Abstract
Collections of cells called engrams are thought to represent memories. Although there has been progress in identifying and manipulating single engrams, little is known about how multiple engrams interact to influence memory. In lateral amygdala (LA), neurons with increased excitability during training outcompete their neighbors for allocation to an engram. We examined whether competition based on neuronal excitability also governs the interaction between engrams. Mice received two distinct fear conditioning events separated by different intervals. LA neuron excitability was optogenetically manipulated and revealed a transient competitive process that integrates memories for events occurring closely in time (coallocating overlapping populations of neurons to both engrams) and separates memories for events occurring at distal times (disallocating nonoverlapping populations to each engram).
Memory of an event is thought to be represented by an ensemble of neurons, referred to as its memory trace or engram (1, 2). Despite recent advances in localizing and manipulating single engrams, little is known about how multiple engrams interact to influence memory function. Two engrams may engage nonoverlapping neuronal populations, thus minimizing interference between distinct memory representations. Alternatively, engrams may engage overlapping neuronal populations to functionally link those memories. Here, we examined the rules governing engram interaction.
In lateral amygdala (LA), a region critical for conditioned fear memory (3–5), eligible neurons compete for engram allocation. Neurons with relatively higher function of transcription factor CREB or increased excitability at training preferentially win this competition and are allocated to an engram (6–10). Silencing these neurons prevents memory expression, whereas their activation artificially elicits memory expression, indicating that these allocated neurons are both necessary and sufficient for expression of that memory (7–10). Similar competition governs engram allocation in mice without experimental manipulation of CREB or excitability (11).
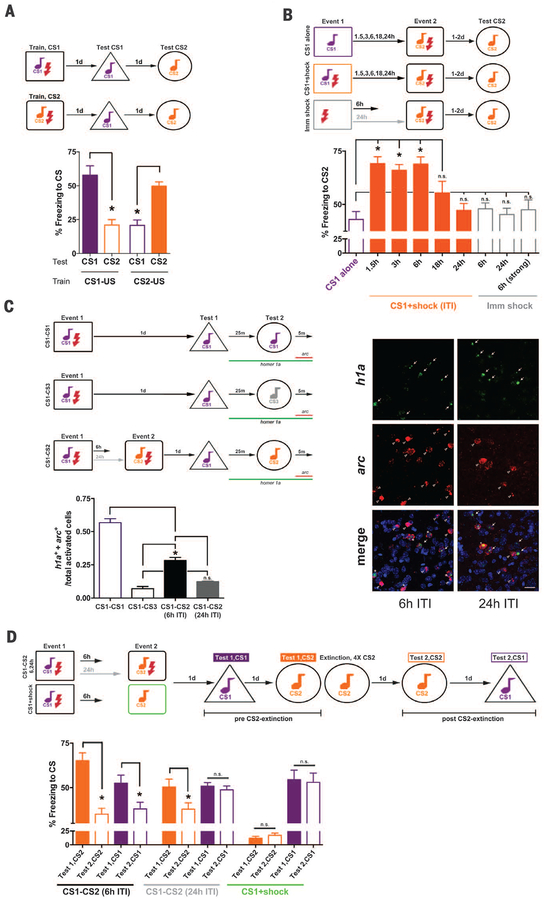
We asked if two fear-conditioning events that occur closely in time are coallocated to overlapping populations of neurons, thereby functionally linking these memories. Mice received two events (event 1, event 2) featuring distinct auditory conditioned stimuli (CS1 or CS2, Fig. 1A; see supplementary materials) separated by varying intertraining intervals (ITIs; 1.5, 3, 6, 18, or 24 hours). Event 2 (CS2+footshock) was the same across groups, but event 1 content and timing differed. Memory for event 1 was stable across ITIs (fig. S1B). However, event 2 memory was enhanced if event 1 (CS1+footshock) occurred shortly before (1.5 to 6 hours, but not 18 to 24 hours; Fig. 1B and fig. S2, B and C), even if event 1 consisted of light-footshock pairing (fig. S2A). Event 2 memory enhancement depended on previous fear conditioning (not sensitization); mice failed to show enhanced event 2 memory if event 1 consisted of CS1 alone or immediate footshock.
Fig. 1. Events occurring closely in time are coallocated to overlapping engrams, and memories become linked.
(A) Mice distinguish conditioned stimuli (CS1, CS2), freezing more in response to trained, than untrained, auditory CS. ANOVA: Train-CS × Test-CS, F(1,12) = 44.29, *P < 0.001. Means ± SEM. n = 7 mice per group. *P < 0.05. (B) Event 2 memory was enhanced if event 1 (CS1+shock) occurred within a short intertraining interval (ITI). Enhancement was not due to CS1 alone or immediate shock (Imm shock) during event 1; F(8,142) = 17.03, *P < 0.001. n = 56 for CS1 alone (see fig. S1A), n = 5 to 16 for other groups. n.s., not statistically different. (C) Engrams for events with a 6-hour (but not 24-hour) ITI coallocated to overlapping LA neurons; F(3,8) = 119.01, *P < 0.001. (Right) Coallocation in 6-hour, but not 24-hour, ITI. arc+ (CS2 reexposure, red), homer1a+ (h1a+, CS1 reexposure, green) neurons, 4´,6-diamidino-2-phenylindole (DAPI; blue, nuclear stain). Scale bar, 20 μm. n = 3 mice per group. (D) Extinguishing event 2 memory also decreased event 1 memory if a 6-hour (but not 24-hour) ITI was used. Group × CS-Freezing, F(2,50) = 12.53, *P < 0.001. n = 12 mice, 6-hour, 24-hour groups; n = 4 mice, control group.
To examine if neural representations of events separated by short (not long) intervals are co-allocated to overlapping neuronal populations, we used fluorescent in situ hybridization (FISH) for two activity-dependent genes. Nuclear arc mRNA labels neurons that were active in the preceding 5 min, whereas nuclear homer1a (h1a) mRNA labels neurons that were active in the preceding 30 to 40 min (12). Mice received event 1 and event 2 separated by 6 or 24 hours. During the memory test, CS1 was presented; then, 25 min later, CS2 was presented. Brains were removed 5 min later. arc+ neurons were activated by CS2, h1a+ neurons were activated by CS1, and arc+/h1a+ neurons were activated by both CSs (part of both engrams). Two control groups trained on event 1 only and tested under two conditions provided upper and lower overlap boundaries. During the test, the CS1-CS1 group was reexposed to CS1 twice (h1a+ = first CS1 reexposure, arc+ = second CS1 reexposure, “overlap ceiling”), whereas the CS1-CS3 group was reexposed to CS1 and novel CS3 (“floor”). The overlap observed in mice trained on event 1 and event 2 separated by 24 hours was not different from floor conditions, whereas the overlap observed in mice similarly trained with a 6-hour interval was higher, indicating coallocation (Fig. 1C and fig. S3, A to C).
To test whether coallocation functionally links memories, we extinguished event 2 memory (repeatedly presented CS2 without footshock) and examined the effect on event 1 memory. After CS2 extinction, all groups froze less to CS2. However, mice trained with short (6 hours), but not long (24 hours), ITIs also froze less in response to CS1, even though CS1 was not explicitly extinguished (Fig. 1D).
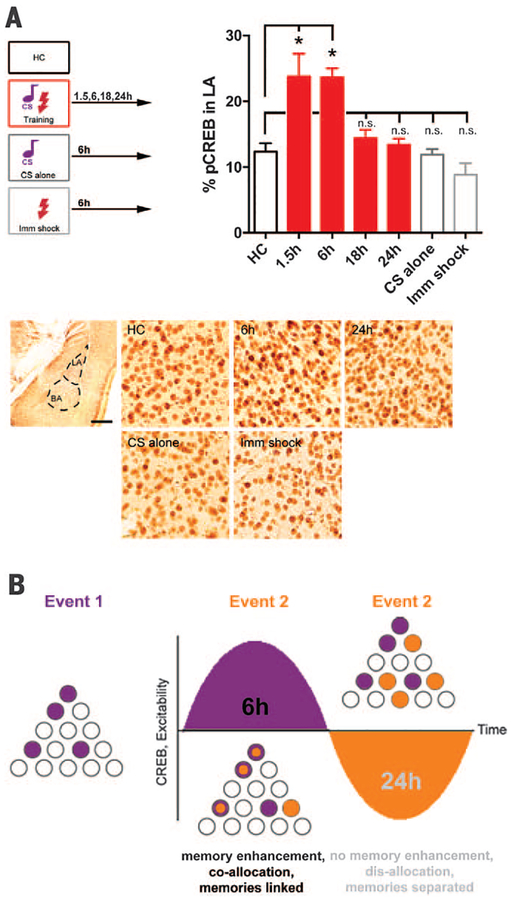
We next examined potential molecular and local circuit mechanisms governing engram interaction. Fear conditioning transiently (1.5 to 6 hours, but not 18 to 24 hours) increased the number of LA principal neurons with activated CREB (Fig. 2A), in agreement with reports showing that learning transiently increases neuronal excitability (13, 14). CREB function cycles over time, in part, due to autoregulatory feedback (15) and, consistent with this, we observed that a depolarizing stimulus in cultured neurons first activated CREB, then increased an inhibitory CREB isoform (fig. S4).
Fig. 2. Engram interaction may be governed by neuronal CREB function and excitability.
(A) (Top) Fear conditioning transiently increases the percentage of LA neurons with activated CREB (pCREB) relative to home-cage (HC), CS alone (6h), or immediate shock (6h, Imm shock); F(6,21) = 12.53, *P < 0.001. (Bottom) Left panel: LA. Scale bar, 400 μm. Right panels: pCREB staining, Scale bar, 25 μm. n = 3 to 5 mice per group. (B) Schematic of hypothesized engram interaction. Event 1 transiently increases CREB function and excitability in a population of LA neurons (purple). If event 2 occurs when these neurons have elevated CREB and excitability (6h), then engrams are coal-located, memories are linked (purple+orange neurons), and event 2 memory is enhanced. If event 2 occurs later (24h), neurons activated by event 1 are no longer more excitable (perhaps they are in a “refractory-like period”), engrams are disallocated to nonoverlapping neurons, and memories are distinct.
The oscillation in CREB function and neuronal excitability provides a potential mechanism by which engrams supporting events occurring closely in time are coallocated, whereas events occurring at more remote time points are disallocated (Fig. 2B) (16). Although the size of an LA engram supporting a fear memory remains stable, a competitive process determines which neurons comprise each engram (7–10). If event 1 transiently increases CREB function and neuronal excitability in a population of LA neurons and event 2 occurs while these neurons are more excitable than their neighbors (within 6 hours), then overlapping neurons would be coallocated to both events. However, if event 2 occurs when event 1 neurons are no longer more excitable (e.g., 24 hours after event 1), the engram underlying event 2 is disallocated, thus functionally disambiguating these memories. We tested these predictions by manipulating LA neuronal excitability to control allocation. We sought to force coallocation to artificially link normally separated memories (24-hour interval) and force disallocation to artificially separate normally linked memories (6-hour interval).
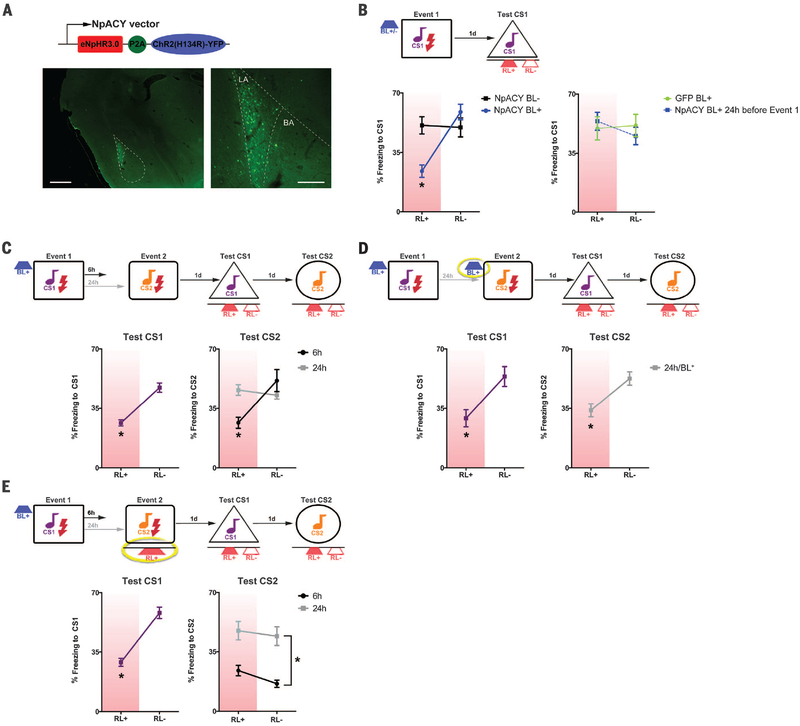
To bidirectionally manipulate excitability in the same neuron, we constructed a replication-defective herpes simplex viral vector (HSV) expressing both channelrhodopsin-2 (ChR2, excitatory opsin) and halorhodopsin (NpHR3.0, inhibitory opsin) [HSV-NpACY (17, 18)]. Blue light (via ChR2) increases excitability, whereas red light (via NpHR3.0) inhibits activity (Fig. 3A and figs. S5 and S7). HSV infects ~15% of LA principal neurons (10) (Fig. 3A and fig. S6). We verified this approach using one fear-conditioning event. To allocate HSV-infected neurons, we excited them (blue light, BL+) before training (10). To confirm their allocation, we tested mice under basal conditions (no red light, RL–) and while inhibiting infected neurons (red light, RL+). Mice trained immediately after activation of infected neurons froze less when these neurons were inhibited (Fig. 3B), indicating that optogenetically exciting neurons before training was sufficient for allocation. Inhibiting a similar number of neurons in control groups (BL−, BL+ 24 hours before training, BL+ but no opsin) did not affect memory expression, nor did BL+ affect training variables or function as a cue during the test (fig. S8B).
Fig. 3. Bidirectionally manipulating excitability to force coallocation or disallocation reveals competition governing engram interaction.
(A) HSV-NpACY was used to excite (ChR2) and inhibit (NpHR3.0) the same neurons. (Bottom) Yellow fluorescent protein (YFP) expression. LA, lateral amygdala; BA, basal amygdala. Scale bars, 400 μm (left), 150 μm (right). (B) Exciting HSV-NpACY neurons immediately (but not 24 hours) before event 1 [blue light (BL+)] allocates them to the engram; inhibiting them [red light (RL+)] during the test decreased CS freezing versus no red light (RL−). There was no effect of inhibiting random neurons (BL−). Group × Test condition, F(3,28) = 17.41, *P < 0.001. n = 7 to 9 mice for all groups. (C) Exciting neurons before event 1 (BL+) allocates them to event 1 engram (*P < 0.001) and coallocates them to event 2, if event 2 occurs 6 hours, but not 24 hours, later. (D) Exciting the same neurons before event 2 (24h/BL+, yellow circle) forces coallocation, F(2,20) = 17.91, *P < 0.001. n = 8 for 6-hour, n = 7 for 24-hour, and n = 8 for 24-hour/BL+ groups. (E) Exciting neurons before event 1, but inhibiting them during event 2 (RL+, yellow circle), impairs event 2 memory, if event 2 occurred 6 hours, but not 24 hours, later. F(1,16) = 5.63, *P < 0.05. n = 7 for 24-hour and n = 11 for 6-hour groups.
We confirmed time-limited coallocation of two events. Principal neurons expressing HSV-NpACY were excited immediately before event 1. Infected neurons were coallocated to event 2, if event 2 occurred 6 hours, but not 24 hours, later (Fig. 3C). To artificially link normally separated memories (24-hour interval), we excited the same population of HSV-infected neurons before event 1 and event 2, forcing coallocation. Inhibiting infected neurons impaired both event 1 and event 2 memory expression (Fig. 3D), indicating overlapping engrams of infected neurons. Similarly, we artificially linked normally separated memories (24-hour interval) by virally overexpressing CREB (vCREB) to excite the same neurons during event 1 and event 2. Silencing vCREB-infected neurons with the inhibitory DREADD receptor hM4Di [designer receptors exclusively activated by designer drug (19)] disrupted expression of both memories (fig. S9, A to D).
To artificially separate normally linked memories (6-hour interval), we excited infected neurons before event 1, then inhibited (RL+) these neurons during event 2. Surprisingly, event 2 memory was impaired, even when mice were tested without red light (Fig. 3E). Event 2 memory was also disrupted if we used vCREB to allocate neurons before event 1 and DREADDs to inhibit these neurons during event 2 (fig. S9E). In contrast, event 2 memory was intact in optogenetic experiments if the interval between events was increased to 24 hours (Fig. 3E). These results indicate that initially allocated neurons transiently prevent nonallocated neurons from being allocated to another engram (winning neurons inhibit losing neurons).
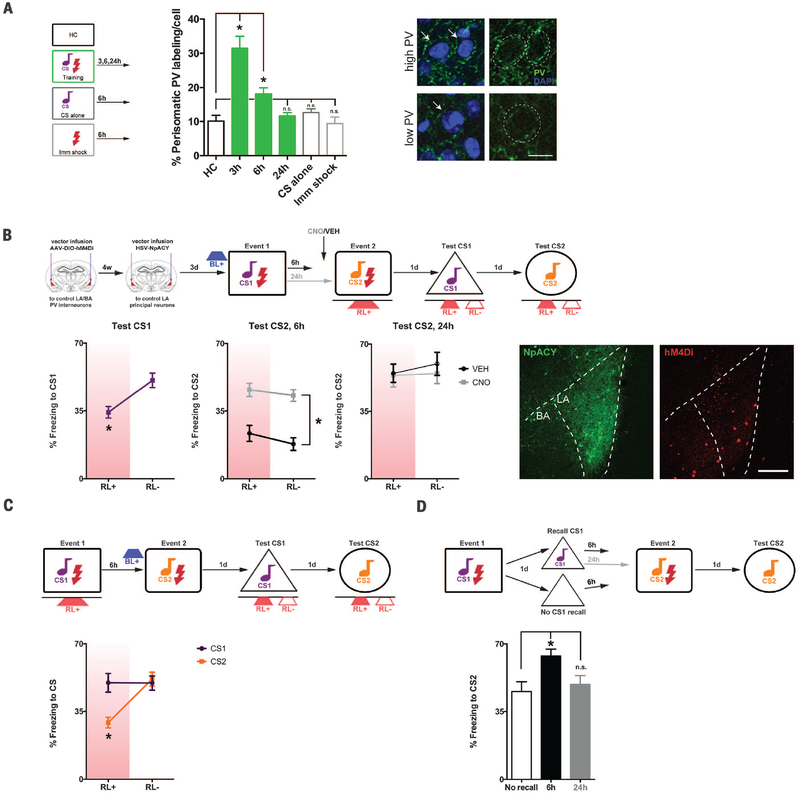
In neural networks, winner-take-all competition maintains sparse coding and is regulated by excitation and inhibition (20). Local LA inhibition is mediated predominantly by γ-aminobutyric acid (GABA)–releasing parvalbumin (PV) interneurons (21, 22). PV interneurons in LA and basal amygdala (BA) form an inhibitory network receiving inputs from, and regulating, LA principal neuron activity by forming perisomatic synapses (23, 24). Fear conditioning induced a transient increase in perisomatic PV immunolabeling (24) surrounding LA principal neurons (1.5 to 6 hours but not 18 to 24 hours, Fig. 4A).
Fig. 4. Competition-mediating engram interaction depends on excitation-inhibition.
(A) Fear conditioning transiently increases perisomatic PV immunolabeling (green) surrounding LA principal neurons relative to home-cage (HC), CS alone (6h), or immediate shock (6h, Imm shock); *P < 0.001. (Right) High or low perisomatic PV, with or without DAPI-labeled nuclei (blue, perisomatic region outlined). Scale bar, 10 μm. n = 3 mice per group. Arrows indicate perisomatic PV immunolabel in example cells designated “high PV”or “low PV.”(B) Event 2 memory is impaired (Fig. 3E) if events are separated by 6 hours and neurons allocated to event 1 are silenced (RL+) during event 2. Inhibiting PV interneurons (hM4Di+CNO) permits event 2 memory formation, group difference only, F(3,23) = 22.61, *P < 0.001. (Right) HSV-NpACY (YFP, green), AAV-hM4Di (mCherry, red). Scale bar, 200 μm. n = 4 to 8 mice for all groups. (C) HSV-infected neurons inhibited (RL+) during event 1 are not allocated to event 1 engram; *P > 0.05. Exciting these neurons before event 2 (BL+) allocates them to event 2 engram, F(1,7) = 51.02, *P < 0.001. n = 8 mice. (D) Event 2 memory is enhanced if CS1 is recalled 6 hours, but not 24 hours, before event 2; F(2,29) = 5.28, *P < 0.05. n = 8 to 12 mice for all groups.
To test whether PV interneurons suppress non-allocated principal neurons, we released LA and BA PV inhibition. Transgenic mice expressing Cre recombinase in PV interneurons (PV-Cre mice) were microinjected with adeno-associated virus (AAV) expressing Cre-dependent hM4Di. Control PV-Cre mice with intact PV function (vehicle-treated) similarly showed impaired event 2 memory (Fig. 3E) if HSV-infected neurons allocated to event 1 were optogenetically silenced during event 2 (6-hour interval, Fig. 4B). However, inhibiting PV neurons before event 2 [clozapine-N-oxide (CNO) to activate hM4Di receptors] permitted event 2 memory formation. Therefore, if principal neurons not allocated to the engram supporting event 1 (losers) are released from PV inhibition, they may be allocated to an engram for an event occurring shortly after (become winners). Similarly, relieving PV inhibition before event 2 had no effect when events were separated by 24 hours, showing the time dependency of this competition (Fig. 4B).
We examined whether directly manipulating LA principal neuron excitability, rather than indirectly relieving PV inhibition, overcomes these competition effects. HSV-infected principal neurons were optogenetically inhibited during event 1 (these neurons were not allocated to event 1 engram; Fig. 4C). Event 2 occurred 6 hours later when neurons allocated to event 1 engram would also be coallocated to event 2 engram. However, immediately before event 2, we optogenetically excited HSV-infected neurons. These “loser” HSV-infected neurons became critical components of the engram supporting event 2, outcompeting previous winning neurons.
These results reveal that, in the LA, a transient competitive process governs the interaction between engrams to integrate memories for events occurring closely in time and distinguish memories for events occurring farther apart in time. Coal-location is not limited to linking memories at encoding. Memory recall may engage a similar process to link new with old memories. We trained mice on event 1, 2 days before event 2. Event 2 memory was enhanced if event 1 was recalled 6 hours, not 24 hours, before event 2 (Fig. 4D). Here, we find that excitatory-inhibitory balance determines whether memories are bound or, alternately, segregated in the LA. More broadly, these principles provide a foundation for understanding how memories are organized within associative networks.
Note added in proof: During final preparation of this manuscript, a notable study showing time-limited coallocation of hippocampal memory traces was published (25).
Supplementary Material
ACKNOWLEDGMENTS
We thank B. Roth for reagents, N. Insel and B. Richards for comments on previous versions of this manuscript, and the Josselyn and Frankland labs for general comments. This work was supported by Canadian Institutes of Health Research (S.A.J., P.W.F.), Natural Sciences and Engineering Research Council of Canada (S.A.J., P.W.F.), Brain Canada (S.A.J., P.W.F.), and Brain and Behavior Research Foundation (S.A.J.) grants. K.D. is supported by the National Institute of Mental Health, National Institute on Drug Abuse, the Wiegers Family Fund, Howard Hughes Medical Institute, and the U.S. Army Research Laboratory and Defense Advanced Research Projects Agency (Cooperative Agreement no. W911NF-14-2-0013). Data are archived in the Dryad repository http://dx.doi.org/10.5061/dryad.5tp75.
Footnotes
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Josselyn SA, Köhler S, Frankland PW, Nat. Rev. Neurosci 16, 521–534 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Tonegawa S, Liu X, Ramirez S, Redondo R, Neuron 87, 918–931 (2015). [DOI] [PubMed] [Google Scholar]
- 3.LeDoux JE, Annu. Rev. Neurosci 23, 155–184 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Maren S, Ann. N. Y. Acad. Sci 985, 106–113 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Duvarci S, Pare D, Neuron 82, 966–980 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim D, Paré D, Nair SS, J. Neurosci 33, 14354–14358 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han JH et al. , Science 316, 457–460 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Han JH et al. , Science 323, 1492–1496 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y et al. , Nat. Neurosci 12, 1438–1443 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yiu AP et al. , Neuron 83, 722–735 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Gouty-Colomer LA et al. , Mol. Psychiatry 21, 364–375 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzowski JF, Hippocampus 12, 86–104 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Oh MM, Kuo AG, Wu WW, Sametsky EA, Disterhoft JF, J. Neurophysiol 90, 2171–2179 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Thompson LT, Moyer JR Jr., Disterhoft JF, J. Neurophysiol 76, 1836–1849 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Monaco L, Foulkes NS, Sassone-Corsi P, Proc. Natl. Acad. Sci. U.S.A 92, 10673–10677 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silva AJ, Zhou Y, Rogerson T, Shobe J, Balaji J, Science 326, 391–395 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mei Y, Zhang F, Biol. Psychiatry 71, 1033–1038 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gradinaru V et al. , Cell 141, 154–165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL, Proc. Natl. Acad. Sci. U.S.A 104, 5163–5168 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shoemaker PA, Front. Comput. Neurosci 9, 12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sah P, Faber ES, Lopez De Armentia M, Power J, Physiol. Rev 83, 803–834 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Wolff SB et al. , Nature 509, 453–458 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Woodruff AR, Sah P, J. Neurosci 27, 553–563 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trouche S, Sasaki JM, Tu T, Reijmers LG, Neuron 80, 1054–1065 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai DJ et al. , Nature 534, 115–118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.