Abstract
OBJECTIVE:
Endometrioid ovarian carcinomas (EOCs) comprise 5–10% of all ovarian cancers and commonly co-occur with synchronous endometrioid endometrial cancer (EEC). We sought to examine the molecular characteristics of pure EOCs in patients without concomitant EEC.
METHODS:
EOCs and matched normal samples were subjected to massively parallel sequencing targeting 341–468 cancer-related genes (n=8) or whole-genome sequencing (n=28). Mutational frequencies of EOCs were compared to those of high-grade serous ovarian cancers (HGSOCs; n=224) and EECs (n=186) from The Cancer Genome Atlas, and synchronous EOCs (n=23).
RESULTS:
EOCs were heterogeneous, frequently harboring KRAS, PIK3CA, PTEN, CTNNB1, ARID1A and TP53 mutations. EOCs were distinct from HGSOCs at the mutational level, less frequently harboring TP53 but more frequently displaying KRAS, PIK3CA, PIK3R1, PTEN and CTNNB1 mutations. Compared to synchronous EOCs and pure EECs, pure EOCs less frequently harbored PTEN, PIK3R1 and ARID1A mutations. Akin to EECs, EOCs could be stratified into the four molecular subtypes: 3% POLE (ultramutated), 19% MSI (hypermutated), 17% copy-number-high (serous-like) and 61% copy-number-low (endometrioid). In addition to microsatellite instability, a subset of EOCs harbored potentially targetable mutations, including AKT1 and ERBB2 hotspot mutations. EOCs of MSI (hypermutated) subtype uniformly displayed a good outcome.
CONCLUSIONS:
EOCs are heterogeneous at the genomic level and harbor targetable genetic alterations. Despite the similarities in the repertoire of somatic mutations between pure EOCs, synchronous EOCs and EECs, the frequencies of mutations affecting known driver genes differ. Further studies are required to define the impact of the molecular subtypes on the outcome and treatment of EOC patients.
Keywords: endometrioid ovarian cancer, massively parallel sequencing, molecular subtypes, somatic mutations, heterogeneity, molecular subtypes
1. INTRODUCTION
Epithelial ovarian cancer is the fourth commonest cause of female cancer death in the developed world, due to its advanced stage at presentation and progressive development of chemo-resistance. ‘Epithelial’ is a collective term that encompasses all histologies historically thought to originate in the ovary. This classification, however, insufficiently accounts for the distinct clinical, pathologic and biologic features of ovarian carcinomas. More recently, ovarian cancers have been classified into type I and II tumors [1]. Type I tumors are low-grade (low-grade serous, endometrioid, mucinous, and clear cell types) and harbor mutations in BRAF, KRAS and PTEN, often with microsatellite instability, and are thought to arise from ovarian or pelvic endometriosis [1,2]. Type II tumors are high-grade serous and carcinosarcomas, which are believed to arise in the fimbriae of the fallopian tube and frequently contain mutations in TP53, BRCA1 and BRCA2 [2]. Although this type I/II classification better encompasses the molecular diversity of epithelial ovarian cancer, it is not employed clinically and does not account for the molecular and biological heterogeneity seen within type I and type II tumors.
Endometrioid ovarian cancers (EOCs) comprise 5–10% of all epithelial ovarian cancers and are thought to arise from endometriosis, commonly co-occur with synchronous endometrial cancer and tend have a better prognosis than other histologic subtypes [3]. In some patients with pure EOC, disease relapse has been recorded, and, in that context, the outcome of EOCs is poor [4]. Microarray gene expression profiling identified different transcriptomic subtypes within EOCs [5], and genome sequencing revealed distinct pathways to tumor progression in EOCs [6], providing evidence to suggest that EOC may be heterogeneous at the molecular level.
In endometrial cancer, massively parallel sequencing analysis led to the identification of multiple molecular subtypes, including the POLE (ultramutated), MSI (hypermutated), copy-number low (endometrioid), copy-number high (serous-like) subtypes, which have prognostic and predictive implications [7,8]. In ovarian cancer, whole-exome (WES) and whole-genome (WGS) sequencing studies have not only revealed the genetic heterogeneity of ovarian cancers, including that of high-grade serous ovarian cancer (HGSOC), clear cell carcinoma, mucinous carcinoma and carcinosarcoma [6,9–12], but also identified the genomic consequences of aberrant DNA repair processes within and between high-grade serous, endometrioid and clear cell ovarian cancers [6]. Furthermore, EOCs synchronously diagnosed with EECs have been studied at the genetic level, and suggested to be of uterine origin [13,14]. Little is known, however, about the landscape of somatic genetic alterations in pure EOCs arising in the ovary. Here, we sought to characterize the repertoire of somatic mutations in key cancer genes in pure EOCs in patients without a concomitant endometrioid endometrial cancer (EEC). In addition, we aimed to define whether the molecular subtypes identified in EECs would also be present in EOCs.
2. METHODS
2.1. Case selection
After retrieving approval from Memorial Sloan Kettering Cancer Center’s (MSKCC’s) institutional review board, we retrospectively identified all patients treated for EOC at our institution between January 2002 and September 2017. We included 8 patients who were diagnosed with ovarian endometrioid adenocarcinoma without a synchronous endometrial carcinoma and which were subjected to targeted massively parallel sequencing (MSK-IMPACT) testing [15,16], none of which had been previously reported. Data were abstracted from medical records. All cases underwent pathologic review at MSKCC by expert gynecologic pathologists. In addition, we obtained the whole-genome sequencing and clinico-pathologic data, including microsatellite instability (MSI) status, from 28 pure EOCs previously described in Wang et al [6], which were reviewed by at least two expert gynecopathologists [6].
2.2. Massively parallel sequencing
Targeted massively parallel sequencing was performed in the Clinical Laboratory Improvement Amendments (CLIA)-certified MSKCC Molecular Diagnostics Service Laboratory on DNA extracted from tumor and matched normal from blood. The MSK Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) sequencing assay was employed, targeting all coding exons of 341 (n=1), 410 (n=5) or 468 (n=2) key cancer-related genes, as described previously [15,16]. Tumor and normal DNA were sequenced to a median of 827x (range 312x-1062x) and 469x (range 319x-830x) sequence coverage, respectively (Supplementary Table 1). Sequencing data were mapped onto the reference human genome GRCh37 using the Burrows-Wheeler Aligner (BWA, v0.7.15)[17], and sequencing data analysis was performed as previously described [18,19]. In addition, somatic single nucleotide variants (SNVs) and small insertions and deletions (indels) of 28 EOCs subjected to whole-genome sequencing were obtained from Wang et al [6]. All somatic SNVs and indels were re-annotated using vcf2maf (https://github.com/mskcc/vcf2maf), and manually curated on the Integrated Genomics Viewer (IGV) [20]. Mutations affecting hotspot codons were annotated according to Chang et al. [21].
2.3. Comparison with EECs, synchronous EOCs and high-grade serous ovarian cancers
The mutational frequencies of pure EOCs were compared to those of high-grade serous ovarian cancers (HGSOCs) from The Cancer Genome Atlas (TCGA, n=224)[9], EECs from TCGA (n=186)[8] and EOCs with concurrent EECs (i.e. synchronous EOCs; Schultheis et al, n=23)[14]. The recently updated WES-derived somatic mutation MC3 data of the HGSOCs and EECs from TCGA were retrieved from the Genomic Data Commons (GDC; https://gdc.cancer.gov/aboutdata/publications/mc3-2017)[22]. We restricted the comparison to the 341 genes targeted by the smallest MSK-IMPACT panel employed in this study. Given the lack of hypermutated HGSOCs due to microsatellite-instability (MSI-high) or POLE exonuclease domain mutations (EDMs), the MSI-high and POLE-mutant EOCs were removed from the comparisons with HGSOCs. To ensure consistency across datasets, all somatic SNVs and indels were re-annotated as described above, and mutations affecting hotspot codons were annotated according to Chang et al. [21].
2.4. MSISensor
For the quantification of MSI in the 8 EOCs subjected to MSK-IMPACT sequencing, MSIsensor was employed [23], and samples with an MSIsensor score ≥10 were deemed MSI-high, as previously described [24].
2.5. Molecular subtyping
To classify the EOCs into the molecular subtypes described for endometrioid and serous endometrial carcinomas by TCGA [8], we employed a surrogate model [25,26], where 1) EOCs with POLE EDMs were classified as of POLE (ultramutated) subtype; 2) Of the remaining cases, EOCs being MSI-high by panel testing (Wang et al [6]) or by MSISensor (MSK-IMPACT cases) were classified as MSI (hypermutated) subtype; 3) Of the remaining cases, EOCs harboring TP53 mutations were classified as of copy-number high-like (serous-like) subtype; and 4) those with wild-type TP53 were classified as of copy-number low-like (endometrioid-like) subtype.
2.6. Statistical analysis
Comparisons of mutation frequencies between different cancer types were performed using Fisher’s exact tests. Resulting p-values were corrected for multiple testing using the Benjamini-Hochberg false discovery rate [27], and two-tailed adjusted p-values <0.05 were deemed statistically significant. Progression-free survival (PFS) was calculated from the date of initial diagnosis to the date of radiologic progression, or death/last follow-up, respectively. Survival was estimated using the Kaplan–Meier method, and estimates compared with the log-rank test. Survival analyses/ statistical tests were performed using SPSS 25.0 (IBM).
3. RESULTS
3.1. EOCs are phenotypically heterogeneous
The median age of EOC diagnosis in this patient series (n=36) was 54.3 years (range, 32–88). At presentation, 61% (22/36) of patients were of FIGO stage I, 11% (4/36) of FIGO stage II, 22% (8/36) of FIGO stage III, and 6% (2/36) were of FIGO stage IV (Table 1). The median follow-up was 55.5 months (range 12.6 – 265.9 months). The median progression-free survival (PFS) for the entire cohort (n=34; two patients excluded due to lack of follow up) was 110.6 months (95% CI: 29.9–191.3). Five patients died of disease during the follow-up period. The 5-year overall-survival was 90.3% (95% CI: 72.7–96.7).
Table 1.
Clinico-pathologic characteristics of the endometrioid epithelial ovarian cancers included in this study.
| Characteristic | N(36) |
|---|---|
| Median age at diagnosis (years) | |
| Median (range) | 54.3 (32–88) |
| FIGO (2009) stage at diagnosis | |
| I | 22 (61%) |
| II | 4 (11%) |
| III | 8 (22%) |
| IV | 2 (6%) |
| Median PFS (months) (95% CI) | 110.6 (29.9–191.3) |
| Median OS (months) (95% CI) | not reached |
| 5-year OS (months) (95% CI) | 90% (72.7–96.7) |
Due to lack of follow-up data on two patients, survival statistics were calculated on 34 patients. CI, confidence interval; FIGO, International Federation of Gynecology and Obstetrics; PFS, progression-free survival; OS, overall survival.
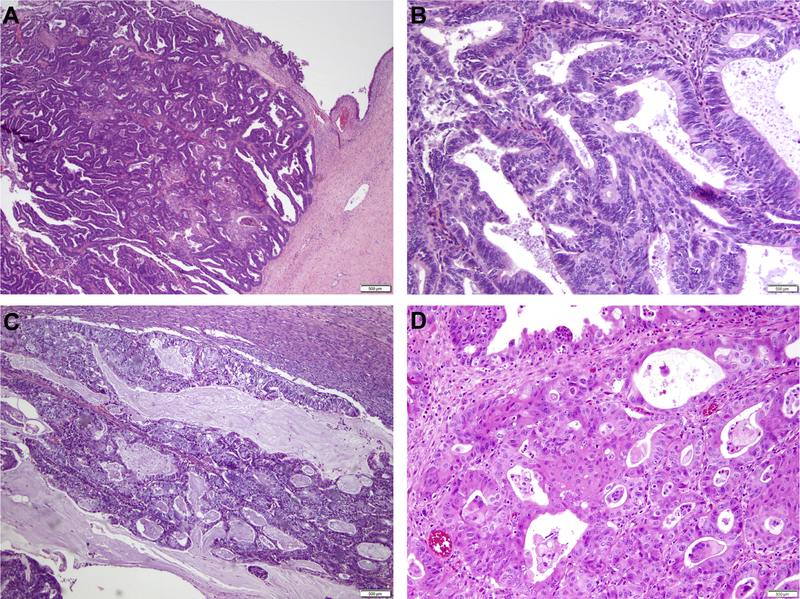
In the MSK cohort (n=8), one patient had a clinical history of endometriosis; however, 50% (4/8) of patients had endometriosis seen on final pathology (Supplementary Table 2). In addition, mucinous or squamous differentiation was present in the well-differentiated EOCs studied (Figure 1). The FIGO tumor grades varied, and EOCs were of grade 1 (2/8), grade 2 (3/8) and grade 3 (3/8) at diagnosis (Supplementary Table 2).
Figure 1. Histologic features of endometrioid adenocarcinomas of the ovary.
(A) Well-differentiated endometrioid adenocarcinoma of the ovary arising in a background of endometriosis. (B) Higher power micrograph of a well-differentiated endometrioid ovarian carcinoma. (C) Well-differentiated endometrioid adenocarcinoma of the ovary with mucinous differentiation. (D) Well-differentiated endometrioid ovarian carcinoma with squamous differentiation. Scale bars, 500 μm.
3.2. The repertoire somatic mutations and copy number alterations in EOCs
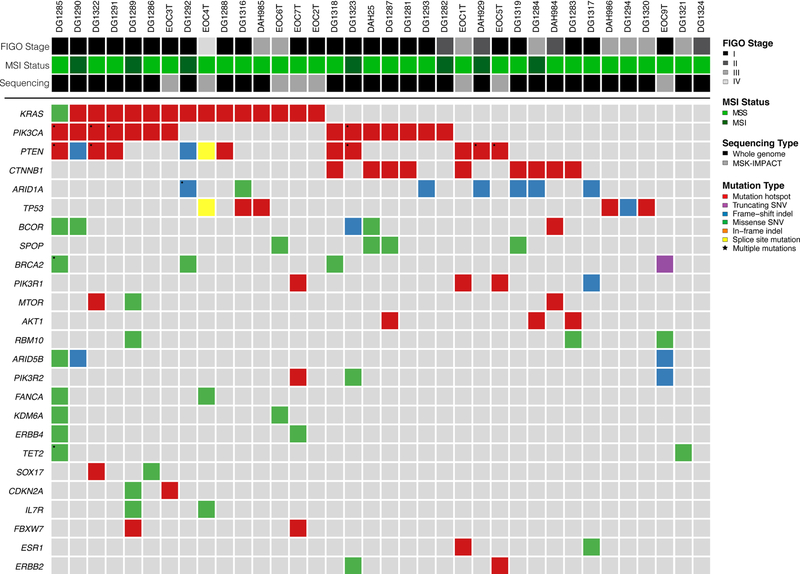
EOCs harbored a median of 7 non-synonymous somatic mutations (range 1–120) in 341 cancer genes tested (smallest gene panel; see Methods; Supplementary Table 3). KRAS was found to be the most frequently mutated gene (15/36; 42%), and all but one mutation were G12 or G13 KRAS hotspot mutations (Figure 2; Supplementary Table 3). In addition, 22/36 (61%) of the EOCs studied harbored mutations in the PI3K pathway, including PIK3CA hotspot mutations (14/36), PTEN mutations (12/36), PIK3R1 mutations (4/36), PIK3R2 mutations (3/36) and AKT1 E17K hotspot mutations (3/36; Figure 2). Nine and seven EOCs harbored CTNNB1 hotspot mutations (25%) and ARID1A mutations (19%), respectively. Cell cycle-related genes, including TP53, CDKN2A, RB1 and MYC, were mutated in six (17%), two (6%), one (3%) and one (3%) cases, respectively (Figure 2; Supplementary Table 3). Four EOCs harbored mutations affecting SPOP and two affecting FBXW7, genes described to be recurrently mutated in serous endometrial cancers [8]. Seven EOCs were found to be MSI-high and one (DG1285) harbored a POLE V411L hotspot mutation (Figure 2). Finally, EOC1 had an ESR1 D538G hotspot mutation and EOC5 harbored an ERBB2 D769H hotspot mutation (Figure 2); both mutations have been shown to have treatment implications in other disease types. In fact, based on the OncoKB annotation [28], three EOCs harbored a level 3A genetic alteration (i.e. AKT1 E17K) and 16 a level 3B genetic alteration (i.e. ERBB2 D769H, ESR1 D538G and PIK3CA hotspot mutations), which are associated with promising/compelling clinical evidence supportive of response prediction to a drug in gynecologic cancers (AKT1)/ in another indication (i.e. investigational therapeutic implications, possibly directed to clinical trials)[28].
Figure 2. Non-synonymous somatic mutations identified in endometrioid ovarian carcinomas using targeted massively parallel sequencing.
Recurrent (n≥2) non-synonymous somatic mutations identified in 36 endometrioid epithelial ovarian cancers by massively parallel sequencing targeting 341 cancer-related genes. Mutation types are color-coded according to the legend. The phenobar provides information on FIGO stage, microsatellite instability (MSI) status and sequencing modality. Indel, small insertion/ deletion; SNV, single nucleotide variant.
3.3. Comparison of the repertoire of somatic mutations of EOCs, HGSOCs, EECs and synchronous EOCs
We next sought to define whether pure EOCs would differ from HGSOCs, synchronous EOCs and endometrioid cancers of the uterus at the mutational level. This comparison was restricted to the smallest sequencing panel (i.e. 341 cancer-related genes, MSK-IMPACT).
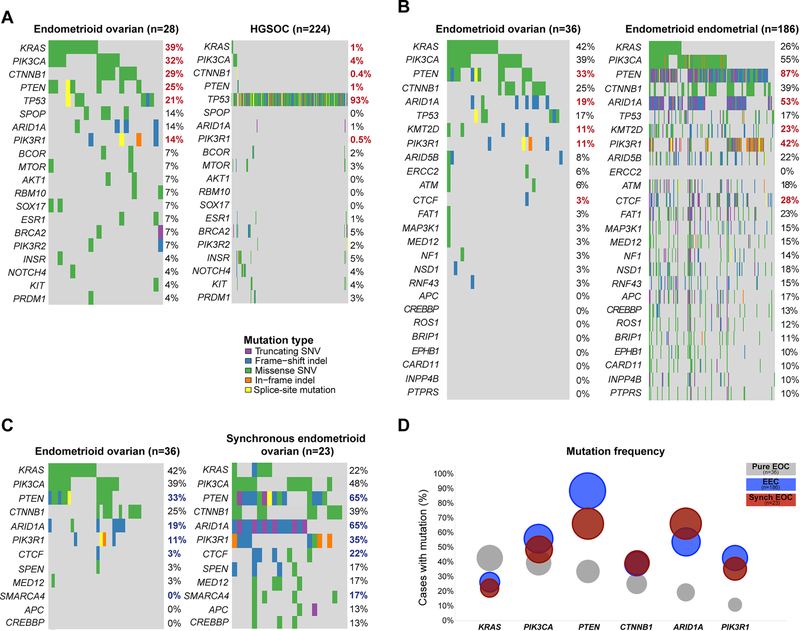
For the comparison with HGSOCs, we removed the MSI-high and POLE EDM EOCs, given the lack of hyper-/ultra-mutated HGSOCs [9]. Consistent with previous reports, pure EOCs were distinct from HGSOCs at the mutational level [29]. The comparison of 28 non-hypermutated pure EOCs and 224 HGSOCs from TCGA [9] revealed that PI3K pathway mutations, including those affecting KRAS (39% vs 1%), PIK3CA (32% vs 4%), PTEN (25% vs 1%) and PIK3R1 (14% vs 0.5%), were significantly more frequent in EOCs than in HGSOCs, as were CTNNB1 mutations (29% vs 0.4%) (all adjusted p<0.05, Fisher’s exact test; Figure 3A). On the other hand, TP53 was significantly more frequently mutated in HGSOCs (93%) than in EOCs (21%, adjusted p<0.05, Fisher’s exact test).
Figure 3. Comparison of the mutational profiles of pure endometrioid ovarian cancers with high-grade serous ovarian cancers, endometrioid endometrial cancers and synchronous endometrioid ovarian cancers.
The repertoire of somatic mutations affecting the 341 genes included in the MSK-IMPACT assay (smallest gene panel) in (A) pure microsatellite-stable/ POLE wild-type endometrioid epithelial ovarian cancers (EOCs) (n=28) and in high-grade serous ovarian cancers from The Cancer Genome Atlas (TCGA; n=224)[9] (B) all pure EOCs (n=36) and endometrioid endometrial cancers (EECs; TCGA; n=186)[8], and (C) all pure EOCs (n=36) and EOCs synchronously diagnosed with EECs (n=23)[14]. (D) Mutation frequencies of KRAS, PIK3CA, PTEN, CTNNB1, ARID1A and PIK3R1 in the pure EOCs studied here (n=36), in EECs (n=186, TCGA)[8] and EOCs synchronously diagnosed with EECs (synchronous EOCs; n=23)[14]. Comparisons were performed using Fisher’s exact test. (A and B) Benjamini-Hochberg adjusted statistically significant p-values (adjusted p<0.05) are shown in red bold font, (C) unadjusted statistically significant p-values (p<0.05) are shown in blue bold font.
The genes mutated in EOCs (n=36) and EECs (n=186) from TCGA [8] were found to be overall similar, however the mutations occurred at different frequencies [30]. Whilst KRAS (42% EOCs vs 26% EECs) and PIK3CA (39% EOCs vs 55% EECs) mutation rates were similar between EOCs and EECs, mutations affecting other PI3K pathway genes, including PTEN (33% vs 87%), PIK3R1 (11% vs 42%), as well as chromatin remodeling genes, including ARID1A (19% vs 53%), KMT2D (11% vs 23%) and CTCF (3% vs 28%), were significantly less frequent in EOCs than in the EECs (all adjusted p<0.05, Fisher’s exact test; Figure 3B). It should be noted, however, that in the endometrial cancer TCGA there was a likely overrepresentation of FIGO grade 2 and grade 3 EECs as well as of MSI-high EECs [8]. When comparing the mutational frequencies of EOCs (n=36) matched by molecular subtype (see below) at a 1:2 ratio with preferentially grade 1 and grade 2 EECs from TCGA (n=72), only PTEN mutation rates were statistically different between EOCs (33%) and EECs (82%) after correction for multiple comparisons (Fisher’s exact test, adjusted p<0.001; Supplementary Table 4).
We have previously shown that sporadic synchronous EECs and EOCs are clonally related [13,14]. When comparing pure EOCs (n=36) with EOCs synchronously diagnosed with EECs (henceforth named synchronous EOCs; n=23)[14], we observed that, akin to EECs, mutations affecting PTEN (33% vs 65%), PIK3R1 (11% vs 35%), ARID1A (19% vs 65%) and CTCF (3% vs 22%) were numerically less frequent in pure EOCs than in EOCs synchronously diagnosed with EECs (p<0.05, Fisher’s exact test), however did not reach statistical significance after Benjamini-Hochberg correction for multiple comparisons (Figure 3C), possibly due to the small sample size. The mutational frequencies of the most frequently mutated genes in pure EOCs (n=36), including KRAS, PIK3CA, PTEN, CTNNB1 and ARID1A, are generally more similar between EECs (n=186; TCGA) and synchronous EOCs than in pure EOCs (Figure 3D). These observations are consistent with the notion that synchronous EOCs likely constitute dissemination from the EEC to the ovary [13,14].
3.4. Molecular subtyping of EOCs
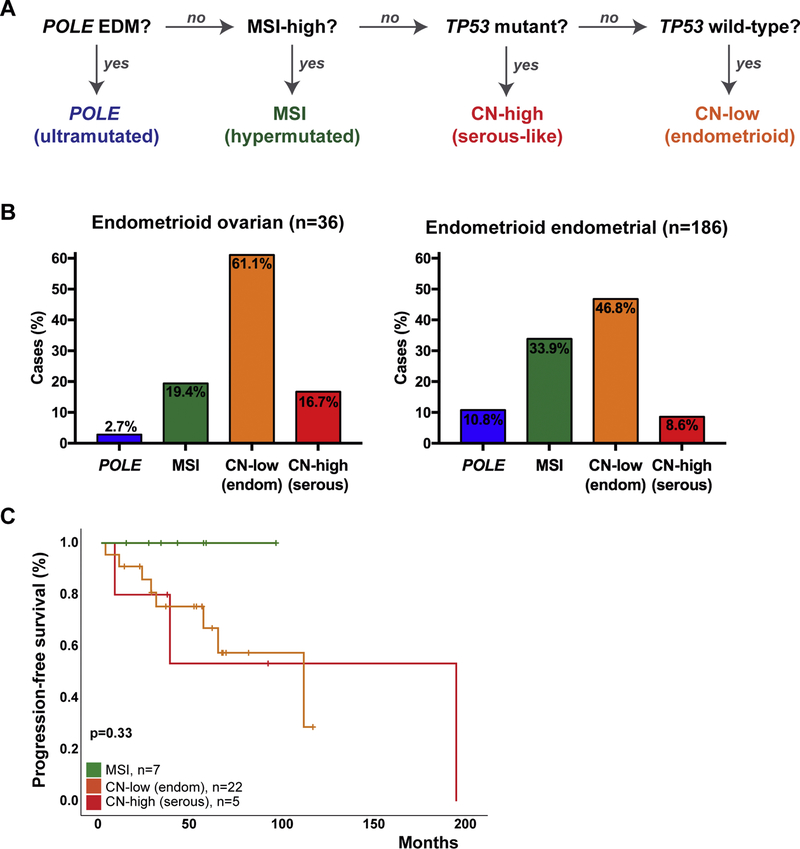
Given the diversity of the repertoire of somatic mutations identified in EOCs, and the similarity between EOCs and EECs at the mutational level, we sought to define whether EOCs could be classified into the molecular subtypes described for endometrial cancers [8]. For this, we employed a surrogate model for the molecular-based TCGA classification [8,26], integrating the POLE mutation status, the MSI status (based on MSI panel testing or MSISensor), and TP53 mutation status (see Methods; Figure 4A). We noted that all four molecular subtypes identified in EECs were also present in EOCs. The majority of EOCs (22/36; 61%) were classified as of copy-number low (endometrioid) subtype, of which 8 (36%) harbored a CTNNB1 hotspot mutation (D32, n=1; S33, n=2; S37, n=3; G34, n=1; T41, n=1 [21]) (Figure 4B). One (2.7%) of the 36 EOCs harbored a very high mutation rate (3757 non-synonymous somatic mutations, WGS), a POLE EDM V411L somatic hotspot mutation, and was classified as of POLE (ultramutated) subtype (Figure 4B). In addition, seven (19%) EOCs were MSI-high based on MSI panel testing, had a high mutation rate (median 562, range 146–828 non-synonymous somatic mutations, WGS) and were classified as of MSI (hypermutated) subtype. The remaining 6 (17%) EOCs were microsatellite stable, harbored TP53 mutations, and, therefore, were classified as of copy-number high (serous-like) subtype (Figure 4B).
Figure 4. Molecular subtyping of endometrioid ovarian cancers.
(A) Surrogate classifier employed for the molecular subtyping of endometrioid epithelial ovarian cancers (EOCs), based on the molecular subtypes described for endometrial cancers by The Cancer Genome Atlas [8,26]. (B), Molecular subtype distribution in EOCs from this study (left) and in endometrioid ovarian cancers from TCGA (right) [8]. (C) Kaplan-Meier progression-free survival curve for EOC patients stratified according to the endometrial cancer molecular subtypes defined with a surrogate model. Two cases (one of POLE subtype and one of CN-high (serous-like) subtype) were excluded due to limited/ lack of follow-up information. P-values of the log-rank test are shown. CN, copy-number.
To compare directly the distribution of the molecular subtypes in EOCs and in EECs, we classified the EECs (n=186) from TCGA [8] into the molecular subtypes using the same surrogate as for the EOCs (Figure 4A). This led to the re-classification of 5/186 (2.7%) EECs; one copy-number high (TCGA) EEC was now classified as copy-number low given the lack of a TP53 mutation, one microsatellite stable copy-number low (TCGA) EEC was now classified as copy-number high given the presence of a C238F TP53 hotspot mutation; and one microsatellite stable copy-number low and two MSI (hypermutated) EEC were now classified as POLE (ultramutated) subtype given the presence of POLE EDM mutations (L242V, A465V, Q453R, Supplementary Table 5). Compared to EECs, EOCs of POLE (ultramutated) and MSI (hypermutated) subtypes are numerically less common (3% vs 11% POLE; 19% vs 34%, MSI) and copy-number low (endometrioid) and copy-number high (serous-like) numerically more common (61% vs 47% copy-number low; 17% vs 9% copy-number high) in EOCs, however none of these differences were statistically significant (all p>0.1 Fisher’s exact test, Figure 4B).
As a hypothesis-generating exploratory aim, we assessed whether, akin to EECs [8], the molecular subtypes in EOCs were associated with distinct outcomes. Although in univariate analysis, MSI (ultramuted), copy-number low (endometrioid) and copy-number high (serous-like) EOCs were not different in their progression-free survival (p=0.33), the MSI (ultramutated) EOCs were found to be associated with a favorable outcome (Figure 4C). One POLE case and one copy-number high (serous-like) case were removed from this analysis due to limited/ lack of follow-up. The POLE EDM EOC patient (DG1285) was alive without disease at last follow-up (160 days) [6]. Larger datasets may be required to assess the impact of the molecular subtypes in EOC on outcome.
4. DISCUSSION
Here we demonstrate that pure EOCs are heterogeneous at the mutational level, and that these tumors are underpinned by mutations described in EECs and in EOCs occurring synchronously with EECs, however at different frequencies. In addition, the heterogeneity of pure EOCs is further reflected by the detection of all four molecular subtypes described originally for endometrioid and serous endometrial cancers.
TGCA first reported in 2013 on a molecular classification of endometrioid and serous endometrial carcinomas [8], and, since then, these molecular subtypes and the associated molecular heterogeneity were also found in other histologic types of endometrial cancer, including clear cell carcinomas and de-/un-differentiated carcinomas of the uterus [25,31]. Here we observed that all four molecular subtypes described in EECs are also present in EOCs. Further studies are warranted to assess not only the frequency of the molecular subtypes in EOCs but also the impact of the molecular subtypes on outcome of patients with this disease, given the association of these subtypes with progression-free survival in EECs [8]. Although we did not see a statistically significant difference in progression-free survival between the molecular subtypes, EOCs of MSI (hypermutated) subtype had favorable outcomes.
EOCs have been reported to generally have a better prognosis than other histologic subtypes of epithelial ovarian cancer [3], however in a subset of EOCs, in particular those with disease relapse, the outcome is poor [4]. Our massively parallel sequencing analysis of EOCs not only identified MSI-high EOCs, which may be eligible for checkpoint blockage immunotherapy [32], but also other potentially targetable alterations. One EOC was found to harbor an ERBB2 hotspot mutation, which has been associated with resistance to Trastuzumab but response to HER2 irreversible inhibitors [33–36]. Three EOCs harbored an AKT1 E17K hotspot mutation, and there is promising clinical data in patients with AKT1 E17K-mutant gynecological cancer treated with the pan-AKT kinase inhibitor AZD5363 [37]. On the other hand, one EOC harbored a D538 hotspot mutation in the ligand-binding domain of ESR1, which is associated with acquired resistance to endocrine therapy [38]. In the era of precision medicine, the assessment of DNA mismatch repair proteins and/ or alterations affecting targetable cancer-related genes and likely mechanisms of resistance may guide treatment stratification of patients with EOCs.
Our study has several limitations, including the small sample size, given the rarity of EOCs. In addition, our data provide evidence to suggest that EOCs are molecularly heterogeneous, however, based on the small numbers in each of the molecular subtypes, further studies are warranted to define whether the distinct EOCs molecular subtypes are associated with distinct outcomes and therapeutic responses. Nonetheless, our findings demonstrate that pure EOCs are heterogenous at the genetic level, harbor potentially actionable alterations, including MSI, and AKT1 and ERBB2 hotspot mutations, and that the mutational spectrum bears resemblance to that of EECs and synchronous EOCs, albeit at different frequencies.
Supplementary Material
HIGHLIGHTS.
Endometrioid eptithelial ovarian cancers (EOCs) are genetically heterogeneous
The four endometrial cancer molecular subtypes can be detected in EOCs
EOCs may be hyper-or ultramutated due to MSI or POLE mutations
A subset of EOCs harbors targetable genetic alterations
ACKNOWLEDGEMENTS
Funding support
Research reported in this publication was supported in part by a Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute (Grant No. P30CA008748). B. Weigelt is funded in part by Cycle for Survival.
We gratefully acknowledge the members of the Molecular Diagnostics Service in the Department of Pathology. We thank YK Wang for assistance with the sequencing data, and J.S. Reis-Filho for critically reading the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
M.M. Leitao Jr is an ad hoc speaker for Intuitive Surgical, Inc. C. Aghajanian reports personal fees from Tesaro, Immunogen, Clovis, Mateon Therapeutics, Cerulean Pharma, and grants from Clovis, Genentech, AbbVie and Astra Zeneca, outside the submitted work. N.R. Abu-Rustum reports grants from Stryker/Novadaq, Olympus and GRAIL paid to the institution, outside the submitted work. S.P. Shah is founder, shareholder and consultant of Contextual Genomics Inc., outside the submitted work. The remaining authors have no conflicts of interest to declare.
REFERENCES
- [1].Shih IM, Kurman RJ. Ovarian tumorigenesis - A proposed model based on morphological and molecular genetic analysis. American Journal of Pathology. 2004;164:1511–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Banerjee S, Kaye SB. New strategies in the treatment of ovarian cancer: current clinical perspectives and future potential. Clin Cancer Res. 2013;19:961–8. [DOI] [PubMed] [Google Scholar]
- [3].Pearce CL, Templeman C, Rossing MA, Lee A, Near AM, Webb PM, et al. Association between endometriosis and risk of histological subtypes of ovarian cancer: a pooled analysis of case-control studies. Lancet Oncology. 2012;13:385–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Soovares P, Pasanen A, Butzow R, Lassus H. L1CAM expression associates with poor outcome in endometrioid, but not in clear cell ovarian carcinoma. Gynecol Oncol. 2017;146:615–22. [DOI] [PubMed] [Google Scholar]
- [5].Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–208. [DOI] [PubMed] [Google Scholar]
- [6].Wang YK, Bashashati A, Anglesio MS, Cochrane DR, Grewal DS, Ha G, et al. Genomic consequences of aberrant DNA repair mechanisms stratify ovarian cancer histotypes. Nat Genet. 2017;49:856–65. [DOI] [PubMed] [Google Scholar]
- [7].Murali R, Soslow RA, Weigelt B. Classification of endometrial carcinoma: more than two types. Lancet Oncology. 2014;15:e268–78. [DOI] [PubMed] [Google Scholar]
- [8].Cancer Genome Atlas Research N, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhao S, Bellone S, Lopez S, Thakral D, Schwab C, English DP, et al. Mutational landscape of uterine and ovarian carcinosarcomas implicates histone genes in epithelial-mesenchymal transition. Proc Natl Acad Sci U S A. 2016;113:12238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jones S, Stransky N, McCord CL, Cerami E, Lagowski J, Kelly D, et al. Genomic analyses of gynaecologic carcinosarcomas reveal frequent mutations in chromatin remodelling genes. Nat Commun. 2014;5:5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ryland GL, Hunter SM, Doyle MA, Caramia F, Li J, Rowley SM, et al. Mutational landscape of mucinous ovarian carcinoma and its neoplastic precursors. Genome Med. 2015;7:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Anglesio MS, Wang YK, Maassen M, Horlings HM, Bashashati A, Senz J, et al. Synchronous Endometrial and Ovarian Carcinomas: Evidence of Clonality. J Natl Cancer Inst. 2016;108:djv428. [DOI] [PubMed] [Google Scholar]
- [14].Schultheis AM, Ng CK, De Filippo MR, Piscuoglio S, Macedo GS, Gatius S, et al. Massively Parallel Sequencing-Based Clonality Analysis of Synchronous Endometrioid Endometrial and Ovarian Carcinomas. J Natl Cancer Inst. 2016;108:djv427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cheng DT, Prasad M, Chekaluk Y, Benayed R, Sadowska J, Zehir A, et al. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med Genomics. 2017;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Geyer FC, Li A, Papanastasiou AD, Smith A, Selenica P, Burke KA, et al. Recurrent hotspot mutations in HRAS Q61 and PI3K-AKT pathway genes as drivers of breast adenomyoepitheliomas. Nat Commun. 2018;9:1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Weigelt B, Bi R, Kumar R, Blecua P, Mandelker DL, Geyer FC, et al. The Landscape of Somatic Genetic Alterations in Breast Cancers From ATM Germline Mutation Carriers. J Natl Cancer Inst. 2018;110:1030–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chang MT, Bhattarai TS, Schram AM, Bielski CM, Donoghue MTA, Jonsson P, et al. Accelerating Discovery of Functional Mutant Alleles in Cancer. Cancer Discov. 2018;8:174–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst. 2018;6:271–81 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30:1015–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Middha S, Zhang L, Nafa K, Jayakumaran G, Wong D, Kim HR, et al. Reliable Pan-Cancer Microsatellite Instability Assessment by Using Targeted Next-Generation Sequencing Data. JCO Precis Oncol. 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].DeLair DF, Burke KA, Selenica P, Lim RS, Scott SN, Middha S, et al. The genetic landscape of endometrial clear cell carcinomas. J Pathol. 2017;243:230–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Murali R, Delair DF, Bean SM, Abu-Rustum NR, Soslow RA. Evolving Roles of Histologic Evaluation and Molecular/Genomic Profiling in the Management of Endometrial Cancer. J Natl Compr Canc Netw. 2018;16:201–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–84. [DOI] [PubMed] [Google Scholar]
- [28].Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Teer JK, Yoder S, Gjyshi A, Nicosia SV, Zhang C, Monteiro ANA. Mutational heterogeneity in non-serous ovarian cancers. Sci Rep. 2017;7:9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].McConechy MK, Ding J, Senz J, Yang W, Melnyk N, Tone AA, et al. Ovarian and endometrial endometrioid carcinomas have distinct CTNNB1 and PTEN mutation profiles. Mod Pathol. 2014;27:128–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Heong V, Ngoi N, Tan DS. Update on immune checkpoint inhibitors in gynecological cancers. J Gynecol Oncol. 2017;28:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ma CX, Bose R, Gao F, Freedman RA, Telli ML, Kimmick G, et al. Neratinib Efficacy and Circulating Tumor DNA Detection of HER2 Mutations in HER2 Nonamplified Metastatic Breast Cancer. Clin Cancer Res. 2017;23:5687–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ng CK, Martelotto LG, Gauthier A, Wen HC, Piscuoglio S, Lim RS, et al. Intra-tumor genetic heterogeneity and alternative driver genetic alterations in breast cancers with heterogeneous HER2 gene amplification. Genome Biol. 2015;16:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Weigelt B, Reis-Filho JS. Activating mutations in HER2: neu opportunities and neu challenges. Cancer Discov. 2013;3:145–7. [DOI] [PubMed] [Google Scholar]
- [36].Hyman DM, Piha-Paul SA, Won H, Rodon J, Saura C, Shapiro GI, et al. HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature. 2018;554:189–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hyman DM, Smyth LM, Donoghue MTA, Westin SN, Bedard PL, Dean EJ, et al. AKT Inhibition in Solid Tumors With AKT1 Mutations. J Clin Oncol. 2017;35:2251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chandarlapaty S, Chen D, He W, Sung P, Samoila A, You D, et al. Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer: A Secondary Analysis of the BOLERO-2 Clinical Trial. JAMA Oncol. 2016;2:1310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rosa-Rosa JM, Leskela S, Cristobal-Lana E, Santon A, Lopez-Garcia MA, Munoz G, et al. Molecular genetic heterogeneity in undifferentiated endometrial carcinomas. Mod Pathol. 2016;29:1390–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.