Abstract
Background
Gastric cancer (GC) is a common malignant cancer in the worldwide, especially in China. Patients with GC have poor prognosis, which is mainly due to lack of early diagnosis. Up to now, there is no good biomarker to detect GC at early stage. Apolipoprotein C1 (APOC1), a component of both triglyceride-rich lipoproteins and high-density lipoproteins, is reported to be involved in numerous biological processes.
Methods
Expression of APOC1 mRNA was analyzed by in silicon assay. Concentration of APOC1 in serum was measured by ELISA assay. Expression of APOC1 protein in GC tissue array was checked by immunohistochemistry.
Results
It was firstly found that concentration of APOC1 in serum was significantly higher in GC than that in control. Expression of APOC1 protein was also higher in GC than that in adjacent issues of GC and normal tissues using tissues array by immunohistochemistry. In addition, the expression of APOC1 is significantly associated with clinical stage (P=0.011), tumor classification (P=0.010), as well as with the lymph node metastasis (P=0.048). Area under the curve (AUC) of receiver operating characteristic (ROC) curve of APOC1 was 0.803. Furthermore, elevated APOC1 expression in GC was found to be correlated with decreased overall survival (P=0.00214).
Conclusions
All these results suggested that APOC1 might be a potential serum biomarker to diagnose GC and a potential prognostic marker for GC.
Keywords: Apolipoprotein C1 (APOC1), gastric cancer (GC), diagnosis, prognosis, biomarker
Introduction
Gastric cancer (GC) is a common malignant cancer which ranks fifth in incidence and third in mortality of cancers in the world (1). GC is the second most common cause of cancer deaths in China (2). Surgery is the primary treatment for GC now. Since the early stages of GC are easily neglected, the majorities of cases are diagnosed at the later stages when it is very hard to treat. Specific and sensitive tests will facilitate early screening and monitoring of cancerous states (3). Hence, the identification of the ideal biomarker is imperative and necessary to diagnose GC at early stage and predict prognosis of GC patients.
APOC1, the smallest apolipoprotein (only 6.6 KDa) and a component of both triglyceride-rich lipoproteins and high-density lipoproteins. APOC1 plays an important role in the metabolism of plasma lipoproteins (4,5). APOC1 was reported to play many important roles in numerous biological processes including membrane remodeling, cholesterol catabolism, and dendritic reorganization (6,7). Additionally, a few studies have shown that APOC1 is related to the progression of multiple diseases, such as type 1 or type 2 diabetes, diabetic nephropathy, Alzheimer's disease and glomerulosclerosis (8-11). Moreover, recent studies have shown that APOC1 may be associated with the development of cancers. APOC1 was overexpressed in pancreatic cancer and high level of APOC1 in preoperative serum was significantly correlated with poor prognosis of patients. Knockdown of APOC1 expression inhibited cell proliferation and induced apoptosis of pancreatic cancer cells (12). Overexpression of APOC1 had diagnostic ability in distinguishing between triple negative breast cancer (TNBC) and non-TNBC (NTNBC). APOC1 presented as a potential prognostic factor for TNBC (13). APOC1 expression was increased and played an oncogenic role in AML (14).
The expressions of APOC1 mRNA and protein were upregulated in prostate tissue and the levels of APOC1 were increased in the serum of prostate adenocarcinoma patients (15). The mRNA and protein levels of APOC1 were also highly expressed in late stage lung cancer tissues, whereas no prognostic value of APOC1 could be identified in serum samples of lung cancer (16).
Some researchers found that APOC1 was down-regulated in serum of NSCLC and could be a potential biomarker of NSCLC (17). The serum levels of APOC1 were significantly decreased in colorectal cancer (18), papillary thyroid carcinoma (19) and child nephroblastoma (20). However, the role of APOC1 in GC has never been reported.
In the study, we detected APOC1 concentration in serum of GC, expression of APOC1 protein in tissues of GC and found that APOC1 concentration was higher in serum of GC than that in control. We also found that expression of APOC1 protein was higher in tissues than that in adjacent issues of GC and normal tissues. APOC1 expression was found to be associated with many clinical characters and prognosis of patients with GC. APOC1 may be a potential biomarker to diagnose GC and predict prognosis of GC.
Methods
Patients and healthy control characteristics
Serum of 65 GC patients and 40 healthy individuals were collected from clinical laboratory of Peking Union Medical College Hospital (PUMCH) from October 2018 to January 2019. GCs were selected according to histological examination in this study and all serum samples were collected from GC patients before surgery and any therapy. Healthy individuals, who received health checks in PUMCH, had no history of malignancy previously or at the time of the health check. The study had been approved by PUMCH Ethics Committee (S-424).
In silicon analysis
GC Database used in silicon assay was from Oncomine (www.oncomine.org) which is very useful for investigating genes expression in many cancers and normal control.
The Cancer Genome Atlas (TCGA) was done by the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI), which provides many kinds of cancer data sets, such as RNA-Seq data, microarray data, sequencing data et al. APOC1 expression in GC was also assayed by using data from website (http://ualcan.path.uab.edu/).
APOC1 concentration in serum was checked by ELISA analysis
Serum samples collected from patients with GC and healthy individuals were stored at -80°C. Concentrations of APOC1 in serum samples were measured by commercially available colorimetric sandwich ELISA kits (Elabscience Biotechnology Co., Ltd) according to the manufacturer’s protocols. The sensitivity of the assays was 5.63 ng/mL and intraassay or interassay coefficient of variation (CV) were always less than 10%. Concentrations of carcinoembryonic antigen (CEA), CA19-9, CA242 and CA72-4 in serum samples were measured by commercially available chemiluminescent immunoanalysis kits (Architect, Abbott, USA).
Expression of APOC1 in GC tissue microarray by immunohistochemistry (IHC) analysis
The GC tissue array (ST2091a) was purchased from company Alenabio (Shaanxi, China). The corresponding clinical details were available online and were also provided by the company. IHC was performed as previously described (21). In brief, paraffin section of GC tissue microarray was deparaffinized, subjected to antigen retrieval, and incubated with primary APOC1 antibody, then followed by development with the LSAB+ kit (DAKO). Finally, the slide was counterstained with hematoxylin. As a negative control, normal rabbit was used instead of the primary antibodies.
The antibody of APOC1 (ab198288) was purchased from AbCam. Images of the sections were independently examined and differentially quantified by two pathologists. The staining intensity of APOC1 expression in GC tissue array by IHC was scored as 0 (negative), 1 (weak brown), 2 (moderate brown), or 3 (strong brown). The extent of staining was scored as 0 (≤10%), 1 (11–25%), 2 (26–50%), 3 (51–75%), or 4 (>75%). Immunoreactive score of Remmele and Stegner (IRS) was determined by the formula: intensity score × extent score. A final score >1 was defined as high expression; otherwise, it was defined as low expression (22,23).
Statistical analysis
Student’s t-test was used for comparison of two groups. Correlation between the APOC1 expression and the clinical characteristics in GC patients were assessed by Spearman’s correlation coefficient test. Correlation between serum CEA, CA125, CA19-9, CA72-4 and the expression of APOC1 was analyzed with Pearson’s correlation coefficient test. The survival analyses were conducted according to the Kaplan-Meier method. P<0.05 (*), P<0.001 (**) and P<0.0001 (***) were considered statistically significant. All statistical analyses were performed using GraphPad Prism 7.0 and SPSS17.0.
Results
APOC1 expression in GC was significantly higher than that in adjacent tissues and normal control
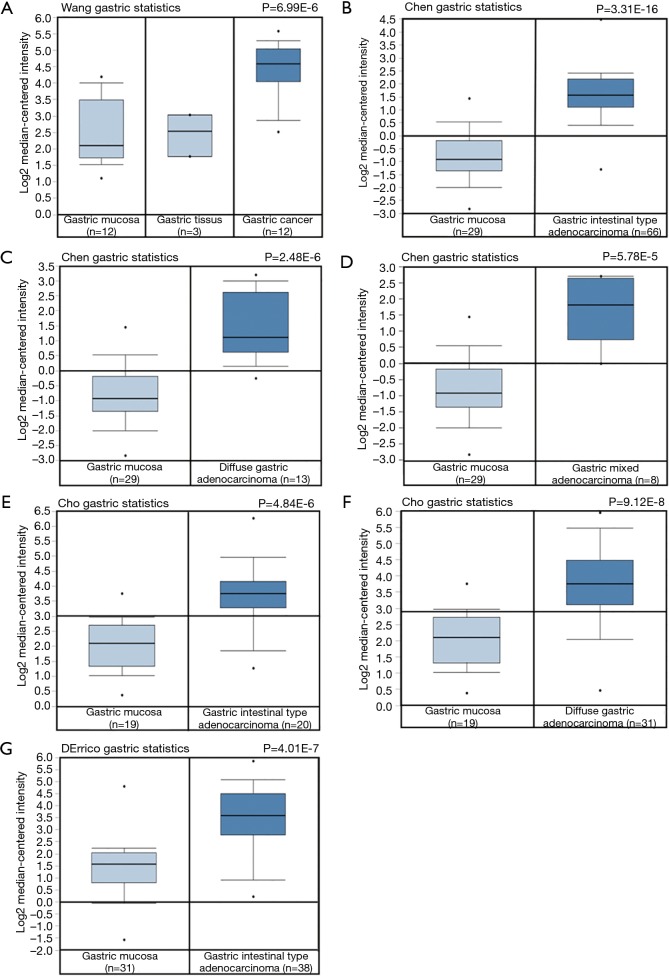
To investigate expression level of APOC1 mRNA in GC, adjacent tissues of GC and normal control, we firstly carried out in silicon assay using data from the Oncomine (www.oncomine.org). It was shown in Figure 1 that there was significantly higher APOC1 expression in GC than that in adjacent tissues of GC and normal control.
Figure 1.
There is significantly higher expression APOC1 mRNA in GC than that in normal control. Expression APOC1 mRNA is higher in GC than control. Both data and statistical value were obtained from www.oncomine.org. APOC1, apolipoprotein C1; GC, gastric cancer.
We also did in silicon assay to check APOC1 expression in GC and normal control by using TCGA (The Cancer Genome Atlas) data which was provided by National Cancer Institute and National Human Genome Research Institute. The similar results were obtained (Figure S1). Furthermore, it was found that APOC1 expression was increased as development of GC (Figure S2).
Figure S1.
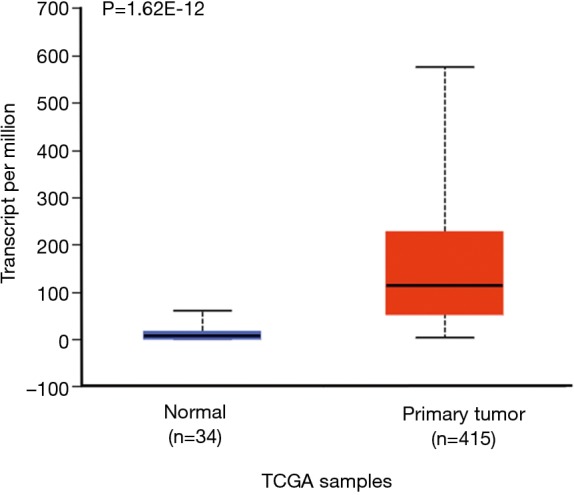
Data from TCGA showed that there was significantly higher APOC1 expression in GCs than that in normal tissues. APOC1, apolipoprotein C1; GC, gastric cancer.
Figure S2.
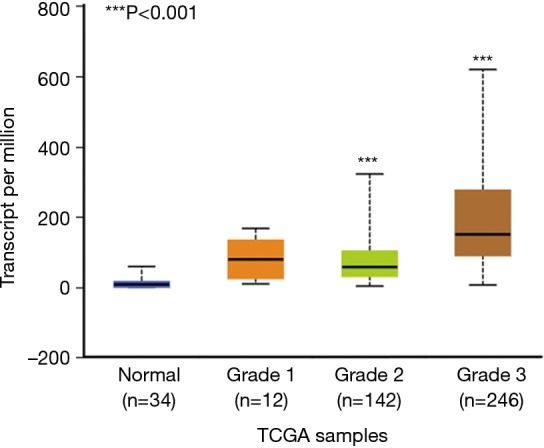
Data from TCGA showed that expression of APOC1 mRNA was increased as GC developed. APOC1, apolipoprotein C1; GC, gastric cancer.
Serum level of APOC1 and histological types of GC
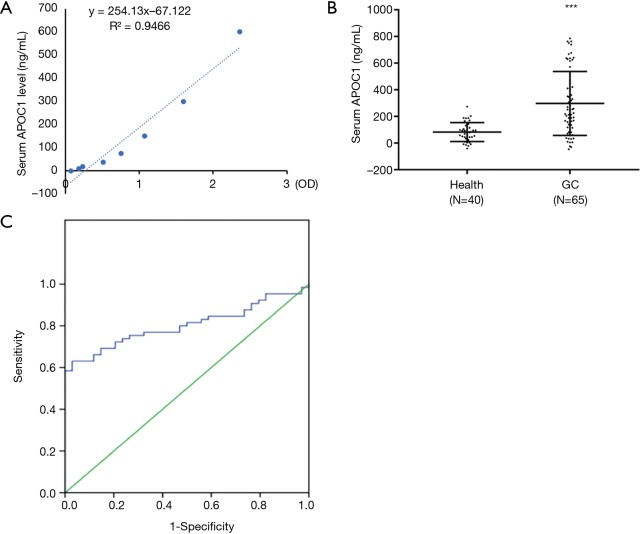
To evaluate if APOC1 could be used as a biomarker for GC, we checked concentration of APOC1 in serum of 65 GC patients and 40 healthy individuals by ELISA assay. Standard curve and concentration of APOC1 in serum were shown in Figure 2A,B, respectively. APOC1 concentration in serum of GC patients were significantly higher than that in healthy individuals (Figure 2B). To determine the measure accuracy of APOC1 in serum, receiving operating characteristics (ROC) analysis was performed. Area under curve (AUC) for APOC1 was 0.803 (Figure 2C). Based on these results, the cut-off value of APOC1 was 0.19 ug/mL. With this cut-off, the sensitivity and the specificity of APOC1 were 63.0% and 93.0%, respectively.
Figure 2.
Concentration of APOC1 in serum of GCs is higher than that in healthy controls. (A) Standard curve of ELISA assay for APOC1 concentration. (B) Concentrations of APOC1 in serum are higher GC than healthy controls. (C) AUC for APOC1. APOC1, apolipoprotein C1; GC, gastric cancer. ***, P<0.0001.
Expression of APOC1 protein was higher in GC than that in normal and is associated with clinical features of GC
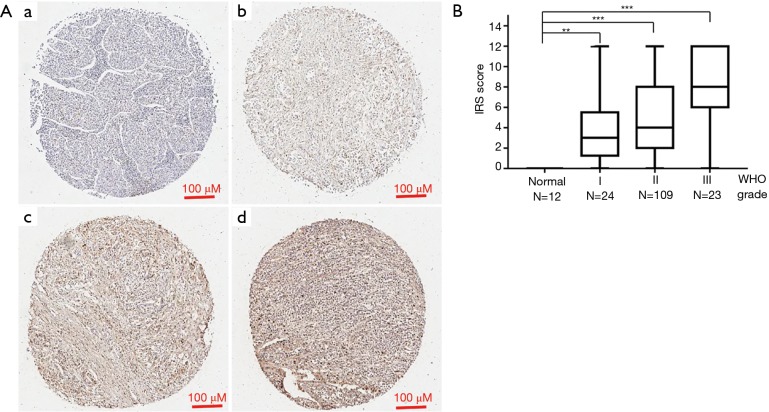
To check expression of APOC1 protein in GC and normal control, tissue microarray of GC with the clinical profiles was purchased and IHC was performed on it. Representative pictures were shown in Figure 3A. IRS statistics for APOC1 are presented in Figure 3B. IRS was shown to increase with clinical stage (P<0.0001). Expression of APOC1 protein is higher in GC than that in normal (Figure 3B). Associations between APOC1 expression and the clinical characteristics of 156 patients in GC tissue arrays including 24 (15.4%) stage I cases, 109 (69.9%) stage II cases and 23 (14.7%) stage III patients. As shown in Table 1, there were significant associations between expression of APOC1 and clinical stage (P=0.011), tumor (T) classification (P=0.010) and lymph node metastasis (P=0.048). However, it was not shown to be associated with patient age (P=0.132), gender (P=0.352). Taken together, these results indicated that expression of APOC1 was associated with many key clinical features of GC.
Figure 3.
Expression of APOC1 protein is much higher in GC than normal tissues. (A) Representative pictures of GC in tissue microarray by IHC, a (negative), b (weak brown), c (moderate brown), d (strong brown). (B) Expression of APOC1 protein was higher in GC than normal tissues. Expression APOC1 protein was increased as GC developed. APOC1, apolipoprotein C1; GC, gastric cancer; IHC, immunohistochemistry. **, P<0.001; ***, P<0.0001.
Table 1. Basic characteristics of patients and association between the expression of APOC1 and clinicopathologic characteristics in GC tissues.
| Characteristics | APOC1 | P value | |
|---|---|---|---|
| Low (score ≤1), N=26 | High (>1), N=130 | ||
| Age (year, mean ± SD) | 0.132 | ||
| <60 | 20 | 74 | |
| ≥60 | 6 | 56 | |
| Gender | 0.352 | ||
| Male | 16 | 91 | |
| Female | 10 | 39 | |
| Stage | 0.011 | ||
| I | 6 | 18 | |
| II | 18 | 91 | |
| III | 2 | 21 | |
| IV | – | – | |
| T-stage | 0.010 | ||
| T1 | 1 | 3 | |
| T2 | 6 | 19 | |
| T3 | 18 | 98 | |
| T4 | 1 | 10 | |
| Lymph node metastasis | 0.048 | ||
| With | 5 | 46 | |
| Without | 21 | 84 | |
APOC1, apolipoprotein C1; GC, gastric cancer.
Spearman association coefficients between APOC1 expression and clinical stage, tumor classification, and lymph node metastasis were 0.479 (P<0.001), 0.236 (P=0.003), 0.187 (P=0.019), respectively (Table 2). These results indicated that expression of APOC1 was associated with many key clinical features of GC.
Table 2. Spearman association analysis between APOC1 and clinical pathologic factors.
| Variables | APOC1 expression level | |
|---|---|---|
| Spearman association | P value | |
| Clinical stage | 0.479 | <0.001 |
| Tumor classification | 0.236 | 0.003 |
| Lymph node metastasis | 0.187 | 0.019 |
APOC1, apolipoprotein C1.
Correlation between APOC1 and CEA, CA19-9, CA242 and CA72-4
The correlation of serum CEA, CA19-9, CA242 CA72-4 and the expression of APOC1 were analyzed using Pearson’s correlation coefficient. None of the serum CEA/CA19-9/CA242/CA72-4 level was correlated with the level of APOC1 expression (Table 3).
Table 3. Association between serum levels of APOC1 and CEA, CA19/9, CA242 and CA72-4 expression.
| Marker | Pearson’s coefficient | P value |
|---|---|---|
| CEA | 0.106 | 0.591 |
| CA19/9 | 0.043 | 0.829 |
| CA242 | −0.041 | 0.844 |
| CA72-4 | −0.080 | 0.722 |
APOC1, apolipoprotein C1; CEA, carcinoembryonic antigen.
Univariate analysis of 5-year survival rate of patients with GC
To further explore the relationship between APOC1 and clinical prognosis in patients with GC, we evaluated the prognostic value of APOC1 using TCGA database (http://www.oncolnc.org/). It was shown in Figure 4 that expression of APOC1 was associated with prognosis of GC patients (P=0.00214). The higher APOC1 expression is, the lower survival rate of GC patients is. On the contrary, the lower APOC1 expression is, the higher survival rate is.
Figure 4.
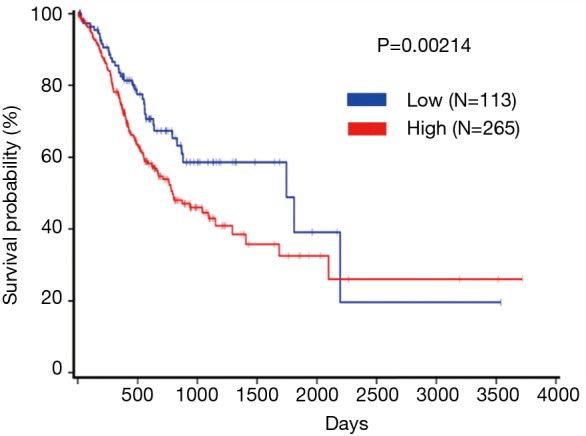
Expression of APOC1 was associated with poor prognosis of GC. APOC1, apolipoprotein C1; GC, gastric cancer.
Discussion
GC is a highly aggressive cancer associated with high mortality in China (24,25). Most GC patients are diagnosed with advanced-stage disease for failure of early diagnosis for treatment options of GC at advanced stage are limited, resulting in an overall 5-year survival rate of 10–28% (26). Serum-based biomarkers are of considerable importance in the early diagnosis of various diseases including cancers (27,28). In recent years, the public availability of cancer gene expression data, such as Oncomine databases and TCGA databases, has provided a unique opportunity to explore initial hypotheses and concepts that can subsequently be validated by additional experiments (29-31). There was much higher APOC1 expression in serum of GC than that in adjacent tissues of GC and normal control. Expression of APOC1 protein was increased as development of GC. Expression of APOC1 was associated with clinical characters and prognosis of patients with GC. APOC1 may be a promising biomarker for diagnosis and prognosis of GC.
In recent years, the relationship between apolipoprotein C and cancers has been highlighted. Borgquist et al. reported that overall cancer risk is associated with the circulatory content of apolipoproteins in males (32). APOC1 is a 57 amino acid residue polypeptide primarily synthesized in the liver (33) and was secreted into serum in an autocrine manner (12). Several studies have found that APOC1 are involved in the development of various cancers (12,13,16), but not serum APOC1. In this study, we firstly reported that the level of APOC1 was increased in serum of patients with GC than healthy controls (P<0.0001), which was in accord with our primary hypothesis. The diagnostic ability of the APOC1 determined based on ROC analysis revealed that APOC1 had distinctly superior ability to distinguish patients with GC with healthy controls (AUC =0.803). Consistent with this result, Kaplan-Meier survival analyses confirmed that the APOC1 could be a novel prognostic biomarker for GC.
In this study, we demonstrated that expression of APOC1 was significantly higher in GC tissues compared with that in adjacent non-cancerous tissues. We also found that the high expression level of APOC1 was associated with clinical stage (P=0.011), tumor classification (P=0.010), as well as with the lymph node metastasis (P=0.048). These findings suggest that the high expression of APOC1 was correlated with unfavorable prognosis of GC patients.
There are numerous cancer markers, such as CEA, cancer antigen125 (CA125), cancer antigen 72-4 (CA72-4) and cancer antigen 19-9 (CA19-9), have been widely used for the diagnosis of GC (34-36). However, these proteins are neither sensitive nor specific tumor biomarker for GC. The sensitivities of CA72-4, CEA, CA125 and CA19-9 at the optimal cut-off level were 67.9%, 73.6%, 63.2% and 60.4%, respectively, and the sensitivity of all four markers used in combination increased to 75.5%. The specificity of CA72-4, CEA, CA125 and CA19-9 at the optimal cut-off level were 83.9%, 75.8%, 77.9% and 83.2%, respectively, and the specificity of all four markers used in combination was75.7% (37). Youden Index is a frequently used summary measure of the ROC curve. It both, measures the effectiveness of a diagnostic marker and enables the selection of an optimal threshold value (cutoff point) for the marker. The corresponding value of the truncation point corresponding to the most approximate index (Youden's index) is regarded as the cut off value (38). With the cut-off value of APOC1 for GC, the sensitivity of APOC1 was 63.0%, though a little low to the combination of four markers (63% vs. 75.5%), the specificity was higher than the combination of four markers (93.0% vs. 75.7%). In addition, APOC1 is an independent marker without correlation with CEA, CA125, CA19-9 and CA72-4.
Conclusions
All in all, our study firstly demonstrated that there was much higher APOC1 concentration in serum of GC than that in health control. We also found that there was much higher expression of APOC1 protein in tissues of GC patients than that in normal. Besides, APOC1 expression was associated with clinical characters of GC patients, such as clinical stage, tumor classification and the lymph node metastasis. Furthermore, APOC1 was found to be associated with prognosis of GC patients. In conclusion, APOC1 may be a potential serum biomarker to diagnose GC and a potential prognostic marker for GC.
Acknowledgments
Funding: This work was supported in part by National Natural Science Foundation of China (81573454 for Jinhua Wang) and supported by Beijing Natural Science Foundation (7172142). This work was also supported by CAMS Innovation Fund for Medical Sciences (2016-I2M-3-007) and Technology Major Projects for “Major New Drugs Innovation and Development” (2018ZX09711001-005-025).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All authors declare no conflict of interest. The study had been approved by PUMCH Ethics Committee (S-424).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Fitzmaurice C, Allen C, Barber RM, et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol 2017;3:524-48. 10.1001/jamaoncol.2016.5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 3.Roth AD. Curative treatment of gastric cancer: towards a multidisciplinary approach. Crit Rev Oncol Hematol 2003;46:59-100. 10.1016/S1040-8428(02)00160-9 [DOI] [PubMed] [Google Scholar]
- 4.Xu Y, Berglund L, Ramakrishnan R, et al. A common Hpa I RFLP of apolipoprotein C-I increases gene transcription and exhibits an ethnically distinct pattern of linkage disequilibrium with the alleles of apolipoprotein E. J Lipid Res 1999;40:50-8. [PubMed] [Google Scholar]
- 5.Su M, Zhou Y, Wang D, Xu T, et al. Expression and purification of recombinant human apolipoprotein C-I in Pichia pastoris. Protein Expr Purif 2011;78:22-6. 10.1016/j.pep.2011.03.003 [DOI] [PubMed] [Google Scholar]
- 6.Leduc V, Jasmin-Bélanger S, Poirier J. APOE and cholesterol homeostasis in Alzheimer's disease. Trends Mol Med 2010;16:469-77. 10.1016/j.molmed.2010.07.008 [DOI] [PubMed] [Google Scholar]
- 7.Poirier J, Hess M, May PC, et al. Cloning of hippocampal poly(A) RNA sequences that increase after entorhinal cortex lesion in adult rat. Brain Res Mol Brain Res 1991;9:191-5. 10.1016/0169-328X(91)90002-F [DOI] [PubMed] [Google Scholar]
- 8.Mooyaart AL, Valk EJ, van Es LA, et al. Genetic associations in diabetic nephropathy: a meta-analysis. Diabetologia 2011;54:544-53. 10.1007/s00125-010-1996-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKay GJ, Savage DA, Patterson CC, et al. Association analysis of dyslipidemia-related genes in diabetic nephropathy. PLoS One 2013;8:e58472. 10.1371/journal.pone.0058472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ki CS, Na DL, Kim DK, et al. Genetic association of an apolipoprotein C-I (APOC1) gene polymorphism with late-onset Alzheimer's disease. Neurosci Lett 2002;319:75-8. 10.1016/S0304-3940(01)02559-9 [DOI] [PubMed] [Google Scholar]
- 11.Bus P, Pierneef L, Bor R, et al. Apolipoprotein C-I plays a role in the pathogenesis of glomerulosclerosis. J Pathol 2017;241:589-99. 10.1002/path.4859 [DOI] [PubMed] [Google Scholar]
- 12.Takano S, Yoshitomi H, Togawa A, et al. Apolipoprotein C-1 maintains cell survival by preventing from apoptosis in pancreatic cancer cells. Oncogene 2008;27:2810-22. 10.1038/sj.onc.1210951 [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, Zhang J, Guo F, et al. Identification of Apolipoprotein C-I Peptides as a Potential Biomarker and its Biological Roles in Breast Cancer. Med Sci Monit 2016;22:1152-60. 10.12659/MSM.896531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X, Lu B, Sun X, et al. ANP32A regulates histone H3 acetylation and promotes leukemogenesis. Leukemia 2018;32:1587-97. 10.1038/s41375-018-0010-7 [DOI] [PubMed] [Google Scholar]
- 15.Su WP, Sun LN, Yang SL, et al. Apolipoprotein C1 promotes prostate cancer cell proliferation in vitro. J Biochem Mol Toxicol 2018:e22158. 10.1002/jbt.22158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko HL, Wang YS, Fong WL, et al. Apolipoprotein C1 (APOC1) as a novel diagnostic and prognostic biomarker for lung cancer: A marker phase I trial. Thorac Cancer 2014;5:500-8. 10.1111/1759-7714.12117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin Y, Yang Y, Su Y, et al. Identification a novel clinical biomarker in early diagnosis of human non-small cell lung cancer. Glycoconj J 2019;36:57-68. 10.1007/s10719-018-09853-z [DOI] [PubMed] [Google Scholar]
- 18.Engwegen JY, Depla AC, Smits ME, et al. Detection of Colorectal Cancer by Serum and Tissue Protein Profiling: A Prospective Study in a Population at Risk. Biomark Insights 2008;3:375-85. 10.4137/BMI.S790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan Y, Shi L, Liu Q, et al. Discovery and identification of potential biomarkers of papillary thyroid carcinoma. Mol Cancer 2009;8:79. 10.1186/1476-4598-8-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Wang J, Dong R, et al. Identification of novel serum biomarkers in child nephroblastoma using proteomics technology. Mol Biol Rep 2011;38:631-8. 10.1007/s11033-010-0149-4 [DOI] [PubMed] [Google Scholar]
- 21.Yi J, Gao R, Chen Y, et al. Overexpression of NSUN2 by DNA hypomethylation is associated with metastatic progression in human breast cancer. Oncotarget 2017;8:20751-65. 10.18632/oncotarget.10612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaemmerer D, Peter L, Lupp A, et al. Comparing of IRS and Her2 as immunohistochemical scoring schemes in gastroenteropancreatic neuroendocrine tumors. Int J Clin Exp Pathol 2012;5:187-94. [PMC free article] [PubMed] [Google Scholar]
- 23.Specht E, Kaemmerer D, Sänger J, et al. Comparison of immunoreactive score, HER2/neu score and H score for the immunohistochemical evaluation of somatostatin receptors in bronchopulmonary neuroendocrine neoplasms. Histopathology 2015;67:368-77. 10.1111/his.12662 [DOI] [PubMed] [Google Scholar]
- 24.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 25.Zong L, Abe M, Seto Y, et al. The challenge of screening for early gastric cancer in China. Lancet 2016;388:2606. 10.1016/S0140-6736(16)32226-7 [DOI] [PubMed] [Google Scholar]
- 26.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer 2013;49:1374-403. 10.1016/j.ejca.2012.12.027 [DOI] [PubMed] [Google Scholar]
- 27.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol 2008;5:588-99. 10.1038/ncponc1187 [DOI] [PubMed] [Google Scholar]
- 28.Coghlin C, Murray GI. Progress in the development of protein biomarkers of oesophageal and gastric cancers. Proteomics Clin Appl 2016;10:532-45. 10.1002/prca.201500079 [DOI] [PubMed] [Google Scholar]
- 29.Akaogi K, Nakajima Y, Ito I, et al. KLF4 suppresses estrogen-dependent breast cancer growth by inhibiting the transcriptional activity of ERalpha. Oncogene 2009;28:2894-902. 10.1038/onc.2009.151 [DOI] [PubMed] [Google Scholar]
- 30.Chabanais J, Labrousse F, Chaunavel A, et al. POFUT1 as a Promising Novel Biomarker of Colorectal Cancer. Cancers (Basel) 2018;10. doi: . 10.3390/cancers10110411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeanquartier F, Jean-Quartier C, Holzinger A. Use case driven evaluation of open databases for pediatric cancer research. BioData Min 2019;12:2. 10.1186/s13040-018-0190-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borgquist S, Butt T, Almgren P, et al. Apolipoproteins, lipids and risk of cancer. Int J Cancer 2016;138:2648-56. 10.1002/ijc.30013 [DOI] [PubMed] [Google Scholar]
- 33.Berbee JF, van der Hoogt CC, Kleemann R, et al. Apolipoprotein CI stimulates the response to lipopolysaccharide and reduces mortality in gram-negative sepsis. FASEB J 2006;20:2162-4. 10.1096/fj.05-5639fje [DOI] [PubMed] [Google Scholar]
- 34.Haglund C, Kuusela P, Roberts P, et al. Tumour marker CA 125 in patients with digestive tract malignancies. Scand J Clin Lab Invest 1991;51:265-70. 10.3109/00365519109091613 [DOI] [PubMed] [Google Scholar]
- 35.Bornschein J, Selgrad M, Wex T, Kuester D, et al. Serological assessment of gastric mucosal atrophy in gastric cancer. BMC Gastroenterol 2012;12:10. 10.1186/1471-230X-12-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ychou M, Duffour J, Kramar A, et al. Clinical significance and prognostic value of CA72-4 compared with CEA and CA19-9 in patients with gastric cancer. Dis Markers 2000;16:105-10. 10.1155/2000/595492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang AP, Liu J, Lei HY, et al. CA72-4 combined with CEA, CA125 and CAl9-9 improves the sensitivity for the early diagnosis of gastric cancer. Clin Chim Acta 2014;437:183-6. 10.1016/j.cca.2014.07.034 [DOI] [PubMed] [Google Scholar]
- 38.Fluss R, Faraggi D, Reiser B. Estimation of the Youden Index and its associated cutoff point. Biom J 2005;47:458-72. 10.1002/bimj.200410135 [DOI] [PubMed] [Google Scholar]