Abstract
Supplementation with NAD precursors such as nicotinamide riboside (NR) has been shown to enhance mitochondrial function in the liver and to prevent hepatic lipid accumulation in high-fat diet (HFD)-fed rodents. Hepatocyte-specific knockout of the NAD+-synthesizing enzyme nicotinamide phosphoribosyltransferase (NAMPT) reduces liver NAD+ levels, but the metabolic phenotype of Nampt-deficient hepatocytes in mice is unknown. Here, we assessed Nampt's role in maintaining mitochondrial and metabolic functions in the mouse liver. Using the Cre-LoxP system, we generated hepatocyte-specific Nampt knockout (HNKO) mice, having a 50% reduction of liver NAD+ levels. We screened the HNKO mice for signs of metabolic dysfunction following 60% HFD feeding for 20 weeks ± NR supplementation and found that NR increases hepatic NAD+ levels without affecting fat mass or glucose tolerance in HNKO or WT animals. High-resolution respirometry revealed that NR supplementation of the HNKO mice did not increase state III respiration, which was observed in WT mice following NR supplementation. Mitochondrial oxygen consumption and fatty-acid oxidation were unaltered in primary HNKO hepatocytes. Mitochondria isolated from whole-HNKO livers had only a 20% reduction in NAD+, suggesting that the mitochondrial NAD+ pool is less affected by HNKO than the whole-tissue pool. When stimulated with tryptophan in the presence of [15N]glutamine, HNKO hepatocytes had a higher [15N]NAD+ enrichment than WT hepatocytes, indicating that HNKO mice compensate through de novo NAD+ synthesis. We conclude that NAMPT-deficient hepatocytes can maintain substantial NAD+ levels and that the Nampt knockout has only minor consequences for mitochondrial function in the mouse liver.
Keywords: NAMPT, nicotinamide adenine dinucleotide (NAD), mitochondrial metabolism, hepatocyte, NAD biosynthesis, fatty-acid metabolism, respiration, tryptophan metabolism
Introduction
Mitochondrial dysfunction is thought to contribute to the development of nonalcoholic fatty liver disease (NAFLD)5 and nonalcoholic steatohepatitis (NASH) (1, 2). Lipid accumulation increases mitochondrial fatty-acid oxidation capacity, but impaired capacity of the mitochondrial respiratory chain complexes results in increased reactive oxygen species formation, which may contribute to the liver damage observed in NASH (3, 4). Hence, improving mitochondrial function in patients with NAFLD and NASH represents a potential treatment strategy.
In several animal models of NAFLD, hepatic levels of NAD+ are reduced (5–10). NAD+ precursor supplementation in a rodent model for NAFLD increases the abundance of enzymes regulating β-oxidation and mitochondrial respiratory complexes (7). Moreover, this treatment improves mitochondrial function and decreases hepatic lipid accumulation (7). NAD+ precursor supplementation has also been shown to efficiently improve mitochondrial function in multiple organs and cell types (9, 11–15), and it has been used to treat rodent models of type 2 diabetes (16, 17). In mitochondria, NAD+ is central for ATP synthesis through oxidative phosphorylation, where NADH serves as an electron donor (18). NAD+ is also used as a precursor for the synthesis of NADP+, which is an important cofactor for biosynthesis and antioxidative defenses (19). However, beneficial effects of NAD+ precursor supplementation may be mediated by increased activity of NAD+-dependent sirtuins (20). Sirtuins (SIRT1–7) comprise a family of deacylase enzymes, several of which regulate mitochondrial biogenesis and function (21).
Sirtuins break down NAD+ to nicotinamide (NAM) and O-acetyl ADP-ribose. As NAM inhibits sirtuin activity, the NAD+ generated from recycled NAM is important for maintaining sirtuin activity (22). NAM and phosphoribosyl pyrophosphate are turned into nicotinamide mononucleotide (NMN) by nicotinamide phosphoribosyltransferase (NAMPT) (23), and nicotinamide mononucleotide adenylyltransferases (NMNATs) further convert NMN to NAD+ using ATP as a substrate. Supplementation with NMN reduces the adverse effects of obesity, diabetes, and aging in rodents (9, 17, 24). In the liver, NAD+ can also be generated de novo from tryptophan or from nicotinic acid (NA) through the Preiss-Handler pathway (25). Synthesis of NAD+ from tryptophan is a multistep process requiring eight separate reactions (26). Synthesis from NA begins with the conversion of NA to nicotinic acid mononucleotide (NAMN) by the action of NA phosphoribosyltransferase (27). NAMN is then further converted to NA adenine dinucleotide and finally to NAD+ by the action of NMNAT and NAD synthase, respectively (27). These final two steps are also required for NAD+ synthesis from tryptophan (26). As Nadsyn1 expression is restricted to liver, kidney, and small intestine, NAD+ synthesis from NA and tryptophan is limited to these tissues (28). Additionally, NAD+ can be synthesized from nicotinamide riboside (NR), which is converted to NMN by the nicotinamide riboside kinases (NRK) (29). NR has been widely applied in NAD+ supplementation studies in both rodents (7, 8, 12, 15, 16, 30) and humans (31–35). NMN and NR can both be delivered orally to increase NAD+ levels in peripheral tissues (7, 8, 12, 24). Interestingly, NMN appears to be converted to NR for cellular uptake, indicating that NR is the “transport form” of these NAD+ precursors (29).
It is not known whether decreased liver NAD+ content observed in rodent models for NAFLD is due to increased activity of NAD+-consuming enzymes or is due to decreased NAD+ biosynthesis. Furthermore, it is not known whether low NAD+ levels are a consequence of hepatic lipid accumulation and mitochondrial dysfunction or whether low NAD+ levels precede steatosis development. The NAMPT inhibitor FK866 aggravates the development of steatosis in high-fat diet–fed mice (36), and FK866 increases the susceptibility of human cancer cell lines to oxidative stress (37). Furthermore, hepatocyte-specific knockout of Nampt decreases hepatic NAD+ content, impairs fatty-acid oxidation, and decreases mitochondrial oxygen consumption (38). Enzymatically-inactive NAMPT transgenic mice are also more susceptible toward NAFLD development, and inducible hepatocyte-specific knockdown of Nampt impairs liver regeneration (8, 39). Previously, low hepatic NAMPT levels were observed in obese animals (5, 8, 10, 17). This suggests that impaired NAD+ salvage may be an important factor for disease development. However, we and others have observed increased NAD+ and NAMPT levels in livers of high-fat diet (HFD)-fed mice (40) or little or no change in hepatic NAD+ content following long-term HFD feeding (41–43). This suggests that hepatic lipid accumulation per se may not cause a shortage of NAD+ or impair NAD+ salvage capacity. Hence, further investigation into the relationship between mitochondrial function, hepatic lipid accumulation, and NAD+ salvage capacity is warranted.
As the metabolic phenotype of hepatocyte-specific Nampt knockout mice has not been thoroughly investigated (38), we hypothesized that disruption of salvage capacity would impact mitochondrial function and increase the susceptibility of the liver to lipid accumulation following a HFD. To this end, we generated stable Nampt knockdown Hepa1c1c7 cells and hepatocyte-specific Nampt knockout (HNKO) mice with the aim to determine the role of Nampt for maintaining mitochondrial function in liver cells under normal and excess-energy states. Our data suggest that the liver is capable of maintaining part of the NAD+ pool in the absence of NAMPT and that deletion of Nampt per se has minor effects on hepatic mitochondrial function.
Results
Nampt knockdown in Hepa1c1c7 cells reduces NAD+ levels without affecting mitochondrial respiratory function
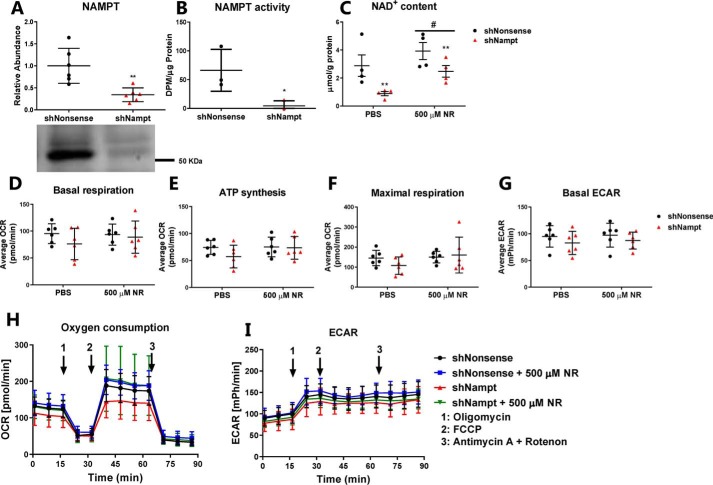
As embryonic deletion of Nampt from hepatocytes reduces hepatic NAD+ content and mitochondrial oxygen consumption rate (38), we investigated whether this could be replicated in a hepatocarcinoma cell line. Stable Nampt knockdown (shNampt) in Hepa1c1c7 cells reduced NAMPT protein abundance (Fig. 1A, p < 0.01) and ablated NAMPT activity (Fig. 1B, p < 0.05). Knockdown of Nampt reduced NAD+ levels by 60% (Fig. 1C, p < 0.01), confirming that salvage of NAM contributes to the maintenance of NAD+ levels in hepatocarcinoma cells. NR treatment increased NAD+ levels in both control and shNampt cells (p < 0.01), but NAD+ levels were still lower in shNampt cells after NR treatment. To determine whether reduced cellular NAD+ content affected mitochondrial oxygen consumption, we performed extracellular flux analysis on control and shNampt cells ± supplementation with NR. Neither knockdown of Nampt nor NR treatment affected basal respiration, oxygen consumption for ATP synthesis, maximal respiratory capacity, or the glycolytic capacity (basal extracellular acidification rate, ECAR) (Fig. 1, D–I). Hence, NAMPT contributes to maintaining NAD+ levels in Hepa1c1c7 cells but appears dispensable for maintaining mitochondrial respiratory function.
Figure 1.
NAMPT is important for maintaining NAD+ levels in Hepa1c1c7 cells but is not essential for mitochondrial function. A, NAMPT abundance (n = 6); B, NAMPT activity (n = 3); and C, NAD+ levels (n = 4) in Hepa1c1c7 cells, expressing either a shNonsense or shNampt construct. D–I, mitochondrial oxygen consumption and extracellular acidification rate as measured by extracellular flux analysis (n = 6). n indicates number of experiments. */** indicate effects of genotype, p < 0.05/0.01, respectively. # indicates effects of NR treatment, p < 0.05.
Hepatocyte-specific knockout of Nampt does not affect body composition or glucose tolerance
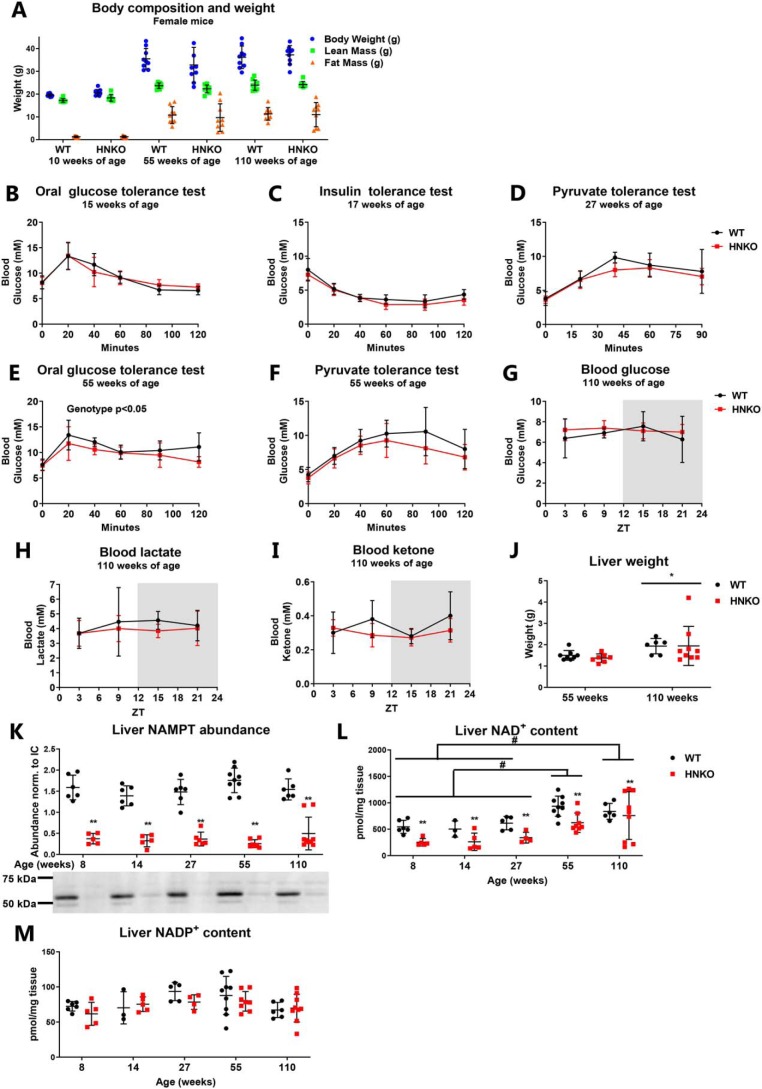
Although mitochondrial respiratory capacity was maintained in Hepa1c1c7 cells, previous reports observed impaired mitochondrial function in hepatocytes from hepatocyte-specific Nampt KO mice (38). We next investigated whether this defect in mitochondrial function would make HNKO mice prone to develop obesity and glucose intolerance. Because male mice often fight when group-housed, and because prolonged single-housing is also associated with stress and depression (44), the metabolic characterization of HNKO mice was performed in female mice. HNKO mice did not become obese with age and had a comparable body composition with WT animals at 10, 55, and 110 weeks of age (Fig. 2A). In young adult female mice (15–27 weeks of age), no significant changes in glucose, insulin, or pyruvate tolerance were detected (Fig. 2, B–D), indicating that glucose homeostasis was maintained. The 55-week-old HNKO mice had a slight shift in glucose tolerance (Fig. 2E, main effect of genotype, p < 0.05), but pyruvate tolerance remained unaltered in these mice (Fig. 2F). As the 110-week-old mice were too frail for tolerance tests, we measured circadian oscillations in blood glucose, lactate, and ketone levels. No differences were observed between WT and HNKO mice (Fig. 2, G–I). Liver weight increased independently of genotype with age (Fig. 2J, p < 0.05). Protein abundance of NAMPT was equally reduced in HNKO mice of all ages (Fig. 2K), and at most time points, hepatic NAD+ levels were ∼50% lower in HNKO mice (Fig. 2L, p < 0.01). Hepatic NAD+ levels increased with age regardless of genotype (p < 0.05), which could indicate an increased reliance on NAD+ synthesis from tryptophan or NA. NADP+ levels were not significantly affected at any of the time points (Fig. 2M). Collectively, these data confirm the importance of NAMPT for maintaining hepatic NAD+ levels in vivo. However, the lack of Nampt does not predispose HNKO mice to the development of obesity, and glucose homeostasis is similarly affected with age between genotypes.
Figure 2.
HNKO causes no whole-body phenotype. A, body composition of female mice at 10, 55, and 110 weeks of age, n = 9. B, oral glucose tolerance. C, insulin tolerance. D, pyruvate tolerance tests performed on HNKO and WT mice of 15, 17, and 27 weeks of age. E, oral glucose tolerance, and F, pyruvate tolerance tests performed on 55-week-old HNKO mice and WT littermates, n = 8–9. Circadian oscillations of blood glucose (G), lactate (H), and ketone bodies (I) in 110-week-old HNKO and WT mice (n = 6–9) are shown. J, liver weight from HNKO and WT mice at 55 and 110 weeks of age, n = 7–9. K, hepatic NAMPT abundance. L, NAD+ content. M, NADP+ content in HNKO and WT mice from 8 to 110 weeks of age, n = 3–9. n indicates number of mice per genotype. */** indicate effects of genotype, p < 0.05/0.01, respectively; #, indicates effect of time, p < 0.05.
HFD-induced fat mass accumulation and glucose intolerance develop independently of hepatic Nampt expression
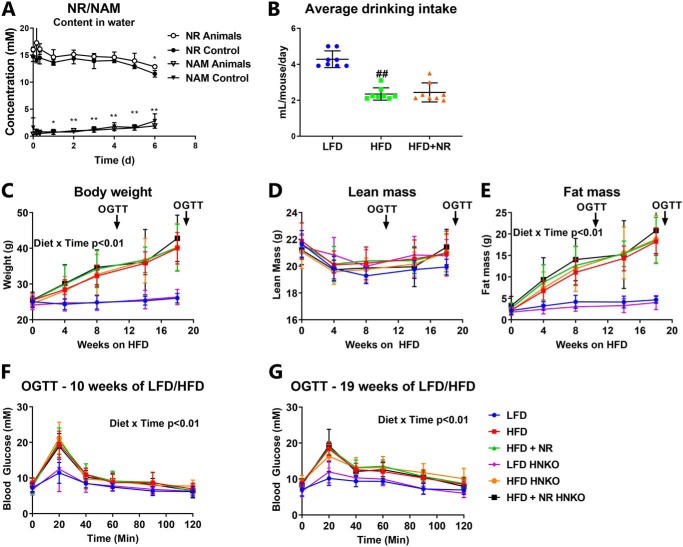
As mice with enzymatically inactive NAMPT in the liver were found to develop hepatic steatosis (8), we challenged HNKO mice with a HFD. Furthermore, we included a group of mice receiving NR in the drinking water. To determine whether NR was stable in solution, we measured NR stability over 1 week. NAM content in the drinking water increased significantly over time (p < 0.01, Fig. 3A), but a significant decrease in NR levels was only detected after 6 days. Even then, more than 75% of the NR remained in the solution (p < 0.01, Fig. 3A). NR stability was not affected by placing the water bottle in a cage with mice, demonstrating that potential bacterial contamination did not accelerate breakdown. Thus, NR appears stable in drinking water. Female WT and HNKO mice were fed a HFD (60% of total energy content from fat) or a matched low-fat control diet (LFD, 10% of total energy content from fat) for 20 weeks. The presence of NR in the drinking water did not affect mouse water intake (Fig. 3B). Body weight increased rapidly in mice fed a HFD (Fig. 3C, time × diet interaction p < 0.01), while body weight was unaltered in LFD-fed mice over time (Fig. 3C). Lean mass was not significantly altered between groups throughout the study (Fig. 3D). Fat mass accumulation increased significantly throughout the study in the HFD groups (Fig. 3E, time × diet interaction p < 0.01), and HNKO mice accumulated fat mass to the same degree as WT mice. Glucose tolerance was impaired regardless of genotype after both 10 and 19 weeks of HFD feeding (Fig. 3, F and G, time × diet interaction p < 0.01). No changes in fat mass accumulation or glucose tolerance were detected in NR-supplemented mice regardless of genotype. Hence, neither Nampt knockout nor NR supplementation affected HFD-induced body weight gain or fat mass accumulation.
Figure 3.
HFD-induced obesity in HNKO mice is indistinguishable from WT littermates. A, NR stability and NAM formation in drinking water from cages with or without mice (n = 2–4). Female HNKO mice at ∼15 weeks of age were fed a LFD or a 60% HFD ± 400 μg of NR/g of body weight/day for 20 weeks. B, average drinking intake per mouse per day after 9 weeks of HFD feeding and NR supplementation, measured over 7 days. C–E, throughout the experiment, mice were weighed and NMR scanned to assess body composition. F and G, glucose tolerance was evaluated after 10 and 19 weeks of HFD-feeding. n = 6–8 per group. */** indicate effects of time, p < 0.05/0.01, respectively. #/## indicate effects of diet, p < 0.05/0.01, respectively.
Liver mitochondrial function is unaffected following 20 weeks of HFD, but NR supplementation increases state III respiration in a genotype-specific manner
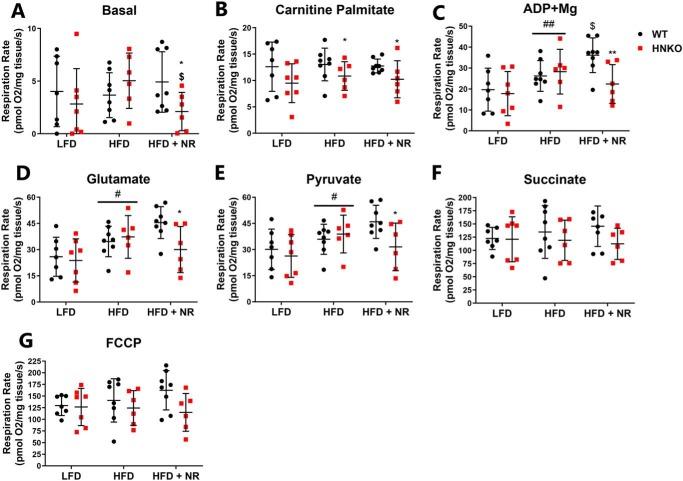
To determine whether mitochondrial respiratory capacity in the liver was affected in HFD/NR-treated HNKO and WT mice, hepatic mitochondrial function was evaluated by high-resolution respirometry. An interaction was observed between genotype and NR for basal respiration (Fig. 4A, interaction p < 0.05), and post hoc testing showed decreased oxygen consumption in the NR-treated HNKO mice (p < 0.05). When stimulated with carnitine palmitate, the HFD-fed HNKO groups had lower respiration rates, indicating an impairment of fatty-acid utilization during uncoupled respiration (Fig. 4B, p < 0.05). HFD caused an increase in state III respiration compared with LFD groups (respiration rate in response to ADP addition, Fig. 4C, p < 0.01). Interestingly, NR treatment increased state III respiration in WT mice but not in HNKO animals (interaction p < 0.05, post hoc test p < 0.05). The differences in oxygen consumption persisted following the addition of glutamate and pyruvate (Fig. 4, D and E). No change in respiration rate was observed between groups after succinate and FCCP stimulation (Fig. 4, F and G), although a borderline effect of genotype was observed between HFD versus HFD + NR following FCCP stimulation (p = 0.06). No change was detected in the cytochrome c/pyruvate ratio between groups, indicating that mitochondria were not damaged (data not shown). Thus, although mitochondrial oxygen consumption was largely maintained in HNKO mice, HNKO mice have minor impairments in fatty-acid handling and a decreased ability to increase respiration in response to NR supplementation.
Figure 4.
Hepatocyte-specific knockout of Nampt has no effect on mitochondrial respiratory function, but reduces the ability of NR to increase state III respiration. Mitochondrial function was evaluated by high-resolution respirometry in liver tissue, where mitochondrial substrates were added in succession. Following basal stimulation (A), liver tissues were stimulated with carnitine-palmitate (B), ADP + Mg (C), glutamate (D), pyruvate (E), succinate (F), and FCCP (G). n = 6–8 per group. */** indicate effects of genotype, p < 0.05/0.01, respectively. #/## indicate effects of diet, p < 0.05/0.01, respectively. $/$$ indicate effects of NR within the HFD groups, p < 0.05 and p < 0.01, respectively.
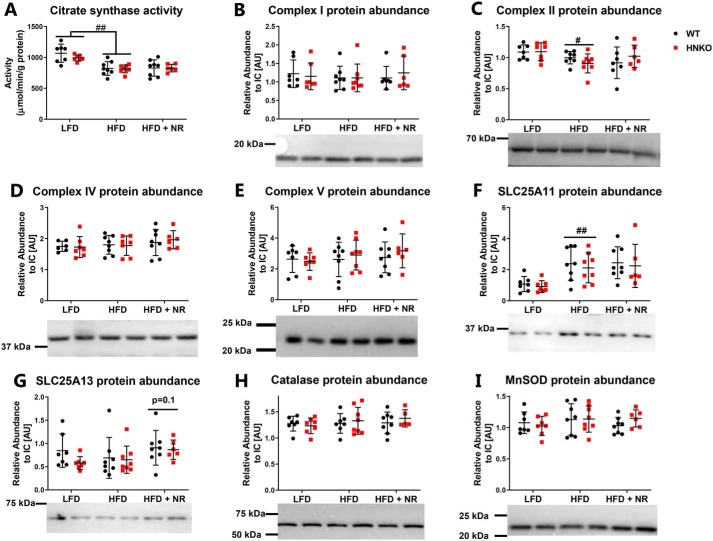
HFD feeding does not affect mitochondrial protein abundance in HNKO mice
To investigate whether the difference in state III respiration between genotypes following NR stimulation was associated with an altered number of mitochondria, we measured citrate synthase activity in the liver. Citrate synthase activity correlates with mitochondrial content in skeletal muscle (45). We observed decreased citrate synthase activity in HFD-fed mice (Fig. 5A, p < 0.01). However, activity was not affected by NR or genotype. NR was shown to increase the abundance of the oxidative phosphorylation (OXPHOS) complexes (7), but in our hands the levels of OXPHOS complexes I, II, IV, and V were not affected by either genotype or NR (Fig. 5, B–E). However, complex II abundance was decreased in HFD-fed mice compared with LFD-fed mice (Fig. 5C, p < 0.05). Reduced NAD+ content in the cytosol may affect transport into the mitochondria of electrons generated through glycolysis (46). We therefore measured the abundance of the malate–aspartate shuttle proteins SLC25A11 and SLC25A13. HFD feeding increased hepatic abundance of SLC25A11, but the abundance was not affected by genotype or NR treatment (Fig. 5F, p < 0.01). SLC25A13 abundance was not affected by diet, genotype, or NR (Fig. 5G). NR has previously been demonstrated to decrease hepatic lipid peroxidation, indicating an attenuation of oxidative stress (7). The abundance of catalase and MnSOD was not affected by diet, genotype, or NR (Fig. 5, H and I), indicating no changes in oxidative stress from HFD or Nampt knockout. Together, these data indicate that HFD decreases citrate synthase activity and alters the abundance of specific mitochondrial proteins. However, these changes were not affected by NR supplementation, and the genotype differences in state III respiration appear not to be associated with alterations in mitochondrial protein abundances.
Figure 5.
HNKO mice have no alterations in abundance of the mitochondrial OXPHOS complexes. WT and HNKO mice were fed a LFD or HFD with or without NR supplementation for 20 weeks. To determine whether NR affected mitochondrial function, citrate synthase activity (A), and abundance of complex I (B), complex II (C), complex IV (D), complex V (E), SLC25A11 (F), and SLC25A13 (G) was determined. To determine whether redox defenses were affected by NR treatment, abundance of catalase (H) and MnSOD (I) was determined. n = 6–8 per group. */** indicate effects of genotype, p < 0.05/0.01, respectively. #/## indicates effects of diet, p < 0.05/0.01, respectively. $/$$ indicates effects of NR within the HFD groups, p < 0.05/0.01, respectively.
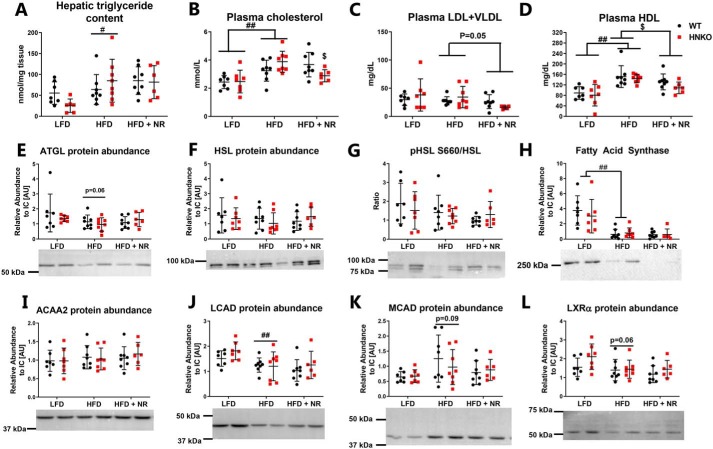
NR decreases LDL + VLDL and HDL content regardless of genotype but does not affect hepatic triglyceride content
To investigate whether the impaired oxygen consumption observed in HNKO liver following palmitate stimulation was associated with changes in hepatic lipid metabolism, we quantified hepatic triglyceride levels. A significant increase in triglyceride was observed following HFD feeding independent of genotype (Fig. 6A, p < 0.05). Interestingly, NR did not decrease hepatic triglyceride content in this model as described previously (12). HFD feeding significantly increased plasma cholesterol levels regardless of genotype (Fig. 6B, p < 0.01). An interaction was observed between NR treatment and genotype (p < 0.05), where NR significantly decreased plasma cholesterol levels in HNKO mice (p < 0.05) but not in WT mice. NR tended to decrease plasma LDL + VLDL levels (Fig. 6C, p = 0.05) and resulted in a small but significant decrease in plasma HDL levels regardless of genotype (Fig. 6D, p < 0.05). We observed a tendency toward decreased hepatic ATGL abundance in HFD-fed mice regardless of genotype (Fig. 6E, p = 0.06), but NR had no significant effects on ATGL abundance. HSL abundance was unaltered between groups (Fig. 6F), and no significant change was observed in pSer-660, indicating a similar regulation of lipolysis between groups (Fig. 6G). HFD-fed mice had low abundance of hepatic fatty-acid synthase (FASN) compared with LFD-fed mice (Fig. 6H, p < 0.01), indicating that control of hepatic lipid synthesis was maintained. No differences in FASN abundance were observed between genotypes or with NR treatment. We found no changes in the abundance of ACAA2, which mediates the final step in β-oxidation (Fig. 6I), but HFD feeding decreased hepatic abundance of LCAD (Fig. 6J, p < 0.01) and borderline increased abundance of MCAD (Fig. 6K, p = 0.09). However, no differences were observed for either protein between genotypes or with NR treatment. HFD tended to decrease abundance of the hepatic cholesterol sensor liver X receptor α (Fig. 6L, p = 0.06), but no change was observed following NR treatment, even though NR lowered plasma cholesterol levels in HNKO mice. Collectively, our data indicate that the decreased respiration following palmitate stimulation in HNKO mice does not result in severe lipid accumulation or altered abundance of proteins associated with lipogenesis or lipolysis. Furthermore, we demonstrate that NR reduces plasma HDL content independent of genotype.
Figure 6.
HNKO mice have no alterations in abundance of proteins associated with oxidative defenses or lipid metabolism. A, to determine whether NR supplementation affected hepatic lipid metabolism, hepatic triglyceride content was determined. To determine the effects of NR supplementation on plasma cholesterol composition, total plasma cholesterol levels (B), plasma LDL + VLDL levels (C), and plasma HDL levels (D) were determined. To investigate whether NR treatment increased abundance of lipases, hepatic ATGL abundance (E), HSL abundance (F), and phosphorylation of Ser-660 on HSL (G) were determined. Hepatic content of fatty-acid synthase (H), ACAA2 (I), LCAD (J), MCAD (K), and LXRα (L) were also determined. n = 6–8 per group. */** indicate effects of genotype, p < 0.05/0.01, respectively. #/## indicate effects of diet, p < 0.05/0.01, respectively. $/$$ indicate effects of NR within the HFD groups, p < 0.05/0.01, respectively.
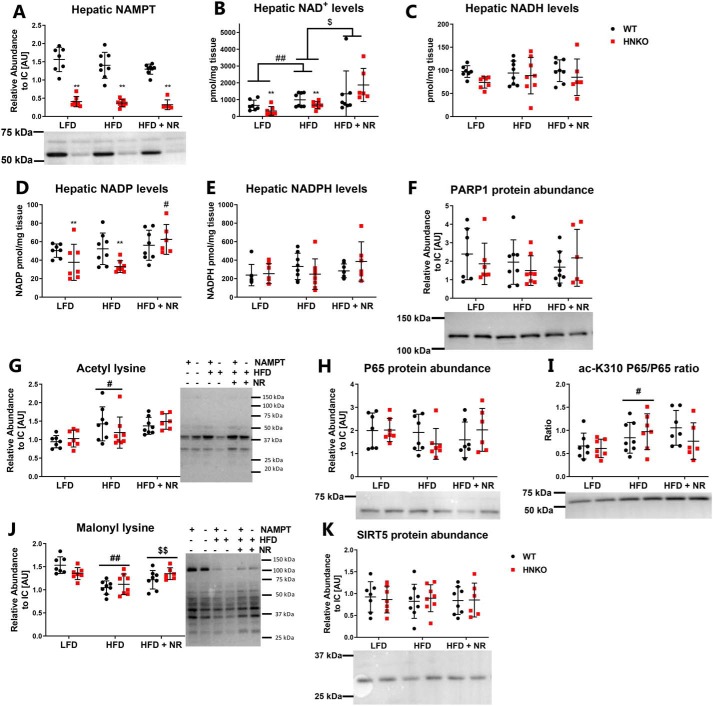
NR supplementation increases hepatic NAD+ content and rescues HFD-induced decrease in protein malonylation
The general lack of effect of NR on fat mass accumulation and hepatic triglyceride content could be due to the lack of effects on hepatic NAD+ metabolism. As expected, NAMPT protein abundance was significantly decreased in HNKO mice (Fig. 7A, p < 0.01) but was not altered by diet or NR treatment in WT mice. Similarly to chow-fed mice, hepatic NAD+ levels were decreased in HNKO mice from both the LFD and HFD groups (Fig. 7B, p < 0.01). NR increased hepatic NAD+ levels compared with HFD-fed mice (Fig. 7B, p < 0.01), but HFD feeding also increased hepatic NAD+ levels regardless of genotype compared with LFD-fed mice (Fig. 7B, p < 0.01). NADH levels were not affected by NR or genotype (Fig. 7C). NADP+ levels decreased in HNKO mice on both LFD and HFD (Fig. 7D, p < 0.01), and NR increased NADP+ levels in HNKO mice compared with HFD-fed mice (Fig. 7D, interaction p < 0.05, KO HFD versus KO HFD + NR, p < 0.01). No alterations in NADPH levels could be detected between groups (Fig. 7E). To identify whether knockout of Nampt or NR supplementation affected the activity of the NAD+-consuming processes, we first attempted to detect formation of poly(ADP)-ribose (PAR) polymers in the liver samples as a measure of PARP activity, but no PAR polymers could be detected in the samples (data not shown). PARP1 abundance was unaltered between groups (Fig. 7F), although a high degree of variance was observed between samples. NR has been demonstrated to increase SIRT1- and SIRT3-mediated deacetylation in skeletal muscle, liver, and brown adipose tissue (12), and a HFD causes hyperacetylation of mitochondrial proteins (47). We found increased lysine acetylation following HFD feeding (Fig. 7G, p < 0.05), but no effect of genotype or NR supplementation. We also measured the acetylation levels of P65, a subunit of NF-κB, as an indirect measure of SIRT1 activity (48). P65 content was similar between all groups (Fig. 7H), but HFD increased P65 acetylation (Fig. 7I, p < 0.01), and acetylation status was not affected by either genotype or by NR. Hence, although NR increased hepatic NAD+ content, this did not decrease acetylation of this SIRT1 target. In addition to deacetylation, other sirtuins can remove different acyl groups such as succinyl and malonyl from lysine residues (49). In line with previous observations (42), HFD feeding caused a significant decrease in lysine malonylation compared with LFD feeding (Fig. 7J, p < 0.01). Interestingly, no differences were observed between malonylation in HNKO and WT mice, but NR supplementation caused a small increase in lysine malonylation (Fig. 7J, p < 0.01), indicating either increased malonylation or decreased demalonylation. No change was observed in SIRT5 abundance (Fig. 7K). Collectively, knockout of Nampt has the expected effects on NAD+ levels, and NR is able to increase hepatic NAD+ concentrations. However, NR affects only lysine malonylation and not lysine acetylation.
Figure 7.
NR treatment increases hepatic NAD+ levels and increases lysine malonylation. WT and HNKO mice were fed a LFD or HFD with or without NR supplementation for 20 weeks. To determine the effects of HFD feeding and NR supplementation in HNKO mice, hepatic NAMPT abundance (A), NAD+ content (B), NADH content (C), NADP+ content (D), and NADPH content (E) levels were determined. F, PARP1 abundance was quantified to investigate whether HFD feeding or NAMPT knockout affected poly(ADP-ribose) polymer formation. G, to investigate indications of altered sirtuin activity, total lysine acetylation was determined. Total hepatic P65 content (H), and acetylation of lysine 310 on P65 (I) were determined as an indication of SIRT1 activity. Total lysine malonylation (J) and SIRT5 abundance (K) were also determined to investigate whether SIRT5 activity was affected. n = 6–8 per group. */** indicate effects of genotype, p < 0.05/0.01, respectively. #/## indicate effects of diet, p < 0.05/0.01, respectively. $/$$ indicate effects of NR within the HFD groups, p < 0.05/0.01, respectively.
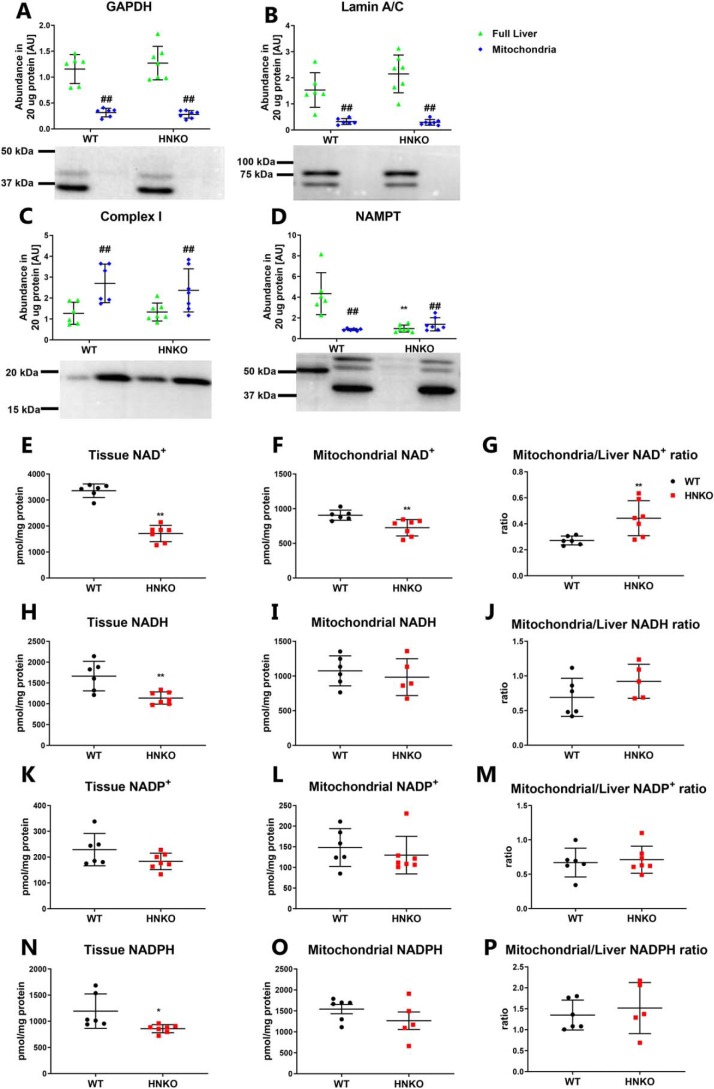
Hepatic mitochondrial NAD+ pool is less sensitive to NAMPT ablation than the whole-tissue NAD+ pool
The respirometry analysis showed only minor alterations in mitochondrial oxygen consumption in HNKO mice, and our data suggest that mitochondrial function is maintained in the absence of Nampt. A recent report suggested that mitochondria contain no NAMPT and instead import NAD+ from the cytosol (50). We therefore hypothesized that the mitochondrial NAD+ pool could be less sensitive to Nampt ablation, which would allow mitochondrial function to be maintained. To this end, we isolated mitochondria from livers of male HNKO mice and WT littermates. Mitochondrial fractions contained no GAPDH (Fig. 8A, p < 0.01) or lamin A/C (Fig. 8B, p < 0.01), indicating that the mitochondrial fraction was free of cytosolic and nuclear proteins. In contrast, the mitochondrial fraction contained an increased abundance of OXPHOS complex I (Fig. 8C, p < 0.01). NAMPT was not detected in the livers of HNKO mice (Fig. 8D), and the mitochondrial fraction from WT livers did not contain any NAMPT as expected (50). Although total NAD+ content was reduced by 50% in livers from HNKO mice (Fig. 8E, p < 0.01), NAD+ levels in mitochondria from HNKO mice were only reduced by 20% (Fig. 8F, p < 0.01). The ratio between mitochondrial NAD+ and total liver NAD+ content was increased in HNKO mice (Fig. 8G, p < 0.01), indicating that a larger proportion of the total NAD+ pool of HNKO hepatocytes resides in the mitochondria. Tissue NADH levels were reduced by 40% in HNKO livers (Fig. 8H, p < 0.01), although levels were unaltered in HNKO mitochondria (Fig. 8I). However, the ratio between mitochondrial and total NADH levels was not significantly increased in HNKO mice (Fig. 8J). NADP+ levels were not altered in HNKO mice (Fig. 8, K–M). Interestingly, NADPH levels were significantly decreased in HNKO liver tissue (Fig. 8N, p < 0.05), but they were not significantly altered in the HNKO mitochondria (Fig. 8O). The ratio between mitochondrial NADPH and whole-liver NADPH was not significantly altered (Fig. 8P). Collectively, our data show that the mitochondrial NAD+ and NADP pools are less sensitive to Nampt ablation compared with whole-tissue levels. This provides a possible explanation for how mitochondrial function can be maintained in the absence of Nampt.
Figure 8.
Mitochondrial NAD pool is prioritized in response to hepatocyte-specific knockout of Nampt. To verify the purity of the mitochondrial fraction, the mitochondrial fraction was investigated for the presence of GAPDH (A), lamin A/C (B), and complex I (C) by immunoblotting (n = 6–7). Furthermore, the mitochondrial fraction was investigated for the presence of NAMPT (n = 6–7) (D). NAD+ content was determined for whole liver tissue (E) and the mitochondrial fraction (F), and it was used to calculate the ratio between the mitochondrial NAD+ pool and the total NAD+ pool (n = 6–7) (G). Likewise, NADH content was determined for whole-liver tissue (H) and the mitochondrial fraction (I) and was used to calculate the ratio between the mitochondrial NADH pool and the total NADH pool (n = 5–6) (J). This was also done for NADPH (K–M) and NADP+ (N–P). n indicates number of animals. */** indicate effects of genotype, p < 0.05/0.01, respectively. #/## indicate effects of compartment, p < 0.05/0.01, respectively.
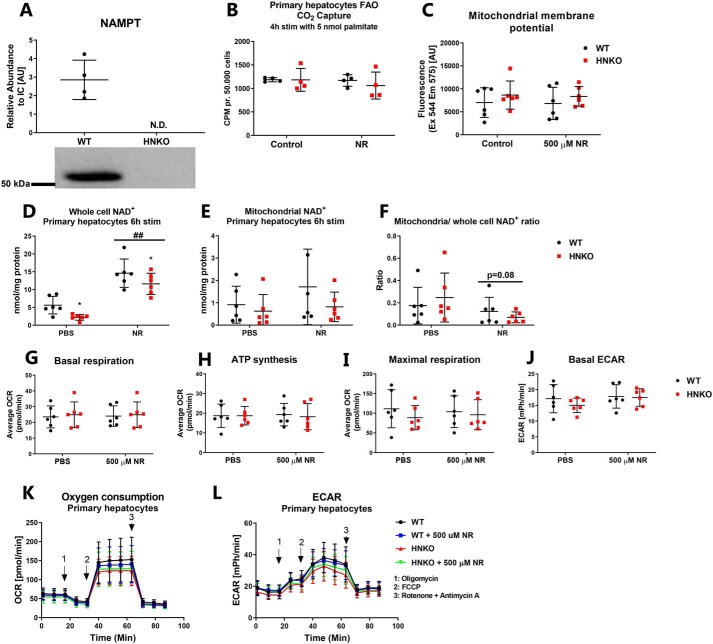
Mitochondrial function is maintained in primary hepatocytes with knockout of Nampt
We next investigated whether mitochondrial function was preserved in primary mouse hepatocytes from HNKO mice. As several reports have observed enhanced mitochondrial function following NR treatment (12, 13, 15), we also treated cells with NR. No NAMPT protein was detected in primary hepatocytes from HNKO mice (Fig. 9A). Decreased fatty-acid oxidation in liver homogenates from hepatocyte-specific Nampt knockout mice has been reported previously (38). However, fatty-acid oxidation was not different between WT and HNKO primary hepatocytes (Fig. 9B). Primary hepatocytes were incubated with TMRM dye to measure mitochondrial membrane potential, but no difference in fluorescence intensity was detected between genotypes (Fig. 9C). Following 6 h of NR treatment, NAD+ content increased in primary hepatocytes regardless of genotype (Fig. 9D, p < 0.01); however, no increase was observed in the mitochondrial NAD+ pool (Fig. 9E). Thus, there was a borderline decrease in the mitochondrial/whole-cell NAD+ ratio after NR treatment compared with untreated cells (Fig. 9F, p = 0.08), suggesting that NAD+ repletion may primarily increase cytosolic and nuclear NAD+ pools. Finally, we found no differences in basal respiration rate, oxygen consumption for ATP synthesis, maximal respiration rate, or basal ECAR with or without 500 μm NR (Fig. 9, G–L). Thus, in primary hepatocytes, mitochondrial respiratory function, fatty-acid oxidation, and mitochondrial membrane potential are not affected by Nampt knockout or NR stimulation.
Figure 9.
Mitochondrial respiratory capacity is maintained in HNKO hepatocytes. A, NAMPT abundance in primary hepatocytes from WT and HNKO mice, n = 4. B, CO2 release from primary mouse hepatocytes from WT and HNKO mice following 4 h of stimulation with [14C]palmitate (n = 4). C, TMRM fluorescence was used as a measure of mitochondrial membrane potential, n = 6. NAD+ content was quantified in primary mouse hepatocytes (D) and in hepatocyte mitochondria (E) following 6 h of NR stimulation, n = 5. F, ratio of mitochondrial to whole-cell NAD+ content following 6 h of NR stimulation. G–L, oxygen consumption and respiratory capacity of primary mouse hepatocytes, measured by the SeaHorse XF system (n = 6). n indicates number of experiments. */** indicate effects of genotype, p < 0.05/0.01, respectively. #/## indicate effects of NR treatment, p < 0.05/0.01, respectively.
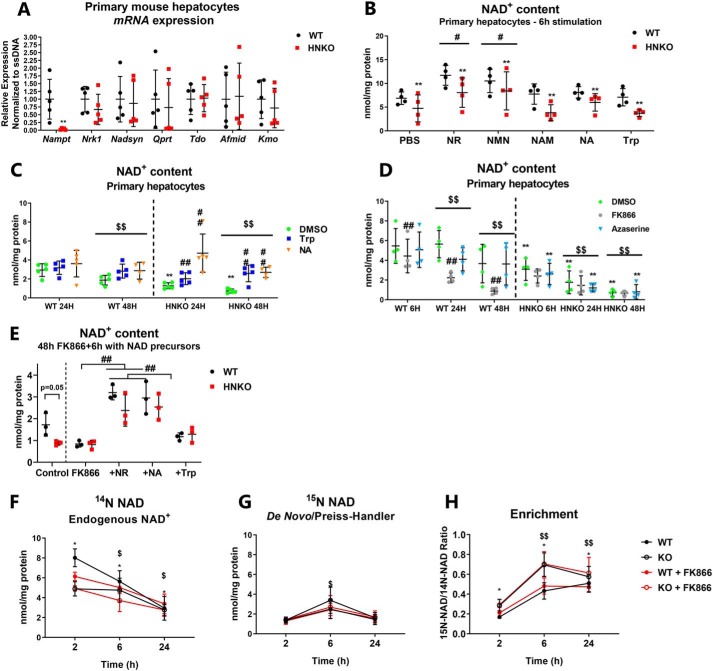
NAMPT is the major contributor for maintaining NAD+ levels in primary mouse hepatocytes
Across experiments, HNKO mice maintained 50% of liver NAD+ levels. To determine whether HNKO mice were more efficient in utilizing other NAD+ precursors, we measured expression of genes from different NAD+-synthesizing pathways. Primary hepatocytes from HNKO mice expressed very low levels of Nampt (Fig. 10A). HNKO mice expressed Tdo2, Afmid, Kmo, or Qprt to WT levels (Fig. 10A), suggesting a maintained expression level of de novo synthesis genes. Nrk1 expression was also not affected in HNKO hepatocytes (Fig. 10A), and no change was observed in expression of Nadsyn (Fig. 10A). To test the efficiency of NAD+ synthesis from NAD+ precursors, we incubated HNKO and WT hepatocytes with different substrates for 6 h. Across treatments there was an overall reduction in NAD+ in primary HNKO hepatocytes as assessed by a two-way ANOVA testing genotype versus treatment (Fig. 10B, main effect p < 0.01). NR and NMN significantly increased NAD+ levels regardless of genotype, although NAD+ content was still lower in HNKO hepatocytes (Fig. 10B, p < 0.05). Treatment with NAM, NA, or Trp did not affect NAD+ levels after 6 h (Fig. 10B). However, when HNKO hepatocytes were incubated with NA or Trp for 24 and 48 h, both precursors prevented the decline in NAD+ observed in the control group (Fig. 10C, p < 0.01). These data suggest the different NAD+ precursors affect the hepatocyte NAD+ pool in distinct ways. WT and HNKO hepatocytes were subsequently incubated for 48 h with the NAMPT inhibitor, FK866, or the NADSYN inhibitor, azaserine, which blocks synthesis from Trp and NA (51). Regardless of genotype, NAD+ content declined over time (Fig. 10D, p < 0.01). NAD+ levels were reduced in HNKO hepatocytes at all investigated time points (Fig. 10D, p < 0.01). FK866 treatment significantly reduced NAD+ levels in WT hepatocytes over time (Fig. 10D, p < 0.01), but not in HNKO hepatocytes. After 48 h NAD+ levels in FK866-treated WT hepatocytes were similar to NAD+ levels in HNKO hepatocytes, suggesting that NAMPT is the primary contributor to the hepatic NAD+ pool in vitro. Azaserine did not significantly affect NAD+ levels regardless of genotype, indicating that the contribution of the de novo pathway was reduced in culture. When WT and HNKO hepatocytes were stimulated with NAD+ precursors following 48 h of FK866 treatment, NR and NA caused an almost 3-fold increase in NAD+ content independent of genotype (Fig. 10E, p < 0.01). Trp caused a minor increase in NAD+ content when tested alone against FK866 with a two-way ANOVA (p < 0.01), but the increase was much smaller than for both NA and NR. No genotype-specific differences were observed in response to stimulation with the various precursors, indicating that HNKO hepatocytes do not utilize NA or Trp for NAD+ synthesis to a greater extent compared with WT cells. To directly demonstrate whether HNKO hepatocytes rely on de novo synthesis for maintaining NAD+ levels, we incubated HNKO and WT hepatocytes with 500 μm tryptophan and [15N]glutamine for up to 24 h. This results in 15N-labeling of NAD+ produced through NADSYN1 (27). To prevent the confounding factor of NAD+ salvage in WT hepatocytes, the experiment was also carried out in the presence of FK866. A main effect of genotype was observed for endogenous [14N]NAD+ levels (Fig. 10F, p < 0.05), although [14N]NAD+ levels declined throughout the experiment (p < 0.05). [15N]NAD+ could be detected at all time points, but levels peaked after 6 h (Fig. 10G, p < 0.05). Enrichment of [15N]NAD+ increased over time (Fig. 10H, p < 0.01) and was significantly higher in HNKO mice (p < 0.05). No effect of FK866 was observed, indicating that salvage of [15N]NAD+ was not a major factor in WT mice. Through MS/MS analysis of NAD+, enrichment occurred at the carboxamide nitrogen of the NAM ring of NAD+ (data not shown) as expected for de novo synthesis of NAD+. Collectively, these data indicate that hepatocytes from HNKO mice can compensate for the lack of NAMPT by increasing the amount of NAD+ produced from Trp.
Figure 10.
NAMPT is an important contributor to the maintenance of NAD+ levels in primary mouse hepatocytes. A, mRNA levels of enzymes associated with NAD+ synthesis were determined in primary mouse hepatocytes, n = 4. B, NAD+ content in primary mouse hepatocytes from HNKO mice and WT littermates stimulated for 6 h with various NAD+ precursors (NMN, nicotinamide mononucleotide; Trp, tryptophan, n = 4). C, NAD+ content in primary mouse hepatocytes from WT and HNKO mice stimulated for 24 or 48 h with DMSO, 500 μm Trp, or 500 μm NA, n = 4–5. D, NAD+ content in primary mouse hepatocytes from WT and HNKO mice following 6, 24, and 48 h of incubation with the NAMPT inhibitor FK866 (10 μm), the NAD synthase inhibitor azaserine (10 μm), or DMSO, n = 4. E, NAD+ content in primary hepatocytes from WT and HNKO mice, following 48 h of 10 μm FK866 treatment and 6 h of stimulation with 500 μm of NR, NA, or Trp, n = 3. To determine the contribution of de novo synthesis, primary hepatocytes from WT and HNKO mice were stimulated with 500 μm Trp in the presence of [15N]glutamine to label NAD+ produced by NAD+ synthase. Cells were incubated with DMSO or 10 μm FK866 to prevent recycling of [15N]NAD+. F, [14N]NAD+ is endogenous NAD+ or NAD+ produced independently of NADSYN1; G, [15N]NAD+ is NAD+ produced by amidation through NADSYN1. H, enrichment indicates the ratio of [15N]NAD+ to [14N]NAD+, n = 3. n indicates number of experiments. */** indicate effects of genotype, p < 0.05/0.01, respectively. #/## indicate effects of treatment, p < 0.05/0.01, respectively. $/$$ indicates effects of time, p < 0.05/0.01 respectively.
Discussion
As dietary NAD+ precursor supplementation can alleviate symptoms of a wide range of diseases and conditions, it is essential to investigate the underlying mechanisms associated with NAD+ deficiency. We demonstrate that while NAMPT contributes significantly to the hepatic NAD+ pool, hepatocytes are capable of maintaining a substantial fraction of NAD+ in the absence of Nampt through the de novo pathway. We also show that deletion of Nampt in the liver causes no obvious phenotype in the adult animal and that HNKO mice are not susceptible to diet-induced fat mass accumulation. Hence, under normal culturing/feeding conditions, ablation of NAMPT-mediated NAD+ salvage appears to have only minor metabolic consequences.
NAD+ levels were significantly decreased in young and adult HNKO mice. These results are in line with previously published results in hepatocyte-specific Nampt knockout mice and mice expressing enzymatically inactive NAMPT in the liver (8, 38). In contrast, inducible knockout of Nampt in liver does not affect NAD+ levels (39). HNKO mice resembled WT littermates in both body composition and in the various tolerance tests. At 110 weeks of age, mice maintained normoglycemia, indicating that HNKO mice are not prone to age-induced loss of glucose control. A surprising finding was that hepatic NAD+ content increased with age for both HNKO and WT mice, which is in contrast to several reports (8, 52, 53). This may be a consequence of the continuous backcrossing of our model onto the C57BL/6JBomTac background, done to avoid the Nnt mutation present in other C57BL6/J strains (54). We have previously reported that the hepatic NAMPT/NAD+ levels in C57BL/6JBomTac mice are resistant to prolonged high-fat diet feeding (42), and this strain may therefore also be resistant to aging-induced decreases in hepatic NAD+ levels. In HNKO mice, the increased NAD+ levels may reflect an increased efficiency of the de novo synthesis pathway with time compared with young mice, adapting to a life-long shortage of NAD+ in the liver.
The mitochondrial NAD+ pool of HNKO mice was less affected than the total NAD+ pool, suggesting that cytosolic and nuclear NAD+ pools are primary sites of NAD+ depletion, but also that the mitochondrial NAD+ pool is prioritized over the cytosolic/nuclear NAD+ pool. A recent study showed that mitochondria are unable to utilize NAM for NAD+ synthesis (50). Instead, mitochondria were found to import NAD+ from the cytosol. It would be of interest to determine whether the mitochondrial NAD+ pool can be maintained if NAD+ levels in the cytosol/nucleus are further depleted. Interestingly, decreased oxygen consumption was observed in isolated mitochondria from liver-specific Nampt knockout mice following stimulation with palmitoyl carnitine (38). We also observed decreased oxygen consumption following palmitate- carnitine stimulation of whole liver from HNKO mice. Our data therefore suggest that a decrease in hepatocyte NAD+ content may be associated with a minor impairment of fatty-acid oxidation. However, no decrease in fatty-acid oxidation was observed in HNKO primary hepatocytes. Moreover, no HNKO-specific reductions in abundance of proteins associated with fatty-acid oxidation were observed, indicating that decreased oxidation might be a result of decreased pathway activity rather than decreased protein abundance.
Stimulation with different NAD+ precursors showed differential effects on hepatocyte NAD+ levels. NR and NMN induced a rapid increase in cellular NAD+ levels. The ability of NR to increase cellular NAD+ levels has been demonstrated across multiple cell lines (12) and in primary mouse hepatocytes (29). NA and Trp were not efficient precursors for NAD+ after 6 h, but prevented a decline in NAD+ content in HNKO mice after 24 and 48 h of incubation. NA and Trp may not be capable of increasing NAD+ levels after 6 h due to the culturing media used, as the media contain 50 μm Trp and 200 nm NAM and NA, respectively. The presence of these metabolites may mask true precursor utilization capacities in the stimulated wells. When cells were stimulated with NA and tryptophan for 6 h following 48 h of NAD+ depletion and without replacing the media, NA was as potent as NR and caused a much higher increase in NAD+ content than Trp. Moreover, we found that tryptophan contributes to the maintenance of the NAD+ pool but that it is not efficient at increasing total NAD+ content. This is in line with a recent report, demonstrating that primary hepatocytes can utilize Trp to generate NAD+, but to a much lower degree than full liver (55). However, the labeling experiment also demonstrates a higher enrichment of NADSYN-produced NAD+ in HNKO hepatocytes, indicating that this pathway may be the primary way of compensation for the lack of NAM salvage in HNKO mice.
The NADSYN inhibitor azaserine did not affect NAD+ content (51), suggesting that an acute inhibition of the de novo pathway causes no immediate decline in NAD+ content. We demonstrate the importance of the NAD+ salvage pathway in primary hepatocytes by showing that FK866 treatment depleted NAD+ levels in WT hepatocytes after 48 h, whereas no effect was observed in HNKO hepatocytes. This is in contrast to human primary hepatocytes that demonstrated resistance to FK866-mediated NAD+ depletion (56). In a previous study, mouse hepatocytes maintained 50% of NAD+ following 30 h of FK866 stimulation (29). This resistance to full depletion could arise from efficient “detoxification” of FK866 in primary hepatocytes or from a lower turnover of NAD+. Primary mouse hepatocytes may require little new NAD+ synthesis in the absence of stress. However, we found that FK866 treatment accelerated the loss of NAD+, demonstrating the importance of NAMPT for maintaining NAD+ levels over time.
NAD+ levels increased independent of genotype in response to HFD feeding, and this was associated with a significant increase in triglyceride levels in both genotypes. Thus, in contrast to several published studies (5–10, 17), fat accumulation was positively associated with NAD+ levels. This was especially true for the HNKO mice that increased hepatic triglyceride content by 3-fold in response to HFD feeding. Female mice on a C57BL/6JBomTac background have been shown to only develop hepatic steatosis when housed under thermo-neutral conditions (57). The choice of mouse strain for hepatic steatosis models should therefore be carefully considered. HFD feeding did, however, induce severe fat mass accumulations in our experiment indicating that this model can be used to mimic obesity development.
The HFD induced changes in liver protein abundance and modified the levels of specific post-translational modifications. HFD feeding decreased citrate synthase activity and OXPHOS complex II abundance and increased lysine acetylation and decreased lysine malonylation. Both of the latter effects have been reported before (42, 47). NR supplementation did not decrease hepatic lysine acetylation, suggesting that sirtuin activity may not have been increased despite increased NAD+ levels. Furthermore, NR supplementation did not correct the HFD-induced increase in P65 acetylation, suggesting SIRT1 activity was not increased. However, NR supplementation partially corrected the HFD-induced decrease in lysine malonylation. Future investigations should determine how HFD affects the protein-specific malonylation patterns in the liver and define the mechanism by which NR causes increased malonylation. As SIRT5 is a NAD+-dependent demalonylase (49), it seems counterintuitive that increased NAD+ levels would cause increased malonylation. Currently, it is not known whether increased malonylation may be related to putative beneficial effects of NR.
NR was stable in the drinking water, and oral NR administration significantly increased hepatic NAD+ and NADP+ levels. However, NR did not prevent fat mass accumulation or improve glucose tolerance, as reported previously (7, 12), but decreased plasma HDL and LDL/VLDL levels as reported earlier (12). However, dietary NR supplementation for 12 weeks in obese, older, and dyslipidemic men was recently shown to not affect plasma levels of LDL, HDL, or total cholesterol (32). Intriguingly, our data revealed that although NR increased oxygen consumption following induction of state III respiration in WT mice, this was not observed in HNKO mice. No genotype effects were observed following FCCP stimulation, demonstrating that maximal uncoupled respiratory capacity was not compromised in the HNKO liver. At present, we have no explanation for this observation, but since NR increased NAD+ levels in both genotypes, the underlying cause may be unrelated to reduced NAD+ content induced by knockout of Nampt.
In conclusion, NAMPT plays an important role in maintaining NAD+ levels in hepatocytes. However, the lack of NAMPT has only minor effects on mitochondrial function measured either in vitro or ex vivo. Although NR was able to increase hepatic NAD+ levels in both cell cultures and whole liver and showed positive effects on the blood lipid profile in response to HFD feeding, it did not significantly affect hepatic triglyceride content, glucose tolerance, or whole-body fat mass accumulation in HFD-fed mice. Future studies will be directed toward clarifying the contribution of individual hepatic NAD+ salvage pathways as well as the de novo synthesis pathway in response to dietary interventions that exert a more profound metabolic stress on the liver.
Experimental procedures
Ethical approval
Animal experiments were performed in accordance with the European directive 2010/63/EU of the European Parliament and of the Council for the Protection of Animals Used for Scientific Purposes. Ethical approval was given by the Danish Animal Experiments Inspectorate (numbers 2012-15-2934-307 and 2015-15-0201-00796).
Chemicals and reagents
Unless otherwise noted, all chemicals and reagents were purchased from Merck (Kenilworth, NJ).
Cell culture experiments
Generation and culturing of Nampt knockdown cell line
Hepa1c1c7 cells were transfected with lentivirus carrying an shRNA construct to generate a stable Nampt KD cell line (shNampt, catalog no. RMM3981-201824136, Open Biosystems Dharmacon, Lafayette, CO) or a nonsense shRNA control cell line (sh-Nonsense, catalog no. SHC202, Sigma), as described previously (58). Cells were cultured in minimum essential medium Eagle's α (MEM-α, catalog no. 12561, Thermo Fisher Scientific, Waltham, MA) with 1% penicillin/streptomycin (P/S) (catalog no. 15070063, Thermo Fisher Scientific), 10% fetal bovine serum (FBS, catalog no. F7524), and 2.5 μg/ml puromycin for selection, at 37 °C with 5% CO2. Cells were transfected in passage 4. Experiments were performed from passage 7 to 20.
Nicotinamide riboside supplementation
For NAD+ measurement experiments, 500,000 cells/well of each cell line were seeded in 6-well plates and allowed to attach overnight. The medium was changed to serum-free MEMα (12561, Thermo Fisher Scientific) with 1% P/S ± 500 μm NR (Chromadex, Irvine, CA) dissolved in PBS. After ∼24 h of incubation, media were removed, and cells were rinsed once in PBS. Subsequently, 200 μl of trypsin (trypsin-EDTA, Gibco) were added to cells and then incubated at 37 °C for 10 min. Trypsin was neutralized with MEMα with 10% serum, and cells were centrifuged for 5 min at 200 × g. The media were removed, and the pellets were rinsed once in PBS and centrifuged for 5 min at 200 × g. NAD+ levels were measured using resazurin conversion (described below). NAD+ concentration was normalized to protein concentration, determined from the cell pellets following NAD+ extraction. Proteins were extracted by adding 100 μl of 2% SDS followed by heating to 95 °C for 5 min, after which protein concentration was determined using the bicinchoninic acid assay (BCA) (23227, Thermo Fisher Scientific).
Extracellular flux analysis
Mitochondrial function was assessed using the XF96 extracellular flux analyzer (Seahorse Bioscience, Agilent, Santa Clara, CA). SeaHorse plates were coated with Corning Matrigel Basement Membrane Matrix (catalog no. 356234, BD Biosciences), diluted 1:10 in MEMα with 10% FBS. 10 μl were added to each well for 5 min at room temperature, before the plates were dried for 20 min at 37 °C. 30,000 cells/well of shNampt and shNonsense cells (22–24 wells per group) were seeded in MEMα with 10% FBS, 1% P/S, and puromycin (2.5 μg/ml), ±500 μm NR dissolved in PBS, for 20 h at 37 °C with 5% CO2. Four wells containing only media were included for background measurement. The SeaHorse XF96 Sensor Cartridge was hydrated with Seahorse XF Calibrant Solution (100840–000, Agilent) and was incubated overnight at 37 °C without CO2 in the XF Prep Station. On the day of measurement, 5.6 mm glucose and 1 mm pyruvate were added to the Seahorse XF assay medium (catalog no. 102365-100, Agilent), heated to 37 °C, and pH-adjusted to 7.4 using 1 m HCl. Following incubation, each well was washed in 180 μl of Seahorse XF assay medium and incubated for 1 h in 180 μl of assay medium ± 500 μm NR in the XF Prep Station at 37 °C without CO2. Cells were stimulated sequentially with 1 μm oligomycin, 0.2 μm FCCP (optimized for maximum oxygen consumption rate), and finally 1 μm antimycin A + 1 μm rotenone, delivered through the sensor cartridge injection ports in XF assay medium. All oxygen consumption rates (OCR) were subtracted nonmitochondrial respiration, measured as average OCR following rotenone and antimycin A addition. Basal respiration was calculated as an average of OCR prior to oligomycin addition. Oxygen consumption for ATP synthesis was calculated by subtracting OCR following oligomycin addition from basal respiration rate. Maximal respiration was calculated as an average of OCR following FCCP addition. Basal ECAR was calculated as an average of ECAR prior to oligomycin addition.
Animal experiments
Mouse generation and maintenance
HNKO mice were generated by breeding homozygous floxed animals (15) with mice heterozygous for the cre recombinase gene that was driven by the serum albumin promoter (Alb-cre). Both lines were continuously backcrossed to the C57BL/6JBomTac background, and breeding was maintained by pairing Namptfl/fl, cre+/− (HNKO) males with Namptfl/fl (wildtype (WT)) littermate females. For the aging cohort, female HNKO mice or WT littermates between 10 and 110 weeks of age were used. Mice were group-housed and had access to ad libitum chow (1310, Altromin, Lage, Germany) and water. Mice were housed in a temperature-controlled environment (22 ± 1 °C) with 12-h light/dark cycles. In the “55-week-old” group, mice were between 53 and 58 weeks of age, and in the “110-week-old” group, mice were between 99 and 113 weeks of age. Body compositions were obtained by NMR scanning (EchoMRI 4–1, EchoMRI, Houston, TX). Before oral glucose tolerance tests (OGTT), mice were fasted for 5.5 h, from Zeitgeber time (ZT) 2.5 to 8. Basal blood glucose levels were measured using Contour Blood Glucose Meter (Bayer, Leverkusen, Germany). Mice were given an oral gavage of 2 g of glucose per kg of body weight. Blood glucose levels were measured after 20, 40, 60, 90, and 120 min following the gavage. For the insulin tolerance test, mice were fasted similarly as for the OGTT and were given an intraperitoneal dose of 0.75 units of insulin per kg of body weight, diluted in Gelofusine® (B. Braun, Melsungen, Germany). Blood glucose levels were measured immediately before injection, and 20, 40, 60, 90, and 120 min after. For the pyruvate tolerance test, mice were fasted for 18 h, beginning at ZT 10. Mice were given an intraperitoneal injection dosed as 20 μl/g body weight of a pyruvate/lactate solution in saline (0.1 g of lactate/ml and 10 mg of pyruvate/ml) (59). Blood glucose levels were measured before and 20, 40, 60, and 90 min after the injection. For the 55-week-old group, a 120-min time point was included. The 110-week-old group was deemed too frail for tolerance tests, so blood glucose, lactate, and ketone bodies were measured at ZT 3, 9, 15, and 21 during the light/dark cycle over 2 weeks. Lactate levels were measured using a Lactate Pro 2 (HaB Direct, Southam, UK), and ketone bodies were measured using Freestyle Optimum Neo (Abbott). All blood measurements were performed on blood from the tail vein, obtained by making a small incision with a 25-gauge needle. Mice were sedated with pentobarbital (0.1 mg/g body weight, diluted in saline) and killed by cardiac puncture, and livers were snap-frozen in liquid nitrogen and stored at −80 °C until used for NAD+ measurements and Western blot analysis.
Mitochondria isolation
Mitochondria were isolated from the livers of female WT and HNKO mice (11–15 weeks of age) sacrificed by cervical dislocation. The right liver lobe was transferred to a buffer containing 100 mm sucrose, 100 mm KCl, 50 mm Tris-HCl, 1 mm KH2PO4, 0.1 mm EGTA, and 0.2% BSA. Samples were placed on wet ice throughout the isolation protocol, and mitochondria were isolated following a procedure adapted from muscle (60). Following isolation, the mitochondrial fraction was distributed into four tubes and centrifuged at 7000 × g for 4 min. The supernatant was discarded, and the pellets were stored at −80 °C.
High-fat diet experiments
Female HNKO and WT mice (14–18 weeks of age) were randomized into three diet groups. Mice were fed a 60% HFD (D12492, Research Diets, New Brunswick, NJ) or a sucrose-matched low-fat diet (LFD, D12450J, Research Diets). Half of the HFD-fed mice received NR (Chromadex, Irvine, CA) in the drinking water, which was changed every 2–3 days. NR was supplied to deliver 400 mg/kg body weight/day, dosed as 3.33 g/liter drinking water. This was based on previous experiments measuring water intake in HFD-fed mice (data not shown). Mice were fed this diet for 20 weeks, and body composition was assessed every 4th week by NMR (EchoMRITM). The experiment was repeated twice to obtain a sufficient number of animals in each group. After 9 weeks, water intake was measured for a subset of mice. The water bottle was weighed five times over the course of 1 week, and the average water intake per mouse/day was calculated from these measurements. After 10 and 19 weeks of HFD/LFD feeding, mice were subjected to an OGTT as described above. On the day of sacrifice, mice were sedated with pentobarbital and killed by cardiac puncture. Part of the liver was transferred to BIOPS buffer (as described below) for respirometry measurement, and the rest was snap-frozen in liquid nitrogen. Before further processing, frozen livers were crushed in liquid nitrogen using a mortar and pestle.
Respirometry measurement
High-resolution respirometry was performed as described previously (45), with minor modifications. Following dissection, one piece of liver was transferred to ice-cold BIOPS buffer (containing 2.77 mm CaK2EGTA, 7.23 mm K2EGTA, 5.77 mm Na2ATP, 6.56 mm MgCl2·6H2O, 20 mm taurine, 15 mm Na2 phosphocreatine, 20 mm imidazole, 0.5 mm DTT, 50 mm MES, pH 7.1). The liver piece was cut into smaller pieces, ∼0.5 × 0.5 mm, in ice-cold MiR05 buffer (containing 0.5 mm EGTA, 3 mm MgCl2·6H2O, 60 mm K-lactobionate, 20 mm taurine, 10 mm KH2PO4, 20 mm HEPES, 110 mm sucrose, 1 g/liter BSA, pH 7.1). Tissues were not treated with permeabilization reagents as this was previously demonstrated to be unnecessary for liver tissue (61). The liver pieces were transferred to fresh respiration buffer on ice for 10 min before being weighed and transferred to the Oxygraph-2k (Oroboros Instruments, Innsbruck, Austria). Respirometry measures were performed in MiR05 buffer at 37 °C in hyper-oxygenated conditions in triplicates. Oxygen flux was processed by DatLab software (Oroboros Instruments). Mitochondrial respiratory function was assessed by the sequential addition of mitochondrial substrates. State II respiration was assessed by the addition of 2 mm malate. Then 25 μm palmitate/carnitine was added to determine fatty-acid oxidation. State III respiration was achieved by the addition of ADP-Mg (5 mm) followed by the addition of 10 mm glutamate and 5 mm pyruvate (complex I substrates). 10 μm cytochrome c was added to assess the integrity of the outer mitochondrial membrane. 10 mm succinate was added to assess maximal complex I + II-linked respiration, and finally 0.25 μm FCCP was added to achieve maximal uncoupled respiration.
Primary hepatocyte experiments
Isolation and culturing of primary mouse hepatocytes
Isolation of primary mouse hepatocytes was based on two protocols (62, 63). Male HNKO or WT mice (3–7 months of age) were sedated using avertin, prepared from a stock of 1 g/ml tribromoethanol in T-amyl alcohol, diluted 1:20 in saline, and dosed as 10 μl/g body weight. An incision was made into the abdomen, and the cava vein was localized. A 24-gauge catheter (1962025, Mediq Danmark, Denmark) was inserted into the inferior vena cava and fixed with a suture and a vein clamp. The catheter was connected to a peristaltic pump (520s, Watson Marlow, MA). The portal vein was cut, and the liver was perfused with 50 ml of Hanks' buffered saline solution without calcium and magnesium (HBSS, catalog no. 14170112, Thermo Fisher Scientific), supplemented with 76 mg/liter EGTA and 10 mm HEPES, and heated to 42 °C at 3.5 ml/min. After this, the liver was perfused with 50 ml of Williams E medium (catalog no. 32551020, Thermo Fisher Scientific) supplemented with 400 mg/liter collagenase (C5138, Sigma). Hepatocytes were dispensed in a Petri dish containing plating medium (Minimum essential medium Eagle-199 (MEM-199) (catalog no. 41150, Thermo Fisher Scientific), 10% FBS, 1% P/S), and the cell suspension was passed through a 70-μm strainer (352350, Corning, Corning, NY). Cells were centrifuged three times at 50 × g for 2 min; between each step, the pellet was washed in plating medium. Following the washes, the cell pellet was dissolved in plating medium, and viability was assessed (NucleoCounter NC-200, ChemoMetec). 500,000 living cells/well were seeded in 12-well plates; 1 million. living cells/well were seeded in 6-well plates, and 50,000 living cells/well were seeded in 96-well plates. All plates were collagen-coated. Cells were allowed a minimum of 2 h to attach. Medium was changed to culture medium (MEM-199 with 0.5% FBS, 1% P/S, 1 μm dexamethasone, and 1 nm insulin), and the cells were incubated overnight at 37 °C with 5% CO2.
NAD+ precursor stimulation experiments
The day following seeding in a 6-well plate, the medium was changed to fresh culture medium and PBS or 500 μm either of NR, nicotinamide mononucleotide (NMN), NAM, tryptophan, NA, 10 μm FK866, or 10 μm azaserine was added. Cells were stimulated for 6, 24, or 48 h, after which cells were harvested as described for Hepa1c1c7 cells. NAD+ levels were quantified using resazurin conversion, as described below. For the FK866 NAD+-depletion experiment, media were removed on the day following isolation and replaced with culture medium supplemented with 10 μm FK866. Cells were incubated for 48 h, after which to each well was added NR, NA, or Trp to a final concentration of 500 μm (or an equal volume of DMSO for the control wells), still in the presence of FK866. Cells were stimulated for an additional 6 h, after which cells were harvested as described for Hepa1c1c7 cells.
Fatty-acid oxidation detection
Primary mouse hepatocytes from HNKO and WT mice were plated at a density of 50,000 cells/well in a collagen-coated 96-well plate. Then 14C-labeled palmitate/BSA complexes were prepared by evaporating the ethanol from a 0.1 μCi/μl palmitic acid solution (55 μCi/mmol, MC 121, Moravek, Brea, CA) under a nitrogen stream, using 1.4 μl/well. The dried palmitate stock was subsequently added 5 μl/well of 1 mm palmitate/BSA prepared as described previously (64), and the solution was incubated for 30 min at 37 °C. 100 μl/well of glucose-free Dulbecco's modified Eagle's medium was added to the vial, and part of the media were supplemented with 500 μm NR. The media were subsequently added to the hepatocytes. 30 μl of 1 m NaOH/well was added to a glass-fiber filter 96-well plate (catalog no. 6005199, PerkinElmer Life Sciences), and clamped on top of the culture plate, separated by a custom-made perforated rubber insert. The back of the filter plate was sealed with a back cover seal (catalog no. 6005199, PerkinElmer Life Sciences), and the plates were wrapped in parafilm. The cells were incubated at 37 °C for 4 h, and the filter plate was recovered to detect 14CO2 release. The filter plate was dried at 50 °C for 1 h. To each well of the filter plate was added 50 μl of Microscint-20 (catalog no. 6013621, PerkinElmer Life Sciences). The plate was sealed with a Topseal (catalog no. 6050185, PerkinElmer Life Sciences), and the plate was counted on a TopCount NXT scintillation counter (PerkinElmer Life Sciences). Counts from wells with cells stimulated with media without [14C]palmitate were subtracted as background. Wells with 14C-media but without cells were included as an internal control and showed the background level of counts/min on the filter (data not shown).
TMRM staining of primary hepatocytes
Two days following seeding in a 12-well plate, medium was changed to culture medium with 500 μm NR. Cells were stimulated for 1 h. TMRM (T668, Thermo Fisher Scientific) was added from a 1 μm stock in PBS to a final concentration of 50 nm, and cells were incubated for 20 min at 37 °C. Cells were washed in PBS, and fluorescence was detected using a Hidex Sense plate reader (excitation 544 nm and emission 575 nm; Hidex, Finland). Values were corrected by subtracting fluorescence from hepatocytes without TMRM addition.
Oxygen consumption measurements in primary hepatocytes
Oxygen consumption was measured using the XF96 Extracellular Flux Analyzer as described for Hepa1c1c7 cells, with minor modifications. Plates were collagen-coated by adding 50 μl of collagen solution (0.1 mg/ml in PBS) per well, and the plate was incubated for 5 min at room temperature. Liquid was discarded, and the plate was left to dry overnight at room temperature. 40,000 cells per well were seeded in plating medium. After 2 h, medium was changed to culture medium, ±500 μm NR. Cells were incubated overnight, and on the day of the experiment they were incubated for 1 h in SeaHorse medium, prepared as described for Hepa1c1c7 cells, ±500 μm NR. Measurement and data analysis were performed as described for Hepa1c1c7 cells.
Mitochondria isolation
Mitochondria were isolated from primary mouse hepatocytes seeded in 6-well plates, following 6 h of stimulation in culture medium with 500 μm NR or PBS. 500,000 cells per condition were recovered (for total NAD+ measurement), and mitochondria were isolated from the remaining 500,000 cells, following the protocol described above for full liver. The final mitochondrial fraction was split into two vials for further analysis.
Measurement of NR stability in solution
Sample collection
1.7 g of NR was dissolved in water bottles used for drinking water in mouse experiments. Two bottles were placed in empty cages, and four were placed in separate cages each containing three female C57BL/6JBomTac mice to account for possible increased NR breakdown due to bacterial contamination. NR water was sampled from bottles after 0, 4, and 8 h and once every other day for a total of 6 days. Immediately after collection, samples were transferred to −80 °C where they were stored prior to analysis.
Materials for LC-MS analysis
Unless otherwise stated, all reagents were HPLC grade or higher. Nicotinamide-d4 (NAM-d4) (catalog no. D-3457) and nicotinic acid-d4 (NA-d4) (catalog no. D-4368) were purchased from C/D/N Isotopes (Pointe-Claire, Quebec, Canada). NR-13C,d was synthesized as described (33) and kindly provided by Prof. Marie Migaud (University of South Alabama). N1-Methylnicotinamide-d3 (MeNAM-d3) was prepared as described using nonlabeled NAM (33). Nonlabeled NAM (catalog no. 72340), NA (catalog no. 72309), LCMS grade water (catalog no. 1.15333), 2-propanol (catalog no. 1.02781), acetonitrile (catalog no. 1.00029), ammonium acetate (catalog no. 5.33004), and formic acid (catalog no. 5.33002) were purchased from Merck (Darmstadt, Germany). Hexakis(1H,1H,2H-perfluorethoxy) phosphazene (catalog no. PC1165) was purchased from Apollo Scientific Ltd. (Stockport, UK).
Sample and standard preparation
Samples were thawed on wet ice after storage at −80 °C and diluted 1:10,000 in LCMS grade water. 12 μl of diluted sample was combined with internal standard solution (290 pmol of NR-13C,d, NAM-d4, NA-d4, and 15 pmol of MeNAM-d3) in 48 μl of LCMS grade water in a Verex autosampler vial (catalog no. AR0-3920-12, Phenomenex ApS, Denmark). Vials were capped with Verex Cert + MSQ PTFE caps (catalog no. AR0-8952-12, Phenomenex ApS, Denmark). Standards were composed such that the final volume and internal standard concentrations were the same as for samples. Final standard concentrations were 0.001, 0.003, 0.01, 0.03, 0.1, 0.3, 1, 3, and 10 μm. Quality controls (QCs) were composed in triplicate at final concentrations of 0.063, 0.63, and 6.25 μm in the same manner as standards. Samples were randomized prior to analysis. All samples, standards, and QCs were placed in a Dionex Ultimate 3000 autosampler pre-chilled to 6 °C and injected (10 μl) for analysis.
LC-MS
Analytes were separated in a similar fashion as described (65) with slight modification. These modifications include a faster flow rate (0.55 ml/min) and a modified gradient between mobile phase A (10 mm ammonium acetate + 0.1% (v/v) formic acid) and B (0.1% formic acid in acetonitrile). In the modified gradient, the mobile phase was held at 5% B for 1.8 min followed by a linear increase to 64.1% B over 1.2 min. The column was washed at 90% B for 2 min and then re-equilibrated to starting conditions (5% B) for 1.3 min. Analytes eluted in the following order at the designated retention times: MeNAM (0.87 min), NA (3.22 min), NR (3.31 min), and NAM (3.53 min). Before beginning analysis, the instrument was calibrated by direct infusion of sodium formate. Calibration was modeled with HPC mode. Calibrant was also infused at the beginning of each analysis. A lock mass (hexakis(1H,1H,2H-perfluorethoxy) phosphazene in 2-propanol (0.1 mg/ml)) was employed to improve mass accuracy over the duration of analysis. Analytes were ionized via electrospray and detected in positive, full scan (m/z: 50–1000) ion mode with a Bruker Impact II Q-TOF. The instrument was operated using the following conditions: cone voltage = 45 kV, collision energy = 7 eV, nebulizer gas flow = 10 liters/h, nebulizer gas temperature = 220 °C, nebulizer pressure = 2 bar, and scan rate = 3 Hz.
Quantitation and accuracy
Analytes were identified based upon exact m/z ratio (±0.01 Da) and retention times (±0.2 min). Analyte m/z ratios were 137.0709, 124.0393, 255.0975, and 123.0553 for MeNAM, NA, NR, and NAM, respectively. QCs were injected at equal intervals during the analysis to test precision and accuracy with deviations from theoretical concentrations of ≤20% deemed acceptable. QCs at all concentration levels in all analytes except NA passed. The lowest QC was undetectable for NA; all other concentration levels passed tolerance for NA. Samples were quantified using an internally controlled standard curve (1/x weighted). Micromolar amounts were corrected for dilution.
15N-enrichment of NAD+
On the day following primary hepatocyte isolation, medium was removed and replaced with glutamine-free MEM-199 (M2154, Sigma) supplemented with 0.1 g/liter [15N]glutamine (490024, Sigma) and 500 μm tryptophan. Cells were incubated and harvested as described for Hepa1c1c7 cells after 2, 6, and 24 h. Cell pellets were stored at −80 °C prior to analysis. Prior to extraction, internal standard (heavy-labeled yeast) was composed by incubating yeast in the presence of d-[U-13C]glucose and 0.3 mm adenine and the absence of tryptophan as described previously (65). 0.125% of the total preparation was dosed into each sample, standard, and quality control sample in a 10-μl volume while samples rested on dry ice. Cells were extracted following Ref. 55 with slight modification. Cells were suspended in 1 ml of dry ice-chilled methanol/water (80:20% v/v) (both LCMS grade) with constant shaking (highest energy setting) at 4 °C using a Heidolph Multi Reax system (Heidolph Instruments GmbH and Co. KG, Schwabach, Germany) for 15 min. Any nonsuspended samples were suspended with vigorous, repeated pipetting. All samples were allowed to rest on dry ice for 20 min. The supernatants of centrifuged samples (10,000 × g, 4 C, 5 min) were transferred to fresh tubes and dried via speed vacuum with a Gene Vac EZ 2 system for no more than 4 h. Samples were resuspended in 50 μl of LCMS grade water and centrifuged (20,238 × g, 22 °C, 10 min). Clarified supernatants were transferred to autosampler vials and placed in a pre-chilled (6 °C) autosampler in a random order. A standard curve with nine calibration points (see Ref. 65 for concentrations) and nine quality control samples (final concentrations of 0.225, 7.5, and 75 μm, prepared in triplicate at each concentration) were composed by extracting 10 μl after freezing in tubes on dry ice in the same manner as samples. An internal standard blank containing heavy labeled yeast and a blank containing only extraction solvent were also prepared.
Analytes were separated using LC conditions as described (33) for the alkaline separation. MS conditions were exactly as described above except that the nebulizer gas temperature was 350 °C rather than 220 °C. 14N-Labeled NAD+ was detected using its theoretical m/z ratio of 664.1164 ± 0.005 Da and its retention time (5.9 ± 0.2 min). 14N-Labeled NAD+ was quantified as described (65). Accuracy and precision were determined as described above. 15N-Labeled NAD+ was detected using its theoretical m/z ratio of 665.11405 ± 0.005 Da and co-elution with 14N-labeled NAD+. 15N-Labeled NAD+ was corrected for natural isotopic distribution, isotope skewing (15N abundance of 0.004), and the purity of [15N]glutamine (2/98% [14N1]glutamine/[15N1]glutamine) (66). The concentration of labeled NAD was then determined by multiplying the ratio of [15N]NAD+/[14N]NAD+ (enrichment) by the concentration of nonlabeled NAD+. The location of the labeled 15N on the molecule was determined by fragmenting doubly-labeled NAD+ (m/z: 332.5618) using MS/MS (quadrupole resolution = 3 Da, collision energy = 20 eV). The enrichment of [15N]NAM produced by fragmenting NAD+ was identical to intact NAD+ (data not shown).
Molecular and biochemical analyses
Western blot analysis
To determine protein abundance in cell cultures, cells were harvested and centrifuged as for the NAD+ measurement experiments. Pellets were lysed in 200 μl of lysis buffer (42). For tissue samples, ∼20 mg of pulverized tissue was homogenized in 500 μl of lysis buffer using a Tissuelyser II (Qiagen, Germany), two times for 30 s at 30 Hz. Lysates were incubated end–over–end for 45 min at 4 °C, and were centrifuged for 10 min at 16,000 × g at 4 °C. Following freezing at −80 °C, protein concentration was determined using BCA. 20 μg of protein lysate was loaded on SDS-polyacrylamide gels, subjected to SDS-PAGE, and transferred as described previously (42). On each gel, an internal control of mixed samples was loaded on each side for normalization and for evaluation of transfer quality. Proteins were subjected to immunoblotting with antibodies against the following: NAMPT (catalog no. 372A, Bethyl Lab, Montgomery, TX); MnSOD (catalog no. 06-984, Millipore, Burlington, MA); acetyl-lysine (catalog no. 9441s, Cell Signaling); malonyl lysine (catalog no. 14942, Cell Signaling); fatty-acid synthase (catalog no. 3189, Cell Signaling); ATGL (catalog no. 2138s, Cell Signaling); HSL (catalog no. 4107, Cell Signaling); pHSL Ser-660 (4126, Cell Signaling); catalase (AF3398, R&D Systems, Minneapolis, MN); P65 (catalog no. 8242, Cell Signaling); Ac-P65 Lys-310 (catalog no. 3045, Cell Signaling); complex I (catalog no. 459210, Invitrogen); complex II (catalog no. 459200, Invitrogen); complex IV (catalog no. 459600, Invitrogen); complex V (catalog no. sc74786, Santa Cruz Biotechnology, Dallas, TX); lamin A/C (catalog no. 2032s, Cell Signaling); GAPDH (catalog no. 2118, Cell Signaling); SIRT5 (catalog no. 8782, Cell Signaling); PARP1 (catalog no. 9542, Cell Signaling); poly(ADP-ribose) (catalog no. ALX-210-890A-0100, Enzo Life Sciences, Farmingdale, NY); SLC25A11 (catalog no. Ab80464, Abcam); SLC25A13 (catalog no. Ab156010, Abcam). Bands were developed and quantified as described previously (42). For each gel, band volumes were normalized to the average band volume of the internal controls to account for differences in transfer efficiency.
NAMPT activity assay
The conversion of 14C-labeled nicotinamide to [14C]NMN can be measured by precipitation of [14C]NMN in acetone on filter paper, through scintillation counting. Cell pellets were harvested with trypsin as described above, and 100 μl of phosphate buffer (0.01 m Na2HPO4/NaH2PO4 buffer, pH 7.4, containing protease inhibitor (Sigma)) was added to each tube. Samples were further processed, and NAMPT activity was determined on 30 μg of protein as described previously (15, 67, 68). Background disintegrations/min from scintillation fluid was subtracted from each measurement.
Citrate synthase activity
Liver lysates, prepared in lysis buffer, were diluted 1:10 in 0.1 m Tris-HCl, pH 8.1. Diluted lysate (10 μl) was added in duplicates in a 96-well plate, along with 150 μl of reaction mix containing 44 μg/ml 5,5′-dithiobis(2-nitrobenzoic acid), and 333.3 μg/ml acetyl-CoA in 0.1 m Tris-HCl. Absorbance at 412 nm was measured for 5 min in a Hidex Sense plate reader (Hidex, Finland) to assess background activity (one measurement per 30 s) and for an additional 5 min after the addition of 10 μl of 10 mm oxaloacetate. Citrate synthase activity was calculated in the linear range of absorbance increase and was normalized to protein content of the lysate.
NAD(H)/NADP(H) measurement
NAD+, NADH, NADP+, and NADPH levels were determined using an enzymatic cycling assay, as described previously (42). 10–20 mg of frozen liver was homogenized in 400 μl of either 0.6 m perchloric acid (for NAD+) or 0.1 m NaOH (for NADH) with a Tissuelyser II (Qiagen, Germany). The NADH extracts were incubated at 70 °C for 10 min, and both fractions were centrifuged for 3 min at 13,000 × g. Supernatants were recovered. The NAD+ extracts were diluted 1:1500 in 100 mm Na2HPO4 buffer, pH 8, and the NADH extracts were diluted in 1:500 in 10 mm Tris, pH 6. For NADP+, measurements extracts were diluted 1:100 and for NADPH 1:1200 in the same buffers as above. 100 μl of the diluted extracts were pipetted into a white 96-well plate, and NAD+/NADH levels were determined as described previously (42) and normalized to tissue weight. For NAD+/NADH measurements on cells and mitochondria, one pellet was homogenized in 100 μl of 0.6 m perchloric acid or 0.1 m NaOH, and NAD+/NADH was extracted and measured as for liver tissue. NAD+/NADH levels were normalized to protein concentration extracted from the cell pellet. Total protein was extracted by heating the pellet to 95 °C in 100 μl of 0.2 m NaOH for 5 min. The solution was then centrifuged at 16,000 × g for 5 min, and protein concentration of the supernatant was determined by the BCA assay. For mitochondrial pellets isolated from whole liver, 400 μl of 0.6 m perchloric acid, 0.1 m NaOH was added to each pellet and homogenized using a TissueLyser II (Qiagen, Germany). NAD+/NADH was extracted and measured as for liver tissue. Total protein was extracted from the mitochondrial pellet by adding 100 μl of 2% SDS and 200 μl of 0.2 m NaOH to each pellet for 5 min at 95 °C followed by 5 min of centrifugation at 16,000 × g. For comparison, NAD+ and NADH content from whole tissue from the mitochondria isolation experiment were also normalized to protein content, extracted by the same protocol.
Quantitative real-time PCR
RNA from 15 mg of liver was isolated using TRIzol (Invitrogen). Samples were homogenized using a Tissuelyser II (Qiagen). RNA concentration was determined on a Nanodrop 8000 (Thermo Fisher Scientific). RNA quality was assessed by 260 nm/280 nm ratio, and for all samples the ratio was above 1.8. RNA was reverse-transcribed to cDNA using iScript cDNA synthesis kit (catalog no. 170-8891, Bio-Rad). Reverse transcription was performed on C1000 Thermal Cycler (Invitrogen), using the following protocol: 5 min at 25 °C, 40 min at 42 °C, and 5 min at 85 °C. Real-time PCR was performed on a CFX96 real-time system (Invitrogen) using SYBR Green PCR master mix (600882, AH Diagnostics, Aarhus, Denmark). The qPCR was run according to the following protocol: 3 min at 95 °C for denaturation, 40 cycles of 95 °C for 5 s and 60 °C for 10 s for amplification. A melting curve was generated by gradual heating from 65 to 95 °C and was used to verify the specificity of the reaction. The reaction was run on a 2-μl cDNA template and 300 nm sense and antisense oligonucleotides in a 10-μl reaction. Relative expression was determined from the Cq value and from a standard curve made by serial dilution of a mixture of samples. Expression was normalized to ssDNA, and concentration ws determined using Qubit ssDNA assay kit (qQ10212, Invitrogen). The Nampt primer sequences were used as described previously (69). The remaining primer sequences were obtained from PrimerBank (70). The following primer sequences were used: Nampt: forward, 5′-TGCCGTGAAAAGAAGACAGA-3′, and reverse, 5′-ACTTCTTTGGCCTCCTGGAT-3′; Nrk1 (PrimerBank ID: 21703978a1): forward, 5′-TCATTGGAATTGGTGGTGTGAC-3′, and reverse, 5′-CAACAGGAAACTGCTGACATCAT-3′; Tdo2 (PrimerBank ID: 31982697a1): forward, 5′-ATGAGTGGGTGCCCGTTTG-3′, and reverse, 5′-GGCTCTGTTTACACCAGTTTGAG-3′; Afmid (PrimerBank ID: 21746157a1): forward, 5′-TTGGGAACTTCGTGCAGATAGG-3′, and reverse, 5′-CAGTTTCTCCCCTTCGCCATC-3′; Kmo (PrimerBank ID: 19527030a1): forward, 5′-ATGGCATCGTCTGATACTCAGG-3′, and reverse, 5′-CCCTAGCTTCGTACACATCAACT-3′; Qprt (PrimerBank ID 19526852a1): forward, 5′-CCGGGCCTCAATTTTGCATC-3′, and reverse, 5′-GGTGTTAAGAGCCACCCGTT-3; Nadsyn (PrimerBank ID: 26338135a1): forward, 5′-AATATGCGGCTATGGATGTTGG-3′, and reverse, 5′-GGAGCGAATGCAGGAGAGT-3.
Biochemical measurements
Hepatic triglyceride content was quantified using a commercial kit (KA0847, Abnova, Taipei, Taiwan). Triglycerides were extracted from 40 to 50 mg of pulverized tissue, added 350 μl of ethanolic KOH (2 parts ethanol and 1 part 30% KOH). Tissues were incubated overnight at 55 °C. Each sample was added 50% ethanol up to a final volume of 1 ml. Samples were centrifuged for 5 min at 13,000 × g. 200 μl was recovered and diluted 1:50 in water prior to measurement as per the manufacturer's instructions. Absorbance at 570 nm was measured using a Hidex Sense (Hidex, Finland). HDL/LDL plasma levels were determined by a kit (catalog no. ab65390, Abcam, Cambridge, UK) according to the manufacturer's instructions. Total plasma cholesterol levels were determined by a kit (CH200, Randox, Crumlin, UK).
Statistics
Data are presented as means ± S.D. Data sets were tested for equal variance using either F-test (for comparing two groups) or Brown-Forsythe test (for more than two groups). In cases of unequal variance, data were log-transformed prior to further analysis. Data were analyzed using unpaired t test (when comparing two groups), one-way analysis of variance (ANOVA, when comparing more than two groups) or two- or three-way ANOVAs (when comparing two or more groups subjected to two or more treatments) with Sidak's multiple comparison test post hoc. Data from the study with NR supplementation was analyzed by two, two-way ANOVAs; one analysis to determine genotype differences in response to the HFD, and one analysis to determine genotype differences in response to NR treatment. All statistical analyses were performed using SigmaPlot 13.0 (Systat Software Inc, San Jose, CA), Graphpad Prism 8 (Graphpad Software, San Diego), and IBM SPSS Statistics 25 (SPSS, Chicago). Statistical significance was set at p < 0.05. All figures were produced using Graphpad Prism 8.
Author contributions
M. D. and J. T. T. conceptualization; M. D., S. A. J. T., M. Asping, A. S. H., M. Agerholm, S. G. V., M. P. G., S. L., and J. T. T. data curation; M. D., S. A. J. T., A. S. H., M. Agerholm, and S. L. formal analysis; M. D., S. A. J. T., M. Asping, A. S. H., S. L., and J. T. T. validation; M. D., S. A. J. T., M. Asping, A. S. H., M. Agerholm, S. G. V., S. L., and J. T. T. investigation; M. D. visualization; M. D., S. A. J. T., M. Asping, M. Agerholm, S. G. V., M. P. G., and S. L. methodology; M. D. writing-original draft; M. D., A. S. H., M. P. G., and J. T. T. writing-review and editing; S. G. V., M. P. G., S. L., and J. T. T. supervision; J. T. T. resources; J. T. T. funding acquisition; J. T. T. project administration.
Acknowledgments
Novo Nordisk Foundation Center for Basic Metabolic Research is an independent Research Center at the University of Copenhagen and is partially funded by an unrestricted donation from the Novo Nordisk Foundation. 13C,2H-NR (33) was kindly provided by Prof. Marie Migaud (University of South Alabama) for use as chemical standard in mass spectrometry measurements, and mice expressing Cre recombinase under the albumin promoter were kindly provided by Prof. Johan Auwerx (École Polytechnique Fédérale, Lausanne, Switzerland). Profs. Zachary Gerhart-Hines and Birgitte Holst provided access to the Seahorse XF96 Extracellular Flux Analyzer. Regitze Kraunsøe is acknowledged for skilled technical assistance during the oxygraph analyses, and Signe Marie Borch Nielsen, Hanne Cathrine Bisgaard, and Sabina Chubanava provided valuable advice during primary mouse hepatocyte isolation and liver mitochondria isolation, respectively. Jeremie Boucher and AstraZeneca are acknowledged for providing skilled technical assistance in setting up the fatty-acid oxidation assay in primary hepatocytes. We acknowledge the generous donation of NR from Chromadex (Irvine, CA).
This work was supported in part by Novo Nordisk Foundation Excellence Project Award NNF14OC0009315, Danish Council for Independent Research Grant DFF 4004-00235) (to J. T. T.), and by the Novo Nordisk Foundation Center for Basic Metabolic Research. The authors declare that they have no conflicts of interest with the contents of this article.
- NAFLD
- nonalcoholic fatty liver disease
- NASH
- nonalcoholic steatohepatitis
- HNKO
- hepatocyte-specific knockout of Nampt
- LFD
- low-fat diet
- HFD
- high-fat diet
- NR
- nicotinamide riboside
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- TMRM
- tetramethylrhodamine
- NAM
- nicotinamide
- NA
- nicotinic acid
- NAMPT
- nicotinamide phosphoribosyltransferase
- NMNAT
- nicotinamide mononucleotide adenylyltransferase
- NAMN
- nicotinic acid mononucleotide
- NMN
- nicotinamide mononucleotide
- OXPHOS
- oxidative phosphorylation
- OGTT
- oral glucose tolerance test
- FBS
- fetal bovine serum
- P/S
- penicillin/streptomycin
- OCR
- oxygen consumption rate
- ECAR
- extracellular acidification rate
- MEMα
- α-minimal essential medium
- ssDNA
- single-stranded DNA
- ATGL
- adipose triglyceride lipase
- FASN
- fatty-acid synthase
- PAR
- poly(ADP)-ribose
- PARP
- poly(ADP)-ribose polymerase
- ANOVA
- analysis of variance
- QC
- quality control
- MeNAM-d3
- N1-methylnicotinamide-d3
- MnSOD
- Mn-superoxide dismutase
- HDL
- high-density lipoprotein
- LCAD
- long chain acyl CoA dehydrogenase
- MCAD
- medium chain acyl CoA dehydrogenase
- LDL
- low-density lipoprotein
- VLDL
- very-low-density lipoprotein
- HSL
- hormone sensitive lipase.
References
- 1. Begriche K., Massart J., Robin M. A., Bonnet F., and Fromenty B. (2013) Mitochondrial adaptations and dysfunctions in nonalcoholic fatty liver disease. Hepatology 58, 1497–1507 10.1002/hep.26226 [DOI] [PubMed] [Google Scholar]
- 2. Pessayre D., and Fromenty B. (2005) NASH: a mitochondrial disease. J. Hepatol. 42, 928–940 10.1016/j.jhep.2005.03.004 [DOI] [PubMed] [Google Scholar]
- 3. Pérez-Carreras M., Del Hoyo P., Martín M. A., Rubio J. C., Martín A., Castellano G., Colina F., Arenas J., and Solis-Herruzo J. A. (2003) Defective hepatic mitochondrial respiratory chain in patients with nonalcoholic steatohepatitis. Hepatology 38, 999–1007 10.1002/hep.1840380426 [DOI] [PubMed] [Google Scholar]
- 4. Sanyal A. J., Campbell-Sargent C., Mirshahi F., Rizzo W. B., Contos M. J., Sterling R. K., Luketic V. A., Shiffman M. L., and Clore J. N. (2001) Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology 120, 1183–1192 10.1053/gast.2001.23256 [DOI] [PubMed] [Google Scholar]
- 5. Choi S.-E., Fu T., Seok S., Kim D.-H., Yu E., Lee K.-W., Kang Y., Li X., Kemper B., and Kemper J. K. (2013) Elevated microRNA-34a in obesity reduces NAD+ levels and SIRT1 activity by directly targeting NAMPT. Aging Cell 12, 1062–1072 10.1111/acel.12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang Z.-F., Fan S.-H., Zheng Y.-L., Lu J., Wu D.-M., Shan Q., and Hu B. (2014) Troxerutin improves hepatic lipid homeostasis by restoring NAD+-depletion–mediated dysfunction of lipin 1 signaling in high-fat diet–treated mice. Biochem. Pharmacol. 91, 74–86 10.1016/j.bcp.2014.07.002 [DOI] [PubMed] [Google Scholar]
- 7. Gariani K., Menzies K. J., Ryu D., Wegner C. J., Wang X., Ropelle E. R., Moullan N., Zhang H., Perino A., Lemos V., Kim B., Park Y. K., Piersigilli A., Pham T. X., Yang Y., et al. (2016) Eliciting the mitochondrial unfolded protein response by nicotinamide adenine dinucleotide repletion reverses fatty liver disease in mice. Hepatology 63, 1190–1204 10.1002/hep.28245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou C. C., Yang X., Hua X., Liu J., Fan M. B., Li G. Q., Song J., Xu T. Y., Li Z. Y., Guan Y. F., Wang P., and Miao C. Y. (2016) Hepatic NAD+ deficiency as a therapeutic target for non-alcoholic fatty liver disease in ageing. Br. J. Pharmacol. 173, 2352–2368 10.1111/bph.13513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Uddin G. M., Youngson N. A., Sinclair D. A., and Morris M. J. (2016) Head to head comparison of short-term treatment with the NAD+ precursor nicotinamide mononucleotide (NMN) and 6 weeks of exercise in obese female mice. Front. Pharmacol. 7, 258 10.3389/fphar.2016.00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang X., Zhang Z.-F., Zheng G.-H., Wang A.-M., Sun C.-H., Qin S.-P., Zhuang J., Lu J., Ma D.-F., and Zheng Y.-L. (2017) The inhibitory effects of purple sweet potato color on hepatic inflammation is associated with restoration of NAD+ levels and attenuation of NLRP3 inflammasome activation in high-fat diet–treated mice. Molecules 22, 1315 10.3390/molecules22081315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khan N. A., Auranen M., Paetau I., Pirinen E., Euro L., Forsström S., Pasila L., Velagapudi V., Carroll C. J., Auwerx J., and Suomalainen A. (2014) Effective treatment of mitochondrial myopathy by nicotinamide riboside, a vitamin B3. EMBO Mol. Med. 6, 721–731 10.1002/emmm.201403943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cantó C., Houtkooper R. H., Pirinen E., Youn D. Y., Oosterveer M. H., Cen Y., Fernandez-Marcos P. J., Yamamoto H., Andreux P. A., Cettour-Rose P., Gademann K., Rinsch C., Schoonjans K., Sauve A. A., and Auwerx J. (2012) The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 15, 838–847 10.1016/j.cmet.2012.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang H., Ryu D., Wu Y., Gariani K., Wang X., Luan P., D'Amico D., Ropelle E. R., Lutolf M. P., Aebersold R., Schoonjans K., Menzies K. J., and Auwerx J. (2016) NAD+ repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352, 1436–1443 10.1126/science.aaf2693 [DOI] [PubMed] [Google Scholar]
- 14. Cerutti R., Pirinen E., Lamperti C., Marchet S., Sauve A. A., Li W., Leoni V., Schon E. A., Dantzer F., Auwerx J., Viscomi C., and Zeviani M. (2014) NAD(+)-dependent activation of Sirt1 corrects the phenotype in a mouse model of mitochondrial disease. Cell Metab. 19, 1042–1049 10.1016/j.cmet.2014.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Agerholm M., Dall M., Jensen B. A. H., Prats C., Madsen S., Basse A. L., Graae A.-S., Risis S., Goldenbaum J., Quistorff B., Larsen S., Vienberg S. G., and Treebak J. T. (2018) Perturbations of NAD+ salvage systems impact mitochondrial function and energy homeostasis in mouse myoblasts and intact skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 314, E377–E395 10.1152/ajpendo.00213.2017 [DOI] [PubMed] [Google Scholar]
- 16. Trammell S. A., Weidemann B. J., Chadda A., Yorek M. S., Holmes A., Coppey L. J., Obrosov A., Kardon R. H., Yorek M. A., and Brenner C. (2016) Nicotinamide riboside opposes type 2 diabetes and neuropathy in mice. Sci. Rep. 6, 26933 10.1038/srep26933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoshino J., Mills K. F., Yoon M. J., and Imai S. (2011) Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 14, 528–536 10.1016/j.cmet.2011.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DiMauro S., and Schon E. A. (2003) Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 348, 2656–2668 10.1056/NEJMra022567 [DOI] [PubMed] [Google Scholar]
- 19. Agledal L., Niere M., and Ziegler M. (2010) The phosphate makes a difference: cellular functions of NADP. Redox Rep. 15, 2–10 10.1179/174329210X12650506623122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bonkowski M. S., and Sinclair D. A. (2016) Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nat. Rev. Mol. Cell Biol. 17, 679–690 10.1038/nrm.2016.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Giblin W., Skinner M. E., and Lombard D. B. (2014) Sirtuins: guardians of mammalian health span. Trends Genet. 30, 271–286 10.1016/j.tig.2014.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Avalos J. L., Bever K. M., and Wolberger C. (2005) Mechanism of sirtuin inhibition by nicotinamide: altering the NAD+ cosubstrate specificity of a Sir2 enzyme. Mol. Cell 17, 855–868 10.1016/j.molcel.2005.02.022 [DOI] [PubMed] [Google Scholar]
- 23. Cantó C., Menzies K. J., and Auwerx J. (2015) NAD(+) Metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab. 22, 31–53 10.1016/j.cmet.2015.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mills K. F., Yoshida S., Stein L. R., Grozio A., Kubota S., Sasaki Y., Redpath P., Migaud M. E., Apte R. S., Uchida K., Yoshino J., and Imai S. (2016) Long-term administration of nicotinamide mononucleotide mitigates age-associated physiological decline in mice. Cell Metab. 24, 795–806 10.1016/j.cmet.2016.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Preiss J., and Handler P. (1958) Biosynthesis of diphosphopyridine nucleotide: I. Identification of intermediates. J. Biol. Chem. 233, 488–492 [PubMed] [Google Scholar]
- 26. Bender D. A. (1983) Biochemistry of tryptophan in health and disease. Mol. Aspects Med. 6, 101–197 10.1016/0098-2997(83)90005-5 [DOI] [PubMed] [Google Scholar]
- 27. J P, and Handler P. (1958) Biosynthesis of diphosphopyridine nucleotide. II. Enzymatic aspects. J. Biol. Chem. 233, 493–500 [PubMed] [Google Scholar]
- 28. Mori V., Amici A., Mazzola F., Di Stefano M., Conforti L., Magni G., Ruggieri S., Raffaelli N., and Orsomando G. (2014) Metabolic profiling of alternative NAD biosynthetic routes in mouse tissues. PLoS ONE 9, e113939 10.1371/journal.pone.0113939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ratajczak J., Joffraud M., Trammell S. A., Ras R., Canela N., Boutant M., Kulkarni S. S., Rodrigues M., Redpath P., Migaud M. E., Auwerx J., Yanes O., Brenner C., and Cantó C. (2016) NRK1 controls nicotinamide mononucleotide and nicotinamide riboside metabolism in mammalian cells. Nat. Commun. 7, 13103 10.1038/ncomms13103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ryu D., Zhang H., Ropelle E. R., Sorrentino V., Mázala D. A., Mouchiroud L., Marshall P. L., Campbell M. D., Ali A. S., Knowels G. M., Bellemin S., Iyer S. R., Wang X., Gariani K., Sauve A. A., et al. (2016) NAD+ repletion improves muscle function in muscular dystrophy and counters global PARylation. Sci. Transl. Med. 8, 361ra139 10.1126/scitranslmed.aaf5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dellinger R. W., Santos S. R., Morris M., Evans M., Alminana D., Guarente L., and Marcotulli E. (2017) Repeat dose NRPT (nicotinamide riboside and pterostilbene) increases NAD+ levels in humans safely and sustainably: a randomized, double-blind, placebo-controlled study. NPJ Aging Mech. Dis. 3, 17 10.1038/s41514-017-0016-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dollerup O. L., Christensen B., Svart M., Schmidt M. S., Sulek K., Ringgaard S., Stødkilde-Jørgensen H., Møller N., Brenner C., Treebak J. T., and Jessen N. (2018) A randomized placebo-controlled clinical trial of nicotinamide riboside in obese men: safety, insulin-sensitivity, and lipid-mobilizing effects. Am. J. Clin. Nutr. 108, 343–353 10.1093/ajcn/nqy132 [DOI] [PubMed] [Google Scholar]
- 33. Trammell S. A., Schmidt M. S., Weidemann B. J., Redpath P., Jaksch F., Dellinger R. W., Li Z., Abel E. D., Migaud M. E., and Brenner C. (2016) Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat. Commun. 7, 12948 10.1038/ncomms12948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martens C. R., Denman B. A., Mazzo M. R., Armstrong M. L., Reisdorph N., McQueen M. B., Chonchol M., and Seals D. R. (2018) Chronic nicotinamide riboside supplementation is well-tolerated and elevates NAD+ in healthy middle-aged and older adults. Nat. Commun. 9, 1286 10.1038/s41467-018-03421-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Airhart S. E., Shireman L. M., Risler L. J., Anderson G. D., Nagana Gowda G. A., Raftery D., Tian R., Shen D. D., and O'Brien K. D. (2017) An open-label, non-randomized study of the pharmacokinetics of the nutritional supplement nicotinamide riboside (NR) and its effects on blood NAD+ levels in healthy volunteers. PLoS ONE 12, e0186459 10.1371/journal.pone.0186459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang L.-F., Wang X.-N., Huang C.-C., Hu L., Xiao Y.-F., Guan X.-H., Qian Y.-S., Deng K.-Y., and Xin H.-B. (2017) Inhibition of NAMPT aggravates high fat diet-induced hepatic steatosis in mice through regulating Sirt1/AMPKα/SREBP1 signaling pathway. Lipids Health Dis. 16, 82 10.1186/s12944-017-0464-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu R., Yuan Z., Yang L., Li L., Li D., and Lv C. (2017) Inhibition of NAMPT decreases cell growth and enhances susceptibility to oxidative stress. Oncol. Rep. 38, 1767–1773 10.3892/or.2017.5793 [DOI] [PubMed] [Google Scholar]
- 38. Peek C. B., Affinati A. H., Ramsey K. M., Kuo H.-Y., Yu W., Sena L. A., Ilkayeva O., Marcheva B., Kobayashi Y., Omura C., Levine D. C., Bacsik D. J., Gius D., Newgard C. B., Goetzman E., Chandel N. S., et al. (2013) Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 342, 1243417 10.1126/science.1243417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mukherjee S., Chellappa K., Moffitt A., Ndungu J., Dellinger R. W., Davis J. G., Agarwal B., and Baur J. A. (2017) Nicotinamide adenine dinucleotide biosynthesis promotes liver regeneration. Hepatology 65, 616–630 10.1002/hep.28912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Penke M., Larsen P. S., Schuster S., Dall M., Jensen B. A., Gorski T., Meusel A., Richter S., Vienberg S. G., Treebak J. T., Kiess W., and Garten A. (2015) Hepatic NAD salvage pathway is enhanced in mice on a high-fat diet. Mol. Cell. Endocrinol. 412, 65–72 10.1016/j.mce.2015.05.028 [DOI] [PubMed] [Google Scholar]
- 41. Drew J. E., Farquharson A. J., Horgan G. W., and Williams L. M. (2016) Tissue-specific regulation of sirtuin and nicotinamide adenine dinucleotide biosynthetic pathways identified in C57Bl/6 mice in response to high-fat feeding. J. Nutr. Biochem. 37, 20–29 10.1016/j.jnutbio.2016.07.013 [DOI] [PubMed] [Google Scholar]
- 42. Dall M., Penke M., Sulek K., Matz-Soja M., Holst B., Garten A., Kiess W., and Treebak J. T. (2018) Hepatic NAD+ levels and NAMPT abundance are unaffected during prolonged high-fat diet consumption in C57BL/6JBomTac mice. Mol. Cell. Endocrinol. 473, 245–256 10.1016/j.mce.2018.01.025 [DOI] [PubMed] [Google Scholar]
- 43. Shi W., Hegeman M. A., van Dartel D. A. M., Tang J., Suarez M., Swarts H., van der Hee B., Arola L., and Keijer J. (2017) Effects of a wide range of dietary nicotinamide riboside (NR) concentrations on metabolic flexibility and white adipose tissue (WAT) of mice fed a mildly obesogenic diet. Mol. Nutr. Food Res. 61, 1600878 10.1002/mnfr.201600878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kalliokoski O., Teilmann A. C., Jacobsen K. R., Abelson K. S., and Hau J. (2014) The lonely mouse–single housing affects serotonergic signaling integrity measured by 8-OH-DPAT-induced hypothermia in male mice. PLoS ONE 9, e111065 10.1371/journal.pone.0111065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Larsen S., Nielsen J., Hansen C. N., Nielsen L. B., Wibrand F., Stride N., Schroder H. D., Boushel R., Helge J. W., Dela F., and Hey-Mogensen M. (2012) Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J. Physiol. 590, 3349–3360 10.1113/jphysiol.2012.230185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gutiérrez-Aguilar M., and Baines C. P. (2013) Physiological and pathological roles of mitochondrial SLC25 carriers. Biochem. J. 454, 371–386 10.1042/BJ20121753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hirschey M. D., Shimazu T., Jing E., Grueter C. A., Collins A. M., Aouizerat B., Stančáková A., Goetzman E., Lam M. M., Schwer B., Stevens R. D., Muehlbauer M. J., Kakar S., Bass N. M., Kuusisto J., et al. (2011) SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol. Cell 44, 177–190 10.1016/j.molcel.2011.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., and Mayo M. W. (2004) Modulation of NF-κB–dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23, 2369–2380 10.1038/sj.emboj.7600244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Du J., Zhou Y., Su X., Yu J. J., Khan S., Jiang H., Kim J., Woo J., Kim J. H., Choi B. H., He B., Chen W., Zhang S., Cerione R. A., Auwerx J., Hao Q., and Lin H. (2011) Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809 10.1126/science.1207861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Davila A., Liu L., Chellappa K., Redpath P., Nakamaru-Ogiso E., Paolella L. M., Zhang Z., Migaud M. E., Rabinowitz J. D., and Baur J. A. (2018) Nicotinamide adenine dinucleotide is transported into mammalian mitochondria. Elife 7, e33246 10.7554/eLife.33246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Narrod S. A., Bonavita V., Ehrenfeld E. R., and Kaplan N. O. (1961) Effect of azaserine on the biosynthesis of diphosphopyridine nucleotide in mouse. J. Biol. Chem. 236, 931–935 [PubMed] [Google Scholar]
- 52. Braidy N., Guillemin G. J., Mansour H., Chan-Ling T., Poljak A., and Grant R. (2011) Age related changes in NAD+ metabolism oxidative stress and Sirt1 activity in Wistar rats. PLoS ONE 6, e19194 10.1371/journal.pone.0019194 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53. Mouchiroud L., Houtkooper R. H., Moullan N., Katsyuba E., Ryu D., Cantó C., Mottis A., Jo Y. S., Viswanathan M., Schoonjans K., Guarente L., and Auwerx J. (2013) The NAD(+)/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell 154, 430–441 10.1016/j.cell.2013.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Toye A. A., Lippiat J. D., Proks P., Shimomura K., Bentley L., Hugill A., Mijat V., Goldsworthy M., Moir L., Haynes A., Quarterman J., Freeman H. C., Ashcroft F. M., and Cox R. D. (2005) A genetic and physiological study of impaired glucose homeostasis control in C57BL/6J mice. Diabetologia 48, 675–686 10.1007/s00125-005-1680-z [DOI] [PubMed] [Google Scholar]
- 55. Liu L., Su X., Quinn W. J. 3rd., Hui S., Krukenberg K., Frederick D. W., Redpath P., Zhan L., Chellappa K., White E., Migaud M., Mitchison T. J., Baur J. A., and Rabinowitz J. D. (2018) Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab. 27, 1067–1080.e5 10.1016/j.cmet.2018.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schuster S., Penke M., Gorski T., Gebhardt R., Weiss T. S., Kiess W., and Garten A. (2015) FK866-induced NAMPT inhibition activates AMPK and downregulates mTOR signaling in hepatocarcinoma cells. Biochem. Biophys. Res. Commun. 458, 334–340 10.1016/j.bbrc.2015.01.111 [DOI] [PubMed] [Google Scholar]
- 57. Giles D. A., Moreno-Fernandez M. E., Stankiewicz T. E., Graspeuntner S., Cappelletti M., Wu D., Mukherjee R., Chan C. C., Lawson M. J., Klarquist J., Sünderhauf A., Softic S., Kahn C. R., Stemmer K., Iwakura Y., et al. (2017) Thermoneutral housing exacerbates nonalcoholic fatty liver disease in mice and allows for sex-independent disease modeling. Nat. Med. 23, 829–838 10.1038/nm.4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brandauer J., Vienberg S. G., Andersen M. A., Ringholm S., Risis S., Larsen P. S., Kristensen J. M., Frøsig C., Leick L., Fentz J., Jørgensen S., Kiens B., Wojtaszewski J. F. P., Richter E. A., Zierath J. R., et al. (2013) AMP-activated protein kinase regulates nicotinamide phosphoribosyl transferase expression in skeletal muscle. J. Physiol. 591, 5207–5220 10.1113/jphysiol.2013.259515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gray L. R., Sultana M. R., Rauckhorst A. J., Oonthonpan L., Tompkins S. C., Sharma A., Fu X., Miao R., Pewa A. D., Brown K. S., Lane E. E., Dohlman A., Zepeda-Orozco D., Xie J., Rutter J., et al. (2015) Hepatic mitochondrial pyruvate carrier 1 is required for efficient regulation of gluconeogenesis and whole-body glucose homeostasis. Cell Metab. 22, 669–681 10.1016/j.cmet.2015.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tonkonogi M., and Sahlin K. (1997) Rate of oxidative phosphorylation in isolated mitochondria from human skeletal muscle: effect of training status. Acta Physiol. Scand. 161, 345–353 10.1046/j.1365-201X.1997.00222.x [DOI] [PubMed] [Google Scholar]
- 61. Lund M. T., Kristensen M., Hansen M., Tveskov L., Floyd A. K., Støckel M., Vainer B., Poulsen S. S., Helge J. W., Prats C., and Dela F. (2016) Hepatic mitochondrial oxidative phosphorylation is normal in obese patients with and without type 2 diabetes. J. Physiol. 594, 4351–4358 10.1113/JP272105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gandin V., Marchisio P. C., and Biffo S. (2008) Isolation of murine hepatocytes to measure protein synthesis ex vivo upon stimulation. Protoc. Exch. 10.1038/nprot.2008.209 [DOI] [Google Scholar]
- 63. Bahjat F. R., Dharnidharka V. R., Fukuzuka K., Morel L., Crawford J. M., Clare-Salzler M. J., and Moldawer L. L. (2000) Reduced susceptibility of nonobese diabetic mice to TNF-α and d-galactosamine-mediated hepatocellular apoptosis and lethality. J. Immunol. 165, 6559–6567 10.4049/jimmunol.165.11.6559 [DOI] [PubMed] [Google Scholar]
- 64. Pike L. S., Smift A. L., Croteau N. J., Ferrick D. A., and Wu M. (2011) Inhibition of fatty acid oxidation by etomoxir impairs NADPH production and increases reactive oxygen species resulting in ATP depletion and cell death in human glioblastoma cells. Biochim. Biophys. Acta 1807, 726–734 10.1016/j.bbabio.2010.10.022 [DOI] [PubMed] [Google Scholar]
- 65. Trammell S. A., and Brenner C. (2013) Targeted, LCMS-based metabolomics for quantitative measurement of NAD(+) metabolites. Comput. Struct. Biotechnol. J. 4, e201301012 10.5936/csbj.201301012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Midani F. S., Wynn M. L., and Schnell S. (2017) The importance of accurately correcting for the natural abundance of stable isotopes. Anal. Biochem. 520, 27–43 10.1016/j.ab.2016.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schuster S., Penke M., Gorski T., Petzold-Quinque S., Damm G., Gebhardt R., Kiess W., and Garten A. (2014) Resveratrol differentially regulates NAMPT and SIRT1 in hepatocarcinoma cells and primary human hepatocytes. PLoS ONE 9, e91045 10.1371/journal.pone.0091045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Elliott G. C., Ajioka J., and Okada C. Y. (1980) A rapid procedure for assaying nicotinamide phosphoribosyltransferase. Anal. Biochem. 107, 199–205 10.1016/0003-2697(80)90512-6 [DOI] [PubMed] [Google Scholar]
- 69. Nielsen K. N., Peics J., Ma T., Karavaeva I., Dall M., Chubanava S., Basse A. L., Dmytriyeva O., Treebak J. T., and Gerhart-Hines Z. (2018) NAMPT-mediated NAD+ biosynthesis is indispensable for adipose tissue plasticity and development of obesity. Mol. Metab. 11, 178–188 10.1016/j.molmet.2018.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Spandidos A., Wang X., Wang H., and Seed B. (2010) PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 38, D792–D799 10.1093/nar/gkp1005 [DOI] [PMC free article] [PubMed] [Google Scholar]