SUMMARY
The cell wall of Gram-positive bacteria has been shown to mediate environmental stress tolerance, antibiotic susceptibility, host immune evasion and overall virulence. The majority of these traits have been demonstrated for the well-studied system of wall teichoic acid (WTA) synthesis, a common cell wall polysaccharide among Gram-positive organisms. Streptococcus mutans, a Gram-positive odontopathogen that contributes to the enamel-destructive disease dental caries, lacks the capabilities to generate WTA. Instead, the cell wall of S. mutans is highly decorated with rhamnose-glucose polysaccharides (RGP), for which functional roles are poorly defined. Here, we demonstrate that the RGP has a distinct role in protecting S. mutans from a variety of stress conditions pertinent to pathogenic capability. Mutant strains with disrupted RGP synthesis failed to properly localize cell division complexes, suffered from aberrant septum formation and exhibited enhanced cellular autolysis. Surprisingly, mutant strains of S. mutans with impairment in RGP side chain modification grew into elongated chains and also failed to properly localize the presumed cell wall hydrolase, GbpB. Our results indicate that fully mature RGP has distinct protective and morphogenic roles for S. mutans, and these structures are functionally homologous to the WTA of other Gram-positive bacteria.
Keywords: Streptococcus mutans, Rhamnose, RGP, cell wall, cell division, morphogenesis
Graphical Abstract
The cell wall of Streptococcus mutans is richly decorated by the rhamnose-glucose polysaccharides (RGP). Here, we show that loss of mature RGP causes undercrosslinking of the peptidoglycan, leading to heightened cell wall stress sensitivity for S. mutans. In addition, the RGP structures were shown to have an important functional role in maintaining proper cellular morphology and coordinating events during cell division.
INTRODUCTION
The outermost surface-exposed layer of Streptococcus mutans is distinctively decorated by rhamnose-glucose polysaccharides (RGP) (Van de Rijn & Bleiweis, 1973). These RGP structures become covalently linked to the peptidoglycan (PG) to complete the cell envelope of S. mutans via activity of the LytR-CpsA-Psr (LCP) family of proteins in a manner reminiscent of the formation of wall teichoic acids (WTA) in many other Gram-positive bacteria (Kawai et al., 2011, De et al., 2017). Notably, S. mutans lacks the ability to produce WTA, while the RGP that is produced makes up approximately half of the total weight of the cell wall (Mistou et al., 2016).
The pathogenic capability of S. mutans is tightly linked to the bacterium’s physiology. As an efficient sugar importer, this odontopathogen can rapidly ferment available carbohydrates into lactic acid, generating acidic microenvironments within the multispecies dental plaque biofilms in which it resides (Ajdic & Pham, 2007). Frequent sugar consumption by the human host promotes recurring acidification, leading to S. mutans dominance over acid-susceptible neighboring organisms within these biofilms, as well as the destruction of tooth enamel and progression of dental caries. Oxidative stress, in the form of reactive oxygen species produced by competing bacteria, or the presence of diffusible oxygen within the oral cavity, compound the challenges S. mutans routinely faces and must overcome (Kreth et al., 2005, Derr et al., 2012, Baker et al., 2014). Previous work by our group and others has elucidated a robust network of genetic and physiological changes that S. mutans undergoes in order to tolerate and withstand these otherwise inhospitable conditions (Baker et al., 2017). Recently, we have also demonstrated that the RGP structures themselves contribute to acid and oxidative stress tolerance in S. mutans (Kovacs et al., 2017). Mutant strains impaired in their ability to generate mature RGP also lacked the capacity to grow into robust biofilms and were severely attenuated in virulence (Kovacs et al., 2017). Beyond this, however, elucidation of the biological roles for RGP remains understudied.
Various polysaccharide structures are known to incorporate into the cell envelope, providing support for shape and overall cellular integrity for Gram-positive bacteria. By far the most widely studied include the anionic ribitol-phosphate or glycerol-phosphate WTAs of model Gram-positive organisms such Staphylococcus aureus and Bacillus subtilis, respectively. Although different in composition, the stepwise construction is common among these polymers, and both depend on the activity of TarO/TagO (teichoic acid ribitol/teichoic acid glycerol), responsible for linking N-acetylglucosamine-1-phosphate (GlcNAc) onto a membrane-anchored bactoprenol lipid carrier, which is required to initiate WTA synthesis (Brown et al., 2013). In S. mutans, the UDP-GlcNAc transferase enzyme RgpG shares 40% amino acid homology with TagO and facilitates initiation of RGP synthesis by linking GlcNAc onto bactoprenol (Yamashita et al., 1999). The poly-rhamnose chain that makes up the backbone of RGP is built via specific rhamnosyltransferase enzymes, first by RgpA, followed by alternating actions of RgpB and RgpF, which in tandem elongate the structure (Shibata et al., 2002). Side chain modification is then achieved by glucose linkage, carried out by activities of the glycosyltransferase enzymes RgpE, RgpH and RgpI (Ozaki et al., 2002, Shibata et al., 2003, Nakano et al., 2005).
While functional relevance for RGP is unclear, much insight can be gained from studies interrogating the roles of WTAs and other cell wall polysaccharides (CWPs). In tarO mutants of S. aureus, loss of WTA has been shown to restore sensitivity to β-lactam antibiotics in methicillin-resistant S. aureus (MRSA) strains (Wang et al., 2013). Physiologically, ∆tarO mutants experience drastic morphological changes and cell envelope defects (Campbell et al., 2011). When WTA formation is inactivated in B. subtilis, the bacterium loses its ability to transition to rod shape and becomes sensitive to a number of cell wall stresses, including temperature and salinity (Pollack & Neuhaus, 1994). The WTA of S. aureus has also been shown to be implicitly required for efficient cell division, by coordinating the localization of penicillin binding proteins (PBP) for crosslinking the newly-formed PG and also directing autolysin enzymes to the septum for cell separation (Atilano et al., 2010). Similar features have additionally been described for the role of GBC (Group B Carbohydrate) in Streptococcus agalactiae, which upon disruption leads to severely under-crosslinked PG and aberrant localization of the murein hydrolase PcsB (Caliot et al., 2012). Characterization of this cysteine- and histidine-dependent aminohydrolase/peptidase (CHAP)-domain-containing protein in Streptococcus pneumoniae has revealed direct interaction of PcsB with the FtsEX cell division complex (Sham et al., 2011, Sham et al., 2013, Bajaj et al., 2016). Biochemical analysis of PcsB from S. pneumoniae has shown hydrolyzing activity of its CHAP domain on PG, a requirement for the splitting of daughter cells to complete cell division (Bartual et al., 2014). The glucan binding protein B (GbpB) of S. mutans has been identified as an ortholog to PcsB, yet despite its association with the cell membrane, specific hydrolase activity of GbpB has not been detected. However, mutation of GbpB leads to cell morphology defects, and the enzyme also contains a CHAP domain, suggesting that its role in cell division is not yet fully understood (Mattos-Graner et al., 2001, Mattos-Graner et al., 2006).
The present study interrogated the roles of RGP on overall physiology and morphogenesis in S. mutans. By examining the consequences of disrupting RGP at various steps along the structure’s maturation, we demonstrate that fully-formed RGP is required to protect the cell and tolerate various cell wall-targeting insults. Further, mature RGP is necessary to efficiently coordinate cell division in S. mutans. Surprisingly, not only complete inhibition of RGP, but also inhibition of its side chain formation, caused irregular cellular morphology and inability to properly localize the GbpB enzyme, which we propose acts as a murein hydrolase required for cell separation.
RESULTS
Disruption of RGP formation causes growth impairment in S. mutans.
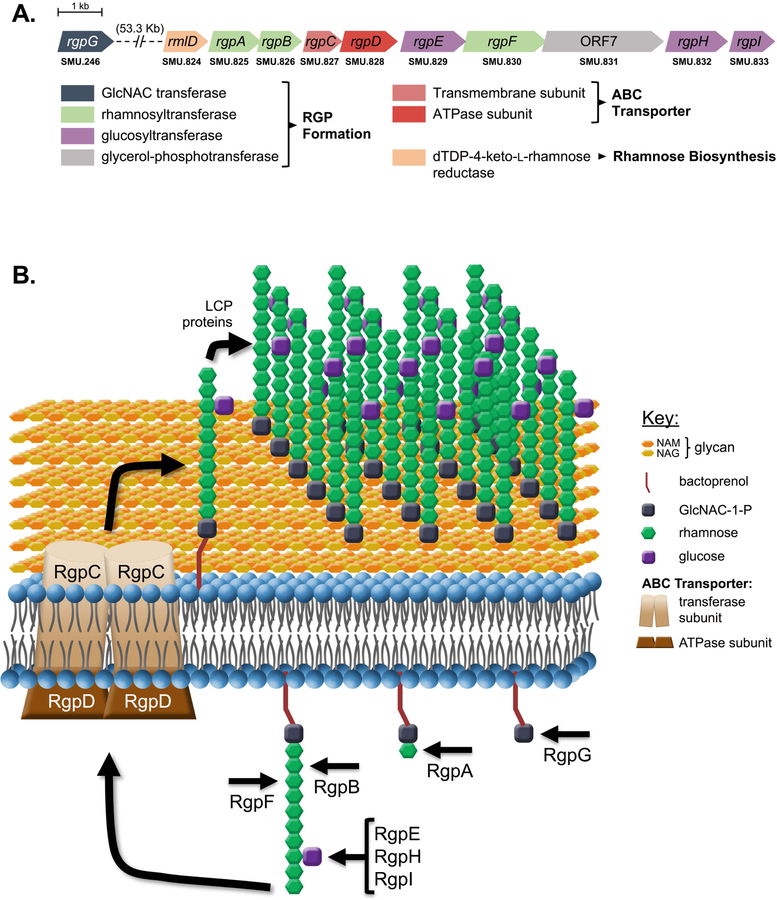
RGP biosynthesis is a multi-factorial process (summarized in Figure 1) reliant on the enzymatic products encoded in the ten-gene rgp operon (Figure 1A). In order to characterize the functional roles of RGP, specific genes within the rgp operon were deleted to inhibit different steps along RGP maturation in the S. mutans UA159 parent strain. We predicted that loss of RgpE (∆rgpE), responsible for glucose side chain linkage, would cause the least severe defects, since formation of the core poly-rhamnose backbone should remain intact. The ∆rgpE mutant experienced a significantly reduced growth rate, compared to the UA159 parent strain, but was capable of growing to the same maximal cell density and displayed little overall fitness defects (Figure S1A, Table S1) (Quivey et al., 2015). We have previously demonstrated that deletion of rgpF causes substantial fitness defects, with significantly attenuated stress tolerance, biofilm and virulence phenotypes (Kovacs et al., 2017, Quivey et al., 2015). As such, this mutant was chosen to represent more severe RGP disruption (Figure S1A, Table S1). RgpG, the enzyme that initiates RGP formation, was also deleted in order to investigate complete loss of the polysaccharide, and the ∆rgpG mutant was associated with the greatest impact on growth and fitness of the three mutant strains (Figure S1A, Table S1).
FIGURE 1. Rhamnose-glucose polysaccharide (RGP) formation in S. mutans.
The genetic loci (A) and formation steps (B) corresponding to RGP biosynthesis are displayed. Enzymes encoded by each gene in Panel A are color-coordinated based on functional classes.
RGP structure influences response to cell wall stress.
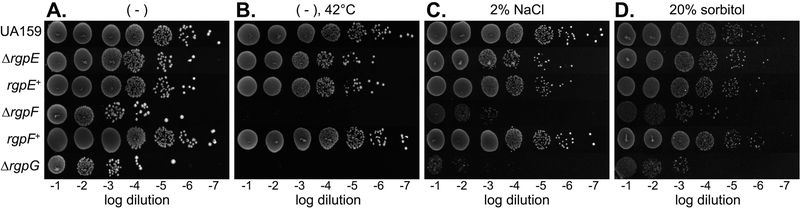
As part of the outermost layer of the S. mutans cell envelope, RGPs are presumably the initial point of contact for the cell with its surroundings. As such, tolerance of environmental insults is likely facilitated by intact, mature RGP. With this in mind, overnight cultures of UA159 and the various rgp mutant strains were serially diluted and spot plated onto BHI agar medium, or agar medium containing 2% NaCl (saline stress) or 20% sorbitol (osmotic stress) and incubated for recovery of surviving organisms. The effect of elevated temperature was also analyzed by growing cultures on BHI agar at 42°C. In general, the trends in sensitivity followed the degree of RGP disruption caused by each mutation. Comparator conditions, where cells were grown on standard BHI agar at 37°C allowed for maximal outgrowth of cells from UA159 and mutant strains (Figure 2A). Despite normalization of culture optical densities, both the ∆rgpF and ∆rgpG mutants displayed outgrowth that was reduced by several orders of magnitude, highlighting a basal growth impairment for those strains that exhibited the most severe inhibition of RGP synthesis. The greatest impact on ∆rgpF and ∆rgpG growth was caused by elevated temperature, as both of these mutants failed to grow at 42°C, while ∆rgpE and the parent strain maintained normal growth, that is, similar to growth on BHI agar medium (Figure 2B compared to Figure 2A). Exposure of cultures to 2% NaCl did not affect the ability of either WT or ∆rgpE to grow, whereas both the ∆rgpF and ∆rgpG mutant strains failed to grow in conditions of elevated salinity (Figure 2C). While osmotic pressure caused by the presence of 20% sorbitol negatively affected all strains tested, the impact on the ∆rgpG strain was most evident. All cultures grown on agar medium containing 20% sorbitol were faint in appearance with smaller colony morphology, while the ∆rgpG strain also displayed reduced outgrowth compared to its control condition (Figure 2D versus Figure 2A). Growth kinetics were also evaluated in liquid BHI medium for all strains under similar conditions of stress (Figure S1, Table S1). Overall, the trends on growth rates were reminiscent of those seen with dilution spot plating. Growth at elevated temperature was completely inhibitory to both the ∆rgpF and ∆rgpG strains (Figure S1B, Table S1). These two mutant strains also experienced the most significant reduction in growth rates when exposed to 1% NaCl (Figure S1C, Table S1) and 20% sorbitol (Figure S1D, Table S1), although significant effects on growth rate were also observed for all strains, compared to growth in BHI medium alone. Calculated doubling times also revealed significant improvement for the rgpE+ and rgpF+ complement strains versus their respective isogenic mutants (Table S1). Of note, none of the strains tested were capable of growing in liquid medium supplemented with 2% NaCl. Taken together, these results indicate that the presence of RGP structures on the surface of S. mutans contribute to the ability of the bacterium to tolerate a variety of environmental stresses, and that cell wall integrity is of paramount importance.
FIGURE 2. Outgrowth of rgp mutants is impaired under conditions of cell wall stress.
Overnight cultures of S. mutans UA159, ∆rgpE, rgpE+, ∆rgpF, rgpF+ and ∆rgpG were serially diluted ten-fold and spotted onto (A) BHI agar medium, (B) BHI agar medium and incubated at 42°C, (C) BHI agar medium supplemented with 2% NaCl, or (D) BHI agar medium supplemented with 20% sorbitol. Plates for panels A, C and D were incubated at 37°C. Images are representative of three independent experiments each plated in duplicate.
Antibiotic susceptibilities of rgp-mutants reveal impairment in peptidoglycan synthesis.
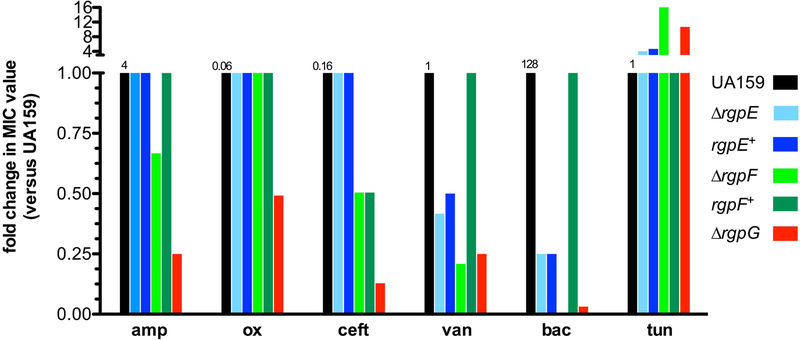
The observation that RGP disruption impaired the ability of S. mutans to tolerate cell wall stress led us to investigate more targeted treatments using antibiotics that inhibit various steps in cell wall synthesis. While revealing susceptibilities of mutant strains, this approach also allows for insight into specific processes where RGP formation may be involved. Proper maturation of PG on the extracellular surface has been shown to rely on the presence of CWP in Gram-positive bacteria, thus several β-lactam antibiotics were tested against the rgp mutant strains to determine if RGP is similarly involved (Atilano et al., 2010, Caliot et al., 2012). The most substantial effects were seen in the ∆rgpG background, where RGP formation was completely inhibited. When treated with ampicillin, oxacillin, and ceftriaxone, the ∆rgpG mutant was consistently 4-fold, 2-fold and 8-fold, respectively, more susceptible than UA159 (Figure 3). The ∆rgpF strain also displayed enhanced sensitivity to β-lactam treatment, especially ampicillin (2-fold) and ceftriaxone (2-fold). These susceptibilities were consistently reproducible for ∆rgpF, indicating a bona fide response to this class of antibiotics influenced by loss of RgpF. In comparison, the ∆rgpE mutant was as sensitive to β-lactam treatment as WT cells.
FIGURE 3. Disruption of RGP causes enhanced sensitivities to antibiotics that target peptidoglycan transpeptidation.
Antibiotic susceptibility testing was performed using cultures of S. mutans UA159 (black), ∆rgpE (light blue), rgpE+ (dark blue), ∆rgpF (light green), rgpF+ (dark green) and ∆rgpG (red) according to CLSI standards and as outlined in Experimental Procedures section. Data are represented as fold-change in consensus MIC value versus UA159 for each drug tested. Numbers above black bars indicate MIC value (μg ml−1) for UA159 for each drug tested. Data are derived from consensus results from three independent experiments each performed in triplicate. amp = ampicillin, ox = oxacillin, ceft = ceftriaxone, van = vancomycin, bac = bacitracin, tun = tunicamycin.
Vancomycin, a PG cross-linking inhibitor, was 2, 4, and 4-fold more potent against ∆rgpE, ∆rgpF and ∆rgpG, respectively, compared to the parent strain (Figure 3). Bacitracin, to which S. mutans is relatively resistant (MIC value 128 μg ml−1) was particularly effective upon RGP disruption, with a 256-fold decrease in MIC for ∆rgpF and 32-fold reduction for ∆rgpG. As bacitracin targets recycling of the lipid carrier onto which the RGP is built, a disruption in the structure’s synthesis would disturb the balance of available bactoprenol, causing heightened sensitivity. Susceptibility to tunicamycin was also tested, as it is an inhibitor of GlcNAc transferases, the class of enzymes to which RgpG belongs. When RGP synthesis was ablated genetically in ∆rgpG, the mutant strain became 8-fold more resistant to treatment with tunicamycin. Similarly, the ∆rgpF strain displayed a 16-fold increase in resistance to the antibiotic, whereas ∆rgpE was 4-fold more resistant (Figure 3). Because RGP formation is dependent upon the target of tunicamycin, it stands to reason that strains carrying mutations affecting the RGP would be able to circumvent activity of the antibiotic as the target becomes less readily available for inhibition. Altogether, the sensitivity profile demonstrated by these mutant strains signifies that the RGP structures may play an important role in the process of PG transpeptidation, given that RGP disruption further sensitized S. mutans to treatment with β-lactam and vancomycin antibiotics.
Additional antibiotics were also tested that target various intracellular events (Table S2). Use of d-cycloserine, a d-alanine analog that disrupts formation of the PG pentapeptide, was completely ineffective against all strains. The S. mutans WT and ∆rgpE strains were also highly resistant to exposure to the pore-forming lipid II inhibitor nisin (MIC value of 512 μg ml−1 for both strains), while the MIC values for ∆rgpF and ∆rgpG were reduced to 32 μg ml−1 (16-fold change) and 64 μg ml−1 (8-fold change), respectively. These results are more difficult to interpret as the susceptibility values remained relatively high in the mutant strains. Interestingly, sensitivity to kanamycin was also drastically enhanced in both ∆rgpF and ∆rgpG (32-fold and 8-fold, respectively). It is unlikely that the RGP is in any way connected to protein synthesis in S. mutans; however, these non-specific changes in susceptibilities may arise from perturbations in membrane permeability, as we have previously demonstrated to be the case in the ∆rgpF mutant strain (Kovacs et al., 2017).
The relationship between RGP and PG was further investigated using combined antibiotic treatments of tunicamycin plus the β-lactam ampicillin to measure for potentiating, or drug synergy, effects. Synergistic benefits for the combination, as determined by FIC values < 0.5, were observed for the UA159 WT strain, as well as for ∆rgpE and the rgpE+ and rgpF+ complement control strains (all of which displayed equivalent MIC values against ampicillin alone) (Table 1). In contrast, synergy was not achieved using these drug combinations against either the ∆rgpF or ∆rgpG mutant strains. Isobolograms were also used to analyze drug-partnering interactions against all strains, recapitulating the calculated FIC values (Figure S2).
TABLE 1.
Fractional Inhibitory Concentration (FIC) testing for antibiotic potentiation using combinatorial treatment of ampicillin and tunicamycin with cultures of rgp mutant strains. Synergistic combinations (FIC < 0.5) are highlighted in green.
| MIC of drug (μg ml−1) |
|||||
|---|---|---|---|---|---|
| Alone |
In combination |
||||
| Strain | Tun (A) | Amp (B) | Tun (AB) | Amp (BA) | FIC |
| UA159 | 4 | 8 | 1 | 1 | 0.375 |
| ∆rgpE | 4 | 8 | 0.5 | 2 | 0.375 |
| rgpE+ | 4 | 8 | 1 | 1 | 0.375 |
| ∆rgpF | 16 | 4 | 8 | 2 | 1 |
| rgpF+ | 4 | 8 | 1 | 1 | 0.375 |
| ∆rgpG | 16 | 4 | 8 | 4 | 1.5 |
Tun = tunicamycin
Amp = ampicillin
FIC defined as sum of (MICAB/MICA) and (MICBA/MICB)
These results, using chemical inhibition of various stages of RGP formation, support the susceptibilities described above with genetic ablation and point to a particular reliance of S. mutans on the RGP system to allow for tolerance against PG-targeting antibiotics and, thus, proper PG maturation.
Polar effects of rgpE, rgpF and rgpG deletion.
The rgpG gene is co-transcribed with mecA (SMU.245), while both rgpE and rgpF are transcribed as a polycistronic message along with additional rgp genes within the operon (De et al., 2018). To determine if loss of these genes caused polar effects on downstream expression, qRT-PCR was performed to measure mRNA produced in each mutant background for the appropriate surrounding genes. Loss of rgpG did not alter expression of mecA, or that of the downstream gene, SMU.247 (Figure S3A). Similarly, expression of downstream genes in the ∆rgpE background was unaffected (Figure S3B). For the ∆rgpF strain, expression of ORF7 (SMU.831), immediately downstream of rgpF, was reduced by ~2.5-fold (p ≤ 0.05), and no changes were observed for the next downstream gene, rgpH (Figure S3C).
RGP is required for proper, localized septum formation in dividing S. mutans.
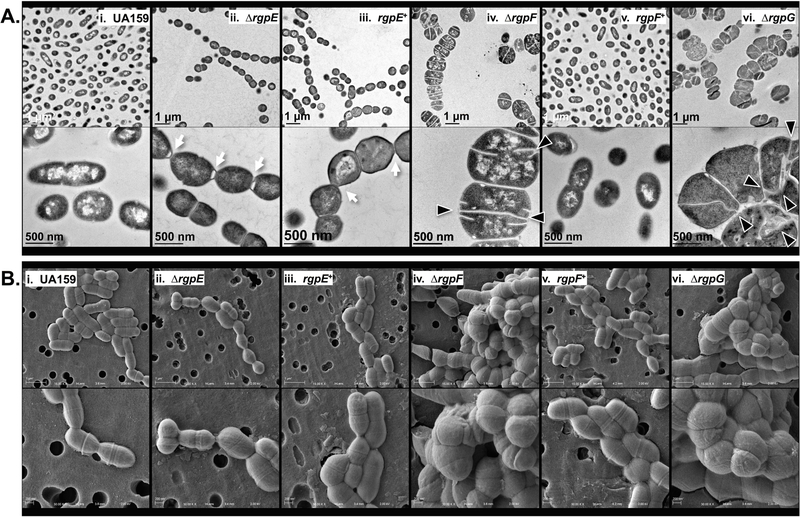
Detailed examination of cellular morphologies caused by RGP mutagenesis was employed using transmission and scanning electron microscopy (TEM and SEM; Figures 4A and 4B, respectively). S. mutans UA159 displayed predominantly as single cells with classic ovococcal cell shape and elongation occurring perpendicular to a centralized septal plane, as captured in dividing cells (Figure 4A and B, panel i). Images of the ∆rgpE strain revealed elongated chains of cells that failed to efficiently separate from one another post-division (Figure 4A and B, panel ii). The most striking morphologies were observed in both the ∆rgpF and ∆rgpG mutant strains, which both appeared as swollen, distended cells in long chains or immense clusters (Figure 4A and B, panels iv and vi, respectively). Additionally, septation in these mutant strains occurred at seemingly random locales along the cell, with apparent dysregulation of new crosswall progression, as evidenced by the bifurcation of individual septa. Interestingly, morphologic complementation was not seen in the rgpE+ strain (Figure 4A and B, panel iii), while the rgpF+ strain (Figure 4A and B, panel v) is similar in appearance to the parent strain, UA159. Additionally, WT S. mutans treated with sub-MIC amounts of tunicamycin to chemically inhibit RGP (specifically RgpG) also demonstrated irregular septation and bifurcated crosswalls (Figure 5A).
FIGURE 4. Electron microscopy reveals morphological abnormalities that arise upon RGP disruption.
Cultures of S. mutans UA159 (panel i), ∆rgpE (panel ii), rgpE+ (panel iii), ∆rgpF (panel iv), rgpF+ (panel v) and ∆rgpG (panel vi) were grown to mid-log phase (OD600 ~0.4) and imaged using (A) transmission and (B) scanning electron microscopy as described in Experimental Procedures section. Images in Panel A were taken at 15,000X (upper) and 60,000X (lower) magnification, while those in Panel B were taken at 15,000X (upper) and 30,000X (lower) magnification. Excessive chains of cells (white arrow) and aberrant cellular morphologies are apparent in each mutant strain, with malformed and bifurcated septa highlighted (black triangles).
FIGURE 5. Chemical perturbation of S. mutans cell wall.
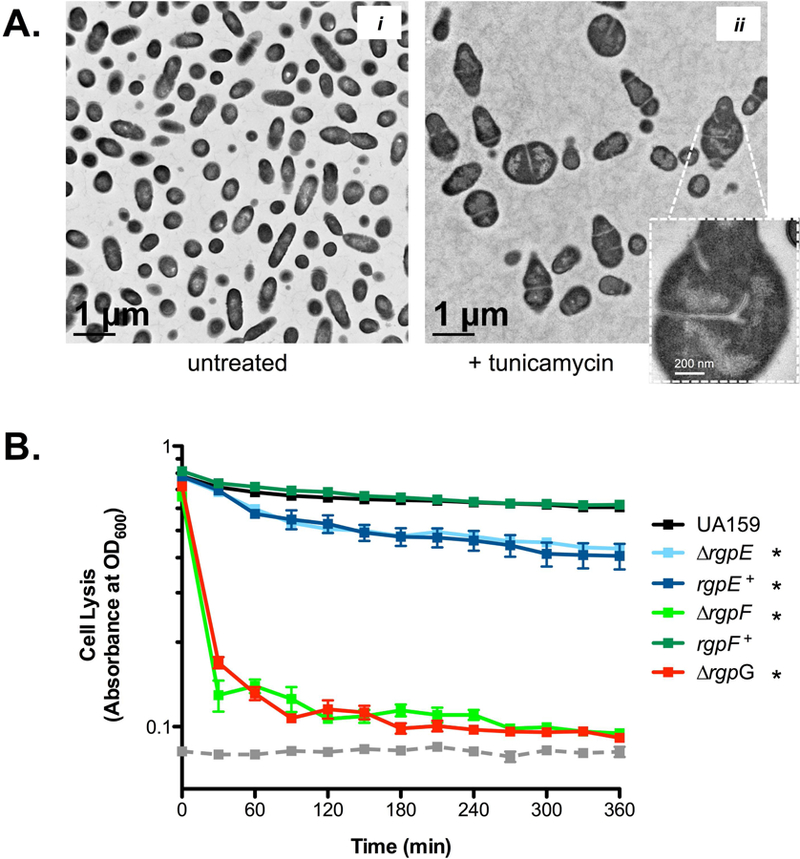
(A) Untreated (image i) S. mutans UA159 or UA159 exposed to 0.5 μg ml−1 tunicamycin for 3 hours (image ii) were imaged using TEM. Inset in image ii highlights morphological defects, depicting malformed and bifurcated septum. (B) Cultures of S. mutans UA159 (black), ∆rgpE (light blue), rgpE+ (dark blue), ∆rgpF (light green), rgpF+ (dark green) and ∆rgpG (red) were grown to stationary phase, washed and suspended in buffer containing 50 U ml−1 mutanolysin. Rates of cellular lysis were assessed by measuring optical density at 600 nm in a Bioscreen C plate reader (as described in Experimental Procedures). The hashed gray line indicates optical density readings of buffer alone. Data is representative of two independent experiments each performed with 10 replicates and expressed as mean ± SD. Statistical significance (* p ≤ 0.005; versus UA159) was determined by pairwise comparison of terminal optical density readings using Student’s t-test.
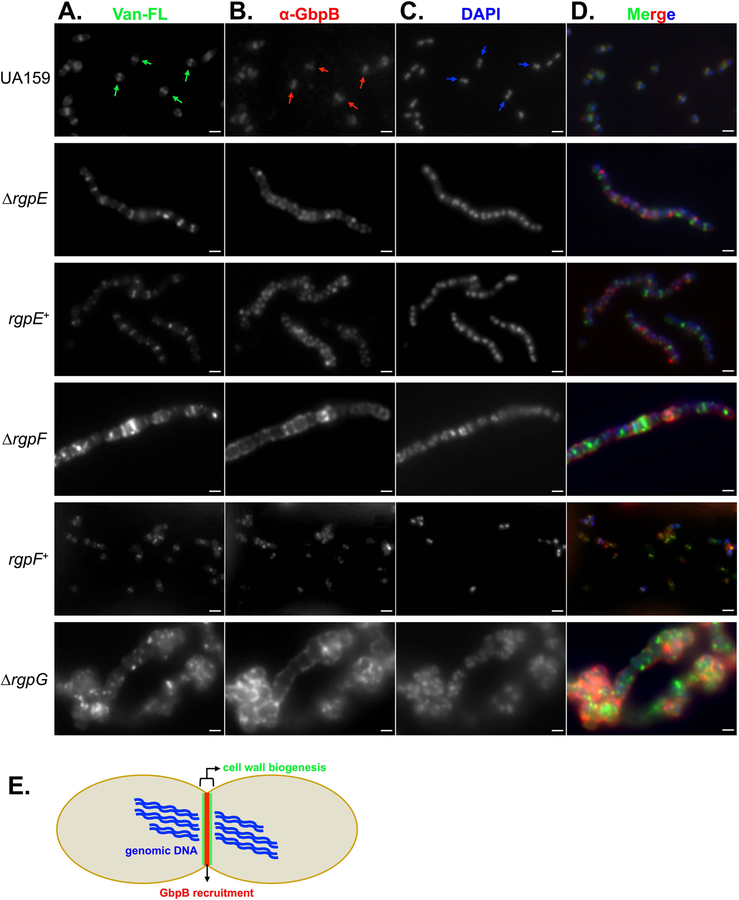
Further confirmation of septal dysregulation was shown using BODIPY-vancomycin (Van-FL), a fluorescently-conjugated derivative of the antibiotic that incorporates into sites of newly-synthesized cell wall. Cells from cultures of UA159 grown to early-log phase displayed distinct septum formation, as demonstrated by localized fluorescence that only appeared across the midline of dividing cells and perpendicular to the DAPI-stained DNA (Figure 6A and C, respectively). The ∆rgpE mutant strain also displayed midline staining with Van-FL, indicating crosswall formation proceeded normally in this mutant, although chains of cells were still apparent, as in TEM imaging. Again, the rgpE+ strain retained the same morphological pattern as its corresponding mutant. Both the ∆rgpF and ∆rgpG mutant strains exhibited random staining patterns, with regions of intense fluorescence coupled with diffuse signal across individual cells, while the rgpF+ complement strain resembled the pattern observed in the WT. Altogether, these data are demonstrative of a loss in coordinated cell division that arises upon disruption of RGP synthesis.
FIGURE 6. Mature RGP is required for proper localization of cell division complexes.
Cultures of S. mutans UA159, ∆rgpE, rgpE+, ∆rgpF, rgpF+ and ∆rgpG were grown to early log phase (OD600 ~ 0.2) and treated with 2 μg ml−1 fluorescently-conjugated vancomycin (Van-FL; A) for ten minutes. Cells were collected, fixed and stained with α-GbpB polyclonal antibody (B) and mounted onto glass slides with mounting media containing DAPI stain (C). Imaging was performed as described in the Experimental Procedures section with a 100X objective using oil overlay. Arrows in the UA159 panel indicate position and orientation of staining patterns. Merged fluorescent images of all three channels are displayed in panel D. Scale bars = 1 μM. (E) Model of fluorescent staining pattern in a dividing S. mutans UA159 cell, depicting DNA (blue) separating horizontally, with cell wall biogenesis occurring perpendicularly to form a septum (green), and localization of the cell wall hydrolase, GbpB (red), distinctly at septal plane.
Recruitment of the presumed cell wall hydrolase, GbpB, to the septum of dividing cells is reliant upon mature RGP.
The highly-crosslinked PG of S. mutans confers a degree of resistance for the organism towards autolysis-inducing muramidases, such as mutanolysin (Chatfield et al., 2005, Ahn & Burne, 2006). Each of the rgp mutant strains were sensitive to mutanolysin, with near-complete lysis occurring for both ∆rgpF and ∆rgpG (Figure 5B). The enhanced autolysis observed upon loss of rgpE may suggest that beyond aiding in the crosslinking of PG, fully mature RGP structures may possibly provide physical protection against cell wall-degrading enzymes. In comparison, no differences in lysis were observed when subjecting the rgp mutant strains to either lysozyme or Triton-X100 (data not shown).
Previous work investigating the role of CWP during morphogenesis in Gram-positive bacteria has shown a clear dependency on the placement of these structures and the ability of cells to effectively divide in S. aureus, S. pneumoniae and S. agalactiae (Atilano et al., 2010, Ng et al., 2004, Caliot et al., 2012). In the latter, cell division was severely altered in mutants lacking GBC, resultant from mislocalization of the cell wall hydrolase enzyme, PcsB (protein for cell separation in Group B Streptococcus). Given the phenotypes of the S. mutans rgp mutant strains described above, we sought to explore the role of RGP in recruitment of additional cell division machinery in these backgrounds. S. mutans possesses an ortholog to PcsB named GbpB (glucan binding protein B), although specific hydrolase function has not yet been demonstrated for this protein (Mattos-Graner et al., 2001). Using a polyclonal antibody raised against S. mutans GbpB, we tracked localization of the protein by immunofluorescence. In WT UA159 cells, GbpB fluorescence overlaid distinctly with the site of septum formation (as identified by Van-FL staining), indicating that, indeed, GbpB becomes localized to areas of cell wall synthesis (Figure 6B and D). Markedly, the localization pattern of GbpB was completely lost in each of the rgp mutant strains examined, with staining occurring instead around the perimeter of these cells. Further, this staining pattern was not in accordance with Van-FL staining, suggesting that GbpB was no longer able to localize to areas where cell wall was being synthesized when RGP formation was disrupted.
DISCUSSION
While the importance of WTAs has been demonstrated for certain organisms, it has become increasingly appreciated that these structures are not entirely ubiquitous among Gram-positive bacteria. Notably, members of the order Lactobacillales can generate alternative CWP that utilize a variety of carbohydrates and are abundant within the cell envelope (Mistou et al., 2016). Of these, the monosaccharide l-rhamnose is particularly relied upon. These rhamnosylated structures have been shown to mitigate important cellular functions including capsule formation, bacteriophage uptake and immune cell evasion in Streptococcal and Lactococcal species (James & Yother, 2012, Shibata et al., 2009, Tsuda et al., 2000, Sadovskaya et al., 2017). As a prototypical member of the lactic acid bacteria group, S. mutans possesses a cell wall with a distinct polysaccharide composed of a poly-rhamnose core and glucose side chains. Formation of these RGP structures was originally described in a heterologous expression system using Escherichia coli and quantification of the rhamnosyl-lipopolysaccharide formed (Shibata et al., 2002). Here, we have shown that a number of cellular processes – from protection to cell division – are incumbent upon fully mature RGP structures.
Disruption of RGP formation in S. mutans resulted in attenuated growth responses to ionic and osmotic stresses, as well as temperature-dependent growth inhibition. These phenotypes were most evident in the ∆rgpF and ∆rgpG mutant strains where the greatest degree of RGP inhibition was achieved. Survival of the ∆rgpF strain, in addition to non-lethal phenotypes for ∆rgpA and ∆rgpB, highlights the most substantial differences between the RGP system of S. mutans versus WTA formation in many other Gram-positive bacteria (Quivey et al., 2015, Kovacs et al., 2017). In the later, inactivation of genes responsible for early steps in WTA formation is lethal due to accumulation of lipid-linked undecaprenol without downstream processing, a phenotype that can be rescued upon loss of the WTA-initiating gene, tagO (D’Elia et al., 2006a, D’Elia et al., 2006b). Non-lethality of ∆rgpF in S. mutans (likewise for ∆rgpA and ∆rgpB) suggests that perhaps additional cell wall polysaccharides utilizing undecaprenol may continue to mature and in turn, help to replenish available pools of the molecule. In support of this hypothesis, when cell walls from a S. mutans deletion strain of rgpG were probed for total cell wall antigen content, the mutant strain retained 29.5% of reactivity (De et al., 2017).
The ∆rgpE strain, while slightly attenuated in growth, as compared to the parent strain, continued to persist in the face of the stress conditions tested in this study, suggesting that the RGP backbone structure is most important for tolerance of difficult environmental circumstances. RgpE has been shown to contribute to formation of glucose side chains extending off the rhamnose stalk on the RGP structure and is thought to function after the core poly-rhamnose chain is assembled. Of note, the genes downstream of rgpE, rgpH and rgpI, encode the glucosyltransferases RgpH and RgpI, which function to control the branching frequency of side chain linkage in S. mutans UA159 (Ozaki et al., 2002). RgpE and RgpI are required for the addition of the α1,2-glucose moieties defining serotype c, the most prevalent clinical isolate associated with dental caries (Shibata et al., 2003, Nomura et al., 2005). Serotype e and f strains harbor β1,2- and α1,3- glucosidic linkages, respectively, although the precise enzymes involved in creating these bonds is poorly understood (Shibata et al., 2003, Nomura et al., 2005). RgpH and RgpI can exert control over one another at the protein level to control the frequency and branching of side-chain addition in a non-serotype specific way, as loss of either alters RGP formation, yet does not impact gene expression (Ozaki et al., 2002).
Considering the varied stress tolerance outcomes displayed by the different rgp mutant strains, a hierarchical dependence upon the RGP can be inferred, whereby side chain modification (as the terminal maturation step) can be dispensable for the bacterium during challenging growth conditions, despite impacting its overall growth. Clinical evidence may support this conclusion as well. S. mutans serotype k lacks glucose side chains naturally due to an insertional mutation within rgpE, and this particular strain is least common (<1% reported in carious lesions) amongst the various infective serotypes of S. mutans (Nomura et al., 2005). Given the clinical distribution of this particular serotype, it is interesting to speculate on how side chain modification of RGP can influence pathogenicity. Similar conclusions have been reached for the Group A polysaccharide of S. pyogenes, which exhibited significant attenuation in virulence upon side chain disruption of the bacterium’s poly-rhamnose CWP (van Sorge et al., 2014).
Detailed examination of the rgp mutant strains via a combination of TEM and SEM imaging revealed striking cellular morphology defects, compared to the parent strain. Cells of the mutant strains appeared in longer chains, while the ∆rgpF and ∆rgpG strains both revealed distended and irregularly shaped bacteria. Septation was also disrupted in ∆rgpF and ∆rgpG, causing crosswall formation at random locations within cells. Previous work has also examined cell wall abnormalities as a consequence of rgpG deletion in S. mutans, demonstrating altered cellular morphology and a poor biofilm growth phenotype (De et al., 2017). Similar morphological abnormalities have been observed with mutant strains incapable of producing WTA in other Gram-positive bacteria. Deletion of tagO in Lactobacillus plantarum causes aberrant cell shape and decreased surface roughness (Andre et al., 2011). Loss of WTA formation in B. subtilis prevents transition of spherical cells into their destined rod shape (Pollack & Neuhaus, 1994). Deletion of the RgpG ortholog in S. agalactiae, GbcO, also resulted in septum malformations, and the mutation was alleviated by ectopic expression of the S. aureus tarO gene in the ∆gbcO background, demonstrating a shared functional role across genera (Caliot et al., 2012). In S. aureus, septum formation and subsequent cellular division were severely altered with both genetic ablation of tarO and by chemical inhibition of WTA formation using tunicamycin (Brown et al., 2008, Campbell et al., 2011, Lee et al., 2016). The irregularities seen here upon RGP disruption suggest that, similar to the functions of WTA and other CWP, the RGP plays a major role in coordinating efficient cell division in S. mutans. Further insight into this topic was revealed by the enhanced sensitivities of the ∆rgpF and ∆rgpG strains to treatment with ampicillin and vancomycin, and also to the muramidase enzyme mutanolysin, together suggesting reduced PG crosslinking resultant from disrupted RGP formation. Similarly, inhibition of WTA formation in S. aureus prohibited the recruitment of penicillin binding proteins to newly-synthesized glycan strands, causing under-crosslinking of the PG (Atilano et al., 2010). This, in turn, has broader impact on overall fitness, as loss of WTA also causes re-sensitization of MRSA strains to β-lactam antibiotics (Wang et al., 2013, Lee et al., 2016). Thus, the RGP of S. mutans likely serves a function homologous to that of WTA during cell division, facilitating the proper localization of enzyme complexes within the cell envelope.
In order for dividing Gram-positive bacteria to efficiently separate and produce daughter cells, cell wall lytic enzymes must be recruited to the newly-formed crosswall and initiate hydrolysis at distinct locations to allow for cell splitting. Several such enzymes have been described in a variety of bacteria, including PcsB of Streptococcal species. PcsB contains a CHAP domain, a feature common in cell wall hydrolases that target the murein sacculus (Reinscheid et al., 2001, Reinscheid et al., 2003). Specific activity of PcsB has been eloquently described in Streptococcus pneumoniae, where the C-terminal end of the enzyme was shown to interact with the extracellular loops of the transmembrane ABC transporter complex FtsEX (Sham et al., 2011, Sham et al., 2013). Intracellularly, FtsX interacts with FtsE, an ATPase that causes conformational changes in FtsX upon ATP hydrolysis (Bartual et al., 2014). In turn, PcsB becomes activated, allowing for its specific hydrolytic activity upon the PG (Bartual et al., 2014, Bajaj et al., 2016). This extends to S. agalactiae as well, where PcsB was shown to localize specifically to the septum (Caliot et al., 2012). In S. mutans, GbpB shares 60% sequence homology with PcsB, and the GbpB enzyme similarly contains a CHAP domain (Mattos-Graner et al., 2001, Mattos-Graner et al., 2006). In this study, immunofluorescence using a GbpB polyclonal antibody demonstrated that the enzyme perfectly aligns with regions of active PG synthesis in the wild-type UA159 strain. This localization pattern was completely disturbed in each of the rgp mutant strains tested, highlighting that not only is the RGP structure itself required, but also a fully mature form of the polymer is necessary for efficient GbpB recruitment. Furthermore, the significantly enhanced rates of autolysis revealed in the rgp mutant strains suggests that hydrolytic enzymes, such as GbpB, have a greater affinity for cell wall binding upon RGP disruption (represented visually by the more intense α-GbpB staining for each rgp mutant strain, compared to UA159). These data also help to explain the excessive chaining phenotype observed with the ∆rgpE mutant strain, in that the presumed hydrolase activity of GbpB at very discrete sites in the crosswall likely contributes to separation of newly-divided cells. As these cells still appeared similar in size and shape to the parent strain, we suspect that the immature form of RGP (that is, lacking side chains) is sufficient for the majority of cell wall remodeling steps to occur since properly formed septa were observed in the ∆rgpE mutant strain. This is in accordance with loss of PcsB in S. pneumoniae, where crosswalls also formed normally, suggesting that the enzyme is activated following septum formation (Sham et al., 2011). The absence of glucose side chains on S. mutans RGP may simply alter the spatial landscape within the cell wall, thus promoting mislocalization of enzymes that are otherwise recruited to particular locations extracellularly. Upon more substantial ablation of RGP formation in the ∆rgpF and ∆rgpG mutant strains, GbpB was not only mislocalized, but septation was no longer distinct, and instead appeared to form randomly, further evidenced by diffuse Van-FL staining. Again, these findings are reminiscent of the consequences of the loss of WTA formation, which results in increased binding of cell wall lytic enzymes across the cell, promoting autolysis in both S. aureus and B. subtilis (Schlag et al., 2010, Biswas et al., 2012, Kasahara et al., 2016). Mature WTA densely populates the cell wall in regions apart from the septum, while within the crosswall these structures are yet immature and less populous. It has been suggested that these WTA-rich regions restrict autolytic enzymes from interacting with their murein targets, directing them instead to the septal plane, thus protecting the rest of the cell from non-specific lysis (Schlag et al., 2010). This spatial exclusion phenomenon may also be attributed to mature RGP of S. mutans, since the defined staining pattern of GbpB became completely aberrant in each of the rgp mutant strains.
Recently, the previously unannotated S. mutans gene ORF7 (SMU.831) was shown to encode an additional RGP-modifying transferase responsible for linkage of glycerol-phosphate side chains, conferring a negative charge to the structure (Edgar et al., 2018). The full implications of this tailoring mechanism on S. mutans morphogenesis, or the possibility of other alterations affecting RGP charge remains to be fully understood. That said, failure to recruit GbpB to the septum in the glucosyltransferase deletion mutant strain, ∆rgpE, implies that additional factors, such as potential charge interactions, may also be at play in governing hydrolytic activity in S. mutans. In fact, autolysin trafficking has been shown to rely on the proton-binding capacity of immature WTA in S. aureus where the activity of the major autolysin AtlA correlates to regions of higher pH within the cell wall (Biswas et al., 2012). Altogether, these data emphasize that proper localization of cell division enzymes and complexes is necessitated by mature RGP formation, and demonstrate an essential role for these polymers during S. mutans growth.
Finally, it is worth mentioning the difficulty encountered with attempts to generate a genetic complement strain of the ∆rgpG mutant, which was entirely recalcitrant to our efforts. A number of conditions were tested, including the use of cultures at different stages of growth for transformation, addition of competence stimulating peptide (CSP), electroporation and direct, markerless transformation using WT chromosomal rgpG DNA. Notably, control experiments using shuttle vectors to transform the ∆rgpG strain were unsuccessful as well. When the complementation construct was introduced first into wild-type UA159, termed “UA159+[rgpG+],” followed by deletion of the native rgpG gene (“UA159+[rgpG+/∆rgpG]”), cells were viable; however, they remained phenotypically similar to the ∆rgpG strain (Figure S4). In addition, gene expression of rgpG in the UA159+[rgpG+/∆rgpG] background was markedly lower than that of WT (albeit detectable above background; p ≤ 0.01) suggesting that the genomic context of rgpG location is important in governing its proper expression/function (Figure S5A). The rgpG gene is co-transcribed with its upstream neighbor, mecA (SMU.245) and separated by 74 bases from the next downstream gene, SMU.247 (De et al., 2018). When expression of these genes was measured via qRT-PCR in the different rgpG mutant strains, no changes were observed (Figure S3A). MecA has been directly implicated in negatively regulating competency in S. mutans, facilitating the proteolysis of the sigma factor SigX (Tian et al., 2013). SigX activity is, in turn, promoted by the accumulation of CSP extracellularly. We posit that deletion of rgpG diminishes a major component of the cell wall, the RGP, affecting the competency-signaling network and overall transformability of S. mutans. How these physiological properties affect transformation remains to be fully understood, and defining the possible regulatory networks that govern proper rgpG gene expression in its native locus is an area of current research interest.
Behavior of the rgpG deletion mutant provides possible insight into the lack of complete complementation observed with the rgpE+ strain, which was constructed using standard conditions whereby the construct was inserted ectopically into the chromosome. We suspect that, similarly, native regulatory mechanisms exist within the rgp operon that are required for proper post-translational expression/modification of these gene products, and this hypothesis is currently being investigated, particularly since expression of rgpE was fully restored in the rgpE+ complementation strain (Figure S6). Since loss of rgpG results in complete inhibition of RGP formation, we, therefore, conclude that the RGP structures may also serve a critical function in DNA uptake; however, this is an area not yet fully explored. It is known that binding and absorption of bacteriophage M102 to S. mutans is mediated by RGP, lending merit to this idea (Shibata et al., 2009). Furthermore, transcriptomic analysis in S. mutans has revealed that mutagenesis of the rgpI gene, associated with side chain tailoring on RGP, can impair efficiency of the alternative sigma factor, SigX, in turn reducing genetic competency (Shields et al., 2018). Whether the RGP structures themselves are directly involved with DNA interaction and uptake, or if the loss of competency in ∆rgpG results from gross morphological alterations of the cell surface remains to be determined.
EXPERIMENTAL PROCEDURES
Bacterial strains
Streptococcus mutans strain UA159, the genomic type strain and a clinical isolate of serotype c, was used as the parent strain for all experiments (Ajdic et al., 2002). Isogenic mutants of S. mutans were created via single deletions of rgpE, rgpF and rgpG by replacement of the target open reading frames with an erythromycin resistance cassette, as previously described (Kovacs et al., 2017, Quivey et al., 2015). All S. mutans strains, were maintained on either brain heart infusion (BHI; BD/Difco, Franklin Lakes, NJ) solid agar medium or liquid BHI medium at 37°C in a 5% (v/v) CO2/95% air atmosphere, unless otherwise noted. Escherichia coli Stellar (Clonetech Laboratories, Inc., Mountain View, CA) was grown on LB agar medium at 37°C or in liquid LB medium with continuous shaking at 37°C.
Cloning methodologies
Genetic complementation of ∆rgpF has been described previously (Kovacs et al., 2017). A similar approach was used to complement both ∆rgpE and ∆rgpG. Briefly, appropriate open reading frames were PCR amplified from UA159 genomic DNA with primers (rgpE: 5’-GAGCTCGAATAGATCTGACTCTG CCAGTGGACTTATTC-3’ (forward) and 5’-ATTTAAAAATAGATCTCATGGGAAC TTACCCGATTGTA-3’ (reverse); rgpG: 5’- GAGCTCGAATAGATCTCATGCTGG TAGAGGAACAAAGA-3’ (forward) and 5’-ATTTAAAAATAGATCTTTGATTTC CCAGTCCCGTTAG-3’), designed to contain a BglII restriction site (underlined) (Kovacs et al., 2017). The resulting amplicons contained 15 bp overhangs on each end, complementary to the integration site of the cloning vector, pSUGK-Bgl, which had been similarly linearized with BglII (New England Biolabs, Ipswich, MA) (Derr et al., 2012). Linearized vector and the rgpE or rgpG amplicons were combined using the In-Fusion® HD Cloning Kit (Clontech Laboratories, Inc., Mountain View, CA) as per the manufacturer’s recommendations. The ligation reaction was transformed into E. coli Stellar competent cells (Clontech Laboratories) and transformants were selected on LB agar medium containing 50 µg ml−1 kanamycin. Positive transformants were verified by colony PCR and nucleotide sequencing using gene-specific primers (rgpE: 5’-GACTCTGCCAGTGGACTTATTC-3’ (forward) and 5’-CATGGGAACTT ACCCGATTGTA-3’ (reverse); rgpG: 5’-CATGCTGGTAGAGGAACAAAGA-3’ (forward) and 5’-TTGATTTCCCAGTCCCGTTAG-3’ (reverse)). Plasmid DNA was isolated from a positive construct (pSUGKrgpE and pSUGKrgpG) and used to transform ∆rgpE or ∆rgpG, respectively, for integration into the gtfA (SMU.881) locus using previously published methods (Derr et al., 2012). Transformants were selected on BHI agar medium containing 1 mg ml−1 kanamycin. Positive transformants complemented in this fashion were only recovered for the ∆rgpE strain, which were verified by colony PCR and direct nucleotide sequencing, and one such strain was named rgpE+. Antibiotic selection was reduced to 50 μg ml−1 kanamycin for transformations involving the ∆rgpG strain. This concentration was permissive for growth of the UA159 parent strain, but not for either ∆rgpF or ∆rgpG. No colonies were recovered following transformation of ∆rgpG even when selection was reduced. Further commentary on lost competency upon RGP disruption is provided in the Discussion section.
Cell wall stress dilution plating
Overnight cultures of S. mutans UA159, ∆rgpE, rgpE+, ∆rgpF, rgpF+ and ∆rgpG were adjusted to OD600 ~1.0 and serially diluted ten-fold in liquid BHI medium. An aliquot of 5 μl from each dilution was spotted onto BHI agar medium, BHI agar medium supplemented with 2% NaCl (saline stress) and BHI agar medium supplemented with 20% sorbitol (osmotic stress) and incubated at 37°C in a 5% (v/v) CO2/95% air atmosphere. In addition, 5 μl from each dilution was spotted onto BHI agar medium and incubated at 42°C (heat stress) in a 5% (v/v) CO2/95% air atmosphere. Data shown are representative images from three independent experiments, each plated in duplicate.
Minimum inhibitory concentration (MIC) testing
MIC testing was performed as per Clinical and Laboratory Standards Institute guidelines to assess antimicrobial efficacy of ampicillin, oxacillin, ceftriaxone, vancomycin, bacitracin, tunicamycin, nisin, cycloserine and kanamycin against S. mutans UA159, ∆rgpE, rgpE+, ∆rgpF, rgpF+ and ∆rgpG (Clinical Laboratory Standards Institute, 2018). Briefly, two-fold serial dilutions of test article were made across wells of a 96-well microtiter plate in 100 μl of BHI medium, to which 100 μl of BHI containing ~3.0 × 105 cells were added. Plates were incubated overnight at 37°C in a 5% (v/v) CO2/95% air atmosphere and MIC values were determined as the lowest concentration within a dilution scheme that lacked evident growth. Data are derived from consensus MIC values of three independent experiments, performed in triplicate.
Fractional inhibitory concentration (FIC) testing
Synergistic interactions between ampicillin and tunicamycin were determined by FIC testing in culture medium (Odds, 2003). Two-fold serial dilutions of ampicillin were made across wells of a 96-well microtiter plate in 50 μl of BHI medium. Separately, two-fold serial dilutions of tunicamycin were made in BHI medium and 50 μl of this was added to ampicillin dilutions to create a checkerboard of ampicillin/tunicamycin concentration. Then, 100 μl of BHI medium containing ~3.0 × 105 cells of S. mutans UA159, ∆rgpE, rgpE+, ∆rgpF, rgpF+ or ∆rgpG were added to all wells and plates were incubated overnight at 37°C in a 5% (v/v) CO2/95% air atmosphere. The FIC of combinatorial treatment was determined by the sum of FICa (defined as the MIC of ampicillin in combination, divided by MIC of ampicillin alone) and FICb (defined as the MIC of tunicamycin in combination, divided by MIC of tunicamycin alone). Synergy correlates to a FIC value of ≤0.5, while >0.5 – 4.0 indicates no interaction and >4.0 is antagonistic.
Transmission electron microscopy
Cultures of S. mutans UA159, ∆rgpE, rgpE+, ∆rgpF, rgpF+ and ∆rgpG were grown to mid-log phase (OD600 ~ 0.4) in BHI medium. For chemical inhibition of RGP, UA159 was grown in BHI medium containing either 0.25 or 0.5 μg ml−1 tunicamycin for three hours. Cultures were washed three times in PBS, then fixed (2.5% glutaraldehyde in 0.1M sodium cacodylate buffer) for 24 hours. Cells were then rinsed in buffer, post-fixed for 30 minutes in buffered 1.0% osmium tetroxide, rinsed in dH2O and trapped in agarose. Dehydration was carried out in a graded series of ethanol to 100%, where after samples were transitioned into propylene oxide, then EPON/Araldite epoxy resin overnight and the next day embedded into molds and polymerized at 60°C. One micron sections were stained with toluidine blue to confirm the presence of bacteria by light microscopy and then thin-sectioned at 70nm with a diamond knife onto formvar/carbon nickel slot grids. The grids were stained with aqueous uranyl acetate and lead citrate, then photographed using a Hitachi 7650 TEM (Hitachi High Technologies, Clarksburg, MD) with an attached Gatan Erlangshen 11 megapixel digital camera (Gatan Inc, Pleasanton, CA).
Scanning electron microscopy
Cultures of S. mutans UA159, ∆rgpE, rgpE+, ∆rgpF, rgpF+ and ∆rgpG were grown to mid-log phase (OD600 ~ 0.4), washed three times in PBS and fixed in 3% glutaraldehyde (in phosphate buffer) for one hour at room temperature. Cells were vacuum filtered and collected onto Nuclepore Track-Etched Membrane (0.4 micron pore size) filters (GE Healthcare, Chicago, IL) and subsequently washed with sterile dH2O. Sample filters were left to air dry, mounted onto pegs and coated with platinum for 210s at 800V and 10 mA. Imaging was performed using a Zeiss Auriga Scanning Electron Microscope/Focused Ion Beam Tool (Carl Zeiss Microscopy, Thornwood, NY).
Fluorescence microscopy and immunohistochemistry
Early-log phase (OD600 ~ 0.2) cultures of S. mutans UA159, ∆rgpE, rgpE+, ∆rgpF, rgpF+ and ∆rgpG were treated with BODIPY FL Vancomycin (Thermo Fisher Scientific, Waltham, MA) mixed with equal volume vancomycin to yield a final concentration of 2 μg ml−1 for ten minutes. Cells were washed in PBS, fixed in 4% paraformaldehyde for one hour at room temperature, and washed again with PBS. Cell pellets were resuspended in PBS and treated with a 1:500 dilution of polyclonal α-GbpB antibody raised in chicken (generous gift from M. Duncan, The Forsyth Institute, Cambridge, MA), followed by treatment with Cy3-conjugated secondary IgY antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) (Mattos-Graner et al., 2006). Stained cells were then mounted onto glass microscope slides with ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific, Waltham, MA). Samples were imaged with a 100X objective and oil immersion using a Zeiss Axio Observer inverted microscope and the AxioVision digital image processing software (Carl Zeiss Microscopy, Thornwood, NY).
Autolysis assay
S. mutans UA159, ∆rgpE, rgpE+, ∆rgpF, rgpF+ and ∆rgpG were grown to late-log phase (OD600 ~ 0.8), washed, and resuspended in autolysis buffer (50 mM Tris, pH 8.0). The muramidase enzyme mutanolysin (Sigma Chemical Company, St. Louis, MO) was added to each cell suspension at 50 U ml−1 and 0.3 ml was aliquoted to wells of a microtiter plate. Cellular lysis was measured turbidometrically at OD600 every 30 minutes using a Bioscreen C Plate Reader at 37°C. Assays were performed in three independent experiments with 5 replicates each and are displayed as mean OD600 values ±SD. Statistical significance (p ≤ 0.001) was determined by pairwise comparison using Student’s t-test.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully thank Dr. Margaret Duncan (Forsyth Institute, Boston, MA) for generously providing the GbpB polyclonal antibody. We also thank Dr. Wei Hsu, and especially Dr. Heng Lin (Center for Oral Biology, URMC, Rochester, NY), for use of fluorescence microscopy equipment. Electron microscopy and image acquisition was provided by Karen Bentley (Electron Microscopy Shared Resource, URMC, Rochester, NY) with assistance from Brian McIntyre (Integrated Nanosystems Center, UR, Rochester, NY), and their input was greatly appreciated. We acknowledge the many helpful discussions and suggestions shared with Drs. Martin Pavelka and Brian Ward (URMC, Rochester, NY).
This study was supported by NIH/NIDCR DE013683 and DE017425 (to R.G.Q.), as well as the Training Program in Oral Sciences DE021985 (to C.J.K. and A.P.B.).
Footnotes
The authors declare no conflicts of interest.
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Ahn SJ & Burne RA, (2006) The atlA operon of Streptococcus mutans: role in autolysin maturation and cell surface biogenesis. J Bacteriol 188: 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA & Ferretti JJ, (2002) Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A 99: 14434–14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdic D & Pham VT, (2007) Global transcriptional analysis of Streptococcus mutans sugar transporters using microarrays. J Bacteriol 189: 5049–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre G, Deghorain M, Bron PA, van S II, Kleerebezem M, Hols P & Dufrene YF, (2011) Fluorescence and atomic force microscopy imaging of wall teichoic acids in Lactobacillus plantarum. ACS Chem Biol 6: 366–376. [DOI] [PubMed] [Google Scholar]
- Atilano ML, Pereira PM, Yates J, Reed P, Veiga H, Pinho MG & Filipe SR, (2010) Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc Natl Acad Sci USA 107: 18991–18996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj R, Bruce KE, Davidson AL, Rued BE, Stauffacher CV & Winkler ME, (2016) Biochemical characterization of essential cell division proteins FtsX and FtsE that mediate peptidoglycan hydrolysis by PcsB in Streptococcus pneumoniae. Microbiologyopen 5: 738–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JL, Derr AM, Karuppaiah K, MacGilvray ME, Kajfasz JK, Faustoferri RC, Rivera-Ramos I, Bitoun JP, Lemos JA, Wen ZT & Quivey RG Jr., (2014) Streptococcus mutans NADH oxidase lies at the intersection of overlapping regulons controlled by oxygen and NAD+ levels. J Bacteriol 196: 2166–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JL, Faustoferri RC & Quivey RG Jr., (2017) Acid-adaptive mechanisms of Streptococcus mutans-the more we know, the more we don’t. Mol Oral Microbiol 32: 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartual SG, Straume D, Stamsas GA, Munoz IG, Alfonso C, Martinez-Ripoll M, Havarstein LS & Hermoso JA, (2014) Structural basis of PcsB-mediated cell separation in Streptococcus pneumoniae. Nat Commun 5: 3842. [DOI] [PubMed] [Google Scholar]
- Biswas R, Martinez RE, Gohring N, Schlag M, Josten M, Xia G, Hegler F, Gekeler C, Gleske AK, Gotz F, Sahl HG, Kappler A & Peschel A, (2012) Proton-binding capacity of Staphylococcus aureus wall teichoic acid and its role in controlling autolysin activity. PLoS One 7: e41415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Santa Maria JP Jr. & Walker S, (2013) Wall teichoic acids of Gram-positive bacteria. Annu Rev Microbiol 67: 313–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Zhang YH & Walker S, (2008) A revised pathway proposed for Staphylococcus aureus wall teichoic acid biosynthesis based on in vitro reconstitution of the intracellular steps. Chem Biol 15: 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliot E, Dramsi S, Chapot-Chartier MP, Courtin P, Kulakauskas S, Pechoux C, Trieu-Cuot P & Mistou MY, (2012) Role of the Group B antigen of Streptococcus agalactiae: a peptidoglycan-anchored polysaccharide involved in cell wall biogenesis. PLoS Pathog 8: e1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J, Singh AK, Santa Maria JP Jr., Kim Y, Brown S, Swoboda JG, Mylonakis E, Wilkinson BJ & Walker S, (2011) Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem Biol 6: 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatfield CH, Koo H & Quivey RG Jr., (2005) The putative autolysin regulator LytR in Streptococcus mutans plays a role in cell division and is growth-phase regulated. Microbiology 151: 625–631. [DOI] [PubMed] [Google Scholar]
- Clinical Laboratory Standards Institute (2018) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—10th ed. M07-A11 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- D’Elia MA, Millar KE, Beveridge TJ & Brown ED, (2006a) Wall teichoic acid polymers are dispensable for cell viability in Bacillus subtilis. J Bacteriol 188: 8313–8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Elia MA, Pereira MP, Chung YS, Zhao W, Chau A, Kenney TJ, Sulavik MC, Black TA & Brown ED, (2006b) Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J Bacteriol 188: 4183–4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De A, Jorgensen AN, Beatty WL, Lemos J & Wen ZT, (2018) Deficiency of MecA in Streptococcus mutans Causes Major Defects in Cell Envelope Biogenesis, Cell Division, and Biofilm Formation. Front Microbiol 9: 2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De A, Liao S, Bitoun JP, Roth R, Beatty WL, Wu H & Wen ZT, (2017) Deficiency of RgpG causes major defects in cell division and biofilm formation, and deficiency of LCP proteins leads to accumulation of cell wall antigens in culture medium by Streptococcus mutans. Appl Environ Microbiol 83: e00928–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr AM, Faustoferri RC, Betzenhauser MJ, Gonzalez K, Marquis RE & Quivey RG Jr., (2012) Mutation of the NADH oxidase gene (nox) reveals an overlap of the oxygen- and acid-mediated stress responses in Streptococcus mutans. Appl Environ Microbiol 78: 1215–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RJ, van Hensbergen VP, Ruda A, Turner AG, Deng P, Le Breton Y, El-Sayed NM, Belew AT, McIver KS, McEwan AG, Morris AJ, Lambeau G, Walker MJ, Rush JS, Korotkov KV, Widmalm G, van Sorge NM & Korotkova N, (2019) Discovery of glycerol phosphate modification on streptococcal rhamnose polysaccharides. Nat Chem Biol 15: 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James DB & Yother J, (2012) Genetic and biochemical characterizations of enzymes involved in Streptococcus pneumoniae serotype 2 capsule synthesis demonstrate that Cps2T (WchF) catalyzes the committed step by addition of beta1–4 rhamnose, the second sugar residue in the repeat unit. J Bacteriol 194: 6479–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara J, Kiriyama Y, Miyashita M, Kondo T, Yamada T, Yazawa K, Yoshikawa R & Yamamoto H, (2016) Teichoic acid polymers affect expression and localization of dl-endopeptidase LytE required for lateral cell wall hydrolysis in Bacillus subtilis. J Bacteriol 198: 1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y, Marles-Wright J, Cleverley RM, Emmins R, Ishikawa S, Kuwano M, Heinz N, Bui NK, Hoyland CN, Ogasawara N, Lewis RJ, Vollmer W, Daniel RA & Errington J, (2011) A widespread family of bacterial cell wall assembly proteins. EMBO J 30: 4931–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs CJ, Faustoferri RC & Quivey RG Jr., (2017) RgpF is required for maintenance of stress tolerance and virulence in Streptococcus mutans. J Bacteriol 199: e00497–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Shi W & Qi F, (2005) Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol 187: 7193–7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Wang H, Labroli M, Koseoglu S, Zuck P, Mayhood T, Gill C, Mann P, Sher X, Ha S, Yang SW, Mandal M, Yang C, Liang L, Tan Z, Tawa P, Hou Y, Kuvelkar R, DeVito K, Wen X, Xiao J, Batchlett M, Balibar CJ, Liu J, Xiao J, Murgolo N, Garlisi CG, Sheth PR, Flattery A, Su J, Tan C & Roemer T, (2016) TarO-specific inhibitors of wall teichoic acid biosynthesis restore beta-lactam efficacy against methicillin-resistant staphylococci. Sci Transl Med 8: 329ra332. [DOI] [PubMed] [Google Scholar]
- Mattos-Graner RO, Jin S, King WF, Chen T, Smith DJ & Duncan MJ, (2001) Cloning of the Streptococcus mutans gene encoding glucan binding protein B and analysis of genetic diversity and protein production in clinical isolates. Infect Immun 69: 6931–6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattos-Graner RO, Porter KA, Smith DJ, Hosogi Y & Duncan MJ, (2006) Functional analysis of glucan binding protein B from Streptococcus mutans. J Bacteriol 188: 3813–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistou MY, Sutcliffe IC & van Sorge NM, (2016) Bacterial glycobiology: rhamnose-containing cell wall polysaccharides in Gram-positive bacteria. FEMS Microbiol Rev 40: 464–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Nomura R, Nakagawa I, Hamada S & Ooshima T, (2005) Role of glucose side chains with serotype-specific polysaccharide in the cariogenicity of Streptococcus mutans. Caries Res 39: 262–268. [DOI] [PubMed] [Google Scholar]
- Ng WL, Kazmierczak KM & Winkler ME, (2004) Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol Microbiol 53: 1161–1175. [DOI] [PubMed] [Google Scholar]
- Nomura R, Nakano K & Ooshima T, (2005) Molecular analysis of the genes involved in the biosynthesis of serotype specific polysaccharide in the novel serotype k strains of Streptococcus mutans. Oral Microbiol Immunol 20: 303–309. [DOI] [PubMed] [Google Scholar]
- Odds FC, (2003) Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother 52: 1. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Shibata Y, Yamashita Y, Nakano Y, Tsuda H & Koga T, (2002) A novel mechanism for glucose side-chain formation in rhamnose-glucose polysaccharide synthesis. FEBS letters 532: 159–163. [DOI] [PubMed] [Google Scholar]
- Pollack JH & Neuhaus FC, (1994) Changes in wall teichoic acid during the rod-sphere transition of Bacillus subtilis 168. J Bacteriol 176: 7252–7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivey RG Jr., Grayhack EJ, Faustoferri RC, Hubbard CJ, Baldeck JD, Wolf AS, MacGilvray ME, Rosalen PL, Scott-Anne K, Santiago B, Gopal S, Payne J & Marquis RE, (2015) Functional profiling in Streptococcus mutans: construction and examination of a genomic collection of gene deletion mutants. Mol Oral Microbiol 30: 474–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinscheid DJ, Ehlert K, Chhatwal GS & Eikmanns BJ, (2003) Functional analysis of a PcsB-deficient mutant of Group B streptococcus. FEMS Microbiology Letters 221: 73–79. [DOI] [PubMed] [Google Scholar]
- Reinscheid DJ, Gottschalk B, Schubert A, Eikmanns BJ & Chhatwal GS, (2001) Identification and molecular analysis of PcsB, a protein required for cell wall separation of Group B streptococcus. J Bacteriol 183: 1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovskaya I, Vinogradov E, Courtin P, Armalyte J, Meyrand M, Giaouris E, Palussiere S, Furlan S, Pechoux C, Ainsworth S, Mahony J, van Sinderen D, Kulakauskas S, Guerardel Y & Chapot-Chartier MP, (2017) Another brick in the wall: a rhamnan polysaccharide trapped inside peptidoglycan of Lactococcus lactis. MBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlag M, Biswas R, Krismer B, Kohler T, Zoll S, Yu W, Schwarz H, Peschel A & Gotz F, (2010) Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol Microbiol 75: 864–873. [DOI] [PubMed] [Google Scholar]
- Sham LT, Barendt SM, Kopecky KE & Winkler ME, (2011) Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. Proc Natl Acad Sci U S A 108: E1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham LT, Jensen KR, Bruce KE & Winkler ME, (2013) Involvement of FtsE ATPase and FtsX extracellular loops 1 and 2 in FtsEX-PcsB complex function in cell division of Streptococcus pneumoniae D39. MBio 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Ozaki K, Seki M, Kawato T, Tanaka H, Nakano Y & Yamashita Y, (2003) Analysis of loci required for determination of serotype antigenicity in Streptococcus mutans and its clinical utilization. J Clin Microbiol 41: 4107–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Yamashita Y, Ozaki K, Nakano Y & Koga T, (2002) Expression and characterization of streptococcal rgp genes required for rhamnan synthesis in Escherichia coli. Infect Immun 70: 2891–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Yamashita Y & van der Ploeg JR, (2009) The serotype-specific glucose side chain of rhamnose-glucose polysaccharides is essential for adsorption of bacteriophage M102 to Streptococcus mutans. FEMS Microbiol Lett 294: 68–73. [DOI] [PubMed] [Google Scholar]
- Shields RC, Zeng L, Culp DJ & Burne RA, (2018) Genomewide identification of essential genes and fitness determinants of Streptococcus mutans UA159. mSphere 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian XL, Dong G, Liu T, Gomez ZA, Wahl A, Hols P & Li YH, (2013) MecA protein acts as a negative regulator of genetic competence in Streptococcus mutans. J Bacteriol 195: 5196–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda H, Yamashita Y, Toyoshima K, Yamaguchi N, Oho T, Nakano Y, Nagata K & Koga T, (2000) Role of serotype-specific polysaccharide in the resistance of Streptococcus mutans to phagocytosis by human polymorphonuclear leukocytes. Infect Immun 68: 644–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Rijn I & Bleiweis AS, (1973) Antigens of Streptococcus mutans. I. Characterization of a serotype-specific determinant from Streptococcus mutans. Infect Immun 7: 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sorge NM, Cole JN, Kuipers K, Henningham A, Aziz RK, Kasirer-Friede A, Lin L, Berends ETM, Davies MR, Dougan G, Zhang F, Dahesh S, Shaw L, Gin J, Cunningham M, Merriman JA, Hutter J, Lepenies B, Rooijakkers SHM, Malley R, Walker MJ, Shattil SJ, Schlievert PM, Choudhury B & Nizet V, (2014) The classical lancefield antigen of Group A Streptococcus is a virulence determinant with implications for vaccine design. Cell Host Microbe 15: 729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Gill CJ, Lee SH, Mann P, Zuck P, Meredith TC, Murgolo N, She X, Kales S, Liang L, Liu J, Wu J, Santa Maria J, Su J, Pan J, Hailey J, McGuinness D, Tan CM, Flattery A, Walker S, Black T & Roemer T, (2013) Discovery of wall teichoic acid inhibitors as potential anti-MRSA beta-lactam combination agents. Chem Biol 20: 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Shibata Y, Nakano Y, Tsuda H, Kido N, Ohta M & Koga T, (1999) A novel gene required for rhamnose-glucose polysaccharide synthesis in Streptococcus mutans. J Bacteriol 181: 6556–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.