Abstract
Listeria monocytogenes is a foodborne Gram-positive bacterium causing listeriosis in both animals and humans. It can persist and grow in various environments including conditions countered during saprophytic or intra-host lifestyles. Sigma (σ) subunit of RNA polymerase is a transcriptional factor responsible for guiding the core RNA polymerase and initiating gene expression under normal growth or physiological changes. In L. monocytogenes, there is one housekeeping sigma factor, σA, and four alternative sigma factors σB, σC, σH, and σL. Generally, σA directs expression of genes required for normal growth while alternative σ factors alter gene expression in response to specific conditions (e.g., stress). In this study, we aimed to determine the exclusive role of σA in L. monocytogenes by comparing a wild type strain with its isogenic mutant lacking genes encoding all alternative sigma factors (i.e., sigB, sigC, sigH, and sigL). We further investigated their survival abilities in 6% porcine bile (pH 8.2) mimicking gallbladder bile and their transcriptomics profiles in rich medium (i.e., BHI) and 1% porcine bile. Surprisingly, the results showed that survival abilities of wild type and ΔsigBΔsigCΔsigHΔsigL (or ΔsigBCHL) quadruple mutant strains in 6% bile were similar suggesting a compensatory role for σA. RNA-seq results revealed that bile stimulon of L. monocytogenes wild type contained 66 genes (43 and 23 genes were up- and down-regulated, respectively); however, only 29 genes (five up- and 24 down-regulated by bile) were differentially expressed in ΔsigBCHL. We have shown that bile exposure mediates increased transcription levels of dlt and ilv operons and decreased transcription levels of prfA and heat shock genes in wild type. Furthermore, we identified σA-dependent bile inducible genes that are involved in phosphotransferase systems, chaperones, and transporter systems; these genes appear to contribute to L. monocytogenes cellular homeostasis. As a result, σA seemingly plays a compensatory role in the absence of alternative sigma factors under bile exposure. Our data support that the bile stimulon is prone to facilitate resistance to bile prior to initiated infection.
Keywords: RNA-seq, Listeria monocytogenes, housekeeping sigma σA, sigma factor, bile, stress
Introduction
The foodborne pathogen Listeria monocytogenes is a facultative Gram-positive intracellular bacterium that is able to adapt to a broad range of environments such as soil and waste water. It can survive and grow in a variety of temperature ranging from −1.5°C to up to 45°C, a wide range of pH and hypertonic conditions (Hudson et al., 1994; Membre et al., 2005; Goulet et al., 2013). This pathogen is the causative agent of listeriosis in human and animals. Immunocompromised individuals, including the elderly and pregnant women, are considered to be high-risk populations (Southwick and Purich, 1996). While L. monocytogenes infections are rare, the mortality rate is 15.6% (EFSA, 2015). It ranks third among foodborne pathogens causing death in the United States (Scallan et al., 2011) and results in approximately 23,000 cases of listeriosis each year worldwide (de Noordhout et al., 2014).
Sigma factors (σ) are the dissociable subunit of bacterial RNA polymerase (RNAP) enzyme; they recognize specific promoter sequences and initiate gene/operon transcription. Unbound sigma factors can bind to core RNAP to form the holoenzyme and enhance interaction between RNAP and promoter consensus sequences (Burgess, 2001). This results in conformational changes in the recognized promoter regions upstream of the transcription starting site. Most gene expression in the bacterial cells is dependent on the housekeeping sigma factor σ70 in Bacillus subtilis (Gruber et al., 2001) or σA in L. monocytogenes. σ70 is encoded by rpoD in E. coli and it is evident that the intracellular concentrations of RNAP and RpoD remain constant under several growth conditions (Jishage et al., 1996; Piper et al., 2009). σ70 is required for cell growth related transcription, while alternative sigma factors regulate gene expression in response to stress (Mauri and Klumpp, 2014). For example, the alternative sigma factor σB is known to respond to heat stress in L. monocytogenes and B. subtilis (Schumann, 2003) as well as σF, σE, σG, and σK are known for developmental programing such as sporulation in B. subtilis (Losick and Stragier, 1992). The concentration of alternative sigma factors [e.g., σH (σ32), σS (σ38) in E. coli] vary considerably within altered physiological states (Jishage et al., 1996; Grigorova et al., 2006).
Under different environmental conditions, L. monocytogenes regulates gene expression through the use of four alternative sigma factors: σB, σC, σH, and σL (Chaturongakul et al., 2011). Co-regulation between σA and each alternative sigma factor have been evaluated. A number of genes have σA- and σB-dependent promoters such as prfA (encoding a master regulator of virulence genes), qoxABCD (encoding quinol oxidase important for oxidative stress response), and cggR (encoding central glycolytic gene regulator) (Liu et al., 2017). Promoter consensus sequences of σA and σH have been found in competence genes such as comG (Medrano Romero and Morikawa, 2016). Genes co-regulated by housekeeping σA and other alternative sigma factors have also been identified in other bacteria including B. subtilis and Escherichia coli (Wade et al., 2006). The extracytoplasmic function (ECF) sigma factor is also shown to co-regulate with σA in B. subtilis (Kingston et al., 2011). In addition to the concentration of sigma factors inside the cell, the affinity between each sigma factor and RNAP determines the probability of associating with the core RNAP. In E. coli, σ70 has been shown to have the highest affinity with core RNAP (Sharma and Chatterji, 2010). Previous studies have demonstrated that sigma factors can compensate for each other as explored in in vitro transcriptional assays (Maeda et al., 2000; Jishage et al., 2002) and competition experiments (Mauri and Klumpp, 2014).
A L. monocytogenes quadruple deletion mutant (ΔsigBΔsigCΔsigHΔsigL or ΔsigBCHL) with only σA remaining grew as well as wild type in phosphotransferase system (PTS)-dependent carbon sources (e.g., mannose, cellobiose, and glucose) (Wang et al., 2014) suggesting that having the housekeeping σA alone is sufficient to maintain transcription. This raised our interest in the potential of up-regulation of genes by σA in stress conditions in L. monocytogenes. For instance, following consumption of contaminated food, L. monocytogenes encounters the low pH of the stomach and the bile salt and high osmolality in intestinal fluid during gastric passage. In order to colonize the intestine and successfully establish infection, L. monocytogenes needs to survive bile exposure, an important antimicrobial component in gastrointestinal fluid (Olier et al., 2004). It has also been reported that L. monocytogenes may utilize unique bile resistance mechanisms to survive in the gallbladder (Hardy et al., 2004). A number of studies have evaluated L. monocytogenes proteins that affect bile resistance including bile salt hydrolase (Bsh) (Dussurget et al., 2002; Begley et al., 2005b), the bile exclusion system (BilE) (Sleator et al., 2005), and multidrug resistance (MDR) efflux pump MdrT (Quillin et al., 2011). Previously, it was determined that bile salt hydrolase as well as bile exclusion system are regulated by σB (Dussurget et al., 2002; Begley et al., 2005b). However, the bile stimulon defined recently by RNA-seq analyses was not identified as σB-dependent (Guariglia-Oropeza et al., 2018). This is surprising, however, they suggested that it is possibly due to comparing transcriptional profiles of L. monocytogenes strains exposed to pH 5.5 with and without bile. The σB regulon is induced under acidic condition (Sue et al., 2004); no induction has been observed in the presence of bile. It is more likely that σB-dependent gene expression plays a more crucial role in earlier stage of gastrointestinal infection (e.g., stomach) and prime the bacteria for the establishment in the small intestine. We hypothesized that under bile stress exposure, ΔsigBCHL will have similar survival ability as wild type due to the compensatory role of housekeeping σA in ΔsigBCHL. To test the hypothesis, we phenotypically and transcriptomically compared ΔsigBCHL with solely functional σA with its isogenic parental wild type under bile exposure. Genes under σA regulation in L. monocytogenes during gallbladder bile exposure were identified.
Materials and Methods
Bacterial Strains and Growth Conditions
Listeria monocytogenes 10403S and its isogenic quadruple alternative sigma factor mutant strain, ΔsigBCHL [FSL C3-135 (Mujahid et al., 2013)] were used in this study. Both strains were maintained in Brain Heart Infusion broth (BHI; DifcoTM, BD, United States) and 50% glycerol stocks at −80°C. They were streaked onto BHI agar plates prior to each experiment, and plates were incubated at 37°C overnight. A single colony inoculated into five ml BHI broth for 37°C overnight (16–18 h) incubation with shaking (200 rpm). Overnight culture was diluted 1:100 into a fresh five ml BHI broth and incubated at 37°C with shaking to an OD600 of approximately 0.4, representing mid-log phase. An aliquot of 500 μl of the culture was subsequently passaged in 50 ml of BHI broth to generate synchronized cells at mid-log phase before exposure to the simulated bile stress.
Bile Stress Survival Assay
Once mid-log phase was reached, cells were immediately challenged with simulated gallbladder bile. The in vitro bile fluid was prepared as previously described (Versantvoort et al., 2005). Briefly, 0.5 ml of 2 × 6% porcine bile (Sigma, United States), pH 8.2 ± 0.2 (30 ml of 175.3 g/l NaCl, 68.3 ml of 84.7 g/l NaHCO3, 4.2 ml of 89.6 g/l KCl, 150 μl of 37% g/g HCl, 10 ml of 25 g/l urea, 10 ml of 22.2 g/l CaCl2.2H2O, and 1.8 g BSA to 500 ml distilled water) was added to 0.5 ml of mid-log phase culture. The challenged cultures were incubated at 37°C with shaking for 10 and 20 min, respectively. Survival ability was determined at 10 and 20 min after stress (t = 10 and t = 20). A 100 μl aliquot of the pre-treated control, untreated control (culture with additional 20 min incubation) along with t = 10 and t = 20 cultures were ten-fold serially diluted in PBS (pH 7.4); 10 μl of each dilution was plated onto BHI agar plates for subsequent enumeration. Experiments were conducted in triplicate on separate days.
Statistical Analysis for Survival Assay
The significant differences in survival between L. monocytogenes wild type and quadruple mutant were determined by t-test (SPSS v. 23, IBM, United States). p < 0.05 was considered statistically significantly different.
RNA Isolation and DNase Treatment
RNA was extracted from mid-log phase cultures after 10 min exposure to 1% porcine bile or BHI (as a control) as previously described with minor modifications (Pleitner et al., 2014). Briefly, all experiments were conducted in three biological replicates on different days. For each strain and condition, one ml of BHI or bile-exposed culture was collected and immediately added to ice-cold stop solution of 10% acid-phenol chloroform pH 4.5 (InvitrogenTM, United States) in ethanol, and followed as outlined previously (Pleitner et al., 2014). DNase treatment was performed using TURBO DNA-freeTM DNase treatment kit (Invitrogen, United States) according to the manufacturer’s protocol. Total RNA concentration was quantified using Nanodrop (DeNovix, United States).
rRNA Depletion, Library Preparation, and RNA Sequencing
Ribosomal RNA (rRNA) was depleted by using Ribo-Zero rRNA removal kit (Epicentre, Madison, WI, United States) following manufacturer’s instruction. Quality of RNA was assessed using the 2100 Bioanalyzer (Agilent Technology, Santa Clara, CA, United States). Sample with RNA Integrity Number (RIN) score greater than 8.0 was considered acceptable for further library preparation and RNA-sequencing.
cDNA library preparation was constructed using ScriptSeq v2 RNA-seq Library Preparation kit for Bacteria (Epicentre, Madison, WI, United States). Purification of cDNA and indexed RNA-seq libraries were performed using Agencourt® AMPure® XP kit (Beckman Coulter Inc., Brea, CA, United States). Quantity and quality of the libraries were determined using the 2100 Bioanalyzer. All experiments were performed in three biological replicates. Sequencing was carried out on a HiSeq 2 × 100 High Output paired-end, 100 bp read at the Purdue University genomics core facility.
RNA-Seq Analysis
Sequencing reads were mapped against L. monocytogenes 10403S (NCBI accession number: NC_017544.1). The average coverage per base on both sense and anti-sense strands of L. monocytogenes 10403S wild type and quadruple mutant (ΔsigBCHL) as well as rRNA match rates in BHI and bile are shown in Supplementary Table S1. The paired-end RNA-seq data were submitted to the SRA database (SRA accession number: PRJNA544468). Reads were aligned and mapped with Bowtie2 and TopHat2 (Langmead and Salzberg, 2012; Trapnell et al., 2012). Reads were counted by HTSeq-count (Anders et al., 2015). Differential expression (DE) was compared and analyzed in R version 3.3.3 using the package DESeq2 (Love et al., 2014). The analyses were conducted from three replicates of each sample, except wild type sample in BHI (two replicates were used in analysis). Genes were considered differentially expressed when log2 fold-change <−1 or >1 (representing down- or up-regulation) and adjusted p-values < 0.05.
Gene Set Enrichment Analysis
GOseq package 1.34.1 package for R (Young et al., 2010) available from Bioconductor was used to evaluate whether differential expressed genes identified by DESeq2 were enriched for Gene Ontology (GO) terms. This tool allowed us to statistically confirm the specific metabolic pathways that L. monocytogenes utilized under BHI and/or bile conditions.
Quantitative PCR (qPCR) Validation of RNA-Seq Data
Selected differentially expressed genes from RNA-seq results were validated using qPCR as previously described with minor modifications (Sue et al., 2004; Pleitner et al., 2014). Target genes used for RNA-seq data validation were (i) rpoB and bglA as housekeeping genes and (ii) gadT2, plcA, LMRG_00091, LMRG_01149, LMRG_01669, and LMRG_02283 for sigma A function. A list of TaqMan primers and probes designed by PrimerQuest (IDT DNA, Coralville, IA, United States) are shown in Table 1. TaqMan probes were synthesized with a 5′ 6-carboxyfluorescein (6-FAM) reporter dye and a 3′ ZENTM dark quencher dye. All reactions were run via Rotor-Gene Q (Qiagen, Germany). cDNA synthesis was performed at 48°C for 30 min followed by PCR setting of 1 cycle at 95°C for 10 min, 40 cycles at 95°C for 15s, and 55°C for 1 min. Transcripts were normalized by a geometric mean of housekeeping genes, rpoB and bglA. The transcript levels of each strain in BHI and bile were compared statistically using unpaired t-test, SPSS version 23 (IBM, United States). Statistical comparison of gene expression levels among L. monocytogenes strains in each condition were analyzed by unpaired t-test.
TABLE 1.
TaqMan primers and probes used for RNA-seq data validation.
| Target gene | Forward primer∗ | Probe∗ | Reverse primer∗ |
| bglA | TTCATGAGCGGCGGTATTT | TACCAAGCTGTCCACCACGAACTT | TGTGCTTCGGGCATGATT |
| gadT2 | TGGCAAGAAGGCGGTATTT | TTCTTGGGTGGGAAATACACTCGGG | TCACGAATCCGACCGTTATTT |
| malG | GCAGCCCTAACAGCTTTCT | AGCAATGCTACTAGGTGCGCTTGA | GAATCCCACCACCGATGTAAA |
| plcA Kazmierczak et al., 2006 | GATTTATTTACGACGCACATTAGTTT | CCCATTAGGCGGAAAAGCATATTCGC | GAGTTCTTTATTGGCTTATTCCAGTTATT |
| rpoB Pleitner et al., 2014 | CGATCTTGGAGAGCCGAAATA | CGGTAGAAGAATCTAAGAAC | GAGCCGCATAGTTTGCATCAC |
| LMRG_01149 | GAGCATCTATCCCTCCAGAAATTA | TAGGAACTGCCTTCGCGATTTCGA | AATCCCAAGAGCTAACGCTAC |
| LMRG_01669 | GATTTCGCAAAGACTCGGATTAC | TTGCACCAAGTTTGCGAGAATGGC | ACGACGACGAATCGCTTT |
| LMRG_02283 | GTCATTTACCAGCCCGTATCTC | AGTACCGCTTGTTTGGTCAATTTGGT | CGGACTCATCATGTTCCAATCA |
∗ All sequences are written 5′–3′.
Results
L. monocytogenes Lacking All Alternative Sigma Factors Has Similar Phenotype as Its Parental Strain
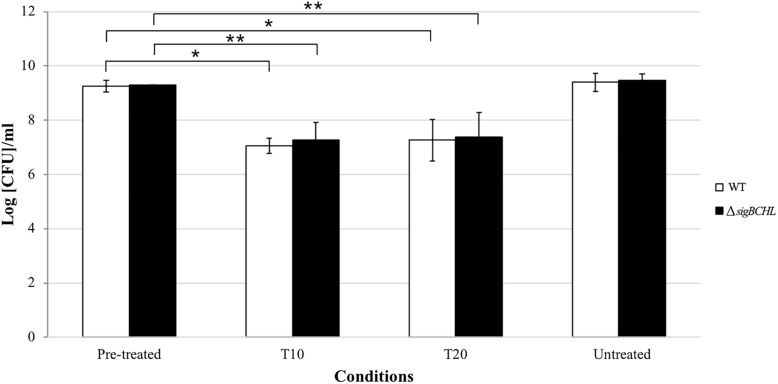
To examine the survival ability under bile exposure, L. monocytogenes wild type and the quadruple mutant (ΔsigBCHL) were exposed to simulated gallbladder bile for 20 min. Viability of each culture was determined at 10 and 20 min after bile treatment; two log CFU/ml reductions were observed in both wild type and ΔsigBCHL mutant when compared to untreated controls (p < 0.05) (Figure 1). No significant difference was observed between wild type and ΔsigBCHL mutant suggesting housekeeping sigma factor σA alone is sufficient to coordinate expression of genes responsible for survival under bile stress.
FIGURE 1.
Survival of Listeria monocytogenes 10403S wild type and its quadruple mutant in simulated gallbladder bile at various time points: pre-treated control, T10, T20, untreated control; WT (white), ΔsigBCHL (black). Asterisks indicate significant difference when compared to pre-treated controls using t-test (p < 0.05).
Sixty-Six Significantly Differentially Expressed Genes Were Identified in Wild Type L. monocytogenes Exposed to Bile
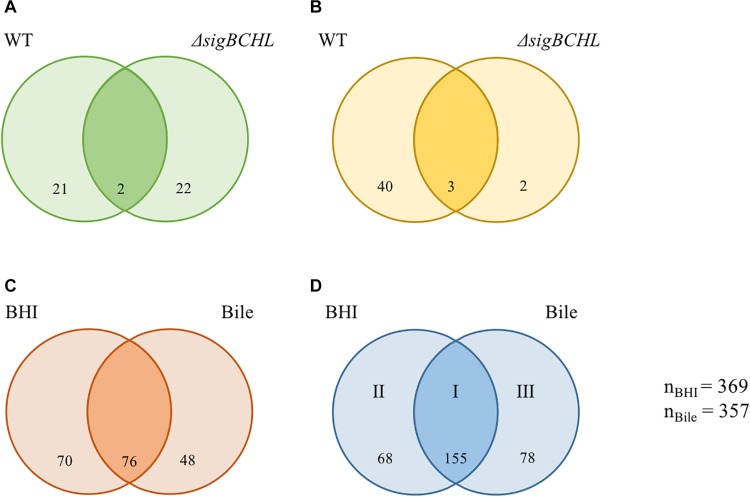
We evaluated wild type and ΔsigBCHL mutant transcriptomic profiles in both BHI and bile conditions to characterize the bile stimulon and σA regulon. To define the bile response genes of L. monocytogenes 10403S, we compared RNA-seq data from wild type grown in BHI (control) to 1% bile for 10 min. Genes with FC either >2 or <−2 and adjusted p-value < 0.05 were considered significant difference. We found 66 genes significantly differentially expressed upon bile exposure of which 23 and 43 genes were down- and up-regulated, respectively (Figures 2A,B and Table 2). Twenty-three genes with statistically significant reduced transcript levels were decreased by two–three folds. Among these genes, purE, LMRG_02276, and groL were the most down-regulated with FC of 3.3, 3.2, and 3.2, respectively. purE encodes a phosphoribosylaminoimidazole carboxylase catalytic subunit involved in inosine-5′-phosphate biosynthesis II. LMRG_02276 and groL encode QacE family quaternary ammonium compound efflux SMR transporter and heat shock protein 60 family chaperone (GroL), respectively. Surprisingly, the master positive virulence regulator, PrfA, was down-regulated (FC = 2.14) in bile exposure conditions. The chaperone encoding gene dnaK was also shown to have lower transcript levels after bile exposure. opuCA and opuCD involved in L-carnitine/choline ABC transporter were also down-regulated. On the other hand, among the up-regulated genes, LMRG_01622 had the highest fold-change at 18.59; its function is not yet characterized. We also observed that inlC2 (encoding internalin C2), dltC (encoding D-alanyl carrier protein), ilvBCD (encoding valine biosynthesis associated proteins), and uspA2 (encoding universal stress protein) were up-regulated under bile exposure. GO terms associated with biological process involved in defense response, antigenic variation, and interspecies interaction between organisms were enriched in bile exposure condition (Supplementary Table S2).
FIGURE 2.
Venn diagrams showing the overlaps of differentially expressed genes (DEGs) of L. monocytogenes wild type and ΔsigBCHL under bile exposure compared to BHI. (A,B) Numbers of genes with significantly decreased and increased transcripts after exposure to bile, respectively. (C,D) Numbers of genes under σA regulation with significantly lower and higher levels in ΔsigBCHL in comparison to wild type, respectively. Differentially expressed genes in BHI and bile conditions are shown in left and right circles, respectively. The number indicated in the circle represents the number of differentially expressed genes in each strain and condition. I, II, and III represent groups of genes positively regulated by σA. The total numbers of differentially expressed genes under BHI and bile calculated from either lower or higher level are 369 and 357, respectively.
TABLE 2.
Differentially expressed genes under bile exposure in L. monocytogenes 10403S wild type.
| LMRG Locus tag | lmo Locus tag | Gene name | Operon | Annotation | FC | Adjusted p-value |
| LMRG_00151 | LMRG_02887, LMRG_00151- LMRG_00152 | Hypothetical protein | 3.08 | 0.00 | ||
| LMRG_00152 | Hypothetical protein | 2.39 | 0.01 | |||
| LMRG_00222 | lmo0540 | Penicillin-binding protein | 2.04 | 0.00 | ||
| LMRG_00293 | lmo0610 | Internalin-like protein | 2.4 | 0.00 | ||
| LMRG_00311 | lmo0628 | LMRG_00311-LMRG_00312 | Hypothetical protein | 4.56 | 0.00 | |
| LMRG_00334 | lmo0647 | Hypothetical protein | 2.83 | 0.00 | ||
| LMRG_00341 | lmo0654 | LMRG_00341-LMRG_00342 | Hypothetical protein | 3.11 | 0.00 | |
| LMRG_00404 | lmo0715 | LMRG_00402-LMRG_00407 | Hypothetical protein | 2.02 | 0.00 | |
| LMRG_00482 | lmo0794 | Putative Rrf2-linked NADH-flavin reductase | 2.41 | 0.00 | ||
| LMRG_00529 | lmo1067 | GTP-binding protein TypA/BipA | 2.23 | 0.00 | ||
| LMRG_00530 | lmo1068 | Fomain-containing protein | 2.05 | 0.01 | ||
| LMRG_00706 | lmo1257 | Hypothetical protein | 2.53 | 0.00 | ||
| LMRG_00826 | lmo1375 | Peptidase M20 | 2.07 | 0.00 | ||
| LMRG_00942 | lmo1489 | LMRG_00945-LMRG_00939 | RNA binding protein | 2.22 | 0.00 | |
| LMRG_01131 | lmo1983 | ilvD | ilv-leu | Dihydroxy-acid dehydratase | 2.62 | 0.01 |
| LMRG_01132 | lmo1984 | ilvB | Acetolactate synthase large subunit | 3.12 | 0.00 | |
| LMRG_01134 | lmo1986 | ilvC | Ketol-acid reductoisomerase | 2.91 | 0.00 | |
| LMRG_01453 | lmo1517 | LMRG_01454-LMRG_01453 | Nitrogen regulatory protein P-II | 2.07 | 0.02 | |
| LMRG_01561 | lmo2269 | Hypothetical protein; | 6.18 | 0.00 | ||
| LMRG_01602 | lmo2230 | arsC | Ars | Arsenate reductase | 2.56 | 0.01 |
| LMRG_01609 | lmo2223 | Putative transcriptional regulator | 2.09 | 0.04 | ||
| LMRG_01622 | lmo2210 | Hypothetical protein | 18.59 | 0.00 | ||
| LMRG_01630 | lmo2202 | fabH | LMRG_01630-LMRG_01631 | 3-oxoacyl-[acyl-carrier-protein] synthase, KASIII | 2.05 | 0.00 |
| LMRG_01761 | lmo2487 | Hypothetical protein | 2.14 | 0.00 | ||
| LMRG_01976 | lmo2720 | Acyl-coenzyme A synthetases/AMP-(fatty) acid ligases, YtcI homolog | 4.32 | 0.00 | ||
| LMRG_02011 | lmo0911 | Hypothetical protein | 2.43 | 0.00 | ||
| LMRG_02071 | lmo0972 | dltC | dltABCD, LMRG_02074 | D-alanyl carrier protein | 2.04 | 0.00 |
| LMRG_02074 | Teichoic acid D-Ala incorporation-associated protein DltX | 2.28 | 0.01 | |||
| LMRG_02191 | lmo2646 | LMRG_02193-LMRG_02190 | Hypothetical protein | 2.51 | 0.03 | |
| LMRG_02218 | lmo2673 | uspA2 | uspA2-rpiB | Universal stress protein UspA | 2.37 | 0.02 |
| LMRG_02232 | lmo2686 | Hypothetical protein | 2.23 | 0.00 | ||
| LMRG_02304 | lmo0880 | Putative peptidoglycan bound protein (LPXTG motif) | 4.47 | 0.00 | ||
| LMRG_02364 | lmo0115 | lmaD | lmaDCBA | Listeria protein LmaD, associated with virulence | 3.79 | 0.00 |
| LMRG_02365 | lmo0116 | lmaC | LmaC, associated with virulence in Listeria | 3.34 | 0.00 | |
| LMRG_02366 | lmo0117 | lmaB | Listeria protein LmaB, associated with virulence | 2.04 | 0.02 | |
| LMRG_02372 | lmo0123 | LMRG_02369-LMRG_02378 | Putative tail or base plate protein gp18 [Bacteriophage A118] | 2.1 | 0.04 | |
| LMRG_02423 | lmo2852 | ASCH domain-containing protein | 2.02 | 0.02 | ||
| LMRG_02427 | lmo2856 | rpmH | 50S ribosomal protein L34 | 2.06 | 0.00 | |
| LMRG_02611 | lmo0265 | dapE | Succinyl-diaminopimelate desuccinylase | 2.87 | 0.00 | |
| LMRG_02646 | lmo0263 | inlC2, inlH | Internalin C2 | 2.22 | 0.04 | |
| LMRG_02700 | lmo2568 | LMRG_02700-LMRG_02701 | Hypothetical protein | 9.41 | 0.00 | |
| LMRG_02768 | lmo1694 | CDP-abequose synthase, Putative sugar nucleotide epimerase | 3.68 | 0.00 | ||
| LMRG_02808 | lmo2132 | Cyclic nucleotide-binding protein | 2.52 | 0.00 | ||
| LMRG_00273 | lmo0591 | LMRG_00271-LMRG_00273 | Hypothetical protein | −2.12 | 0.01 | |
| LMRG_00501 | lmo1040 | LMRG_00501-LMRG_00499 | molybdenum ABC transporter permease | −2.57 | 0.04 | |
| LMRG_00852 | lmo1400 | LMRG_00852-LMRG_00854 | Phosphinothricin N-acetyltransferase | −2.20 | 0.00 | |
| LMRG_00877 | lmo1425 | opuCD | opuC | Osmotically activated L-carnitine/choline ABC transporter, permease protein OpuCD, subunit of predicted ATP-driven transporter complex of CARNITINE/choline | −2.29 | 0.00 |
| LMRG_00880 | lmo1428 | opuCA | Osmotically activated L-carnitine/choline ABC transporter, ATP-binding protein OpuCA, subunit of predicted ATP-driven transporter complex of CARNITINE/choline | −2.05 | 0.00 | |
| LMRG_00926 | lmo1473 | dnaK | hrcA-grpE-dnaK-dnaJ | Chaperone protein, DnaK | −2.39 | 0.00 |
| LMRG_01023 | lmo1877 | fhs | Formate–tetrahydrofolate ligase | −2.55 | 0.00 | |
| LMRG_01218 | lmo2068 | groL | groSL | Heat shock protein 60 family chaperone GroEL | −3.20 | 0.00 |
| LMRG_01219 | lmo2069 | groS | Heat shock protein 60 family co-chaperone GroES | −2.58 | 0.00 | |
| LMRG_01286 | lmo1681 | LMRG_01286-LMRG_01288 | 5-methyltetrahydropteroyltriglutamate–homocysteine methyltransferase | −2.03 | 0.03 | |
| LMRG_01399 | lmo1568 | LMRG_01399-LMRG_01401 | Membrane protein | −2.46 | 0.00 | |
| LMRG_01585 | lmo2247 | LMRG_01585-LMRG_01587 | Xidoreductase of aldo/keto reductase family, subgroup 2, glyoxal reductase | −2.09 | 0.01 | |
| LMRG_01842 | lmo2406 | Hypothetical protein | −2.38 | 0.00 | ||
| LMRG_02144 | lmo2600 | LMRG_02142-LMRG_02144 | ATPase component of general energizing module of ECF transporters | −2.40 | 0.00 | |
| LMRG_02241 | lmo2694 | Arginine decarboxylase/Lysine decarboxylase | −2.14 | 0.00 | ||
| LMRG_02276 | lmo0853 | LMRG_02275-LMRG_02277 | QacE family quaternary ammonium compound efflux SMR transporter | −3.22 | 0.02 | |
| LMRG_02326 | lmo0075 | LMRG_02326-LMRG_02327 | Phosphonomutase, probable carboxyvinyl-carboxyphosphonate phosphorylmutase | −2.03 | 0.04 | |
| LMRG_02358 | lmo0109 | LMRG_02358-LMRG_02359 | AraC family transcriptional regulator | −2.18 | 0.00 | |
| LMRG_02386 | lmo0137 | LMRG_02385-LMRG_02386 | Oligopeptide ABC transporter, permease protein | −2.29 | 0.03 | |
| LMRG_02496 | lmo1775 | purE | LMRG_02496-LMRG_02507 | Phosphoribosylaminoimidazole carboxylase catalytic subunit, 5-(carboxyamino)imidazole ribonucleotide mutase | −3.30 | 0.00 |
| LMRG_02510 | lmo1761 | Sodium-dependent transporter | −2.78 | 0.00 | ||
| LMRG_02622 | lmo0200 | prfA | prfA-plcA | Listeriolysin regulatory protein, PrfA | −2.14 | 0.00 |
| LMRG_02716 | lmo2371 | LMRG_02716-LMRG_02717 | ABC transporter, permease protein | −2.59 | 0.00 |
Twenty-Nine Significantly Differentially Expressed Genes Were Identified in L. monocytogenes Quadruple Mutant Exposed to Bile
In the ΔsigBCHL strain, we were able to assess the role σA alone plays in response to bile. To assess this assumption, ΔsigBCHL RNA-seq data from exposed and non-exposed to bile were compared for differentially expressed genes. Remarkably, we found a small number of differentially expressed genes in this comparison; in total, 29 genes were identified. Of these, 24 and five genes were down- and up-regulated under bile treatment, respectively. Overall, 2–3 FC was observed (Table 3). No specific GO terms were enriched among these genes. Among the five bile up-regulated genes, four encode hypothetical proteins and only LMRG_02808 is annotated as a cyclic nucleotide-binding protein.
TABLE 3.
Differentially expressed genes under bile exposure in L. monocytogenesΔsigBCHL quadruple mutant.
| LMRG Locus tag | lmo Locus tag | Gene name | Operon | Annotation | FC | Adjusted p-value |
| LMRG_00151 | LMRG_02887, LMRG_00151-LMRG_00152 | Hypothetical protein | 2.23 | 0.00 | ||
| LMRG_01622 | lmo2210 | Hypothetical protein | 4.25 | 0.00 | ||
| LMRG_01645 | lmo2187 | Hypothetical protein | 2.22 | 0.02 | ||
| LMRG_01919 | lmo2778 | Hypothetical protein | 2.05 | 0.03 | ||
| LMRG_02808 | lmo2132 | Cyclic nucleotide-binding protein | 2.08 | 0.00 | ||
| LMRG_00091 | lmo0398 | LMRG_00091-LMRG_00095 | PTS system, IIA component | −2.51 | 0.01 | |
| LMRG_00092 | lmo0399 | PTS system, IIB component | −3.52 | 0.00 | ||
| LMRG_00352 | lmo0665 | LMRG_00351-LMRG_00353 | Hypothetical protein | −2.32 | 0.01 | |
| LMRG_00353 | lmo0666 | Domain-containing protein | −2.12 | 0.01 | ||
| LMRG_00369 | lmo0681 | Flagellar biosynthesis regulator FlhF | −2.01 | 0.00 | ||
| LMRG_00373 | lmo0685 | motA | motAB | Flagellar motor rotation protein MotA | −2.43 | 0.00 |
| LMRG_00852 | lmo1400 | LMRG_00852-LMRG_00854 | phosphinothricin N-acetyltransferase | −2.06 | 0.00 | |
| LMRG_01073 | lmo1926 | LMRG_01075-LMRG_01070 | Chorismate mutase II | −2.2 | 0.01 | |
| LMRG_01156 | lmo2008 | LMRG_01157-LMRG_01155 | ABC transporter, permease protein | −2.71 | 0.02 | |
| LMRG_01228 | lmo2077 | LMRG_01229-LMRG_01228 | Inactive homolog of metal-dependent proteases, Putative molecular chaperone tRNA (adenosine(37)-N6)-threonylcarbamoyltransferase complex dimerization subunit type 1 TsaB | −2.61 | 0.00 | |
| LMRG_01283 | lmo2129 | Hypothetical protein | −2.03 | 0.00 | ||
| LMRG_01286 | lmo1681 | metE | LMRG_01286-LMRG_01288 | 5-methyltetrahydropteroyl-triglutamate–homocysteine methyltransferase | −2.23 | 0.00 |
| LMRG_01287 | lmo1680 | Cystathionine gamma-synthase | −2.59 | 0.04 | ||
| LMRG_01333 | lmo1633 | trpE | trp | Anthranilate synthase, aminase component | −2.21 | 0.00 |
| LMRG_01428 | lmo1542 | rplU | LMRG_01428-LMRG_01430 | 50S ribosomal protein L21 | −3.22 | 0.00 |
| LMRG_01435 | lmo1535 | LMRG_01435-LMRG_01436 | YebC/PmpR family DNA-binding transcriptional regulator | −2.25 | 0.00 | |
| LMRG_01591 | lmo2241 | LMRG_01591-LMRG_01593 | Transcriptional regulator, GntR family | −2.25 | 0.00 | |
| LMRG_01669 | lmo2163 | LMRG_01669-LMRG_01673 | Myo-inositol 2-dehydrogenase 1, Oxidoreductase | −2.21 | 0.00 | |
| LMRG_01810 | lmo2438 | ClbS/DfsB family four-helix bundle protein | −2.97 | 0.00 | ||
| LMRG_01955 | lmo2741 | LMRG_01955-LMRG_01957 | Multidrug-efflux transporter, major facilitator superfamily (MFS);Efflux pump Lde | −2.62 | 0.00 | |
| LMRG_02234 | lmo2688 | LMRG_02235-LMRG_02233 | Cell division protein FtsW | −2.56 | 0.01 | |
| LMRG_02481 | lmo0052 | LMRG_02481-LMRG_02483 | Phosphoesterase, DHH family protein | −2.07 | 0.00 | |
| LMRG_02665 | lmo0241 | LMRG_02938, LMRG_02667-LMRG_02664 | TrmH family tRNA/rRNA methyltransferase YacO | −2.67 | 0.00 | |
| LMRG_02671 | lmo0235 | ispD1 | LMRG_02672-LMRG_02669 | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | −2.27 | 0.00 |
LMRG_01669 and LMRG_00091 Genes Were Differentially Expressed in ΔsigBCHL Compared to Wild Type Under BHI and Bile Exposure Conditions, Respectively
To further determine the role of σA, the transcriptional profiles of the ΔsigBCHL mutant in BHI (control) and bile was used to compare to its wild type. We identified 369 and 357 genes that were differentially expressed in BHI or after bile exposure, respectively (Figures 2C,D). A total of 194 genes had statistically significantly lower transcripts levels in ΔsigBCHL mutant under BHI and/or bile conditions (Figure 2C). These genes were considered positively regulated by alternative sigma factors since we compared them with wild type. The gadT2 gene, a member of gadT2D2 operon encoding glutamate/gamma-aminobutyrate antiporter, was chosen to represent the genes in lower transcript group. The qPCR data showed that the level of gadT2 expression was decreased in ΔsigBCHL. We identified 301 genes that were up-regulated in both or either conditions in ΔsigBCHL; 68 and 78 genes had higher transcripts in BHI alone or bile alone, respectively (Figure 2D). The up-regulated genes were defined as σA-dependent genes since they showed higher expression levels in the quadruple mutant compared to wild type. Certainly, regulatory networks are more complicated. Lower expression levels in wild type could imply that these genes are negatively regulated in the presence of alternative sigma factors e.g., by the negative regulators that are not transcribed by σA. For this study, we intentionally minimize discussions on negative roles of alternative sigma factors as de-repression of these genes (or enhanced levels of expression in ΔsigBCHL) was still σA-dependent.
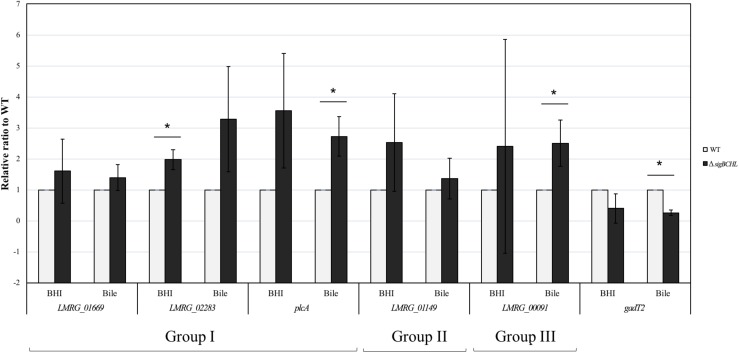
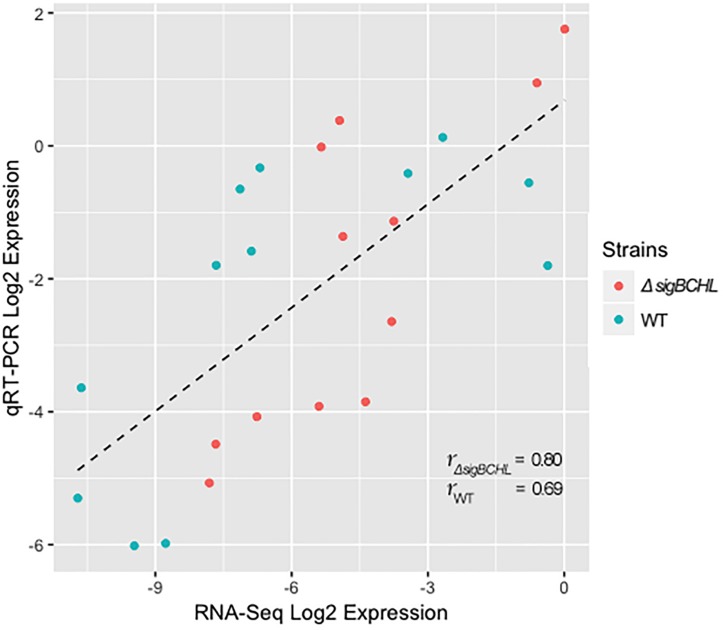
We classified σA-dependent genes into three groups: group I σA-dependent genes expressed in both conditions, group II σA-dependent genes expressed in BHI, and group III σA-dependent bile inducible genes (Figure 2D). Among the 155 genes in group I, three of these genes (LMRG_00092- LMRG_00094) with higher FC are part of an operon that includes genes encoding a PTS fructose transporter subunit and alpha-mannosidase. LMRG_02456 and LMRG_02567 encode PTS system beta-glucoside-specific components. LMRG_01933 encodes PTS system cellobiose-specific IIB component. LMRG_01669-LMRG_01673 is an operon encoding genes involved in myo-inositol degradation and LMRG_01535 is a phage capsid scaffolding encoded gene. LMRG_02283 encodes membrane-spanning permease protein, MsmF, and a member of multiple sugar ABC transporter showed up to six FC. Known σA-regulated genes in prfA-plcA operon are also present in this cluster (Supplementary Table S3). We then confirmed differential expression of LMRG_01669, LMRG_02283, and plcA in both BHI and bile conditions using qPCR (Figure 3). The RNA-seq and qPCR data correlate well with the Pearson correlation coefficients (r) of expression levels in the wild type and the quadruple mutant at 0.69 and 0.80, respectively (Figure 4). Expression of these genes are shown with higher transcripts in ΔsigBCHL than in WT in both BHI and bile conditions. Gene set enrichment analysis identified 48 GO terms that were overrepresented among group I members (Supplementary Table S4). They were mostly involved with cellular process, molybdopterin bioprocess, transmembrane transporter, and other transports.
FIGURE 3.
Confirmation of DEGs by TaqMan qPCR. Transcript levels were quantified in up-regulated sets of genes i.e., Group I (LMRG_01669, LMRG_02283, and plcA), Group II (LMRG_01149), and Group III (LMRG_00091) and in down-regulated gene (gadT2). Wild type transcript levels are in gray and ΔsigBCHL transcript levels are in black. Transcript levels are expressed relative to WT after normalization to geometric mean of housekeeping genes, rpoB and bglA (Supplementary Table S7). Experiments were performed in biological triplicates. Asterisk indicates significance fold difference relative to wild type.
FIGURE 4.
Pearson correlation between RNA-seq and qPCR for DEGs of L. monocytogenes wild type (blue) and quadruple mutant (red). All expression data were transformed to Log2 and normalized by geometric mean of housekeeping genes (rpoB and bglA).
Among the 68 genes that were up-regulated in the BHI control (group II), we found that LMRG_01516 encoding for unknown protein showed the highest FC, as high as 52.6 (Supplementary Table S5). Other significantly up-regulated genes include an operon of ferrous iron transport genes (LMRG_00057-LMRG_00059), members of the ilv operon (i.e., ilvB and ilvC), and LMRG_01149 encoding mannose specific IIC component in a PTS system. Higher transcript levels in LMRG_01149 in BHI was validated by qPCR (Figure 3). Functional groups were enriched and significant specific GO terms associated with group II were not identified.
We identified 78 genes that were up-regulated only in bile exposure (group III; bile-stimulon) (Supplementary Table S6). LMRG_00091 encoding PTS fructose transporter subunit IIA was up-regulated 27.7 FC. Beta-glucosidase encoding gene LMRG_01934 as well as triosephosphate isomerase encoding gene tpiA2 were up-regulated up to six FC. Interestingly, several bacteriophage-associated genes such as LMRG_01524-LMRG_01525, LMRG_01536, LMRG_01540, and LMRG_01543 showed higher expression levels ranging from five to 14 FC. Virulence associated genes (clpB, clpP, groL, inlB, and dnaK) also had statistically increased their expression levels. However, GO terms associated with group III are not enriched.
Discussion
A number of studies revealed the role of sigma factors in various stress responses using bioinformatic approaches including RNA-sequencing and protein level analyses. In this study, we define the exclusive role of housekeeping σA in BHI and under gallbladder bile (pH ∼8.2) stress exposure. In addition, we defined the bile stimulon, which includes genes involved mainly in stress defense mechanisms.
Bile Mediates Up-Regulation of dlt and ilv Operons but Down-Regulated prfA and Heat Shock Genes
Approximately 2% of the total transcriptome was differentially expressed genes in L. monocytogenes 10403S wild type under bile exposure compared to BHI control. This finding showed less differentially expressed genes compared to a previous study (Guariglia-Oropeza et al., 2018) that showed approximately 16% of differentially expressed genes in L. monocytogenes 10403S exposed to bile pH 5.5. It has been shown that the toxicity of bile is pH-dependent; the lower the pH, the higher the toxicity (Begley et al., 2005b; White et al., 2015). As a result, the genes that respond to bile at lower pH would likely be different due to dual stresses. Nevertheless, the key differentially expressed genes are similar. For instance, dltC encoding D-alanyl carrier protein and LMRG_02074 encoding teichoic acid D-Ala incorporation-associated protein DltX were induced under bile exposure. This finding supports that of others in that dltABCD operon is induced under bile exposure in both L. monocytogenes 10403S (lineage II) and H7858 (lineage I) (Guariglia-Oropeza et al., 2018) and Lactobacillus rhamnosus (Koskenniemi et al., 2011). DltABCD proteins are involved in D-alanylation of teichoic acid, which facilitates resistance to antimicrobial peptides (Revilla-Guarinos et al., 2014). These Dlt proteins have been shown to contribute to resistance to various cell wall disruption stress such as that induced by nisin (Kang et al., 2015) and lysozyme (Guariglia-Oropeza and Helmann, 2011). We also observed that ilvBCD genes were highly expressed in our bile exposure condition. The ilv operon is involved in branched-chain amino acid (BCAA) isoleucine or valine biosynthesis, which is essential for intracellular survival (Chatterjee et al., 2006). Recently, BCAA synthesis of isoleucine, leucine, and valine were shown to directly and indirectly affect PrfA (Lobel et al., 2012, 2015). Particularly, isoleucine is found to be a crucial signal for L. monocytogenes intra-host gene expression (Lobel et al., 2012; Brenner et al., 2018) and low concentrations of isoleucine can enhance virulence (Brenner et al., 2018). In addition to acting as signaling molecules, isoleucine and valine have been shown to serve as osmolytes that accumulate in the cytosol of plant cells under salt stress (Parida and Das, 2005). Bacteria accumulate osmoprotective solutes to manage hyperosmotic stress (Yancey et al., 1982). L. monocytogenes has been shown earlier to accumulate glycine, alanine, and proline that increase growth rate under high osmolarity (Patchett et al., 1992). Therefore, the induction of L. monocytogenes ilv operon under bile exposure may function as a signaling molecule pathway for accumulation of osmoprotectants that increase growth rate under high osmolarity.
We also found that prfA was down-regulated under bile exposure. This finding is consistent with previous microarray analysis showing that the entire PrfA regulon including plcA, hly, mpI, actA, plcA, inlA, and inlB was repressed in response to bile (Quillin et al., 2011). However, it conflicted with previous RNA-seq data in which some PrfA regulon genes were induced under bile exposure (Guariglia-Oropeza et al., 2018). This could be explained by the pH difference of the bile used in these studies. The microarray study used neutral bile while the RNA-seq used acidic bile pH 5.5. The bile components used in this study mimics gallbladder bile which is more similar to the condition used in the microarray study. The down-regulation of virulence associated genes such as inlA and inlB might allow the bacterium to initiate penetration after successful establishment (Prouty and Gunn, 2000), therefore conserving resources until primed for infection.
In agreement with previous proteomic analysis of L. monocytogenes under bile exposure, gene expression of chaperone proteins DnaK and DnaJ were lower upon bile treatment (Wright et al., 2016). Our data revealed that the expression of chaperone genes (i.e., dnaK, groL, and groS) were down-regulated under bile exposure in L. monocytogenes 10403S wild type strain. DnaK (heat-shock protein 70 family) and DnaJ (heat-shock protein 40 family) are involved in protein folding and survival under stress conditions (Genevaux et al., 2007). GroL exists in a double heptameric ring structure, which facilitates newly synthesized proteins folding. Deletions of dnaK in Campylobacter jejuni and Salmonella enterica serovar Typhimurium lead to diminish growth in macrophages and reduced ability to colonize mice (Konkel et al., 1998; Takaya et al., 2004). Heat shock proteins are important for bacterial survival in host niches (Neckers and Tatu, 2008). However, inactivation of hspR genes (encoding heat shock protein receptor and transcription factor) in Mycobacterium tuberculosis yielded high production of DnaK resulting in significantly impaired persistence in the mouse model (Stewart et al., 2001; Das Gupta et al., 2008). Findings in other organisms suggest that increased DnaK protein concentrations could boost the immune response in early stages of infection and possibly lead to expedited clearance of the bacteria. These data suggest that overexpression of the heat shock proteins might not increase the virulence of the bacteria. Deviant production of the heat shock proteins is possibly disadvantageous to the pathogen and regulating the magnitude and the timing of the production are important. Moreover, in E. coli and, DnaK inhibits the σ32 or σH by binding to and targeting it for degradation via the FtsH, a membrane-bound protease (Tomoyasu et al., 1995; Blaszczak et al., 1999). They found that the induction of σ32 in DnaKJ depleted-cell induces broad changes in proteomes and re-organization in cellular function in which, the majority of repair and maintenance functions are up-regulated, while the proliferation and metabolic process go down. Therefore, it suggests that DnaK is not only involved in the DNA replication or misfolded protein catalytic, but also essential for regulatory function when its folding activity is dispensable (Schramm et al., 2017). Collectively, the down-regulation of dnaK, groL, and groS genes after bile exposure in our study may support avoidance of the immune system in early stage of infection as well as play a part in a regulatory complex for maintenance activity.
σA Compensates Functions of Other Alternative Sigma Factors
As the quadruple alternative sigma factor mutant had a comparable phenotype to wild type under bile exposure, we queried differentially expressed genes in the quadruple mutant under BHI and bile exposure. Surprisingly, only 29 genes were differentially expressed between the L. monocytogenes quadruple mutant exposed to bile compared to BHI control. Although it has no alternative sigma factors, the number of differentially expressed genes was limited. It is feasible that σA compensated for the alternative sigma factors, thus resulting in a small number of differentially expressed genes. One possible explanation could be the competition among sigma factors. The concept of “competition” sets in when the concentrations of sigma factors are in excess of core RNAP (Mauri and Klumpp, 2014). The competition is more complicated when the affinities of different sigma factors are varied. The stronger-binding sigma factor dislocates the lower affinity sigma factors even under concentration equilibrium of alternative sigma factors. Previous study has measured E. coli sigma factors dissociation constants and revealed that the housekeeping sigma factor σ70 is found to have the strongest-binding affinity to core RNAP (Maeda et al., 2000). In addition, the affinity of the housekeeping sigma factor binding to core RNAP can be modulated by alarmone ppGpp (guanosine tetraphosphate), which is induced in bacteria or plants by several stresses (Srivatsan and Wang, 2008). If the ppGpp can specifically modulate housekeeping sigma factors, but not other alternative sigma factors, then during stress ppGpp can enhance successful competition of alternative sigma factors over the housekeeping (Mauri and Klumpp, 2014). Therefore, in the absence of alternative sigma factors in the quadruple mutant, no competition can be observed leading to enhanced function of the housekeeping sigma factor σA.
Moreover, it has previously been shown that strong binding between promoter and RNAP could lead to relatively lower transcription initiation rates since the promoter is occupied by RNAP most of the time (Hatoum and Roberts, 2008). For example, in E. coli, σN which is structurally and mechanically different from other sigma factors (Mauri and Klumpp, 2014) together with core RNAP can occupy a promoter sequence and, upon binding to RNAP holoenzyme, the promoter remains inactive in a closed transcription initiation complex. The complex becomes active upon binding of the ATPase activator, which typically occupy a site at a distance from the promoter and contacts the holoenzyme via DNA looping (Friedman and Gelles, 2012). In L. monocytogenes, σL is classified as a member of RpoN (σ54), which is structurally similar to σN in E. coli. Thus, existence of alternative sigma factor such as σL could passively prevent transcription that would otherwise be initiated by other sigma factors resulting in a temporary stop in transcription. In addition, the cross-talk among alternative sigma factors take place in various regulatory networks. The quadruple mutant with no alternative sigma factors would not have the insulation effect, therefore, it drives transcription more conveniently.
σA-Dependent Genes
Most of the σA-dependent genes are considered as housekeeping genes that are important for the growth of the bacteria. It is equally important to better understanding the role of σA during stress exposures. Here, we report the list of σA-dependent genes that were higher expressed in both BHI and bile conditions to provide the information of σA-dependent and housekeeping genes for further analyses. The functional categories of σA-dependent genes in this study using gene set enrichment analysis (GOseq), were found that these genes are involved in many crucial biological processes including carbohydrate metabolic process, pentose phosphate shunt, NADP metabolic process, transport, cell communication, signal transduction and regulation, which are important for cell growth and survival.
The bglA gene encoding beta-glucosidase is used as a reference housekeeping gene in several studies (Tasara and Stephan, 2007; Kwong et al., 2016; Hilliard et al., 2018) and is shown in our list. The level of bglA was constantly expressed in both conditions. In addition to bglA, the entire five genes in a σA-dependent operon LMRG_01669-LMRG_01673 are shown in our list as constantly expressed at high levels in both conditions. This operon is involved in myo-inositol degradation. In the Gram-negative bacterium, Legionella pneumophila, myo-inositol promotes its growth and virulence for infection of amoeba and macrophage (Manske et al., 2016). Our qPCR confirmed that the expression level of the representative gene in this operon, LMRG_01669 was constantly expressed in both conditions. Therefore, this gene can be used as a σA-dependent gene in further study.
σA-Dependent Bile Inducible Genes
Since acidic bile (pH 5.5) does not induce the σB regulon, we further focused on the housekeeping sigma factor σA regulation upon basic bile exposure (pH 8.2). Consistent with previous bile stimulon findings, we confirmed that LMRG_01119 encoding a PTS system enzyme in the LMRG_01117 – LMRG_01121 operon is a bile responsive gene. Here, we reported that this lineage II specific gene is σA-dependent and induced by bile. This operon is suggested to facilitate bacterial survival during gastrointestinal stages of infection (Guariglia-Oropeza et al., 2018). Besides LMRG_01119, LMRG_00091 encoding for PTS fructose transporter subunit is σA-dependent bile inducible gene with high FC ∼ 27. In contrast, the PTS associated proteins in either glucose or mannose systems from previous proteomic study have decreased upon exposure to bile (Wright et al., 2016). To better understand the link between them, further investigation is warranted.
The up-regulation of clpP, a gene involved in degradation of misfolded proteins and required for growth subsequent to stress exposure, was consistent with previous reports demonstrating the level of ClpP protein increased in L. monocytogenes HCC23 strain under bile exposure in aerobic condition (Gaillot et al., 2000; Wright et al., 2016). This data suggested that clpP is induced in response to damaged proteins; bile contributes to conformational changes of proteins resulting in misfolding and denaturation (Begley et al., 2005a). Importantly, the induction of inlB, encoding the surface protein of L. monocytogenes that partially mediates entry into host cell by binding to the host receptor Met (Bierne and Cossart, 2002), was observed. It can implicate that bile triggers expression of bacterial genes involved in host cell invasion.
In contrast to differentially expressed genes in L. monocytogenes wild type, we found that the chaperone genes, dnaK and groL were σA-dependent and induced under bile exposure. Due to the lesser complicated regulatory network in quadruple mutant, regulated by σA, the negative effects of other alternative sigma factors such as insulation effects on promoter sequences were removed; therefore, the up-regulation of these chaperone genes was noticed. It could suggest that σA drives the expression of dnaK and groL in response to bile, hence, we conclude that these genes are σA-dependent bile inducible genes.
Conclusion
In this study, we used RNA-seq to define transcriptomes of L. monocytogenes 10403S wild type and its isogenic quadruple mutant (ΔsigBCHL) in BHI and under 1% gallbladder bile (pH 8.2) exposure. This study underscores that σA is a housekeeping sigma factor that can compensate for the absence of all other sigma factors. Our data suggest σA is sufficient to survive in gallbladder bile. The σA-dependent bile inducible genes are represented by PTS systems, chaperones, and transporters, which contribute to cellular homeostasis of the bacteria. We proposed a compensatory role of σA in the absence of alternative sigma factors.
Data Availability
The datasets generated or analyzed for this study can be found in the sequence read archive (SRA): https://www.ncbi.nlm.nih.gov/sra/PRJNA544468.
Author Contributions
HO and SC conceived the study. AB performed the experiments and analyzed the data. All authors drafted and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the Department of Statistics at Purdue University for statistical advice.
Footnotes
Funding. AB was supported by the Science Achievement Scholarship of Thailand (SAST), Ministry of Education (Thailand). The study was supported by the Office of the Higher Education Commission (Thailand) for Talent Mobility Program (to SC).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02070/full#supplementary-material
References
- Anders S., Pyl P. T., Huber W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley M., Gahan C. G., Hill C. (2005a). The interaction between bacteria and bile. FEMS Microbiol. Rev. 29 625–651. 10.1016/j.femsre.2004.09.003 [DOI] [PubMed] [Google Scholar]
- Begley M., Sleator R. D., Gahan C. G., Hill C. (2005b). Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect. Immun. 73 894–904. 10.1128/IAI.73.2.894-904.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H., Cossart P. (2002). InlB, a surface protein of Listeria monocytogenes that behaves as an invasin and a growth factor. J. Cell Sci. 115 3357–3367. [DOI] [PubMed] [Google Scholar]
- Blaszczak A., Georgopoulos C., Liberek K. (1999). On the mechanism of FtsH-dependent degradation of the sigma 32 transcriptional regulator of Escherichia coli and the role of the Dnak chaperone machine. Mol. Microbiol. 31 157–166. 10.1046/j.1365-2958.1999.01155.x [DOI] [PubMed] [Google Scholar]
- Brenner M., Lobel L., Borovok I., Sigal N., Herskovits A. A. (2018). Controlled branched-chain amino acids auxotrophy in Listeria monocytogenes allows isoleucine to serve as a host signal and virulence effector. PLoS. Genet. 14:e1007283. 10.1371/journal.pgen.1007283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess R. R. (2001). “Sigma Factors,” in Encyclopedia of Genetics, eds Brenner S., Miller J. H. (Cambridge, MA: Academic Press; ), 1831–1834. 10.1006/rwgn.2001.1192 [DOI] [Google Scholar]
- Chatterjee S. S., Hossain H., Otten S., Kuenne C., Kuchmina K., Machata S., et al. (2006). Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74 1323–1338. 10.1128/IAI.74.2.1323-1338.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturongakul S., Raengpradub S., Palmer M. E., Bergholz T. M., Orsi R. H., Hu Y., et al. (2011). Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors sigmaB, sigmaC, sigmaH, and sigmaL in Listeria monocytogenes. Appl. Environ. Microbiol. 77 187–200. 10.1128/AEM.00952-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Gupta T., Bandyopadhyay B., Das Gupta S. K. (2008). Modulation of DNA-binding activity of Mycobacterium tuberculosis HspR by chaperones. Microbiol 154 484–490. 10.1099/mic.0.2007/012294-0 [DOI] [PubMed] [Google Scholar]
- de Noordhout C. M., Devleesschauwer B., Angulo F. J., Verbeke G., Haagsma J., Kirk M., et al. (2014). The global burden of Listeriosis: a systematic review and meta-analysis. Lancet. Infect. Dis. 14 1073–1082. 10.1016/S1473-3099(14)70870-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussurget O., Cabanes D., Dehoux P., Lecuit M., Buchrieser C., Glaser P., et al. (2002). Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of Listeriosis. Mol. Microbiol. 45 1095–1106. 10.1046/j.1365-2958.2002.03080.x [DOI] [PubMed] [Google Scholar]
- EFSA (2015). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J. 13:3991 10.2903/j.efsa.2015.3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L. J., Gelles J. (2012). Mechanism of transcription initiation at an activator-dependent promoter defined by single-molecule observation. Cell 148 679–689. 10.1016/j.cell.2012.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillot O., Pellegrini E., Bregenholt S., Nair S., Berche P. (2000). The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35 1286–1294. 10.1046/j.1365-2958.2000.01773.x [DOI] [PubMed] [Google Scholar]
- Genevaux P., Georgopoulos C., Kelley W. L. (2007). The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol. Microbiol. 66 840–857. 10.1111/j.1365-2958.2007.05961.x [DOI] [PubMed] [Google Scholar]
- Goulet V., King L. A., Vaillant V., de Valk H. (2013). What is the incubation period for Listeriosis? BMC. Infect. Dis. 13:11. 10.1186/1471-2334-13-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorova I. L., Phleger N. J., Mutalik V. K., Gross C. A. (2006). Insights into transcriptional regulation and sigma competition from an equilibrium model of RNA polymerase binding to DNA. Proc. Natl. Acad. Sci. U.S.A. 103 5332–5337. 10.1073/pnas.0600828103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber T. M., Markov D., Sharp M. M., Young B. A., Lu C. Z., Zhong H. J., et al. (2001). Binding of the initiation factor sigma (70) to core RNA polymerase is a multistep process. Mol. Cell. 8 21–31. 10.1016/S1097-2765(01)00292-1 [DOI] [PubMed] [Google Scholar]
- Guariglia-Oropeza V., Helmann J. D. (2011). Bacillus subtilis sigma (V) confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and D-alanylation of teichoic acids. J. Bacteriol. 193 6223–6232. 10.1128/JB.06023-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guariglia-Oropeza V., Orsi R. H., Guldimann C., Wiedmann M., Boor K. J. (2018). The Listeria monocytogenes bile stimulon under acidic conditions is characterized by strain-specific patterns and the upregulation of motility, cell wall modification functions, and the prfa regulon. Front. Microbiol. 9:120. 10.3389/fmicb.2018.00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J., Francis K. P., DeBoer M., Chu P., Gibbs K., Contag C. H. (2004). Extracellular replication of Listeria monocytogenes in the murine gall bladder. Science 303 851–853. 10.1126/science.1092712 [DOI] [PubMed] [Google Scholar]
- Hatoum A., Roberts J. (2008). Prevalence of RNA polymerase stalling at Escherichia coli promoters after open complex formation. Mol. Microbiol. 68 17–28. 10.1111/j.1365-2958.2008.06138.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard A., Leong D., O’Callaghan A., Culligan E. P., Morgan C. A., DeLappe N., et al. (2018). Genomic characterization of Listeria monocytogenes isolates associated with clinical Listeriosis and the food production environment in Ireland. Genes 9:171. 10.3390/genes9030171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson J. A., Mott S. J., Penney N. (1994). Growth of Listeria monocytogenes, Aeromonas hydrophila, and Yersinia enterocolitica on vacuum and saturated carbon dioxide controlled atmosphere-packaged sliced roast beef. J. Food. Prot. 57 204–208. 10.4315/0362-028X-57.3.204 [DOI] [PubMed] [Google Scholar]
- Jishage M., Iwata A., Ueda S., Ishihama A. (1996). Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 178 5447–5451. 10.1128/jb.178.18.5447-5451.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M., Kvint K., Shingler V., Nystrom T. (2002). Regulation of sigma factor competition by the alarmone ppGpp. Genes. Dev. 16 1260–1270. 10.1101/gad.227902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Wiedmann M., Boor K. J., Bergholz T. M. (2015). VirR-mediated resistance of Listeria monocytogenes against food antimicrobials and cross-protection induced by exposure to organic acid salts. Appl. Environ. Microbiol. 81 4553–4562. 10.1128/AEM.00648-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak M. J., Wiedmann M., Boor K. J. (2006). Contributions of Listeria monocytogenes sigmaB and PrfA to expression of virulence and stress response genes during extra- and intracellular growth. Microbiology 152 1827–1838. 10.1099/mic.0.28758-0 [DOI] [PubMed] [Google Scholar]
- Kingston A. W., Subramanian C., Rock C. O., Helmann J. D. (2011). A sigmaW-dependent stress response in Bacillus subtilis that reduces membrane fluidity. Mol. Microbiol. 81 69–79. 10.1111/j.1365-2958.2011.07679.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel M. E., Kim B. J., Klena J. D., Young C. R., Ziprin R. (1998). Characterization of the thermal stress response of Campylobacter jejuni. Infect. Immun. 66 3666–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskenniemi K., Laakso K., Koponen J., Kankainen M., Greco D., Auvinen P., et al. (2011). Proteomics and transcriptomics characterization of bile stress response in probiotic Lactobacillus rhamnosus GG. Mol. Cell. Proteomics 10:M110002741. 10.1074/mcp.M110.002741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong J. C., Mercoulia K., Tomita T., Easton M., Li H. Y., Bulach D. M., et al. (2016). Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J. Clin. Microbiol. 54 333–342. 10.1128/JCM.02344-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Orsi R. H., Boor K. J., Wiedmann M., Guariglia-Oropeza V. (2017). Home alone: elimination of all but one alternative sigma factor in Listeria monocytogenes allows prediction of new roles for sigma (B). Front. Microbiol. 8:1910. 10.3389/fmicb.2017.01910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel L., Sigal N., Borovok I., Belitsky B. R., Sonenshein A. L., Herskovits A. A. (2015). The metabolic regulator CodY links Listeria monocytogenes metabolism to virulence by directly activating the virulence regulatory gene prfA. Mol. Microbiol. 95 624–644. 10.1111/mmi.12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel L., Sigal N., Borovok I., Ruppin E., Herskovits A. A. (2012). Integrative genomic analysis identifies isoleucine and CodY as regulators of Listeria monocytogenes virulence. PLoS Genet. 8:e1002887. 10.1371/journal.pgen.1002887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R., Stragier P. (1992). Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature 355 601–604. 10.1038/355601a0 [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H., Fujita N., Ishihama A. (2000). Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 28 3497–3503. 10.1093/nar/28.18.3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manske C., Schell U., Hilbi H. (2016). Metabolism of myo-Inositol by Legionella pneumophila promotes infection of amoebae and macrophages. Appl. Environ. Microbiol. 82 5000–5014. 10.1128/AEM.01018-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri M., Klumpp S. (2014). A model for sigma factor competition in bacterial cells. PLoS Comput. Biol. 10:e1003845. 10.1371/journal.pcbi.1003845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano Romero V., Morikawa K. (2016). Listeria monocytogenes sigmaH contributes to expression of competence genes and intracellular growth. J. Bacteriol. 198 1207–1217. 10.1128/JB.00718-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Membre J. M., Leporq B., Vialette M., Mettler E., Perrier L., Thuault D., et al. (2005). Temperature effect on bacterial growth rate: quantitative microbiology approach including cardinal values and variability estimates to perform growth simulations on/in food. Int. J. Food Microbiol. 100 179–186. 10.1016/j.ijfoodmicro.2004.10.015 [DOI] [PubMed] [Google Scholar]
- Mujahid S., Orsi R. H., Boor K. J., Wiedmann M. (2013). Protein level identification of the Listeria monocytogenes sigma H, sigma L, and sigma C regulons. BMC Microbiol. 13:156. 10.1186/1471-2180-13-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckers L., Tatu U. (2008). Molecular chaperones in pathogen virulence: emerging new targets for therapy. Cell Host Microbe 4 519–527. 10.1016/j.chom.2008.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olier M., Rousseaux S., Piveteau P., Lemaitre J. P., Rousset A., Guzzo J. (2004). Screening of glutamate decarboxylase activity and bile salt resistance of human asymptomatic carriage, clinical, food, and environmental isolates of Listeria monocytogenes. Int. J. Food Microbiol. 93 87–99. 10.1016/j.ijfoodmicro.2003.10.010 [DOI] [PubMed] [Google Scholar]
- Parida A. K., Das A. B. (2005). Salt tolerance and salinity effects on plants: a review. Ecotoxicol Environ. Saf. 60 324–349. 10.1016/j.ecoenv.2004.06.010 [DOI] [PubMed] [Google Scholar]
- Patchett R. A., Kelly A. F., Kroll R. G. (1992). Effect of sodium chloride on the intracellular solute pools of Listeria monocytogenes. Appl. Environ. Microbiol. 58 3959–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper S. E., Mitchell J. E., Lee D. J., Busby S. J. (2009). A global view of Escherichia coli Rsd protein and its interactions. Mol. Biosyst. 5 1943–1947. 10.1039/B904955j [DOI] [PubMed] [Google Scholar]
- Pleitner A. M., Trinetta V., Morgan M. T., Linton R. L., Oliver H. F. (2014). Transcriptional and phenotypic responses of Listeria monocytogenes to chlorine dioxide. Appl. Environ. Microbiol. 80 2951–2963. 10.1128/AEM.00004-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouty A. M., Gunn J. S. (2000). Salmonella enterica serovar typhimurium invasion is repressed in the presence of bile. Infect. Immun. 68 6763–6769. 10.1128/IAI.68.12.6763-6769.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillin S. J., Schwartz K. T., Leber J. H. (2011). The novel Listeria monocytogenes bile sensor BrtA controls expression of the cholic acid efflux pump MdrT. Mol. Microbiol. 81 129–142. 10.1111/j.1365-2958.2011.07683.x [DOI] [PubMed] [Google Scholar]
- Revilla-Guarinos A., Gebhard S., Mascher T., Zuniga M. (2014). Defence against antimicrobial peptides: different strategies in Firmicutes. Environ. Microbiol. 16 1225–1237. 10.1111/1462-2920.12400 [DOI] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R. M., Angulo F. J., Tauxe R. V., Widdowson M. A., Roy S. L., et al. (2011). Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 17 7–15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm F. D., Heinrich K., Thuring M., Bernhardt J., Jonas K. (2017). An essential regulatory function of the DnaK chaperone dictates the decision between proliferation and maintenance in Caulobacter crescentus. PLoS Genet. 13:e1007148. 10.1371/journal.pgen.1007148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann W. (2003). The Bacillus subtilis heat shock stimulon. Cell Stress Chaperones 8 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U. K., Chatterji D. (2010). Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity. FEMS Microbiol. Rev. 34 646–657. 10.1111/j.1574-6976.2010.00223.x [DOI] [PubMed] [Google Scholar]
- Sleator R. D., Wemekamp-Kamphuis H. H., Gahan C. G., Abee T., Hill C. (2005). A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol. Microbiol. 55 1183–1195. 10.1111/j.1365-2958.2004.04454.x [DOI] [PubMed] [Google Scholar]
- Southwick F. S., Purich D. L. (1996). Intracellular pathogenesis of Listeriosis. N. Engl. J. Med. 334 770–776. 10.1056/NEJM199603213341206 [DOI] [PubMed] [Google Scholar]
- Srivatsan A., Wang J. D. (2008). Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 11 100–105. 10.1016/j.mib.2008.02.001 [DOI] [PubMed] [Google Scholar]
- Stewart G. R., Snewin V. A., Walzl G., Hussell T., Tormay P., O’Gaora P., et al. (2001). Overexpression of heat-shock proteins reduces survival of Mycobacterium tuberculosis in the chronic phase of infection. Nat. Med. 7 732–737. 10.1038/89113 [DOI] [PubMed] [Google Scholar]
- Sue D., Fink D., Wiedmann M., Boor K. J. (2004). sigmaB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiol 150 3843–3855. 10.1099/mic.0.27257-0 [DOI] [PubMed] [Google Scholar]
- Takaya A., Tomoyasu T., Matsui H., Yamamoto T. (2004). The DnaK/DnaJ chaperone machinery of Salmonella enterica serovar Typhimurium is essential for invasion of epithelial cells and survival within macrophages, leading to systemic infection. Infect. Immun. 72 1364–1373. 10.1128/IAI.72.3.1364-1373.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasara T., Stephan R. (2007). Evaluation of housekeeping genes in Listeria monocytogenes as potential internal control references for normalizing mRNA expression levels in stress adaptation models using real-time PCR. FEMS Microbiol. Lett. 269 265–272. 10.1111/j.1574-6968.2007.00633.x [DOI] [PubMed] [Google Scholar]
- Tomoyasu T., Gamer J., Bukau B., Kanemori M., Mori H., Rutman A. J., et al. (1995). Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor sigma 32. EMBO J. 14 2551–2560. 10.1002/j.1460-2075.1995.tb07253.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versantvoort C. H., Oomen A. G., Van de Kamp E., Rompelberg C. J., Sips A. J. (2005). Applicability of an in vitro digestion model in assessing the bioaccessibility of mycotoxins from food. Food Chem. Toxicol. 43 31–40. 10.1016/j.fct.2004.08.007 [DOI] [PubMed] [Google Scholar]
- Wade J. T., Castro Roa D., Grainger D. C., Hurd D., Busby S. J., Struhl K., et al. (2006). Extensive functional overlap between sigma factors in Escherichia coli. Nat. Struct. Mol. Biol. 13 806–814. 10.1038/nsmb1130 [DOI] [PubMed] [Google Scholar]
- Wang S., Orsi R. H., Tang S., Zhang W., Wiedmann M., Boor K. J. (2014). Phosphotransferase system-dependent extracellular growth of Listeria monocytogenes is regulated by alternative sigma factors sigmaL and sigmaH. Appl. Environ. Microbiol. 80 7673–7682. 10.1128/AEM.02530-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S. J., McClung D. M., Wilson J. G., Roberts B. N., Donaldson J. R. (2015). Influence of pH on bile sensitivity amongst various strains of Listeria monocytogenes under aerobic and anaerobic conditions. J. Med. Microbiol. 64 1287–1296. 10.1099/jmm.0.000160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright M. L., Pendarvis K., Nanduri B., Edelmann M. J., Jenkins H. N., Reddy J. S., et al. (2016). The effect of oxygen on bile resistance in Listeria monocytogenes. J. Proteomics Bioinformatics 9 107–119. 10.4172/jpb.1000396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey P. H., Clark M. E., Hand S. C., Bowlus R. D., Somero G. N. (1982). Living with water stress: evolution of osmolyte systems. Science 217 1214–1222. 10.1126/science.7112124 [DOI] [PubMed] [Google Scholar]
- Young M. D., Wakefield M. J., Smyth G. K., Oshlack A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated or analyzed for this study can be found in the sequence read archive (SRA): https://www.ncbi.nlm.nih.gov/sra/PRJNA544468.