Summary
The development and adoption of hybrid seed technology have led to dramatic increases in agricultural productivity. However, it has been a challenge to develop a commercially viable platform for the production of hybrid wheat (Triticum aestivum) seed due to wheat's strong inbreeding habit. Recently, a novel platform for commercial hybrid seed production was described. This hybridization platform utilizes nuclear male sterility to force outcrossing and has been applied to maize and rice. With the recent molecular identification of the wheat male fertility gene Ms1, it is now possible to extend the use of this novel hybridization platform to wheat. In this report, we used the CRISPR/Cas9 system to generate heritable, targeted mutations in Ms1. The introduction of biallelic frameshift mutations into Ms1 resulted in complete male sterility in wheat cultivars Fielder and Gladius, and several of the selected male‐sterile lines were potentially non‐transgenic. Our study demonstrates the utility of the CRISPR/Cas9 system for the rapid generation of male sterility in commercial wheat cultivars. This represents an important step towards capturing heterosis to improve wheat yields, through the production and use of hybrid seed on an industrial scale.
Keywords: CRISPR/Cas9, genome editing, hybrid wheat, Ms1, Triticum aestivum, male sterility
Introduction
Global demand for food crops is projected to double between 2005 and 2050 (Tilman et al., 2011). In order to meet future demand and limit the environmental impact associated with doing so, new breeding technologies must be developed to increase crop yields (Tester and Langridge, 2010). Capturing heterosis through hybrid breeding is one of few crop improvement technologies that offers rapid and significant yield gains across diverse production environments. Hybrid seed has long been widely used for the production of major cereal crops such as maize and rice, but it has been a challenge to develop a commercially viable hybridization platform for bread wheat (Triticum aestivum) due to wheat's strong inbreeding features, and the absence of a simple, inexpensive and reliable system for hybrid seed production. Heterotic yield gains of more than 10% and enhanced yield stability have been observed in experimental wheat hybrids (Longin et al., 2013; Mühleisen et al., 2014), underscoring the potential of this breeding method. Given that wheat provides approximately one‐fifth of dietary calories and protein for the human population (Shiferaw et al., 2013), it is clear that the development of a viable wheat hybridization platform could have a substantial positive impact on global food security.
Commercial hybrid seed production requires efficient cross‐pollination between genetically distinct parental inbred lines. In addition, self‐pollination of the female parent must be avoided. In wheat, this has been difficult to achieve on a large scale due to a lack of efficient and reliable methods for separating the sexes and forcing outcrossing (Whitford et al., 2013). Recently, a novel hybridization platform that utilizes nuclear male sterility was described for maize (Wu et al., 2016). A key component of the platform is seed production technology (SPT), a process that enables the propagation of non‐transgenic nuclear male‐sterile inbred lines for use as female parents. This hybridization platform has since been extended to rice, a development that was made possible by the identification and isolation of the rice male fertility gene OsNP1 (Chang et al., 2016). Recently, we identified the wheat male fertility gene Ms1 by map‐based cloning and demonstrated its function via complementation of the EMS‐derived mutation ms1d (Tucker et al., 2017). In our previous report, we also described how Ms1 could be used to establish SPT in wheat, and we highlighted the potential of genome editing for rapidly introducing highly penetrant recessive ms1 alleles into elite wheat lines.
The CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 (CRISPR‐associated protein 9) system is currently the most widely used genome editing technology, largely due to its simplicity and flexibility. The system has two components that together form a ribonucleoprotein complex: the Cas9 endonuclease and a small guide RNA (gRNA). The gRNA contains a 20 nucleotide guide sequence that is designed to target a specific site (protospacer) in the genome via Watson–Crick base pairing. The protospacer must be located immediately 5′ to a protospacer adjacent motif (PAM), whose canonical form is 5′‐NGG‐3′ (Jinek et al., 2012). Following target site recognition, Cas9 creates a DNA double‐strand break (DSB). Repair of the DSB through the error‐prone non‐homologous end‐joining (NHEJ) pathway often leaves a lesion in the form of a small insertion/deletion (indel) mutation. Such mutations can shift the open reading frame of a coding sequence or introduce a pre‐mature stop codon, resulting in gene knockout.
Here, we used the CRISPR/Cas9 system to generate Ms1 knockout wheat lines that exhibit male sterility in the first generation. One of the recessive ms1 alleles was highly penetrant and stably transmitted to the T1, T2 and T3 generations. Our results demonstrate the utility of the CRISPR/Cas9 system for the rapid generation of nuclear male sterility in hexaploid wheat. We anticipate that this approach will facilitate the development of a commercially viable wheat hybridization platform.
Results
gRNA and vector design
Using partial Ms1 (chromosome 4BS), Ms‐A1 (4AL homoeolog) and Ms‐D1 (4DS homoeolog) sequences derived from T. aestivum cultivars Fielder and Gladius (NCBI GenBank accessions MK039721, MK039722, MK039723, MK039724, MK039725, MK039726), we designed three gRNAs (LTPG1‐1, LTPG1‐2 and LTPG1‐4) that target exon 1 of Ms1 (Figure 1a). All three gRNAs targeted a predicted signal peptide. LTPG1‐1 was mismatched on chromosomes 4AL (positions +8 and +10 upstream of the PAM in the protospacer) and 4DS (position +8). LTPG1‐2 was mismatched on chromosomes 4BS (position +20), 4AL (positions +2, +4 and +20) and 4DS (positions +4, +15 and +20). LTPG1‐4 was mismatched on chromosomes 4AL (positions +2, +11, +14 and +20) and 4DS (position +20). Given that single mismatches at the PAM‐distal end of the guide sequence (i.e. position +20) are well‐tolerated unlike mismatches at the PAM‐proximal end (Hsu et al., 2013), we expected that LTPG1‐1 and LTPG1‐2 would specifically target Ms1 on 4BS, while LTPG1‐4 would target both Ms1 and Ms‐D1. A schematic of the Agrobacterium T‐DNA binary vector used for transformation is shown in Figure 1b.
Figure 1.
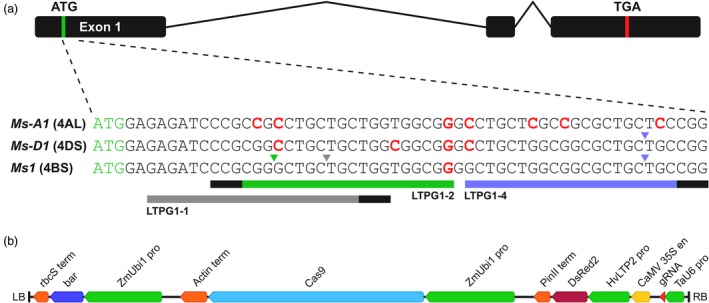
Target sites for gRNAs targeting Ms1, and the vector used for transformation. (a) The gene structure of Ms1 and its homoeologs is shown. The detail underneath shows partial sequences for Ms1 and its homoeologs, as well as the target sites for the three gRNAs (LTPG1‐1, LTPG1‐2 and LTPG1‐4) targeting exon 1 of Ms1 on chromosome 4BS. Target sites are indicated by coloured bars (grey, green and purple). PAM sites (5′‐NGG‐3′) are indicated by black bars at the ends of the coloured bars. Downward‐pointing coloured arrow heads indicate the position of the canonical cut site, and the predicted specificity of the gRNA based on the number and distribution of gRNA mismatches (red text). (b) T‐DNA (12.8 kb) of the Agrobacterium binary vector used for transformation. LB, left border; RB, right border.
Identification of transgenic T0 plants with targeted mutations in Ms1
We transformed 444 immature embryos of cv. Gladius and 352 immature embryos of cv. Fielder. We produced a total of 74 T0 transgenic lines (36 of cv. Gladius and 38 of cv. Fielder) carrying gRNA LTPG1‐1 (18 lines), LTPG1‐2 (40 lines) or LTPG1‐4 (16 lines). Transgene copy numbers ranged from 1 to 18 Table (S1). Two edited lines, both carrying LTPG1‐2, were identified by capillary separation of fluorescently labelled Ms1‐specific amplicons (Figure 2a). Line GL353‐119 (cv. Gladius, transgene copy number = 12) was a biallelic heterozygous mutant (+1/−3), and FL353‐19 (cv. Fielder, transgene copy number = 7) was a biallelic mutant (+1/+1). TIDE (Tracking of Indels by DEcomposition) analysis (Brinkman et al., 2014) of Sanger sequence traces (Data [Link], [Link], [Link], [Link], [Link], [Link], [Link], [Link], [Link], [Link], [Link], [Link], [Link], [Link]) confirmed the presence of targeted mutations in GL353‐119 (+1/−3) and FL353‐19 (+1/+1; Figure 2b). CRISPResso analysis (Pinello et al., 2016) of NGS reads (NCBI BioProject PRJNA495044) revealed that the targeted mutations in GL353‐119 and FL353‐19 were located precisely at the canonical cut site for LTPG1‐2 (Figure 3). In GL353‐119, the +1 insertion was an A nucleotide that created a new AluI restriction site, and the −3 deletion removed the Leu7 residue in the predicted signal peptide. In FL353‐19, the +1 insertions were A and T nucleotides. The CRISPResso analysis also identified several chimeric T0 lines in which <4% of genomic DNA was mutated at the target site (Figure 3). Of the three gRNAs, LTPG1‐2 induced targeted mutations in Ms1 at the highest frequency, and it also had the highest WU‐CRISPR score (Wong et al., 2015) based on in silico predicted on‐target gRNA activity (Table 1). By contrast, LTPG1‐1 induced targeted mutations at very low frequencies, despite having the highest sgRNA Designer score (Doench et al., 2016; Table 1). None of the T0 lines contained off‐target mutations in Ms‐A1 or Ms‐D1 at a frequency above that observed in the wild‐type negative control (0.1%). GL353‐119 was partially male‐sterile (Figure S1), and FL353‐19 was fully male‐sterile, whereas all other T0 lines and the wild‐type negative controls were fully fertile.
Figure 2.
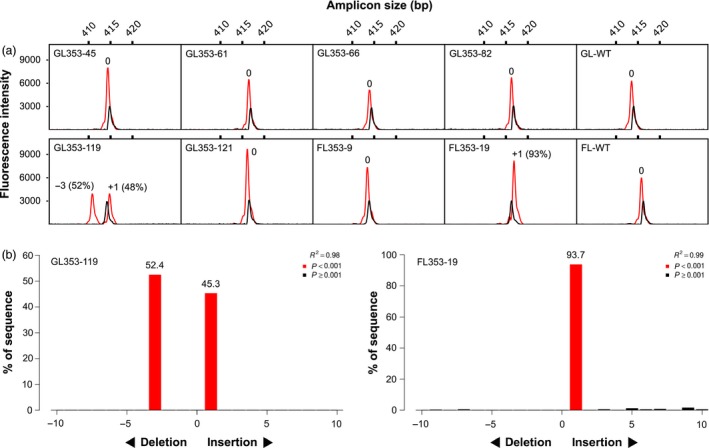
Screening of transgenic T0 wheat lines for targeted mutations. (a) Capillary separation of fluorescently labelled Ms1‐specific (chromosome 4BS) amplicons derived from eight representative lines carrying gRNA LTPG1‐2. A wild‐type control is also shown for each cultivar (GL‐WT, cv. Gladius; FL‐WT, cv. Fielder). Black peaks, wild‐type spike‐in (size reference); red peaks, transgenic or wild‐type (negative control) line. (b) TIDE analysis of Sanger sequence traces for the two putative biallelic mutants identified in (a).
Figure 3.

NGS reads from transgenic T0 mutant wheat lines carrying gRNA LTPG1‐2 targeting Ms1 on chromosome 4BS. The number of reads is shown in the first column. The per cent of total reads is shown in the second column. Green text, start codon; bold text, PAM; blue text, target site. Downward‐pointing arrow heads indicate the position of the canonical cut site. AluI restriction sites are highlighted in grey.
Table 1.
Summary of editing frequencies in T0 wheat lines carrying gRNAs targeting Ms1 on 4BS
| gRNA | Number of transgenic lines | In silico predicted gRNA activity | ||||
|---|---|---|---|---|---|---|
| Total | >0.1% NHEJ | ≥1.0% NHEJ | ≥99% NHEJ | sgRNA Designer score* | WU‐CRISPR score† | |
| LTPG1‐1 | 18 | 4 (22%) | 0 (0%) | 0 (0%) | 0.4 | <50 |
| LTPG1‐2 | 40 | 28 (70%) | 10 (25%) | 2 (5%) | 0.3 | 70 |
| LTPG1‐4 | 16 | 0 (0%) | N/A | N/A | – | <50 |
| None | N/A | 0 (0%) | N/A | N/A | N/A | N/A |
Potential range of gRNA activity score = 0–1.
Potential range of gRNA activity score = 0–100 (scores < 50 are not output); –, no score; N/A, not applicable. Editing frequencies (%NHEJ) are based on CRISPResso analysis of NGS reads. Wild‐type cv. Gladius was used as the negative control (bottom row).
Inheritance of targeted mutations and male‐sterile phenotypes in the T1, T2 and T3 generations
To determine whether the targeted mutations were heritable, we tracked the inheritance of the +1 mutant allele in the progeny of the partially male‐sterile line GL353‐119 (+1/−3). As the +1 insertion of an A nucleotide created a new AluI restriction site, we were able to easily detect the mutant allele via AluI restriction enzyme assay.
By crossing GL353‐119 (+1/−3) with wild‐type cv. Gladius (Figure 4a), we obtained T1 progeny (+1/WT or −3/WT), all of which were fully fertile (Figure 4b). Line T1‐1 (+1/WT) was selfed to produce 94 T2 seeds that were either DsRed‐positive (63 seeds) or DsRed‐negative (31 seeds), based on fluorescence microscopy (Figure 4c). The Cas9 transgene was detected by PCR in 59 of the 63 DsRed‐positive T2 plants (94%), while it was detected in only 5 of the 31 DsRed‐negative T2 plants (16%). The 26 DsRed/Cas9‐negative T2 plants were genotyped for the +1 allele and phenotyped for fertility. All of the WT/WT and +1/WT plants were fully fertile, whereas all of the +1/+1 plants were male‐sterile and produced no seed (Figure 4d and Table 2). The recessive +1 allele was inherited in a Mendelian fashion (Figure 4e and Table 2). Thus, we obtained four male‐sterile T2 plants that were apparently non‐transgenic (Table 2). Line T2‐14 (+1/WT) was randomly selected and selfed to produce T3 progeny. Fifty T3 plants were genotyped and phenotyped. All of the WT/WT and +1/WT plants were fully fertile, whereas all of the +1/+1 plants were male‐sterile, and the targeted mutations were inherited in a Mendelian fashion (Table 3). Additionally, the partially male‐sterile GL353‐119 line produced a T1 seed through selfing. The T1 plant grown from this seed had the parental genotype (+1/−3), and it too was partially male‐sterile.
Figure 4.
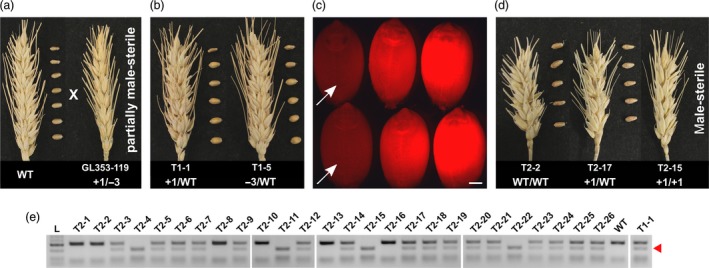
Inheritance and segregation of targeted mutations and male sterility generated with gRNA LTPG1‐2. (a) Crossing of the transgenic T0 mutant line GL353‐119 with wild‐type cv. Gladius. (b) Representative examples of T1 progeny derived from the cross shown in (a). The T1 progeny were selfed to produce T2 seeds. (c) Selection of DsRed‐negative (presumed non‐transgenic) T2 seeds produced by line T1‐1. Scale bar = 1 mm. (d) Representative examples of T2 progeny grown from the selected DsRed‐negative seeds. (e) Genotyping of the 26 DsRed/Cas9‐negative T2 progeny. The +1 mutant allele contains an AluI restriction site that is not present in the WT allele. Cleavage at the AluI restriction site results in a 210 bp band (red arrow head).
Table 2.
Summary of genotypes and phenotypes of DsRed/Cas9‐negative T2 progeny from line T1‐1
| Genotype | Number of lines | Male‐sterile |
|---|---|---|
| +1/WT | 16 (61.5%) | 0 (0%) |
| +1/+1 | 4 (15.4%) | 4 (100%) |
| WT/WT | 6 (23.1%) | 0 (0%) |
| Chi‐squared | 1.69* |
Segregation conforms to a Mendelian 1:2:1 ratio based on the chi‐squared test (P = 0.43).
Table 3.
Summary of genotypes and phenotypes of T3 progeny from line T2‐14
| Genotype | Number of lines | Male‐sterile |
|---|---|---|
| +1/WT | 22 (44%) | 0 (0%) |
| +1/+1 | 11 (22%) | 11 (100%) |
| WT/WT | 17 (34%) | 0 (0%) |
| Chi‐squared | 2.16* |
Segregation conforms to a Mendelian 1:2:1 ratio based on the chi‐squared test (P = 0.34).
Discussion
The CRISPR/Cas9 system is a powerful tool for studying gene function and generating genetic diversity in crops. In this study, our aim was to demonstrate the utility of the CRISPR/Cas9 system for the rapid generation of nuclear male sterility in hexaploid wheat. We hypothesized that knockout of Ms1 through the introduction of targeted biallelic frameshift mutations would result in male sterility, a trait that has high agronomic value for hybrid seed production.
Five per cent (2 of 40) of T0 plants carrying gRNA LTPG1‐2 were biallelic mutants, while the majority of the plants (26 of 40) were chimeras in which only a small proportion of cells were edited. Similar editing efficiencies have been reported for gRNAs targeting other wheat genes such as TaMLO‐A1 (Wang et al., 2014) and TaGW2 (Zhang et al., 2016). We observed substantial variation in editing efficiencies between different gRNAs; of the three gRNAs tested, only one (LTPG1‐2) had sufficient activity to generate heritable targeted mutations in Ms1. In silico prediction of gRNA activity was carried out using the sgRNA Designer (Doench et al., 2016) and WU‐CRISPR (Wong et al., 2015) tools. These tools use different algorithms to calculate an activity score for each gRNA, where higher scores indicate higher predicted gRNA activity. WU‐CRISPR has a score range of 0–100, but only gRNAs with scores ranging from 50 to 100 are displayed in the database as they are considered ‘good’ candidates (Xiaowei Wang, personal communication). LTPG1‐2 was predicted by WU‐CRISPR to be a good candidate (activity score = 70), whereas LTPG1‐1 and LTPG1‐4 (activity scores < 50) were not. Our experimental results were in agreement with these predictions. sgRNA Designer has a score range of 0–1, and there is no threshold for what is considered a ‘good’ candidate. LTPG1‐1 (activity score = 0.4) was predicted by sgRNA Designer to be the most active of the three gRNAs, but our experimental results were not in agreement with this prediction. The lack of correlation between prediction and experiment in this case is not surprising, as we tested only three gRNAs. Given the variability in gRNA efficacy, and considering the time‐consuming and laborious nature of wheat transformation and tissue culture, it is recommended that gRNAs be validated through transient expression in wheat protoplasts prior to commencing plant transformation (Shan et al., 2014).
We did not observe any off‐target editing of Ms‐A1 or Ms‐D1 in T0 lines. In the case of LTPG1‐2, the presence of multiple mismatches (including at least one mismatch in the critical PAM‐proximal ‘seed’ region of the protospacer) was sufficient to abolish gRNA activity, as expected (Hsu et al., 2013; O'Geen et al., 2015; Sternberg et al., 2014; Wu et al., 2014). Thus, LTPG1‐2 shows high specificity for Ms1 on chromosome 4BS. In the case of LTPG1‐1 and LTPG1‐4, on‐target activity was very low or undetectable, and therefore, the lack of off‐target activity was also expected.
We used three different methods for detecting targeted mutations in transgenic T0 plants: capillary separation of fluorescently labelled amplicons, TIDE analysis of Sanger sequence traces and CRISPResso analysis of NGS reads. Only NGS had sufficient sensitivity to detect the low‐frequency targeted mutations in the chimeras, but the three methods produced very similar results for the two biallelic mutants. Therefore, capillary separation of fluorescently labelled homoeolog‐specific amplicons and TIDE analysis of homoeolog‐specific Sanger sequence traces are rapid, reliable and cost‐effective options for initial screening of T0 plants for targeted mutations that are likely to be heritable.
As expected, biallelic knockout of Ms1 in the T0 line FL353‐19 (+1/+1) resulted in complete male sterility. By contrast, incomplete knockout of Ms1 in the biallelic mutant T0 line GL353‐119 (+1/−3) resulted in partial male sterility. The targeted mutations carried by GL353‐119 were inherited in a Mendelian fashion, and completely male‐sterile (+1/+1) mutants were recovered in the T2 and T3 generations, along with fully fertile +1/WT and WT/WT plants. These results are consistent with previous reports of male sterility in EMS‐derived Ms1 knockout mutants (Tucker et al., 2017; Wang et al., 2017). Furthermore, we obtained four T2 knockout mutants that were apparently non‐transgenic based on fluorescence microscopy and PCR assays. The identification of these mutants was streamlined by employing a fluorescence‐based seed sorting strategy, similar to that which has been developed for Arabidopsis (Gao et al., 2016). Interestingly, the partial male‐sterile phenotype, observed in GL353‐119, was inherited by a +1/−3 T1 plant. This suggests that the Leu7 residue (deleted in the −3 allele) is required for proper functioning of the signal peptide in Ms1.
Genome editing has been successfully applied for the generation of male‐sterile rice and sorghum lines (Chang et al., 2016; Cigan et al., 2017; Li et al., 2016; Zhou et al., 2016). Initial attempts to use (meganuclease‐based) genome editing for the generation of male‐sterile wheat lines (Cigan et al., 2017) were met with limited success, as editing of only one of the three Ms26 homoeologs (A, B or D‐genome) did not confer male sterility due to functional redundancy of the gene. However, conventional crossing of single‐genome biallelic Ms26 mutants was successfully carried out to produce triple biallelic Ms26 mutants exhibiting male sterility (Singh et al., 2017). More recently, the CRISPR/Cas9 system was used to edit the wheat male fertility gene Ms45, and by selfing a triple monoallelic mutant T1 plant, male‐sterile triple biallelic mutant T2 plants were recovered (Singh et al., 2018). Thus, gene functional redundancy can slow the process of recovering edited wheat lines with the desired phenotype, as the targeted mutations often need to be combined and/or made homozygous via conventional breeding.
Ms1 is a single copy gene located on chromosome 4BS (Tucker et al., 2017), and the homoeologs Ms‐A1 and Ms‐D1 are epigenetically silenced (Wang et al., 2017). This lack of functional redundancy among homoeologs makes Ms1 a particularly attractive target for genome editing. Indeed, our results demonstrate that the CRISPR/Cas9 system can be used to generate Ms1 knockout wheat lines that exhibit male sterility in the first generation. We also report here the sequence for LTPG1‐2, an active gRNA that specifically targets Ms1 in cv. Fielder and cv. Gladius. Further studies will be needed to determine the efficacy of LTPG1‐2 in other wheat cultivars, including elite germplasm from different breeding pools. If LTPG1‐2 is found to be ineffective in a particular target cultivar, for example due to gene functional redundancy, then a different gRNA or multiple gRNAs may be needed. Furthermore, as we only tested three gRNAs, it is likely that screening of additional gRNAs would lead to the identification of a gRNA(s) that exhibits even higher activity than that observed for LTPG1‐2.
Our study provides a methodological foundation and molecular tools for the rapid development of nuclear male‐sterile wheat lines. This represents a significant step toward the establishment of a commercially viable hybrid wheat platform. Thus, we anticipate that the adoption of genome editing technologies for precision wheat breeding, together with a better understanding of wheat floral architecture and the flower opening process (Okada et al., 2018), will ultimately lead to increased yield gains through capturing heterosis.
Experimental procedures
gRNA design
Partial Ms1, Ms‐A1 and Ms‐D1 sequences derived from T. aestivum cultivars Fielder and Gladius were used for gRNA design. The gRNAs were designed to target exon 1 of Ms1 on chromosome 4BS, in a region immediately downstream of the start codon, based on the presence of the canonical PAM (5′‐NGG‐3′). Guide sequences were 20 nucleotides in length. To ensure efficient transcription from the TaU6 promoter, all gRNAs had a G nucleotide at position +20 (PAM‐distal end) of the guide sequence (Sander and Joung, 2014).
Vector design and construction
All vectors were designed using Vector NTI software. The gRNA expression cassette (Shan et al., 2013) consisting of the TaU6 promoter and a non‐targeting (random guide sequence) gRNA was synthesized (GenScript) and cloned into pUC57, resulting in pUC57‐gRNA. Annealed oligos containing the Ms1‐targeting guide sequence (Table S2) were cloned into pUC57‐gRNA by simultaneous digestion/ligation with BbsI and T4 DNA ligase. Positive clones were identified by diagnostic restriction digest and validated by Sanger sequencing (Australian Genome Research Facility).
The rice codon‐optimized SpCas9 gene with N‐ and C‐terminal nuclear localization signals (Shan et al., 2013) was synthesized (GenScript) and inserted into PHP62407M as an NcoI–AscI fragment between the maize Ubi1 promoter and the Sorghum bicolor actin terminator, resulting in the entry vector pCas9‐NB.
The Agrobacterium T‐DNA binary vector pMDC123 (Curtis and Grossniklaus, 2003) was modified by replacing the original selection cassette with an intron‐containing bar gene regulated by the maize Ubi1 promoter and the wheat rbcS Class II terminator. In addition, an aleurone‐specific fluorescent reporter (DsRed2) cassette (Wu et al., 2016) was inserted between the Gateway cassette and the right border, resulting in the destination vector pMDC‐Bar‐DsRed. The Cas9 expression cassette from pCas9‐NB was Gateway cloned into pMDC‐Bar‐DsRed to produce the intermediate vector pNB1.
Finally, the gRNA expression cassettes from three different pUC57‐gRNA vectors pre‐loaded with Ms1‐targeting guide sequences (LTPG1‐1, LTPG1‐2 and LTPG1‐4, respectively) were individually cloned into pNB1 as AsiSI‐PmeI fragments between the DsRed2 expression cassette and the right border. The resulting T‐DNA binary vectors (pNB‐LTPG1‐1, pNB‐LTPG1‐2 and pNB‐LTPG1‐4) were used for Agrobacterium‐mediated transformation.
Agrobacterium‐mediated transformation
Transformation of cv. Fielder and cv. Gladius was carried out as described (Ishida et al., 2015), with minor modifications. Briefly, immature embryos were isolated from spikes harvested at 14 days post‐anthesis. Isolated embryos were transferred to WLS‐liq solution, centrifuged at 16 000 g for 10 min, incubated in WLS‐inf solution containing Agrobacterium (strain AGL1) for 5 min and then transferred to WLS‐AS media for 2 days of co‐cultivation. After co‐cultivation, embryo axes were removed, and then, scutella were transferred to WLS‐Res media for 5 days of resting culture. After the resting culture, scutella were transferred to WLS‐P5 callus induction media (selection with 5 mg/L phosphinothricin) for 2 weeks, followed by WLS‐P10 callus induction media (selection with 10 mg/L phosphinothricin) for 3 weeks. Calli were then transferred to LSZ‐P5 regeneration media (selection with 5 mg/L phosphinothricin) for 2 weeks under a cycle of 12 h dark/12 h light (~70 μmol/m2/s). Regenerants were transferred to LSF‐P5 rooting media (selection with 10 mg/L phosphinothricin) for 2 weeks, before being transferred to potted soil in the greenhouse. Timentin was substituted for cefotaxime in all tissue culture media.
Detection of targeted mutations by capillary separation of fluorescently labelled amplicons
Genomic DNA was extracted from the second leaves of transgenic T0 plants at the vegetative stage, using a freeze‐dried method (Kovalchuk, 2014). The target site was amplified by PCR using Phusion High‐Fidelity DNA Polymerase (New England BioLabs), Phusion GC Buffer, 5% DMSO, 1 m betaine and a pair of Ms1‐specific 6‐FAM‐labelled primers Table (S2). To generate wild‐type amplicons for spike‐in (size reference), the same primer pair labelled with HEX was used. Touchdown PCR cycling conditions were as follows: initial denaturation at 98 °C for 3 min, denaturation at 98 °C for 15 s, annealing at 70–65 °C for 20 s, extension at 72 °C for 15 s and final extension at 72 °C for 5 min. The starting annealing temperature was decreased by 0.5 °C each cycle for 10 cycles, followed by 25 cycles at the final annealing temperature. A sample of the PCR product was run on an agarose gel to confirm the presence of a single band of the expected size (432 bp). The fluorescently labelled amplicons were diluted and subjected to capillary electrophoresis (Australian Genome Research Facility) on an AB3730 DNA Analyzer (Applied Biosystems, Foster City, CA). The results were analysed using PeakScanner Software 2 (Applied Biosystems). In PeakScanner, the peak for the HEX‐labelled wild‐type size reference was adjusted to a fluorescence intensity of approximately 3000, to improve visual clarity.
Detection of targeted mutations by Sanger sequencing and TIDE
Genomic DNA was extracted from transgenic T0 plants as described above. The target site was amplified by PCR using Phusion High‐Fidelity DNA Polymerase, Phusion GC Buffer, 5% DMSO, 1 m betaine and a pair of Ms1‐specific primers Table (S2). Touchdown PCR cycling conditions were as follows: initial denaturation at 95 °C for 8 min, denaturation at 94 °C for 10 s, annealing at 62–57 °C for 30 s, extension at 72 °C for 30 s and final extension at 72 °C for 5 min. The starting annealing temperature was decreased by 0.5 °C each cycle for 10 cycles, followed by 30 cycles at the final annealing temperature. A sample of the PCR product was run on an agarose gel to confirm the presence of a single band of the expected size (577 bp). The amplicons were then column‐purified and Sanger sequenced (Australian Genome Research Facility) on a 3730xl DNA Analyzer (Applied Biosystems). Bases were called with KB Basecaller v1.4.1.8, and the AB1 files were uploaded to the online TIDE analysis tool (Brinkman et al., 2014). In TIDE, the indel size range was set at 10, and the other settings were adjusted based on information provided on the online TIDE analysis tool Troubleshooting webpage. The proportion of edited DNA in the sampled tissue was calculated as the sum of all significant indels (P < 0.001) detected by TIDE.
Detection of targeted mutations by NGS and CRISPResso analysis
Genomic DNA was extracted from transgenic T0 plants as described above. To generate amplicons for NGS, two rounds of PCR were carried out. The PCR mixtures contained Phusion High‐Fidelity DNA Polymerase, Phusion GC Buffer, 5% DMSO and 1 m betaine. In the first round of PCR, the target site was amplified using conserved primers flanked by 5′ universal tail sequences Table (S2). PCR cycling conditions were as follows: initial denaturation at 98 °C for 3 min, followed by 30 cycles of 98 °C for 20 s, 72 °C for 20 s, with a final extension at 72 °C for 2 min. A sample of the PCR product was run on an agarose gel to confirm the presence of bands of the expected sizes (300 bp, 313 bp, 327 bp). In the second round of PCR, barcodes and adapters were added using Illumina Nextera XT primers that anneal to the tail sequences of the primers used in the first round of PCR. Cycling conditions in the second round of PCR were as follows: initial denaturation at 98 °C for 3 min, followed by six cycles of 98 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, with a final extension of 72 °C for 5 min. The barcoded PCR products were purified using Agencourt AMPure XP beads (Beckman Coulter), quantified by qPCR, pooled in equimolar amounts, spiked with 10% PhiX Control v3 and then sequenced (Australian Genome Research Facility) on the Illumina MiSeq platform using the MiSeq Reagent Kit v3 300 cycle. The raw reads from each sample were filtered using Ms1, Ms‐A1 and Ms‐D1‐specific tag sequences Table (S2) and assigned to their respective homoeologs. FASTQ files containing the homoeolog‐specific reads were used as input for the CRISPResso analyses (Pinello et al., 2016). In CRISPResso, the following parameters were used: –w 20 –hide_mutations_outside_window_NHEJ –save_also_png –trim_sequences ‐q 30 –exclude_bp_from_left 5 –exclude_bp_from_right 5 –ignore_substitutions. Allele frequencies in Figure 3 were calculated by summing the values in the %Reads column of the CRISPResso allele frequency table, after filtering out aligned sequences that did not contain the partial allele sequence shown. Editing frequencies in Table 1 were calculated using data from the CRISPResso pie charts.
Pollen viability assay
Pollen viability was assessed by Lugol (1% I3K solution) staining. Pollen grains were mounted on glass microscope slides and imaged using a Nikon Ni‐E microscope equipped with a DS‐Ri1‐U3 camera (Adelaide Microscopy Waite Facility). Images were captured with NIS‐Elements software.
DsRed expression assay
Mature seeds were imaged with a Leica MZ FLIII microscope equipped with a Leica DFC450 C digital camera and a DsRed filter set. Images were captured with Leica Application Suite v4.10.0 software.
Detection of the Cas9 transgene in T2 plants
Genomic DNA was extracted from T2 plants as described above. The Cas9 transgene was detected by PCR using Phusion High‐Fidelity DNA Polymerase, Phusion HF Buffer and a pair of Cas9‐specific primers Table (S2). Touchdown PCR cycling conditions were as follows: initial denaturation at 98 °C for 2 min, denaturation at 98 °C for 15 s, annealing at 72–69 °C for 20 s, extension at 72 °C for 15 s and final extension at 72 °C for 5 min. The starting annealing temperature was decreased by 0.5 °C each cycle for 6 cycles, followed by 24 cycles at the final annealing temperature. A sample of the PCR product was run on an agarose gel to check for the presence/absence of the expected 281 bp band.
AluI assay for genotyping T2 and T3 plants
Genomic DNA was extracted from T2 and T3 plants as described above. The target site was amplified by PCR using Phusion High‐Fidelity DNA Polymerase, Phusion GC Buffer, 5% DMSO, 1 m betaine and pair of Ms1‐specific primers Table (S2). Touchdown PCR cycling conditions were as follows: initial denaturation at 95 °C for 8 min, denaturation at 94 °C for 10 s, annealing at 70–65 °C for 30 s, extension at 72 °C for 20 s and final extension at 72 °C for 5 min. The starting annealing temperature was decreased by 0.5 °C each cycle for 10 cycles, followed by 27 cycles at the final annealing temperature. A sample of the PCR product was run on an agarose gel to confirm the presence of a single band of the expected size (432 bp). Five μL of unpurified amplicons was digested with 2 units of AluI in a 7 μL reaction and then run on a 2% agarose gel.
Statistics
To test for Mendelian inheritance of edited alleles in the T2 and T3 generations, Pearson's chi‐squared test was used, as described (Montoliu, 2012).
Post hoc in silico prediction of gRNA activity
gRNA on‐target activity was predicted using the sgRNA Designer (Doench et al., 2016) and WU‐CRISPR (Wong et al., 2015) tools, according to the developers’ guidelines.
Conflict of interest
ET, UB and RW have filed a patent on previous published work on Ms1. The remaining authors declare no conflict of interest.
GenBank accession numbers
Partial Ms1 sequence obtained from cultivar Fielder: MK039721; Partial Ms‐A1 sequence obtained from cultivar Fielder: MK039722; Partial Ms‐D1 sequence obtained from cultivar Fielder: MK039723; Partial Ms1 sequence obtained from cultivar Gladius: MK039724; Partial Ms‐A1 sequence obtained from cultivar Gladius: MK039725; Partial Ms‐D1 sequence obtained from cultivar Gladius: MK039726.
Supporting information
Table S1 Transgene copy numbers for T0 lines.
Table S2 Primers, oligonucleotides and sequence tags used in this study.
Figure S1 Pollen viability assay based on detection of starch via iodine‐potassium iodide staining. Wild type cv. Gladius (left) and the partially male‐sterile edited line GL353‐119 (right) are shown. Scale bar = 100 μm.
Data S1 Ms1 Sanger sequence trace for cv. Fielder transgenic T0 line FL353‐9.
Data S2 Ms1 Sanger sequence trace for cv. Fielder transgenic T0 line FL353‐14.
Data S3 Ms1 Sanger sequence trace for cv. Fielder transgenic T0 line FL353‐19.
Data S4 Ms1 Sanger sequence trace for wild type cv. Fielder.
Data S5 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐45.
Data S6 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐48.
Data S7 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐61.
Data S8 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐69.
Data S9 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐82.
Data S10 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐119.
Data S11 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐121.
Data S12 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐123.
Data S13 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐124.
Data S14 Ms1 Sanger sequence trace for wild type cv. Gladius.
Acknowledgements
We are grateful to Matthew Tucker (The University of Adelaide) for critical discussions and reading of the manuscript, Adam Croxford (The University of Adelaide) for providing helpful advice and technical assistance with NGS, Yuan Li (The University of Adelaide) for copy number analysis, Ursula Langridge‐Reimold and Yuriy Onyskiv (The University of Adelaide) for glasshouse management and plant care, Iryna Mazonka, Sheridan Morton and Christine Kastner (The University of Adelaide) for assistance with plant transformation and tissue culture, and Mathieu Baes (The University of Adelaide) for technical support. This research was funded by DuPont Pioneer Hi‐Bred International Inc. We are grateful for the support provided by The University of Adelaide, Australian Research Council, Grains Research and Development Corporation and the South Australian State Government.
References
- Brinkman, E.K. , Chen, T. , Amendola, M. and van Steensel, B. (2014) Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Z. , Chen, Z. , Wang, N. , Xie, G. , Lu, J. , Yan, W. , Zhou, J. et al (2016) Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc. Natl. Acad. Sci. USA, 113, 14145–14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan, A.M. , Singh, M. , Benn, G. , Feigenbutz, L. , Kumar, M. , Cho, M.‐J. , Svitashev, S. et al (2017) Targeted mutagenesis of a conserved anther‐expressed P450 gene confers male sterility in monocots. Plant Biotechnol. J. 15, 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M.D. and Grossniklaus, U. (2003) A Gateway cloning vector set for high‐throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench, J.G. , Fusi, N. , Sullender, M. , Hegde, M. , Vaimberg, E.W. , Donovan, K.F. , Smith, I. et al (2016) Optimized sgRNA design to maximize activity and minimize off‐target effects of CRISPR‐Cas9. Nat. Biotechnol. 34, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X. , Chen, J. , Dai, X. , Zhang, D. and Zhao, Y. (2016) An effective strategy for reliably isolating heritable and Cas9‐free Arabidopsis mutants generated by CRISPR/Cas9‐mediated genome editing. Plant Physiol. 171, 1794–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, P.D. , Scott, D.A. , Weinstein, J.A. , Ran, F.A. , Konermann, S. , Agarwala, V. , Li, Y. et al (2013) DNA targeting specificity of RNA‐guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida, Y. , Tsunashima, M. , Hiei, Y. and Komari, T. (2015) Wheat (Triticum aestivum L.) transformation using immature embryos. Methods Mol. Biol. 1223, 189–198. [DOI] [PubMed] [Google Scholar]
- Jinek, M. , Chylinski, K. , Fonfara, I. , Hauer, M. , Doudna, J.A. and Charpentier, E. (2012) A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk, N. (2014) High‐throughput analysis pipeline for achieving simple low‐copy wheat and barley transgenics. Methods Mol. Biol. 1145, 239–252. [DOI] [PubMed] [Google Scholar]
- Li, Q. , Zhang, D. , Chen, M. , Liang, W. , Wei, J. , Qi, Y. and Yuan, Z. (2016) Development of japonica photo‐sensitive genic male sterile rice lines by editing Carbon Starved Anther using CRISPR/Cas9. J. Genet. Genom. 43, 415–419. [DOI] [PubMed] [Google Scholar]
- Longin, C. , Gowda, M. , Mühleisen, J. , Ebmeyer, E. , Kazman, E. , Schachschneider, R. , Schacht, J. et al (2013) Hybrid wheat: quantitative genetic parameters and consequences for the design of breeding programs. Theoret. Appl. Genet. 126, 2791–2801. [DOI] [PubMed] [Google Scholar]
- Montoliu, L. (2012) Mendel: a simple excel workbook to compare the observed and expected distributions of genotypes/phenotypes in transgenic and knockout mouse crosses involving up to three unlinked loci by means of a χ2 test. Transgenic Res. 21, 677–681. [DOI] [PubMed] [Google Scholar]
- Mühleisen, J. , Piepho, H.‐P. , Maurer, H.P. , Longin, C.F.H. and Reif, J.C. (2014) Yield stability of hybrids versus lines in wheat, barley, and triticale. Theoret. Appl. Genet. 127, 309–316. [DOI] [PubMed] [Google Scholar]
- O'Geen, H. , Henry, I.M. , Bhakta, M.S. , Meckler, J.F. and Segal, D.J. (2015) A genome‐wide analysis of Cas9 binding specificity using ChIP‐seq and targeted sequence capture. Nucleic Acids Res. 43, 3389–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, T. , Jayasinghe, J.E.A.R.M. , Nansamba, M. , Baes, M. , Warner, P. , Kouidri, A. , Correia, D. et al (2018) Unfertilized ovary pushes wheat flower open for cross‐pollination. J. Exp. Bot. 69, 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinello, L. , Canver, M.C. , Hoban, M.D. , Orkin, S.H. , Kohn, D.B. , Bauer, D.E. and Yuan, G.‐C. (2016) Analyzing CRISPR genome‐editing experiments with CRISPResso. Nat. Biotechnol. 34, 695–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander, J.D. and Joung, J.K. (2014) CRISPR‐Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32, 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, Q. , Wang, Y. , Li, J. , Zhang, Y. , Chen, K. , Liang, Z. , Zhang, K. et al (2013) Targeted genome modification of crop plants using a CRISPR‐Cas system. Nat. Biotechnol. 31, 686–688. [DOI] [PubMed] [Google Scholar]
- Shan, Q. , Wang, Y. , Li, J. and Gao, C. (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. Nature Publishing Group 9, 2395–2410. [DOI] [PubMed] [Google Scholar]
- Shiferaw, B. , Smale, M. , Braun, H.‐J. , Duveiller, E. , Reynolds, M. and Muricho, G. (2013) Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Security, 5, 291–317. [Google Scholar]
- Singh, M. , Kumar, M. , Thilges, K. , Cho, M.‐J. and Cigan, A.M. (2017) MS26/CYP704B is required for anther and pollen wall development in bread wheat (Triticum aestivum L.) and combining mutations in all three homeologs causes male sterility. PLoS ONE, 12, e0177632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, M. , Kumar, M. , Albertsen, M.C. , Young, J.K. and Cigan, A.M. (2018) ‘Concurrent modifications in the three homeologs of Ms45 gene with CRISPR‐Cas9 lead to rapid generation of male sterile bread wheat (Triticum aestivum L.). Plant Mol. Biol. Springer, Netherlands 97, 371–383. [DOI] [PubMed] [Google Scholar]
- Sternberg, S.H. , Redding, S. , Jinek, M. , Greene, E.C. and Doudna, J.A. (2014) DNA interrogation by the CRISPR RNA‐guided endonuclease Cas9. Nature, Nature Publishing Group 507, 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester, M. and Langridge, P. (2010) Breeding technologies to increase crop production in a changing world. Science, 327, 818–822. [DOI] [PubMed] [Google Scholar]
- Tilman, D. , Balzer, C. , Hill, J. and Befort, B.L. (2011) Global food demand and the sustainable intensification of agriculture. Proc. Natl Acad. Sci. USA, 108, 20260–20264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, E.J. , Baumann, U. , Kouidri, A. , Suchecki, R. , Baes, M. , Garcia, M. , Okada, T. et al (2017) Molecular identification of the wheat male fertility gene Ms1 and its prospects for hybrid breeding. Nat. Commun. 8, 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Cheng, X. , Shan, Q. , Zhang, Y. , Liu, J. , Gao, C. and Qiu, J.‐L. (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Li, J. , Chen, S. , Heng, Y. , Chen, Z. , Yang, J. , Zhou, K. et al (2017) Poaceae‐specific MS1 encodes a phospholipid‐binding protein for male fertility in bread wheat. Proc. Natl. Acad. Sci. USA, 114, 12614–12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford, R. , Fleury, D. , Reif, J.C. , Garcia, M. , Okada, T. , Korzun, V. and Langridge, P. (2013) Hybrid breeding in wheat: technologies to improve hybrid wheat seed production. J. Exp. Bot. 64, 5411–5428. [DOI] [PubMed] [Google Scholar]
- Wong, N. , Liu, W. and Wang, X. (2015) WU‐CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 16, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X. , Scott, D.A. , Kriz, A.J. , Chiu, A.C. , Hsu, P.D. , Dadon, D.B. , Cheng, A.W. et al (2014) Genome‐wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. Nature Publishing Group 32, 670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , Fox, T.W. , Trimnell, M.R. , Wang, L. , Xu, R. , Cigan, A.M. , Huffman, G.A. et al (2016) Development of a novel recessive genetic male sterility system for hybrid seed production in maize and other cross‐pollinating crops. Plant Biotechnol. J. 14, 1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Liang, Z. , Zong, Y. , Wang, Y. , Liu, J. , Chen, K. , Qiu, J.‐L. et al (2016) Efficient and transgene‐free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H. , He, M. , Li, J. , Chen, L. , Huang, Z. , Zheng, S. , Zhu, L. et al (2016) Development of commercial thermo‐sensitive genic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9‐mediated TMS5 editing system. Sci. Rep. 6, 37395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Transgene copy numbers for T0 lines.
Table S2 Primers, oligonucleotides and sequence tags used in this study.
Figure S1 Pollen viability assay based on detection of starch via iodine‐potassium iodide staining. Wild type cv. Gladius (left) and the partially male‐sterile edited line GL353‐119 (right) are shown. Scale bar = 100 μm.
Data S1 Ms1 Sanger sequence trace for cv. Fielder transgenic T0 line FL353‐9.
Data S2 Ms1 Sanger sequence trace for cv. Fielder transgenic T0 line FL353‐14.
Data S3 Ms1 Sanger sequence trace for cv. Fielder transgenic T0 line FL353‐19.
Data S4 Ms1 Sanger sequence trace for wild type cv. Fielder.
Data S5 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐45.
Data S6 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐48.
Data S7 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐61.
Data S8 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐69.
Data S9 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐82.
Data S10 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐119.
Data S11 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐121.
Data S12 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐123.
Data S13 Ms1 Sanger sequence trace for cv. Gladius transgenic T0 line GL353‐124.
Data S14 Ms1 Sanger sequence trace for wild type cv. Gladius.


