Abstract
Twelve carbapenem-resistant Escherichia coli strains were obtained from goose farms in Jiangsu, China. These isolates were resistant to multiple antimicrobials, and positive for the blaNDM–5. The carbapenem-resistance of all strains mediated by blaNDM–5 were successfully conjugated to E. coli J53. S1-PFGE and WGS results showed blaNDM–5 was located on IncX3 conjugative plasmids with a size of ca. 46 kb. All blaNDM–5-bearing IncX3 plasmids shared the same genetic context almost identical to pNDM_MGR194-blaNDM–5 and pNDM-QD28-blaNDM–5 reported in India and China, respectively. The twelve strains belonged to three STs, in which the dominant type of E. coli isolated from breeding goose farm carrying blaNDM–5 was ST48. The emergence of blaNDM–5-bearing strains in goose farms and the clonal transmission of E. coli within the breeding goose farm highlighted the potential reservoir of carbapenemase genes in waterfowl farming system, which may further contaminate environments and pose a threat to public health. Comprehensive surveillance of carbapenem-resistant bacteria in goose farms warrants further study to evaluate the underlying risks.
Keywords: carbapenemase genes, bla NDM–5 , E. coli, long-read sequencing, plasmids
Introduction
Enterobacteriaceae such as Escherichia coli, Klebsiella pneumoniae and Salmonella spp. are important pathogens that cause human infections. Carbapenemase-producing Enterobacteriaceae (CPE) is constantly reported worldwide and has become an urgent public health threat (Potter et al., 2016). So far, CPE has been detected in animals, environment and vegetable samples that are closely related to humans (Walsh et al., 2011; Wang J. et al., 2018; Wang R.B. et al., 2018). As one main type of carbapenemases, New Delhi metallo-β-lactamase (NDM) is able to confer resistance to almost all β-lactams. Since the first report of blaNDM–1 in 2009, 21 variants of NDM enzymes (NDM-1 to NDM-21) have been reported (Yong et al., 2009; Liu et al., 2018). Among them, the NDM-5-encoding gene, blaNDM–5, was first reported in an E. coli recovered from a patient in the United Kingdom (Hornsey et al., 2011). In China, a lot of studies have reported the blaNDM–5 in Enterobacteriaceae of animal origin. Regarding poultry origin, there was one report of the presence of NDM in Enterobacteriaceae in the poultry production environment, which may indicated that blaNDM–5 has been widely distributed among Enterobacteriaceae in poultry farms (Zhang et al., 2019). Especially, the coexistence of blaNDM–5 and mcr-1 in E. coli of single duck origin has also been reported, which raised the public concern about the antibiotic resistance of waterfowl (Yang et al., 2016).
In addition to duck origin, goose breeding industry is particularly developed in Jiangsu, China. The breeding mode of geese in farms is different from other poultry farms, because many pools are built in goose farms to support their growth. Owing to the use of antibiotics and possible dissemination of resistant bacteria via water system in goose farms, the antibiotic resistance may be extremely complicated. However, there are few reports of antibiotic resistance in goose farms. Therefore, enhancing the surveillance of antibiotic resistance in goose farms is necessary. To cover this gap, we investigated the prevalence of blaNDM–5-positive E. coli isolates in goose farms and probed the genomic features of these isolates with cutting-edge nanopore long-read sequencing technology.
Materials and Methods
Identification of Bacterial Strains and Detection of Carbapenemase Genes
A total of 117 samples, including anal swab, feed, feces, water and soil, were collected within one day from a breeding goose farm (n = 68) and a goose hatchery (n = 49) in Jiangsu Province, China in 2018 (Table S1). There are about 10 goose houses in the breeding farm where farming scale approaches to one thousand. We randomly selected three goose houses as sample sites. It was worth pointing out that we considered the samples collected from the three goose houses as related samples due to the same feed and water used in different goose houses, the open environment and the free movement of the breeders. The hatchery farm which isolated from outside had some distance away from the breeding goose farm (Figure S7). Both farms belonged to the same goose producing chain meaning that the goslings from hatchery were transported to the breeding farm for feeding. To be note, the breeding goose were mainly for food and the water supply of the goose farm was from the river outside the farm. Besides, there were no hospitals, factories or other source of pollutions near the farm. Isolation of carbapenem-resistant E. coli was performed as following method. Briefly, each sample was added to 3 mL of buffer peptone water (BPW) and incubated for 6–8 h in 37°C, followed by inoculating into Mossel Enterobacteria enrichment broth (MEE broth) with 0.5 mg/L meropenem for 6 h. The carbapenem-resistant bacteria were isolated by streaking on MacConkey plates containing 2 mg/L meropenem and identified by MALDI-TOF MS and 16S rDNA sequencing (Kim et al., 2010). In this way, significant number of well-formed carbapenem-resistant colonies were grown on MacConkey plates, but only one colony was selected for further investigations. The carbapenem-resistant isolates were screened for the most found β-lactamases genes including the OXA-1-like broad-spectrum β-lactamases, extended-spectrum β-lactamases (ESBLs), plasmid-mediated AmpC β-lactamases and class A, B and D carbapenemases using multiplex PCR assays reported previously (Dallenne et al., 2010). The complete coding sequence of blaNDM–5 was amplified with the primers (NDM-up, 5′-CTTCCAACGGTTTGATCGTC; NDM-dw, 5′-ATTGGCATAAGTCGCAATCC) and confirmed by ABI3730 sequencing.
Antimicrobial Susceptibility Testing, Conjugation Assay and S1-PFGE
The MICs of blaNDM–5-positive E. coli isolates against antibiotics (Table 1), as well as their transconjugants, were determined using the microdilution broth method and interpreted according to CLSI (2014) guidelines. E. coli strain ATCC 25922 was used as the quality control. To investigate transferability of blaNDM–5, conjugation assay was performed for the twelve blaNDM–5 positive E. coli isolates with the sodium azide-resistant E. coli J53 as the recipient strain. Overnight culture of donor strains and E. coli J53 were mixed (ratio of 1:4) in LB broth, then subjected to overnight incubation on LB agar plates. The mixture culture was then diluted and spread on a selective LB agar plate supplemented with meropenem (2 mg/L) and sodium azide (200 mg/L) to recover transconjugants. Carriage of blaNDM–5 in the transconjugant was confirmed by PCR and MICs. The frequencies of conjugation transfer of five blaNDM–5 plasmids were conducted and expressed as transconjugants per donor cell (T/D) as previously described (Zhong et al., 2012). S1-PFGE was performed to obtain plasmid profiles in donor strains and transconjugants, and the Salmonella enterica serotype Braenderup H9812 was used as the standard size marker.
TABLE 1.
Antibiotic susceptibility profiles of the carbapenem-resistant E. coli strains and the corresponding transconjugants to different antibiotics (mg/L).
| Isolates |
Antibioticsa
|
||||||||
| MEM | STR | ATM | AMX | CQM | FFC | DOX | ENR | CL | |
| L33 | 128 | 128 | 256 | >64 | >128 | >64 | 32 | 16 | 1 |
| L37 | 128 | 64 | 32 | >64 | >128 | >64 | 16 | 16 | 1 |
| L41-1 | 128 | 16 | 4 | >64 | >128 | >64 | 32 | ≤0.125 | 1 |
| L43-1 | 256 | 32 | 8 | >64 | >128 | >64 | 32 | 0.25 | 1 |
| L53 | 128 | 128 | 256 | >64 | >128 | >64 | 32 | 4 | 1 |
| L56 | 128 | >128 | >256 | >64 | >128 | >64 | 16 | 8 | 1 |
| L65 | 128 | 1 | ≤ 0.5 | >64 | >128 | >64 | 32 | 1 | 1 |
| L99 | 128 | >128 | >256 | >64 | >128 | >64 | 32 | 4 | 1 |
| L100 | 128 | 64 | 32 | >64 | >128 | >64 | 32 | ≤0.125 | 0.5 |
| L102 | 128 | 32 | 8 | >64 | 128 | 4 | 16 | ≤0.125 | 1 |
| L103-1 | 64 | 128 | >256 | >64 | >128 | >64 | 16 | 8 | 0.125 |
| L103-2 | 128 | 64 | 256 | >64 | >128 | >64 | 16 | ≤0.125 | 1 |
| L33T | 128 | 1 | ≤0.5 | >64 | 128 | 8 | 4 | ≤0.125 | 0.5 |
| L37T | 32 | 1 | ≤0.5 | >64 | 128 | 8 | 4 | ≤0.125 | 0.5 |
| L41-1T | 64 | 0.5 | 2 | >64 | >128 | 4 | 8 | ≤0.125 | 0.5 |
| L43-1T | 64 | 1 | 1 | >64 | >128 | 8 | 8 | ≤0.125 | 0.25 |
| L53T | 128 | 1 | 64 | >64 | 128 | 8 | 4 | ≤0.125 | 1 |
| L56T | 64 | 1 | ≤0.5 | >64 | 128 | 4 | 4 | ≤0.125 | 0.5 |
| L65T | 64 | 1 | ≤0.5 | >64 | 128 | 4 | 2 | ≤ 0.25 | 0.5 |
| L99T | 64 | 2 | ≤ 0.5 | >64 | 64 | 4 | 2 | ≤ 0.125 | 1 |
| L100T | 64 | 1 | 2 | >64 | 128 | 4 | 2 | ≤0.125 | 1 |
| L102T | 128 | 1 | 1 | >64 | >128 | 4 | 4 | ≤ 0.125 | 0.5 |
| L103-1T | 128 | 1 | ≤ 0.5 | >64 | 128 | 8 | 4 | ≤ 0.125 | 1 |
| L103-2T | 64 | 1 | 64 | >64 | 128 | 4 | 8 | ≤0.125 | 1 |
| J53b | ≤0.25 | 1 | ≤0.5 | 1 | ≤0.25 | 4 | 4 | ≤0.125 | 0.5 |
| ATCC25922c | ≤0.25 | 0.5 | ≤0.5 | 1 | ≤0.25 | 2 | 0.5 | ≤0.125 | 0.5 |
aMEM, meropenem; STR, streptomycin; ATM, aztreonam; AMX, amoxicillin; CQM, cefquinome; FFC, florfenicol; DOX, doxycycline; ENR, enrofloxacin; CL, colistin. All susceptibility tests were repeated at least three times according to CLSI method. The results of colistin susceptibility were interpreted according to EUCAST breakpoints. brecipient strain. cquality control strain.
DNA Extractions, WGS and Bioinformatics Analysis
Genomic DNA of the twelve carbapenem-resistant strains were prepared using the TIANamp Bacteria DNA Kit (Tiangen, China) and subjected to WGS using the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, United States) generating 2 × 150 bp paired-end reads (1 Gbp per sample). The software SPAdes (v3.11.1) was utilized to assemble the genomes. Furthermore, genomes of six strains (L37, L41-1, L53, L65, L100, and L103-2) belonging to different clonal groups based on Illumina data were selected for MinION long-read sequencing (400 Mbp per sample) with the Rapid Barcoding Kit RBK004 without size selection to obtain the complete genome sequences according to the published method (Li R. et al., 2018). Briefly, de novo assembly with hybrid strategy combining Illumina short-read data and MinION long-read data was performed with unicycler (v0.4.4) as the reported method (Wick et al., 2017; Li R. et al., 2018). Sequence Types (STs) and antimicrobial resistance genes were determined using the GoSeqIt tool1. Plasmid replicon type and plasmid multi-locus STs were determined using the PlasmidFinder and pMLST tools2. The complete genome sequences were annotated using the RAST3 automatically and modified manually. Comparisons between blaNDM–5-bearing plasmids and homologous plasmid sequences available in NCBI database were performed using the BRIG tool and Easyfig (Alikhan et al., 2011; Sullivan et al., 2011). The phylogenetic tree based on SNPs was generated as previously reported (Li et al., 2017).
The complete genome sequences of the six strains were deposited in NCBI database with the accession numbers listed in Table 2. The de novo assembled results of another six strains were deposited in figshare database4 for reference.
TABLE 2.
Basic information of the twelve blaNDM–5 positive E. coli strains revealed by WGS data.
| Number of | ||||||||||
| MLST | Conjugation | MinION | Plasmid names | Size | resistance | Accession | ||||
| Strains | Species | types | Source | Transferabilitya | Resistance genes | sequencing | (or replicons) | (bp) | genes | numbers |
| L37 | Escherichia coli | ST-48 | Anal swab | + | fosA, sul2, aac(3)-IId, strA, strB, qnrS1, dfrA14, tet(A), floR, blaCTX–M–55, blaNDM–5, blaTEM–1B, aph(6)-Id, aph(3″)-Ib | + | pL37-2 (IncFIB, p0111) | 145704 | 9 | CP034590 |
| pL37-3 (IncX3)d | 45650 | 1 | CP034591 | |||||||
| pL37-4 (IncFII) | 76559 | 3 | CP034592 | |||||||
| L41-1 | Escherichia coli | ST-48 | Feed | + | strA, strB, sul2, floR, blaCTX–M–64, blaNDM–5, blaTEM–1A, tet(A) | + | pL41-1-2 (IncFIB, IncHIB, p0111) | 201021 | 8 | CP034728 |
| pL41-1-3 (IncFIB) | 111458 | 1 | CP034729 | |||||||
| pL41-1-4 (IncX3)d | 46238 | 1 | CP034730 | |||||||
| L53 | Escherichia coli | ST-48 | Water | + | fosA, sul2, strA, strB, aac(3)-IId, qnrS1, dfrA14, tet(A), floR, blaTEM–1B, blaNDM–5, blaCTX–M–55 | + | pL53-2 (IncFIB, p0111) | 137848 | 9 | CP034745 |
| pL53-3 (IncFII) | 76642 | 3 | CP034756 | |||||||
| pL53-4 (IncX3)d | 46259 | 1 | CP034757 | |||||||
| L65 | Escherichia coli | ST-3076 | Feces | + | tet(A), qnrS1, dfrA14, floR, blaTEM–1C, blaNDM–5 | + | pL65-2 (IncFIB, IncFII) | 145346 | 6 | CP034739 |
| pL65-9 (IncX3)d | 46161 | 1 | CP034744 | |||||||
| L100 | Escherichia coli | ST-48 | Anal swab | + | strA, strB, sul2, floR, blaCTX–M–64, blaTEM–1A, blaNDM–5, tet(A) | + | pL100-2 (IncFIB, IncHIB, p0111) | 202387 | 8 | CP034746 |
| pL100-3 (IncFIB) | 111458 | 1 | CP034747 | |||||||
| pL100-4 (IncX3)d | 46261 | 1 | CP034748 | |||||||
| L103-2 | Escherichia coli | ST8809 | Anal swab | + | arr-2, rmtB, aph(3′)-IIa, aadA1, fosA, dfrA14, tet(A), cmlA1, floR, blaTEM–1B, blaNDM–5, blaOXA–10, blaCTX–M–55 | + | pL103-2-2 (IncFIB) | 110210 | 0 | CP034844 |
| pL103-2-3 (IncFIB) | 102295 | 1 | CP034845 | |||||||
| pL103-2-4 (IncFII) | 100667 | 6 | CP034846 | |||||||
| pL103-2-5 (IncX3)d | 46163 | 1 | CP034847 | |||||||
| pL103-2-6 (IncX1) | 13362 | 1 | CP034848 | |||||||
| pL103-2-7 (unknown) | 12606 | 6 | CP034849 | |||||||
| L33 | Escherichia coli | ST-48 | Anal swab | + | strB, strA, aac(3)-IId, sul2, fosA, qnrS1, dfrA14, tet(A), floR, blaNDM–5, blaCTX–M–55, blaTEM–1B | – | IncX3d, p0111 | 46kbc | 12 | Onlineb |
| L43-1 | Escherichia coli | ST-48 | Feces | + | strA, strB, sul2, floR, blaCTX–M–64, blaTEM–1A, blaNDM–5, tet(A) | – | IncX3d, p0111, colRNAI | 46kb | 8 | Online |
| L56 | Escherichia coli | ST-48 | Water | + | fosA, sul2, strA, strB, aac(3)-IId, qnrS1, dfrA14, tet(A), floR, blaTEM–1B, blaNDM–5, blaCTX–M–55 | – | IncX3d, p0111 | 46kb | 12 | Online |
| L99 | Escherichia coli | ST-48 | Anal swab | + | fosA, sul2, strA, strB, aac(3)-IId, qnrS1, dfrA14, tet(A), floR, blaNDM–5, blaCTX–M–55, blaTEM–1B | – | IncX3d, p0111 | 46kb | 12 | Online |
| L102 | Escherichia coli | ST-48 | Anal swab | + | tet(A), sul2, blaCTX–M–64, blaNDM–5, strB, strA | – | IncX3d, p0111 | 46kb | 6 | Online |
| L103-1 | Escherichia coli | ST-48 | Anal swab | + | strB, strA, aac(3)-IId, sul2, fosA, qnrS1, dfrA14, tet(A), floR, blaNDM–5, blaTEM–1B, blaCTX–M–55 | – | IncX3d, p0111, colRNAI | 46kb | 12 | Online |
aTransference of plasmids carrying blaNDM–5. bThe assembly results of six strains without complete genome sequences were deposited in figshare database for reference (https://figshare.com/articles/L33-L103-1/8044847). cThe length of blaNDM–5-bearing IncX3 plasmids in samples without MinION sequencing were infered from S1-PFGE. dblaNDM–5-bearing plasmid.
Results and Discussion
Characterization of Carbapenem-Resistant E. coli Strains
Twelve carbapenem-resistant E. coli strains were isolated from the 117 samples. The twelve carbapenem-resistant E. coli strains included seven strains (58.3%) from anal swab samples (L33, L37, L99, L100, L102, L103-1, L103-2), two strains (16.7%) from water samples (L53, L56), two isolates (16.7%) from fecal samples (L43-1, L65) and one isolate (8.3%) from a feed sample (L41-1). Among them, L65 was isolated from goose hatchery, while the other eleven strains were isolated from the breeding goose farm indicating the occurrence of carbapenem-resistant strains of breeding goose farm was more prevalent. These strains were resistant to multiple antibiotics including meropenem, streptomycin, amoxicillin, cefquinome, florfenicol and doxycycline, but most of them were susceptible to enrofloxacin (Table 1).
All the carbapenem-resistant E. coli strains harbored blaNDM–5. The strains were also found to carry multiple resistance genes conferring resistance to β-lactam (blaCTX–M, blaTEM–1, blaOXA), aminoglycoside (strA, strB, aac(3)-IId, aph(6)-Id, aph(3″)-Ib, rmtB) fosfomycin (fosA), tetracycline (tet(A)), phenicols (cmlA1, floR), quinolone (qnrS1), sulfonamide(sul2), rifampicin (arr-2) and trimethoprim (dfrA14) (Table 2). Moreover, the twelve E. coli isolates were categorized into two STs and one novel ST according to WGS data. Ten strains belonged to ST48 and one novel ST (ST8809) strain (L103-2) was isolated from the breeding goose farm, whereas only one strain L65 isolated from hatchery pertained to ST3076, which indicated that resistant strains between the two farms was unrelated phylogenetically. Two clonal groups within ST48 were identified in breeding goose farm confirmed by SNP analysis (Figure S1).
Transferability of blaNDM–5 Gene and the Underlying Mechanisms
Conjugation assay was performed to investigate transferability of blaNDM–5 gene, and transconjugants were successfully recovered from twelve donor strains (Table 1). It is noteworthy that the conjugation frequencies of blaNDM–5-bearing plasmids were stable (∼10–6 per donor) (Table S2). S1-PFGE demonstrated that plasmids of ca. 46 kb in size could be observed in both donor strains and recipient strains except L53T, the transconjugant of L53, in which a plasmid of ca. 120 kb was found (Figure S2). The underlying molecular mechanism warrants further study. Two donor strains can transfer two plasmids into receipt strains (L103-1 and L103-2), enabling the two transconjugants resistant to multiple antibiotics (Table 1). The fact that all blaNDM–5-bearing transconjugants carried a ca. 46 kb plasmid indicated blaNDM–5 was located on a conjugative plasmid with a similar structure. To probe this hypothesis, complete genomes of six strains belonging to different clonal groups were obtained through short-read and long-read sequencing methods.
All blaNDM–5-bearing plasmids from L37, L41-1, L53, L65, L100, L103-2 were almost identical (99% query coverage and 99% nucleotide identity) (Figure 1), with sizes ranging from 45,650 to 46,261 bp (Table 2). The blaNDM–5 gene was in the typical structure ISAba125-IS5-blaNDM–5- bleMBL-trpF-dsbC-IS26 found in IncX3 type plasmids. No other antimicrobial resistance genes were detected in these plasmids. These plasmids belonged to IncX3 type and showed 99% nucleotide identity with the first identified blaNDM–5-harboring plasmid pNDM-MGR194 of K. pneumoniae MGR-K194 in India (KF220657) (Krishnaraju et al., 2015) and pNDM-QD28 obtained from E. coli QD28 in China (KU167608) (Figure 1). The pNDM-MGR194-like plasmids were reported in China frequently, which indicated IncX3 type plasmids were the epidemic vehicles mediating dissemination of the blaNDM–5 in China (Krishnaraju et al., 2015; Li X. et al., 2018). The QD28 was the first identification of blaNDM–5-carrying E. coli in the neonatal infection (Zhu et al., 2016). The blaNDM–5-bearing IncX3 plasmid has been shown to be carried in several clinical isolates (Wailan et al., 2015; Zhang et al., 2016). The discovery of these blaNDM–5-bearing plasmids in goose farm demonstrated the IncX3 plasmids accounted for widespread of blaNDM–5 in this goose farm.
FIGURE 1.
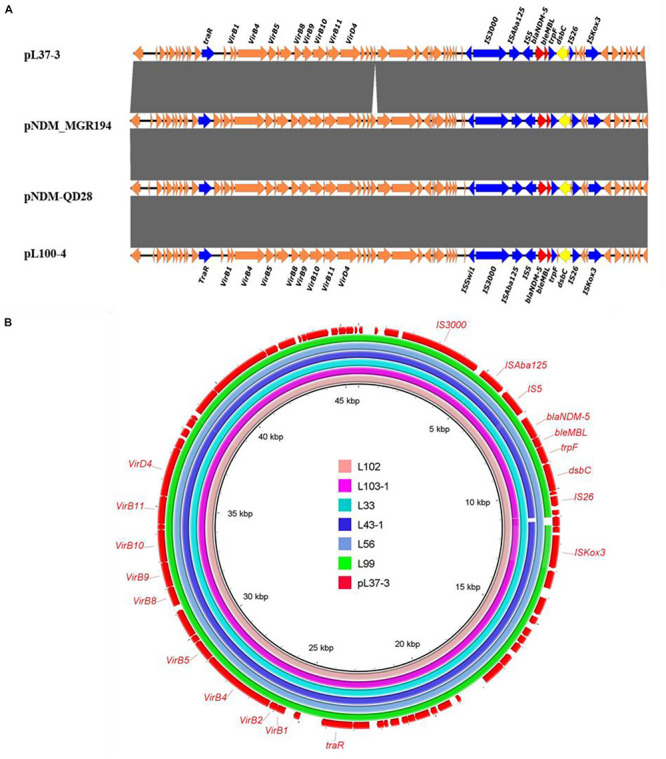
Sequence alignment analysis of IncX3 blaNDM–5 plasmids. (A) Linear alignment of four blaNDM–5-bearing E. coli strains. pNDM-QD28 (GenBank accession no. KU167608.1) and pNDM_MGR194-blaNDM–5 (GenBank accession KF220657.1) were the reported blaNDM–5-bearing plasmids. Plasmids pL41-1-4, pL53-4, pL65-9 and pL103-2-5 shown identical structure with pL100-4. (B) Circular comparison between the blaNDM–5-bearing IncX3 plasmid pL37-3 and the assembled contigs of other six blaNDM–5 positive strains without MinION data and based on Illumina data.
Whole Genome Sequencing Analysis
Analysis of whole genomes of the strains revealed the distribution of antibiotic resistance genes, replicon types and STs (Table 2). The twelve strains were divided into three clonal groups based on SNP analysis, and strains belonging to ST48 from both clonal group A and C were isolated from various sources including water, anal swabs and feces collected from the breeding goose farm, demonstrating the potential clonal spread of these blaNDM–5-bearing strains and transmission of blaNDM–5-bearing plasmid among subtypes of ST48 in breeding goose farm (Figure S1). However, L65 (ST3076) isolated from goose hatchery and L103-2 (ST8809) obtained from breeding goose farm showed great difference from other strains (ST48) isolated from the breeding goose farm, suggesting that there was spread of blaNDM–5 mediated by the conjugative plasmid among different E. coli strains (Figure S1).
Apart from the blaNDM–5-bearing plasmids, other MDR plasmids were identified from the six complete genomes. L37 and L53 belonging to the same clonal group also carried other two plasmids, the IncFII plasmid (pL37-4, pL53-3) and the IncFIB/p0111 hybrid plasmid (pL37-2, pL53-2), respectively. A mosaic MDR region consisting of ISs and various resistance gene including qnrS1, floR, blaTEM–1B, sul2, aac(3)-IId, aph(6)-Id, aph(3″)-Ib, dfrA14 and tet(A) was identified in IncFIB/p0111 hybrid plasmids (Figure S3). Two large similar plasmids (pL41-1-2 and pL100-2) over 200 kb were found in both L41-1 and L100 belonging to the same clonal group, and these two hybrid plasmids composing of IncFIB, IncHIB and p0111 replicons contained a MDR region (blaTEM–1A, sul2, floR, tet(A), aph(6)-Id and aph(3″)-Ib) dispersed among many insertion sequences. High insertion sequences abundance dispersed in plasmids may account for plasmid plasticity resulting in generation of hybrid plasmids. IS26 was proved to be involved in plasmid recombination (Wong et al., 2017). The formation of these two hybrid plasmids (pL41-1-2 and pL100-2) was probably related to insertion sequences (Figures S4, S5). A hybrid plasmid pL65-2 was found in L65, and pL65-2 was composed of IncFIB and IncFII replicons and the typical characteristic of this plasmid was a transfer region containing conjugative transfer genes, but no resistance genes were found (Figure S6).
Carbapenems are currently only approved for usage in humans and prohibited in animals. The emergence of blaNDM–5 positive E. coli in animals and surrounding environments posed a great public health concern. Previously, there were reports of blaNDM–5 positive strains found in various chicken and swine farms (Ho et al., 2018; Xiang et al., 2018). However, to the best of our knowledge, this was the first report of blaNDM–5 positive strains of goose origin which was the important waterfowl in Jiangsu breeding industry. Further research revealed that ST48 blaNDM–5 positive E. coli, as dominant type, underwent clone spread within goose farm. ST48 type E. coli is frequently associated to various β-lactamases, including ESBL and NDM (Liu et al., 2017; Said et al., 2017), thus we assumed that goose farming system could be a reservoir of β-lactamases genes. The occurrence of carbapenem-resistant strains in goose breeding farm was higher than that in goose hatchery, which may be due to the layout of the two farms. The breeding goose farm is an open space suitable for goose activities, facilitating transmission of MDR strains. The goose hatchery adopts a completely closed layout to protect the goslings from external environments, and few MDR bacteria contaminated the goose hatchery and prevalence of resistant strains was low.
IncX3 plasmids carrying various of blaNDM variants have been increasingly reported all over the world in recent years (Paskova et al., 2018; Xu and He, 2019). Previous study has proved the ability of blaNDM-bearing IncX3 plasmid transfer to different Enterobacterial species at a wide range of temperatures (Wang Y. et al., 2018). Based on these facts, we believed that once the IncX3 plasmid carrying blaNDM–5 was detected in goose farms, the risk of its widespread dissemination will be greatly increased.
Water is an essential element during goose growth, and water samples were collected from the river around the goose farm. The goose farm received and discharged river water without any purification measures, which may accelerate transfer of MDR bacteria. Bacteria encoding blaNDM–5 was found in river (Almakki et al., 2017). Thus, river water has become a reservoir of drug-resistant bacteria. Apart from the feces and feed samples, water is another environmental sample detected positive for NDM-5-producing E. coli in this study. This indicated that contamination of the NDM-5 producing E. coli between feces and water occurred in goose farm. Comprehensive surveillance of MDR bacteria in water surrounding the goose farm must be strengthened to understand the accurate contamination route. Since similar blaNDM–5-bearing plasmids were always found in clinical settings and no carbapenem exposure existed in goose farms, this blaNDM–5-bearing plasmid may derive from humans, further transfer and persist in goose farms to constitute a potential risk.
In conclusion, this study first identified the blaNDM–5-bearing E. coli isolates in goose farms and its surroundings. Prevalence of NDM-5 producing E. coli posed a potential risk to public health. It is extremely urgent to reinforce the surveillance of resistance in the waterfowl system to curb the transmission or persistence of MDR bacteria.
Data Availability
Publicly available datasets were analyzed in this study. This data can be found here: Refer to Table 2 for all accession numbers.
Author Contributions
ZW and RL conceived and designed the study. ZL, YLi, and XX performed the study, analyzed the data, and prepared the manuscript. XX, YLiu, and RL revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jiayu Xiao and Xiaoyu Lu in Wang’s Lab for sample collection.
Funding. This work was supported by the National Key Research and Development Program of China (2016YFD0501310), National Natural Science Foundation of China (31872523 and 31872526), the Natural Science Foundation of Jiangsu Province (BK20180900), and a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The Postgraduate Research and Practice Innovation Program of Jiangsu Province (Yangzhou University), XKYCX18_117 to ZL.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02002/full#supplementary-material
References
- Alikhan N. F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almakki A., Maure A., Pantel A., Romano-Bertrand S., Masnou A., Marchandin H., et al. (2017). NDM-5-producing Escherichia coli in an urban river in montpellier, France. Int. J. Antimicrob. Agents 50 123–124. 10.1016/j.ijantimicag.2017.04.003 [DOI] [PubMed] [Google Scholar]
- CLSI (2014). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-fourth Informational Supplement M100-S24. Wayne, PA: CLSI. [Google Scholar]
- Dallenne C., Da Costa A., Decre D., Favier C., Arlet G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65 490–495. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- Ho P. L., Wang Y., Liu M. C., Lai E. L., Law P. Y., Cao H., et al. (2018). IncX3 epidemic plasmid carrying blaNDM-5 in Escherichia coli from swine in multiple geographic areas in China. Antimicrob. Agents Chemother. 62:e02295-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornsey M., Phee L., Wareham D. W. (2011). A novel variant, NDM-5, of the new delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 55 5952–5954. 10.1128/AAC.05108-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. W., Kim Y. H., Kim S. E., Lee J. H., Park C. S., Kim H. Y. (2010). Identification and distribution of bacillus species in doenjang by whole-cell protein patterns and 16S rRNA gene sequence analysis. J. Microbiol. Biotechnol. 20 1210–1214. 10.4014/jmb.1002.02008 [DOI] [PubMed] [Google Scholar]
- Krishnaraju M., Kamatchi C., Jha A. K., Devasena N., Vennila R., Sumathi G., et al. (2015). Complete sequencing of an IncX3 plasmid carrying bla(NDM-5) allele reveals an early stage in the dissemination of the bla(NDM) gene. Indian J. Med. Microbiol. 33 30–38. 10.4103/0255-0857.148373 [DOI] [PubMed] [Google Scholar]
- Li R., Xie M., Dong N., Lin D., Yang X., Wong M. H. Y., et al. (2018). Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience 7 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Fu Y., Shen M. Y., Huang D. Y., Du X. X., Hu Q. F., et al. (2018). Dissemination of bla(NDM-5) gene via an IncX3-type plasmid among non-clonal Escherichia coli in China. Antimicrob. Resist. Infect. Control 7:59. 10.1186/s13756-018-0349-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. C., Ye L. W., Wong M. H. Y., Zheng Z., Chan E. W. C., Chen S. (2017). Evolution and comparative genomics of pAQU-like conjugative plasmids in vibrio species. J. Antimicrob. Chemother. 72 2503–2506. 10.1093/jac/dkx193 [DOI] [PubMed] [Google Scholar]
- Liu L., Feng Y., Mcnally A., Zong Z. Y. (2018). bla(NDM-21), a new variant of bla(NDM) in an Escherichia coli clinical isolate carrying bla(CTX-M-55) and rmtB. J. Antimicrob. Chemother. 73 2336–2339. 10.1093/jac/dky226 [DOI] [PubMed] [Google Scholar]
- Liu Z., Wang Y., Walsh T. R., Liu D., Shen Z., Zhang R., et al. (2017). Plasmid-mediated novel blaNDM-17 gene encoding a carbapenemase with enhanced activity in a sequence type 48 Escherichia coli strain. Antimicrob. Agents Chemother. 61:e002233-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paskova V., Medvecky M., Skalova A., Chudejova K., Bitar I., Jakubu V., et al. (2018). Characterization of NDM-encoding plasmids from Enterobacteriaceae recovered from Czech Hospitals. Front. Microbiol. 9:1549. 10.3389/fmicb.2018.01549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter R. F., D’souza A. W., Dantas G. (2016). The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist. Updat. 29 30–46. 10.1016/j.drup.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said L. B., Hamdaoui M., Jouini A., Boudabous A., Slama K. B., Torres C., et al. (2017). First detection of CTX-M-1 in extended-spectrum beta-lactamase-producing Escherichia coli in seafood from Tunisia. J. Food Pro. 17 1877–1881. 10.4315/0362-028X.JFP-16-324 [DOI] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27 1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wailan A. M., Paterson D. L., Caffery M., Sowden D., Sidjabat H. E. (2015). Draft genome sequence of NDM-5-producing Escherichia coli sequence type 648 and genetic context of blaNDM-5 in Australia. Genome Announc. 3:e00194-15. 10.1128/genomeA.00194-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T. R., Weeks J., Livermore D. M., Toleman M. A. (2011). Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11 355–362. 10.1016/S1473-3099(11)70059-7 [DOI] [PubMed] [Google Scholar]
- Wang J., Yao X., Luo J., Lv L. C., Zeng Z. L., Liu J. H. (2018). Emergence of Escherichia coli co-producing NDM-1 and KPC-2 carbapenemases from a retail vegetable, China. J. Antimicrob. Chemother. 73 252–254. 10.1093/jac/dkx335 [DOI] [PubMed] [Google Scholar]
- Wang R. B., Liu Y. Q., Zhang Q., Jin L. Y., Wang Q., Zhang Y. W., et al. (2018). The prevalence of colistin resistance in Escherichia coli and Klebsiella pneumoniae isolated from food animals in China: coexistence of mcr-1 and bla(NDM) with low fitness cost. Int. J. Antimicrob. Agents 51 739–744. 10.1016/j.ijantimicag.2018.01.023 [DOI] [PubMed] [Google Scholar]
- Wang Y., Tong M. K., Chow K. H., Cheng V. C., Tse C. W., Wu A. K., et al. (2018). Occurrence of highly conjugative IncX3 epidemic plasmid carrying bla NDM in Enterobacteriaceae isolates in geographically widespread areas. Front. Microbiol. 9:2272. 10.3389/fmicb.2018.02272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R. R., Judd L. M., Gorrie C. L., Holt K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. H. Y., Chan E. W. C., Chen S. (2017). IS26-mediated formation of a virulence and resistance plasmid in Salmonella enteritidis. J. Antimicrob. Chemother. 72 2750–2754. 10.1093/jac/dkx238 [DOI] [PubMed] [Google Scholar]
- Xiang R., Zhang A. Y., Ye X. L., Kang Z. Z., Lei C. W., Wang H. N. (2018). Various sequence types of Enterobacteriaceae isolated from commercial chicken farms in china and carrying the blaNDM-5 gene. Antimicrob. Agents Chemother. 62:e00779-18. 10.1128/AAC.00779-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., He F. (2019). Characterization of a NDM-7 carbapenemase-producing Escherichia coli ST410 clinical strain isolated from a urinary tract infection in China. Infect. Drug Resist. 12 1555–1564. 10.2147/IDR.S206211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R. S., Feng Y., Lv X. Y., Duan J. H., Chen J., Fang L. X., et al. (2016). Emergence of NDM-5- and MCR-1-producing Escherichia coli clones ST648 and ST156 from a single muscovy duck (Cairina moschata). Antimicrob. Agents Chemother. 60 6899–6902. 10.1128/AAC.01365-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong D., Toleman M. A., Giske C. G., Cho H. S., Sundman K., Lee K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a Unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53 5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Xie L., Wang X., Han L., Guo X., Ni Y., et al. (2016). Further spread of bla NDM-5 in Enterobacteriaceae via IncX3 plasmids in shanghai, China. Front. Microbiol. 7:424. 10.3389/fmicb.2016.00424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Li J., Wang Y., Shen J., Shen Z., Wang S. (2019). Presence of NDM in non-E. coli Enterobacteriaceae in the poultry production environment. J. Antimicrob. Chemother. 74 2209–2213. 10.1093/jac/dkz193 [DOI] [PubMed] [Google Scholar]
- Zhong X., Droesch J., Fox R., Top E. M., Krone S. M. (2012). On the meaning and estimation of plasmid transfer rates for surface-associated and well-mixed bacterial populations. J. Theor. Biol. 294 144–152. 10.1016/j.jtbi.2011.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. Q., Zhao J. Y., Xu C., Zhao H., Jia N., Li Y. N. (2016). Identification of an NDM-5-producing Escherichia coli sequence type 167 in a neonatal patient in China. Sci. Rep. 6:29934. 10.1038/srep29934 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: Refer to Table 2 for all accession numbers.


