Antibiotic resistance poses an alarming and ever-increasing threat to modern health care. Although the current antibiotic crisis is widely acknowledged, actions taken so far have proved insufficient to slow down the rampant spread of resistant pathogens. Problematically, routine screening methods and strategies to restrict therapy failure almost exclusively focus on genetic resistance, while evidence for dangers posed by other bacterial survival strategies is mounting.
KEYWORDS: antibiotic resistance, antibiotics, evolution, persistence
ABSTRACT
Antibiotic resistance poses an alarming and ever-increasing threat to modern health care. Although the current antibiotic crisis is widely acknowledged, actions taken so far have proved insufficient to slow down the rampant spread of resistant pathogens. Problematically, routine screening methods and strategies to restrict therapy failure almost exclusively focus on genetic resistance, while evidence for dangers posed by other bacterial survival strategies is mounting. Antibiotic tolerance, occurring either population-wide or in a subpopulation of cells, allows bacteria to transiently overcome antibiotic treatment and is overlooked in clinical practice. In addition to prolonging treatment and causing relapsing infections, recent studies have revealed that tolerance also accelerates the emergence of resistance. These critical findings emphasize the need for strategies to combat tolerance, not only to improve treatment of recurrent infections but also to effectively address the problem of antibiotic resistance at the root.
ANTIBIOTICS AND THE RESISTANCE CRISIS
Antibiotics have transformed medicine and saved millions of lives since the introduction of penicillin in the 1940s (1). However, either by de novo mutation or horizontal gene transfer, bacteria can acquire mechanisms to degrade or inactivate the antibiotic, pump the antibiotic out of the cell, or modify the drug target. These resistance mechanisms allow bacteria to grow at elevated concentrations of antibiotics. Reports on resistance to antibiotics appeared even before their first therapeutic use (2). Initially, the emergence of resistant pathogens was not a matter of significant public concern, as novel effective antibiotics were being discovered regularly. However, after the golden years of antibiotic discovery from the 1940s through the 1960s, the development of new and effective antibiotics steadily declined (3). The decreasing supply of novel compounds, together with the massive increase in antibiotic consumption, including nontherapeutic use for growth promotion in agriculture and aquaculture, has led to the antibiotic resistance crisis that we are facing today (4). Each year, infections with antibiotic-resistant bacteria are estimated to result in more than 23,000 and 33,000 deaths in the United States and Europe, respectively (5, 6). Infections with antibiotic-resistant pathogens cause an extra 10,000 to 40,000 U.S. dollars in hospital costs per patient (7). If adequate measures are not taken, the toll of antimicrobial resistance in Europe, North America, and Australia is estimated to rise to 2.4 million casualties per year at an annual cost of up to 3.5 billion U.S. dollars (8).
ANTIBIOTIC TOLERANCE AND PERSISTENCE
Overshadowed by the more prominent narrative of antibiotic resistance, scientists have also warned of the dangers of another strategy by which bacteria can overcome antibiotic treatment. Antibiotic tolerance allows bacteria to temporarily withstand or slow down the lethal consequences of high doses of bactericidal antibiotics but without being able to grow in their presence (9). Increased tolerance can result from mutations, but it can also result from environmental conditions, for example conditions that slow down growth (10). The ability of a whole bacterial population to survive longer treatments with bactericidal antibiotics is denoted as “tolerance” throughout the text. Tolerance can also occur in a subpopulation of phenotypic variants called “persister cells.” This specific type of tolerance is referred to as “persistence” (11). As was the case with resistance, tolerance and persistence were first observed shortly after the introduction of penicillin (12–15).
Increasing evidence suggests that tolerance and persistence play a considerable and currently underappreciated role in the recalcitrance and relapse of bacterial infections (11, 16–19). This evidence includes the following. (i) High levels of persisters are present in biofilms and inside host cells (20–23). (ii) A well-known persistence gene is frequently mutated in clinical isolates of uropathogenic Escherichia coli (24). (iii) Increased tolerance or persistence evolves in the face of periodical antibiotic treatment both in vitro and in vivo (25–29). (iv) Recurrent episodes of infection can be due to relapse caused by the original, nonresistant founder strain rather than reinfection with a new strain (30–32). (v) Several studies demonstrated that nongrowing or slowly growing bacteria tolerate antibiotics and cause therapy failure in in vivo infection models (23, 33–37). The increasing awareness of the clinical consequences of tolerance and persistence has strongly encouraged research on these phenomena in the past decade. However, different experimental setups are often adopted in different studies, resulting in inconsistencies in the available data. A set of standardized and generally approved guidelines were recently proposed to measure antibiotic persistence and should facilitate the comparison of experimental results (38).
TOLERANCE AND PERSISTENCE AS DRIVERS OF RESISTANCE EVOLUTION
Three decades ago, Moreillon and Tomasz found that cyclic exposure of Streptococcus pneumoniae to high concentrations of penicillin selects for tolerant mutants, while resistant mutants evolve during exposure to sustained, low levels of penicillin (39). On the basis of this finding, they hypothesized that tolerant cells constitute a reservoir of viable cells from which resistant mutants can emerge (“Resistant mutants would then be recruited from this pool of lysis- and kill-defective bacteria […]”), implying that tolerance facilitates the evolution of resistance (“selection for resistance […] may be secondary to selection for increased survival.”). In further agreement with this hypothesis, a tolerant S. pneumoniae mutant consistently displays greater efficiency in transformation with DNA from streptomycin- or penicillin-resistant clinical isolates compared to a wild-type S. pneumoniae strain (40). Mathematical simulation of a bacterial infection similarly showed that tolerant cells may play an important role in the emergence of therapy resistance (41). However, strong experimental evidence in favor of this hypothesis was published only recently. Nguyen et al. used a murine infection model to demonstrate that resistance development is abolished in a strain with strongly decreased antibiotic tolerance (21). Sebastian et al. showed that persisters are a source of de novo resistant mutants during long-term rifampin exposure in Mycobacterium tuberculosis (42). The group of Nathalie Balaban found that the evolution of increased tolerance or persistence precedes the emergence and spread of resistance-conferring mutations in E. coli populations under intermittent ampicillin exposure (43). Recently, we discovered a strong, positive correlation between persister levels and the likelihood to evolve resistance in natural isolates and lab strains of E. coli (44). Importantly, these findings hold for different types of antibiotics, in different setups or conditions, suggesting a widespread link between persistence and the evolution of resistance.
Apart from constituting a viable reservoir from which resistant clones can emerge, tolerance and persistence may also facilitate the development of resistance in a more intricate fashion. Stress responses play a central role in the formation of persisters (11, 16, 21, 45), and are also known to cause a temporal increase in cellular mutation rates (46–49). Hence, high persister levels and high mutation rates may act synergistically in stressed bacteria and increase the likelihood for resistance-conferring mutations to occur in the persister reservoir (50). Evidence for stress responses being a major determinant of the persistence-resistance link was provided by Sebastian et al., who reported that resistance mutations occurring in Mycobacterium persisters are provoked by elevated levels of oxidative stress (42). Moreover, high-persistence and mutator phenotypes are both known to thrive in fluctuating environments (25, 51–53), suggesting that these traits could evolve under similar environmental conditions. A recent study from our lab confirmed that persistence is pleiotropically linked with mutation rates. Using Luria-Delbrück fluctuation assays, we found increased mutation rates in two high-persistence mutants and a lower mutation rate in a low-persistence strain. In populations plated on supra-MIC antibiotic concentrations, the number of resistant mutants that emerged per surviving persister shows the same trend (44). Spontaneous or antibiotic-induced DNA damage may still inflict mutations in nongrowing persisters, because these cells display DNA turnover even in the absence of chromosomal replication (54–56). Indeed, Barrett et al. recently observed a strong induction of the SOS response in persisters, indicative of DNA damage (57). These persisters exhibit accelerated resistance development, a process in which error-prone DNA polymerases are involved (57). Similarly, Yaakov et al. demonstrated that Saccharomyces cerevisiae persisters are characterized by an increased load of DNA damage, pointing at an increased mutation rate and evolvability of these cells (58). A correlation between persistence and mutation rates was also recently confirmed by El Meouche and Dunlop (59). Their data suggest that heterogeneity in the expression of the multidrug efflux pump AcrAB-TolC gives rise to a subpopulation that is not only transiently resistant to several drugs but also exhibits a lower growth rate and a reduced expression of the DNA mismatch repair enzyme MutS, resulting in an increased spontaneous mutation rate (59).
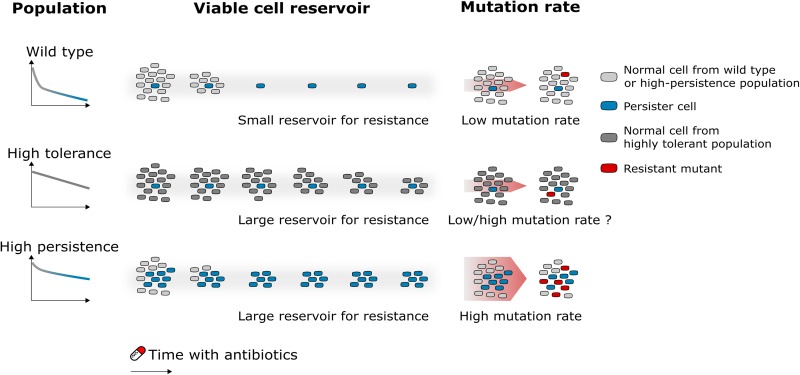
On the basis of this accumulating experimental evidence, we propose a framework in which two factors constitute the link between tolerance or persistence and resistance (Fig. 1). On the one hand, high levels of persistence or tolerance lead to a higher number of viable cells during antibiotic treatment, which results in an increased statistical probability for the occurrence of resistance-conferring mutations (Fig. 1). On the other hand, increased persistence, and possibly also tolerance, is pleiotropically linked with increased mutation rates both in growing cells (when antibiotic concentrations are low) and in persisters (when antibiotic concentrations are high) (Fig. 1). A mathematical model that simulates the course of antibiotic treatment in a patient with a bacterial infection confirmed that persistence contributes to the emergence of resistance in clinical settings by an interplay of increased survival and increased adaptability through elevated mutation rates (44).
FIG 1.
Framework for how antibiotic tolerance and persistence accelerate the evolution of genetic resistance. First, in populations displaying high levels of tolerance or persistence, an increased number of viable cells is available for mutation, increasing the likelihood for a resistant mutant to arise. Second, persistence is linked with higher mutation rates, again causing an increased likelihood for the occurrence of resistance-conferring mutations. Future research will reveal if a similar link exists between tolerance and mutation rates.
COMBATTING TOLERANCE TO STOP RESISTANCE
Continuous improvements in medical care increase the life span of immunocompromised patients, and the use of indwelling medical devices, prone to biofilm-related infections that shield bacteria from the activity of the immune system, is ever growing. Therefore, the clinical manifestation of antibiotic tolerance can be expected to increase considerably in the future. Given the recent studies that unravelled a link between tolerance or persistence and the evolution of genetic resistance, this will lead not only to an increased prevalence of chronic and recurrent infections but likely also to a further increase in the emergence and spread of antibiotic resistance. Therefore, to avoid the devastating consequences of the imminent postantibiotic era, we argue that the clinical focus should not lie exclusively with battling antibiotic resistance.
In clinical settings, screening for resistance by disk diffusion assays or MIC measurements has become routine practice to guide antibiotic therapy of bacterial infections (60). On the other hand, antibiotic tolerance and persistence are overlooked, mainly due to the lack of a similarly reliable, quantitative, and easily measurable parameter. As a consequence, data on the clinical manifestation of tolerance and persistence are rather scarce (9). On the basis of the decreased killing rate characterizing antibiotic-tolerant cells, the Balaban group introduced the “minimum duration for killing” (MDK) as a quantitative parameter to identify tolerance and persistence (28, 61). As time-kill assays are labor-intensive, the same group also described a modification of the standard disk diffusion assay that allows a semiquantitative evaluation of tolerance and persistence (62). However, further validation of these proposed metrics in species other than E. coli and in clinical isolates is necessary before they can be implemented by clinical microbiology labs. Moreover, it is important to note that tolerance and persistence strongly depend on the environmental conditions, necessitating their detection to occur in a context that resembles the in vivo infection environment (63). Current attempts to investigate this in vivo context include the use of engineered strains reporting on the environmental conditions and their impact on bacterial physiology (64, 65). This knowledge should be used to move away from conventional culture conditions toward model systems that mimic the infection environment (65).
Therapeutic choices could be further supported by sequence-based prediction of resistance and tolerance. The ever-decreasing costs of sequencing make the identification of tolerance- and persistence-associated genetic markers increasingly feasible (18). Nevertheless, knowledge gaps regarding the genetic basis of tolerance and persistence are currently hampering the use of DNA-based diagnostic tools. On the other hand, expression-based and physiology-based markers have been proposed as tools to diagnose antibiotic tolerance and persistence (18, 66). For example, Stokes et al. recently developed the “solid media portable cell killing” (SPOCK) assay, a method that allows high-throughput screening of antibiotic killing of tolerant bacteria (67). In this assay, antibiotic lethality against cells residing in colonies is monitored with a redox-sensitive dye (67). However, this method does not assess the capacity for regrowth of surviving cells, and therefore does not discriminate between antibiotic-tolerant cells and viable but nonculturable cells (VBNCs). As the existing methods to diagnose tolerance and persistence are clearly still in their infancy, we advocate that more research should be focused on the further development and valorization of these tools. A better understanding of the genetic basis and physiology of antibiotic-tolerant cells would improve the prospect of marker-based diagnostics. Furthermore, an expeditious introduction of these tools in screenings for novel antibiotics and clinical decision-making will become indispensable.
In addition to the development of diagnostic tools for tolerance and persistence, considerable efforts should be devoted to the development of strategies able to eliminate tolerant cells, as these have the potential to preclude the evolution of resistance. The effectiveness of this approach was illustrated in a proof-of-concept experiment where E. coli cultures were treated with mannitol. This compound is known to sensitize persister cells, and consequently slows down resistance development (44). Similarly, it has been proposed that inhibitors of the AcrAB-TolC efflux pump might not only sensitize cells to antibiotics but also restore their mutation rates, potentially leading to an improved treatment outcome (68).
Strikingly, drug-tolerant persisters are also present in cancer cell populations, where they are implicated in tumor recurrence (69, 70). An increasing body of evidence suggests that this subpopulation of phenotypic variants acts as a substantial reservoir for the emergence of therapy resistance (71, 72). This intriguing parallel between cancer and infections, two seemingly distinct types of disease, indicates that antipersister strategies may also help to improve the treatment outcome of cancer (73–75). Indeed, inhibition of the lipid hydroperoxidase GPX4, which is necessary for survival of persisters, results in persister cell death and consequently prevents the acquisition of drug resistance by cancerous cells (76). Understanding, detecting, and targeting tolerance and persistence will require joint efforts of microbiologists and clinicians and should eventually lead to reduced therapy failure in both infectious diseases and cancer.
ACKNOWLEDGMENTS
E.M.W. and B.V.D.B. are Research Foundation-Flanders (FWO) fellows, and J.E.M. is a fellow of the Agency for Innovation by Science and Technology (IWT). This research was funded by the KU Leuven Research Council (PF/10/010 and C16/17/006), FWO (G047112N, G0B2515N, and G055517N), and the Flemish Institute for Biotechnology (VIB). B.V.D.B. was also funded by a fellowship of the Belgian American Education Foundation (BAEF) and The European Molecular Biology Organization (EMBO).
We declare that we have no conflicts of interest.
E.M.W. and J.E.M. wrote the paper (contributed equally). B.V.D.B., M.F., and J.M. edited the paper.
Footnotes
Citation Windels EM, Michiels JE, Van den Bergh B, Fauvart M, Michiels J. 2019. Antibiotics: combatting tolerance to stop resistance. mBio 10:e02095-19. https://doi.org/10.1128/mBio.02095-19.
Contributor Information
Slava Epstein, Northeastern University.
Eric J. Rubin, Harvard School of Public Health.
REFERENCES
- 1.Lobanovska M, Pilla G. 2017. Penicillin’s discovery and antibiotic resistance: lessons for the future? Yale J Biol Med 90:135–145. [PMC free article] [PubMed] [Google Scholar]
- 2.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies J. 2006. Where have all the antibiotics gone? Can J Infect Dis Med Microbiol 17:287–290. doi: 10.1155/2006/707296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martens E, Demain AL. 2017. The antibiotic resistance crisis, with a focus on the United States. J Antibiot (Tokyo) 70:520–526. doi: 10.1038/ja.2017.30. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 6.Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, Colomb-Cotinat M, Kretzschmar ME, Devleesschauwer B, Cecchini M, Ouakrim DA, Oliveira TC, Struelens MJ, Suetens C, Monnet DL, Burden of AMR Collaborative Group. 2019. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis 19:56–66. doi: 10.1016/S1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cecchini M, Langer J, Slawomirski L. 2015. Resistance in G7 countries and beyond: economic issues, policies and options for action. Organisation for Economic Co-operation and Development, Paris, France. [Google Scholar]
- 8.OECD. 2018. Stemming the superbug tide: just a few dollars more. In OECD Health Policy Studies. OECD Publishing, Paris, France. [Google Scholar]
- 9.Brauner A, Fridman O, Gefen O, Balaban NQ. 2016. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 10.Tuomanen E, Cozens R, Tosch W, Zak O, Tomasz A. 1986. The rate of killing of Escherichia coli by β-lactam antibiotics is strictly proportional to the rate of bacterial growth. J Gen Microbiol 132:1297–1304. doi: 10.1099/00221287-132-5-1297. [DOI] [PubMed] [Google Scholar]
- 11.Michiels JE, Van den Bergh B, Verstraeten N, Michiels J. 2016. Molecular mechanisms and clinical implications of bacterial persistence. Drug Resist Updat 29:76–89. doi: 10.1016/j.drup.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Hobby G, Meyer K, Chaffee E. 1942. Observations on the mechanism of action of penicillin. Exp Biol Med 50:281–285. doi: 10.3181/00379727-50-13773. [DOI] [Google Scholar]
- 13.Bigger J. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244:497–500. doi: 10.1016/S0140-6736(00)74210-3. [DOI] [Google Scholar]
- 14.Sabath LD, Laverdiere M, Wheeler N, Blazevic D, Wilkinson BJ. 1977. A new type of penicillin resistance of Staphylococcus aureus. Lancet 309:443–447. doi: 10.1016/S0140-6736(77)91941-9. [DOI] [PubMed] [Google Scholar]
- 15.McDermott W. 1958. Microbial persistence. Yale J Biol Med 30:257–291. [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher RA, Gollan B, Helaine S. 2017. Persistent bacterial infections and persister cells. Nat Rev Microbiol 15:453–464. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- 17.Conlon BP. 2014. Staphylococcus aureus chronic and relapsing infections: evidence of a role for persister cells. Bioessays 36:991–996. doi: 10.1002/bies.201400080. [DOI] [PubMed] [Google Scholar]
- 18.Van den Bergh B, Michiels JE, Fauvart M, Michiels J. 2016. Should we develop screens for multidrug tolerance? Expert Rev Anti Infect Ther 14:613–616. doi: 10.1080/14787210.2016.1194754. [DOI] [PubMed] [Google Scholar]
- 19.Van den Bergh B, Fauvart M, Michiels J. 2017. Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol Rev 41:219–251. doi: 10.1093/femsre/fux001. [DOI] [PubMed] [Google Scholar]
- 20.Spoering AL, Lewis K. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol 183:6746–6751. doi: 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, Mckay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK, Youngman E, Wolberger C, Boeke JD. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K. 2013. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW. 2014. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schumacher MA, Balani P, Min J, Chinnam NB, Hansen S, Vulic M, Lewis K, Brennan RG. 2015. HipBA-promoter structures reveal the basis of heritable multidrug tolerance. Nature 524:59–66. doi: 10.1038/nature14662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van den Bergh B, Michiels JE, Wenseleers T, Windels EM, Vanden Boer P, Kestemont D, De Meester L, Verstrepen KJ, Verstraeten N, Fauvart M, Michiels J. 2016. Frequency of antibiotic application drives rapid evolutionary adaptation of Escherichia coli persistence. Nat Microbiol 1:16020. doi: 10.1038/nmicrobiol.2016.20. [DOI] [PubMed] [Google Scholar]
- 26.Mechler L, Herbig A, Paprotka K, Fraunholz M, Nieselt K, Bertram R. 2015. A novel point mutation promotes growth phase-dependent daptomycin tolerance in Staphylococcus aureus. Antimicrob Agents Chemother 59:5366–5376. doi: 10.1128/AAC.00643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulcahy LR, Burns JL, Lory S, Lewis K. 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol 192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. 2014. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513:418–421. doi: 10.1038/nature13469. [DOI] [PubMed] [Google Scholar]
- 29.Grahn E, Holm SE, Roos K. 1987. Penicillin tolerance in beta-streptococci isolated from patients with tonsillitis. Scand J Infect Dis 19:421–426. doi: 10.3109/00365548709021674. [DOI] [PubMed] [Google Scholar]
- 30.Okoro CK, Kingsley RA, Quail MA, Kankwatira AM, Feasey NA, Parkhill J, Dougan G, Gordon MA. 2012. High-resolution single nucleotide polymorphism analysis distinguishes recrudescence and reinfection in recurrent invasive nontyphoidal Salmonella Typhimurium disease. Clin Infect Dis 54:955–963. doi: 10.1093/cid/cir1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ejrnaes K, Sandvang D, Lundgren B, Ferry S, Holm S, Monsen T, Lundholm R, Frimodt-Moller N. 2006. Pulsed-field gel electrophoresis typing of Escherichia coli strains from samples collected before and after pivmecillinam or placebo treatment of uncomplicated community-acquired urinary tract infection in women. J Clin Microbiol 44:1776–1781. doi: 10.1128/JCM.44.5.1776-1781.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bingen E, Denamur E, Lambert-Zechovsky N, Braimi N, El Lakany M, Elion J. 1992. DNA restriction fragment length polymorphism differentiates recurrence from relapse in treatment failures of Streptococcus pyogenes pharyngitis. J Med Microbiol 37:162–164. doi: 10.1099/00222615-37-3-162. [DOI] [PubMed] [Google Scholar]
- 33.Kaiser P, Regoes RR, Dolowschiak T, Wotzka SY, Lengefeld J, Slack E, Grant AJ, Ackermann M, Hardt W. 2014. Cecum lymph node dendritic cells harbor slow-growing bacteria phenotypically tolerant to antibiotic treatment. PLoS Biol 12:e1001793. doi: 10.1371/journal.pbio.1001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claudi B, Spröte P, Chirkova A, Personnic N, Zankl J, Schürmann N, Schmidt A, Bumann D. 2014. Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell 158:722–733. doi: 10.1016/j.cell.2014.06.045. [DOI] [PubMed] [Google Scholar]
- 35.Adams KN, Takaki K, Connolly LE, Wiedenhoft H, Winglee K, Humbert O, Edelstein PH, Cosma CL, Ramakrishnan L. 2011. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 145:39–53. doi: 10.1016/j.cell.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manina G, Dhar N, McKinney JD. 2015. Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe 17:32–46. doi: 10.1016/j.chom.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 37.Brennan RO, Durack DT. 1983. Therapeutic significance of penicillin tolerance in experimental streptococcal endocarditis. Antimicrob Agents Chemother 23:273–277. doi: 10.1128/aac.23.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balaban NQ, Helaine S, Lewis K, Ackermann M, Aldridge B, Andersson DI, Brynildsen MP, Bumann D, Camilli A, Collins JJ, Dehio C, Fortune S, Ghigo J-M, Hardt W-D, Harms A, Heinemann M, Hung DT, Jenal U, Levin BR, Michiels J, Storz G, Tan M-W, Tenson T, Van Melderen L, Zinkernagel A. 2019. Definitions and guidelines for research on antibiotic persistence. Nat Rev Microbiol 17:441–448. doi: 10.1038/s41579-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreillon P, Tomasz A. 1988. Penicillin resistance and defective lysis in clinical isolates of pneumococci: evidence for two kinds of antibiotic pressure operating in the clinical environment. J Infect Dis 157:1150–1157. doi: 10.1093/infdis/157.6.1150. [DOI] [PubMed] [Google Scholar]
- 40.Novak R, Henriques B, Charpentier E, Normark S, Tuomanen E. 1999. Emergence of vancomycin tolerance in Streptococcus pneumoniae. Nature 399:590–593. doi: 10.1038/21202. [DOI] [PubMed] [Google Scholar]
- 41.Levin BR, Rozen DE. 2006. Non-inherited antibiotic resistance. Nat Rev Microbiol 4:556–562. doi: 10.1038/nrmicro1445. [DOI] [PubMed] [Google Scholar]
- 42.Sebastian J, Swaminath S, Nair RR, Jakkala K, Pradhan A, Ajitkumar P. 2017. De novo emergence of genetically resistant mutants of Mycobacterium tuberculosis from the persistence phase cells formed against antituberculosis drugs in vitro. Antimicrob Agents Chemother 61:e01343-16. doi: 10.1128/AAC.01343-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. 2017. Antibiotic tolerance facilitates the evolution of resistance. Science 355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 44.Windels EM, Michiels JE, Fauvart M, Wenseleers T, Van den Bergh B, Michiels J. 2019. Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J 13:1239–1251. doi: 10.1038/s41396-019-0344-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poole K. 2012. Bacterial stress responses as determinants of antimicrobial resistance. J Antimicrob Chemother 67:2069–2089. doi: 10.1093/jac/dks196. [DOI] [PubMed] [Google Scholar]
- 46.Cirz RT, Chin JK, Andes DR, de Crécy-Lagard V, Craig WA, Romesberg FE. 2005. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol 3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foster PL. 2007. Stress-induced mutagenesis in bacteria. Crit Rev Biochem Mol Biol 42:373–397. doi: 10.1080/10409230701648494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Al Mamun AAM, Lombardo M-J, Shee C, Lisewski AM, Gonzalez C, Lin D, Nehring RB, Saint-Ruf C, Gibson JL, Frisch RL, Lichtarge O, Hastings PJ, Rosenberg SM. 2012. Identity and function of a large gene network underlying mutagenic repair of DNA breaks. Science 338:1344–1348. doi: 10.1126/science.1226683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pribis JP, García-Villada L, Zhai Y, Lewin-Epstein O, Wang AZ, Liu J, Xia J, Mei Q, Fitzgerald D, Bos J, Austin RH, Herman C, Bates D, Hadany L, Hastings PJ, Rosenberg SM. 2019. Gamblers: an antibiotic-induced evolvable cell subpopulation differentiated by reactive-oxygen-induced general stress response. Mol Cell 74:785–800. doi: 10.1016/j.molcel.2019.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cohen NR, Lobritz MA, Collins JJ. 2013. Microbial persistence and the road to drug resistance. Cell Host Microbe 13:632–642. doi: 10.1016/j.chom.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kussell E, Kishony R, Balaban NQ, Leibler S. 2005. Bacterial persistence: a model of survival in changing environments. Genetics 169:1807–1814. doi: 10.1534/genetics.104.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carja O, Liberman U, Feldman MW. 2014. Evolution in changing environments: modifiers of mutation, recombination, and migration. Proc Natl Acad Sci U S A 111:17935–17940. doi: 10.1073/pnas.1417664111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishii K, Matsuda H, Iwasa Y, Sasaki A. 1989. Evolutionarily stable mutation rate in a periodically changing environment. Genetics 121:163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bridges BA. 1997. DNA turnover and mutation in resting cells. Bioessays 19:347–352. doi: 10.1002/bies.950190412. [DOI] [PubMed] [Google Scholar]
- 55.Loewe L, Textor V, Scherer S. 2003. High deleterious genomic mutation rate in stationary phase of Escherichia coli. Science 302:1558–1560. doi: 10.1126/science.1087911. [DOI] [PubMed] [Google Scholar]
- 56.Bjedov I, Tenaillon O, Gérard B, Souza V, Denamur E, Radman M, Taddei F, Matic I. 2003. Stress-induced mutagenesis in bacteria. Science 300:1404–1409. doi: 10.1126/science.1082240. [DOI] [PubMed] [Google Scholar]
- 57.Barrett TC, Mok WWK, Murawski AM, Brynildsen MP. 2019. Enhanced antibiotic resistance development from fluoroquinolone persisters after a single exposure to antibiotic. Nat Commun 10:1177. doi: 10.1038/s41467-019-09058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yaakov G, Lerner D, Bentele K, Steinberger J, Barkai N. 2017. Coupling phenotypic persistence to DNA damage increases genetic diversity in severe stress. Nat Ecol Evol 1:16. doi: 10.1038/s41559-016-0016. [DOI] [PubMed] [Google Scholar]
- 59.El Meouche I, Dunlop MJ. 2018. Heterogeneity in efflux pump expression predisposes antibiotic resistant cells to mutation. Science 690:686–690. doi: 10.1126/science.aar7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jenkins SG, Schuetz AN. 2012. Current concepts in laboratory testing to guide antimicrobial therapy. Mayo Clin Proc 87:290–308. doi: 10.1016/j.mayocp.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brauner A, Shoresh N, Fridman O, Balaban NQ. 2017. An experimental framework for quantifying bacterial tolerance. Biophys J 112:2664–2671. doi: 10.1016/j.bpj.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gefen O, Chekol B, Strahilevitz J, Balaban NQ. 2017. TDtest: easy detection of bacterial tolerance and persistence in clinical isolates by a modified disk-diffusion assay. Sci Rep 7:1–9. doi: 10.1038/srep41284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang JH, Bening SC, Collins JJ. 2017. Antibiotic efficacy — context matters. Curr Opin Microbiol 39:73–80. doi: 10.1016/j.mib.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Certain LK, Way JC, Pezone MJ, Collins JJ. 2017. Using engineered bacteria to characterize infection dynamics and antibiotic effects in vivo. Cell Host Microbe 22:263–268.e4. doi: 10.1016/j.chom.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 65.Meylan S, Andrews IW, Collins JJ. 2018. Targeting antibiotic tolerance, pathogen by pathogen. Cell 172:1228–1238. doi: 10.1016/j.cell.2018.01.037. [DOI] [PubMed] [Google Scholar]
- 66.Haaber J, Friberg C, McCreary M, Lin R, Cohen SN, Ingmer H. 2015. Reversible antibiotic tolerance induced in Staphylococcus aureus by concurrent drug exposure. mBio 6:e02268-14. doi: 10.1128/mBio.02268-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stokes JM, Gutierrez A, Lopatkin AJ, Andrews IW, French S, Matic I, Brown ED, Collins JJ. 2019. A multiplexable assay for screening antibiotic lethality against drug-tolerant bacteria. Nat Methods 16:303–306. doi: 10.1038/s41592-019-0333-y. [DOI] [PubMed] [Google Scholar]
- 68.Frimodt-Møller J, Løbner-Olesen A. 2019. Efflux-pump upregulation: from tolerance to high-level antibiotic resistance? Trends Microbiol 27:291–293. doi: 10.1016/j.tim.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 69.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, Wong KK, Brandstetter K, Wittner B, Ramaswamy S, Classon M, Settleman J. 2010. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Knoechel B, Roderick JE, Williamson KE, Zhu J, Lohr JG, Cotton MJ, Gillespie SM, Fernandez D, Ku M, Wang H, Piccioni F, Silver SJ, Jain M, Pearson D, Kluk MJ, Ott CJ, Shultz LD, Brehm MA, Greiner DL, Gutierrez A, Stegmaier K, Kung AL, Root DE, Bradner JE, Aster JC, Kelliher MA, Bernstein BE. 2014. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet 46:364–370. doi: 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, Maruvka YE, Ji F, Bhang H-E, Krishnamurthy Radhakrishna V, Siravegna G, Hu H, Raoof S, Lockerman E, Kalsy A, Lee D, Keating CL, Ruddy DA, Damon LJ, Crystal AS, Costa C, Piotrowska Z, Bardelli A, Iafrate AJ, Sadreyev RI, Stegmeier F, Getz G, Sequist LV, Faber AC, Engelman JA. 2016. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med 22:262–269. doi: 10.1038/nm.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramirez M, Rajaram S, Steininger RJ, Osipchuk D, Roth MA, Morinishi LS, Evans L, Ji W, Hsu C-H, Thurley K, Wei S, Zhou A, Koduru PR, Posner BA, Wu LF, Altschuler SJ. 2016. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat Commun 7:1–8. doi: 10.1038/ncomms10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oxnard GR. 2016. The cellular origins of drug resistance in cancer. Nat Med 22:232–234. doi: 10.1038/nm.4058. [DOI] [PubMed] [Google Scholar]
- 74.Vallette FM, Olivier C, Lézot F, Oliver L, Cochonneau D, Lalier L, Cartron P-F, Heymann D. 2019. Dormant, quiescent, tolerant and persister cells: four synonyms for the same target in cancer. Biochem Pharmacol 162:169–176. doi: 10.1016/j.bcp.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 75.Glickman MS, Sawyers CL. 2012. Converting cancer therapies into cures: lessons from infectious diseases. Cell 148:1089–1098. doi: 10.1016/j.cell.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hangauer MJ, Viswanathan VS, Ryan MJ, Bole D, Eaton JK, Matov A, Galeas J, Dhruv HD, Berens ME, Schreiber SL, McCormick F, McManus MT. 2017. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 551:247–250. doi: 10.1038/nature24297. [DOI] [PMC free article] [PubMed] [Google Scholar]