Abstract
Junctophilins (JPH1–JPH4) are expressed in excitable and nonexcitable cells, where they tether endoplasmic/sarcoplasmic reticulum (ER/SR) and plasma membranes (PM). These ER/SR–PM junctions bring Ca-release channels in the ER/SR and Ca as well as Ca-activated K channels in the PM to within 10–25 nm. Such proximity is critical for excitation–contraction coupling in muscles, Ca modulation of excitability in neurons, and Ca homeostasis in nonexcitable cells. JPHs are anchored in the ER/SR through the C-terminal transmembrane domain (TMD). Their N-terminal Membrane-Occupation-Recognition-Nexus (MORN) motifs can bind phospholipids. Whether MORN motifs alone are sufficient to stabilize JPH–PM binding is not clear. We investigate whether S-palmitoylation of cysteine (Cys), a critical mechanism controlling peripheral protein binding to PM, occurs in JPHs. We focus on JPH2 that has four Cys residues: three flanking the MORN motifs and one in the TMD. Using palmitate–alkyne labeling, Cu(I)-catalyzed alkyne-azide cycloaddition reaction with azide-conjugated biotin, immunoblotting, proximity–ligation–amplification, and various imaging techniques, we show that JPH2 is S-palmitoylatable, and palmitoylation is essential for its ER/SR–PM tether function. Palmitoylated JPH2 binds to lipid–raft domains in PM, whereas palmitoylation of TMD-located Cys stabilizes JPH2's anchor in the ER/SR membrane. Binding to lipid–raft domains protects JPH2 from depalmitoylation. Unpalmitoylated JPH2 is largely excluded from lipid rafts and loses the ability to form stable ER/SR–PM junctions. In adult ventricular myocytes, native JPH2 is S-palmitoylatable, and palmitoylated JPH2 forms distinct PM puncta. Sequence alignment reveals that the palmitoylatable Cys residues in JPH2 are conserved in other JPHs, suggesting that palmitoylation may also enhance ER/SR–PM tethering by these proteins.
Keywords: endoplasmic reticulum (ER), post-translational modification, lipid raft, plasma membrane, excitation–contraction coupling (E-C coupling), ER–PM junction, palmitoylation
Introduction
Endoplasmic reticulum–plasma membrane (ER–PM)2 junctions, also called junctional membrane complexes, are specialized cellular domains, where ER and PM membranes come very close to each other (10–25 nm apart), and critical cellular functions such as Ca release from ER, Ca entry through PM, lipid transfer, and phosphoinositide signaling take place (1). ER–PM junctions require protein tethers for their formation and maintenance. A growing list of ER–PM protein tethers has been identified: extended synaptotagmins (E-Syt1 to E-Syt3); vesicle-associated membrane protein–associated proteins A and B (VAP-A and VAP-B) (1); Kv2.1 (2), and junctophilins (JPHs) (3). Malfunctioning of ER–PM junctions can lead to human diseases (4). Understanding the molecular mechanisms for these ER–PM protein tethers, and how their function is regulated or modulated by cellular context and post-translational modifications, is important for identifying therapeutic targets to ameliorate these diseases.
The focus of this study is junctophilin-2 (JPH2), the major sarcoplasmic reticulum (SR)–PM tether in cardiac myocytes (3, 4). The JPH2 knockout mouse is embryonically lethal, and the myocytes manifest abnormalities in junctional membrane complexes and Ca transients (3). Acute JPH2 knockdown in adult mice leads to disruption of junctional membrane complexes and heart failure (5). Overexpressing JPH2 in mouse hearts can protect the hearts from failure in the presence of cardiac stress (6). In a canine model of chronic cardiac stress, down-regulation of JPH2 is a major factor for impairment in Ca-induced Ca release (CICR) (7). These observations support an important role played by JPH2 in maintaining SR–PM contacts and proper CICR.
Similar to other members of the junctophilin family, JPH2 is anchored in the ER/SR membrane by its C-terminal transmembrane domains (TMDs) and binds PM with its N-terminal Membrane Occupation and Recognition Nexus (MORN) motifs (MORN1–MORN8) (3). Given the essential role of JPH2–PM binding in its function as the major SR–PM tether in cardiac myocytes, are the MORN motifs alone sufficient to ensure stable JPH2–PM binding? Lipidation is a common strategy to stabilize binding of peripheral proteins to the phospholipid bilayer. There are three forms of protein lipidations (8), among which S-palmitoylation (attaching saturated long-acyl chains, most commonly palmitoyl or C16:0, to the thiol group of cysteine) provides the strongest stabilizing force for protein–PM binding (9). Furthermore, because of the labile nature of thioester bonds that can be cleaved by depalmitoylating enzymes (8, 10), S-palmitoylation is the only reversible form of lipidation, thus affording a dynamic control of protein–membrane association. Human JPH2 has four Cys residues: three of them flank the MORN region (Cys-15 and Cys-29 in MORN1 (aa 17–36), and Cys-328 in MORN8 (aa 314–336)), and the fourth one (Cys-678) is at the beginning of the C-terminal TMD (aa 675–695). In this study, we addressed the following questions. 1) Can JPH2 be S-palmitoylated? 2) If so, which Cys side chain(s) is(are) involved? 3) What is the subcellular distribution pattern of palmitoylated JPH2? 4) What are the functional consequences of JPH2 palmitoylation? We used heterologous expression of JPH2 in COS-7 cells to validate experimental approaches. We then applied these approaches to study native JPH2 in adult ventricular myocytes.
Results
JPH2 is S-palmitoylatable and all four cysteine side chains are involved
We expressed JPH2 in COS-7 cells and used metabolic labeling with palmitate–alkyne, followed by Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAC) reaction with azide-conjugated biotin to biotinylate palmitoylated proteins. Specificity in the detection of palmitoylated (i.e. biotinylated) JPH2 was achieved by JPH2 immunoprecipitation followed by immunoblot experiments and by proximity ligation amplification (PLA (11)) followed by imaging experiments. These experimental procedures are presented in Fig. S1. The degree of metabolic labeling depends on the kinetics of de- and re-palmitoylation cycles during palmitate–alkyne incubation. In the following experiments we incubated cells with palmitate–alkyne at 100 μm for 12 h (except where noted), to ensure a sufficient level of metabolic labeling of JPH2 for detection. We detected a strong biotinylation signal in the CuAAC-reacted JPH2 immunoprecipitate (Fig. 1A, right, IP (CuAAC)). This signal was absent in the WCL lane (not reacted with biotin-azide), despite the much stronger JPH2 band intensity in WCL than IP lanes (Fig. 1A, left). Similar findings were obtained in five independent experiments, indicating that JPH2 is palmitoylatable.
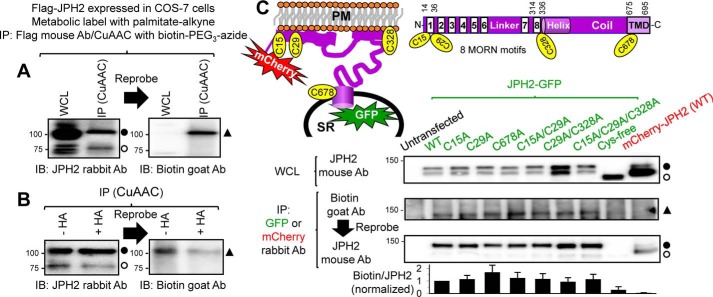
Figure 1.
JPH2 can be S-palmitoylated, and all four Cys side chains are involved. A and B, FLAG–JPH2 expressed in COS-7 cells was labeled with palmitate–alkyne and immunoprecipitated (IP) from WCL with FLAG mouse Ab and protein A/G magnetic beads. Protein-bound beads were reacted with biotin–PEG3–azide in CuAAC. WCL and CuAAC-reacted immunoprecipitate were analyzed by sequential immunoblot (IB), first with JPH2 rabbit Ab and then with biotin goat Ab. A, FLAG–JPH2 migrated as 75- and 100-kDa bands in both WCL and IP lanes. Biotinylation (i.e. palmitoylation) signal was detected at 100-kDa position in CuAAC-reacted IP but not in WCL. The experiment shown in B was similar to A, except that CuAAC-reacted beads were divided into two halves: one treated with neutral hydroxylamine (+HA, cleaving thioester bonds), and the other treated with Tris (−HA, preserving thioester bonds). Densitometry quantification shows that HA treatment reduced palmitoylation signal (biotin band intensity divided by JPH2 band intensity) by 62%, indicating that JPH2 was S-palmitoylated. C, top right, domain structure of JPH2, showing eight MORN motifs at the N terminus, followed by helical and coil domains, and a TMD at the C terminus. Domains are not drawn to the scale. The locations of four Cys side chains are noted. Top left, JPH2 topology, depicting SR, PM, JPH2 domains, including four Cys side chains, and fluorescent protein reporters (mCherry or GFP, fused to the N and C termini, respectively). Cys to Ala mutations in the JPH2–GFP background are noted as C#A, where # is position number, except when all four Cys side chains were mutated (Cys-free). JPH2–GFP, WT or mutants, and mCherry–JPH2 were expressed in COS-7 cells, labeled with palmitate–alkyne, and immunoprecipitated with GFP or mCherry rabbit Ab. Untransfected COS-7 cells served as negative control for IP with GFP Ab. First row: whole-cell lysates probed with JPH2 mouse Ab. Second and third rows: CuAAC-reacted immunoprecipitates analyzed by sequential immunoblot with biotin goat Ab, and then JPH2 mouse Ab. Fourth row: histogram of palmitoylation signal (biotin band intensity divided by JPH2 band intensity) normalized to that of JPH2–GFP WT. Data are pooled from three to six independent experiments. In this figure and Figs. 3B, 4B, and 7B, Reprobe and arrow between adjacent immunoblot images indicate that the PVDF membrane was probed with the first Ab, stripped, and reprobed with the second Ab. The open and closed circles next to JPH2 immunoblot images denote the expected JPH2 band based on its molecular weight and the putative palmitoylated JPH2 band (increase in molecular mass by 25 kDa). The closed triangles next to biotin Ab immunoblot images (or streptavidin ECL image in Fig. 7B) denote the band of palmitoylated JPH2 biotinylated through the CuAAC reaction.
Palmitoyl chain can be attached to the amine group of an N-terminal glycine by an amide bond (N-palmitoylation), hydroxyl group of serine by an ester bond (O-palmitoylation), or thiol group of Cys by a thioester bond (S-palmitoylation). Among the three, only S-palmitoylation is reversible, allowing a dynamic control of protein palmitoylation. To check whether palmitoylation of JPH2 occurs at Cys side chain(s), we used neutral hydroxylamine (HA) to cleave thioester bonds. Fig. 1B shows that the JPH2 biotinylation signal was markedly reduced if CuAAC-reacted IP were incubated with HA, indicating that JPH2 was S-palmitoylated.
JPH2 has four Cys side chains (Fig. 1C, top). To check which one(s) may be involved in S-palmitoylation, we mutated these Cys residues to Ala, one at a time, two or three in combination, and eventually all four. These were done in a JPH2–GFP background. Fig. 1C shows that WT JPH2–GFP and mutants with up to three Cys side chains replaced by Ala could all generate biotinylation signals above that of background (in the untransfected lane). Removing all four Cys (Cys-free JPH2–GFP) reduced the biotinylation signal to a level similar to background, suggesting that all four Cys side chains in JPH2 are involved in S-palmitoylation. However, there is no quantitative relationship between the intensity of palmitoylation signal and the number of Cys side chains present in JPH2–GFP. This will be discussed (see “Discussion”).
The size of FLAG–JPH2 is 75 kDa. However, it migrated as 75- and 100-kDa bands in SDS-PAGE (Fig. 1, A and B, WCL and IP lanes probed with JPH2 Ab). The palmitoylation signal of FLAG–JPH2 was exclusively detected in the 100-kDa band (IP lanes probed with biotin Ab). Similarly, the size of JPH2–GFP is 100 kDa, but it migrated as 100- and 125-kDa bands, and the palmitoylation signal was exclusively detected in the 125-kDa band (Fig. 1C). This apparent increase in the size of palmitoylated JPH2 was a consistent finding (see below). Its possible causes will be presented under the “Discussion.”
Palmitoylated JPH2 is distributed as distinct puncta close to the plasma membrane
We used Palm–PLA (11) to monitor the subcellular distribution pattern of palmitoylated JPH2 (palm*–JPH2). Validation of the Palm–PLA protocol is presented in Fig. S2. In the same experiments, we also monitored the distribution pattern of JPH2 by immunofluorescence, JPH2 (IF), and the ER morphology by coexpressing a fluorescently labeled ER marker (dsRed2–ER-5). Fig. 2 depicts images of palm*–JPH2, JPH2 (IF), and ER and their merges obtained using three imaging modalities: confocal (diffraction-limited); total internal reflection fluorescence (TIRF, detecting signals <200 nm from the areas of plasma membrane in contact with coverslip); and structured illumination (doubling XY plane resolution) microscopy.
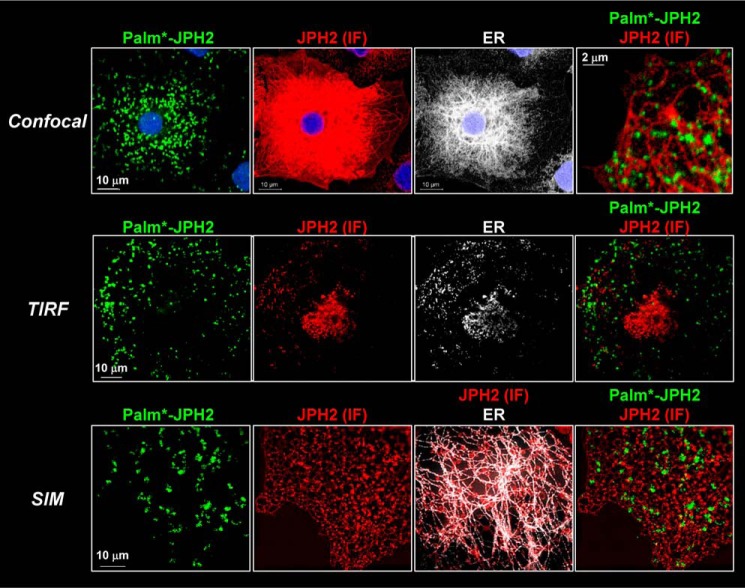
Figure 2.
palm*–JPH2 forms distinct puncta close to the plasma membrane, although the majority of JPH2 is in cytoplasmic ER. COS-7 cells expressing FLAG–JPH2 and ER markers (dsRed2–ER-5) were subject to metabolic labeling with palmitate–alkyne, CuAAC reaction with biotin–PEG3–azide, Palm–PLA, IF labeling of JPH2, and viewed by confocal, TIRF, and SIM. Protocol for Palm–PLA and its validation are presented in Figs. S1 and S2, respectively. In all cases, palm*–JPH2 was detected by green fluorophore, and JPH2 (IF) and ER marker were pseudo-colored red and white, respectively.
In confocal images, palm*–JPH2 manifested as distinct puncta, whereas JPH2 (IF) displayed an ER morphology overlapping with the ER marker. The differences in their distribution patterns are best seen in the merged view of palm*–JPH2 and JPH2 (IF). TIRF imaging indicated that the bright, distinct puncta of palm*–JPH2 were within the evanescent field (i.e. close to the cell surface). However, the signals of JPH2 (IF) were largely missing in the TIRF view, similar to the signals of the ER marker. This indicates that the majority of JPH2 (IF) was in the cytoplasmic ER away from the cell surface. Some TIRF images showed JPH2 (IF) as small and sparse puncta that did not overlap with the larger and more abundant palm*–JPH2 puncta, as is seen in the merged TIRF view of palm*–JPH2 and JPH2 (IF). The distinctly different patterns of palm*–JPH2 and JPH2 (IF) were confirmed in structured illumination images. The higher resolution with structured illumination enabled the detection of puncta in JPH2 (IF) signals. Again, they showed little overlap with the puncta of palm*–JPH2. Together, these observations indicate that although the majority of JPH2 stayed in the cytoplasmic ER, palmitoylated JPH2 preferentially bound to the plasma membrane in distinct puncta. There were small and sparse JPH2 (IF) puncta close to the cell surface, but they appeared to bind to different membrane domains from those of palm*–JPH2.
Fig. 1C shows that the biotinylation signal of mCherry–JPH2 was at the background level despite the presence of four Cys side chains, suggesting that mCherry–JPH2 was under-palmitoylated. This was corroborated by Palm–PLA data in Fig. S3: Palm–PLA applied to mCherry–JPH2 did not produce signals of palm*–JPH2, although the clear mCherry fluorescence and JPH2 immunofluorescence confirmed strong mCherry–JPH2 expression. Furthermore, the same reagents applied to FLAG–JPH2 in a parallel experiment detected prominent palm*–JPH2 signals, ruling out the possibility of experimental errors.
JPH2 in lipid–raft domains is enriched with palmitoylated form
It has been shown that palmitoylation promotes binding of many peripheral proteins to lipid–raft domains in the cell membrane (9). We performed two sets of experiments to test whether this is the case for JPH2. First, we compared the degrees of palmitoylation of JPH2 in raft versus nonraft domains. We used a detergent-free procedure (12) to purify raft and nonraft components from COS-7 cells expressing FLAG–JPH2 labeled with palmitate–alkyne (Fig. 3A). FLAG–JPH2 was immunoprecipitated from these two components, reacted with biotin-PEG3-azide, and analyzed by sequential immunoblots, first with JPH2 and then with biotin Abs (Fig. 3B). The degree of FLAG–JPH2 palmitoylation was quantified by the ratio of the biotin band intensity to the palmitoylated JPH2 (100 kDa) band intensity. Fig. 3C shows that the degree of FLAG–JPH2 palmitoylation in the raft component is four times that in the nonraft component, indicating that palmitoylated JPH2 preferentially clustered to lipid rafts of the cell membrane.
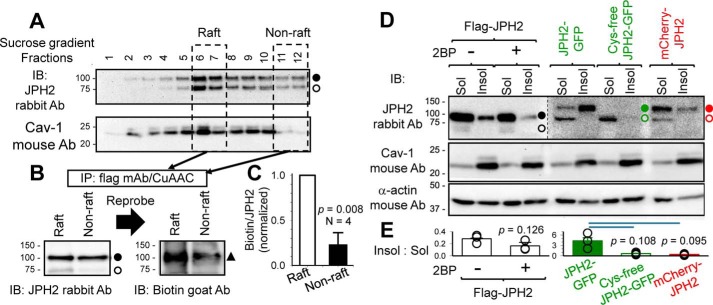
Figure 3.
JPH2 in lipid–raft domain is enriched with palmitoylated form, and inhibiting JPH2 palmitoylation reduces its presence in the lipid–raft domains. A, raft and nonraft components of cell membranes from COS-7 cells expressing FLAG–JPH2 labeled with palmitate–alkyne were separated by detergent-free purification/sucrose-gradient fractionation. Fractions were analyzed by immunoblots (IB) with JPH2 and caveolin-1 (Cav-1, raft marker) Abs. Fractions 6 and 7 with high Cav-1 contents were combined as “Raft” component, and fractions 11 and 12 with the lowest Cav-1 contents were combined as “Non-raft” component. Both components were solubilized, and FLAG–JPH2 was immunoprecipitated and reacted with biotin–PEG3–azide in the CuAAC reaction. B, immunoblot images of CuAAC reacted IPs from raft and nonraft components probed with JPH2 Ab and then biotin Ab. C, degree of JPH2 palmitoylation was quantified by dividing the biotinylation band intensity (100 kDa) by the total JPH2 band intensity at 100 kDa. D, Triton-soluble and -insoluble fractions (Sol and Insol) were isolated from COS-7 cells expressing FLAG–JPH2 (without or with 2BP pretreatment), WT, and Cys-free JPH2–GFP or mCherry–JPH2, and analyzed by immunoblot with JPH2, Cav-1, and α-actin Abs (the latter as loading control). FLAG–JPH2 migrated as 100- and 75-kDa bands in Sol lanes (closed and open circles), but as a single 100-kDa band in Insol lanes. JPH2–GFP and mCherry–JPH2 migrated as 125- and 100-kDa bands in Sol lanes, but as single 125 kDa in Insol lanes. Cys-free JPH2–GFP migrated as a single 100-kDa band in the Sol lane and was barely detectable in the Insol lane. Image of the right six lanes from the same membrane is shown in high contrast to better illustrate the differential banding pattern described above. E, bar plot of distribution of JPH2 variants between Insol and Sol components, estimated by dividing the JPH2 band intensity in the Insol lane by the combined JPH2 band intensities in the Sol lane.
Second, we monitored the effect of reducing or preventing JPH2 palmitoylation on its distribution between raft and nonraft domains. In this case, we separated the two domains as Triton-insoluble and Triton-soluble fractions (Insol and Sol, respectively) (13). The average ratio of band intensity of caveolin-1 (Cav-1, lipid raft marker) in the Insol lane to the corresponding Sol lane was 7.5 ± 0.6 (Fig. 3D, middle panel, Cav-1 immunoblot image), confirming the separation. Fig. 3D, top panel (JPH2 immunoblot), shows that FLAG–JPH2 migrated as 100- and 75-kDa bands in the Sol lanes but as a single 100-kDa band in the Insol lanes. JPH2–GFP and mCherry–JPH2 migrated as 100- and 125-kDa bands in the Sol lanes but as a single 125-kDa band in the Insol lanes. The banding patterns in the Insol lanes are consistent with those of palmitoylated JPH2 (Fig. 1). Inhibiting palmitoylation by incubating cells expressing FLAG–JPH2 with 2-bromopalmitate (2BP, simultaneously with palmitate–alkyne, both at 100 μm, overnight) (14) or interfering with palmitoylation in mCherry–JPH2 shifted JPH2 from Triton-insoluble to Triton-soluble fractions. This is quantified by the reduction of the Insol/Sol JPH2 band intensity ratio (Fig. 3E). The most dramatic case is Cys-free JPH2–GFP, which migrated as a single 100-kDa band (unpalmitoylated form) in the Sol lane and was barely detectable in the Insol lane. Together, these data show that palmitoylation promotes JPH2 binding to lipid rafts in the cell membrane, whereas unpalmitoylated JPH2 is largely excluded from the raft domain.
Residence in lipid–raft domains sustains the palmitoylation status of JPH2
If palmitoylated JPH2 preferentially binds to lipid rafts of the cell membrane, does disrupting lipid rafts simply disperse the pool of palmitoylated JPH2, or does it induce JPH2 depalmitoylation? We incubated cells with methoxy-β-cyclodextrin (MβCD, 2 mm at 36 °C for 2 h) to extract cholesterol from the cell membrane and disrupt cholesterol-rich lipid–raft domains. Fig. S4 confirmed the effectiveness of MβCD treatment in two tests. First, signals of a membrane lipid environment–sensitive dye, Nile Red 12S (15), indicated that the liquid-order phase of cell membrane was reduced after MβCD treatment. Second, there was a dramatic change in the distribution pattern of the lipid-raft marker, cholera toxin subunit B (15), suggesting a disruption of lipid rafts after MβCD treatment.
We used Palm–PLA to monitor the distribution pattern of palmitoylated JPH2 in conjunction with JPH2 immunofluorescence. Fig. 4A, top, depicts confocal images focused on the footprint of COS-7 cells. MβCD treatment greatly reduced the number of palm*–JPH2 puncta, without reducing the JPH2 (IF) signal intensity. The ratio of palm*–JPH2/JPH2 (IF) was dramatically reduced by MβCD treatment (Fig. 4A, bottom, from 1.0 ± 0.08 to 0.17 ± 0.02, p < 0.001). Fig. 4B, top, shows that MβCD treatment markedly reduced the palmitoylation signal in CuAAC-reacted IP (biotin Ab immunoblot), without altering the JPH2 band intensities in either WCL or IP (JPH2 Ab immunoblot). Data summarized from three independent experiments (Fig. 4B, bottom) shows that MβCD treatment caused ∼50% decrease in JPH2 palmitoylation. These observations indicate that disrupting lipid rafts dislodged palm*–JPH2 without altering the total JPH2 pool. Furthermore, the degree of JPH2 palmitoylation was reduced by MβCD treatment, suggesting that JPH2 dislodged from lipid rafts was prone to de-palmitoylation.
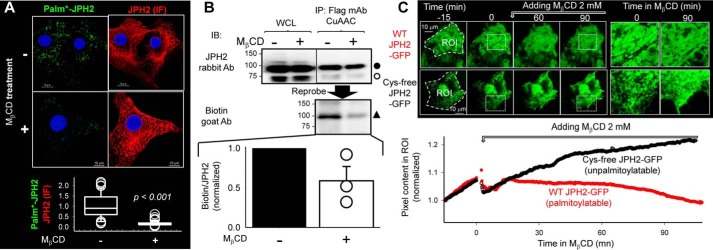
Figure 4.
Disrupting cholesterol-rich lipid rafts in the plasma membrane by extracting cholesterol with MβCD reduces JPH2 palmitoylation and shrinks the cortical ER compartment. A and B, COS-7 cells expressing FLAG–JPH2 were incubated with palmitate–alkyne overnight and then treated with MβCD (2 mm, 36 °C, 2 h) or not before experiments. A depicts distribution of palmitoylated JPH2 by Palm–PLA. Top: representative fluorescence images of palm*–JPH2 and JPH2 IF in cells without or with MβCD treatment. Bottom, data summary. Degree of JPH2 palmitoylation was quantified by ratio of palm*–JPH2 to JPH2 (IF). Data were pooled from three experiments, t test: p < 0.001. B depicts quantification of JPH2 palmitoylation by immunoblotting (IB) of immunoprecipitated and CuAAC-reacted JPH2. Top, immunoblot images of FLAG–JPH2 in WCL (not CuAAC-reacted) and IP (CuAAC-reacted), probed first with JPH2 Ab (100 and 75 kDa, closed and open circles), and then with biotin Ab (100 kDa, closed triangle). Bottom, densitometry summary from three experiments. Degree of JPH2 palmitoylation was quantified by ratio of biotin band intensity to the 100-kDa JPH2 band intensity (biotin/JPH2). A and B together show that MβCD treatment markedly reduced the degree of JPH2 palmitoylation without affecting the total JPH2 protein level. C, direct observation of effect of MβCD on juxtamembrane WT JPH2–GFP and Cys-free JPH2–GFP using live cell TIRF imaging. Top left: selected images before and at 60 and 90 min after MβCD application. ROI indicates the regions of interest where pixel contents were measured. Top right, enlarged views of rectangle areas in the left panels marked by white dotted lines. Bottom, time courses of changes in ROI pixel contents normalized by signals at the beginning of imaging.
To directly observe the effects of MβCD treatment on the juxtamembrane JPH2 pool, we applied TIRF imaging to live COS-7 cells expressing WT (palmitoylatable) or Cys-free (unpalmitoylatable) JPH2–GFP (Fig. 4C). The GFP signals reported juxtamembrane JPH2 pool in the cortical ER compartment, which was stabilized by ER–PM junctions. During TIRF imaging, we observed a gradual increase in the GFP signals, similar to the spontaneous formation of ER–PM junctions in Kv2.1-expressing cells during laser-scanning imaging (2). In the case of WT JPH2–GFP, this upward trend of GFP signals stopped after MβCD application followed by a gradual decline. For Cys-free JPH2–GFP, the upward trend continued for >90 min after MβCD application. To check whether the difference between WT and Cys-free JPH2–GFP was due to the mutations or due to the difference in palmitoylation, we compared WT JPH2–GFP without or with 2BP pretreatment. Fig. S5 shows that inhibiting WT JPH2–GFP palmitoylation by 2BP also prevented the decline of juxtamembrane GFP signals after MβCD application. These observations show that disrupting lipid rafts dislodged palmitoylated JPH2 from the raft domains and decreased the cortical ER compartment. In contrast, unpalmitoylated JPH2–GFP was not affected by lipid–raft disruption, suggesting that it might bind to nonraft domains of the cell membrane.
JPH2 palmitoylation stabilizes ER–PM junctions
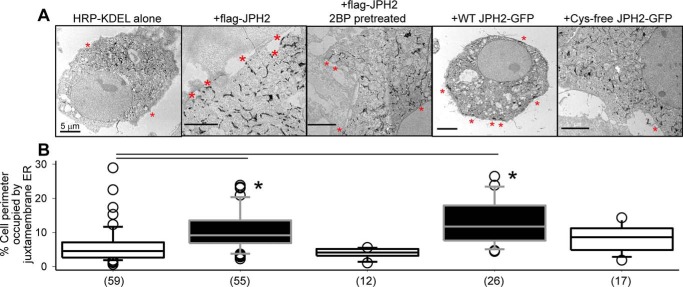
TIRF imaging showed that juxtamembrane WT JPH2–GFP often distributed as large elongated patches of 5–10 μm in long axis and 2–3 μm in short axis, whereas juxtamembrane Cys-free JPH2–GFP manifested a combination of ER network and small patches (Fig. 4C, top right, enlarged view). These observations suggest that palmitoylated JPH2 could stabilize ER–PM junctions, so that more ER elements stayed close to the PM enlarging the cortical ER compartment. To directly test whether this was the case, we used transmission EM (TEM) to monitor the distribution of ER elements relative to the plasma membrane. ER was marked by an electron-dense reaction product in the ER lumen from coexpressed HRP–KDEL (16). Labeled ER elements in transfected cells could be unequivocally identified in TEM images (Fig. 5A). ER at the ER–PM junction (juxtamembrane ER) was defined as ER elements abutting on the PM (Fig. 5A, red asterisks) (16). The degree of abundance in ER–PM junctions was quantified by the percentage of cell perimeter occupied by juxtamembrane ER (16).
Figure 5.
Palmitoylation of JPH2 stabilizes ER–PM junctions assessed by quantifying juxtamembrane ER elements using transmission EM. COS-7 cells were transfected with horseradish peroxidase (HRP)-conjugated-KDEL, alone or with specified JPH2 variants. In the case of FLAG–JPH2, cells were incubated under the control conditions or in the presence of 2BP (100 μm) overnight before experiment. The ER lumen was marked by amplification of HRP–KDEL followed by peroxidase reaction with DAB in 0.01% H2O2 (16). A, representative TEM images. Red asterisks mark juxtamembrane ER. All scale bars refer to 5 μm. B, summary of percent cell perimeter occupied by juxtamembrane ER. Data were pooled from three independent experiments; numbers of cells analyzed are listed in parentheses. One-way ANOVA of all five groups, p < 0.001, followed by Dunn's tests versus HRP–KDEL alone; *, p < 0.05. Both FLAG–JPH2 and JPH2–GFP increased juxtamembrane ER elements. Inhibiting FLAG–JPH2 palmitoylation by 2BP or replacing all four Cys side chains with Ala in Cys-free JPH2–GFP nullified this effect.
In cells expressing HRP–KDEL alone, native ER–PM tether proteins in COS-7 cells maintained the ER–PM junctions. Coexpression with FLAG–JPH2 or JPH2–GFP significantly increased the abundance of ER–PM junctions. Inhibiting FLAG–JPH2 palmitoylation by 2BP pretreatment or preventing JPH2–GFP palmitoylation in the Cys-free version reduced the abundance of juxtamembrane ER, to a degree not different from that in cells expressing HRP–KDEL alone. These TEM data corroborated live-cell TIRF data. Importantly, they indicate that palmitoylation of JPH2 was crucial for its ability to stabilize ER–PM junctions. Preventing JPH2 palmitoylation, despite the presence of intact phospholipid-binding MORN motifs, abolished its ability to stabilize ER–PM junctions.
Palmitoylation of Cys-678 stabilizes JPH2 anchor in the ER membrane
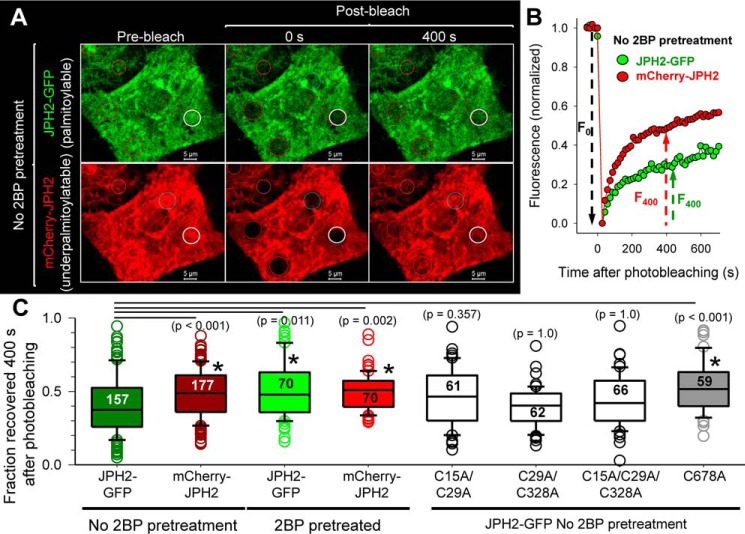
ER membrane proteins can move laterally in the ER network. Their mobility within the ER network can be assessed by fluorescence recovery after photobleaching (FRAP) (17), where fluorescence in a photobleached area recovers due to random motions of fluorescent molecules moving into the bleached area. We coexpressed JPH2–GFP (palmitoylatable) and mCherry–JPH2 (under-palmitoylated) in COS-7 cells and monitored their fluorescence recovery in small circular areas (∼5 μm in diameter) in the ER network where fluorescence had been bleached to 10–30% of pre-bleach levels. Fig. 6A depicts representative fluorescence images. The time courses of fluorescence recovery in the areas specified by white circles are shown in Fig. 6B. Fluorescence recovery for both JPH2–GFP and mCherry–JPH2 followed a bi-exponential time course. To simplify comparison, we used the fraction of fluorescence recovered 400 s after photobleach as a measure of JPH2 mobility. The recovery of JPH2–GFP signal was slower than that of mCherry–JPH2 (Fig. 6, B and C). Inhibiting protein palmitoylation by 2BP pretreatment significantly accelerated JPH2–GFP mobility, without changing the mobility of mCherry–JPH2. As a result, there was no difference in their mobility after 2BP treatment (Fig. 6C). These data indicate that JPH2 palmitoylation slowed its lateral mobility in the ER membrane.
Figure 6.
Palmitoylation of JPH2 at Cys-678 slows its lateral mobility in the ER network assessed by FRAP experiments. A, representative fluorescence images of coexpressed JPH2–GFP (palmitoylatable) and mCherry–JPH2 (under-palmitoylatable) before and at 0 and 400 s after photobleaching of small circular areas (marked by circles). White circles denote the areas where the recovery time courses are plotted in B. B, time courses of fluorescence recovery, with signals normalized so that prebleach level equals 1, and first scan after bleach equals 0. The fraction of fluorescence recovered at 400 s after photobleaching (F400/F0) is used for quantification as shown in C. C, box plots of summary data. Data were pooled from four independent experiments. Numbers of ROIs analyzed are listed inside the boxes. One-way ANOVA, p < 0.001, followed by Dunn's tests against “JPH2–GFP without 2BP pretreatment.” All p values are listed in parentheses, with p < 0.05 marked with *. JPH2 mobility in the ER was accelerated by reducing palmitoylation: 2BP pretreatment of JPH2–GFP or fusing mCherry to the N terminus. In the case of mCherry–JPH2, 2BP pretreatment did not cause a further increase in its mobility in the ER. Replacing Cys-678 by Ala accelerated JPH2–GFP mobility to that of JPH2–GFP after 2BP pretreatment or mCherry–JPH2. However, double or triple mutations of the other three Cys side chains did not increase JPH2–GFP mobility.
To test which Cys was responsible for this effect, we compared the mobilities of Cys–to–Ala mutants. Preventing Cys flanking the MORN motifs from palmitoylation (C15A/C29A, C29A/C328A, and C15A/C29A/C328A) did not accelerate JPH2 mobility. However, the single C678A mutation that prevented palmitoylation at the ER TMD significantly accelerated JPH2 mobility, to the same level as WT JPH2–GFP after 2BP pretreatment or mCherry–JPH2 (Fig. 6C).
Native JPH2 in ventricular myocytes is concentrated in the lipid–raft domain in palmitoylated state
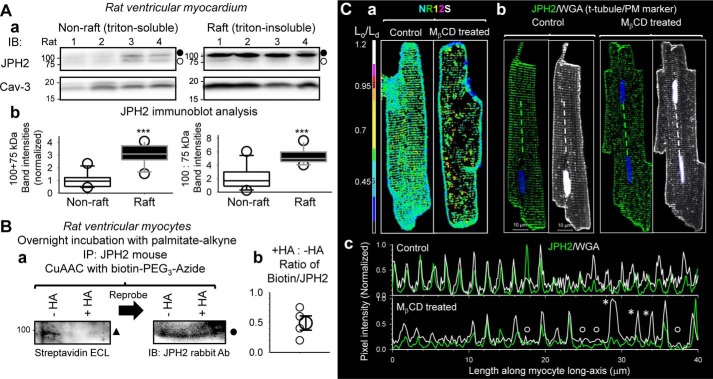
We used rat heart as our model. Rat JPH2 has three Cys residues, corresponding to Cys-15, Cys-29, and Cys-678 in human JPH2 (sequence alignment in supporting information). We separated Triton-soluble and -insoluble fractions from left ventricles of 11 animals, and we used immunoblots to quantify JPH2 protein levels and banding pattern in the two fractions. The latter was motivated by the distinctly different banding pattern of palmitoylated versus unpalmitoylated JPH2 in COS-7 experiments (Figs. 1, 3, and 4). Rat JPH2 is 74 kDa in size. We expected to see palmitoylated and unpalmitoylated rat JPH2 migrated as 74- and 99-kDa bands, respectively. Immunoblot images from 4 of the 11 animals are shown in Fig. 7A, panel a. Immunoblot with caveolin-3 Ab (Cav-3, lipid–raft marker) confirmed the enrichment of the lipid–raft domain in the Triton-insoluble fraction.
Figure 7.
Native JPH2 in rat ventricular myocytes is concentrated in lipid–raft domains in palmitoylated form and stabilizes dyads. A, panel a, immunoblot images of JPH2 and caveolin-3 (Cav-3) in nonraft and raft domains (Triton-soluble and -insoluble, respectively) isolated from left ventricular myocardium of rats. Loading was 45 μg/lane. The two fractions were run on separate SDS-polyacrylamide gels. After the proteins were transferred to PVDF membranes, for each of the immunoblots, the two membranes were incubated with the same Ab solutions and subject to ECL reaction/imaging side–by–side to allow quantitative comparison of immunoreactive band intensities between the nonraft and raft domains. Immunoblot of Cav-3 (raft marker) confirms enrichment of raft domains in Triton-insoluble fraction. Filled and open circles next to the JPH2 immunoblots denote putative palmitoylated and unpalmitoylated forms. Panel b, box plots of densitometry analysis of JPH2 immunoblot. Left, combined 100- and 75-kDa band intensities normalized by the mean value of nonraft lanes. Right, ratio of 100:75-kDa band intensities in nonraft and raft domains. t test, nonraft versus raft, ***, p < 0.001. B, testing whether native JPH2 in rat ventricular myocytes is S-palmitoylatable. The experimental procedures and graph format are similar to those of Fig. 1B. Native JPH2 was immunoprecipitated from WCL with JPH2 mouse Ab. On-bead CuAAC reaction linked biotin–PEG3–azide to palmitate–alkyne. Beads were divided into two halves, one reacted with hydroxylamine (+HA) to cleave thioester bonds, and the other incubated with Tris as control (−HA). Proteins were eluted from beads, fractionated by SDS-PAGE, transferred to PVDF membrane, incubated with JPH2 rabbit Ab, and analyzed by sequential ECL reactions as shown in panel a, HRP-conjugated streptavidin (left, to detect biotinylated proteins) and, after stripping, HRP-conjugated secondary Ab targeting rabbit Ab (right, to detect JPH2 bands). Solid triangle and circle denote the 100-kDa palmitoylated JPH2 bands. The PVDF membrane was cut above the 75-kDa size marker because of a strong biotin-positive band at 75-kDa position unrelated to palmitoylated JPH2 (seen in WCL that was not reacted with biotin-azide). Panel b, scatter plot of densitometry analysis. The degree of JPH2 palmitoylation was quantified by dividing biotinylated band intensity to JPH2 band intensity (biotin/JPH2), and the values of +HA/−HA from four independent experiments are plotted as small symbols. The large symbol denotes average (0.49 ± 0.12). C, disrupting lipid rafts by MβCD treatment (2 mm, 36 °C, 2 h) caused disarray of JPH2 and t-tubule organization in rat ventricular myocytes. Panel a, confirming the effectiveness of MβCD treatment in disrupting lipid rafts using a membrane lipid environment-sensitive fluorescent dye, Nile Red 12S (NR12S) (18). Live myocytes were incubated with NR12S (50 nm, in normal Tyrode's solution) at room temperature for 7 min, rinsed, and imaged by confocal microscopy. NR12S was excited by 514-nm laser. Its emission was sensitive to the lipid environment (18): we defined emission in the 523–581-nm range as “liquid-ordered, Lo, channel” and emission in the 591–698 nm range as “liquid-disordered, Ld, channel.” Shown are Lo/Ld ratio images of NR12S in myocytes without (control) or with MβCD treatment. Color scale of Lo/Ld is shown on the left. Panel b, fluorescence images of JPH2 (detected by rabbit Ab/Alexa488 anti-rabbit) and Alexa647-wheat germ agglutinin (WGA, marker of t-tubules and PM, pseudo-colored white) in control and Mβd-treated myocytes. Dashed lines mark where pixel profiles were determined. Panel c, pixel profiles of JPH2 and WGA in the control and MβCD-treated myocytes. Alexa488 and Alexa647 fluorescence signals were background-subtracted and normalized by respective maxima in the profile. Open circles mark missing JPH2 and t-tubules, and asterisks mark missing JPH2 where t-tubule was present. Images in panels a and b are shown at the same magnification as the one with the scale bars.
JPH2 migrated as ∼100- and ∼75-kDa bands, consistent with palmitoylated and unpalmitoylated forms. The combined JPH2 band intensity was much higher in the raft than the nonraft domains (3.02 ± 0.23 versus 1.0 ± 0.16, n = 11 each, p < 0.001) (Fig. 7A, panel b, left). Furthermore, the ratio of 100:75-kDa band intensities was much higher in the raft than nonraft domains (5.35 ± 0.33 versus 2.06 ± 0.49, n = 11 each, p < 0.001) (Fig. 7A, panel b, right). These data show that native JPH2 in rat ventricles is concentrated in the lipid–raft domain in palmitoylated form.
We then tested whether native JPH2 is S-palmitoylated, using an approach similar to that described for Fig. 1B. Isolated rat ventricular myocytes were incubated with palmitate–alkyne (100 μm) for 12 h. Native JPH2 was immunoprecipitated from whole-cell lysate with JPH2 mouse or rabbit Ab and reacted with biotin–PEG3–azide in the CuAAC reaction. The palmitoylation (i.e. biotinylation) signal was detected by either HRP-conjugated streptavidin or biotin goat Ab. Repeated attempts showed that the efficiency of immunoprecipitating rat JPH2 by either rabbit or mouse JPH2 Ab was low, and the biotinylation signal by either detection method was dim (one example depicted in Fig. 7B, panel a), likely reflecting the extremely slow palmitoyl turnover of native JPH2 in rat heart (see below). The intensity of JPH2 palmitoylation signal appeared to be lower after hydroxylamine treatment (Fig. 7B, panel b, biotin/JPH2 ratio in +HA samples decreased to 0.49 ± 0.12 relative to the ratio in −HA samples, n = 4). This is consistent with S-palmitoylation of native JPH2 in rat ventricular myocytes.
To further probe the functional role of native JPH2 palmitoylation in rat ventricular myocytes, we tested how MβCD treatment affected the distribution pattern of JPH2 and the organization of t-tubules. This was inspired by the COS-7 experiments shown in Fig. 4: disrupting lipid rafts by MβCD treatment reduced JPH2 palmitoylation and dislodged JPH2 from ER–PM junctions. Fig. 7C, panel a, confirms that the liquid-ordered raft domains present along the t-tubules in control myocytes (revealed by a lipid environment-sensitive fluorescent dye, Nile Red 12S (18)) were disrupted after MβCD treatment (2 mm, 36 °C, 2 h). This was accompanied by a disruption of the regularity in JPH2 distribution and in t-tubule organization (Fig. 7C, panel b). This finding was consistent among control and MβCD-treated myocytes (20 and 19, respectively) analyzed in the same manner. The disruption in JPH2 and t-tubule regularity is emphasized by their pixel profiles in myocyte center along the longitudinal axis (Fig. 7C, panel c). In the control myocyte, the peaks of fluorescence signals of JPH2 and t-tubule marker overlap at a regular spacing of ∼2 μm. In MβCD-treated myocytes, we observed missing JPH2 peaks where the t-tubule was present or missing both JPH2 and t-tubules.
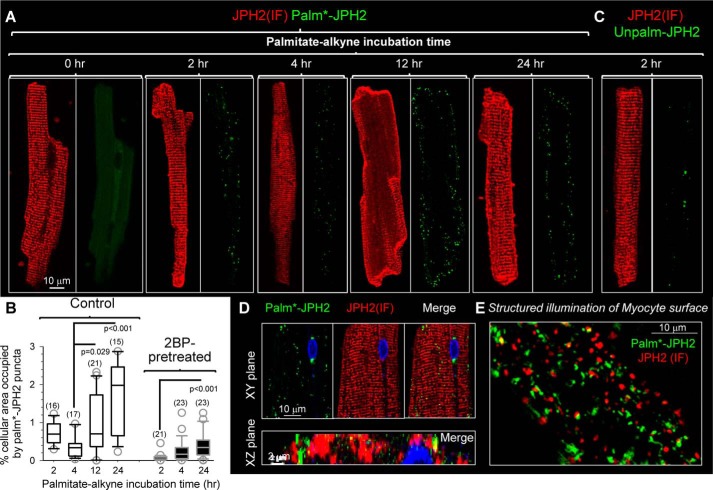
We used the Palm–PLA approach to examine the distribution pattern of palmitoylated JPH2 (palm*–JPH2) in rat ventricular myocytes. Fig. 8A depicts representative images of JPH2 (IF) and Palm–PLA signals in myocytes that had been incubated with palmitate–alkyne for the specified durations. The procedure of quantifying palm*–JPH2 puncta is described in Fig. S6, and the data summary is presented in Fig. 8B. The Palm–PLA approach applied to myocytes was authenticated by two tests. First, myocytes without palmitate–alkyne incubation but processed in parallel were free of Palm–PLA signals (0 h myocyte, in Fig. 8A). Second, pretreating myocytes with 2BP (100 μm, for 2 h) before palmitate–alkyne incubation markedly reduced the Palm–PLA signals, even 24 h after its removal (Fig. 8B, 2BP-pretreated versus control). This is consistent with the irreversible nature of 2BP inhibition of palmitoylation (14).
Figure 8.
Native JPH2 in rat ventricular myocytes has slow palmitoyl turnover in the junctional SR–PM junctions. The procedures of detecting Palm–PLA and Unpalm–PLA are described in Figs. S1 and S7, respectively. A, modest increase in palm*–JPH2 signals after palmitate–alkyne incubation from 2 to 24 h. The palm*–JPH2 signals clustered to the lateral cell surface, although JPH2 (IF) confirmed JPH2 localization along the z-line (jSR–PM junctions). The 0-h time point (no palmitate–alkyne incubation) serves as a negative control. Images are shown at the same magnification as the left one with scale bar. B, quantification of palm*–JPH2 signals after specified palmitate–alkyne incubation times under the control conditions or after 2BP pretreatment (100 μm, 2 h). The quantification procedure is described in Fig. S6. Shown are box plots of % cellular area occupied by palm*–JPH2 puncta. Numbers of myocytes analyzed are listed in parentheses. One-way ANOVA p < 0.001, followed by Dunn's all-pairwise tests. Groups showing significant differences are marked. C, Unpalm–PLA procedure detected very few Unpalm–JPH2 signals in myocytes after a 2-h incubation with palmitate–alkyne. D, detection of palm*–JPH2 in cytoplasm along the z-lines. Top, XY plane images from the myocyte center. Bottom, merged image of palm*–JPH2 and JPH2 (IF) in an XZ plane along a z-line. E, structured illumination image of myocyte surface. Both palm*–JPH2 and JPH2 (IF) manifested as distinct puncta with little overlap.
Fig. 8, A and B (quantification of palm*–JPH2 puncta in “control” myocytes), shows that palmitate–alkyne incubation led to the appearance of Palm–PLA puncta in a time-dependent manner, confirming that native JPH2 can be palmitoylated. However, the palm*–JPH2 signals after a 2- and 4-h incubation with palmitate–alkyne were very sparse, despite the abundant JPH2 (IF) signals. To check whether this was because the majority of native JPH2 in rat ventricular myocytes was unpalmitoylated, we designed an approach to monitor this population of JPH2 (Unpalm–PLA, procedure and validation presented in Fig. S7). Fig. 8C shows that in a myocyte incubated with palmitate–alkyne for 2 h, the Unpalm–PLA procedure detected very little signaling of unpalmitoylated JPH2. The same was observed in myocytes incubated with palmitate–alkyne for 4 or 12 h. This is consistent with data presented in Fig. 7A: native JPH2 in rat ventricles was largely in the palmitoylated form.
Prolonging the palmitate–alkyne incubation time to 12 and 24 h only modestly increased the palm*–JPH2 signals. Although we could detect palm*–JPH2 in the cytoplasm along z-lines (Fig. 8D, in a myocyte after 24-h palmitate–alkyne incubation), palm*–JPH2 mainly clustered to the lateral surface of myocytes despite abundant JPH2 (IF) signals along the z-lines. Palmitate–alkyne labeling required palmitoyl turnover during the incubation period. The low level of palmitate–alkyne labeling even after 24 h of incubation, in conjunction with data presented in Fig. 7A, suggests that native JPH2 was stably palmitoylated, i.e. there was very slow palmitoyl turnover resulting in a low degree of palmitate–alkyne labeling. This can explain why we could detect only a low level of native JPH2 palmitoylation in immunoblot experiments (Fig. 7B).
Fig. 8E depicts the distribution pattern of palm*–JPH2 and JPH2 (IF) on a myocyte surface revealed by structured illumination. The two groups of puncta showed very little overlap, suggesting binding to distinctly different PM subdomains.
Discussion
Our major findings can be summarized as follows. 1) JPH2 was S-palmitoylatable, both as heterologously expressed protein encoded by human JPH2 in COS-7 cells (Fig. 1, A and B), and as native protein in adult rat ventricular myocytes (Fig. 7B). 2) Palmitoylation is essential for JPH2's role as an ER–PM tether (Fig. 5). 3) All four Cys side chains in human JPH2 were involved in S-palmitoylation (Fig. 1C). 4) Functionally, palmitoylation promoted JPH2 binding to lipid–raft domains of PM (Fig. 3), which likely involved Cys side chains flanking the MORN motifs based on their proximity to the PM. Palmitoylation of the single Cys at the beginning of the TMD stabilized the JPH2 anchor in the ER/SR membrane (Fig. 6). 5) In both COS-7 cells and myocytes, palmitoylated JPH2 formed distinct puncta close to the PM (Figs. 2 and 8E), likely representing palmitoylated JPH2 at ER/SR–PM junctions. 6) Unpalmitoylated JPH2 was largely excluded from the raft domains (Fig. 3, D and E, comparing WT and Cys-free JPH2–GFP). However, unpalmitoylated JPH2 could form small and sparse puncta close to the PM (suggested by structured illumination images in Figs. 2 and 8E). They overlapped a little with puncta of palmitoylated JPH2, suggesting binding to different subdomains in the PM. 7) In COS-7 cells, disrupting lipid rafts by MβCD treatment not only dislodged JPH2 from the PM (Fig. 4, A and C) but also reduced the degree of JPH2 palmitoylation (Fig. 4B). In rat ventricular myocytes, disrupting lipid rafts by MβCD treatment caused a disarray of JPH2 distribution and t-tubule organization (Fig. 7C).
Complementary roles of S-palmitoylation and MORN motifs in JPH2 function as ER–PM tether
Palmitoylation of JPH2 greatly enhanced the stability of ER–PM contacts. This was manifested by the 5–10-μm-size patches of juxtamembrane JPH2–GFP signals in TIRF images (reflecting large cortical ER compartments stabilized by ER–PM junctions) and by the increase in juxtamembrane ER elements in TEM images. Importantly, these were observed in cells expressing palmitoylatable JPH2 but not unpalmitoylated JPH2 or after 2BP pretreatment, despite the presence of intact MORN motifs in the latter cases. These observations suggest that MORN motifs alone could not provide sufficient binding force to stabilize ER–PM junctions.
Palmitoyltransferases are membrane-embedded enzymes, requiring their substrates to travel to where they are to be palmitoylated (8). Many S-palmitoylatable proteins are dual-lipidated. They are either N-terminal myristoylated or C-terminal isoprenylated, which provide relatively weak binding forces to the lipid bilayer but are sufficient to bring the proteins to the palmitoyltransferases for S-palmitoylation (8). JPH2 does not have myristoylation or isoprenylation signals. Instead, the phospholipid-binding MORN motifs can bring JPH2 close to membrane-embedded palmitoyltransferases and facilitate S-palmitoylation.
S-Palmitoylation of native JPH2 in rat ventricular myocytes
Based on the JPH2 (IF) images as shown in Fig. 8A, we estimate that ∼75% of native JPH2 was in the cytosolic domain of rat ventricular myocytes. This pool of JPH2 was distributed as striations along the z-lines, where JPH2 tethered jSR to the t-tubule membrane. The other 25% of native JPH2 was in the cell periphery, where JPH2 tethered jSR to the peripheral plasma membrane. The palm*–JPH2 detected by the Palm–PLA reaction mainly existed as puncta in the cell periphery, rarely found in cytoplasm along the z-lines (Fig. 8, A and D). The disparity between JPH2 (IF) and palm*–JPH2 signals would suggest that in cardiac myocytes native JPH2 does not need S-palmitoylation to fulfill its function as a jSR–t-tubule tether. However, the following observations offer an alternative scenario. First, densitometry analysis in Fig. 7A, panel b, shows that 75% of native JPH2 in rat ventricles was in lipid–raft domains, where ∼90% of it was palmitoylated. Second, Fig. 8, A–C, shows that the turnover rate of palmitoyl on native JPH2 was very slow, making the degree of palmitate–alkyne labeling essentially a readout of the palmitoyl turnover rate. The fact that we could readily detect palm*–JPH2 in the cell periphery, but only rarely along the z-lines, indicates that palmitoylation of JPH2 along the z-lines was very stable, whereas palmitoylation of JPH2 along the lateral surface was more dynamic.
S-Palmitoylation guides JPH2 to travel to ER/SR–PM junctions
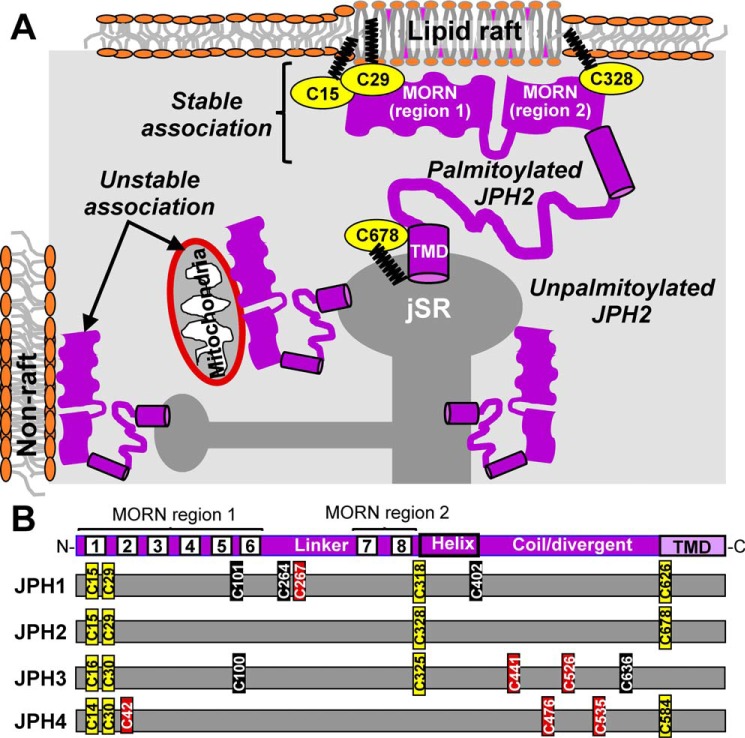
JPH2 translated in the rough ER compartment needs to traffic to ER/SR at the cell periphery to fulfill its function as an ER/SR–PM tether. During this journey, its MORN motifs are exposed to the cytoplasm and can potentially bind cytoplasmic organelles (e.g. mitochondria). How do cells prevent such “unproductive” binding of JPH2 to organelle membranes? Fig. 9A depicts our hypothesis. Our data suggest that palmitoylated JPH2 preferentially binds to lipid–raft domains of the plasma membrane, and binding to lipid–raft domains protects JPH2 from depalmitoylation. These two processes reinforce each other, resulting in stable ER/SR–PM junctions tethered by palmitoylated JPH2. Because lipid rafts mainly exist in the plasma membrane (19), this creates a guiding force favoring palmitoylated JPH2 to localize to junctional membrane complexes, where the ER/SR membrane is in close proximity to the PM, instead of staying in the cytosolic ER/SR. Unpalmitoylated JPH2 can bind to cytoplasmic organelles or nonraft subdomains of PM by the MORN motifs. However, these JPH2/membrane associations are less stable and thus are likely transient.
Figure 9.
A, working hypothesis for how palmitoylation directs JPH2 to SR/ER–PM junctions. Unpalmitoylated JPH2 in SR/ER has its MORN motifs exposed to the cytoplasm and may form unstable/transient associations with nonraft domains in plasma membrane or organelle (e.g. mitochondria). Palmitoylation of JPH2 promotes its binding to lipid–raft domains in PM, and binding to lipid–raft domains protects JPH2 from depalmitoylation. This synergism creates a stable association between palmitoylated JPH2 and PM. Palmitoylation of Cys-678 stabilizes JPH2 anchor in SR/ER membrane. B, Cys residues present in JPH1 (human sequence, accession no. Q9HDC5.2), JPH2 (Q9BR39.2), JPH3 (Q8WXH2.2), and JPH4 (NP_001139500.1) are marked relative to the shared JPH structural domains (top) and color-coded based on sequence alignment with JPH2 (yellow), high score (brown), or low score (black) as S-palmitoylation sites. Sequence alignment of JPH1–4 was done using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) (32, 33) (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.), and prediction of Cys residues as potential S-palmitoylation sites was done using CSS-Palm 4.0 (http://csspalm.biocuckoo.org/online.php (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.)).
Conformation dependence of JPH2 S-palmitoylation
JPH2 has a high-proline content in the linkers between the MORN regions I and II and between MORN II and the C-terminal transmembrane domain (both at 14%). The proline content in JPH2's linkers is more than double the average percentage of proline in human proteins (6.3%) (20). Such a high-proline content is predicted to confer a high degree of conformational flexibility. The conformation of the very flexible linkers in JPH2 may impact the orientation of Cys side chains with respect to palmitoylating enzymes that are embedded in the PM and/or ER/SR membranes (21). Fusing mCherry to the N terminus of JPH2 reduced its palmitoylation, while fusing GFP to the C terminus did not. We suggest that the former modification induced conformational changes in the cytoplasmic domain of JPH2 that reduced the accessibility of its Cys side chains to palmitoylating enzymes, whereas the impact of the latter modification was insulated by the ER/SR membrane.
Sequentially removing the palmitoylation sites by Cys–to–Ala mutations did not lead to a linear reduction in the palmitoylation signals (Fig. 1C). It is possible that preventing palmitoylation at one or more site(s) may increase the efficiency of palmitoylation and/or decrease the efficiency of depalmitoylation of the remaining sites, so that only when all four palmitoylation sites were removed (Cys-free JPH2–GFP), or when the overall palmitoylation efficiency was reduced (mCherry–JPH2), could the palmitoylation signals be reduced to the background level.
S-Palmitoylation of other JPHs in different cell types
Other members of the junctophilin family are expressed in skeletal and smooth muscles, neurons (4), and nonexcitable cells, such as JPH3 in pancreatic β cells (22) and JPH4 in T-cells (23). In all cases, junctophilins tether ER/SR to PM and promote cross-talks between these two compartments. Sequence alignment of JPH1–JPH4 encoded by human genes (Fig. 9B and supporting information) shows that palmitoylatable Cys residues identified in JPH2 are largely conserved in other junctophilins: the two Cys residues in MORN1 are conserved in all JPHs; Cys in MORN8 is conserved in JPH1–JPH3; and Cys in or close to TMD is conserved in JPH1, -2, and -4. This suggests that palmitoylation may also help the other junctophilins stabilize the ER/SR–PM junction.
Predictions by CSS Palm 4.0 server suggest that several other Cys residues in JPH1, JPH3, and JPH4 may also be palmitoylatable (Fig. 9B and supporting information). If confirmed, palmitoylation of JPH1 in the linker between MORN regions 1 and 2 and palmitoylation of JPH3 and JPH4 in the coil region preceding the transmembrane domain are expected to alter the nanodomains between ER/SR and the PM, with functional consequences in muscle contractility, neuronal excitability, and Ca-homeostasis in nonexcitable cells.
JPH2 banding pattern in SDS-PAGE
The amino acid sequences of full-length JPH2 in different species, including human, predict a molecular size of 74–75 kDa. However, we (7, 24) and others (25, 26) routinely observed a major 100-kDa band of native JPH2 in hearts. When JPH2 (with an 8-aa FLAG tag) was expressed in COS-7 cells, we saw both 75- and 100-kDa bands (Fig. 1A). JPH2 fused with GFP (increase in size by 25 kDa) migrated as 100- and 125-kDa bands. Unpalmitoylatable Cys-free JPH2–GFP migrated as a single band just below the 100-kDa marker (Fig. 3D). These observations indicate that the mobility of JPH2 in SDS-PAGE depended on its palmitoylation status. It is possible that palmitoylation of JPH2 conferred certain conformation that promoted tight association with ∼25-kDa protein(s) expressed in both cardiac myocytes and COS-7 cells, and this tight association could resist the denaturing power of SDS during SDS-PAGE. Possible candidates include caveolins (24), i.e. caveolin-3 in cardiac myocytes (18 kDa) and caveolin-1 in COS-7 cells (20 kDa). This possibility, as well as other candidates, should be tested in future experiments.
Experimental procedures
Molecular constructs
Plasmids carrying human JPH2 cDNAs were purchased from GeneCopoeia: isoform 1 (Q9BR39.2, 696 aa) with FLAG or mCherry fused to the N terminus or with GFP fused to the C terminus (termed FLAG–JPH2, mCherry–JPH2, and JPH2–GFP, respectively). Cysteine to alanine mutations in JPH2–GFP were created using Q5 site-directed mutagenesis kit (New England Biolabs) and verified by DNA sequencing. Plasmid encoding HRP–KDEL was a gift from Dr. Alice Ting (Addgene plasmid no. 85582). DsRed2–ER-5 was a gift from Michael Davidson (Addgene plasmid no. 55836).
COS-7 culture and transfection
COS-7 cells are maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, nonessential amino acids, and penicillin/streptomycin at 36 °C in a humidified CO2 incubator. Cells were plated on Matrigel-coated dishes or coverslips, and when reaching ∼70% confluency, they were transfected with cDNA (0.2–0.7 μg/ml) in the presence of Lipofectamine 2000 following the manufacturer's instruction. Cells were cultured for another 16–36 h before experiments.
Adult rat ventricular myocyte isolation and culture
The study of animals was reviewed and approved by the Institutional Animal Care and Use Committee of Virginia Commonwealth University. Ventricular myocytes were isolated from adult (>2 months) male Sprague-Dawley rats as described previously (27). Briefly, the aorta was cannulated, and the heart was mounted on a Langendorff apparatus and perfused sequentially with the following solutions (36 °C, oxygenated): normal Tyrode's (10 min); nominally Ca-free Tyrode's with 0.1% BSA (10 min); the same Ca-free/BSA Tyrode's with collagenase (Worthington type II, 0.5 mg/ml, 10–20 min); and KB medium (28) (30 ml). Afterward, the ventricles were placed in KB medium, minced, and gently triturated to release single myocytes. After recovery in KB medium at room temperature for 1 h, myocytes were plated on Matrigel-coated dishes or coverslips and cultured in medium 199, supplemented with l-carnitine (5 mm), creatine phosphate (5 mm), taurine (5 mm), BSA (0.2%), fetal calf serum (2%), cytochalasin D (0.2 μm), and penicillin/streptomycin in a 36 °C humidified incubator.
Detection of JPH2 palmitoylation
COS-7 cells transfected with JPH2 constructs were incubated with palmitate–alkyne (100 μm) at 36 °C overnight (∼12 h). Cells were harvested and incubated on ice for 1 h in lysis buffer (mm): NaCl 145, MaCl2 0.1, HEPES 15, EGTA 10, pH 7.4, with NaOH, supplemented with phenylmethylsulfonyl fluoride (PMSF, 0.2 mm), protease mixture (Sigma), and 1% Triton X-100. After centrifugation (20,000 × g, 15 min) to remove insoluble debris, the supernatant (whole-cell lysate) was subject to immunoprecipitation with a suitable antibody targeting JPH2 (specified in figure or text) and protein A/G magnetic beads or protein G–agarose beads. Protein-bound beads were subject to CuAAC reaction by incubating with biotin–PEG3–azide, CuSO4, ascorbic acid, tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and PMSF all at 1 mm, plus tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA, 0.1 mm) at room temperature for 1 h (29). In some experiments, protein-bound and CuAAC-reacted beads were incubated with hydroxylamine (HA, 0.75 m, neutralized to pH 7.4 with NaOH) or with Tris at room temperature for 1 h before elution. Proteins were eluted from beads by incubation with 2× SDS sample buffer with mercaptoethanol (5% v/v) at room temperature for 30 min before the immunoblot experiments. The above procedures are diagrammed in Fig. S1A.
In situ PLA (30)
Subcellular distribution pattern of palmitoylated JPH2 was detected by combining palmitate–alkyne labeling, CuAAC reaction with azide-conjugated biotin, and in situ proximity ligation amplification in a process termed “Palm–PLA” (11). COS-7 cells expressing JPH2 constructs or ventricular myocytes expressing native JPH2 were plated on coverslips and incubated with palmitate–alkyne as described above. Cells were fixed (4% paraformaldehyde in PBS at room temperature for 10 min), permeabilized (0.1% Triton X-100 in PBS at room temperature for 10 min), and clicked with biotin–PEG3–azide using Click-it cell reaction buffer kit (Invitrogen) according to the manufacturer's instructions. Cells were incubated with the appropriate primary Ab pair (e.g. biotin goat Ab/JPH2 rabbit Ab, specified in figure or text) at 4 °C overnight. Cells were subject to probe annealing, ligation, and amplification reactions using Duolink fluorescence assay kit (Sigma Millipore), following the manufacturer's instructions. The above procedures are diagrammed in Fig. S1B.
We designed a similar approach to detect unpalmitoylated JPH2 (Unpalm–PLA, Fig. S7). In this case, COS-7 cells or cardiac myocytes were fixed and permeabilized as described above. Cells were incubated with TCEP (100 μm at room temperature for 1 h) to reduce disulfide bonds. This was followed by incubation with thiol-reactive EZ-link BMCC-biotin (160 μm at room temperature for 2 h) to biotinylate free (unpalmitoylated) thiol groups. This was followed by the same PLA procedure as described above.
Disruption and purification of lipid rafts
To disrupt cholesterol-rich lipid–raft domains, cells were incubated with MβCD (2 mm at 36 °C for 2 h) to extract cholesterol before experiments. We purified lipid rafts from cell membranes using detergent-free purification (12) or as Triton-insoluble fraction (13).
Immunoblot experiments
Whole-cell lysates or immunoprecipitates were mixed with 2× SDS sample buffer with 5% v/v mercaptoethanol and rocked at room temperature for 30 min. Proteins were fractionated by 6, 7.5, or 14% SDS-polyacrylamide gels and blotted to PVDF membrane. Membrane was blocked in 1× TBST with 10% nonfat dry milk at room temperature for 3 h +and incubated with primary Ab (specified in figures or text) overnight at 4 °C. Membrane was incubated with HRP-conjugated secondary Ab, and immunoreactive bands were visualized with enhanced chemiluminescence kit (SuperSignal West Femto Maximum Sensitivity Substrate, Thermo Fisher Scientific) using an imager FluorChem E. Band intensities were quantified using AlphaView SA, and numerical data were exported to Excel for processing.
Imaging experiments and data analysis
To image fixed cells, cells were plated on Matrigel-coated no. 1.5 coverslips, transfected with cDNAs and treated with reagents described above or in the text, fixed, permeabilized, labeled, and mounted on glass slides with ProLong Diamond antifade mountant (Molecular Probes). Proteins were detected by their fused fluorescent protein (GFP, mCherry, or dsRed), immunofluorescence (labeled with primary Abs followed by Alexa-conjugated secondary Abs), or by PLA reaction. Nuclei were labeled with DAPI.
To image live cells, cells were plated on Matrigel-coated no. 1.5 glass-bottom microwell dishes (MatTek), transfected with cDNA, and treated with reagents described above. After staining nuclei with Hoechst dye, cells were imaged in normal Tyrode's solution at 37 °C (confocal) or at room temperature (TIRF).
Confocal microscopy using Zeiss 710
Fixed COS-7 cells or ventricular myocytes were imaged with ×63 or ×40 oil immersion objective. Fluorophores were sequentially excited by lasers of 405 nm (DAPI and Hoechst dye), 488 nm (Alexa488 or GFP), 561 nm (Alexa568, dsRed, or mCherry), and 633 nm (Alexa647). The emission lights were collected within appropriate range to avoid “bleed through.”
To image NR12S-stained live cells, we used a 514-nm laser for excitation, and the emission lights were collected at 523–581 nm (defined as liquid-ordered, Lo, channel) and 591–698 nm (defined as liquid-disordered, Ld, channel). To obtain ratiometric images, a plug in for ImageJ, prepared by R. Vauchelles, was used (see Ref. 18).
For FRAP experiments on live COS-7 cells, the regions of interest (ROIs) were specified: small circular areas (∼5 μm diameter) of cytoplasmic ER (to be bleached) and a cell-free area (as background). Emission lights from GFP and/or mCherry in ROIs were measured with low power of 488- and 561-nm lasers (≤1%) once every 15 s. After the control data were collected, fluorescence signals in ROIs of ER regions were bleached with 100% laser power until the fluorescence intensities dropped to 10–30% of pre-bleach levels. Imaging with low-laser power was resumed for another 10–15 min until the fluorescence recovery reached a steady state. Data of pixel values and areas of ROIs were exported to Excel. Background-subtracted pixel contents of ROIs in ER regions were calculated, and the fraction of fluorescence was recovered at 400 s after photobleach was used for comparison.
TIRF microscopy
A Nikon microscope in the TIRF mode was used. Fluorophores were separately excited by lasers of 405 nm (Hoechst dye), 488 nm (GFP), and 561 nm (mCherry). After obtaining epifluorescence images as control, the angle of incident light for each laser was adjusted to just before the fluorescence signal disappeared, i.e. limiting the detection of fluorophores at <200 nm from the plasma membrane in contact with the coverslip. Time-lapse images of fluorophores were recorded before and after application of MβCD (stock solution directly added to the imaging dish, reaching a final concentration of 2 mm), with the “perfect focus system” of the microscope automatically adjusting the focus during imaging. Regions of interest were specified off-line, and the values of pixel contents were exported to Excel for processing.
Structured illumination microscopy (SIM) (31)
Nikon microscopy in the SIM mode was used. Images were obtained using the 3D SIM function and then reconstructed to obtain information at about double the resolution of diffraction-limited microscopy.
TEM using Jeol 1400 TEM
To quantify endoplasmic reticulum at ER–PM junctions, COS-7 cells were transfected with HRP–KDEL alone or with JPH2 variants. After 24 h of culture, cells were fixed with 2% glutaraldehyde in 0.1 m sodium cacodylate buffer (20 min at room temperature). Cells were treated with a tyramide signal amplification (TSA) kit (PerkinElmer Life Sciences) at room temperature for 30 min, which deposited up to 100-fold of biotin molecules at HRP–KDEL. After TSA treatment, cells were reacted with an ABC kit (Vector Laboratories) at room temperature for 30 min, which linked peroxidase to the biotin molecules. After these amplification steps, cells were incubated with 1 mg/ml 3,3′-diaminobenzidine (DAB) at room temperature for 15 min, and then in the presence of 0.01% H2O2 at room temperature for 30 min to produce electron-dense material in the ER lumen (16). Cells were processed for TEM. Images were collected at ×800–2000 magnification. TIFF files of TEM images were analyzed by ImageJ (National Institutes of Health) to quantify percent of cell perimeter occupied by ER elements ≤50 nm from the plasma membrane.
Sources of antibodies and reagents
The following primary antibodies were used: JPH2 mouse and rabbit Abs (Santa Cruz Biotechnology, sc-377086 and sc-134875); mCherry rabbit Ab (Abcam, ab167453); GFP rabbit Ab (Abcam, ab290); FLAG mouse Ab (Sigma, F3165); dsRed rabbit Ab (Clontech, 632496); caveolin-1 and caveolin-3 mouse Abs (BD Transduction Laboratories, 610057 and 610421); biotin goat Ab (Sigma, B3640); Cav1.2 mouse Ab (NeuroMab, 75–257); and α-actin mouse Ab (Sigma, A7811). The following secondary antibodies were used: Alexa-conjugated Abs (Molecular Probes); horseradish peroxidase (HRP)-conjugated Abs (Invitrogen).
The following reagents were used: in situ proximity amplification kit (Sigma, DUO92014); protein A/G magnetic beads (Pierce, 88803); protein G–agarose beads (20398, Pierce); palmitate–alkyne and biotin-PEG3–azide (Click Chemistry Tools, 1165-25 and AZ104-25); EZ-link BMCC-biotin (ThermoFisher Scientific, 21900); TCEP (Sigma, C4706); tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine TBTA (Sigma, 678937); hydroxylamine (Sigma, 159417); 2BP (Sigma 21604); MβCD (Sigma M7439); TSA biotin system (PerkinElmer Life Sciences, NEL700A001KT); ABC kit (Vector Laboratories, PK-6100); Matrigel (Sigma, E1270); Lipofectamine2000 (Thermo Fisher Scientific, 11668019); HRP-conjugated streptavidin (Thermo Fisher Scientific, N100); and Tris-glycine SDS sample buffer (2×) (Novex, LC2676).
Statistical analysis
To compare the two groups, the Excel function “t test: two-sample assuming unequal variances” was used to calculate the p value of the two-tailed t test. For comparison among three or more groups, SigmaStat (version 4.0) was used to perform one-way ANOVA. If the p value was less than 0.01, Dunn's all-pairwise test or test against a reference group was used to detect statistically significant differences among the groups. Data are presented in box plot format: box showing 25th percentile, median, and 75th percentile values, whiskers showing 10th and 90th percentile values, and symbols showing outlying values.
Author contributions
M. J., J. H., A. M., S. W. W., and G.-N. T. data curation; M. J., J. H., S. W. W., and G.-N. T. investigation; M. J., J. H., F. K. H. W., J. W., and G.-N. T. methodology; A. S. K. resources; G.-N. T. conceptualization; G.-N. T. formal analysis; G.-N. T. supervision; G.-N. T. funding acquisition; G.-N. T. writing-original draft; G.-N. T. project administration; G.-N. T. writing-review and editing.
Supplementary Material
Acknowledgments
Confocal, TIRF, and structured illumination microscopy was performed at the Department of Anatomy and Neurobiology Microscopy Facility, Virginia Commonwealth University, which is supported in part by National Institutes of Health–NINDS Center Core Grant 5P30NS047463. We thank Dr. James Smyth (Virginia Tech Carilion, Roanoke, VA) for critical review of an early version of this manuscript.
This work was supported by National Institutes of Health Grants RO1s HL128610 and HL96962 from NHLBI and by Grant-in-aid 16GRNT29920012 from American Heart Association/Mid-Atlantic Affiliate (to G. -N. T.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S7, supporting C–E information, and supporting Refs. 1–4.
- ER–PM
- endoplasmic reticulum–plasma membrane
- JPH
- junctophilin
- MORN
- membrane-occupation-recognition-nexus
- TMD
- transmembrane domain
- CuAAC
- Cu(I)-catalyzed alkyne-azide cycloaddition
- CICR
- Ca-induced Ca release
- palm*–JPH2
- palmitoylated junctophilin 2
- NR12S
- Nile Red 12S
- SR
- sarcoplasmic reticulum
- ROI
- region of interest
- WCL
- whole-cell lysate
- Ab
- antibody
- IP
- immunoprecipitation
- PVDF
- polyvinylidene difluoride
- PMSF
- polyvinylidene difluoride
- HRP
- horseradish peroxidase
- TCEP
- tris(2-carboxyethyl)phosphine hydrochloride
- TEM
- transmission EM
- TSA
- tyramide signal amplification
- DAB
- 3,3′-diaminobenzidine
- TBTA
- tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine
- HA
- hydroxylamine
- ECL
- enhanced chemiluminescence
- IF
- immunofluorescence
- aa
- amino acid
- 2BP
- 2-bromopalmitate
- MβCD
- methoxy-β-cyclodextrin
- SIM
- structured illumination microscopy
- WGA
- wheat germ agglutinin
- DAPI
- 4′,6-diamidino-2-phenylindole
- jSR
- junctional sarcoplasmic reticulum.
References
- 1. Manford A. G., Stefan C. J., Yuan H. L., Macgurn J. A., and Emr S. D. (2012) ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell 23, 1129–1140 10.1016/j.devcel.2012.11.004 [DOI] [PubMed] [Google Scholar]
- 2. Fox P. D., Haberkorn C. J., Akin E. J., Seel P. J., Krapf D., and Tamkun M. M. (2015) Induction of stable ER-plasma-membrane junctions by Kv2.1 potassium channels. J. Cell Sci. 128, 2096–2105 10.1242/jcs.166009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Takeshima H., Komazaki S., Nishi M., Iino M., and Kangawa K. (2000) Junctophilins: a novel family of junctional membrane complex proteins. Mol. Cell 6, 11–22 10.1016/S1097-2765(05)00005-5 [DOI] [PubMed] [Google Scholar]
- 4. Takeshima H., Hoshijima M., and Song L.-S. (2015) Ca2+ microdomains organized by junctophilins. Cell Calcium 58, 349–356 10.1016/j.ceca.2015.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Oort R. J., Garbino A., Wang W., Dixit S. S., Landstrom A. P., Gaur N., De Almeida A. C., Skapura D. G., Rudy Y., Burns A. R., Ackerman M. J., and Wehrens X. H. (2011) Disrupted junctional membrane complexes and hyperactive ryanodine receptors after acute junctophilin knockdown in mice. Circulation 123, 979–988 10.1161/CIRCULATIONAHA.110.006437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guo A., Zhang X., Iyer V. R., Chen B., Zhang C., Kutschke W. J., Weiss R. M., Franzini-Armstrong C., and Song L.-S. (2014) Overexpression of junctophilin-2 does not enhance baseline function but attenuates heart failure development after cardiac stress. Proc. Natl. Acad. Sci. U.S.A. 111, 12240–12245 10.1073/pnas.1412729111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang Y., Eltit J. M., Kaszala K., Tan A., Jiang M., Zhang M., Tseng G.-N., and Huizar J. F. (2014) Cellular mechanism of premature ventricular contraction-induced cardiomyopathy. Heart Rhythm 11, 2064–2072 10.1016/j.hrthm.2014.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chamberlain L. H., and Shipston M. J. (2015) The physiology of protein S-acylation. Physiol. Rev. 95, 341–376 10.1152/physrev.00032.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salaun C., Greaves J., and Chamberlain L. H. (2010) The intracellular dynamic of protein palmitoylation. J. Cell Biol. 191, 1229–1238 10.1083/jcb.201008160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lord C. C., Thomas G., and Brown J. M. (2013) Mammalian αβ-hydrolase domain (ABHD) proteins: lipid metabolizing enzymes at the interface of cell signaling and energy metabolism. Biochim. Biophys. Acta 1831, 792–802 10.1016/j.bbalip.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao X., and Hannoush R. N. (2014) Method for cellular imaging of palmitoylated proteins with clickable probes and proximity ligation applied to Hedgehog, tubulin, and Ras. J. Am. Chem. Soc. 136, 4544–4550 10.1021/ja410068g [DOI] [PubMed] [Google Scholar]
- 12. Song K. S., Li Shengwen, Okamoto T., Quilliam L. A., Sargiacomo M., and Lisanti M. P. (1996) Co-purification and direct interaction of Ras with caveolin, an integral membrane protein of caveolae microdomains. Detergent-free purification of caveolae membranes. J. Biol. Chem. 271, 9690–9697 10.1074/jbc.271.16.9690 [DOI] [PubMed] [Google Scholar]
- 13. Chamberlain L. H. (2004) Detergents as tools for the purification and classification of lipid rafts. FEBS Lett. 559, 1–5 10.1016/S0014-5793(04)00050-X [DOI] [PubMed] [Google Scholar]
- 14. Jennings B. C., Nadolski M. J., Ling Y., Baker M. B., Harrison M. L., Deschenes R. J., and Linder M. E. (2009) 2-Bromopalmitate and 2-(2-hydroxy-5-nitrobenzylidene)-benzo[b]thiophen-3-one inhibit DHHC-mediated palmitoylation in vitro. J. Lipid Res. 50, 233–242 10.1194/jlr.M800270-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klymchenko A. S., and Kreder R. (2014) Fluorescent probes for lipid rafts: from model membranes to living cells. Chem. Biol. Rev. 21, 97–113 10.1016/j.chembiol.2013.11.009 [DOI] [PubMed] [Google Scholar]
- 16. Wu M. M., Buchanan J., Luik R. M., and Lewis R. S. (2006) Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J. Cell Biol. 174, 803–813 10.1083/jcb.200604014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lippincott-Schwartz J., Snapp E., and Kenworthy A. (2001) Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2, 444–456 10.1038/35073068 [DOI] [PubMed] [Google Scholar]
- 18. Darwich Z., Kucherak O. A., Kreder R., Richert L., Vauchelles R., Mély Y., and Klymchenko A. S. (2013) Rational design of fluorescent membrane probes for apoptosis based on 3-hydroxyflavone. Methods Appl. Fluoresc. 1, 025002 10.1088/2050-6120/1/2/025002 [DOI] [PubMed] [Google Scholar]
- 19. Simons K., and Sampaio J. L. (2011) Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 3, a004697 10.1101/cshperspect.a004697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morgan A. A., and Rubenstein E. (2013) Proline: the distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PLoS One 8, e53785 10.1371/journal.pone.0053785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fukata Y., Murakami T., Yokoi N., and Fukata M. (2016) Local palmitoylation cycles and specialized membrane domain organization. Curr. Top. Membr. 77, 97–141 10.1016/bs.ctm.2015.10.003 [DOI] [PubMed] [Google Scholar]
- 22. Li L., Pan Z.-F., Huang X., Wu B.-W., Li T., Kang M.-X., Ge R.-S., Hu X.-Y., Zhang Y.-H., Ge L.-J., Zhu D.-Y., Wu Y.-L., and Lou Y.-J. (2016) Junctophilin 3 expresses in pancreatic beta cells and is required for glucose-stimulated insulin secretion. Cell Death Dis. 7, e2275 10.1038/cddis.2016.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Woo J. S., Srikanth S., Nishi M., Ping P., Takeshima H., and Gwack Y. (2016) Junctophilin-4, a component of the endoplasmic reticulum–plasma membrane junctions, regulates Ca2+ dynamics in T cells. Proc. Natl. Acad. Sci. U.S.A. 113, 2762–2767 10.1073/pnas.1524229113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jiang M., Zhang M., Howren M., Wang Y., Tan A., Balijepalli R. C., Huizar J. F., and Tseng G.-N. (2016) JPH-2 interacts with Cai-handling proteins and ion channels in dyads: contribution to premature ventricular contraction-induced cardiomyopathy. Heart Rhythm 13, 743–752 10.1016/j.hrthm.2015.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo A., Hall D., Zhang C., Peng T., Miller J. D., Kutschke W., Grueter C. E., Johnson F. L., Lin R. Z., and Song L.-S. (2015) Molecular determinants of calpain-dependent cleavage of junctophilin-2 protein in cardiomyocytes. J. Biol. Chem. 290, 17946–17955 10.1074/jbc.M115.652396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quick A. P., Wang Q., Philippen L. E., Barreto-Torres G., Chiang D. Y., Beavers D., Wang G., Khalid M., Reynolds J. O., Campbell H. M., Showell J., McCauley M. D., Scholten A., and Wehrens X. H. (2017) Striated muscle preferentially expressed protein kinase (SPEG) is essential for cardiac function by regulating junctional membrane complex activity. Circ. Res. 120, 110–119 10.1161/CIRCRESAHA.116.309977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang M., Wang Y.-H., and Tseng G.-N. (2017) Adult ventricular myocytes segregate KCNQ1 and KCNE1 to keep the IKs amplitude in check until when larger IKs is needed. Circ. Arrhythm. Electrophysiol. 10, e005084 10.1161/CIRCEP.117.005084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Isenberg G., and Klockner U. (1982) Calcium tolerant ventricular myocytes prepared by preincubation in a “KB medium”. Pflugers Arch. 395, 6–18 10.1007/BF00584963 [DOI] [PubMed] [Google Scholar]
- 29. Kolb H. C., Finn M. G., and Sharpless K. B. (2001) Click chemistry: diverse chemical function from a few good reactions. Agnew. Chem. Int. Ed. Engl. 40, 2004–2021 10.1002/1521-3773(20010601)40:11andlt;2004::AID-ANIE2004andgt;3.0.CO;2-5 [DOI] [PubMed] [Google Scholar]
- 30. Söderberg O., Leuchowius K.-J., Gullberg M., Jarvius M., Weibrecht I., Larsson L.-G., and Landegren U. (2008) Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation. Methods 45, 227–232 10.1016/j.ymeth.2008.06.014 [DOI] [PubMed] [Google Scholar]
- 31. Gustafsson M. G., Shao L., Carlton P. M., Wang C. J., Golubovskaya I. N., Cande W. Z., Agard D. A., and Sedat J. W. (2008) Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94, 4957–4970 10.1529/biophysj.107.120345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y. M., Buso N., and Lopez R. (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43, W580–W584 10.1093/nar/gkv279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.