Abstract
Little is known about the post-transcriptional mechanisms that modulate the genetic effects in the molecular pathways underlying Alzheimer disease (AD), and even less is known about how these changes might differ across diverse populations. RNA editing, the process that alters individual bases of RNA, may contribute to AD pathogenesis due to its roles in neuronal development and immune regulation. Here, we pursued one of the first transcriptome-wide RNA editing studies in AD by examining RNA sequencing data from individuals of both African-American (AA) and non-Hispanic White (NHW) ethnicities. Whole transcriptome RNA sequencing and RNA editing analysis were performed on peripheral blood specimens from 216 AD cases (105 AA, 111 NHW) and 212 gender matched controls (105 AA, 107 NHW). 449 positions in 254 genes and 723 positions in 371 genes were differentially edited in AA and NHW, respectively. While most differentially edited sites localized to different genes in AA and NHW populations, these events converged on the same pathways across both ethnicities, especially endocytic and inflammatory response pathways. Furthermore, these differentially edited sites were preferentially predicted to disrupt miRNA binding and induce nonsynonymous coding changes in genes previously associated with AD in molecular studies, including PAFAH1B2 and HNRNPA1. These findings suggest RNA editing is an important post-transcriptional regulatory program in AD pathogenesis.
Introduction
Alzheimer disease (AD) is a common neurodegenerative disease characterized by progressive loss of cognitive functioning, particularly in the domains of short-term memory, language, orientation, behavior and motor skills, and by pathological findings of amyloid plaques and neurofibrillary tangles leading to atrophy (1). AD is the most common form of dementia and the 6th leading cause of death in the USA (2). It affects all racial and ethnic groups, with African-American (AA) individuals at increased risk (3–6).
Late-onset AD (age at onset >=65) is highly heritable with an estimated 74% of liability explained by genetic factors (7,8), the most significant of which is the ε4 allele of apolipoprotein E (APOE) on Chromosome 19q13 (9–13). More than 30 genetic loci have been implicated in AD risk through large-scale genomic studies (14–17), yet they explain no more than 50% of the heritability of AD (18). While these studies of AD have focused primarily on non-Hispanic white (NHW) individuals, AA individuals are at increased risk for AD due to both environmental and hereditary factors (19). There is growing evidence that the genetic architecture of AD differs across ethnicities. For example, the risk of AD associated with the APOEε4 allele is significantly higher in NHW compared to AA individuals (20), and there are AA specific risk variants in genes such as ABCA7 (17,21). Across both ethnicities, though, genetic loci associated with AD fall within or near genes that are enriched in molecular pathways including lipid metabolism (e.g. APOE, ABCA7), immune regulation (e.g. SORL1, TREM2) and endocytosis (e.g. BIN1), implicating these processes in disease pathogenesis (14,16).
The majority of AD-associated genetic variants are located in non-coding regions, suggesting the importance of gene transcription regulatory function (18), thus several studies have attempted to identify transcriptional alterations in AD. For example, in postmortem cortical tissue, differential gene expression has been observed for several AD risk genes, including ABCA7, BIN1, CD33, CLU, FRMD4A, PTK2B, TREM2 and genes within the HLA and MS4A clusters (22–25). Studies in glial cells have shown differences in gene expression in AD patients versus controls in metabolic and immune-related pathways, particularly acute phase response, cytokine, cell adhesion and interferons (26,27). However, these studies have focused exclusively on NHW samples, highlighting a need for more transcriptomic studies in multi-ethnic populations. Furthermore, little work has been done to characterize how genes involved in AD-related biological processes are affected by post-transcriptional regulatory events like RNA editing, a process that alters not the quantity of RNA that is transcribed in a cell but rather the specific sequence coded by those RNA molecules.
RNA editing is necessary for life due to its role in biological processes including neuronal functioning and immune regulation (28–30). RNA editing is the enzyme-mediated process by which individual bases in double-stranded mRNA are altered. The most common (‘canonical’) type of RNA editing is the deamination of adenosine (A) bases to inosine (I) by the Adenosine Deaminase RNA Specific (ADAR) family of enzymes, and the second is the deamination of cytidine (C) to uridine (U) by the activation induced cytidine deaminase (AID)/apolipoprotein B editing complex (APOBEC) cytidine deaminases (31–35). Editing events can introduce coding changes, alter splice sites, create or disrupt microRNA (miRNA)-binding sites, or alter other RNA processing events (36,37). Ethnic variation in cis polymorphisms influences the rate and location of editing through their effects on RNA secondary structure (38,39).
Altered RNA editing levels at specific sites have been associated with various neurological disorders, such as Aicardi–Goutieres syndrome, amyotrophic lateral sclerosis, epilepsy, major depression, schizophrenia and AD (40–42). The previous studies of RNA editing in AD have shown altered editing in autopsy-derived hippocampal tissue at specific sites (43–45), most prominently at the GluA2 QR site (44). However, the scope of these studies has largely been limited to pre-selected sites within synaptic genes, and genome-wide studies of RNA editing in AD are lacking. In addition, no work has been done to study the role of RNA editing on immune effectors in the context of AD, despite increasing evidence for the role of immunity in mediating disease pathogenesis, nor to study RNA editing in different ethnicities.
Herein, we describe a screen for transcriptome-wide AD-related changes of RNA editing in whole blood from both AA and NHW populations. We chose to study whole blood for multiple reasons, including that immune and endocytic processes, which are implicated in AD but poorly understood, are highly active in blood (46,47). In addition, blood is more accessible than brain while still reflecting many of the known regulatory mechanisms that occur in brain (48). Since there is evidence of racial- and ethnic-specific genetic risk factors in AD, here we tested whether RNA editing in blood differs between AD cases and controls and examine whether those differences generalized across ethnicities or are population-specific. We hypothesize that the specific loci with AD-associated editing changes will differ between ethnicities and yet AD-specific differences in RNA editing will converge on similar AD-related biological processes.
Results
Identifying RNA-edited sites
A total of 44 985 RNA editing events meeting our quality control metrics of ≥10× coverage, minimum average Phred-scaled base quality of 25, and minimum alignment quality of 20 were detected across whole-blood RNAseq data from 105 AA cases, 111 NHW cases, 105 AA controls and 107 NHW controls. Of those, 88% were of the canonical A-to-I (82%) or C-to-U (6%) types (Supplementary Material, Fig. S1a). 83% of all identified editing events and 94% of identified A-to-I events have been reported previously within the RADAR and DARNED databases (Supplementary Material, Fig. S1a). Editing events primarily occurred in noncoding regions, especially introns and 3′ untranslated regions (UTRs), and 86% were in Alu repeat regions which are primate-specific repetitive elements that comprise ∼ 10% of the human genome. (Supplementary Material, Fig. S1b). Overall, the average frequency of editing was 0.28 (SD = 0.10, Supplementary Material, Fig. S1c).
Transcriptome-wide RNA editing analysis
There were no significant differences in the global number of editing events when comparing all AD cases against all controls nor when comparing AD cases and controls stratified by ethnicity. This was also true when examining only A-to-I events alone (Supplementary Material, Fig. S2a). Similarly, there was no significant difference between any of the four groups, AA cases, AA controls, NHW cases and NHW controls, in the average type of substitution or the genomic distribution of editing events (Supplementary Material, Fig. S2b and c). There were no notable differences in the expression of ADAR, the enzyme that mediates most RNA editing events, between AD cases and controls of either race, consistent with previous studies (43,45).
Differentially edited sites
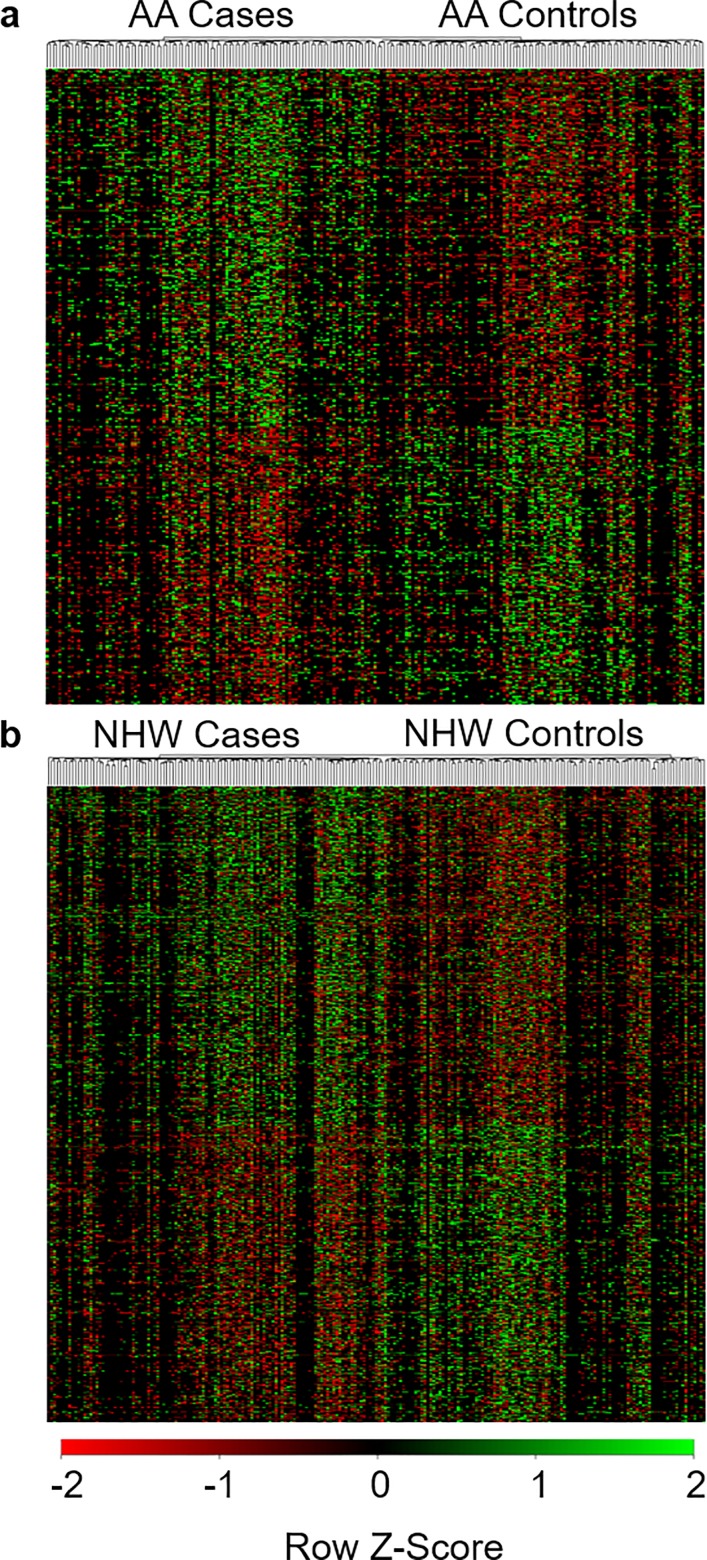
We identified specific sites that were differentially edited between cases and controls using the logistic regression model. AA and NHW samples were analyzed separately to account for potential ethnic differences in overall editing and to identify ethnicity-specific editing signatures. Across both ethnicities, we detected differential editing in a total of 550 genes. In AA, a total of 449 sites in 273 genes were differentially edited (Fig. 1a), and, in NHW, 723 sites in 371 genes were differentially edited (Fig. 1b). Ninety-four genes had at least one differentially edited site in both AA and NHW. This overlap was significantly greater than that expected if sites were randomly distributed across the genome (P = 7 × 10−5). 82% of differentially edited sites were of the canonical A-to-I type and most were noncoding (Supplementary Material, Fig. S3).
Figure 1.
Differentially edited sites. (A, B) Sites that are differentially edited between (A) AA and (B) NHW cases and controls. Increased editing at each site is in green and decreased editing in red.
Editing in AD-associated genes
Of the 24 genes significantly associated with AD in the most recent genome-wide association study (GWAS, 49), 11 contained edited sites identified in our study (Supplementary Material, Table S2). Sites that were differentially edited between AD cases and controls in both ethnicities were located within two of these AD-associated genes, SPI1 and SORL1. INPP5D and ADAM10 contained sites that were differentially edited in AA alone.
Functional annotation of differentially edited sites
Five of the 550 differentially edited genes included differentially edited sites predicted to disrupt miRNA target sites (Table 1). Two of these genes, PAFAH1B2 and HNRNPA1, each had two differentially edited sites within each gene disrupting miRNA binding sites. Only one of these five genes (CTC1) showed decreased editing in AD cases, indicating greater miRNA-binding affinity relative to controls, while the remaining four genes showed increased editing at miRNA target sites in AD cases. Notably, CTC1 was also the only gene in AA with differential editing predicted to affect miRNA binding.
Table 1.
Differentially edited sites disrupting miRNA binding sites
| Race | Position | Feature | Gene | miRNA | Mean Diff. | P-value |
|---|---|---|---|---|---|---|
| NHW | 1:52818232 | 3UTR | CC2D1B | miR-145-5p | 0.130456 | 0.038833 |
| NHW | 11:117039192 | 3UTR | PAFAH1B2 | miR-505-3p.1 | 0.067959 | 0.045713 |
| NHW | 11:117039195* | 3UTR | PAFAH1B2 | miR-505-3p.1 | 0.072055 | 0.03772 |
| NHW | 12:54679031 | intron | HNRNPA1 | miR-582-5p | 0.085919 | 0.009666 |
| NHW | 12:54679032 | intron | HNRNPA1 | miR-582-5p | 0.082697 | 0.019395 |
| NHW | 22:39260256 | 3UTR | CBX6 | miR-129-5p | 0.097871 | 0.040377 |
| AA | 17:8129592* | 3UTR | CTC1 | miR-28-3p | −0.10383 | 0.028583 |
A positive mean difference (Mean Diff.) indicates increased editing in cases, and a negative mean difference indicates decreased editing in cases. *denotes changes affecting the seed region of the miRNA binding site.
In addition, 11 differentially edited sites functionally affected proteins by inducing single amino acid substitutions within coding regions (Table 2). Of the 11 editing-induced amino acid substitutions, 5 were predicted to be damaging to protein function by the SIFT prediction algorithm (50).
Table 2.
Differentially edited sites where edits result in nonsynonymous coding changes
| Race | Position | Gene | Change | Prediction | Mean Diff. | P-value |
|---|---|---|---|---|---|---|
| NHW | 11:93463255 | CEP295 | E > K | Damaging | 0.089847 | 0.024796 |
| NHW | 16:89754057 | CDK10 | S > G | Tolerated | −0.01237 | 0.030494 |
| NHW | 1:117944875 | MAN1A2 | E > K | Tolerated | 0.126331 | 0.036984 |
| NHW | 22:46688132 | TTC38 | N > S | Damaging | 0.030061 | 0.043901 |
| AA | 17:45786136 | TBKBP1 | Q > P | Tolerated | −0.02873 | 0.00488 |
| AA | 13:20222592 | MPHOSPH8 | E > K | Tolerated | 0.011837 | 0.010106 |
| AA | 16:2011667 | NDUFB10 | T > P | Damaging | −0.00519 | 0.018482 |
| AA | 17:43219954 | ACBD4 | T > A | Damaging | −0.03817 | 0.019082 |
| AA | 17:7751162 | KDM6B | L > P | Damaging | −0.03452 | 0.043542 |
Predictions were made using the SIFT algorithm. A positive mean difference (Mean Diff.) indicates increased editing in cases, and a negative mean difference indicates decreased editing in cases.
Pathway analysis
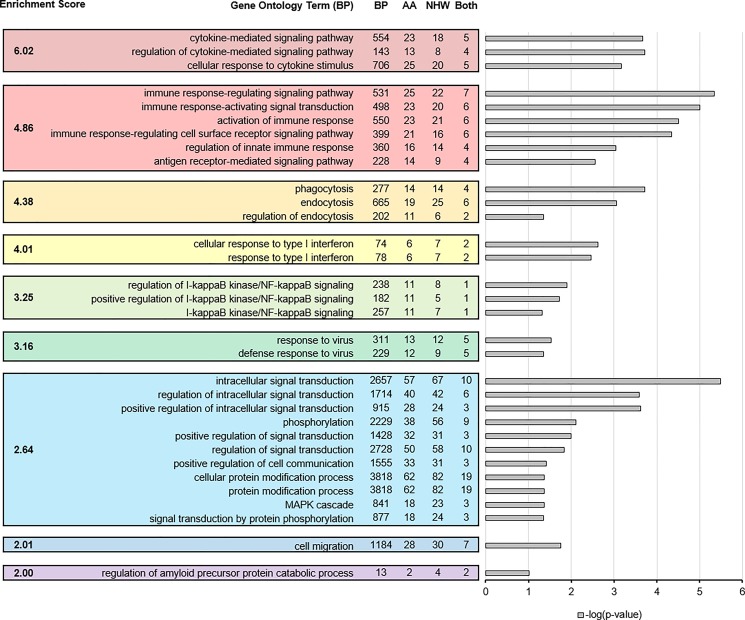
Gene ontology term enrichment for the entire set of differentially edited genes across both ethnicities showed that these differential editing events were enrichened for biological processes involved in immune regulation, inflammation and endocytosis (Fig. 2). Type-1 interferon signaling, the NF-kappa B signaling cascade, and negative regulation of MAP kinase (MAPK) cascade were particularly enriched. Regulation of amyloid precursor protein catabolism was also enriched.
Figure 2.
Functional annotation clusters (‘biological process’, BP) of differentially edited genes (DAVID). Enriched clusters and characteristic examples of their GO Terms are shown, together with their enrichment scores, in the colored boxes. Clusters of enriched pathways that are similar in function were identified using a minimum enrichment score of 2.0, corresponding with an aggregated P < 0.01. Columns next to terms shows total number of genes in pathway (BP) and the number of genes for each enrichment term that were differentially edited in AA, NHW and both. Bars show the negative log of Benjamini–Hochberg corrected P-values.
When looking at the genes uniquely edited in each ethnicity, AA and NHW groups both showed significant enrichment for immune and endocytic processes (Supplementary Material, Fig. S4). However, the set of genes that were differentially edited in only AA showed stronger enrichment for immune-related terms than those in NHW (AA P = 5.72 × 10−6, NHW P = 0.019). The set of genes that was differentially edited in both groups was likewise enriched for immune and endocytic terms, with additional enrichment for protein metabolic processes. In all, 46% of all differentially edited genes in AA and NHW groups were associated with either immune system process or vesicle-mediated transport terms in gene ontology.
Furthermore, we conducted a gene-based association study to identify genes that are significantly enriched with multiple differentially edited sites. This set included 31 genes in AA, including AD candidate gene INPP5D, and 12 genes in NHW (Supplementary Material, Table S3). The 43 total enriched genes also included two APOBEC complex genes. Similar to our single site per gene analysis, these 43 significant genes by gene-based analysis were enriched for gene ontology terms related to immune responses, when the two ethnicities were evaluated together or separately.
Discussion
We explored how RNA editing in AD contributes to the regulation of AD-related processes in blood cells in both AA and NHW populations. Differences between AD cases and controls were identified at the site-specific and gene-specific level in both AA and NHW, and those changes were enriched in genes involved in immune regulatory, inflammatory and endocytic processes across both ethnicities. Notably, SORL1 and SPI1 were differentially edited in both AA and NHW and have previously been associated with AD in both ethnicities by GWAS (51,52). Differential editing was observed in two additional AD-associated genes, INPP5D and ADAM10, in AA alone. Other differentially edited genes were enriched for key AD pathways such as interferon signaling, NF-kappa B pathways, the MAPK cascade, regulation of endocytosis and amyloid precursor protein catabolism (46,47,53,54). In fact, nearly half of all differentially edited genes in AA and NHW whole blood were associated with either immune system process or vesicle-mediated transport gene ontology terms. Lipid metabolism was not enriched in our dataset, despite its role in AD, but differentially edited sites were found within several genes in lipid metabolism pathways including ACBD4, which contained a differentially edited site resulting in a deleterious coding change. Interestingly, while a number of differentially edited sites localized to different genes in AA and NHW, the differential editing events converged on the same pathways across both ethnicities. These different editing sites supports the notion that AA and NHW may have different underlying risk factors, in addition to previously identified differences in heritable risk factors (17,20,21), which lead to the same pathological changes.
Several differentially edited sites are predicted to disrupt miRNA binding or to induce deleterious coding changes, including in genes associated with AD-related pathways. Of particular note, PAFAH1B2 and HNRNPA1 each contained two separate differentially edited sites predicted to increase disruption of miRNA binding in cases, and strikingly both genes have previously been linked to AD-phenotypes in molecular studies (55–57). The only gene in AA with differential editing predicted to affect miRNA binding was CTC1. CTC1 codes a telomere maintenance protein whose function is known to be influenced by ethnicity and to contribute to the longer leukocyte telomere length observed in those of African ancestry versus those of European ancestry (58–60). CTC1 has also been implicated in neurological functioning and disease in previous studies, including a transcriptomic study of AD (61).
Other differentially edited sites resulted in deleterious nonsynonymous coding changes in genes associated with lipid-metabolism (ACBD4) (62,63), the NADH dehydrogenase complex (NDUFB10) (64,65) and inflammation (KDM6B, TTC38) (62,66,67). KDM6B specifically influences neurodegenerative diseases, including the immune pathogenesis of Parkinson’s disease (68), and is involved in modulating the NF-kappa B and MAPK pathways (67,69). This coding site in KDM6B was differentially edited in AA alone, highlighting the need for multiethnic studies to identify novel disease gene candidates. Our findings indicate that altered RNA editing in AD has functional consequences affecting known AD-related genes in key AD pathways.
Here we have demonstrated that AD-associated changes of RNA editing in whole blood affect genes enriched in pathways including endocytosis, inflammation and immune regulation in both AA and NHW. AD-associated RNA editing changes at specific sites provide evidence of a post-transcriptional program contributing to the underlying complexity of AD pathophysiology and suggest that post-transcriptional dysregulation may contribute broadly to the pathogenesis of AD. As such, post-transcriptional modifications of RNA may provide a tool with which to identify new potential therapeutic targets in AD, and AD-associated RNA editing signatures in blood raise the possibility of creating new biomarkers for disease-associated processes. Furthermore, we found that the specific loci of differential editing between AD cases and cognitively intact controls varied by ethnicity, including in genes that are known to be influenced by ethnicity such as CTC1. Despite involving different genes, these events converged on the same pathways. This indicates that these populations have different underlying risk factors contributing to the same pathological processes, a key finding for the development of therapeutics and biomarkers that are effective across populations. Overall, this study is the first multi-ethnic study of RNA editing in AD as well as the first study to demonstrate that RNA editing affects key genes and pathways in AD in whole blood, and the results presented here have important implications for the role of post-transcriptional modifications in modulating pathogenic processes in AD.
Methods and Materials
Sample collection
All participants in this study provided informed consent or the immediate next of kin or legal representative provided written consent on the behalf of the participant prior to their inclusion with oversight by the University of Miami Institutional Review Board protocol #20070307. Participants include 428 individuals (AA—105 with AD, 105 cognitively intact; NHW—111 with AD, 107 cognitively intact) ascertained by the John P. Hussman Institute for Human Genomics at the University of Miami Miller School of Medicine (Miami, FL), North Carolina A&T State University (Greensboro, NC) and Case Western Reserve University (Cleveland, OH) (Supplementary Material, Table S1).
All participants underwent rigorous phenotyping; diagnostic criteria followed the previously described criteria of the National Institute of Neurological and Communicative Disorders and Stroke—Alzheimer’s Disease and Related Disorders Association (17,70,71). The cognitive status of controls was measured with either the Mini-Mental State Examination (72) or with the modified Mini-Mental State (73) combined with the clinical dementia rating scale, which assesses functional decline (74). Individuals self-identified their race and ethnicity; genetic ancestry was confirmed using EIGENSTRAT analysis of existing genome-wide genotyping data.
RNA extraction and sequencing
RNA was isolated from whole blood collected in PAXgene RNA tubes (PreAnalytiX, Hombrechtikon, Switzerland) utilizing automation on the QIAsymphony instrument (Qiagen, Germantown, MD) and quantified on the 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA). RNA was required to have an RNA integrity score (RIN) ≥ 5. 500 ng of total RNA was prepped for sequencing using the NuGEN Universal Plus mRNA-Seq with globin and ribosomal depletion (NuGEN, San Carlos, CA). Libraries were sequenced on paired end 100 bp reactions to generate at least 40 million reads on the Illumina HiSeq3000 (Illumina, San Diego, CA). Raw FASTQ reads were processed through an in house bioinformatics pipeline including quality and adapter trimming with TrimGalore version 0.4.2 (https://github.com/FelixKrueger/TrimGalore), alignment with the STAR aligner 2.5.0a (75) to the human genome hg19/GRCh37, and base quality recalibration. PCR duplicates were removed with Picard 1.103 (http://broadinstitute.github.io/picard).
Identification of RNA editing sites with REDItools
We used the REDItools Denovo software package, version 1.0.4, to create tables of every potential RNA editing site (76). To be included in the analysis, edited sites were required to pass the following QC measures: minimum depth of coverage 10×, minimum base quality of 25, no overlap with known SNPs in the dbSNP138 database (77), mean mapping quality greater than 20 and REDItools confidence in editing call with P < 0.05. Annotation of each editing site in the selected files was achieved by using the hg19 build of the Human RefSeq database (62), the UCSC RepeatMask table (78) and the DARNED (79) and RADAR (80) editing databases (access date for RefSeq and RepeatMask: 7/18/2017; access date for DARNED and RADAR: 7/20/2017). Sites were removed if they were located in a region of low sequence complexity or simple repeat as defined by the UCSC RepeatMask table. To eliminate artefacts and rare SNPs, novel sites were removed if intronic within 4 bp of an intron/exon boundary, if the average editing frequency across the population was less than 0.1 or if the edit was present in <5% of subjects (81).
Global analysis
Global editing frequency is a single value calculated by dividing the total number of edited reads by the total number of reads across all edited positions. A two-tailed student’s t-test was used to compare mean global editing frequencies between cases and controls with a significance level of 0.05. An ANOVA test with a significance level of 0.05 was used to test for differences in global editing frequencies between the four groups, AA cases, AA controls, NHW cases and NHW controls. Because A-to-I edits are the most common and are considered ‘canonical’ edits, we tested for differences in the editing frequency at A-to-I events alone, in addition to testing editing frequency across all detected sites.
Identification of differentially edited sites
Editing frequency at an individual site is given by the number of edited reads divided by the total number of reads mapped to that specific site. Individual sites that were differentially edited between cases and controls were identified using a logistic regression model in R software version 3.3.1 (https://www.r-project.org/). For each edited site, this model included case control status as the outcome variable, editing percentage as an independent variable and age of exam, sex, collection center, sequencing batch and overall coverage (number of sites with reads mapped to those sites for each sample) as covariate variables. AA and NHW were analyzed separately. Because RNA editing in coding regions is more tightly regulated than in non-coding regions, we applied different criteria for these two groups. Differentially edited sites in coding regions were required to have a marginal P < 0.05 and an absolute difference in mean frequency of editing between cases and controls greater than 0.05 to select for sites that were more likely to have a substantial functional effect, in line with established protocols (82). We relaxed our parameters to include marginally differentially edited sites in non-coding regions, which had P < 0.05 and an absolute mean difference in editing frequency of 0.01 between cases and controls. Genes identified in Kunkle et al. (49), were used to identify sites within AD-associated genes.
To determine if the overlap of differentially edited sites between AA and NHW was significantly more than expected by chance, given total number of sites (Ntotal) and observed number of significant sites in each ethnicity (NsigAA, NsigNHW), we conducted a simulation study. Briefly, we randomly sampled NsigAA and NsigNHW sites from Ntotal sites, and computed the number of overlapping sites. This procedure was repeated 100 000 times to obtain the distribution of the number of overlapping sites expected by chance (the null distribution). The P-value was estimated by the number of times the observed number of overlaps was greater than the number of overlaps in null distribution.
Gene ontology enrichment analysis
We analyzed the set of genes with at least one differentially edited site against the set of genes that are expressed in blood for gene ontology term enrichment using the DAVID GO biological process enrichment analysis tool. Significance was determined using Benjamini–Hochberg corrected P < 0.1 (83). The DAVID functional annotation-clustering tool was used to identify clusters of pathways with an enrichment score greater than 2.0, corresponding with an aggregated P < 0.01. Pathway analysis results in DAVID were further confirmed using the GOSeq software (84), which accounts for gene length differences in the genes (longer genes are more likely to include significantly differentially edited sites, simply because they have more sites). In both tests, GTEx genes with transcripts per million > 1 in whole blood were used as the background genes (85). Pathway analysis was conducted on all differentially edited genes and then again, separately, on genes that were differentially edited in AA only, in NHW only or in both.
Gene-based analysis
We conducted a gene-based association test using VEGAS2v02 (86). The nearest SNP was used as a proxy for each editing event for mapping to the appropriate gene. Genes with a P < 0.05 were evaluated using DAVID, as described above, to identify pathways enriched with significant genes.
Protein and miRNA predictions
We predicted the effects of editing events in coding regions using SIFT (50). Differentially edited sites within miRNA binding sites were identified using TargetScan 7.1 (87). Disruptive events were predicted using ImiRP, a tool which allows users to selectively mutate positions and predict the effect on miRNA binding (88).
Supplementary Material
Acknowledgements
Author’s thank the community members who graciously agreed to participate in the study and made this research possible.
Conflict of Interest statement. None declared.
Funding
National Institutes of Health (NIH)/National Institute on Aging (NIA) (grants AG028786, AG027944) and an administrative supplement to NIH/NIA (grant U01AG052410-01).
References
- 1. Vemuri P. and Jack C.R. Jr. (2010) Role of structural MRI in Alzheimer’s disease. Alzheimers Res. Ther., 2, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. 2018 Alzheimer’s Disease Facts and Figures Alzheimer’s dement. J. Alzheimers Assoc., 14, 367–429. [Google Scholar]
- 3. Steenland K., Goldstein F.C., Levey A. and Wharton W. (2016) A meta-analysis of Alzheimer’s disease incidence and prevalence comparing African-Americans and Caucasians. J. Alzheimers Dis., 50, 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weuve J., Barnes L.L., Mendes de Leon C.F., Rajan K.B., Beck T., Aggarwal N.T., Hebert L.E., Bennett D.A., Wilson R.S. and Evans D.A. (2018) Cognitive aging in black and White Americans: cognition, cognitive decline, and incidence of Alzheimer disease dementia. Epidemiology, 29, 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Potter G.G., Plassman B.L., Burke J.R., Kabeto M.U., Langa K.M., Llewellyn D.J., Rogers M.A.M. and Steffens D.C. (2009) Cognitive performance and informant reports in the diagnosis of cognitive impairment and dementia in African Americans and whites. Alzheimers Dement., 5, 445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gurland B.J., Wilder D.E., Lantigua R., Stern Y., Chen J., Killeffer E.H. and Mayeux R. (1999) Rates of dementia in three ethnoracial groups. Int. J. Geriatr. Psychiatry, 14, 481–493. [PubMed] [Google Scholar]
- 7. Gatz M., Pedersen N.L., Berg S., Johansson B., Johansson K., Mortimer J.A., Posner S.F., Viitanen M., Winblad B. and Ahlbom A. (1997) Heritability for Alzheimer’s disease: the study of dementia in Swedish twins. J. Gerontol. A Biol. Sci. Med. Sci., 52, M117–M125. [DOI] [PubMed] [Google Scholar]
- 8. Pericak-Vance M.A., Yamaoka L.H., Haynes C.S., Speer M.C., Haines J.L., Gaskell P.C., Hung W.Y., Clark C.M., Heyman A.L. and Trofatter J.A. (1988) Genetic linkage studies in Alzheimer’s disease families. Exp. Neurol., 102, 271–279. [DOI] [PubMed] [Google Scholar]
- 9. Farrer L.A., Cupples L.A., Haines J.L., Hyman B., Kukull W.A., Mayeux R., Myers R.H., Pericak-Vance M.A., Risch N. and van Duijn C.M. (1997) Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. a meta-analysis. APOE and Alzheimer disease meta analysis consortium. JAMA, 278, 1349–1356. [PubMed] [Google Scholar]
- 10. Pericak-Vance M.A., Bebout J.L., Gaskell P.C.J., Yamaoka L.H., Hung W.Y., Alberts M.J., Walker A.P., Bartlett R.J., Haynes C.A. and Welsh K.A. (1991) Linkage studies in familial Alzheimer disease: evidence for chromosome 19 linkage. Am. J. Hum. Genet., 48, 1034–1050. [PMC free article] [PubMed] [Google Scholar]
- 11. Strittmatter W.J., Weisgraber K.H., Huang D.Y., Dong L.M., Salvesen G.S., Pericak-Vance M., Schmechel D., Saunders A.M., Goldgaber D. and Roses A.D. (1993) Binding of human apolipoprotein E to synthetic amyloid beta peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A., 90, 8098–8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corder E.H., Saunders A.M., Strittmatter W.J., Schmechel D.E., Gaskell P.C., Small G.W., Roses A.D., Haines J.L. and Pericak-Vance M.A. (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science, 261, 921–923. [DOI] [PubMed] [Google Scholar]
- 13. Corder E.H., Saunders A.M., Risch N.J., Strittmatter W.J., Schmechel D.E., Gaskell P.C.J., Rimmler J.B., Locke P.A., Conneally P.M. and Schmader K.E. (1994) Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat. Genet., 7, 180–184. [DOI] [PubMed] [Google Scholar]
- 14. Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B. et al. (2013) Meta-analysis of 74, 046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet., 45, 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jun G.R., Chung J., Mez J., Barber R., Beecham G.W., Bennett D.A., Buxbaum J.D., Byrd G.S., Carrasquillo M.M., Crane P.K. et al. (2017) Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimers Dement., 13, 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. International Genomics of Alzheimer's Disease Consortium (IGAP) (2015) Convergent genetic and expression data implicate immunity in Alzheimer’s disease. Alzheimers Dement., 11(6), 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reitz C., Jun G., Naj A., Rajbhandary R., Vardarajan B.N., Wang L.-S., Valladares O., Lin C.-F., Larson E.B., Graff-Radford N.R. et al. (2013) Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E 4, and the risk of late-onset Alzheimer disease in African Americans. JAMA, 309, 1483–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Naj A.C. and Schellenberg G.D. (2017) Genomic variants, genes, and pathways of Alzheimer’s disease: an overview. Am. J. Med. Genet. B Neuropsychiatr. Genet., 174, 5–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tang M.X., Cross P., Andrews H., Jacobs D.M., Small S., Bell K., Merchant C., Lantigua R., Costa R., Stern Y. et al. (2001) Incidence of AD in African-Americans, Caribbean Hispanics, and Caucasians in northern Manhattan. Neurology, 56, 49–56. [DOI] [PubMed] [Google Scholar]
- 20. Tang M.X., Stern Y., Marder K., Bell K., Gurland B., Lantigua R., Andrews H., Feng L., Tycko B. and Mayeux R. (1998) The APOE-epsilon4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA, 279, 751–755. [DOI] [PubMed] [Google Scholar]
- 21. Cukier H.N., Kunkle B.W., Vardarajan B.N., Rolati S., Hamilton-Nelson K.L., Kohli M.A., Whitehead P.L., Dombroski B.A., Van Booven D., Lang R. et al. (2016) ABCA7 frameshift deletion associated with Alzheimer disease in African Americans. Neurol. Genet., 2, e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Allen M., Zou F., Chai H.S., Younkin C.S., Crook J., Pankratz V.S., Carrasquillo M.M., Rowley C.N., Nair A.A., Middha S. et al. (2012) Novel late-onset Alzheimer disease loci variants associate with brain gene expression. Neurology, 79, 221–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martiskainen H., Viswanathan J., Nykanen N.-P., Kurki M., Helisalmi S., Natunen T., Sarajarvi T., Kurkinen K.M.A., Pursiheimo J.-P., Rauramaa T. et al. (2015) Transcriptomics and mechanistic elucidation of Alzheimer’s disease risk genes in the brain and in vitro models. Neurobiol. Aging, 36, 1221.e15–1221.e28. [DOI] [PubMed] [Google Scholar]
- 24. Karch C.M., Jeng A.T., Nowotny P., Cady J., Cruchaga C. and Goate A.M. (2012) Expression of novel Alzheimer’s disease risk genes in control and Alzheimer’s disease brains. PLoS One, 7, e50976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Humphries C., Kohli M.A., Whitehead P., Mash D.C., Pericak-Vance M.A. and Gilbert J. (2015) Alzheimer disease (AD) specific transcription, DNA methylation and splicing in twenty AD associated loci. Mol. Cell. Neurosci., 67, 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stopa E.G., Tanis K.Q., Miller M.C., Nikonova E.V., Podtelezhnikov A.A., Finney E.M., Stone D.J., Camargo L.M., Parker L., Verma A. et al. (2018) Comparative transcriptomics of choroid plexus in Alzheimer’s disease, frontotemporal dementia and Huntington’s disease: implications for CSF homeostasis. Fluids Barriers CNS, 15, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rangaraju S., Dammer E.B., Raza S.A., Gao T., Xiao H., Betarbet R., Duong D.M., Webster J.A., Hales C.M., Lah J.J. et al. (2018) Quantitative proteomics of acutely-isolated mouse microglia identifies novel immune Alzheimer’s disease-related proteins. Mol. Neurodegener., 13, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Park E., Williams B., Wold B.J. and Mortazavi A. (2012) RNA editing in the human ENCODE RNA-seq data. Genome Res., 22, 1626–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hamilton C.E., Papavasiliou F.N. and Rosenberg B.R. (2010) Diverse functions for DNA and RNA editing in the immune system. RNA Biol., 7, 220–228. [DOI] [PubMed] [Google Scholar]
- 30. Hwang T., Park C.-K., Leung A.K.L., Gao Y., Hyde T.M., Kleinman J.E., Rajpurohit A., Tao R., Shin J.H. and Weinberger D.R. (2016) Dynamic regulation of RNA editing in human brain development and disease. Nat. Neurosci., 19, 1093. [DOI] [PubMed] [Google Scholar]
- 31. Polson A.G., Crain P.F., Pomerantz S.C., McCloskey J.A. and Bass B.L. (1991) The mechanism of adenosine to inosine conversion by the double-stranded RNA unwinding/modifying activity: a high-performance liquid chromatography-mass spectrometry analysis. Biochemistry, 30, 11507–11514. [DOI] [PubMed] [Google Scholar]
- 32. Savva Y.A., Rieder L.E. and Reenan R.A. (2012) The ADAR protein family. Genome Biol., 13, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bass B.L. (2002) RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem., 71, 817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nishikura K. (2010) Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem., 79, 321–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li J.B. and Church G.M. (2013) Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nat. Neurosci., 16, 1518–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bazak L., Haviv A., Barak M., Jacob-Hirsch J., Deng P., Zhang R., Isaacs F.J., Rechavi G., Li J.B., Eisenberg E. et al. (2014) A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res., 24, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang L., Yang C.S., Varelas X. and Monti S. (2016) Altered RNA editing in 3′ UTR perturbs micro RNA-mediated regulation of oncogenes and tumor-suppressors. Sci. Rep., 6:23226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Park E., Guo J., Shen S., Demirdjian L., Wu Y.N., Lin L. and Xing Y. (2017) Population and allelic variation of A-to-I RNA editing in human transcriptomes. Genome Biol., 18, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ramaswami G., Deng P., Zhang R., Anna Carbone M., Mackay T.F. and Li J.B. (2015) Genetic mapping uncovers cis-regulatory landscape of RNA editing. Nat. Commun., 6:8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maas S., Kawahara Y., Tamburro K.M. and Nishikura K. (2006) A-to-I RNA editing and human disease. RNA Biol., 3, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tariq A. and Jantsch M.F. (2012) Transcript diversification in the nervous system: a to I RNA editing in CNS function and disease development. Front. Neurosci., 6, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Slotkin W. and Nishikura K. (2013) Adenosine-to-inosine RNA editing and human disease. Genome Med., 5, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Khermesh K., D’Erchia A.M., Barak M., Annese A., Wachtel C., Levanon E.Y., Picardi E. and Eisenberg E. (2016) Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer’s disease. RNA, 22, 290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gaisler-Salomon I., Kravitz E., Feiler Y., Safran M., Biegon A., Amariglio N. and Rechavi G. (2014) Hippocampus-specific deficiency in RNA editing of GluA2 in Alzheimer’s disease. Neurobiol. Aging, 35, 1785–1791. [DOI] [PubMed] [Google Scholar]
- 45. Annese A., Manzari C., Lionetti C., Picardi E., Horner D.S., Chiara M., Caratozzolo M.F., Tullo A., Fosso B., Pesole G. et al. (2018) Whole transcriptome profiling of late-onset Alzheimer’s disease patients provides insights into the molecular changes involved in the disease. Sci. Rep., 8, 4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heneka M.T., Golenbock D.T. and Latz E. (2015) Innate immunity in Alzheimer’s disease. Nat. Immunol., 16, 229–236. [DOI] [PubMed] [Google Scholar]
- 47. Marsh S.E., Abud E.M., Lakatos A., Karimzadeh A., Yeung S.T., Davtyan H., Fote G.M., Lau L., Weinger J.G., Lane T.E. et al. (2016) The adaptive immune system restrains Alzheimer’s disease pathogenesis by modulating microglial function. Proc. Natl. Acad. Sci., 113, E1316–E1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Qi T., Wu Y., Zeng J., Zhang F., Xue A., Jiang L., Zhu Z., Kemper K., Yengo L., Zheng Z. et al. (2018) Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat. Commun., 9, 2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kunkle B.W., Grenier-Boley B., Sims R., Bis J.C., Damotte V., Naj A.C., Boland A., Vronskaya M., van der Lee S.J., Amlie-Wolf A. et al. (2019) Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet., 51, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kumar P., Henikoff S. and Ng P.C. (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc., 4, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 51. Miyashita A., Koike A., Jun G., Wang L.-S., Takahashi S., Matsubara E., Kawarabayashi T., Shoji M., Tomita N., Arai H. et al. (2013) SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PLoS One, 8, e58618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee J.H., Cheng R., Schupf N., Manly J., Lantigua R., Stern Y., Rogaeva E., Wakutani Y., Farrer L., St George-Hyslop P. et al. (2007) The association between genetic variants in SORL1 and Alzheimer disease in an urban, multiethnic, community-based cohort. Arch. Neurol., 64, 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nixon R.A. (2005) Endosome function and dysfunction in Alzheimer’s disease and other neurodegenerative diseases. Neurobiol. Aging, 26, 373–382. [DOI] [PubMed] [Google Scholar]
- 54. Cataldo A.M., Peterhoff C.M., Troncoso J.C., Gomez-Isla T., Hyman B.T. and Nixon R.A. (2000) Endocytic pathway abnormalities precede amyloid β deposition in sporadic Alzheimer’s disease and down syndrome. Differential effects of APOE genotype and presenilin mutations. Am. J. Pathol., 157, 277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Berson A., Barbash S., Shaltiel G., Goll Y., Hanin G., Greenberg D.S., Ketzef M., Becker A.J., Friedman A. and Soreq H. (2012) Cholinergic-associated loss of hn RNP-A/B in Alzheimer’s disease impairs cortical splicing and cognitive function in mice. EMBO Mol. Med., 4, 730–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Villa C., Fenoglio C., De Riz M., Clerici F., Marcone A., Benussi L., Ghidoni R., Gallone S., Cortini F., Serpente M. et al. (2011) Role of hnRNP-A1 and miR-590-3p in neuronal death: genetics and expression analysis in patients with Alzheimer disease and frontotemporal lobar degeneration. Rejuvenation Res., 14, 275–281. [DOI] [PubMed] [Google Scholar]
- 57. Page R.M., Münch A., Horn T., Kuhn P.-H., Colombo A., Reiner O., Boutros M., Steiner H., Lichtenthaler S.F. and Haass C. (2012) Loss of PAFAH1B2 reduces amyloid-β generation by promoting the degradation of amyloid precursor protein C-terminal fragments. J. Neurosci., 32, 18204–18214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mangino M., Hwang S.-J., Spector T.D., Hunt S.C., Kimura M., Fitzpatrick A.L., Christiansen L., Petersen I., Elbers C.C., Harris T. et al. (2012) Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum. Mol. Genet., 21, 5385–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stewart J.A., Wang F., Chaiken M.F., Kasbek C., Chastain P.D.,. 2nd, Wright W.E. and Price C.M. (2012) Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J., 31, 3537–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hunt S.C., Chen W., Gardner J.P., Kimura M., Srinivasan S.R., Eckfeldt J.H., Berenson G.S. and Aviv A. (2008) Leukocyte telomeres are longer in African Americans than in whites: the National Heart, Lung, and Blood Institute Family Heart Study and the Bogalusa Heart Study. Aging Cell, 7, 451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Qiu W., Guo X., Lin X., Yang Q., Zhang W., Zhang Y., Zuo L., Zhu Y., Li C.-S.R., Ma C. et al. (2017) Transcriptome-wide piRNA profiling in human brains of Alzheimer’s disease. Neurobiol. Aging, 57, 170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al. (2016) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res., 44, D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Di Paolo G. and Kim T.-W. (2011) Linking lipids to Alzheimer’s disease: cholesterol and beyond. Nat. Rev. Neurosci., 12, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Friederich M.W., Erdogan A.J., Coughlin C.R.,. 2nd, Elos M.T., Jiang H., O’Rourke C.P., Lovell M.A., Wartchow E., Gowan K., Chatfield K.C. et al. (2017) Mutations in the accessory subunit NDUFB10 result in isolated complex I deficiency and illustrate the critical role of intermembrane space import for complex I holoenzyme assembly. Hum. Mol. Genet., 26, 702–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Aksenov M.Y., Tucker H.M., Nair P., Aksenova M.V., Butterfield D.A., Estus S. and Markesbery W.R. (1999) The expression of several mitochondrial and nuclear genes encoding the subunits of electron transport chain enzyme complexes, cytochrome c oxidase, and NADH dehydrogenase, in different brain regions in Alzheimer’s disease. Neurochem. Res., 24, 767–774. [DOI] [PubMed] [Google Scholar]
- 66. Wang H., Guo J., Jiang J., Wu W., Chang X., Zhou H., Li Z. and Zhao J. (2017) New genes associated with rheumatoid arthritis identified by gene expression profiling. Int. J. Immunogenet., 44, 107–113. [DOI] [PubMed] [Google Scholar]
- 67. Yu S., Chen X., Xiu M., He F., Xing J., Min D. and Guo F. (2017) The regulation of Jmjd 3 upon the expression of NF-kappaB downstream inflammatory genes in LPS activated vascular endothelial cells. Biochem. Biophys. Res. Commun., 485, 62–68. [DOI] [PubMed] [Google Scholar]
- 68. Tang Y., Li T., Li J., Yang J., Liu H., Zhang X.J. and Le W. (2014) Jmjd3 is essential for the epigenetic modulation of microglia phenotypes in the immune pathogenesis of Parkinson’s disease. Cell Death Differ., 21, 369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ohguchi H., Harada T., Sagawa M., Kikuchi S., Tai Y.-T., Richardson P.G., Hideshima T. and Anderson K.C. (2017) KDM6B modulates MAPK pathway mediating multiple myeloma cell growth and survival. Leukemia, 31, 2661–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McKhann G., Drachman D., Folstein M., Katzman R., Price D. and Stadlan E.M. (1984) Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA work group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology, 34, 939–944. [DOI] [PubMed] [Google Scholar]
- 71. McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R.J., Kawas C.H., Klunk W.E., Koroshetz W.J., Manly J.J., Mayeux R. et al. (2011) The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement., 7, 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Folstein M.F., Folstein S.E. and McHugh P.R. (1975) Mini-mental state’. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res., 12, 189–198. [DOI] [PubMed] [Google Scholar]
- 73. Teng E.L. and Chui H.C. (1987) The modified mini-mental state (3MS) examination. J. Clin. Psychiatry, 48, 314–318. [PubMed] [Google Scholar]
- 74. Morris J.C. (1993) The clinical dementia rating (CDR): current version and scoring rules. Neurology, 43, 2412–2414. [DOI] [PubMed] [Google Scholar]
- 75. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M. and Gingeras T.R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Picardi E., D’Erchia A.M., Montalvo A. and Pesole G. (2015) Using REDItools to detect RNA editing events in NGS datasets. Curr. Protoc. Bioinformatics, 49, 12.12.1–12.12.15. [DOI] [PubMed] [Google Scholar]
- 77. Sherry S.T., Ward M.-H., Kholodov M., Baker J., Phan L., Smigielski E.M. and Sirotkin K. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res., 29, 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M. and Haussler D. (2002) The human genome browser at UCSC. Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kiran A. and Baranov P.V. (2010) DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics, 26, 1772–1776. [DOI] [PubMed] [Google Scholar]
- 80. Ramaswami G. and Li J.B. (2014) RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res., 42, D190–D113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Srivastava P.K., Bagnati M., Delahaye-Duriez A., Ko J.-H., Rotival M., Langley S.R., Shkura K., Mazzuferi M., Danis B., van Eyll J. et al. (2017) Genome-wide analysis of differential RNA editing in epilepsy. Genome Res., 27, 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yablonovitch A.L., Fu J., Li K., Mahato S., Kang L., Rashkovetsky E., Korol A.B., Tang H., Michalak P., Zelhof A.C. et al. (2017) Regulation of gene expression and RNA editing in drosophila adapting to divergent microclimates. Nat. Commun., 8, 1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Huang D.W., Sherman B.T. and Lempicki R.A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc., 4, 44–57. [DOI] [PubMed] [Google Scholar]
- 84. Young M.D., Wakefield M.J., Smyth G.K. and Oshlack A. (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol., 11, R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Carithers L.J., Ardlie K., Barcus M., Branton P.A., Britton A., Buia S.A., Compton C.C., DeLuca D.S., Peter-Demchok J., Gelfand E.T. et al. (2015) A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv. Biobank., 13, 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mishra A. and Macgregor S. (2015) VEGAS2: software for more flexible gene-based testing. Twin Res. Hum. Genet., 18, 86–91. [DOI] [PubMed] [Google Scholar]
- 87. Lewis B.P., Burge C.B. and Bartel D.P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120, 15–20. [DOI] [PubMed] [Google Scholar]
- 88. Ryan B.C., Werner T.S., Howard P.L. and Chow R.L. (2016) ImiRP: a computational approach to microRNA target site mutation. BMC Bioinform., 17, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.