Abstract
Proteins are composed of α-amino acid residues. This consistency in backbone structure likely serves an important role in the display of an enormous diversity of peptides by Class II MHC (MHC-II) products, which make contacts with main chain atoms of their peptide cargo. Peptides that contain residues with an extra carbon in the backbone (derived from β-amino acids) have biological properties that differ starkly from those of their conventional counterparts. How changes in the structure of the peptide backbone affect the loading of peptides onto MHC-II or recognition of the resulting complexes by T-cell receptors has not been widely explored. We prepared a library of analogues of MHC-II-binding peptides derived from ovalbumin, in which at least one α-amino acid residue was replaced with a homologous β-amino acid residue. The latter contain an extra methylene unit in the peptide backbone but retain the original side chain. We show that several of these α/β-peptides retain the ability to bind tightly to MHC-II, activate T-cell receptor signaling and induce responses from T cells in mice. One α/β-peptide exhibited enhanced stability in the presence of an endosomal protease relative to the index peptide. Conjugation of this backbone-modified peptide to a camelid single-domain antibody fragment specific for MHC-II enhanced its biological activity. Our results suggest that backbone modification offers a method to modulate MHC binding and selectivity, T cell stimulatory capacity and susceptibility to processing by proteases such as those found within endosomes where antigen processing occurs.
INTRODUCTION:
The immune system recognizes and responds to proteinaceous antigens through the production and display of short peptide antigens bound to the products of the major histocompatibility complex (MHC). Class I MHC (MHC-I) products that stimulate CD8+T cells typically display peptides of 8–10 residues in length, derived mostly from intracellular proteins, whereas Class II MHC (MHC-II) products that stimulate CD4+T cells display longer peptides, mostly from proteins originating in extracellular space or their topological equivalents. The loading of peptides onto MHC products requires the breakdown of the source proteins into peptide fragments through the action of proteases and peptidases(1, 2). The resulting peptides are then exposed to receptive MHC molecules through a coordinated series of trafficking steps that differ for MHC-I and MHC-II(3). Productive binding of peptide to MHC yields peptide-MHC complexes that are transported for display at the surface of the antigen-presenting cell. These complexes engage T-cell receptor (TCR) complexes, stimulate TCR signaling and drive subsequent T cell-mediated immune responses. Each of these steps (proteolysis, MHC binding, TCR recognition) requires recognition of the polypeptide antigen or some part of it.
Modifications of peptide structure can affect each recognition step required for T cell engagement. Structural alterations that affect recognition by proteases can protect peptides against proteolysis, which not only generates but can also destroy the ligands presented by MHC-II products. For example, synthetic proteins composed entirely of D-amino acids are not degraded by proteases and cannot be processed for loading onto MHC products. Such proteins therefore fail to induce immune responses that require T cell participation(4). Alternatively, the introduction of modifications that inhibit extracellular proteolysis can enhance the efficacy of peptide vaccines(5). In some cases, endosomal proteolysis can destroy potentially antigenic peptides(6), and in this context, inhibition of proteolysis enhances immunogenicity(7, 8). These findings suggest that carefully introduced structural modifications that forestall destruction of antigenic peptides might enhance immunogenicity(9).
MHC-I and MHC-II bind to an enormously diverse array of peptides. The impact of side chain identities at specific sites on binding to specific allomorphs of MHC products and subsequent recognition by T cells has been extensively analyzed(10, 11). The development of compounds with selectivity among these allomorphs could be useful to address autoimmune disorders(12). In contrast, relatively little attention has been paid to changes in the peptide backbone(13). Interactions between MHC-II and the peptide backbone are crucial for affinity and stability of the peptide-MHC-II complex(14, 15). Only a few studies have probed the impact of peptide backbone modifications on MHC binding(16–22), with diverse outcomes. TCR recognition of peptide-MHC complexes and the ensuing T cell responses are sensitive to peptide backbone modifications as well(23–30).
Replacement of α-residues with β-amino acid counterparts has been used to probe the importance of peptide backbone structure in a variety of contexts(31, 32). Each α→β replacement adds a carbon atom to the backbone compared to the analogous conventional peptide. Such replacements can retain the original side chain. The incorporation of a β-amino acid residue provides protection against proteolysis, at least for peptide bonds in proximity to the site of incorporation(33–35). Carefully designed α/β-peptides (i.e., peptides containing a mixture of α- and β-residues) can mimic the conformations and biological properties of α-helical peptides, but the α/β-peptides display greater resistance to proteolysis and have prolonged activity in vivo(36–40).
Only a few studies have probed the impact of incorporating β-residues on recognition by the machinery of the immune system; MHC-I restricted peptide epitopes have been favored(41–43). In general, the MHC-I allomorph and TCR involved in recognition of a conventional peptide epitope can also recognize analogues of this epitope that contain one or two α→β replacements; however, no general trend for tolerance to β-residue substitution was established. It is unclear whether certain positions in MHC-I binding epitopes are especially tolerant to α→β replacements or whether multiple such replacements can be incorporated with retention of MHC binding and TCR activation. Epitope analogues containing a β-residue density sufficient for robust protection from proteolysis were generally unable to bind MHC-I or elicit TCR responses(44).
Since the mode of peptide interaction with MHC-I differs from that of MHC-II, the impact of α→β modifications on MHC-II epitope recognition may differ from that seen for MHC-I epitopes(10). There are two relevant studies on this subject(45, 46). A model peptide derived from histone H4 was the template for a set of peptides in which one or two α-residues were replaced with aza-β3-residue(s). These peptides were then tested for their ability to bind mouse MHC-II (I-Ad and I-AE) and stimulate proliferation or cytokine release by the appropriately specific T cells. Substitution was tolerated only at the three residues at the C-terminus of the peptide. The backbone of aza-β3-residues differs from that of the β3-residues used in this work(44). The second example concerns analogues of an encephalitogenic peptide derived from myelin oligodendrocyte glycoprotein (MOG33–55) binding to I-Ab, in which a single residue known to contact the TCR was replaced with a β3-residue(46). The analogue of MOG33–35 with β3-homoPhe at a TCR contact position was not encephalitogenic and exerted a protective effect against the induction of experimental autoimmune encephalitis. While the mechanism that underlies this protection is unclear, replacement of α-Phe with β3-homoPhe clearly altered the nature of the T cell response. These two precedents suggest that replacement of an α residue with a homologous β3 residue is tolerated, at least in some positions, in the context of the recognition of MHC-II restricted peptides.
Here we performed a more systematic survey of α→β replacements with preservation of the native side chain. These modifications allow us to isolate the impact of backbone alterations. Does the introduction of one or a few homologous β residues impact proteolytic processing in a way that alters immunogenicity in cell-based or animal-based assays? Does the placement of β homologues at specific sites within an MHC-II-restricted peptide provide antigens with altered T cell stimulatory behavior? Our experiments were designed to answer these questions.
MATERIALS AND METHODS:
General.
Cell lines were routinely tested for cell surface marker expression and mycoplasma infection. LC/MS was performed on a Waters Xevo TOF system equipped with a HPLC-C8 column. Calculations and graphing were performed using Microsoft Excel and GraphPad Prism 6. EC50 values were generated using a sigmoidal dose-response with variable slope model (4-paremeter model). Statistical significance was evaluated using a two-tailed, paired t-test.
Flow cytometry and antibodies.
For cytofluorimetry, gating was performed on viable cells based on forward scatter and side scatter profiles. Fluorescently labeled antibodies were purchased from eBioscience (anti-CD45.1, A20; anti-CD4, RM4–5; anti-CD11c, N418) or BioLegend (anti-CD45.2, 104; anti–I-A/I-E, M5/114.15.2). Proliferation was assessed using the dye CellTrace Violet (Life Technologies) according to manufacturer’s instructions. Staining for cell surface markers was performed at 4°C for 30 m. Flow cytometry data were acquired on an LSRFortessa (Becton Dickinson) instrument and analyzed with the FlowJo software package (Tri-Star).
Peptide Synthesis.
Peptides were synthesized as C-terminal carboxylic acids with uncapped N-termini on Wang resin (100–200 mesh, EMD Millipore). Amino acids were activated with 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) and N-hydroxybenzotriazole (HOBt) in the presence of N,N-diisopropylethylamine (DIPEA), except for the C-terminal residue. C-terminal residues were coupled onto resin using 1-(mesitylene-2-sulfonyl)-3-nitro-1H-1,2,4-triazole (MSNT) as a coupling agent. Main chain peptide amino groups were Fmoc-protected. The growing peptide chain was deprotected using 20% piperidine in dimethylformamide (DMF). Protected β3‐homoamino acids were purchased from PepTech and Chem Impex. After synthesis, peptides were cleaved from the resin and side chains were deprotected using reagent K (82.5% TFA, 5% phenol, 5% H2O, 5% thioanisole, 2.5% ethanedithiol) for two hours. The reagent K solution was dripped into cold diethyl ether to precipitate the deprotected peptide. Peptides were purified via reverse-phase HPLC on a prep‐C18 column. Peptide purity was assessed by analytical reverse phase‐HPLC (solvent A: 0.1% TFA in water, solvent B: 0.1% TFA in acetonitrile, C18 analytical column (4.6 X 250 mm), flow rate 1 mL/min) by quantifying the area under the peak corresponding to the desired product. Peptide stocks of known concentrations were prepared by quantifying the absorption at 214 nm(47).
MHC purification and binding assays.
Purification of H-2 I-Ab and I-Ad class II MHC molecules by affinity chromatography, and the performance of assays based on the inhibition of binding of a high affinity radiolabeled peptide to quantitatively measure peptide binding, were performed as detailed elsewhere(48). Briefly, the mouse B cell lymphoma LB27.4 was used as a source of MHC molecules. A high affinity radiolabeled peptide (0.1–1 nM; peptide ROIV, sequence YAHAAHAAHAAHAAHAA) was co-incubated at room temperature with purified MHC in the presence of a cocktail of protease inhibitors and an inhibitor peptide. Following a two-day incubation, MHC bound radioactivity was determined by capturing MHC/peptide complexes on mAb (Y3JP, I-Ab; MKD6, I-Ad) coated Lumitrac 600 plates (Greiner Bio-one, Frickenhausen, Germany), and measuring bound radioactivity using the TopCount (Packard Instrument Co., Meriden, CT) microscintillation counter. The concentration of peptide yielding 50% inhibition of the binding of the radiolabeled peptide was calculated. Under the conditions utilized, where [label]<[MHC] and IC50 ≥ [MHC], the measured IC50 values are reasonable approximations of the true KD values(49, 50). Each competitor peptide was tested at six different concentrations covering a 100,000-fold range, and in three or more independent experiments. As a positive control, the unlabeled version of the radiolabeled probe was also tested in each experiment.
Cell Lines.
The murine A20 B cell lymphoma line presenting I-Ad MHC was acquired from ATCC (TIB-208). The murine DO11.10 T cell hybridoma line was obtained from Dr. Philippa Marrack, National Jewish Health Center, Denver, Co. A20 cells were maintained in RPMI with 10% heat inactivated fetal bovine serum and 0.05 mM beta mercaptoethanol. DO11.10 cells were maintained in DMEM supplemented with 10% FBS, 100 Units penicillin/50 μg streptomycin per mL and 2 mM L-glutamine. B16-FLT3L and primary T cells were maintained in in RPMI supplemented with 10% FBS, 100 Units penicillin/100 μg streptomycin per mL, 0.05 mM beta mercaptoethanol, 1 mM sodium pyruvate, and 2 mM L-glutamine.
Hybridoma stimulation assays.
These assays were performed essentially as previously described(51), with a few exceptions. Each well of a 96-well plate was seeded with 50,000 A20 cells and DO.110 cells along with indicated concentrations of peptide in medium comprised of 50% A20 medium and 50% DO11.10 medium. After a 24-hour incubation at 37°C in a 5% CO2 atmosphere, supernatant was transferred to a new 96 well plate and analyzed with IL-2 ELISA. All peptide doses were performed in duplicate wells and all experiments were replicated the number of times indicated in figure captions. IL-2 levels were assessed using a matched pair ELISA (BD #555148).
ELIspot assays with splenocytes.
Interferon-gamma (IFNγ) ELISpot assays were performed following published procedures(52). Briefly, 96-well ELISpot plates (BD ELISPOT Mouse IFNg ELISPOT Set, BD Biosciences) were coated with an IFNγ capture antibody (BD Biosciences) in PBS overnight at 4° C and plates were blocked with complete medium (10% fetal bovine serum in RPMI with added sodium pyruvate, beta mercaptoethanol, and non-essential amino acids) for 2 hours at room temperature. Splenocytes (5 × 104) from OT-II mice were added in duplicate to wells containing indicated concentration of peptide in complete medium or medium alone as a negative control. Positive control wells for all experiments contained Cell Stimulation Cocktail (Invitrogen). After plating of the splenocytes, ELISpot plates were incubated overnight at 37° C. After 24 hours, plates were washed and incubated with a biotinylated IFNγ detection antibody (BD Biosciences) for 2 hours, followed by streptavidin–horse radish peroxidase (BD Biosciences) for 1 hour at room temperature. The plates were developed with 3-amino-9-ethyl-carbazole substrate (BD ELISPOT AEC Substrate Set) for 5 minutes and dried for at least 24 hours. Spots were enumerated using the KS ELISpot analysis system (Carl Zeiss).
In vitro proliferation assay.
Dendritic cells were purified from spleens and mesenteric lymph nodes of WT mice injected with 1 × 106 B16-FLT3L cells using the Pan Dendritic Cell (DC) Isolation Kit (Miltenyi, Germany). Naïve CD4+ T cells were purified following manufacturer’s instructions (Miltenyi, Germany) from the spleen and mesenteric lymph nodes of OT-II mice and labelled with 3 μM VioletTrace (ThermoFisher). Purity was always greater than 98% for both purified DCs and T cells. 1 ×105 DCs were incubated with peptide (50 nM) for 1 h before adding 1 *105 T cells. Proliferation was assessed using dilution of VioletTrace and analyzed by flow cytometry after 3.5 days of coculture.
In vivo proliferation assay
TCRαβKO mice received 5 * 105 naïve OT-II CD4+ T cells, purified following manufacturer’s instructions (Miltenyi, Germany and ThermoFisher). Mice were immunized sub-cutaneously 24h later, using 2 μg of peptide in PBS, adsorbed in alum (Imject, Thermo Fisher Scientific). Proliferation was assessed by enumerating the total number of OT-II T cells in the draining lymph nodes (inguinal lymph node).
Assessment of peptide proteolysis.
Peptides were incubated with recombinant human Cathepsin S (R&D Systems 1183-CY-010), preactivated according to manufacturer instructions, in pH 4.5 sodium acetate buffer with 5 mM dithiothreitol at 37° C. Proteolysis was quenched at the indicated time points by the addition of an equal volume of 1% trifluoroacetic acid (TFA) in water (vol/vol). A control experiment showed that addition of TFA prior to addition of peptide (pre quenching of protease) prevented degradation of peptide over the timeframe investigated (data not shown). Quenched samples were analyzed by LC/MS. The ion currents corresponding to intact peptide were used to determine peptide concentration. These values were used to create exponential decay plots and calculate half-lives, which were modeled using GraphPad Prism 6.
Expression and purification of proteins.
VHHs were expressed using the pHEN6 vector as previously described(53). Plasmids coding for PelB-VHH-LPETGG-His6 were transformed into WK6 E.coli. Bacteria were grown under ampicillin selection at 37 °C . Protein expression was induced with 1mM IPTG at 30 °C (overnight). Bacterial pellets were collected and resuspended in TES buffer (50 mM Tris, 650 μM EDTA, 2 M sucrose, 15 mL buffer per liter of culture) in preparation for osmotic shock. After incubating for 2 hours at 4 °C, 75 mL distilled H2O was added and the bacterial suspension was incubated overnight at 4 °C. VHHs were purified from the supernatant by Ni-NTA bead batch purification, followed by buffer exchange. Sortase-A pentamutant was expressed and purified as described(53).
Sortase conjugation conditions.
Sortagging reactions were performed in buffer containing 50 mM Tris, 150 mM NaCl, 5 mM CaCl2 pH 7.4 with 10% glycerol at 12° C overnight with a 1 mM concentration of G3-peptide and 20 μM Sortase-A 5M(53). Sortase A-5M and unreacted VHH-LPETGG-His6 were removed by passage over nickel-NTA beads. Unreacted G3-peptide was removed through use of an Amicon Ultra 10k cutoff centrifugal filtration device. The identity of the conjugates and their purity was assessed using liquid chromatography/mass spectrometry.
Animal protocols.
All procedures were performed in accordance with institutional guidelines and approved by the Boston Children’s Hospital Institutional Animal Care and Use Committee (IACUC protocol number 16–12–3328). Specific pathogen-free female (6–8-week-old) OT-II mice were purchased from The Jackson Laboratory. Prior to harvesting spleens, animals were vaccinated with ovalbumin (100 μg) and Polyinosinic:polycytidylic acid (50 μg) to boost T cell responses.
RESULTS:
Our experimental design focused on a model peptide antigen from ovalbumin (OVA323–339, WT; Figure 1). This peptide and its analogues have been extensively characterized with respect to proteolytic processing, MHC-II binding, and TCR activation(54–56). The portion of this peptide thought to be primarily responsible for binding to MHC-II and engaging the TCR, also called the core region, spans residues 329–337(51).
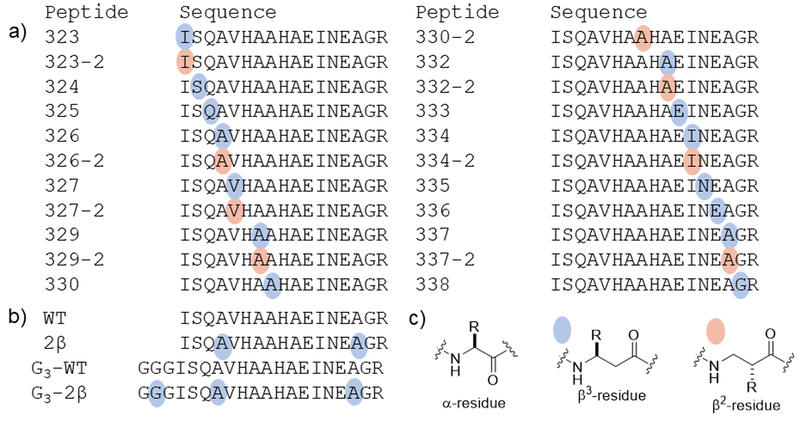
Figure 1. Structures of peptide analogues of ovalbumin 323–339.
(a) Sequences of peptides with a single β residue in this library. The conventional single-letter code is used to indicate α residue identity. Colored dots indicate sites of α-to-β replacement; each β residue bears the side chain of the α residue indicated by the letter. The number of the peptide analogue corresponds to the position of the residue in full-size ovalbumin protein. (b) Sequence of prototype peptide and analogues with two β residues or an N-terminal triglycine extension. (c) Structures of amino acid residues used in this study.
We began by synthesizing a set of analogues in which one of the natural α residues in OVA323–339 was replaced with a homologous β residue (Figure 1a, Supporting Figure 1). Each replacement involved retention of the natural side chain; thus, each α→β replacement simply introduced a methylene group into the peptide backbone. A side chain can be located at either of the two backbone positions between the carbonyl and nitrogen. β3 residues have the side chain adjacent to nitrogen, and β2 residues have the side chain adjacent to the carbonyl. It is straightforward to incorporate both types of β residue into peptides by standard solid-phase synthesis methods. β3 homologues of proteinogenic α-amino acids are commercially available, β3-homohistidine (β3-homoHis) being the only exception. Therefore, we prepared analogues of OVA323–339 containing single α→β3 replacements at 15 of the 17 positions (neither of the His residues was replaced). Fewer protected β2-homoamino acids are commercially available. We thus prepared analogues containing α→β2 replacements at only 8 of the 17 positions in the antigen.
For each of the 22 peptides containing a single α→β replacement, we characterized binding to two allelic products, I-Ad and I-Ab, of mouse MHC-II, both of which are known to bind WT (51, 57). Many sites of α-to-β substitution were tolerated with respect to binding to both MHC-II allelic products (Table 1). Several of the positions in which α-to-β substitution were not tolerated are sensitive to side chain modification(54, 55) and/or make direct contact with MHC-II(58). In general, α-to-β substitutions were tolerated better outside the core of the MHC/TCR-binding region of the peptide. Certain α-to-β substitutions abrogated binding to one MHC-II allelic product while sparing binding to the other. For example, introduction of a β2 or β3 residue at position 330 (330–2, 330) abrogates binding to I-Ab but not to I-Ad. Conversely, placement of a β2 residue at position 329 (329–2) abrogates binding to I-Ab but not to I-Ad. Interpretation of these findings in the absence of a structure of these complexes is difficult because many peptides, including OVA(323–339), can bind to MHC-II in different registers(51). Distinct registers differ in the residues that are crucial for binding to MHC-II and those that contact the TCR. At present we have no information on the effect of α→β replacement in terms of MHC-II binding register.
Table 1: Binding and T cell activation of peptide analogues of OVA323–339 is largely tolerant of single beta amino acid substitutions.
MHC-II binding measurements (I-Ad and I-Ab) represent the mean IC50 values from three independent experiments. Fold-WT is the observed IC50 normalized relative to the IC50 of WT peptide. The responses of T cells (DO11.10 or OT-II) induced by peptides are categorized into strong (+++), medium (++), weak (+) or undetectable (−). ND (not determined) indicates that this peptide was not analyzed in this assay. These categories are defined and the responses enumerated in Supporting Figure 1.
| I-Ad binding | DO11.10 (I-Ad), IL2 response | I-Ab binding | OT-II (I-Ab), IFNγ response | |||||
|---|---|---|---|---|---|---|---|---|
| IC50, nM | Fold WT | 10 μM peptide | IC50, nM | Fold WT | 10 μM peptide, ELISpot | |||
| WT | 2.0 | 1 | +++ | 230 | 1.0 | +++ | ||
| 323 | 5.1 | 3 | +++ | 520 | 2.2 | +++ | ||
| 323–2 | 0.73 | 0.37 | +++ | 370 | 1.6 | +++ | ||
| 324 | 8.0 | 4 | +++ | 610 | 2.6 | +++ | ||
| 325 | 30 | 15 | +++ | 620 | 2.7 | +++ | ||
| 326 | 17 | 8 | +++ | 350 | 1.5 | +++ | ||
| 326–2 | 15 | 7 | ND | 24,000 | 100 | ND | ||
| 327 | 86 | 44 | +++ | 3,500 | 15 | +++ | ||
| 327–2 | 92 | 47 | ++ | 8,900 | 38 | +++ | ||
| 329 | 55 | 28 | + | 21,000 | 89 | + | ||
| 329–2 | 530 | 270 | + | 350 | 1.5 | + | ||
| 330 | 7.1 | 4 | ND | >25,000 | >100 | ND | ||
| 330–2 | 14 | 7 | − | >25,000 | >100 | ++ | ||
| 332 | 100 | 51 | +++ | >25,000 | >100 | − | ||
| 332–2 | 94 | 47 | + | 4,500 | 19 | ++ | ||
| 333 | 171 | 86 | − | 950 | 4.1 | − | ||
| 334 | 43 | 22 | − | >25,000 | >100 | − | ||
| 334–2 | 45 | 23 | − | 10,000 | 43 | − | ||
| 335 | 11 | 6 | ++ | 480 | 2.1 | + | ||
| 336 | 6.9 | 3 | +++ | 550 | 2.3 | − | ||
| 337 | 1.0 | 0.49 | +++ | 270 | 1.1 | +++ | ||
| 337–2 | 6.7 | 3 | +++ | 750 | 3.2 | +++ | ||
| 338 | 0.70 | 0.36 | +++ | 290 | 1.2 | +++ | ||
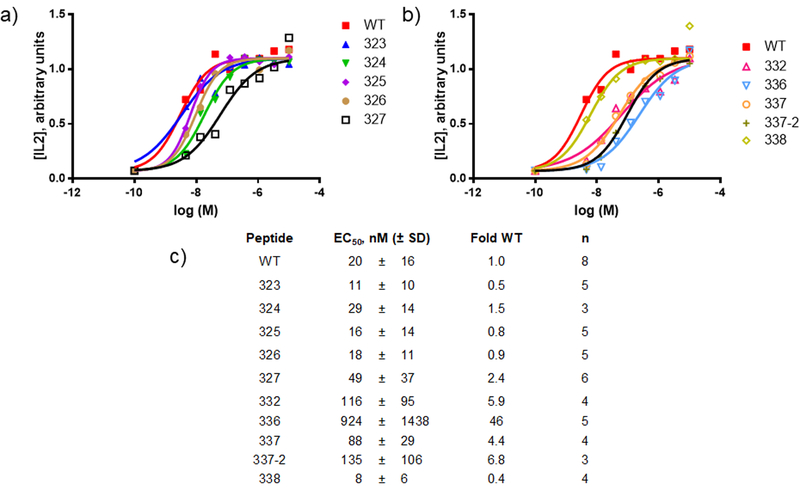
We tested T cell responses to the peptides containing single α→β replacements in the context of both I-Ab and I-Ad . For I-Ad we used the murine B cell lymphoma line (A20 cell) as the antigen presenting cell and the OVA323–339-specific T cell hybridoma DO11.10 as the responder(59). Release of the IL2 by DO11.10 was monitored as an indicator of activation(51). We first surveyed T cell hybridoma activation through incubation of a mixture of A20 and DO11.10 cells with a single concentration of most of the peptides (10 μM, Table 1, Supporting Figure 1). Peptides with β-residues outside the peptide core known to bind MHC-II and engage the TCR were generally efficient at stimulating IL2 production, whereas many with substitutions in the core region showed low or negligible stimulatory activity. The consequences of α-to-β substitution at position 332 depended on side chain location: the β3 substitution (332) was well tolerated, but the β2-substitution (332–2) was not, with respect to retention of T cell stimulatory activity. This difference could not be attributed to differences in I-Ad affinity, which was similar for 332 and 332–2. The finding that β-substitution at position 336 or 337 was well tolerated is surprising, since these residues in WT were predicted to be important for TCR engagement and MHC-II binding(51, 54). For peptides that efficiently induced IL2 release at 10 μM, a more thorough analysis of activity was conducted (Figure 2). The dose-response measurements showed that several analogues matched WT in potency, and one analogue (338) was slightly more potent.
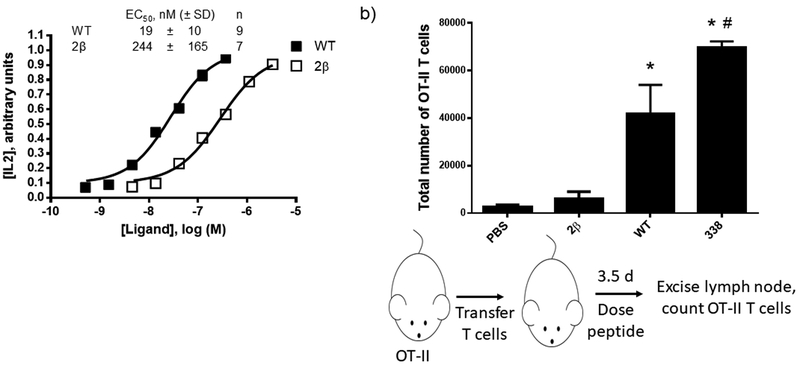
Figure 2. Dose-response for IL2 release from DO11.10 cells following peptide treatment.
Peptide stimulation and measurement of IL2 levels was performed as described in methods. (a-b) Shown are representative dose response curves from a single experiment for selected peptides that stimulate strong IL2 responses in the single dose studies. Both plots in this panel are from the same experiment. WT is included in both plots for comparison. (c) Tabulation of calculated potencies for IL2 production. Data was fit to a 4-parameter model as described in methods. The number of independent replicates (n) is listed.
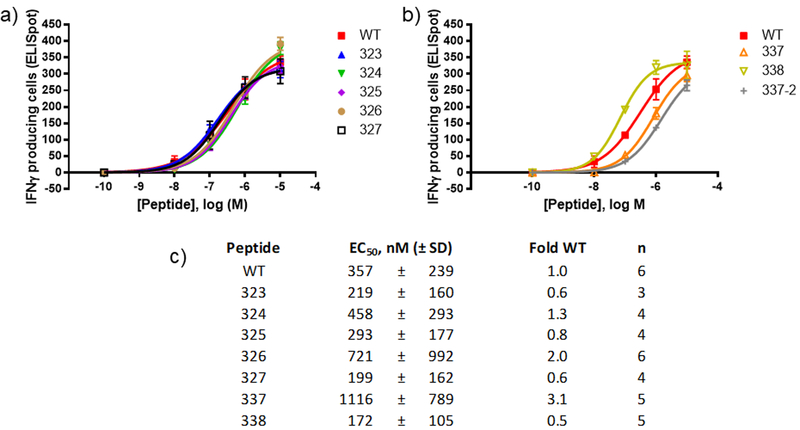
The OT-II mouse model contains CD4+ T cells that respond strongly to WT(54). OT-II mice on a C57/BL6 background were used to assess T cell activation by I-Ab restricted peptides. Total unfractionated splenocyte preparations from ovalbumin-boosted OT-II mice were used as the source of both antigen-presenting cells and antigen-specific T cells. T cell activation was measured by recording the number of interferon gamma (IFNγ) releasing cells in an ELIspot assay. We tested most of the library at a high concentration (10 μM) to compare the efficacies of these peptides for induction of IFNγ production (Table 1, Supporting Figure 2). Most of the peptides with β-substitutions outside of the peptide core were quite active, while peptides with β-substitutions inside the core were only weakly active or inactive in this assay. The exceptions to this latter trend were peptides with α-to-β substitution at position 337. Certain analogues exhibited striking differences in T cell stimulatory activity in the different models. For example, 336 stimulated DO11.10 T cells but not OT-II T cells, behavior that does not correlate with binding to the two MHC-II alleles (strong in both cases). For peptides that at 10 μM displayed efficacy similar to that of WT for stimulating IFNγ release from OT-II T cells, we performed more detailed measurements to estimate potencies (Figure 3). The EC50 values of several analogues were within a few-fold of WT. Analogue 338 was more potent than WT, as observed for IL2 release from DO11–10 cells.
Figure 3. Dose-response curve for interferon gamma release from OT-II splenocyte preparations following peptide treatment.
Splenocyte preparation, treatment and analysis was performed as described in methods. (a-b) Shown are representative dose response curves from a single experiment for selected peptides that stimulate strong interferon-gamma responses in the single dose studies. Both plots in this panel are from the same experiment. WT is included in both plots for comparison. Error bars correspond to standard deviation from technical replicates performed in that experiment. (c) Tabulation of calculated potencies for interferon gamma production. Data was fit to a 4-parameter model as described in methods. The number of independent replicates (n) is listed.
Assessment of T cell proliferation serves as an alternative to the measurement of cytokine release for evaluating peptide-induced T cell activation. We tested whether the WT analogues that contain single α-to-β replacements induced proliferation of OT-II T cells. Purified OT-II T cells were incubated with purified dendritic cells in the presence of each peptide (50 nM). Most peptides efficiently induced proliferation (Supporting Figure 2). Only three peptides (332, 336, 334–2) failed to induce proliferation, and each of them lacked IFNγ stimulatory activity. Peptides that induced IFNγ release only weakly still caused robust T cell proliferation (329, 330–2). T cell proliferation appears to be a more sensitive test of activation than IFNγ production under the conditions tested.
Based on results obtained with WT analogues containing a single α-to-β replacement, we prepared and tested a new analogue designated 2β that contains two α-to-β replacements (Figure 1). The incorporation of two β-residues is expected to improve protection against proteolysis relative to a single substitution. The two sites of β-residue incorporation in 2β were chosen based on tolerance of single α-to-β substitutions (results for 326 and 337). We chose these sites for substitution, and not sites located closer the N- or C-termini of the peptide (such as 338), in an effort to provide protection from proteolysis to the central portion of the peptide. Peptide 2β was ~13-fold less potent than WT in the DO11.10 T cell stimulation assay (Figure 4a).
Figure 4. Characterization of peptide T cell stimulatory activity.
(a) Representative dose-response curve and tabulated data for DO11.10 IL2 assays. Data points represent mean ± standard deviation. Tabulated data is derived from the indicated number of independent replicates (n). Data in this figure are distinct from those in Figure 2. Data was fit to a 4-parameter model as described in methods. (b) Results and schematic for in vivo proliferation assays. OT-II T cells were transferred into recipient mice lacking T cells. Recipient mice were treated with peptide (1 μg) or PBS as a negative control. 3.5 days after peptide treatment inguinal lymph nodes were excised and OT-II T cells were enumerated by flow cytometry. Data represent mean ± standard deviation from 3 (PBS), 4 (WT), or 5 (2β/338) mice, respectively. *Indicates p < 0.05 vs PBS; #indicates p < 0.05 vs 2β.
In order to test whether β-residue-containing analogues of the WT OVA peptide were active in vivo we returned to the OT-II mouse model. T cells from OT-II mice were transferred into mice that lack T cells (TCRα/β knockout mice). Selected peptides were then injected into recipient mice, and the proliferation of the transferred cells was analyzed by determining of the number of transferred T cells in the draining lymph node using flow cytometry (Figure 4b). In the absence of stimulatory peptide (PBS control) few OT-II T cells were recovered. Injection of WT OVA peptide resulted in the recovery of significantly more OT-II T cells, and peptide 338 was even more effective in this regard, in line with findings from cell-based assays (Figures 2, 3). Mice treated with 2β did not differ significantly from mice that did not receive any peptide. This lack of in vivo T cell stimulatory activity for 2β was paralleled by observations made with OT-II cells ex vivo, which showed that 2β only induced weak production of IFNγ according to ELIspot assays (Supporting Figure 3).
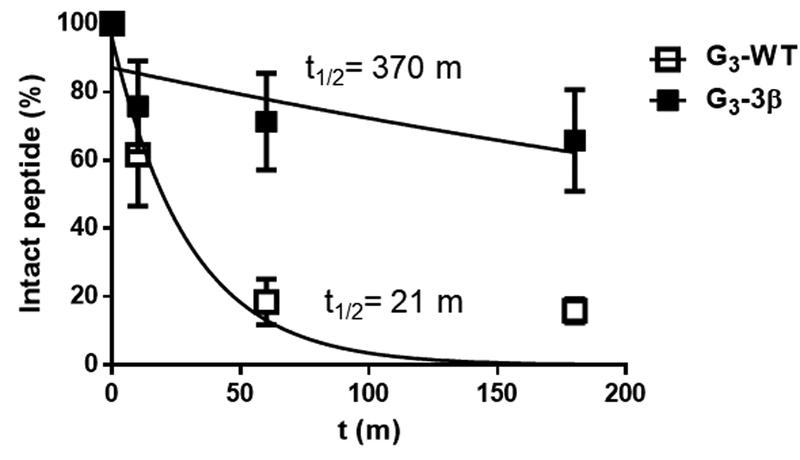
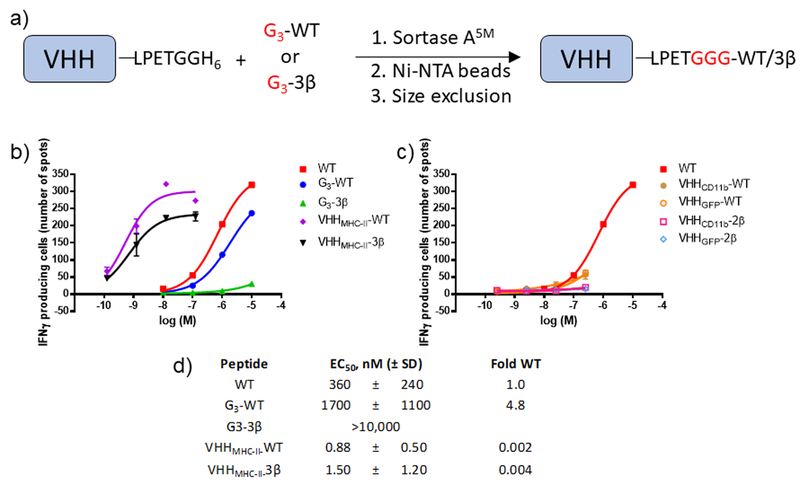
We sought to overcome the negative impact of β-residue incorporation on T cell activation through directed delivery of peptide to antigen presenting cells. Conjugation of an MHC-binding peptide to proteins derived from the variable domain of camelid heavy chain-only antibodies (VHHs or nanobodies) that target cell surface proteins on antigen-presenting cells enhances the immunogenicity of that peptide under immunostimulatory conditions(53, 60, 61). Site-specific and monovalent functionalization of proteins using catalysis by sortase A (sortagging) is our preferred method to generate such adducts(62). We prepared two peptides to pursue this approach. G3-WT is the analogue of WT, bearing a triglycine extension at the N-terminus for sortagging (Figure 1). G3-3β contains the N-terminal extension and three α-to-β replacements, including one in the extension. We examined the susceptibilities of G3-WT and G3-3β to proteolysis by cathepsin S, an endosomal protease important for loading internalized antigens onto MHC-II, (Figure 5, Supporting Figure 4a)(63). As expected, G3-WT was degraded more rapidly than G3-3β, given that β-amino acid residues are known to disfavor proteolytic cleavage of nearby backbone amide bonds(33, 64). Mass spectrometry showed that the principal cleavage by cathepsin S occurred between residues 328 and 329 (Supporting Figure 4b).
Figure 5. A peptide with multiple β-residues is resistant to an endosomal protease.
Time course of degradation of peptides by cathepsin S. Peptides were incubated with cathepsin S for the indicated times and peptide abundance was analyzed by tandem liquid chromatography/mass spectrometry (LC/MS) as described in methods. Data was fit to an exponential decay model to provide indicated half-lives (t1/2). Error bars represent SD.
Sortagging was used to append either G3-WT or G3-3β site-specifically to the C-terminus of VHHs that target MHC-II (VHH7(53)), CD11b (DC13(53)), or green fluorescent protein (GFP, VHH “enhancer”(65)). Delivery of a peptide antigen to MHC-II-expressing cells by conjugation to VHH enhances activation of cognate CD4+ T cells, whereas delivery of antigen to CD11b-expressing cells also promotes activation of CD8+ T cells (53). The GFP-binding VHH serves as a negative control for these experiments. We assessed the VHH-peptide conjugates and the free triglycine-containing peptides used to make them for their ability to stimulate OT-II T cells using an ELISpot assay (Figure 6). G3-WT was less potent than WT, and G3-3β was only weakly active at the highest concentration tested. The latter observation is in line with the lack of activity of 2β in the in vivo OT-II proliferation assay (Figure 5). Conjugates consisting of VHHs that target GFP or CD11b and G3-WT were weakly active, and conjugates of these VHHs with G3-3β were virtually inactive. Because of their weak activity we did not calculate EC50 values. In contrast, conjugates composed of VHH7 (which targets MHC-II) and either G3-WT or G3-3β were more potent than any other compound tested in these assays. This trend was also observed in assays with DO11.10 cells (Supporting Figure 3d–e). The in vivo properties of the various VHH-peptide adducts remain to be explored, although past work has shown that VHHMHC-II-WT is active in mice(53).
Figure 6. Effective stimulation of OT-II T cells by VHHMHC-II-peptide conjugates.
(a) Schematic for synthesis of VHH-peptide conjugates using sortase A catalysis. Reactions were performed as described in methods. (b-c) Representative dose-response curves for stimulation of IFNγ release by peptides or VHH-peptide conjugates. OT-II splenocytes were stimulated overnight and IFNγ production was measured using ELISpot as described in methods. Data points are mean ± SD and curves result from the fitting of four parameter dose-response model. Data in these two panels are split into (b) active conjugates and (c) weakly active conjugates, both taken from the same experiment. (d) Tabulation of dose-response values for peptides and active VHH-peptide conjugates. Data are from ≥ 3 independent experiments with replicates shown in Supporting Figure 3.
DISCUSSION:
These experiments were motivated by the fact that very little is known of how β-residue incorporation affects binding to MHC-II and engagement of the TCR(30, 45, 46). Past studies have probed the impact of reduced amino acid residues(16, 21–24, 26, 27) or N-methylated residues(21, 23) on MHC-II binding and TCR recognition. Reduced analogues of α-amino acid residues replace the carbonyl group with a methylene group. This modification is predicted to alter backbone charge, because the amino group, found in place of the former amide group, should be protonated at physiological pH. In addition, this modification should alter backbone conformational properties. Amide N-methylation is predicted to exert a strong effect on backbone conformation and this change furthermore removes an H-bond donor site. In contrast, replacement of α-amino acid residues with homologous β-amino acid residues may lead to more modest changes, relative to replacements involving reduced residues or N-methyl residues(32, 66).
We prepared a library of analogues of a prototypical MHC-II binding peptide, OVA(323–339), in which a single α-amino acid residue was replaced by the β3 homologue or the β2 homologue (Figure 1). In one case, we examined a double replacement. This study is the first reported systematic evaluation of the impact of incorporating homologous β-residues on MHC-II binding and TCR engagement. We found that α-to-β replacement was tolerated at many positions, mostly those outside the core of the peptide, but at a few within (Figure 2, Table 1). Nonetheless, amino acid substitutions outside the core can affect T cell recognition by changing the register that is predominantly adopted in peptide-MHC-II complexes(51, 67). Substitutions at certain positions imparted selectivity for binding to different allelic variants of MHC-II (329–2, 330–2, 330), while others abrogated T cell stimulatory activity without altering MHC-II binding (330–2, 336) (Table 1). The demonstration that incorporation of a β residue can abrogate TCR engagement without compromising MHC-II binding mirrors past findings with an MHC-II binding peptide derived from MOG(46). Past work has shown that β-residue incorporation can impart selectivity between receptor subtypes(68) and between distinct signaling pathways for a single receptor(69). The replacement of α with β residues might therefore represent a general strategy for creating MHC-II binding peptides with unique functional profiles.
Incorporation of β-residue(s) can stabilize peptides against hydrolysis by proteases, at least near the site of modification(33, 34). Proteolytic processing, predominantly within endosomal compartments, is an important step in transformation of protein antigens into peptides that can be loaded onto MHC-II(3). Whether β-residues can be introduced into MHC-II peptides at a density sufficient to protect against proteolysis while retaining MHC-II binding and the ability to engage the TCR is unknown. In an effort to balance proteolytic susceptibility and antigenic activity, we combined two β-substitutions that were well-tolerated in all assays involving the single α-to-β library. The resulting peptide (2β) effectively stimulated DO11.10 T cells (Figure 4a). An analogue with a short N-terminal extension containing two Gly and one β-homoGly residues (G3-3β) was substantially more resistant to degradation by cathepsin S, an endosomal protease, than the analogous peptide lacking β-residues (G3-WT) (Figure 5). Since full size proteins are often more stable in the presence of proteases than short peptide fragments, the combination of β-residue incorporation and protein conjugation may further improve peptide stability. Whether exogenously added peptides that can bind to MHC-II undergo internalization into protease-containing compartments prior to loading onto MHC-II, or bind directly to peptide-receptive MHC-II at the cell surface is not known(3). We therefore cannot at present prove a direct link between the proteolytic resistance of 2β and the immunological properties of this backbone-modified peptide but speculate that an important connection exists.
Since β-residues disfavor proteolysis at nearby peptide bonds, the α-to-β replacement strategy described here can be readily applied to situations in which sites of cleavage are known. In some cases, discrete proteolytic processing steps are known to be important for the excision of peptide epitopes from full size protein antigens(7, 8). Such processing steps could be modulated by the incorporation of β-residues into larger peptides near sites at which proteolysis occurs. This approach might be useful for guiding the proteolytic excision of peptides with potent immunostimulatory properties, such as cryptic epitopes(6) present in larger proteins.
The observation that 338 is more potent than WT in all assays is striking. Position 338 is outside of the peptide core of OVA(323–339), yet modification at this position enhances MHC-II binding and TCR engagement. The enhanced properties of 338 in MHC-II binding assays are unrelated to stability in the presence of proteases, as these assays were performed with purified proteins or in the presence of protease inhibitors. A more likely explanation for the high potency of 338, compared to all the other β-residue containing analogues tested, relates to the identity of the amino acid at this position. Position 338 of OVA(323–339) contains the only Gly in this peptide, raising the possibility that spatial displacement of the side chain is problematic for α-to-β modifications at non-Gly positions in antigenic peptides. It will be interesting to see whether such modifications at Gly in other antigenic peptides are similarly well tolerated.
While 2β proved effective at stimulating DO11.10 cells, it failed to stimulate proliferation of OT-II T cells in vivo (Figure 4) and its analogue G3-3β did not induce IFNγ release from OT-II splenocyte preparations (Figure 6). To overcome this lack of activity, we made use of past observations that targeting of antigenic peptides to antigen presenting cells by conjugation to VHHs of appropriate specificity can enhance immunogenicity(53, 60, 61). To achieve this goal we needed to introduce β-amino acids in the context of a full-length protein. While the cell’s protein synthesis machinery typically excludes β-amino acids, methods have been developed to incorporate β-amino acids by reprogramming the translational machinery; however, current methods yield relatively modest amounts of product, as mixtures(70). We circumvented this problem by using sortagging to conjugate peptides with β-residues to the C-termini of VHHs. We used sortagging to attach G3-WT or G3-3β to the C-termini of VHHs that target MHC-II, CD11b or GFP. Conjugation of G3-WT or G3-3β to VHHMHC-II resulted in dramatically enhanced T cell stimulatory activity in vitro, whereas conjugation or either peptide to VHHCD11b or VHHGFP did not. This finding is in line with work showing that conjugation of antigenic peptides to VHHMHC-II dramatically enhanced CD4+ T cell and antibody responses(53). It is unknown whether MHC-II can simultaneously bind peptide conjugated to the C-terminus of VHHMHC-II and the VHH itself, either in cis to one and the same MHC-II, or in trans, to an adjoining MHC-II. If simultaneous binding is possible, then peptide conjugated to VHH might be able to productively bind to MHC-II at the cell surface to form a complex that then activates TCR signaling. An alternative and more likely possibility is that the peptide-VHH-MHC-II complex must first be internalized for proteolytic processing, prior to loading of peptide into the MHC-II binding groove. While our results do not differentiate between these possibilities, we note that G3-3β contains a β-residue in the area of the peptide that should be cleaved to release the antigen for recognition by MHC-II. The β-homoGly residue near the N-terminus of G3-3β should inhibit proteolysis of nearby peptide bonds (Figure 5), but this effect does not seem to be detrimental for stimulating T cell responses (Figure 6).
We find that α-to-β replacements at most positions in an MHC-II binding peptide do not abrogate MHC-II or TCR engagement. Because these replacements employ β homologues of the original α residues, only the peptide backbone is modified in these peptides. Our data identify β-residue-containing peptides with a variety of interesting properties, including discrimination between MHC-II allelic variants, MHC-II binding without TCR engagement, enhanced MHC affinity/TCR agonism, and enhanced stability in the presence of a protease. One of our backbone-modified analogues displayed enhanced activity in vivo relative to the index peptide (all α-peptide). The in vitro T cell stimulatory activity of a peptide with two β-residues was substantially enhanced by delivery to MHC-II through conjugation to an MHC-II-specific VHH. These properties highlight the utility of backbone-modified peptides in interrogating antigen presentation and present a strategy for the design of new antigenic peptides with improved properties.
Supplementary Material
KEY POINTS:
Peptides with β-amino acids can bind tightly to MHC-II and activate TCR signaling
Incorporation of β-amino acids enhances resistance to degradation by protease(s)
A selected β- amino acid-containing peptide stimulated T cells in mice
Acknowledgements:
We acknowledge Dr. Philippa Marrack, National Jewish Health Center, Denver, Co. for provision of the DO11.10 hybridoma cell line.
Research was supported by NIH NAIAD contract HHSN272201400045C (AS), NIH grant R01 GM056414 (SHG), Lustgarten Foundation award 388167 (HLP), Melanoma Research Alliance Award 51009 (HLP), and NIH grant R01-AI087879 (H.L.P.). R.W.C. was supported in part by a Biotechnology Training Grant from NIGMS (T32 GM008349) and a Cancer Research Institute Postdoctoral Fellowship. A. W. was supported by a Beckman Institute Postdoctoral Fellowship.
References:
- 1.Villadangos JA, and Ploegh HL. 2000. Proteolysis in MHC class II antigen presentation: Who’s in charge? Immunity 12: 233–239. [DOI] [PubMed] [Google Scholar]
- 2.York IA, Goldberg AL, Mo XY, and Rock KL. 1999. Proteolysis and class I major histocompatibility complex antigen presentation. Immunol. Rev 172: 49–66. [DOI] [PubMed] [Google Scholar]
- 3.Blum JS, Wearsch PA, and Cresswell P. 2013. Pathways of Antigen Processing. Annu. Rev. Immunol 31: 443–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dintzis HM, Symer DE, Dintzis RZ, Zawadzke LE, and Berg JM. 1993. A Comparison of the Immunogenicity of a Pair of Enantiomeric Proteins. Proteins. 16: 306–308. [DOI] [PubMed] [Google Scholar]
- 5.Croft NP, and Purcell AW. 2011. Peptidomimetics: modifying peptides in the pursuit of better vaccines. Expert Rev. Vaccines 10: 211–226. [DOI] [PubMed] [Google Scholar]
- 6.Medd PG, and Chain BM. 2000. Protein degradation in MHC class II antigen presentation: opportunities for immunomodulation. Semin. Cell Dev. Biol 11: 203–210. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez GM, and Diment S. 1995. Destructive Proteolysis of Cysteine Proteases in Antigen Presentation of Ovalbumin. Eur. J. Immunol 25: 1823–1827. [DOI] [PubMed] [Google Scholar]
- 8.ManourySchwartz B, Chiocchia G, Lotteau V, and Fournier C. 1997. Selective increased presentation of type II collagen by leupeptin. Int. Immunol 9: 581–589. [DOI] [PubMed] [Google Scholar]
- 9.Moudgil KD, Sercarz EE, and Grewal IS. 1998. Modulation of the immunogenicity of antigenic determinants by their flanking residues. Immunol. Today 19: 217–220. [DOI] [PubMed] [Google Scholar]
- 10.Rammensee HG 1995. Chemistry of Peptides Associated with MHC Class-I and Class-II Molecules. Curr. Opin. Immunol 7: 85–96. [DOI] [PubMed] [Google Scholar]
- 11.Sidney J, Peters B, Frahm N, Brander C, and Sette A. 2008. HLA class I supertypes: a revised and updated classification. Bmc Immunology 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernando MMA, Stevens CR, Walsh EC, De Jager PL, Goyette P, Plenge RM, Vyse TJ, and Rioux JD. 2008. Defining the role of the MHC in autoimmunity: A review and pooled analysis. Plos Genetics 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Painter CA, and Stern LJ. 2012. Conformational variation in structures of classical and non-classical MHCII proteins and functional implications. Immunol. Rev 250: 144–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McFarland BJ, Katz JF, Beeson C, and Sant AJ. 2001. Energetic asymmetry among hydrogen bonds in MHC class II-peptide complexes. Proc. Natl. Acad. Sci. U.S.A. 98: 9231–9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arneson LS, Katz JF, Liu M, and Sant AJ. 2001. Hydrogen bond integrity between MHC class II molecules and bound peptide determines the intracellular fate of MHC class II molecules. J. Immunol 167: 6939–6946. [DOI] [PubMed] [Google Scholar]
- 16.Benkirane N, Guichard G, Briand JP, and Muller S. 1996. Exploration of requirements for peptidomimetic immune recognition - Antigenic and immunogenic properties of reduced peptide bond pseudopeptide analogues of a histone hexapeptide. J. Biol. Chem 271: 33218–33224. [DOI] [PubMed] [Google Scholar]
- 17.Bergseng E, Xia J, Kim CY, Khosla C, and Sollid LM. 2005. Main chain hydrogen bond interactions in the binding of proline-rich gluten peptides to the Celiac disease-associated HLA-DQ2 molecule. J. Biol. Chem 280: 21791–21796. [DOI] [PubMed] [Google Scholar]
- 18.Bianco A, Brock C, Zabel C, Walk T, Walden P, and Jung G. 1998. New synthetic non-peptide ligands for classical major histocompatibility complex class I molecules. J. Biol. Chem 273: 28759–28765. [DOI] [PubMed] [Google Scholar]
- 19.Bolin DR, Swain AL, Sarabu R, Berthel SJ, Gillespie P, Huby NJS, Makofske R, Orzechowski L, Perrotta A, Toth K, Cooper JP, Jiang N, Falcioni F, Campbell R, Cox D, Gaizband D, Belunis CJ, Vidovic D, Ito K, Crowther R, Kammlott U, Zhang XL, Palermo R, Weber D, Guenot J, Nagy Z, and Olson GL. 2000. Peptide and peptide mimetic inhibitors of antigen presentation by HLA-DR class II MHC molecules. Design, structure-activity relationships, and X-ray crystal structures. J. Med. Chem 43: 2135–2148. [DOI] [PubMed] [Google Scholar]
- 20.McFarland BJ, Katz JF, Sant AJ, and Beeson C. 2005. Energetics and cooperativity of the hydrogen bonding and anchor interactions that bind peptides to MHC class II protein. J. Mol. Biol 350: 170–183. [DOI] [PubMed] [Google Scholar]
- 21.Guichard G, Calbo S, Muller S, Kourilsky P, Briand JP, and Abastado JP. 1995. Efficient Binding of Reduced Peptide-Bond Pseudopeptides to Major Histocompatibility Complex Class-I Molecule. J. Biol. Chem 270: 26057–26059. [DOI] [PubMed] [Google Scholar]
- 22.Quesnel A, Zerbib A, Connan F, Guillet JG, Briand JP, and Choppin J. 2001. Synthesis and antigenic properties of reduced peptide bond analogues of an immunodominant epitope of the melanoma MART-1 protein. J. Pept. Sci 7: 157–165. [DOI] [PubMed] [Google Scholar]
- 23.Cotton J, Herve M, Pouvelle S, Maillere B, and Menez A. 1998. Pseudopeptide ligands for MHC II-restricted T cells. Int. Immunol 10: 159–166. [DOI] [PubMed] [Google Scholar]
- 24.Bastian M, Lozano JM, Patarroyo ME, Pluschke G, and Daubenberger CA. 2004. Characterization of a reduced peptide bond analogue of a promiscuous CD4 T cell epitope derived from the Plasmodium falciparum malaria vaccine candidate merozoite surface protein 1. Mol. Immunol 41: 775–784. [DOI] [PubMed] [Google Scholar]
- 25.de Haan EC, Moret EE, Wagenaar-Hilbers JPA, Liskamp RMJ, and Wauben MHM. 2005. Possibilities and limitations in the rational design of modified peptides for T cell mediated immunotherapy. Mol. Immunol 42: 365–373. [DOI] [PubMed] [Google Scholar]
- 26.Calbo S, Guichard G, Bousso P, Muller S, Kourilsky P, Briand JP, and Abastado JP. 1999. Role of peptide backbone in T cell recognition. J. Immunol 162: 4657–4662. [PubMed] [Google Scholar]
- 27.Stemmer C, Quesnel A, Prevost-Blondel A, Zimmermann C, Muller S, Briand JP, and Pircher H. 1999. Protection against lymphocytic choriomeningitis virus infection induced by a reduced peptide bond analogue of the H-2D(b)-restricted CD8(+) T cell epitope GP33. J. Biol. Chem 274: 5550–5556. [DOI] [PubMed] [Google Scholar]
- 28.Andersson IE, Batsalova T, Haag S, Dzhambazov B, Holmdahl R, Kihberg J, and Linusson A. 2011. (E)-Alkene and Ethylene Isosteres Substantially Alter the Hydrogen-Bonding Network in Class II MHC A(q)/Glycopeptide Complexes and Affect T-Cell Recognition. J. Am. Chem. Soc 133: 14368–14378. [DOI] [PubMed] [Google Scholar]
- 29.Bianco A, Zabel C, Walden P, and Jung G. 1998. N-hydroxy-amide analogues of MHC-class I peptide ligands with nanomolar binding affinities. J. Pept. Sci 4: 471–478. [DOI] [PubMed] [Google Scholar]
- 30.Hart M, and Beeson C. 2001. Utility of azapeptides as major histocompatibility complex class II protein ligands for T-cell activation. J. Med. Chem 44: 3700–3709. [DOI] [PubMed] [Google Scholar]
- 31.Horne WS 2011. Peptide and peptoid foldamers in medicinal chemistry. Expert Opin. Drug Disc. 6: 1247–1262. [DOI] [PubMed] [Google Scholar]
- 32.Checco JW, and Gellman SH. 2016. Targeting recognition surfaces on natural proteins with peptidic foldamers. Curr. Opin. Struct. Biol 39: 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hook DF, Bindschadler P, Mahajan YR, Sebesta R, Kast P, and Seebach D. 2005. The proteolytic stability of ‘designed’ beta-peptides containing alpha-peptide-bond mimics and of mixed alpha,beta-peptides: Application to the construction of MHC-binding peptides. Chem. Biodivers 2: 591–632. [DOI] [PubMed] [Google Scholar]
- 34.Werner HM, Cabalteja CC, and Horne WS. 2016. Peptide Backbone Composition and Protease Susceptibility: Impact of Modification Type, Position, and Tandem Substitution. ChemBioChem 17: 712–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steer DL, Lew RA, Perlmutter P, Smith AI, and Aguilar MI. 2002. beta-amino acids: Versatile peptidomimetics. Curr. Med. Chem 9: 811–822. [DOI] [PubMed] [Google Scholar]
- 36.Horne WS, Johnson LM, Ketas TJ, Klasse PJ, Lu M, Moore JP, and Gellman SH. 2009. Structural and biological mimicry of protein surface recognition by alpha/beta-peptide foldamers. Proc. Natl. Acad. Sci. U.S.A. 106: 14751–14756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horne WS, Boersma MD, Windsor MA, and Gellman SH. 2008. Sequence-based design of alpha/beta-peptide foldamers that mimic BH3 domains. Angew. Chem. Int. Ed 47: 2853–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson LM, Barrick S, Hager MV, McFedries A, Homan EA, Rabaglia ME, Keller MP, Attie AD, Saghatelian A, Bisello A, and Gellman SH. 2014. A Potent alpha/beta-Peptide Analogue of GLP-1 with Prolonged Action in Vivo. J. Am. Chem. Soc 136: 12848–12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheloha RW, Maeda A, Dean T, Gardella TJ, and Gellman SH. 2014. Backbone modification of a polypeptide drug alters duration of action in vivo. Nat. Biotechnol 32: 653–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Checco JW, Kreitler DF, Thomas NC, Belair DG, Rettko NJ, Murphy WL, Forest KT, and Gellman SH. 2015. Targeting diverse protein-protein interaction interfaces with alpha/beta-peptides derived from the Z-domain scaffold. Proc. Natl. Acad. Sci. U.S.A. 112: 4552–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guichard G, Zerbib A, Le Gal FA, Hoebeke J, Connan F, Choppin J, Briand JP, and Guillet JG. 2000. Melanoma peptide MART-1(27–35) analogues with enhanced binding capacity to the human class I histocompatibility molecule HLA-A2 by introduction of a beta-amino acid residue: Implications for recognition by tumor-infiltrating lymphocytes. J. Med. Chem 43: 3803–3808. [DOI] [PubMed] [Google Scholar]
- 42.Reinelt S, Marti M, Dedier S, Reitinger T, Folkers G, de Castro JAL, and Rognan D. 2001. beta-amino acid scan of a class I major histocompatibility complex-restricted alloreactive T-cell epitope. J. Biol. Chem 276: 24525–24530. [DOI] [PubMed] [Google Scholar]
- 43.Webb AI, Dunstone MA, Williamson NA, Price JD, de Kauwe A, Chen WS, Oakley A, Perlmutter P, McCluskey J, Aguilar MI, Rossjohn J, and Purcell AW. 2005. T cell determinants incorporating beta-amino acid residues are protease resistant and remain immunogenic in vivo. J. Immunol 175: 3810–3818. [DOI] [PubMed] [Google Scholar]
- 44.Cheloha RW, Sullivan JA, Wang T, Sand JM, Sidney J, Sette A, Cook ME, Suresh M, and Gellman SH. 2015. Consequences of Periodic alpha-to-beta(3) Residue Replacement for Immunological Recognition of Peptide Epitopes. ACS Chem. Biol 10: 844–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dali H, Busnel O, Hoebeke J, Bi LR, Decker P, Briand JP, Baudy-Floc’h M, and Muller S. 2007. Heteroclitic properties of mixed alpha- and aza-beta(3)-peptides mimicking a supradominant CD4 T cell epitope presented by nucleosome. Mol. Immunol 44: 3024–3036. [DOI] [PubMed] [Google Scholar]
- 46.McDonald CA, Payne NL, Sun GZ, Clayton DJ, Del Borgo MP, Aguilar MI, Perlmutter P, and Bernard CCA. 2014. Single beta(3)-amino acid substitutions to MOG peptides suppress the development of experimental autoimmune encephalomyelitis. J. Neuroimmunol 277: 67–76. [DOI] [PubMed] [Google Scholar]
- 47.Kuipers BJH, and Gruppen H. 2007. Prediction of molar extinction coefficients of proteins and peptides using UV absorption of the constituent amino acids at 214 nm to enable quantitative reverse phase high-performance liquid chromatography-mass spectrometry analysis. J. Agric. Food Chem. 55: 5445–5451. [DOI] [PubMed] [Google Scholar]
- 48.Sidney J, Southwood S, Moore C, Oseroff C, Pinilla C, Grey HM, and Sette A. 2013. Measurement of MHC/peptide interactions by gel filtration or monoclonal antibody capture. Curr. Protoc. Immunol. Chapter 18: Unit 18.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng Y, and Prusoff WH. 1973. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol 22: 3099–3108. [DOI] [PubMed] [Google Scholar]
- 50.Gulukota K, Sidney J, Sette A, and DeLisi C. 1997. Two complementary methods for predicting peptides binding major histocompatibility complex molecules. J. Mol. Biol 267: 1258–1267. [DOI] [PubMed] [Google Scholar]
- 51.Roy BM, Zhukov DV, and Maynard JA. 2012. Flanking Residues Are Central to DO11.10 T Cell Hybridoma Stimulation by Ovalbumin 323–339. Plos One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan LS, Woodham AW, Da Silva DM, and Kast WM. 2015. Functional Analysis of HPV-Like Particle-Activated Langerhans Cells In Vitro. Cervical Cancer: Methods and Protocols 1249: 333–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duarte JN, Cragnolini JJ, Swee LK, Bilate AM, Bader J, Ingram JR, Rashidfarrokhi A, Fang T, Schiepers A, Hanke L, and Ploegh HL. 2016. Generation of Immunity against Pathogens via Single-Domain Antibody-Antigen Constructs. J. Immunol 197: 4838–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robertson JM, Jensen PE, and Evavold BD. 2000. DO11.10 and OT-II T cells recognize a C-terminal ovalbumin 323–339 epitope. J. Immunol 164: 4706–4712. [DOI] [PubMed] [Google Scholar]
- 55.Sette A, Buus S, Colon S, Smith JA, Miles C, and Grey HM. 1987. Structural Characteristics of an Antigen Required for its Interaction with IA and Recognition by T-cells. Nature 328: 395–399. [DOI] [PubMed] [Google Scholar]
- 56.Michalek MT, Benacerraf B, and Rock KL. 1989. 2 Genetically Identical Antigen-Presenting Cell Clones Display Heterogeneity in Antigen Processing. Proc. Natl. Acad. Sci. U.S.A. 86: 3316–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marrack P, Endres R, Shimonkevitz R, Zlotnik A, Dialynas D, Fitch F, and Kappler J. 1983. The Major Histocompatibility Complex-Restricted Antigen Receptor on T-Cells .2. Role of the L3T4 Product. J. Exp. Med 158: 1077–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scott CA, Peterson PA, Teyton L, and Wilson IA. 1998. Crystal structures of two I-A(d)-peptide complexes reveal that high affinity can be achieved without large anchor residues. Immunity 8: 319–329. [DOI] [PubMed] [Google Scholar]
- 59.White J, Haskins KM, Marrack P, and Kappler J. 1983. Use of I Region-Restricted, Antigen-Specific T-Cell Hybridomas to Produce Idiotypically Specific Anti-Receptor Antibodies. J. Immunol 130: 1033–1037. [PubMed] [Google Scholar]
- 60.Woodham AW, Cheloha RW, Ling JJ, Rashidian M, Kolifrath SC, Mesyngier M, Duarte JN, Bader JM, Skeate JG, Da Silva DM, Kast WM, and Ploegh HL. 2018. Nanobody-Antigen Conjugates Elicit HPV-Specific Antitumor Immune Responses. Cancer Immunol. Res 6: 870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fang T, Van Elssen C, Duarte JN, Guzman JS, Chahal JS, Ling JJ, and Ploegh HL. 2017. Targeted antigen delivery by an anti-class II MHC VHH elicits focused alpha MUC1(Tn) immunity. Chem. Sci 8: 5591–5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guimaraes CP, Witte MD, Theile CS, Bozkurt G, Kundrat L, Blom AE, and Ploegh HL. 2013. Site-specific C-terminal and internal loop labeling of proteins using sortase-mediated reactions. Nat. Protoc 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pluger EBE, Boes M, Alfonso C, Schroter CJ, Kalbacher H, Ploegh HL, and Driessen C. 2002. Specific role for cathepsin S in the generation of antigenic peptides in vivo. Eur. J. Immunol 32: 467–476. [DOI] [PubMed] [Google Scholar]
- 64.Johnson LM, Horne WS, and Gellman SH. 2011. Broad Distribution of Energetically Important Contacts across an Extended Protein Interface. J. Am. Chem. Soc 133: 10038–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rothbauer U, Zolghadr K, Tillib S, Nowak D, Schermelleh L, Gahl A, Backmann N, Conrath K, Muyldermans S, Cardoso MC, and Leonhardt H. 2006. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 3: 887–889. [DOI] [PubMed] [Google Scholar]
- 66.Johnson LM, and Gellman SH. 2013. alpha-Helix Mimicry with alpha/beta-Peptides. Methods Enzymol. 523: 407–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Levisetti MG, Suri A, Petzold SJ, and Unanue ER. 2007. The insulin-specific T cells of nonobese diabetic mice recognize a weak MHC-binding segment in more than one form. J. Immunol 178: 6051–6057. [DOI] [PubMed] [Google Scholar]
- 68.Liu S, Cheloha RW, Watanabe T, Gardella TJ, and Gellman SH. 2018. Receptor selectivity from minimal backbone modification of a polypeptide agonist. Proc. Natl. Acad. Sci. U.S.A. 115: 12383–12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hager MV, Johnson LM, Wootten D, Sexton PM, and Gellman SH. 2016. β-Arrestin-Biased Agonists of the GLP-1 Receptor from β-Amino Acid Residue Incorporation into GLP-1 Analogues. J. Am. Chem. Soc 138: 14970–14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Czekster CM, Robertson WE, Walker AS, Soll D, and Schepartz A. 2016. In Vivo Biosynthesis of a beta-Amino Acid-Containing Protein. J. Am. Chem. Soc 138: 5194–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.