SUMMARY
Transporting epithelial cells generate arrays of microvilli, known as a brush border, to enhance functional capacity. To understand brush border formation, we used live cell imaging to visualize apical remodeling early in this process. Strikingly, we found individual microvilli exhibit persistent active motility, translocating across the cell surface at ~0.2 μm/min. Perturbation with inhibitors and photokinetic experiments revealed microvillar motility is driven by actin assembly at the barbed-ends of core bundles, which in turn is linked to robust treadmilling of these structures. Actin regulatory factors IRTKS and EPS8 localize to the barbed-ends of motile microvilli where they control the kinetics and nature of movement. As the apical surface of differentiating epithelial cells is crowded with nascent microvilli, persistent motility promotes collisions between protrusions and ultimately clustering and consolidation into higher order arrays. Thus, microvillar motility represents a previously unrecognized driving force for apical surface remodeling and maturation during epithelial differentiation.
Keywords: epithelium, treadmilling, bundle, protocadherin, adhesion
Graphical Abstract
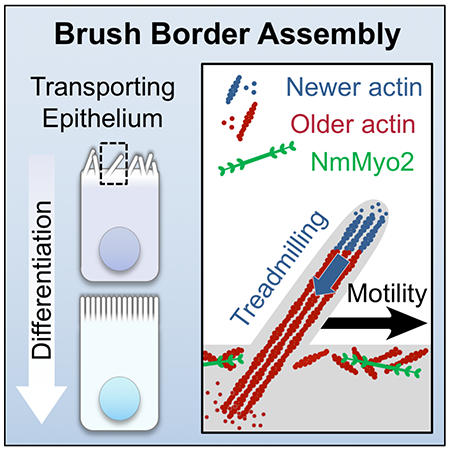
eTOC
Transporting epithelial cells assemble extensive apical arrays of microvilli, known as brush borders. Meenderink et al. use live cell imaging to reveal that nascent microvilli translocate across the cell surface. Cells harness this motility to consolidate microvilli into clusters and tightly packed arrays during differentiation.
INTRODUCTION
Microvilli are evolutionarily ancient cell surface protrusions (Sebe-Pedros et al., 2013) that consist of a core bundle of parallel actin filaments enveloped in plasma membrane. Epithelial cells have adapted this simple structure to fulfill physiologically diverse functions including mechanosensation in the inner ear sensory stereocilia (Schwander et al., 2010), chemosensation in the lung, gut, and urogenital tract (Gerbe and Jay, 2016; Krasteva and Kummer, 2012; Kummer and Deckmann, 2017), and solute uptake in the renal tubules (Coudrier et al., 1988) and intestine (Delacour et al., 2016). Most epithelial cells assemble densely packed apical arrays of microvilli referred to as ‘brush borders’ based on their appearance. Disruption of brush border function through inherited defects (Schneeberger et al., 2018), infectious causes (Vallance et al., 2002), or injury (Emlet et al., 2015) leads to a range of human diseases.
Detailed studies on intestinal epithelial brush borders have revealed that microvillar cores contain 20-30 parallel actin filaments with barbed-ends, the preferred site of monomer addition, oriented toward the distal tips and pointed-ends anchored in a terminal web (Mooseker and Tilney, 1975). Actin cores are stabilized by bundling proteins including villin (Bretscher and Weber, 1979), espin (Bartles et al., 1998), fimbrin (also known as T-plastin) (Bretscher and Weber, 1980), and potentially EPS8 (Hertzog et al., 2010). Additionally, core bundles are linked to the overlying plasma membrane by myosins (Tyska et al., 2005) and ERM proteins (Casaletto et al., 2011). More recent studies indicate that microvillar growth and elongation are regulated by the WH2 domain protein COBL and I-BAR domain protein IRTKS (Grega-Larson et al., 2015; Postema et al., 2018; Wayt and Bretscher, 2014).
Recent work also offers some insight on factors that organize microvilli in mature brush borders. We now know that microvillar packing is driven by an intermicrovillar adhesion complex (IMAC) that forms between the tips of adjacent protrusions (Crawley et al., 2014b). The IMAC is composed of the interacting cadherins CDHR2 (protocadherin-24) and CDHR5 (mucin-like protocadherin), which are positioned at the distal tips through an interaction with the cytoplasmic tripartite assembly of USH1C (also known as harmonin), ANKS4B, and MYO7B (Crawley et al., 2014b; Crawley et al., 2016; Weck et al., 2016). Ultrastructural analysis of Caco-2BBE cells through stages of differentiation also revealed that individual microvilli first form small tepee-shaped clusters, which progressively increase in size (i.e. number of microvilli) until the array fills the entire apical surface, becoming a mature brush border (Crawley et al., 2014b). How adherent clusters of microvilli form and whether adhesion complexes form before or after microvillar growth remains unknown.
Despite growing knowledge and identification of factors that contribute to the assembly of microvilli, how cells organize these structures into mature brush borders remains poorly understood, primarily due to a lack of time-resolved data on apical surface remodeling during this process. Using lattice light-sheet microscopy (LLSM) and spinning disk confocal microscopy (SDCM) to characterize the early steps of brush border maturation, we found that individual microvilli exhibit persistent, active motility. Microvillar movement is driven by actin assembly at microvillar tips, which in turn promotes robust treadmilling that propels protrusions across the cell surface. Although myosin contractility does not drive microvillar motility, it does control the length of motile structures. IRTKS and EPS8 both localize to the barbed ends of motile microvilli and regulate motility via their actin-binding domains. Importantly, we found that microvillar motility promotes collisions and clustering of individual protrusions as well as their consolidation into higher order arrays. This work identifies microvillar motility as a driving force for apical remodeling during epithelial differentiation.
RESULTS
Microvilli exhibit persistent, active motility.
To examine the behavior of microvilli early in differentiation, we imaged the apical surface of LLC-PK1 clone 4 (CL4) cells, which are derived from porcine renal proximal tubules (Hull et al., 1976) and have been used in past studies of microvillar biology (Bartles et al., 1998; Tyska and Mooseker, 2002). Structured illumination microscopy (SIM) of CL4 cells one day post confluence (1 DPC) revealed large numbers of plasma membrane-wrapped linear actin bundles on the apical surface, consistent with nascent microvilli (Figure S1A–C). To gain insight on the dynamics of microvillar growth and brush border maturation, we used LLSM (Chen et al., 2014)(Figure 1A–C) and SDCM (Figure 1C–E) to image cells expressing fluorescent protein-tagged espin, which targets specifically to the parallel actin bundles that support microvilli. Nascent microvilli exhibit a continuum of orientations, ranging from parallel to orthogonal to the cell surface (Figure 1D). Of note, espin was selected for these studies because it localizes along the length of microvilli and has limited impact on actin turnover dynamics (Loomis et al., 2003). However, we did observe microvillar lengthening similar to that reported by others in cells expressing high levels of tagged espin. Therefore, for time-lapse imaging we selected cells expressing low levels of espin, and under these conditions maximum length was no different compared to cells expressing the F-actin probe, EGFP-Lifeact (Figure S1D and Figure 1J).
Figure 1. Microvilli exhibit persistent, active motility.
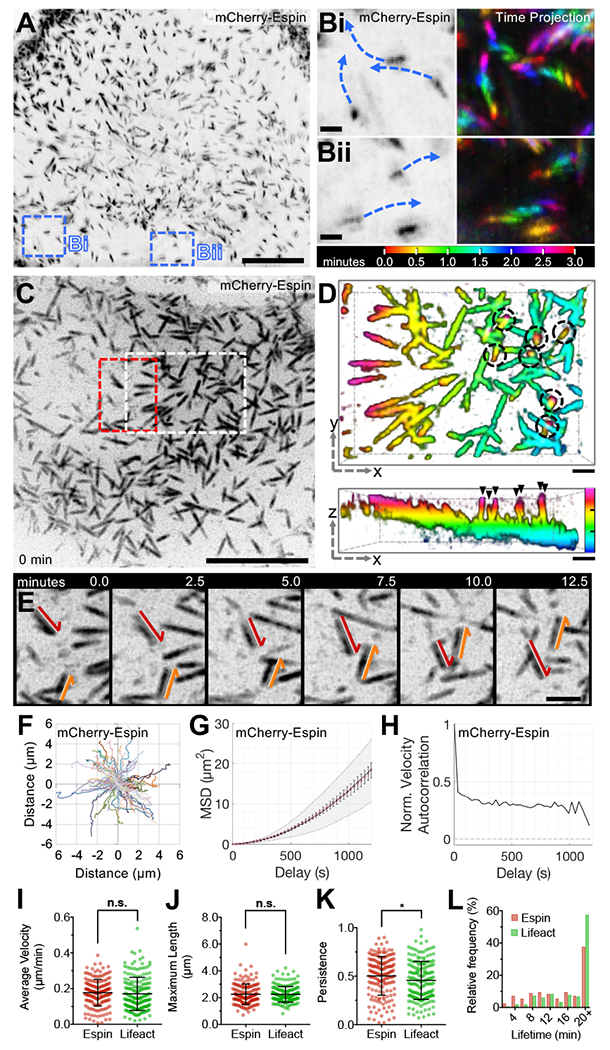
(A) LLSM of a CL4 cell stably expressing mCherry-Espin, reconstructed then viewed as a maximum intensity z-projection. Scale bar 10 μm, blue dashed boxes corresponds to B. (B) Enlarged images from A (left) with time projections (right) showing microvillar movement over 3 minutes. (C) SDCM of the apical surface of a CL4 cell stably expressing mCherry-Espin, viewed as a maximum intensity z-projection. Scale bar 10 μm, white dashed box corresponds to D, red dashed box corresponds to E. (D) 3-dimensional (3D) depth color coded z-stack viewed en face (xy plane, upper panel) or laterally (xz plane, lower panel). Scale bars are 1 μm, z-axis depth color code (lower panel) to scale with tick marks at 1 μm intervals. Microvilli exhibit a range of orientations from parallel (represented by a single color) to perpendicular to the cell surface (spanning multiple color bands, circled—top panel, arrowheads—bottom panel). (E) Time series of microvilli translocating across the cell surface; red and orange arrows highlight the paths of two distinct protrusions. Scale bar 2 μm. (F) Rose plot of trajectories measured from the tips of microvilli (n = 101) for the cell in C. (G) Microvillar trajectories from F were subject to MSD analysis; red open circles represent the mean MSD, error bars indicate standard error of the mean (SEM), grey area marks the weighted standard deviation (SD) over all MSD curves, and the solid line indicates a fit of the data to an active movement model (diffusion coefficient, D = 0.000283 μm2/s and velocity, V = 0.21 μm/min). (H) Trajectories from F were analyzed for normalized velocity autocorrelation, solid line. Dotted line at 0 indicates the velocity autocorrelation of random diffusive movement. (I-K) Average microvillar velocity, maximum microvillar length, and persistence, respectively. (L) Microvillar lifetime frequency histogram. Error bars indicate mean ± SD, (I-L) n = 171 microvilli from 7 cells for mCherry-Espin, n = 183 microvilli from 6 cells for EGFP-Lifeact, * p < 0.05, n.s. = not significant.
Strikingly, time-lapse imaging of CL4s revealed that microvilli are highly motile with actin core bundles translocating across the cell surface (Figure 1B,E and Supplemental Videos 1 and 2) at a mean velocity of 0.18 μm/min (Figure 1I). This rate is similar to microvillar growth rates reported for Xenopus kidney epithelial A6 cells and mouse cells cultured from the inner ear (Gorelik and Gautreau, 2014), but is substantially lower than rates reported for filopodial extension (≥1 μm/min) (Mallavarapu and Mitchison, 1999). To investigate potential patterns of motility, we tracked large numbers of protrusions. Rose plot analysis of the resulting trajectories did not reveal obvious flow patterns or points of nucleation (Figure 1F). However, mean square displacement (MSD) analysis generated a parabolic curve consistent with active motility (Figure 1G). This was further supported by a positive velocity autocorrelation function (Figure 1H), where purely diffusive movement exhibits a decay plateau around 0 and active movement greater than 0. Importantly, microvilli exhibited similar motile properties in CL4 cells expressing EGFP-Lifeact (Figure 1I–L, Figure S1D–H), although our remaining experiments took advantage of the specificity and enhanced signal/noise offered by the espin probe. Importantly, we also observed similar movement of microvilli on the apical surface of CACO-2BBE cells (Figure S1I–L), a human intestinal epithelial cell line used previously in studies of brush border biology (Peterson and Mooseker, 1992).
To further characterize the structure of motile microvilli, we first co-expressed a membrane-targeting GPI-GFP probe to examine the extent of membrane encapsulation in translocating protrusions. Motile microvilli exhibit membrane-wrapping along a significant fraction of their length (Figure S2A,B, see Supplemental Video 3), and scanning EM (SEM) confirmed that these structures were bona fide membrane protrusions (Figure S2C,D). Scanning electron microscopy (SEM) images taken en face and at a 35° angle relative to the sample indicate that most protrusions during this timepoint extend at a low angle relative to the apical surface. Additionally, transmission electron microscopy (TEM) through the long-axis of individual microvilli demonstrates clear membrane wrapping even when microvilli appear parallel to the cell surface (Figure S2E). Motile microvilli were also robustly labeled along their length by a tagged form of Myo1a, a membrane-binding actin-based motor previously implicated in stabilizing membrane-cytoskeleton interactions within brush border microvilli (Figure S2F–H) (Nambiar et al., 2009; Tyska et al., 2005). Together these results indicate that microvilli on differentiating epithelial cells are highly motile, exhibiting directed movement across the apical cell surface.
Microvillar motility is driven by actin assembly.
Motility of subcellular structures can be driven passively by diffusion, or actively by cytoskeletal polymerization or molecular motors. Motion analysis of microvilli (Figure 1F–H) indicates active movement rather than a diffusive process. Interestingly, the rootlets of epithelial microvilli extend from a meshwork of cytoskeletal filaments in the sub-apical cortex known as the terminal web (Fath et al., 1990; Hirokawa and Heuser, 1981; Mooseker and Tilney, 1975). Non-muscle myosin-2 (NmMyo2) was first localized to the terminal web decades ago (Bretscher and Weber, 1978; Mooseker and Tilney, 1975) and more recent proteomic studies indicate that non-muscle myosin-2c (NmMyo2c) is the most abundant isoform (McConnell et al., 2011). Indeed, co-expression of a tagged form of NmMyo2c in CL4 cells revealed robust targeting to the sub-apical cortex beneath motile microvilli (Figure S2I,J). Based on this localization, we evaluated the role of NmMyo2 contractility in microvillar motility. For these experiments, CL4 cells were imaged as described above and, after 5 minutes, we added 20 μM blebbistatin to inhibit NmMyo2 (Supplemental Video 4, left panel). After addition of drug, microvilli continued to translocate across the cell surface (Figure 2A–E). Trajectory analysis of microvilli on blebbistatin-treated cells showed a lower velocity (Figure 2K), but the overall pattern of motion, MSD plots, and autocorrelation outputs were consistent with controls (Figure 2B–E). Interestingly, maximum microvillar length increased significantly during Blebbistatin treatment (Figure 2L), which suggests that under normal conditions, NmMyo2 may contribute to actin core turnover that serves to limit the length of these protrusions.
Figure 2. Microvillar motility is driven by actin assembly and is not dependent on myosin contractility.
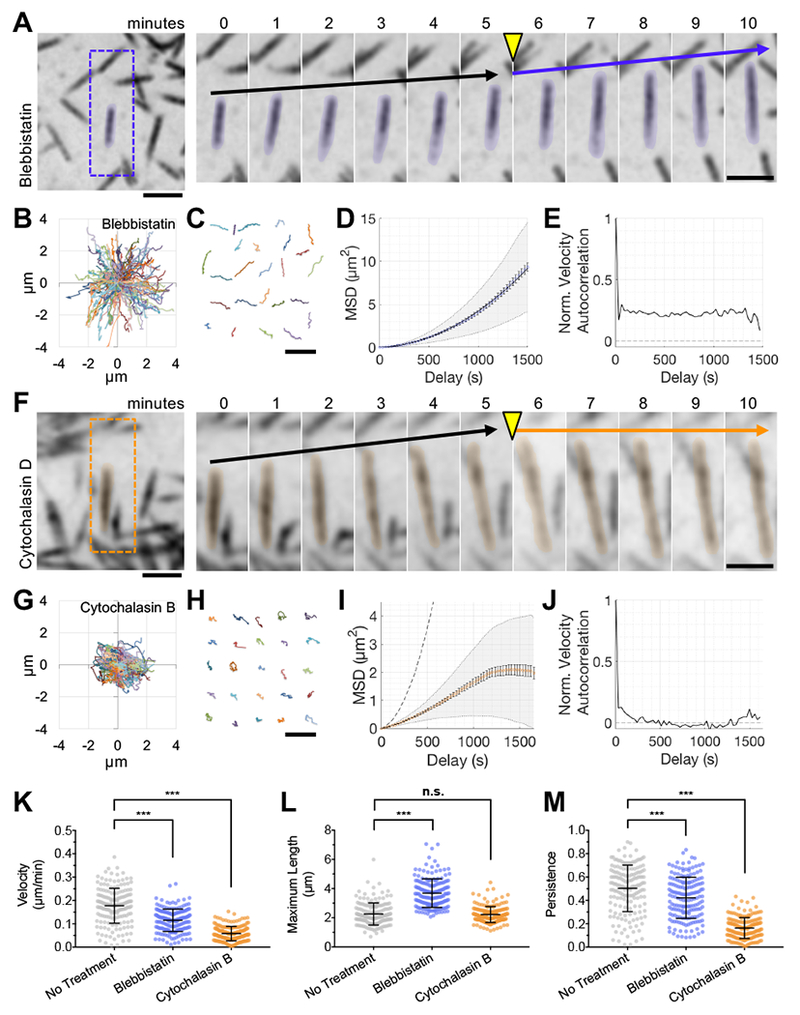
(A) SDCM of the apical surface of CL4 cells stably expressing mCherry-Espin showing the response to 20 μM Blebbistatin. Right, time series montage of a single protrusion highlighted with a 10% pseudo-colored overlay. Scale bars are 2 μm. Drug was added following the 5-minute time interval, yellow arrowhead. Black arrows indicate the baseline rate of microvillar movement. Blue arrow indicates the rate of microvillar movement after the addition of drug. (B) Rose plot shows the microvillar trajectories (n = 100) from a single cell treated with 20 μM Blebbistatin. (C) 25 representative microvillar trajectories are isolated for display. Scale bar is 5 μm. (D) MSD analysis of microvillar trajectories from B. (E) Normalized velocity autocorrelation analysis of microvillar trajectories from B; data were fit to an active movement model with D = 0.000093 μm2/s, V = 0.12 μm/min. (F) SDCM of the apical surface of CL4 cells stably expressing mCherry-Espin showing the response to 30 μM Cytochalasin D. Right, time series montage of a single protrusion highlighted with a 10% pseudo-colored overlay. Scale bars are 2 μm. Drug was added following the 5-minute time interval, yellow arrowhead. Black arrows indicate the baseline rate of microvillar movement. Orange arrow indicates the rate of microvillar movement after the addition of drug. (G) Rose plot shows the microvillar trajectories (n = 100) from a single cell treated with 500 nM Cytochalasin B. (H) 25 representative microvillar trajectories are isolated for display. Scale bar is 5 μm. (I) MSD analysis of microvillar trajectories from G; data could not be fit with an active movement model. The curve for mCherry-Espin with no drug treatment is plotted for comparison (grey dotted line). (J) Normalized velocity autocorrelation analysis of microvillar trajectories from G. (K-M) Average microvillar velocity, maximum microvillar length, and persistence, respectively, measured from untreated cells (from Figure 1), or cells exposed to Blebbistatin or Cytochalasin B. For Blebbistatin and Cytochalasin datasets, n = 178 and 175 microvilli, respectively, from 6-7 cells. Bars represent mean ± SD. *** p < 0.0001, n.s. = not significant.
We next sought to test the possibility that microvillar motion is powered by actin polymerization. Remarkably, microvillar translocation halted immediately after addition of Cytochalasin D (Figure 2F) or B (Figure 2G–J, see Supplemental Video 4, right panel). For detailed trajectory analysis, we used 500 nM Cytochalasin B, which relative to Cytochalasin D had less impact on overall cell shape and facilitated tracking of individual microvilli, although both drugs inhibited microvillar motion. Trajectory analysis showed that these microvilli no longer exhibited persistent motion (Figure 2M), although they continued to exhibit slow, short range displacements over small areas (Figure 2G,H,K). MSD and autocorrelation analysis indicated behavior consistent with constrained diffusion (Figure 2I,J). Together these data indicate that actin assembly at the barbed-ends of microvillar core bundles is required for the active motility of these structures across the apical surface.
Microvillar F-actin cores undergo treadmilling during motility.
The robust inhibition of microvillar movement by Cytochalasin suggests a model where actin incorporation at the distal tips may provide a driving force for the translocation of microvilli. However, because microvilli maintain a relatively stable steady-state length, any barbed-end actin incorporation must be matched by disassembly from the pointed-ends of core bundles. Such a ‘treadmilling’ process has been observed in microvilli before (Loomis et al., 2003; Tyska and Mooseker, 2002), although its relationship to microvillar motility has not been explored. To determine if microvillar core bundles exhibit treadmilling during motility, we expressed mNEON-Green-β-actin in CL4 cells to allow for direct monitoring of actin turnover. Cells were imaged using SDCM as before, however, in this experiment we also photobleached fiduciary marks within translocating microvilli (Figure 3). To visualize the fate of fiduciary marks, images were then processed using Imaris to create three-dimensional (3D) surfaces representing apical microvilli (Figure 3B–D, see Supplemental Video 5). We found that after creating a fiduciary bleach mark at time 0, microvilli continue to translocate over the cell surface. However, we noted a marked expansion of the actin signal at leading end of the protrusion (distal to the bleach mark) up to 1.1 μm/min, which was matched by a gradual loss of signal at the trailing end (Figure 3E). This pattern of turnover indicates that microvillar actin cores treadmill as they translocate across the cell surface with their barbed-ends in the lead.
Figure 3. Microvillar F-actin cores undergo treadmilling during motility.
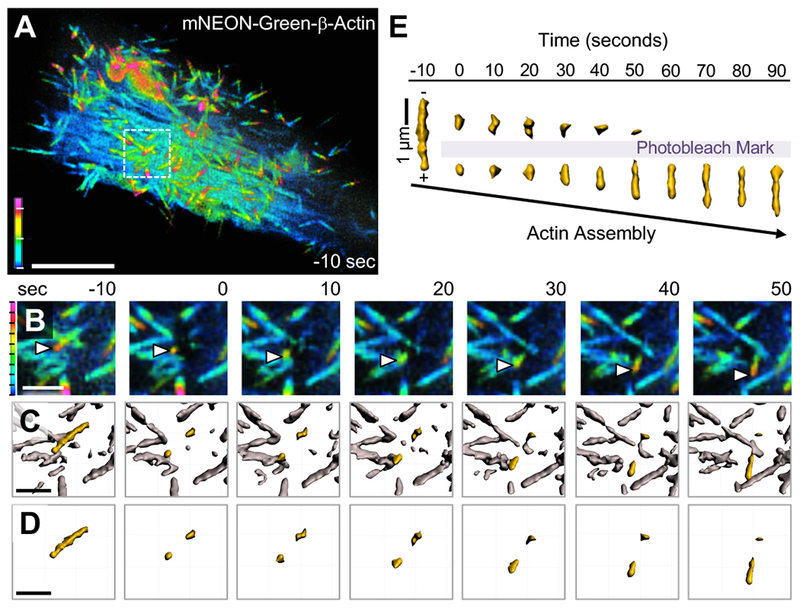
(A) SDCM of a CL4 cell expressing mNEON-Green-β-actin viewed as a depth-coded z-projection. Scale bar is 10 μm, z-axis depth code with tick marks at 1 μm intervals is shown at lower left. (B) Time series montage of area in A highlighted by white dashed box, enlarged, cropped in z to remove cytoplasmic signal, then viewed as a depth-coded z-projection. Time series shows region of interest before photobleaching (−10 seconds), immediately after photobleaching (0 seconds), and during recovery. White arrowheads mark the growing distal tip of a microvillus. (C) Data from B were processed using Imaris to create a 3D surface of the fluorescent actin signal with the microvillus of interest highlighted in yellow. (D) Time series montage showing the isolated microvillus of interest before and after photobleaching. In B-D, scale bar is 2 μm, z-axis depth code (left) with tick marks at 200 nm intervals. (E) The microvillus of interest from D was viewed orthogonally over time and aligned based on the position of the bleached region to highlight treadmilling of the mark through the actin core. Shown here is a representative example of treadmilling observed from n > 20 cells.
Microvillar motility is regulated by barbed-end binding factors.
Given that microvillar motility is inhibited by barbed-end capping (Figure 2F–M) and the barbed-ends of core bundles lead during movement (Figure 3E), we sought to determine if actin regulatory factors that target to the distal tips of microvilli control the motility of these protrusions. Recent work indicates that IRTKS (also known as BAIAP2L1) targets to the distal tips of epithelial microvilli and promotes their elongation (Postema et al., 2018). IRTKS contains an N-terminal I-BAR motif that interacts with outwardly curved membranes, a central SH3 domain, and a C-terminal WH2 domain that binds actin and is needed to promote elongation (Millard et al., 2007; Postema et al., 2018; Yamagishi et al., 2004). When co-expressed with tagged-Espin in CL4 cells, a tagged form of IRTKS localized in dynamic puncta that mapped to the growing tips of motile microvilli (arrowheads in Figure 4A, Supplemental Video 6, top left) and to a lesser extent along the length of core actin bundles. IRTKS overexpression significantly increased microvillar length and velocity (Figure 4E,F), but dramatically reduced overall protrusion lifetime (Figure 4H). A construct lacking the actin-binding WH2 domain (IRTKSΔWH2, Supplemental Video 6, top right) exhibited reduced tip enrichment (Figure 4B) and had little impact on protrusion lifetime relative to cells expressing IRTKS or espin alone (Figure 4H). Interestingly, MSD analysis showed that IRTKSΔWH2 significantly increased the diffusivity of microvillar motion (Figure 4I–L).
Figure 4. Microvillar motility is regulated by barbed-end binding factors.
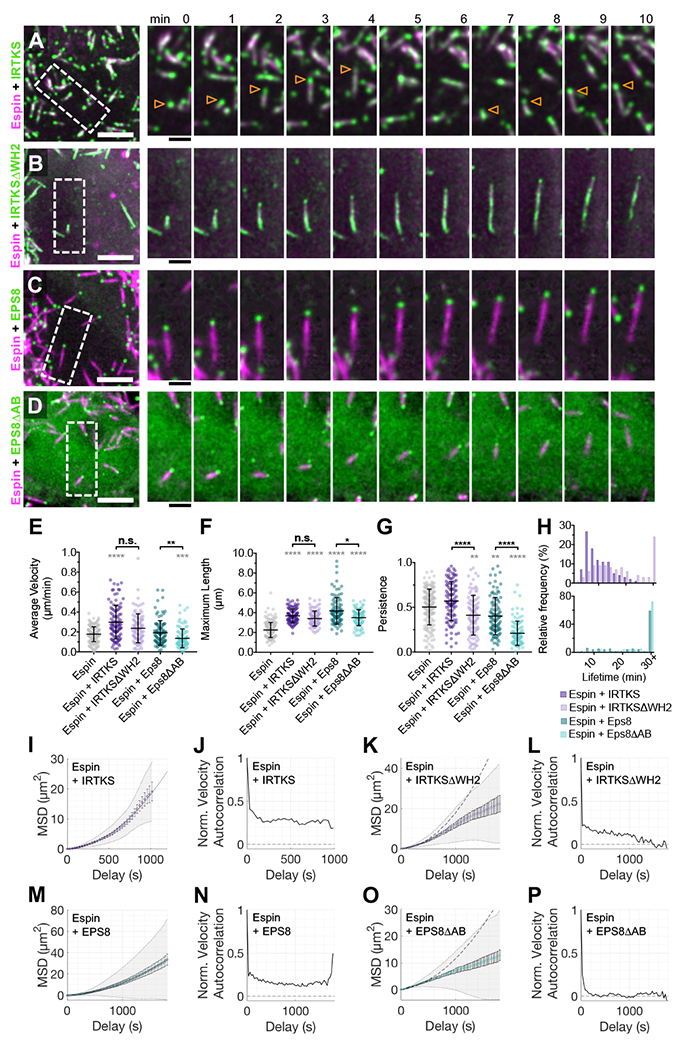
SDCM of the apical surface of CL4 cells stably expressing mCherry-Espin plus either EGFP-IRTKS (A), EGFP-IRTKSΔWH2 (B), EGFP-EPS8 (C), or EGFP-EPS8ΔAB (D). Scale bars are 5 μm. Right, time series montage of individual protrusions. Scale bars are 2 μm. Arrowheads in (A) indicate individual protrusions. (E-G) Average microvillar velocity, maximum microvillar length, and persistence, respectively. (H) Microvillar lifetime frequency histograms. Data for cells expressing mCherry-Espin alone (grey, from Figure 1) shown for comparison. Datasets for dual expression, n = 101 microvilli from 4 cells (Espin+IRTKS), 100 microvilli from 5 cells (Espin+IRTKSΔWH2), 101 microvilli from 5 cells (Espin+EPS8), and 100 microvilli from 7 cells (EPS8ΔAB). Bars represent mean ± SD. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, and n.s. = not significant. Grey asterisks (****) indicate significance compared to expression of mCherry-Espin alone (first column). The microvillar trajectories from E were analyzed for MSD (I, K, M, O) and velocity autocorrelation (J, L, N, P). Normalized velocity autocorrelation shown as a solid line. The dotted line at 0 indicates the velocity autocorrelation of random diffusive movement. MSD data showed a positive curvature and were fit to an active movement model and with D = 0.000276 μm2/s, V = 0.25 μm/min for (I) Espin+IRTKS and D = 0.000715 μm2/s, V = 0.18 μm/min for (M) Espin+EPS8. The curves for wildtype IRTKS (I) and EPS8 (M) are plotted for comparison (grey dotted line) on the graphs for their respective mutants (K and O).
We next examined the involvement of EPS8, which contains an N-terminal split PH domain, a central SH3 domain, and a C-terminal actin binding domain that can bundle and cap filaments in vitro (Hertzog et al., 2010). EPS8 also uses a tandem polyproline motif to bind to the SH3 domain of IRTKS and the resulting complex exerts a potent elongation effect on epithelial microvilli (Postema et al., 2018). Other previous reports also implicate EPS8 in protrusion length control (Croce et al., 2004; Disanza et al., 2006; Manor et al., 2011). A tagged version of EPS8 exhibited highly specific targeting to the tips of motile microvilli (Figure 4C, Supplemental Video 6, lower left), where it increased protrusion length (Figure 4F). Deletion of the C-terminal actin binding motif (EPS8ΔAB, Figure 4D, Supplemental Video 6, lower right) reduced microvillar velocity, length, and most significantly, directional persistence relative to cells expressing full length EPS8 (Figure 4E–H). Co-expression of Espin with EPS8ΔAB also significantly increased the diffusivity of microvillar motion (Figure 4M–P). Together these results indicate that tip-targeted factors IRTKS and EPS8 promote the directed motion of microvilli, and also regulate the directional persistence and lifetime of the underlying actin bundles.
Microvillar motility promotes intermicrovillar collisions, adhesion, and cluster formation.
To understand if microvillar translocation contributes to organizing protrusions during brush border maturation, we studied CL4 cells later in differentiation (2 DPC). Specifically, we selected cells with a mixed population of individual microvilli and small clusters of microvilli. Imaging cells over several hours revealed that microvilli continue to translocate and are highly dynamic, even at this later time point (Figure 5A, Supplemental Video 7). Time-lapse data also revealed that microvillar motility promotes collisions between neighboring protrusions and these encounters can result in the formation of small clusters (arrowheads in Figure 5B, Supplemental Video 8, left panel). Remarkably, even groups of microvilli in adherent clusters retain their ability to translocate across the cell surface, eventually colliding with other clusters and consolidating their numbers (Figure 5C, Supplemental Video 8, right panel). Using a correlative live imaging/fixed immunostaining protocol, we found CDHR5 puncta along the length of motile microvilli and at their distal tips (Figure 5D). Moreover, in live cells, a tagged form of CDHR2 was also enriched in motile microvilli (Figure S3A), suggesting that these structures hold adhesive potential. SEM of these samples confirmed intermicrovillar adhesion link formation between protrusions (Figure S3B,C). Later in differentiation, we also noted that microvilli frequently move across the cell surface until colliding with protrusions at the cell margins (Figure 5H, Supplemental Video 9). Moreover, microvilli at the margins are organized in large and structurally-stable clusters, as they persist in size and relative position for up to 12 hours (Supplemental Video 9). Thus, these live cell observations indicate that microvillar motility provides the driving force for intermicrovillar adhesion and microvillar clustering during brush border maturation.
Figure 5. Microvillar motility promotes intermicrovillar collisions, adhesion, and cluster formation.
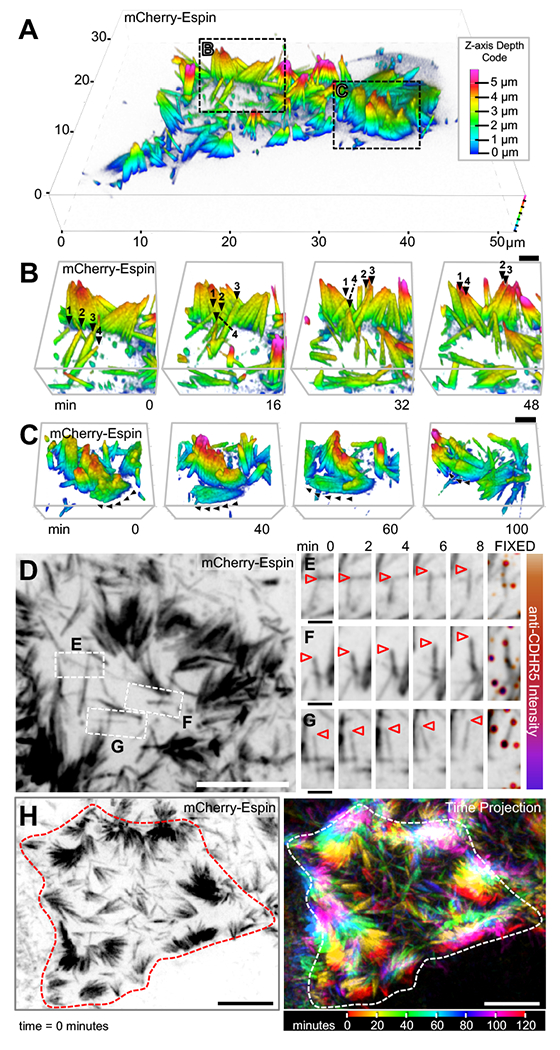
(A) SDCM of the apical surface of a CL4 cell stably expressing mCherry-Espin at 2 DPC, visualized as a depth-coded composite. Image scale is shown on volume frame, depth scale is shown to the right. (B,C) Times series montages of areas highlighted in A, enlarged and rotated to optimize visualization of microvillar movement. In B, individual microvilli of interest are marked at each time point with numbered black arrowheads. In C, black arrowheads mark the path of motion for a cluster of microvilli. Scale bars are 2 μm. Z-axis depth code shown in A also applies to B and C. (D) SDCM of the apical surface of a CL4 cell stably expressing mCherry-Espin at 2 DPC, visualized as a maximum intensity z-projection. Image scale is 10 μm. (E-G) Time series montages of areas highlighted in D, enlarged and rotated to visualize microvillar movement, then fixed and stained for CDHR5 (far right panel). Tips of individual microvilli are marked with arrowheads. Image scales are 2 μm. (H) SDCM of the apical surface of a CL4 cell stably expressing mCherry-Espin at 2 DPC, visualized as a maximum intensity z-projection at time = 0 (left panel) and as a time projection over 120 minutes (right panel). Time scale at lower right. Image scale is 10 μm. Cell edge at time = 0 minutes outlined in red (left panel) or white (right panel).
DISCUSSION
Here we report that microvilli undergo active movement across the apical surface of differentiating epithelial cells. Microvillar motion is driven by actin assembly at the barbed-ends of core bundles and linked to treadmilling of the bundle as structures translocate. Although previously unreported, this robust activity is harnessed by differentiating epithelial cells to organize and consolidate nascent microvilli into tightly packed arrays. Indeed, our time-lapse data provide clear evidence that active motility forces collisions between protrusions, resulting in the formation of larger microvillar clusters (Figure 6A). These data also indicate that nascent microvilli do not emerge from the apical surface in adherent assemblies, but rather elongate from the surface and then interact following their initial growth to form adherent clusters. By identifying a mechanism that serves to initiate and amplify the formation of microvillar clusters, these discoveries provide strong support for the progressive clustering model proposed previously (Crawley et al., 2014a) and refine our understanding of the activities that culminate in a mature brush border (Figure 6A).
Figure 6. A model for microvillar motility in brush border assembly.
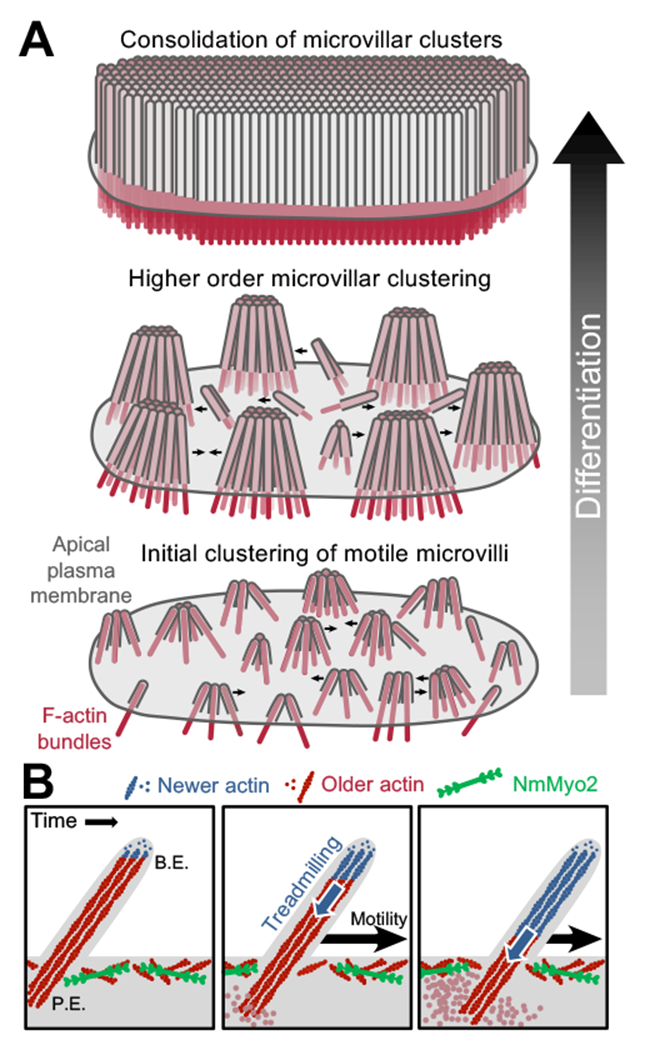
(A) A progressive clustering model for microvillar remodeling and brush border formation during differentiation. Here, nascent microvilli emerge from the apical surface and undergo persistent motility, which promotes collisions between protrusions. These encounters allow adhesion links to form between microvillar tips, resulting in characteristic tepee-shaped structures (originally reported in Crawley et al. Cell 2014). As maturation proceeds, clusters grow by moving across the apical surface, colliding with other clusters and consolidating their numbers until eventually the entire surface is occupied by one continuous large-scale cluster, i.e. a mature brush border. (B) Enlarged view of a single motile microvillus. The microvillar core is comprised of bundled F-actin (red) with the barbed-ends oriented toward the distal tip. New actin monomers incorporate at the barbed-ends (B.E., blue F-actin), which is balanced by monomer disassembly from the pointed-ends (P.E.). This results in ‘treadmilling’ of actin through the microvillar core which provides a pushing force against the membrane that powers microvillar motility. NmMyo2 is localized at the sub-apical cortex and may interact with microvilli rootlets to provide a counterforce translating microvillus actin core treadmilling into movement across the cell surface.
Actin filaments exhibit distinct assembly/disassembly kinetics at their two ends, with the barbed-ends being the preferred site for assembly (Pollard and Cooper, 2009). When monomer incorporation at the barbed-end is balanced by disassembly at the pointed-end, subunits can treadmill through the filament while a steady length is maintained. Polymer treadmilling is a well-established mechanism for generating mechanical force in diverse cellular processes ranging from chromosome separation in mitosis (Mitchison, 1989), to leading edge protrusion in cell motility (Wang, 1985), and pathogen movement during infection (Pantaloni et al., 2001). Actin monomer insertion at the barbed-end of a single filament can produce forces in the piconewton range (Footer et al., 2007; Prass et al., 2006), deforming associated membrane structures (Giardini et al., 2003; Upadhyaya et al., 2003). With their exposed barbed-ends enveloped in plasma membrane, microvillar actin bundles are ideally positioned to generate a protrusive force at microvillar tips (Figure 6B). Our data show clear treadmilling of actin cores in translocating microvilli, linking this mechanism of force generation to microvillar motility.
Forces generated by monomer insertion at the barbed-ends can only be converted to directed motion if they are countered at the pointed-end of the bundle. We propose that interactions between the bundle rootlet and the NmMyo2-rich sub-apical cortex provide these counterforces (Figure 6B). Moreover, while our studies indicate that NmMyo2 is not essential for powering the motility of microvilli, the elongation of these structures after blebbistatin application suggests that this motor plays a role in regulating their steady-state length. Myosin-2 was localized to the terminal web in classic studies (Bretscher and Weber, 1978; Mooseker and Tilney, 1975) and we were able to recapitulate such targeting with a tagged variant of NmMyo2c. Moreover, previous work implicates this motor in regulating microvillar orientation (Temm-Grove et al., 1992). The mechanism driving microvillar elongation under these conditions remains unclear, but myosin may assist with turnover of F-actin at the pointed-ends, which would be reminiscent of contractility-driven actin turnover observed in other systems (Medeiros et al., 2006; Wilson et al., 2010). Future studies examining the organization of terminal web myosin-2 and its attachment to microvillar actin cores will be needed to fully appreciate the role of this motor in microvillar dynamics and brush border assembly.
As proteins that both contain N-terminal membrane-interacting and C-terminal actin-binding domains, IRTKS and EPS8 are uniquely situated to link the barbed-ends of core actin bundles to the overlying plasma membrane. EPS8 has been studied in a variety of biological contexts, including transporting epithelial microvilli, sensory epithelial stereocilia, and crawling cell filopodia (Croce et al., 2004; Disanza et al., 2006; Manor et al., 2011; Postema et al., 2018). In all cases, this factor targets specifically to the distal tips of actin core bundles, and loss-of-function studies indicate a role in protrusion elongation (Croce et al., 2004; Manor et al., 2011; Postema et al., 2018). Recent studies from our group also establish that EPS8 is a binding partner of IRTKS at microvillar tips; together these factors form a complex that promotes the elongation of microvilli (Postema et al., 2018). In the present study, we found that IRTKS and EPS8 both contribute to maintaining the directed motion of nascent microvilli early in differentiation. Indeed, overexpression of dominant negative IRTKS or EPS8 constructs lacking their respective actin binding sites (IRTKSΔWH2 and EPS8ΔAB) reduced directional persistence and markedly increased the diffusivity of motion (Figure 4E–P). One possible explanation of these findings is that these factors help to maintain continuous attachment between the barbed-end of the bundle and the plasma membrane. Alternatively, or in parallel, the IRTKS/EPS8 complex might serve an anti-capping function, or facilitate processive monomer incorporation, similar to that reported for Ena/VASP in filopodia (Winkelman et al., 2014). This latter possibility is consistent with our finding that IRTKS overexpression significantly increases the velocity of microvillar motion, and IRTKS and EPS8 overexpression increases motile bundle length through an actin-binding dependent mechanism (Figure 4E,F).
Finally, the findings we present here hold implications for multiple organ systems. Developing a deeper understanding of molecular mechanisms driving microvillar motility should lead to insight, not only into epithelial brush border formation, but also into the morphogenesis of other complex apical epithelial specializations, such as the inner ear stereocilia. Indeed, early studies of inner ear development showed many years ago that the intricate staircase arrangement of stereocilia on the apical surface of mechanosensory hair cells initially arises from an array of much smaller microvilli (Tilney et al., 1992). Future studies will need to determine if the motile activity reported in this paper plays a role in this remodeling event or the subsequent polarized positioning of the hair bundle on the apical cell surface.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Matthew J. Tyska (matthew.tyska@vanderbilt.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Culture
LLC-PK1-CL4 (CL4), CACO-2BBE, and HEK293FT cells were cultured at 37°C and 5% CO2 in DMEM with high glucose and 2 mM L-glutamine supplemented with 10% fetal bovine serum (FBS) except for CACO-2BBE cells which were supplemented with 20% FBS.
METHOD DETAILS
Constructs
The pmCherry-Espin construct was a kind gift from Dr. James Bartles. pEGFP-Espin was generated by replacing mCherry with EGFP. The pGL-GPI-GFP was provided by the Dr. Anne Kenworthy (University of Virginia). The pEGFP-Myo1a as described in (Tyska and Mooseker, 2002). The EGFP-Lifeact construct was provided by Dr. Irina Kaverina (Vanderbilt University). The pEGFP-NmMyo2c was purchased from Addgene, plasmid #10843. The mNEON-green-β-actin was purchased from Allele Biotechnology. The pEGFP-C1-IRTKS, pEGFP-C1-IRTKSDWH2, and pEGFP-C1-EPS8 as described in (Postema et al., 2018). The pEGFP-C1-EPS8ΔAB construct (amino acids 1-648) was generated via PCR using EGFP-C1-EPS8 as a template. The PCR product was TOPO cloned into the pCR8/GW/TOPO vector (Invitrogen), and then shuttled into the EGFP-C1 backbone (Clontech), adapted for Gateway cloning using the Gateway conversion kit (Invitrogen). CDHR2-EGFP (PCDH24-EGFP as described in (Crawley et al., 2014b).
Stable Cell Line Generation
For generation of stable cell lines CL4 cells were grown to 80-90% confluency in T25 flasks and transfections were performed using Lipofectamine 2000 (Invitrogen) or FuGENE 6 (Promega) according to the manufacturer’s instructions. Selection for stable expression was performed after cells recovered for 2-3 days with addition of 1 mg/ml G418. For cells stably expressing two fluorescently-tagged proteins, the first construct was selected by transient transfection followed by antibiotic selection with G418 (e.g. EGFP-IRTKS, EGFP-IRTKSΔWH2, EGFP-EPS8, and EGFP-EPS8ΔAB). The second construct (mCherry-Espin) was introduced via viral transduction. Lentivirus was generated by co-transfecting HEK293FT cells (Fetal Hs embryonic epithelial cells; T75 flasks at 80% confluency) with 6 mg of pLVX-mCherry-Espin, 4 mg of psPAX2 packaging plasmid, and 0.8 mg of pMD2.G envelope plasmid using Lipofectamine 2000 (Invitrogen). For efficient lentiviral production, cells were incubated for 48 hours, then lentivirus-containing media was collected and concentrated with Lenti-X concentrator (Clontech). To transduce CL4 cells with lentivirus, the media was supplemented with 6 μg/ml polybrene (Sigma) and the lentiviral plasmid. After a 24-hour incubation, the media was changed and supplemented with 6 μg/ml polybrene and lentiviral plasmid for an additional 24 hours. Approximately 72 hours after initial viral transduction, cells were placed under antibiotic selection with Puromycin.
Light Microscopy and Image Processing
For SIM imaging, cells were plated on glass coverslips and allowed to grow to confluence. Cells were washed with warmed phosphate-buffered saline (PBS) and fixed with warm 4% paraformaldehyde/PBS for 15 min at 37°C. Cells were then washed three times with PBS, and blocked for 1 hour at room temperature in 5% bovine serum albumin (BSA) in PBS. Alexa Fluor 568-phalloidin (1:200, A12380; Invitrogen) or WGA-488 (10 μg/ml; W11261 ThermoFischer Scientific) were diluted in blocking solution and incubated for 1 hour at room temperature. Coverslips were washed three times with PBS then mounted on glass slides in ProLong Gold (P36930; Invitrogen). Cells were imaged on a Nikon Structured Illumination Microscope (N-SIM), Andor DU-897 EMCCD camera with four color excitation lasers (405 nm, 488 nm, 561 nm, and 647 nm), and a 100x/1.49 NA TIRF objective. SIM images were reconstructed using the Nikon Elements reconstruction algorithm.
For live-cell SDCM, cells were plated onto plasma-cleaned 35 mm glass bottom dishes (Invitro Scientific, D35-20-1.5-N), then transfected with the appropriate marker construct. Cells were allowed to grow to the appropriate level of confluence. If transfected, cells were imaged within 24 to 72 hours of transfection. Live-cell imaging was performed on a Nikon Ti2 inverted light microscope equipped with a Yokogawa CSU-X1 spinning disk head, Andor DU-897 EMCCD camera or a Photometrics Prime 95B sCMOS camera, 488 nm and 561 nm excitation lasers, a 405 nm photo-stimulation laser directed by a Bruker mini-scanner to enable targeted photoactivation, photoconversion, and photobleaching), and a 100x/1.49 NA TIRF objective. Low density microvilli were imaged when cells were subconfluent (80-90% confluence) or 1 DPC and images were acquired every 30-60 seconds for 20-40 minutes (Figures 1 and 2, and Supplemental Figure 2). During drug treatment, either (−)-Blebbistatin (B592500 Toronto Research Chemicals), Cythochalasin D (C2618 Sigma), or Cytochalasin B (C6762 Sigma) was added after 5 minutes of baseline measurement (Figure 2). For photokinetic studies of actin dynamics, baseline images were obtained for several frames prior to bleaching, then for an additional 3-5 minutes of recovery at 10 second intervals. Bleaching was performed on a line ROI (0.1 μm in width and 3-15 μm in length) using a 405 nm laser at 30% power with a 10 μs dwell time. Higher density microvilli on cells 2 DPC were imaged every 1-2 minutes for up to 4 hours (Figure 4). During imaging, cells were maintained with humidity at 37°C with 5% CO2 using a stage-top incubation system. Image acquisition was controlled with Nikon Elements software. 3D time series images were oversampled in the z-dimension with z-steps ranging from 0.09 μm to 0.18 μm followed by deconvolution (Nikon Elements Automatic or Richardson-Lucy algorithms) for better object resolution. Images were contrast enhanced, cropped, and aligned using Image J software (NIH), Nikon Elements, or Imaris (BITPLANE). Two-dimensional images were viewed as a maximum intensity projection. Three-dimensional depth coding was completed using Nikon Elements with images viewed as alpha-blended 3D composite images. Imaris (BITPLANE) was used to create an initial surface representing the microvillar fluorescence signal (Figure 3) then manually adjusted with fusion/fission of adjacent objects and manual microvillar tracking.
For live-correlative SDCM, cells were imaged as above, then washed with warmed phosphate-buffered saline (PBS) and fixed with warm 4% paraformaldehyde/PBS for 15 min at 37°C. Cells were then washed three times with PBS, and blocked for 1 hour at room temperature in 5% bovine serum albumin (BSA) in PBS. Primary antibody HPA009081 (Sigma) was diluted 1:200 in PBS and incubated with cells at 37°C for 1 hour followed by four washes with PBS. Cells were then incubated with secondary donkey anti-rabbit Alexa Fluor 488 (2 mg/ml, A-21206; Invitrogen) diluted 1:200 at room temperature. Cells were washed four times with PBS then imaged by SDCM.
For live-cell LLSM, a customized version of a lattice light sheet (LLS) microscope was designed and built based on the LLS system originally published by the Betzig group at Janelia Farm Research Campus (JFRC, HHMI) and in accordance with a research license agreement in place between institutions (Chen et al., 2014). In addition to light path modifications to enhance stability, efficiency, and accessibility of the instrument, upgrades particularly relevant for these experiments include a 583nm fiber laser (MPB Communications, Inc.), to excite mCherry at 99% of peak absorbance, and a newer generation sensor (Flash4.0v3, Hamamatsu, Inc.), which together substantially decreased the excitation energy necessary for time lapse imaging. In all cases, a dithered square lattice was utilized for imaging through the use of a spatial light modulator (Fourth Dimension Displays) in combination with an annular mask position corresponding to an annulus with 0.55 outer, and 0.44 inner, numerical apertures. Acquisition of datasets was managed through LabView (National Instruments). Image stacks contained anywhere from 300-500 optical sections per time point, with a step size ranging from 200 nm-320 nm between planes. Camera exposure times ranged from 5 msec - 15 msec per plane, with 25 sec intervals between stacks. Postacquisition, images were deskewed and deconvolved using 10 iterations of Richardson-Lucy Deconvolution. These post-acquisition processing steps were accomplished through LLSpy (Python code developed by Talley Lambert, Harvard University)(Talley, 2019). LLSpy implements cudaDeconv (Rev102), which is CUDA-based deskew and deconvolution developed by the Betzig Lab (JFRC) by Lin Shao and Dan Milkie. Maximum intensity projections were generated from resulting datasets via FIJI (ImageJ) software (Schindelin et al., 2012).
Electron Microscopy
All electron microscopy (EM) reagents were purchased from Electron Microscopy Sciences. To prepare samples for EM, cells were plated on glass coverslips (SEM) or plastic dishes (TEM), washed once with warm SEM buffer (0.1M HEPES, pH 7.3) supplemented with 2 mM CaCl2, then sequentially fixed for 1 hour at room temperature with 2.5% glutaraldehyde and 4% paraformaldehyde in SEM buffer supplemented with 2mM CaCl2, washed with SEM buffer, incubated in 1% tannic acid in SEM buffer for 1 hour, washed with ddH2O, incubated with 1% OsO4 in ddH2O for 1 hour, washed with ddH2O, incubated with 1% uranyl acetate in ddH2O for 30, then washed with ddH2O. Samples were dehydrated in a graded ethanol series. After dehydration, SEM samples were then dried using critical point drying and mounted on aluminum stubs and coated with gold/palladium using a sputter coater. SEM imaging was performed using Quanta 250 Environmental-SEM operated in high vacuum mode with an accelerating voltage of 5-10 kV. After dehydration TEM samples were transitioned to propylene oxide and gradually infiltrated with a Quetol 651 formulation Spurr’s resin (Ellis, 2006) using 1/4th the reported amount of BDMA to reduce viscosity. The resin was polymerized for 48 hours at 60Ό until blocks were firm, but not brittle. Thin sections were cut at nominal thickness of 70 nm and poststained with 2% uranyl acetate and Reynold’s lead citrate. Images were collected with an FEI Technai T-12 transmission electron microscope operating at 100 kV using an AMT CCD camera.
QUANTIFICATION AND STATISTICAL ANALYSIS
Microvillar centroids (x and y coordinates, length, and angle) were manually tracked using ImageJ, and the tip position was calculated using Microsoft Excel (Figure 1F, 2B–C, 2G–H, and Supplemental Figure 2F). The rate of microvillar motility was calculated as the net microvillar tip movement over microvillar lifetime. Maximum microvillar length is the longest length measured for a microvillus during its lifetime. Persistence is calculated as the net microvillar tip displacement divided by the total path traveled. If tips move in a straight line, persistence is equal to 1. All calculations on drug treated samples use the time of drug addition as time = 0. Mean square displacement (MSD) and velocity autocorrelation analysis were completed on microvillar centroids tracked in ImageJ. Data was exported and analyzed with MATLAB using a package specifically developed for MSD analysis (Tarantino et al., 2014), which is publicly available at http://www.mathworks.com/matlabcentral/fileexchange/40692-mean-square-displacement-analysis-of-particles-trajectories. For each individual trajectory, the MSD as a function of time delay (seconds) was calculated using and then averaged overall all trajectories. Random motion is indicated by a linear relationship whereas active motility is represented by convex curvature, and restricted motion by concave curvature. Active motility was fit to MSD (n) = 4Dn + V2n2, where D is the diffusion coefficient, V is the velocity, and n is the time window used for calculation. For MSD plots, open circles represent the mean MSD, error bars are the SEM, and shaded areas represent the weighted SD over all MSD curves. The velocity autocorrelation is likewise calculated for each individual track and then averaged over trajectories. For random motion, displacements are not correlated, thus velocity autocorrelation is 0 except for the initial positive displacement (normalized to 1). For active motion, non-zero velocity autocorrelation is expected. For all figures, error bars indicate SD, and n values are reported in the figure legends. Statistical significance was calculated using the Mann-Whitney-Wilcoxon test for pairwise comparisons (Figure 1I–K), or using one-way ANOVA with Kruskal-Wallis to compare more than 2 conditions (Figure 2K–M, Figure 4E–G). Rose plots and individual tracks were graphed using Excel. MSD and normalized velocity autocorrelation were analyzed and graphed using MATLAB. All other graphs were generated in Prism v.7 (GraphPad). All statistical testing was performed in Prism v.7 (GraphPad) or MATLAB.
DATA AND CODE AVAILABILITY
No large-scale datasets or new code were generated in this study.
Supplementary Material
Supplemental Video 1. Microvilli move across the cell surface, Related to Figure 1A–B. Lattice light-sheet microscopy (LLSM) time lapse images of CL4 cell stably expressing mCherry-Espin (corresponds to Figure 1A). Duration 28 minutes, 30 frames per second (fps). Scale bar is 10 μm. Intensity color code.
Supplemental Video 9. Peripheral clusters of microvilli are stable over up to 12 hours, Related to Figure 5H. Time lapse images of CL4 cells stably expressing mCherry-Espin. Images were deconvolved, processed for extended depth of field (EDF), and visualized as a maximum intensity z-projection with an intensity color code (pictured at left). Individual microvilli (magenta) translocate across the cell surface, merge into small clusters and either dissipate or merge into large clusters of microvilli (yellow/orange) at the cell periphery. These large clusters of microvilli appear to continuously remodel but remain stable over many hours. Duration 12 hours, 30 fps. Movie corresponds to Figure 5H. Scale bar is 10 μm.
Supplemental Video 2. Microvilli move across the cell surface independent of F-actin marker, Related to Figure 1C,E and Figure S1D–E. Time lapse images of CL4 cell stably expressing mCherry-Espin (left panel, corresponds to Figure 1C), or EGFP-Lifeact (right panel, corresponds to Figure S1D). Duration 20 minutes, 12 frames per second (fps). Color-coded arrowheads highlight individual translocating microvilli. Scale bars are 10 μm.
Supplemental Video 3. Early actin structures protruding from the cell surface are wrapped in membrane over time, Related to Figure S1A–B. Time lapse images of CL4 cell stably expressing mCherry-Espin (left panel) and GPI-GFP (right panel), corresponds to Figure S2A–B. The 3D volume was rotated so the microvillus of interest is central and moves vertically across the image, and a depth-code was applied. Z-axis depth color code bottom left with tick marks at 1 μm intervals. Movie contains 6 frames taken at 1 minute intervals.
Supplemental Video 4. Microvillar motility requires actin assembly but not myosin contractility, Related to Figure 2. Time lapse images of CL4 cell stably expressing mCherry-Espin treated with 20 μM Blebbistatin (left panel, corresponds to Figure 2A) or 500 nM Cytochalasin B (representative movie, corresponding Figure 2G–J analysis) with the addition of drug at 5 minutes. Duration 30 minutes, 12 fps. Scale bars are 10 μm.
Supplemental Video 5. Microvillar F-actin core treadmills during microvillar motility, Related to Figure 3. Time lapse images of CL4 cell transiently expressing mNEON-Green-β-actin (top panel), microvillar surface created in Imaris (middle panel), isolated microvillus of interest (bottom panel). Images correspond to Figure 3B–D. Duration 120 seconds, 5 fps. Scale bars are 2 μm.
Supplemental Video 6. IRTKS and EPS8 modulate microvillar motility dependent on actin-binding, Related to Figure 4. Time lapse images of CL4 cells stably expressing mCherry-Espin (magenta) plus either EGFP-IRTKS (top left), EGFP-IRTKSΔWH2 (top right), EGFP-EPS8 (bottom left), or EGFP-EPS8ΔAB (bottom right) in green. Images correspond to Figure 4A–D. Duration 20 minutes, 12 fps. Scale bars ar 5 μm.
Supplemental Video 7. Microvillar translocation drives intermicrovillar collision facilitating cluster formation, Related to Figure 5A. Time lapse images of CL4 cell stably expressing mCherry-Espin. Duration 120 minutes, 10 fps. Movie corresponds to Figure 5A.
Supplemental Video 8. Microvillar translocation drives collision and clustering of individual microvilli as well as movement of large clusters of microvilli across the cell surface, Related to Figure 5C–D. Time lapse images of CL4 cell stably expressing mCherry-Espin. Left Panel Individual microvilli collide and remain connected then translocate across the cell joining with larger clusters of microvilli. Single microvilli are marked by arrowheads, then asterisks once they coalesce with a larger cluster. Duration 120 minutes, 10 fps. Movie corresponds to Figure 5B, zoom of Supplemental Movie 7. Right Panel A large cluster of microvilli (asterisk) moves across the cell surface traveling left then upward. Duration 120 minutes, 10 fps. Movie corresponds to Figure 5C, zoom of Supplemental Movie 7.
Highlights.
Microvilli exhibit persistent, active motility driven by actin assembly.
Microvillar F-actin cores treadmill during motility.
Barbed-end binding factors regulate microvillar motility.
Motility promotes intermicrovillar collisions, adhesion, and cluster formation.
ACKNOWLEDGEMENTS
We thank all members of the Tyska laboratory for feedback and advice. Microscopy was performed in part through the Vanderbilt University Cell Imaging Shared Resource. This work was supported by a Ruth L. Kirschstein National Research Service Award T32-A1007474 (LMM) and NIH grants R01-DK111949 and R01-DK095811 (MJT). The custom LLSM was assembled by BAM and Dr. John Kozub from the BioMIID program, a collaboration between the Vanderbilt Biophotonics Center, Vanderbilt Department of Physics, and the Vanderbilt Cell Imaging Shared Resource, funded by a Trans-Institutional Programs (TIPs) grant from Vanderbilt University (PI: Mahadevan-Jansen). The BioMIID program would like to thank Drs. Eric Betzig, Wes Legant, Jennifer Lippincott-Schwartz, Aubrey Weigel (JFRC), John Heddleston (JFRC), Talley Lambert (Harvard Medical School), and Srigokul Upadhyayula (Harvard Medical School) for technical advice during LLSM development.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Bartles JR, Zheng L, Li A, Wierda A, and Chen B (1998). Small espin: a third actin-bundling protein and potential forked protein ortholog in brush border microvilli. The Journal of cell biology 143, 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, and Weber K (1978). Localization of actin and microfilament-associated proteins in the microvilli and terminal web of the intestinal brush border by immunofluorescence microscopy. The Journal of cell biology 79, 839–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, and Weber K (1979). Villin: the major microfilament-associated protein of the intestinal microvillus. Proceedings of the National Academy of Sciences of the United States of America 76, 2321–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, and Weber K (1980). Fimbrin, a new microfilament-associated protein present in microvilli and other cell surface structures. The Journal of cell biology 86, 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaletto JB, Saotome I, Curto M, and McClatchey AI (2011). Ezrin-mediated apical integrity is required for intestinal homeostasis. Proceedings of the National Academy of Sciences of the United States of America 108, 11924–11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA 3rd, Liu Z, et al. (2014). Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudrier E, Kerjaschki D, and Louvard D (1988). Cytoskeleton organization and submembranous interactions in intestinal and renal brush borders. Kidney international 34, 309–320. [DOI] [PubMed] [Google Scholar]
- Crawley SW, Mooseker MS, and Tyska MJ (2014a). Shaping the intestinal brush border. The Journal of cell biology 207, 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley SW, Shifrin DA Jr., Grega-Larson NE, McConnell RE, Benesh AE, Mao S, Zheng Y, Zheng QY, Nam KT, Millis BA, et al. (2014b). Intestinal brush border assembly driven by protocadherin-based intermicrovillar adhesion. Cell 157, 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley SW, Weck ML, Grega-Larson NE, Shifrin DA Jr., and Tyska MJ (2016). ANKS4B Is Essential for Intermicrovillar Adhesion Complex Formation. Developmental cell 36, 190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce A, Cassata G, Disanza A, Gagliani MC, Tacchetti C, Malabarba MG, Carlier MF, Scita G, Baumeister R, and Di Fiore PP (2004). A novel actin barbed-end-capping activity in EPS-8 regulates apical morphogenesis in intestinal cells of Caenorhabditis elegans. Nat Cell Biol 6, 1173–1179. [DOI] [PubMed] [Google Scholar]
- Delacour D, Salomon J, Robine S, and Louvard D (2016). Plasticity of the brush border - the yin and yang of intestinal homeostasis. Nature reviews Gastroenterology & hepatology 13, 161–174. [DOI] [PubMed] [Google Scholar]
- Disanza A, Mantoani S, Hertzog M, Gerboth S, Frittoli E, Steffen A, Berhoerster K, Kreienkamp HJ, Milanesi F, Di Fiore PP, et al. (2006). Regulation of cell shape by Cdc42 is mediated by the synergic actin-bundling activity of the Eps8-IRSp53 complex. Nat Cell Biol 8, 1337–1347. [DOI] [PubMed] [Google Scholar]
- Ellis EA (2006). Solutions to the Problem of Substitution of ERL 4221 for Vinyl Cyclohexene Dioxide in Spurr Low Viscosity Embedding Formulations. Microscopy Today 14, 32–33. [Google Scholar]
- Emlet DR, Shaw AD, and Kellum JA (2015). Sepsis-associated AKI: epithelial cell dysfunction. Seminars in nephrology 35, 85–95. [DOI] [PubMed] [Google Scholar]
- Fath KR, Obenauf SD, and Burgess DR (1990). Cytoskeletal protein and mRNA accumulation during brush border formation in adult chicken enterocytes. Development 109, 449–459. [DOI] [PubMed] [Google Scholar]
- Footer MJ, Kerssemakers JW, Theriot JA, and Dogterom M (2007). Direct measurement of force generation by actin filament polymerization using an optical trap. Proceedings of the National Academy of Sciences of the United States of America 104, 2181–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F, and Jay P (2016). Intestinal tuft cells: epithelial sentinels linking luminal cues to the immune system. Mucosal immunology 9, 1353–1359. [DOI] [PubMed] [Google Scholar]
- Giardini PA, Fletcher DA, and Theriot JA (2003). Compression forces generated by actin comet tails on lipid vesicles. Proceedings of the National Academy of Sciences of the United States of America 100, 6493–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik R, and Gautreau A (2014). Quantitative and unbiased analysis of directional persistence in cell migration. Nature protocols 9, 1931–1943. [DOI] [PubMed] [Google Scholar]
- Grega-Larson NE, Crawley SW, Erwin AL, and Tyska MJ (2015). Cordon bleu promotes the assembly of brush border microvilli. Molecular biology of the cell 26, 3803–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzog M, Milanesi F, Hazelwood L, Disanza A, Liu H, Perlade E, Malabarba MG, Pasqualato S, Maiolica A, Confalonieri S, et al. (2010). Molecular basis for the dual function of Eps8 on actin dynamics: bundling and capping. PLoS biology 8, e1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, and Heuser JE (1981). Quick-freeze, deep-etch visualization of the cytoskeleton beneath surface differentiations of intestinal epithelial cells. The Journal of cell biology 91, 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull RN, Cherry WR, and Weaver GW (1976). The origin and characteristics of a pig kidney cell strain, LLC-PK. In vitro 12, 670–677. [DOI] [PubMed] [Google Scholar]
- Krasteva G, and Kummer W (2012). “Tasting” the airway lining fluid. Histochemistry and cell biology 138, 365–383. [DOI] [PubMed] [Google Scholar]
- Kummer W, and Deckmann K (2017). Brush cells, the newly identified gatekeepers of the urinary tract. Current opinion in urology 27, 85–92. [DOI] [PubMed] [Google Scholar]
- Loomis PA, Zheng L, Sekerkova G, Changyaleket B, Mugnaini E, and Bartles JR (2003). Espin cross-links cause the elongation of microvillus-type parallel actin bundles in vivo. The Journal of cell biology 163, 1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallavarapu A, and Mitchison T (1999). Regulated actin cytoskeleton assembly at filopodium tips controls their extension and retraction. The Journal of cell biology 146, 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor U, Disanza A, Grati M, Andrade L, Lin H, Di Fiore PP, Scita G, and Kachar B (2011). Regulation of stereocilia length by myosin XVa and whirlin depends on the actin-regulatory protein Eps8. Current biology : CB 21, 167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell RE, Benesh AE, Mao S, Tabb DL, and Tyska MJ (2011). Proteomic analysis of the enterocyte brush border. Am J Physiol Gastrointest Liver Physiol 300, G914–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros NA, Burnette DT, and Forscher P (2006). Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat Cell Biol 8, 215–226. [DOI] [PubMed] [Google Scholar]
- Millard TH, Dawson J, and Machesky LM (2007). Characterisation of IRTKS, a novel IRSp53/MIM family actin regulator with distinct filament bundling properties. J Cell Sci 120, 1663–1672. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ (1989). Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. The Journal of cell biology 109, 637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooseker MS, and Tilney LG (1975). Organization of an actin filament-membrane complex. Filament polarity and membrane attachment in the microvilli of intestinal epithelial cells. The Journal of cell biology 67, 725–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar R, McConnell RE, and Tyska MJ (2009). Control of cell membrane tension by myosin-I. Proceedings of the National Academy of Sciences of the United States of America 106, 11972–11977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaloni D, Le Clainche C, and Carlier MF (2001). Mechanism of actin-based motility. Science 292, 1502–1506. [DOI] [PubMed] [Google Scholar]
- Peterson MD, and Mooseker MS (1992). Characterization of the enterocyte-like brush border cytoskeleton of the C2BBe clones of the human intestinal cell line, Caco-2. J Cell Sci 102 (Pt 3), 581–600. [DOI] [PubMed] [Google Scholar]
- Pollard TD, and Cooper JA (2009). Actin, a central player in cell shape and movement. Science 326, 1208–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postema MM, Grega-Larson NE, Neininger AC, and Tyska MJ (2018). IRTKS (BAIAP2L1) Elongates Epithelial Microvilli Using EPS8-Dependent and Independent Mechanisms. Current biology : CB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prass M, Jacobson K, Mogilner A, and Radmacher M (2006). Direct measurement of the lamellipodial protrusive force in a migrating cell. The Journal of cell biology 174, 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nature methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Roth S, Nieuwenhuis EES, and Middendorp S (2018). Intestinal epithelial cell polarity defects in disease: lessons from microvillus inclusion disease. Disease models & mechanisms 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwander M, Kachar B, and Muller U (2010). Review series: The cell biology of hearing. The Journal of cell biology 190, 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebe-Pedros A, Burkhardt P, Sanchez-Pons N, Fairclough SR, Lang BF, King N, and Ruiz-Trillo I (2013). Insights into the origin of metazoan filopodia and microvilli. Molecular biology and evolution 30, 2013–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talley L (2019). tlambert03/LLSpy: v0.4.0 (Zenodo; ). [Google Scholar]
- Tarantino N, Tinevez JY, Crowell EF, Boisson B, Henriques R, Mhlanga M, Agou F, Israel A, and Laplantine E (2014). TNF and IL-1 exhibit distinct ubiquitin requirements for inducing NEMO-IKK supramolecular structures. The Journal of cell biology 204, 231–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temm-Grove C, Helbing D, Wiegand C, Honer B, and Jockusch BM (1992). The upright position of brush border-type microvilli depends on myosin filaments. J Cell Sci 101 (Pt 3), 599–610. [DOI] [PubMed] [Google Scholar]
- Tilney LG, Tilney MS, and DeRosier DJ (1992). Actin filaments, stereocilia, and hair cells: how cells count and measure. Annual review of cell biology 8, 257–274. [DOI] [PubMed] [Google Scholar]
- Tyska MJ, Mackey AT, Huang JD, Copeland NG, Jenkins NA, and Mooseker MS (2005). Myosin-1a is critical for normal brush border structure and composition. Molecular biology of the cell 16, 2443–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska MJ, and Mooseker MS (2002). MYO1A (brush border myosin I) dynamics in the brush border of LLC-PK1-CL4 cells. Biophysical journal 82, 1869–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya A, Chabot JR, Andreeva A, Samadani A, and van Oudenaarden A (2003). Probing polymerization forces by using actin-propelled lipid vesicles. Proceedings of the National Academy of Sciences of the United States of America 100, 4521–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallance BA, Chan C, Robertson ML, and Finlay BB (2002). Enteropathogenic and enterohemorrhagic Escherichia coli infections: emerging themes in pathogenesis and prevention. Canadian journal of gastroenterology = Journal canadien de gastroenterologie 16, 771–778. [DOI] [PubMed] [Google Scholar]
- Wang YL (1985). Exchange of actin subunits at the leading edge of living fibroblasts: possible role of treadmilling. The Journal of cell biology 101, 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayt J, and Bretscher A (2014). Cordon Bleu serves as a platform at the basal region of microvilli, where it regulates microvillar length through its WH2 domains. Molecular biology of the cell 25, 2817–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weck ML, Crawley SW, Stone CR, and Tyska MJ (2016). Myosin-7b Promotes Distal Tip Localization of the Intermicrovillar Adhesion Complex. Current biology : CB 26, 2717–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CA, Tsuchida MA, Allen GM, Barnhart EL, Applegate KT, Yam PT, Ji L, Keren K, Danuser G, and Theriot JA (2010). Myosin II contributes to cell-scale actin network treadmilling through network disassembly. Nature 465, 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelman JD, Bilancia CG, Peifer M, and Kovar DR (2014). Ena/VASP Enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled F-actin networks with Fascin. Proceedings of the National Academy of Sciences of the United States of America 111, 4121–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi A, Masuda M, Ohki T, Onishi H, and Mochizuki N (2004). A novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J Biol Chem 279, 14929–14936. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Video 1. Microvilli move across the cell surface, Related to Figure 1A–B. Lattice light-sheet microscopy (LLSM) time lapse images of CL4 cell stably expressing mCherry-Espin (corresponds to Figure 1A). Duration 28 minutes, 30 frames per second (fps). Scale bar is 10 μm. Intensity color code.
Supplemental Video 9. Peripheral clusters of microvilli are stable over up to 12 hours, Related to Figure 5H. Time lapse images of CL4 cells stably expressing mCherry-Espin. Images were deconvolved, processed for extended depth of field (EDF), and visualized as a maximum intensity z-projection with an intensity color code (pictured at left). Individual microvilli (magenta) translocate across the cell surface, merge into small clusters and either dissipate or merge into large clusters of microvilli (yellow/orange) at the cell periphery. These large clusters of microvilli appear to continuously remodel but remain stable over many hours. Duration 12 hours, 30 fps. Movie corresponds to Figure 5H. Scale bar is 10 μm.
Supplemental Video 2. Microvilli move across the cell surface independent of F-actin marker, Related to Figure 1C,E and Figure S1D–E. Time lapse images of CL4 cell stably expressing mCherry-Espin (left panel, corresponds to Figure 1C), or EGFP-Lifeact (right panel, corresponds to Figure S1D). Duration 20 minutes, 12 frames per second (fps). Color-coded arrowheads highlight individual translocating microvilli. Scale bars are 10 μm.
Supplemental Video 3. Early actin structures protruding from the cell surface are wrapped in membrane over time, Related to Figure S1A–B. Time lapse images of CL4 cell stably expressing mCherry-Espin (left panel) and GPI-GFP (right panel), corresponds to Figure S2A–B. The 3D volume was rotated so the microvillus of interest is central and moves vertically across the image, and a depth-code was applied. Z-axis depth color code bottom left with tick marks at 1 μm intervals. Movie contains 6 frames taken at 1 minute intervals.
Supplemental Video 4. Microvillar motility requires actin assembly but not myosin contractility, Related to Figure 2. Time lapse images of CL4 cell stably expressing mCherry-Espin treated with 20 μM Blebbistatin (left panel, corresponds to Figure 2A) or 500 nM Cytochalasin B (representative movie, corresponding Figure 2G–J analysis) with the addition of drug at 5 minutes. Duration 30 minutes, 12 fps. Scale bars are 10 μm.
Supplemental Video 5. Microvillar F-actin core treadmills during microvillar motility, Related to Figure 3. Time lapse images of CL4 cell transiently expressing mNEON-Green-β-actin (top panel), microvillar surface created in Imaris (middle panel), isolated microvillus of interest (bottom panel). Images correspond to Figure 3B–D. Duration 120 seconds, 5 fps. Scale bars are 2 μm.
Supplemental Video 6. IRTKS and EPS8 modulate microvillar motility dependent on actin-binding, Related to Figure 4. Time lapse images of CL4 cells stably expressing mCherry-Espin (magenta) plus either EGFP-IRTKS (top left), EGFP-IRTKSΔWH2 (top right), EGFP-EPS8 (bottom left), or EGFP-EPS8ΔAB (bottom right) in green. Images correspond to Figure 4A–D. Duration 20 minutes, 12 fps. Scale bars ar 5 μm.
Supplemental Video 7. Microvillar translocation drives intermicrovillar collision facilitating cluster formation, Related to Figure 5A. Time lapse images of CL4 cell stably expressing mCherry-Espin. Duration 120 minutes, 10 fps. Movie corresponds to Figure 5A.
Supplemental Video 8. Microvillar translocation drives collision and clustering of individual microvilli as well as movement of large clusters of microvilli across the cell surface, Related to Figure 5C–D. Time lapse images of CL4 cell stably expressing mCherry-Espin. Left Panel Individual microvilli collide and remain connected then translocate across the cell joining with larger clusters of microvilli. Single microvilli are marked by arrowheads, then asterisks once they coalesce with a larger cluster. Duration 120 minutes, 10 fps. Movie corresponds to Figure 5B, zoom of Supplemental Movie 7. Right Panel A large cluster of microvilli (asterisk) moves across the cell surface traveling left then upward. Duration 120 minutes, 10 fps. Movie corresponds to Figure 5C, zoom of Supplemental Movie 7.
Data Availability Statement
No large-scale datasets or new code were generated in this study.


