Abstract
Environmental exposures to hazardous chemicals are associated with a variety of human diseases and disorders, including cancers. Phase I metabolic activation and detoxification reactions catalyzed by cytochrome P450 enzymes (CYPs) affect the toxicities of many xenobiotic compounds. Proper regulation of CYP expression influences their biological effects. Noncoding RNAs (ncRNAs) are involved in regulating CYP expression, and ncRNA expression is regulated in response to environmental chemicals. The mechanistic interactions between ncRNAs and CYPs associated with the toxicity and carcinogenicity of environmental chemicals are described in this review, focusing on microRNA-dependent CYP regulation. The role of long non-coding RNAs in regulating CYP expression is also presented and new avenues of research concerning this regulatory mechanism are described.
Keywords: microRNA, long noncoding RNA (lncRNA), cytochrome P450 (CYP), chemical carcinogenesis, epigenomics, epigenetics, biomarker
1. Environmental exposure and impacts of CYPs and their regulation
Humans are exposed throughout their lives to potentially harmful naturally-occurring and man-made organic compounds present in the air, water, and diet. Hazardous xenobiotic (Greek, xenos – stranger; bios – life) environmental chemicals are recognized as important risk factors for cancers and for a variety of other human diseases. Well known examples include the increased risk for lung cancer in humans associated with exposure to nitrosamines and other carcinogens present in tobacco smoke, exposure to the industrial chemical benzene that increases the risk for leukemia, and dietary exposure to aflatoxins that is associated with increased risk for liver cancer.1
Human metabolism influences the acute and chronic biological effects of environmental compounds, for good or for harm, either by catalyzing detoxification reactions or by generating metabolites with enhanced toxicity. Xenobiotic metabolism often involves more than one enzyme-catalyzed step. The complement of xenobiotic metabolizing enzymes (XMEs) in humans includes Phase I cytochrome P450 (CYP) monooxygenase enzymes and Phase II conjugation enzymes, such as UDP-glucuronosyl transferases (UGTs), glutathione S-transferases (GSTs), and sulfotransferases (SULTs). In the ideal case, XME activities convert a toxic environmental compound to a less toxic metabolite; for example, the cytotoxicity of the broad-spectrum antimicrobial agent and environmental contaminant triclosan is partially diminished by CYP-dependent metabolism.2,3 More typically, XMEs facilitate the elimination of hydrophobic xenobiotic compounds by converting them to polar metabolites that are more readily excreted in the urine or bile.4 In other cases, XME activity converts a less toxic compound to a more harmful metabolite, such as the CYP-dependent activation of R-pule-gone, a fragrant monoterpenoid present in pennyroyal oil and mint extracts that is associated with hepatotoxicity, or the metabolism of pyrrolizidine alkaloids that are widely distributed among plant species to generate reactive dehydropyrrolic metabolites that form protein adducts associated with hepatic sinusoidal obstruction syndrome, liver injury, and hepatocarcino-genesis.5,6 XME activity may also generate mixtures of metabolites with varying degrees of toxicity, such as the CYP-dependent activation of the organophosphorus pesticide methyl parathion via desulfuration and its CYP-dependent detoxification via dearylation.7
The CYP enzyme family is noted among XMEs for catalyzing the initial Phase I metabolic steps involved in processing a wide variety of structurally diverse compounds of significant public health interest (Figure 1). The general reaction catalyzed by CYP mixed-function monooxygenase enzymes is given in Reaction (1):
| (Reaction 1) |
Figure 1.
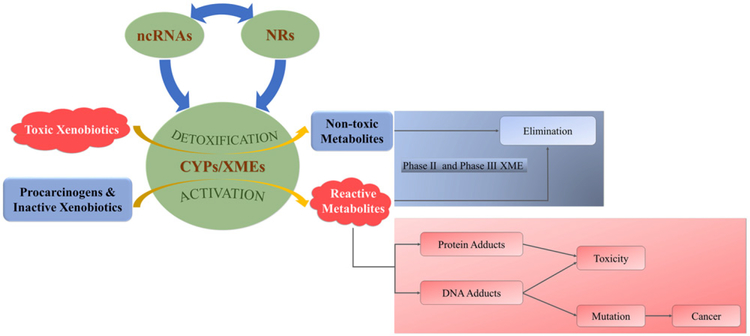
Role of ncRNA-dependent regulation of CYP expression in xenobiotic metabolism. NcRNAs regulate the expression of CYP and NR genes and NRs regulate the expression of ncRNAs and CYPs. CYPs and other Phase I xenobiotic metabolizing enzymes (XMEs) catalyze Phase I metabolism of xenobiotics. Toxic xenobiotics are converted by CYP-mediated Phase I detoxification reactions to polar, nontoxic metabolites that may be processed further by Phase II and Phase III XMEs to enhance elimination. Procarcinogens and inactive xenobiotics are converted by CYP-catalyzed Phase I activation reactions to reactive metabolites that form protein adducts and DNA adducts associated with toxicity or mutations leading to cancer.
In these well-studied reactions, the reduction to water of one oxygen atom provided by O2 and the incorporation of the other oxygen atom into an organic substrate is coupled to NADPH oxidation. The simplicity of Reaction 1 belies the complexity of reaction types catalyzed by different human CYP isoforms. The human CYP superfamily consists of 57 genes that are divided into 18 families, indicating the diversity of metabolic capabilities provided by this enzyme superfamily, including alkyl and aryl hydroxylations, N- and S-oxygenations, epoxidations, N- and S- dealkylations, dehalogenations, desulfurations, ring expansions and closures, aldehyde scissions, dehydrations, desaturations, ester cleavages, cis/trans isomerizations, rearrangements of fatty acid and prostaglandin hydroperoxides, one-electron oxidations, and coupling reactions.8,9
In general, CYP-mediated toxicity is associated with two types of reactions that either (a) produce reactive electrophiles that damage critical bio-molecules or (b) induce oxidative stress via free radical reactions.10 In the first class of toxic reactions, an electrophilic product of a CYP-catalyzed reaction attacks sensitive nucleophilic functional groups on critical biomolecules, such as DNA or proteins, contributing to cancer or toxicity. The formation of DNA adducts, if not repaired, may result in genetic mutations associated with tumor initiation.11,12 The CYP-mediated metabolism of polycyclic aromatic hydrocarbons (PAHs), chemically inert environmental contaminants associated with lung and bladder cancers in humans, is an example of this genotoxic mechanism.13,14 In humans, CYP1A1 and CYP1B1 activities convert benzo[α]pyrene (BaP), a procarcinogenic PAH, to the potent chemical carcinogen (+)-benzo[α]pyrene-7,8-dihydrodiol 9,10-epoxide, an electrophilic metabolite that forms carcinogenic DNA adducts.13,15 Likewise, CYP3A4 and CYP3A5 activities activate aflatoxin B1, a liver carcinogen, to form aflatoxin B1 exo-8,9-epoxide, an electrophilic metabolite that attacks the nucleophilic N7 position of guanine residues.10,16 Both tumor initiation and tumor promotion may be operative in some cases of CYP-dependent toxicity. The organochlorine pesticide heptachlor, metabolized by CYP activity, is linked to genotoxicity in cultured human TK6 lymphoblastoid cells and to the tumor promotion phase of hepatocarcinogenesis in rats.17,18 From the same substrate molecule CYP activity may generate multiple metabolites that mediate different biological effects. CYP2E1 activity creates a mixture of metabolites from benzene, including phenol, 1,2- and 1,4-hydroquinones, 1,2- and 1,4-benzoquinones, and benzene-1,2,4-triol that deplete glutathione reserves, stimulate oxidative stress, generate semiquinone free radicals, produce 8-hydroxy-2′-deoxyguanosine DNA lesions, trigger altered gene expression, and induce chromosomal aberrations associated with leukemia.10,19 CYP-dependent metabolism is also relevant for diseases other than cancer. For example, increased risks of insulin resistance and diabetes are associated with exposure to organochlorine and organophosphorus pesticides, perfluoroalkyl substances, and polychlorobiphenyls, which are environmental contaminants metabolized by CYP activity.20–22
In addition to the role of CYPs in the metabolism of xenobiotic compounds, various hepatic and extrahepatic CYPs are also involved in other normal biological functions, such as steroid and cholesterol biosynthesis and metabolism.23,24 These CYP-mediated functions can affect signaling pathways associated with cell cycle arrest, apoptosis, cell proliferation, and survival in ways that can mediate xenobiotic toxicity.11
The substrate specificities for CYP isoforms, their tissue distributions, and their relative levels of expression influence toxicity. While some CYPs are expressed constitutively, CYP expression may also be induced by endogenous and exogenous chemicals.25 Inter-individual variability in both constitutive and induced CYP expression can cause differential responses to environmental exposure, including susceptibility to diseases. Therefore, the proper regulation of XME expression has a critical bearing on the biochemical toxicology of environmental agents.
A variety of factors influence CYP expression. Genetic variations, including single nucleotide polymorphisms (SNPs), copy number variations (CNVs), and pseudogenes, have been shown to affect CYP expression and CYP-related cancers.26–28 Transcription factors, co-activators and corepressors, and nuclear receptors (NRs) are key transcriptional regulators of CYP expression. Hepatic nuclear factor (HNF) 1 A and HNF4A are important regulators of CYP expression, as are the xenobiotic sensor receptors, such as pregnane X receptor (PXR), constitutive androstane receptor (CAR), aryl hydrocarbon receptor (AhR), and vitamin D receptor (VDR), which can activate CYP expression.29
CYP expression is also affected by many epigenetic factors, including DNA methylation, histone modification, and ncRNA regulation.30,31 DNA methylation at the CpG sites in the promoter region leads to transcriptional repression and has widespread effects on the expression of CYPs in different organs, such as the lung and liver.31–33 Exposure to DNA methylating agents may lead to hypermethylation of CYPs genes by DNA methyltransferases and inhibition of CYP expression.34,35 The amino-terminal tails of histone proteins are accessible for post-translational modifications, including acetylation, methylation, phosphorylation, ubiquitination, and sumoylation.36 Histone modifications can influence gene expression by altering chromatin architecture and recruiting remodeling enzymes and trans-regulatory factors. For example, CYP2E1 upregulation induced by trichostatin A (TSA) is associated with histone H3 acetylation and the recruitment of acetylated histone H3, HNF-1, and HNF-3β to the CYP2E1 promoter.37 DNA methylation and histone modification may work in concert in regulating gene expression activities and are contributing mechanisms to carcinogenesis.38–40
The role of ncRNAs in regulating CYP expression has received increasing attention in the recent decade. It is estimated that 98% of the human genome consists of noncoding genes, producing a large number of ncRNA transcripts of various types.41 NcRNAs comprise a variety of RNA molecules that are not translated into proteins and include transfer RNA (tRNA), ribosomal RNA (rRNA), long noncoding RNA (lncRNA), microRNA (miRNA), small interfering RNA (siRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), piwi-interacting RNA (piRNA), and Y RNA components of the Ro60 ribonucleoprotein essential for DNA replication.42–44 NcRNAs longer than 200 nucleotides are defined as lncRNAs whereas those less than 200 nucleotides long are considered small noncoding RNAs, such as miRNAs, siRNAs, snRNAs, snoRNAs, and piRNAs.42 Of these ncRNA classes, miRNA and lncRNAs have been identified as significant epigenetic modulators of CYP expression.45–49 High-throughput technologies have provided a plethora of genomic and transcriptomic data related to ncRNAs and the analysis of these data using bioinformatics tools has facilitated the characterization of ncRNA functions.50,51
miRNAs are a class of evolutionarily conserved small noncoding RNAs (18–22 nt in length) that are functional in plants and animals. The most recent version of the miRBase database (Release 22, March 2018, http://www.mirbase.org/), an online repository of published miRNAs, contains 38,589 precursor miRNAs that produce 48,885 mature miRNAs in 271 species. Transcription of miRNA encoding genes and cleavage of primary and hairpin-shaped precursor miRNAs give rise to single-stranded mature miRNAs that are proposed to regulate 30–60% of protein-coding genes.52,53
Typically, miRNAs mediate post-transcriptional gene silencing by binding to miRNA recognition elements (MREs) present in the mRNA transcripts of protein-coding genes. Functional interactions between miRNA and mRNA sequences require precise (in plants) or imperfect (in animals) base-pairing between the seed sequence of a miRNA (first 2–8 nucleotides from the 5′-end) and its cognate MRE present in a specific mRNA transcript. Degenerate sequence complementarity in animals involving short stretches of nucleotides allows a single miRNA to regulate more than 100 distinct mRNA transcripts and an individual mRNA to be targeted by multiple miRNAs.54 By recruiting RNA-induced silencing complexes (RISC) to mRNAs, miRNAs downregulate target genes by promoting mRNA degradation or repressing translation.55
ncRNAs and NRs both play regulatory roles in CYP expression, providing opportunities for some ncRNAs and NRs to transregulate the expression of one another. miRNAs may downregulate NR expression, which then suppresses NR-dependent CYP expression downstream. In other cases, NRs may enhance miRNA transcription by acting on their promoters or by interacting with the promoters of miRNA processing factors.45,56
In this review, we describe important ncRNA-dependent regulation of CYP expression that is linked to chemical carcinogenesis and toxicity, namely chemically-induced dysregulation of ncRNA levels, ncRNA-mediated modulation of CYP expression, and toxicity and carcinogenesis caused by CYP-dependent chemical metabolic activation. We discuss mechanisms of CYP regulation by miRNAs, using human CYP2E1, as an example, to illustrate the diversity and complexity in miRNA modulation of CYP expression. We then present current knowledge on the regulation of CYP expression by lncRNAs and present potential directions for future studies.
2. miRNA-dependent regulation of CYP expression in environmental carcinogenesis and toxicity
miRNAs have attracted great interest due to their roles in regulating gene expression associated with nearly all cellular functions, including proliferation, differentiation, and apoptosis, implying significant roles of miRNAs in important human diseases, such as cancer.57–60 miRNAs have also been exploited as potential biomarkers for human diseases. Current investigations are elucidating the roles and molecular mechanisms by which miRNAs regulate CYP expression affecting toxicity and cancer in humans. Table 1 presents recently reported miRNA-dependent regulation of CYP expression that is related to exposures to various types of environmental and endogenous chemical stimuli and the associations of those gene regulatory interactions with toxicity and carcinogenesis.
Table 1.
Mechanistic interactions of miRNAs and CYPs associated with environmental toxicity and carcinogenesis.
| Chemical | Exposure source | miRNA | CYP | Model system | Toxicity | References |
|---|---|---|---|---|---|---|
| DDT | Pesticides | miR-221, −222, −20b ↓ | Predicted to target CYP19A1 | Female rats | Uterus and ovary cancer | 61 |
| BDE47 | Industrial materials | miR-23b ↓ | CYP3A1 (rat orthologue of human CYP3A4) ↑ | H4IIE cells, rats | Cytotoxicity | 62 |
| Aflatoxin Bl | Food contamination | miR-138–1* ↓ | CYP2A13 (not a target of miR-138–1*, but activates aflatoxin B1) | BEAS-2B cells | Malignant cell transformation in lung | 63 |
| NSC-156306 | Chemical | miR-128–3р ↓ | CYP2C9 ↑ | HepG2, HepaRG cells | Associated with hepatocellular carcinoma | 64 |
| NSC-606170 | Chemical | miR-128–3р ↑ | CYP2C9 ↓ | HepG2, HepaRG cells | Associated with hepatocellular carcinoma | 64 |
| Fumonisin B1 | Maize contamination | miR-27b ↓ | CYP1B1 ↑ | HepG2 cells | Liver cancer | 65 |
| NNK | Tobacco | miR-126* ↓ | CYP2A3 ↑ | Male rats | Lung cancer | 66 |
| Benzo(a)pyrene | Coal tar, tobacco smoke, exhaust fume, grilled meat | miR-892a ↓ | CYP1A1 ↑ | MCF-7 cells | Cell viability | 67 |
| lnterleukin-6↑ | miR-27b ↓ | CYP1B1 ↑ | HCT116, SW480 cells | Dietary carcinogen activation; colorectal cancer | 68 |
Various environmental factors are known to influence the expression of miRNAs.69 Modifications in the epigenome due to environmental exposure can have trans-generational effects, i.e. phenotypes or disease states caused by epigenetic alteration from individuals exposed to toxicants can be inherited by their offspring in multiple generations.70 Numerous environmental stressors from various sources, including tobacco, alcohol, food, air pollution, pharmaceuticals, and medical and commercial products, have been shown to alter the expression of miRNAs and lncRNAs.71,72 For example, cigarette smoke can cause dysregulation of many miRNA species, extensive changes in protein expression, and overexpression of a cancer-related lncRNA, SCAL1, in the lung.73,74 BaP treatment downregulates miR-320 and miR-506 but upregulates miR-22, miR-106b, miR-494, miR-638 and lncRNA-DQ786227, which are associated with cancerous transformation of bronchial epithelial cells upon BaP exposure.72 Inorganic arsenic, which may be found in contaminated food and drinking water, induces overexpression of KRAS and RAS oncogenes by inhibiting the expression of multiple oncogene-targeting miRNAs including let-7, miR-134, miR-138, miR-155, miR-181d, miR-205, and miR-373.75 We refer readers to excellent reviews by Marrone et al.72 and Yu and Cho71 for more information on chemically-induced miRNA responses. While the observation of miRNA dysregulation due to environmental exposure is extensively reported, the mechanisms underlying how environmental chemicals alter miRNA expression are less studied. Izzotti and Pulliero have summarized several published mechanisms, including the interconnection of p53 with the miRNA processing machinery affecting miRNA processing in the nucleus, miRNA adduct formation blocking DICER’s access to precursor miRNAs, and the binding between xenobiotic metabolites and DICER inhibiting miRNA maturation.76 It is also possible that xenobiotics may activate transcription factors that regulate the expression of miRNA encoding genes.
Exposures to environmental agents may affect miRNA levels which then affect CYP expression and CYP-dependent chemical bioactivation and carcinogenesis. Lung cancer is a leading cause of cancer death in the world and 80–90% of all lung cancers can be attributed to tobacco smoking.77,78 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) is a tobacco-specific carcinogen that contributes to lung cancer development.79 Kalscheuer et al.66 used male F344 rats treated with NNK for 20 weeks, an early stage of lung carcinogenesis in this model, to identify changes in expression profiles for pulmonary miRNAs. They found that NNK treatment decreased the expression of cancer-associated miRNAs, including miR-34, miR-101, miR-126*, and miR-199; however, the NNK treatment upregulated rat CYP2A3 (an ortholog of human CYP2A13), which is the primary catalyst of NNK bioactivation in the lungs. They demonstrated that miR-126* interacts with an MRE present in the 3′-UTR of CYP2A3 mRNA, suggesting that NNK enhances its oncogenic effects through downregulation of miR-126* to release its block on the expression of CYP2A3 to potentiate further NNK metabolism.
The inhibition of miRNA expression in response to environmental chemicals leading to the upregulation of CYP expression has also been observed in cultured human hepatoma and hepatocyte-like cell models. Fumonisin B1 (FB1), produced by fungi of the genus Fusarium, is a nongenotoxic mycotoxin associated with neural tube defects and esophageal cancer in humans and it causes liver tumors in rats.80 Using HepG2 cells treated with FB1, Chuturgoon and colleagues found that miR-27b expression was decreased while mRNA and protein levels were increased for CYP1B1, a downstream target of this miRNA species, and they showed that transfection with miR-27b molecular mimics reversed the increased expression of CYP1B1 mRNA and protein caused by FB1.65 Although FB1 is not metabolized by CYP enzymes, the induction of CYP1B1 in liver cells by FB1 may increase the probability of CYP1B1-mediated activation of other procarcinogenic substrates; the CYP1B1-mediated hydroxylation of 17β-estradiol to form tumorigenic 4-hydroxyestradiol is an important example.
Yu et al.64 discovered an inverse correlation between CYP2C9 and miR-128–3p expression in hepatocellular carcinoma (HCC) samples and then, using reporter gene constructs, RNA electrophoretic mobility shift assays, and transfection with miR-128–3p mimics, confirmed that miR-128–3p is a novel miRNA regulator of CYP2C9 expression. Furthermore, the chemical NSC-156306 induced miR-128–3p and suppressed CYP2C9 expression in human hepatocyte-like HepaRG cells, while NSC-606170 had the opposite effect on the expression of miR-128–3p and CYP2C9, suggesting that these chemicals influenced CYP2C9 expression through miR-128–3p. Downregulation of CYP2C9 has been reported as a biomarker for liver cancer,81–83 creating the possibility of monitoring miR-128–3p in combination with CYP2C9 as potential diagnostic biomarkers for HCC.64
Endogenous agents can also influence the expression of miRNAs and CYPs. The proinflammatory cytokine interleukin-6 (IL-6) is a key inflammation regulator that is overexpressed in colon cancer tissue and stroma.84 IL-6 is known to induce genome-wide DNA methylation and downregulates miR-27b through a DNA methylation-dependent mechanism in HCT116 and SW480 human colorectal cancer cells, and the inhibition of DNA methylation eliminates IL-6 induced miR-27b downregulation.68 The upregulation of CYP1B1 due to IL-6-induced miR-27b repression can increase the activation of dietary carcinogens and cause DNA damage, leading to colorectal cancer.68 Expression of CYP1B1 is not detectable in most normal tissues but its upregulation is associated with cancers in many organs, including the colon.85 Various environmental factors, such as smoking and diet, have been associated with chronic inflammation in the gastrointestinal tract of patients with inflammatory bowel disease (IBD)86,87 and inflammation is a significant contributor to tumor promotion.88 IL-6-induced CYP1B1 expression, via methylation-dependent miRNA inhibition, provides a novel mechanism for the involvement of IL-6 in inflammation-related colon cancer, suggesting a drug development strategy of preferring drugs that can be activated by CYP1B1 to those that may be deactivated by CYP1B1 for the treatment of colon cancer.68 Moreover, appropriate dietary measures may be taken by IBD patients to reduce the risk of inflammation-associated colon cancer.
The types of miRNA-dependent regulation of CYP expression involved in chemical carcinogenesis are complex. Wang and colleagues reported that the downregulation of miR-138–1* by CYP2A13-dependent aflatoxin B1 activation contributes to malignant transformation of human bronchial epithelial cells.63 They demonstrated that miR-138–1* inhibits cell proliferation, migration, invasion, and colony formation of BEAS-2B cells stably expressing human CYP2A13 by suppressing 3-phosphoinositidedependent protein kinase-1 (PDK1) and its downstream targets in the PI3K/PDK/Akt pathway.63 The metabolism of aflatoxin B1 by CYP2A13 abolishes the tumor suppressive effect of miR-138–1* and promotes lung cancer through PDK1. miRNA-dependent regulation of CYP expression can also affect chemical-induced cytotoxicity. A recent study has shown that BaP treatment leads to decreased miR-892a levels and diminished cell viability concomitantly with increased mRNA and protein levels of CYP1A1, a BaP catalyst, in MCF7 cells.67 In silico and in vitro analyses found that miR-892a targets the CYP1A1 3′-UTR for translational repression, which in turn blocks BaP-induced, CYP1A1-dependent decreased cell viability.
3. The role of miRNAs in CYP expression
3.1. Molecular mechanisms for miRNA-mediated regulation of CYP expression
A recent analysis of the www.microrna.org database predicted that as many as 56 CYP enzymes may be regulated by miRNAs.30 By August 2018, it was known that 17 CYP isoforms, including three CYP1s, seven CYP2s, and CYP3A4, are targeted directly by nearly 40 different miRNAs, with in vitro experimental confirmation in more than 30 studies (Table 2).
Table 2.
CYP isoforms with their mRNAs or promoter as direct binding targets of miRNAs (with experimental confirmation).*
| CYP | miRNA | MRE location† | References | Publication year |
|---|---|---|---|---|
| CYP1A1 | miR-892a | 30-UTR | Choi et al.67 | 2012 |
| CYP1A2 | miR-122–5p | 30-UTR | Gill et al.89 | 2017 |
| miR-132–5p | 30-UTR | Chen et al.90 | 2017 | |
| miR-320 | 30-UTR | Wei et al.91 | 2018 | |
| CYP1B1 | miR-27b | 3”-UTR | Tsuchiya et al.92 | 2006 |
| miR-187–5p | 30-UTR | Mao et al.93 | 2016 | |
| miR-200c | 30-UTR | Chang et al.94 | 2015 | |
| CYP2A6 | miR-126 | 30-UTR | Nakano et al.95 | 2015 |
| CYP2B6 | miR-25–3p | 30-UTR | Jin et al.96 | 2016 |
| CYP2C8 | miR-103 | 30-UTR | Zhang et al.97 | 2012 |
| miR-107 | 30-UTR | Zhang et al.97 | 2012 | |
| CYP2C9 | miR-103 | 30-UTR | Zhang et al.97 | 2012 |
| miR-107 | 30-UTR | Zhang et al.97 | 2012 | |
| miR-128–3p | 30-UTR | Yu et al.64 | 2015 | |
| miR-130b | 30-UTR | Rieger et al.98 | 2015 | |
| CYP2C19 | miR-29a-3p | CDS | Yu et al.99 | 2015 |
| miR-103 | 30-UTR | Zhang et al.97 | 2012 | |
| miR-107 | 30-UTR | Zhang et al.97 | 2012 | |
| CYP2D6 | miR-101 | 30-UTR | Li et al.100 | 2015 |
| miR-128–2 | 30-UTR | Li et al.100 | 2015 | |
| miR-370–3p | CDS | Zeng et al.101 | 2017 | |
| CYP2E1 | miR-214–3p | CDS | Wang et al.102 | 2017 |
| miR-378a-5p‡ | 30-UTR | Mohri et al.103 | 2010 | |
| miR-552 | Promoter, 30-UTR | Miao et al.104 | 2016 | |
| miR-570 | 30-UTR | Nakano et al.105 | 2015 | |
| CYP3A4 | miR-1 | 30-UTR | Wei et al.106 | 2014 |
| miR-27a | 30-UTR | Shi et al.107 | 2015 | |
| miR-27b | 30-UTR | Pan et al.108 | 2009 | |
| miR-122–5p | 30-UTR | Gill et al.89 | 2017 | |
| miR-206 | 30-UTR | Liu et al.109 | 2016 | |
| miR-224–5p | 30-UTR | Yu et al.110 | 2018 | |
| miR-298 | 30-UTR | Pan et al.108 | 2009 | |
| miR-532–3p | 30-UTR | Wei et al.106 | 2014 | |
| miR-577 | 30-UTR | Wei et al.106 | 2014 | |
| miR-627 | 30-UTR | Wei et al.106 | 2014 | |
| miR-628–3p | 30-UTR | Yan et al.111 | 2017 | |
| miR-641 | 30-UTR | Yan et al.111 | 2017 | |
| CYP4Z1 | miR-9 | 30-UTR | Wang et al.112 | 2016 |
| CYP7B1 | miR-17 | 30-UTR | Xi et al.113 | 2016 |
| CYP19A1 | miR-19b | 30-UTR | Kumar et al.114 | 2013 |
| miR-106a | 30-UTR | Kumar et al.114 | 2013 | |
| CYP24 | miR-125b | 30-UTR | Wang et al.115 | 2016 |
| CYP27A1 | miR-204 | 30-UTR | Xin et al.116 | 2018 |
| CYP27B1 | miR-195 | 30-UTR | Singh et al.117 | 2015 |
This list includes relevant studies published until May 2019.
MRE, miRNA recognition element; UTR, untranslated region; CDS, coding sequence.
The miRNA ID “miR-378” was used in Mohri et al. original article and has been curated and annotated as hsa-miR-378a-5p in the latest version of miRBase (Release 22, Mar 2018).
The pioneering work of Tsuchiya et al.92 first identified and validated miR-27b as a post-transcriptional repressor of CYP1B1. CYP1B1 is responsible for the bioactivation of many procarcinogens and CYP1B1 overexpression and increased enzyme activity have been observed in lung, colon, and breast cancers.118 Stable hybridization between miR-27b and its MRE located in the 3′-UTR of the CYP1B1 mRNA transcript was demonstrated using RNase protection assays and the function of this regulatory mechanism was confirmed using luciferase reporter assays upon miR-27b overexpression or introduction of miR-27b inhibitors; the level of miR-27b was inversely correlated with luciferase activity with reporter gene constructs including the CYP1B1 miR-27-MRE. Gain- and loss-of-function assays have also demonstrated that miR-27b binds to CYP1B1 mRNA and decreases CYP1B1 expression and enzymatic activity.92 miRNAs can also affect CYP expression indirectly by targeting transcriptional activators or repressors of CYP expression (Table 3). For instance, the CYP3A4 gene is regulated by multiple NRs, including PXR, farnesoid X receptor (FXR), and CAR.131,132 Takagi et al.123 have shown that CYP3A4 expression is inhibited at both the mRNA and protein levels upon PXR repression by miR-148. Additionally, miR-34a, miR-30c-1–3p, and miR-27b downregulate CYP3A4 by targeting retinoid X receptor α (RXRα), PXR, and VDR, respectively.108,121,122 When a transcriptional repressor of CYPs is targeted by a miRNA, miRNA-dependent silencing of the repressor will lead to an increase of the CYP level. For example, in CYP2D6-humanized mice, feeding cholic acid leads to a significant decrease of small heterodimer partner (SHP) at the protein level by upregulating miR-142–3p, which targets SHP; miR-142–3p-mediated SHP reduction is accompanied by an increase of CYP2D6 mRNA and protein expression, indicating a potential role of bile acid levels in the transcriptional control of CYP2D6.119 Table 3 presents published reports of indirect regulatory pathways by which miRNAs affect the expression of trans-regulatory factors that then regulate CYP genes. It should be noted that additional complexity results from the fact that additional CYP genes are regulated by HNF1A, HNF4A, VDR, and PXR.133,134 Similar miRNA-dependent regulatory pathways involving these CYP genes will require further investigation. Other transcription factors shown to influence the expression of CYP genes, such as the glucocorticoid receptor and FXR, provide additional regulatory targets for miRNA species that could be explored.135,136 miRNAs regulate CYP expression at multiple levels through diverse and novel mechanisms. In the case of CYP2E1, a miRNA species affects transcription by interfering with the assembly of transcription machinery on the promotor region. The multiple mechanisms by which the expression of CYP2E1 is influenced by different miRNAs provides illuminating examples of diverse regulatory systems.
Table 3.
miRNA-dependent regulation of trans-regulatory factors that control CYP expression (with experimental confirmation).
| miRNA | Mediator | Mediator’s role* | CYP | References | Publication year |
|---|---|---|---|---|---|
| miR-142–3p | SHP | Repressor | CYP2D6 | Pan et al.119 | 2017 |
| miR-320a | HNF1A | Activator | CYP2E1 | Yu et al.110 | 2018 |
| miR-877–5p | PXR/NR1I2 | Activator | Yu et al.110 | 2018 | |
| miR-18a-5p | PXR/NR1I2 | Activator | CYP3A4 | Sharma et al.120 | 2017 |
| miR-27b | VDR | Activator | Pan et al.108 | 2009 | |
| miR-30c-1–3p | PXR/NR1I2 | Activator | Vachirayonstien et al.121 | 2016 | |
| miR-34a | RXRα | Activator | Oda et al.122 | 2014 | |
| miR-148a | PXR/NR1I2 | Activator | Takagi et al.123 | 2008 | |
| miR-298 | VDR | Activator | Pan et al.108 | 2009 | |
| miR-449a | HNF4A | Activator | Yu et al.110 | 2018 | |
| miR-320a | RUNX2 | Repressor | CYP11A1 | Zhang et al.124 | 2017 |
| miR-27a-3p | Creb1 | Activator | CYP19A1 | Wang et al.125 | 2017 |
| miR-107 | NR5a1 | Activator | Miao et al.126 | 2016 | |
| miR-133b | Foxl2 | Repressor | Dai et al.127 | 2013 | |
| miR-320a | RUNX2 | Repressor | Zhang et al.124 | 2017 | |
| miR-764–3p | SF-1 | Activator | Wang et al.128 | 2016 | |
| miR-1275 | LRH-1 | Activator | Liu et al.129 | 2018 | |
| miR-34a | RXRα | Activator | CYP26 | Oda et al.122 | 2014 |
| miR-550a | TNF-α | Repressor | CYP27B1 | He et al.130 | 2016 |
The role of mediators in regulating CYP expression.
3.2. Complex miRNA-dependent mechanisms affecting CYP expression – CYP2E1 as an example
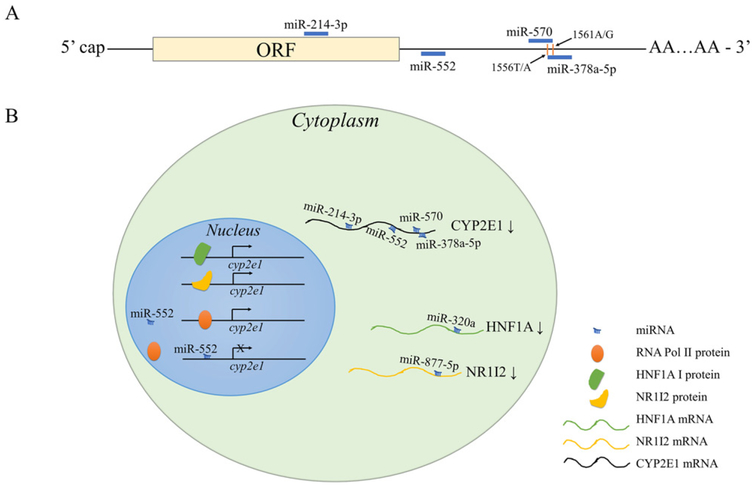
Human CYP2E1 catalyzes the bioactivation of numerous low molecular weight xenobiotics, including N-nitrosodimethylamine, acetaminophen, and ethanol.137 Predominantly expressed in the liver, CYP2E1 constitutes 56% of the total mRNA (the most abundant) and 7% of the total protein (the fourth most abundant) of the hepatic CYPs.138,139 CYP2E1 expression can be induced by its substrates, which potentiates CYP2E1-mediated toxicity. Recent mechanistic studies have expanded our knowledge concerning the roles of miRNAs in the epigenetic regulation of CYP2E1 gene expression. Figure 2 depicts representative miRNA-dependent regulatory mechanisms affecting the expression of CYP2E1.
Figure 2.
Diverse mechanisms of miRNA regulation in CYP2E1 expression. (A) Schematic map of CYP2E1 mRNA with miRNA targeting sites. CYP2E1 mRNA is directly targeted by multiple miRNAs at different regions (miR-214–3p at the coding sequence and miR-552, miR-570, and miR-378a-5p at the 3′-UTR) for post-transcriptional gene repression. 3′ end of the miR-570 MRE and 5′ end of the miR-378a-5p MRE on CYP2E1 mRNA overlap and both encompass two SNPs 1556 T/A and 1561 A/G; however, the SNPs only affect miRNA-mediated gene suppression by miR-570 but not miR-378a-5p. (B) Molecular mechanisms of transcriptional and post-transcriptional regulation of CYP2E1 gene expression in the cell. miR-214–3p, miR-552, miR-570, and miR-378a-5p recognize and bind to CYP2E1 mRNA in the cytoplasm for post-transcription gene suppression. miR-552 can also bind to the promoter region of CYP2E1, which prohibits RNA Pol II from interacting with the promoter and thereby inhibit CYP2E1 transcription. MiR-320a and miR-877–5p downregulates CYP2E1 transactivators HNF1A and NR1I2, respectively, by directly binding to the 3′-UTR of the corresponding transactivator; the decrease of HNF1A and NR1I2 leads to reduced transcription of CYP2E1.
In ways that are analogous to the examples presented above, miRNAs can inhibit CYP2E1 gene expression by targeting its mRNA transcript or by targeting transactivators that regulate its transcription. miR-378a-5p (previously identified as miR-378) binds to the CYP2E1 3′-UTR and decreases the levels of CYP2E1 protein and enzymatic activity via translational repression.103 Interestingly, miR-378–5p downregulates CYP2E1 at the mRNA level without affecting the rate of CYP2E1 mRNA degradation, suggesting the possibility of pre- or co-transcriptional regulation of CYP2E1. The underlying mechanism for this observation is still unclear. Cytoplasmic miR-552 inhibits CYP2E1 translation via binding to its MRE in the 3′-UTR of the CYP2E1 mRNA transcript.104 Meanwhile, miR-552 present in the nucleus hybridizes with the cruciform structure of the CYP2E1 promoter via its nonseed region and prohibits RNA Pol II-dependent CYP2E1 transcription. Thus, the novel dual negative regulatory mechanisms exploited by miR-552 provide evidence of both transcriptional and translational control of CYP2E1 expression. Moreover, miRNAs can reduce CYP2E1 expression indirectly by silencing transactivators of CYP2E1. Utilizing next-generation sequencing (NGS) data from acetaminophen-treated HepaRG cells, Yu et al.110 conducted a thorough search of differentially expressed miRNAs and mRNAs of genes involved in acetaminophen hepatoxicity, including drug metabolizing enzymes and NRs. The expression of HNF1A and NR1I2, well-known transcription factors that stimulate CYP gene expression, was suppressed by acetaminophen in HepaRG cells. These investigators then showed that hsa-miR-320a and hsa-miR-877–5p bind to MREs present in the 3′-UTRs of HNF1A and NR1I2 mRNA transcripts and decrease the mRNA and protein levels of both. The suppressing effects of hsa-miR-320a and hsa-miR-877–5p on CYP2E1 were comparable to those of siRNAs specific for HNF1A and NR1I2. miRNA-dependent mechanisms influence both the constitutive and induced expression of CYP2E1. Hsa-miR-214–3p targets the coding region of CYP2E1 mRNA and suppresses constitutive CYP2E1 expression in HepaRG cells.102 Although ethanol treatment at 200 mM for 24 h had little impact on expression of miR-214–3p in HepG2 cells, overexpression of miR-214–3p upon ethanol induction increased CYP2E1 mRNA and protein levels by 85% and 37% respectively, compared to negative control. This effect is particularly critical as CYP2E1 induction by alcohol enhances the toxicity of acetaminophen overdose and activation of pro-carcinogens to carcinogens in both initiation and promotion stages of alcohol-related liver cancer.140,141 Inter-personal differences in miR-214–3p expression due to genetic variation or drug and environmental exposure may result in differential susceptibility in xenobiotic-induced, CYP2E1-mediated liver toxicity and carcinogenesis; thus, miR-214–3p might serve as a biomarker or as a therapeutic target for such adverse effects.
The expression of CYP2E1 can be influenced by genetic variations in the MREs. In 2015, Nakano et al.105 found that nucleotides 1556 T and 1561 A (haplotype I) residing in the miR-570 MRE located in the CYP2E1 3′-UTR were essential for miR-570/mRNA binding and for miR-570-mediated CYP2E1 translational repression, whereas SNPs 1556 T > A and 1561 A > G (haplotype II) abolished the suppressive effect of miR-570 on CYP2E1 expression. Additionally, miR-570 levels were found to be inversely correlated with CYP2E1 protein levels in human liver for diplo-type I/I but not for diplotypes I/II or II/II. Interestingly, the 3′-portion of the miR-570 MRE (+1519 to +1561) overlaps the 5′-end of miR-378a-5p MRE (+1559 to +1575) and both MREs encompass the SNPs 1556 T/A and 1561 A/G. Although the miR-570-dependent suppression of CYP2E1 expression was found to be sensitive to this genotypic difference, CYP2E1 suppression by miR-378a-5p was not. miRNAs have important roles in CYP2E1-activated acetaminophen toxicity. Upon the discovery of novel miRNAs that represses CYP2E1 expression, it was shown that exogenous introduction of miR-320a, miR-877–5p, and miR-214–3p can reverse the CYP2E1-dependent hepatotoxicity caused by acetaminophen treatment in HepaRG cells.102,110 Elevated levels of CYP2E1-suppressing miRNAs, including miR-320a, miR-877–5p, and miR-378a-5p, have been observed in the serum of acetaminophen overdose pediatric patients, indicating an adaptive response of hepatocytes to acetaminophen-induced liver injury may be operative.89,110
In summary, CYP2E1 expression can be regulated by multiple miRNAs that target distinct DNA and mRNA domains associated with CYP2E1 with different efficiencies of inhibition.102 These miRNAs exert their inhibitory effects on CYP2E1 at transcriptional and post-transcriptional levels via diverse molecular mechanisms. Importantly, modulation of CYP2E1 expression by miRNA interferes with CYP2E1-dependent xenobiotic bioactivation associated with hepatotoxicity. Thus, inter-individual variabilities in miRNA expression and SNPs residing in MREs may also be involved in variations in xenobiotic-induced, CYP-mediated toxicity and susceptibility to cancer linked to other environmental chemicals.
4. The role of lncRNAs in CYP expression
lncRNAs are also recognized as important epigenetic regulatory RNA species and potential biomarkers for human diseases.42 Advancements in high-throughput technologies, such microarray and NGS, have enabled unprecedented detection of novel transcripts, leading to an explosion of data related to lncRNAs. The NONCODE database (version 2016), an online repository of the most complete collection of lncRNAs, has identified 167,150 lncRNAs in humans that are transcribed from 101,700 lncRNA genes.142 Despite the growing amount of data becoming available concerning lncRNAs, much less is known about the properties, functions, and biological significance of individual lncRNAs. Initially considered merely transcriptional noise, lncRNAs were later found to be associated with many human diseases including cancer, metabolic diseases, cardiovascular diseases, autoimmune diseases, and Alzheimer’s disease.42,143–146 Mechanistic studies have demonstrated that lncRNAs can influence gene expression143 that affects developmental147 (e.g. genomic imprinting), physiological148 (e.g. metabolism), and pathological149 (e.g. cancer development and progression) processes. Not only do lncRNAs impact these fundamental processes, they also respond to external stimuli. Indeed, an increasing number of effects of environmental chemicals on lncRNA expression are being reported.46,150,151
4.1. Molecular properties and regulatory mechanisms of lncRNAs
lncRNAs are mainly transcribed by RNA Pol II and are usually 5′ capped and polyadenylated.152 Based on the genomic location of lncRNAs relative to protein-coding genes, lncRNAs are classified into four broad groups: intergenic (residing between two protein-coding genes, also named lincRNAs), intronic (embedded in the intron of a protein-coding gene), and sense and antisense (transcribed from the sense or antisense strand of a protein-coding gene).153 lncRNAs interact with DNA, RNA, and protein molecules and regulate various cellular activities, including chromatin modification and the splicing, stability, and translation of mRNA transcripts.154,155 In contrast to miRNAs and siRNAs, lncRNAs are poorly conserved between different species and are often expressed in low abundance. To date, only a small number of lncRNAs have been functionally characterized; the diverse modes of action for other lncRNAs are evident but remain to be fully understood. Existing studies show that lncRNAs can exert transcriptional control in a cis- or trans- manner and regulate gene expression at the post-transcriptional level. Based on a loose categorization of lncRNA functions in gene regulation, lncRNAs can serve as (1) spatiotemporal signals for developmental stages; (2) molecular decoys to sequester either miRNAs from their target mRNAs or DNA-binding proteins from DNA;(3) guides for RNA binding proteins to their targets, such as promoters and enhancers; and as (4) scaffolds to form large macromolecular complexes.42,152 These mechanisms of action are not mutually exclusive, rather, a lncRNA may adopt multiple functions in different contexts.152
4.2. lncRNA-mediated CYP expression
Current research has identified four regulatory relationships between lncRNAs and CYP expression via either mechanistic or correlational studies (Table 4). The most common type of mechanism underlying lncRNA-mediated gene regulation involves interactions between lncRNA and other regulatory proteins. Depending on the activating or repressive functions of the regulatory proteins, lncRNAs may influence expression of a target gene positively or negatively.
Table 4.
Studies on lncRNA regulation of CYP expression (in chronological order).
| CYP | IncRNA | Subcellular localization* | Type and genomic locus* | Regulatory mechanism | Model system | Comments and references | Publication year |
|---|---|---|---|---|---|---|---|
| CYP8B1 | IncLSTR | Nucleus | Intergenic | Binding to and removing transcriptional suppressor TDP-43 from Cyp8t1 promoter | Mice, primary hepatocytes | In this pioneering work on IncRNA regulation of CYP, Li et al47 delineated a cascade of multi-player molecular events leading to a physiological phenotype in lipid metabolism. | 2015 |
| CYP7A1 | Inc-HC | Nucleus | Sense; overlapping with SIc25a15 3’UTR | Binding to and destabilizing CYP7A1 mRNA in a complex with hnRNPA2B1 | Rats, CBRH- 7919 cells | Lan et al48 presented the first study on post-transcriptional regulation of CYP by IncRNA. | 2016 |
| CYP2B10 | Inc5998 | Undetermined | Anti-sense; near CAR binding site at Cyp2b10 enhancer | Undetermined | Mice | Lodato et al46 identified multiple novel IncRNAs related to CAR targets in chemical-induced, CAR-dependent, sex-based tumorigenesis | 2017 |
| 8 CYPs from CYP1, 2, and 3 families | HNFIa-AS1; HNF4a-AS1 | Undetermined | Antisense; proximal to HNF1a and HNF4a respectively | Undetermined; potential transcriptional regulation in sync with NRs | HepaRG cells | Chen et al49, depict a regulatory network of NRs and their antisense IncRNAs for CYP regulation | 2018 |
| 6 CYPs from CYP 2 and 3 families | HNF1a-ASI | Undetermined | Antisense; proximal to HNF1a | Undetermined | Human liver tissues, Huh 7 cells | Wang et al.156 demonstrated the involvement of HNFIA-ASI in HNFIA’s function as an upstream regulator of NRs and basal and drug-induced CYP expression | 2019 |
Subcellular localization, type, and genomic locus of lncRNAs.
Li et al.47 described a systemic lipid metabolic pathway in mice involving CYP8B1, an important enzyme in bile acid synthesis, and lncLSTR, a liver-specific lncRNA. Identified using the Affymetrix Mouse Genome 430 2.0 Array gene chip, the expression of lncLSTR fluctuated in response to fasting and refeeding, suggesting a role for this lncRNA in energy metabolism. Predominantly expressed in the nucleus, lncLSTR interacts with TDP-43, a transcriptional repressor, to reduce the occupancy of TDP-43 on the Cyp8b1 promoter and abolish TDP-43-dependent CYP8B1 inhibition. In a lncLSTR-knockdown mouse line, depletion of lncLSTR decreased the expression of CYB8B1 and altered the ratio of muricholic acid (MCA)/cholic acid (CA), two major bile acids in mice. This change in the bile acid composition activated the FXR pathway and its downstream gene apoC2, a potent activator of lipoprotein lipase (LPL), which in turn enhanced triglyceride clearance in the plasma. This study depicts a cascade of multi-player molecular events involving lncRNA modulation of CYP expression, which ultimately led to a physiological change in lipid metabolism.
lncRNAs also modulate CYP expression at the post-transcriptional level. A high level of cholesterol in rats activates the C-EBPβ pathway, which promotes the expression of lnc-HC, a recently-identified hepatocyte-specific lncRNA. Lnc-HC interacting with hnRNPA2B1 increases the nuclear localization of hnRNPA2B1. This RNA/protein complex binds to CYP7A1 (cholesterol 7a-hydroxylase) mRNA and decreases its stability, resulting in decreased CYP7A1 protein levels.48 As CYP7A1 has a critical role in converting cholesterol to bile acids, the reduction in CYP7A1 expression leads to cholesterol accumulation. This lncRNA/heterogenous nuclear ribonucleoprotein interaction is essential for CYP7A1 mRNA binding and degradation, even though lncRNAs can hybridize with mRNA directly, highlighting the post-transcriptional control that lncRNAs exert in metabolic systems.
LncRNAs near genomic loci of NRs may modulate CYP expression together with the corresponding NRs. Two anti-sense lncRNAs, HNF1A-AS1, and HNF4A-AS1, are located on chromosomes 12 and 20 near the protein coding genes for the transcription factors HNF1A and HNF4A, respectively, which are important regulators of CYP gene expression. Because anti-sense lncRNAs are often found to have regulatory roles in the expression of neighboring genes, Chen et al.49 hypothesized that these two lncRNAs are involved in a transcription network to control the expression of HNF1A, HNF4A, and the downstream NRs and CYP genes regulated by HNF1A and HNF4A. Chen and coworkers showed that an shRNA-dependent knockdown of HNF1A-AS1 in HepaRG cells suppressed the levels of both lncRNAs, HNF1A-AS1 and HNF4A-AS1, and those of mRNA transcripts for HNF1A, PXR, CAR, AhR, CYP2B6, CYP2C8, CYP2C9, CYP2E1, and CYP3A4, while the mRNA level of CYP1A2 was increased. The level of HNF4A-AS1 was decreased in HepaRG cells treated with an siRNA targeting HNF4A-AS1, yet mRNA levels increased for CAR, AhR, CYP1A2, CYP2B6, CYP2C9, CYP2C19, CYP2E1, and CYP3A4. Moreover, a direct knockdown of HNF1A and HNF4A caused similar effects on mRNA levels of the NRs and CYPs they target as did the knockdown of the corresponding anti-sense lncRNAs, suggesting that the lncRNAs in the neighboring genomic loci of NRs may act as co-regulators of transcription of target genes of HNF1A and HNF4A. Interestingly, a positive correlation was observed in human liver tissue samples between increased mRNA levels for HNF1A, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, PXR, and CAR and upregulation of HNF1A-AS1 lncRNA.156 Although correlations in expression are consistent with this regulatory network, the underlying mechanisms remain elusive.
Another lncRNA, lnc5998, has been found near the enhancer region of Cyp2b10 where CAR binds to activate Cyp2b10 transcription.46 Lnc5998 was found among a number of dysregulated lncRNAs to be in close proximity to CAR target genes, including Cyp2b10, Por, and Alas1, during TCPOBOP (a CAR agonist)-induced tumorigenesis in female mouse liver. Additionally, sex-biased responses of tumor promoting genes were observed in the liver of mice treated with TCPOBOP, indicating sex-based differences in CAR-regulated xenobiotic-induced carcinogenesis.
4.3. Perspectives on unanswered questions related to lncRNA-dependent regulation of CYP expression
lncRNAs provide a sophisticated layer for fine-tuning molecular regulation in the cell. Yet the majority of lncRNAs remain to be functionally annotated and many research issues are open to exploration to address these questions. Which up-stream pathways control lncRNA expression upon environmental exposure? Is altered lncRNA expression a primary or secondary effect of exposures to environmental chemicals?
The ability to interact with other large molecules, including DNA, RNA and proteins, suggests that lncRNA may function in large regulatory networks containing multiple macromolecules. It will be insightful to dissect the interactions between different RNA species (miRNAs/lncRNAs, lncRNA/lncRNA, and lncRNA/mRNA) and their coordination in CYP-dependent metabolic responses. It is also important to investigate the impact of genetic polymorphisms, such as SNPs, on these interactions, since 40% of disease-related SNPs reside in noncoding regions of the human genome.142 Research addressing these issues would further illuminate functional interactions between genetic and epigenetic control of gene expression.
On the organism level, current knowledge on the role of lncRNAs in regulating CYP expression is mainly limited to the liver or hepatocyte models. This is not surprising, given that the liver is the principal site for CYP expression and the primary metabolic center of the body. However, it is also imperative to expand the investigation to extrahepatic, CYP-positive organs, such as the lung, brain, kidney, skin, and adrenal gland. An understanding of the commonalities in lncRNA regulatory mechanisms controlling CYP expression may help construct an informative and predictive platform of lncRNA functions in not only metabolism, but also other biological processes. Another limitation in current studies on lncRNA-dependent modulation of CYP expression is that they mainly focus on the metabolism of endogenous compounds, such as lipid and cholesterol, in the physiological context. There is a great need to explore the role of lncRNA-dependent regulation of CYP expression that is relevant to pathogenesis, in general, and carcinogenesis, specifically, in target organs of environmental chemical exposure, particularly the liver, lung, and kidney. An overarching goal of mechanistic studies is to facilitate the discovery of meaningful biomarkers for disease detection, diagnosis, and prognosis and the development of targeted therapy in the precision medicine era.
Additional important questions include is lncRNA-dependent regulation of CYP expression influenced by other factors such as sex, age, race, diet, and life style? If so, how? Investigating the similarities and disparities in lncRNA-dependent regulation of CYP expression caused by these factors may help us better understand and predict inter-individual variability in environmental chemical induced toxicity and disease susceptibility.
The relatively low expression of lncRNAs complicates research to investigate their regulatory functions that distinguish them from “useless” byproducts of pervasive transcription. Improving methods for detection and bioinformatic analysis may help overcome this obstacle. Additional complications occur when translating findings observed in laboratory species to humans due to the poor conservation of lncRNAs across species. One approach is to identify structurally conserved lncRNA transcripts between animal models and humans, an approach that requires reliable bioinformatic analyses. To simplify the problem, another approach is to focus initial studies on lncRNA genes that are located near protein-coding genes of interest.
5. miRNAs as biomarkers for xenobiotic toxicity and carcinogenesis
A biomarker is defined as an objectively measurable characteristic in biological media (e.g. human tissues, cells, or body fluids) that can indicate biological or pathogenic processes or pharmacological responses.157 Biomarkers have the potential to assist in disease prediction, prevention, diagnosis, prognosis, intervention, and therapeutic efficiency.158 An ideal biomarker features specificity, sensitivity, robustness, validity, actionability, noninvasive detectability, and translatability (from animal models to humans).159 In the era of precision medicine, the development of reliable and robust mechanistic biomarkers will be an important approach to aid clinical practice and regulatory decision-making; their acceptance requires elucidation of underlying mechanisms and advancement of detection technologies. In recent decades, cell free circulating miRNAs have been explored as a new generation of biomarkers for disease, toxicity assessment, and drug development, owing to their conservation, tissue-specificity, stability, and minimally-invasive detectability in human body fluids (e.g. blood, urine, and saliva).69,160–163 miRNAs represent potential biomarkers for detecting exposures to environmental toxicants. Conventional exposure indicators include internal doses of carcinogens and their metabolites, DNA and protein adduct formation, genetic alterations, circulating proteins, and altered gene expression.158,164 The 5-year survival rate for lung cancers overall is only 18%, yet survival increases to 56% when lung tumors are detected before they metastasize; however, only 16% of lung cancer cases are diagnosed at such an early stage.13 The early response of miR-126* in the expression level during lung carcinogenesis indicates miR-126* might be investigated for its potential as an early diagnostic marker for lung cancer.66
Toxicogenomic studies have provided evidence for the association of altered miRNA expression profiles with toxicant exposure and implicated miRNAs as chemical exposure biomarkers, as summarized in other scientific reviews.69,76,158,164–167 For example, Vrijens et al.161 conducted a systematic review on miRNA profile alterations due to personal (e.g. smoking) or environmental (e.g. air pollution) exposure and their roles in exposure-related human diseases and they grouped differentially expressed miRNAs based on exposure types, including smoking, air pollution, nano-particles, and chemicals, in different model systems (in vitro, in vivo, and human studies). Valencia-Quintana and colleagues166 discussed original studies and early reviews on the dysregulation and role of miRNAs in aflatoxin B1 exposure and aflatoxin B1-induced HCC. They stressed the importance of mechanisms of action in biomarker identification and development and summarized miRNAs and their associated cellular events in in vivo and in vitro models exposed aflatoxin B1; they concluded that miR-27a, miR-27b, miR-122, miR-148, miR-155, miR-192, miR-214, miR-221, miR-429, and miR-500 could serve as potential biomarkers of aflatoxin B1 exposure and aflatoxin B1-induced HCC.166 miRNAs can serve as biomarkers for tissue-specific toxicity induced by xenobiotics. Xenobiotic exposure may cause tissue injuries in vital organs, particularly the liver, heart, and kidney; the levels of tissue-specific miRNAs have been found to respond to organ injuries, even prior to detection of other well-established biomarkers. For example, miR-122, a liver-enriched miRNA, shows an elevation in the serum and has been investigated as a potential biomarker for liver injury induced by viral infection, alcohol consumption, chemical exposure, and acetaminophen overdose.168,169 More importantly, increased miR-122 levels in blood can be detected before the elevation of ALT and AST, traditional markers of liver injury, suggesting that it could be used for early detection of acute liver injury.169,170 Heart-specific miR-208 has been shown to be released into the circulation from injured cells and the elevated level of plasma miR-208 has been implicated as a useful biomarker for cardiotoxicity.171,172 We refer readers to recently published review articles describing miRNA species as biomarkers for tissue injury.173–176 miRNAs can also serve as possible biomarkers for cancers. Abundant evidence has demonstrated the regulatory role of miRNAs in various molecular pathways at different stages of cancer, such as initiation, promotion, and progression. Numerous reviews have discussed the potential for using miRNAs as biomarkers for colorectal cancer, lung cancer, hepatocellular carcinoma, breast cancer, ovarian cancer, esophageal cancer, lymphoma, and head and neck cancer.160,177–182
It is a challenging task to establish a mechanistic network that connects xenobiotic exposure, miRNA dysregulation, altered CYP expression, and cancer. Enhanced mechanistic knowledge provides a more secure foundation for the adoption of miRNA-based biomarkers. We emphasize the importance of associating molecular mechanisms involved in a disease with the identification and development of actionable biomarkers. These mechanism-based indicators are likely to be superior to those that merely hallmark the disease state but do not participate in the pathogenic process; the latter are referred to as descriptive biomarkers and, being molecular bystanders, have lower scientific and clinical significance.183 miRNAs as regulators of key genes involved in the activation of environmental chemical toxicants can be explored as informative mechanistic biomarkers because they are rooted in the pathogenesis of human diseases linked to environmental exposures. Thus, the elucidation of such mechanisms will facilitate rational design and development of biomarkers that are useful clinically and for regulatory decision-making.
6. Conclusions
Environmental chemical exposures are major risk factors for many human diseases, including cancers. CYPs and other XMEs are involved both in the detoxification and elimination of xenobiotics and in their molecular activation. Environmental compounds may induce CYP expression by binding to and activating NRs, transcriptional regulators of CYP genes, or alter the level of ncRNAs that regulate CYP genes and influence CYP expression transcriptionally and post-transcriptionally. The interplay between NRs and ncRNAs presents a complex network for finely-regulated CYP expression. Both miRNAs and lncRNAs are able to modulate CYP expression through diverse mechanisms, although they exhibit different biochemical properties, conservation between species, and expression patterns. However, a full understanding of the mechanisms by which ncRNAs affect CYP expression has not yet been attained.
There are also challenges in investigating the role of ncRNA-mediated regulation of CYP expression in human diseases linked to exposures to environmental agents. It can be difficult to determine whether altered ncRNA expression occurs as a direct molecular response to chemical exposure or whether it is a secondary effect of dysregulated cellular pathways involving NRs and/or other transcriptional regulators. Furthermore, although miRNAs are highly conserved, especially in their seed regions, 3′-UTRs of mRNA transcripts often display less similarity across species;52 it is, therefore, difficult to extrapolate findings from the animal models to humans, or to test miRNA-targeted therapies in preclinical models based on results of miRNA functional characterization from human cell lines. This can be partially overcome with the use of xenograft animal models generated by using human cell lines or tissues. Also, most investigations study exposures to individual compounds, which is essential for elucidating the specific mechanisms by which one compound acts, but this approach lacks the ability to mimic the mixture of toxicants present in actual human environments.
Although regulatory interactions between miRNA species and the expression of specific CYP isoforms have been relatively well characterized, studies linking environmental chemicals, ncRNAs, CYPs, and chemical-induced toxicity are still largely lacking. Studies of this type could illuminate cell response pathways activated upon chemical exposures and explore the potential use of miRNAs as biomarkers or as therapeutics for chemical-induced, CYP-activated toxicity. As many current studies are carried out in cell culture or in laboratory rodents, longitudinal clinical studies could help characterize the relationships between the responses of ncRNAs to environmental exposures in humans. Also, it is intriguing to consider potential interplays between ncRNAs and other genetic or other epigenetic factors, such as DNA methylation, in CYP expression. The identification and characterization of miRNA biomarkers for monitoring environmental exposures to specific chemicals in the future will facilitate diagnosis, guide treatments, and promote public health.
Current findings indicate that ncRNAs are important modulators of CYP expression and the CYP-dependent bioactivation of environmental chemicals leading to toxicity or cancer. The increasing significance of ncRNAs in gene regulation marks a new era for the evaluation of ncRNA-dependent modulation of CYP expression in the process of chemical carcinogenesis. Elucidating the mechanisms by which ncRNAs regulate CYP expression could provide valuable insight into xenobiotic-induced carcinogenesis and facilitate investigations leading to novel diagnosis tools for early detection of environmentally-induced cancers and to potential therapeutic targets.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Disclaimer
This review is not an official guidance or policy statement of the U.S. Food and Drug Administration (FDA). No official support or endorsement by the U.S. FDA is intended or should be inferred.
References
- [1].Wogan GN, Hecht SS, Felton JS, Conney AH, Loeb LA. Environmental and chemical carcinogenesis. Semin Cancer Biol. 2004;14(6):473–486. doi: 10.1016/j.sem-cancer.2004.06.010. [DOI] [PubMed] [Google Scholar]
- [2].Wu YF, Chitranshi P, Loukotkova L. Cytochrome P450-mediated metabolism of triclosan attenuates its cytotoxicity in hepatic cells. Arch Toxicol. 2017;91(6): 2405–2423. doi: 10.1007/s00204-016-1893-6. [DOI] [PubMed] [Google Scholar]
- [3].Dhillon GS, Kaur S, Pulicharla R, et al. Triclosan: current status, occurrence, environmental risks and bioaccumulation potential. Int J Environ Res Public Health. 2015;12(5):5657–5684. doi: 10.3390/ijerph120505657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sheweita SA. Drug-metabolizing enzymes: Mechanisms and functions. Curr Drug Metab. 2000;1(2):107–132. doi: 10.2174/1389200003339117. [DOI] [PubMed] [Google Scholar]
- [5].Zarybnicky T, Bousova I, Ambroz M, Skalova L. Hepatotoxicity of monoterpenes and sesquiterpenes. Arch Toxicol. 2018;92(1):1–13. [DOI] [PubMed] [Google Scholar]
- [6].Yang M, Ruan J, Fu PP, Lin G. Cytotoxicity of pyrrolizidine alkaloid in human hepatic parenchymal and sinusoidal endothelial cells: firm evidence for the reactive metabolites mediated pyrrolizidine alkaloid-induced hepatotoxicity. Chem Biol Interact. 2016;243:119–126. doi: 10.1016/j.cbi.2015.09.011. [DOI] [PubMed] [Google Scholar]
- [7].Ellison CA, Tian Y, Knaak JB, Kostyniak PJ, Olson JR. Human hepatic cyto-chrome P450-specific metabolism of the organophosphorus pesticides methyl parathion and diazinon. Drug Metab Dispos. 2012;40(1):1–5. doi: 10.1124/dmd.111.042572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nebert DW, Russell DW. Clinical importance of the cytochromes P450. Lancet. 2002;360(9340):1155–1162. doi: 10.1016/S0140-6736(02)11203-7. [DOI] [PubMed] [Google Scholar]
- [9].Guengerich FP. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol. 2001;14(6):611–650. [DOI] [PubMed] [Google Scholar]
- [10].Guengerich FP. Cytochrome P450s and other enzymes in drug metabolism and toxicity. AAPS J. 2006;8(1):E101–111. doi: 10.1208/aapsj080112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nebert DW, Dalton TP. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat Rev Cancer. 2006;6(12): 947–960. doi: 10.1038/nrc2015. [DOI] [PubMed] [Google Scholar]
- [12].Danielson PB. The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr Drug Metab. 2002;3(6):561–597. [DOI] [PubMed] [Google Scholar]
- [13].Shimada T, Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydro-carbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004;95(1): 1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Guengerich FP. Metabolism of chemical carcinogens. Carcinogenesis 2000;21(3): 345–351. doi: 10.1093/carcin/21.3.345. [DOI] [PubMed] [Google Scholar]
- [15].Karle IL, Yagi H, Sayer JM, Jerina DM. Crystal and molecular structure of a benzo[a]pyrene 7,8-diol 9,10-epoxide N2-deoxyguanosine adduct: absolute configuration and conformation. Proc Natl Acad Sci USA. 2004;101(6):1433–1438. doi: 10.1073/pnas.0307305101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kamdem LK, Meineke I, Godtel-Armbrust U, Brockmoller J, Wojnowski L. Dominant contribution of P450 3A4 to the hepatic carcinogenic activation of aflatoxin B-1. Chem Res Toxicol. 2006;19(4):577–586. doi: 10.1021/tx050358e. [DOI] [PubMed] [Google Scholar]
- [17].Abdo W, Hirata A, Sakai H, El-Sawak A, Nikami H, Yanai T. Combined effects of organochlorine pesticides heptachlor and hexachlorobenzene on the promotion stage of hepatocarcinogenesis in rats. Food Chem Toxicol. 2013;55:578–585. doi: 10.1016/j.fct.2013.01.035. [DOI] [PubMed] [Google Scholar]
- [18].Prado G, Bhalli JA, Marcos R. Genotoxicity of heptachlor and heptachlor epoxide in human TK6 lymphoblastoid cells. Mutat Res. 2009;673(2):87–91. doi: 10.1016/j.mrgentox.2008.12.002. [DOI] [PubMed] [Google Scholar]
- [19].McHale CM, Zhang L, Smith MT. Current understanding of the mechanism of benzene-induced leukemia in humans: implications for risk assessment. Carcinogenesis. 2012;33(2):240–252. doi: 10.1093/carcin/bgr297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shapiro GD, Dodds L, Arbuckle TE, et al. Exposure to organophosphorus and organochlorine pesticides, perfluoroalkyl substances, and polychlorinated biphenyls in pregnancy and the association with impaired glucose tolerance and gestational diabetes mellitus: the MIREC Study. Environ Res. 2016;147:71–81. doi: 10.1016/j.envres.2016.01.040. [DOI] [PubMed] [Google Scholar]
- [21].Lasram MM, Dhouib IB, Annabi A, El Fazaa S, Gharbi N. A review on the molecular mechanisms involved in insulin resistance induced by organophosphorus pesticides. Toxicology. 2014;322:1–13. doi: 10.1016/j.tox.2014.04.009. [DOI] [PubMed] [Google Scholar]
- [22].Li ZM, Guo LH, Ren XM. Biotransformation of 8:2 fluorotelomer alcohol by recombinant human cytochrome P450s, human liver microsomes and human liver cytosol. Environ Sci: Process Impacts. 2016;18(5):538–546. doi: 10.1039/C6EM00071A. [DOI] [PubMed] [Google Scholar]
- [23].Sanderson JT. The steroid hormone biosynthesis pathway as a target for endocrine-disrupting chemicals. Toxicol Sci. 2006;94(1):3–21. doi: 10.1093/toxsci/kfl051. [DOI] [PubMed] [Google Scholar]
- [24].Pikuleva IA. Cholesterol-metabolizing cytochromes P450. Drug Metab Dispos. 2006; 34(4):513–520. doi: 10.1124/dmd.105.008789. [DOI] [PubMed] [Google Scholar]
- [25].Tompkins LM, Wallace AD. Mechanisms of cytochrome P450 induction. J Biochem Mol Toxicol. 2007;21(4):176–181. [DOI] [PubMed] [Google Scholar]
- [26].Agundez JA. Cytochrome P450 gene polymorphism and cancer. Curr Drug Metab. 2004;5(3):211–224. [DOI] [PubMed] [Google Scholar]
- [27].Savolainen VT, Pajarinen J, Perola M, Penttila A, Karhunen PJ. Polymorphism in the cytochrome P450 2E1 gene and the risk of alcoholic liver disease. J Hepatol. 1997;26(1):55–61. [DOI] [PubMed] [Google Scholar]
- [28].Zhou SF, Liu JP, Chowbay B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab Rev. 2009;41(2):89–295. doi: 10.1080/03602530902843483. [DOI] [PubMed] [Google Scholar]
- [29].Honkakoski P, Negishi M. Regulation of cytochrome P450 (CYP) genes by nuclear receptors. Biochem J. 2000;347(Pt 2):321–337. doi: 10.1042/0264-6021:3470321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tang X, Chen S. Epigenetic regulation of cytochrome P450 enzymes and clinical implication. Curr Drug Metab. 2015;16(2):86–96. [DOI] [PubMed] [Google Scholar]
- [31].Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- [32].Dannenberg LO, Edenberg HJ. Epigenetics of gene expression in human hepatoma cells: expression profiling the response to inhibition of DNA methylation and his-tone deacetylation. BMC Genomics. 2006;7(1):181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Habano W, Kawamura K, Iizuka N, Terashima J, Sugai T, Ozawa S. Analysis of DNA methylation landscape reveals the roles of DNA methylation in the regulation of drug metabolizing enzymes. Clin Epigenet. 2015;7(1):105. doi: 10.1186/s13148-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Li Y, Li Y, Zheng G, et al. Cytochrome P450 1A1 and 1B1 promoter CpG island methylation regulates rat liver injury induced by isoniazid. Mol Med Rep. 2018; 17(1):753–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].He L, Gao L, Shi Z, et al. Involvement of cytochrome P450 1A1 and glutathione S-transferase P1 polymorphisms and promoter hypermethylation in the progression of anti-tuberculosis drug-induced liver injury: a case-control study. PLoS One. 2015;10(3):e0119481. doi: 10.1371/journal.pone.0119481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yang H, Nie Y, Li Y, Wan YJ. Histone modification-mediated CYP2E1 gene expression and apoptosis of HepG2 cells. Exp Biol Med (Maywood). 2010;235(1): 32–39. doi: 10.1258/ebm.2009.009252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Esteller M Aberrant DNA methylation as a cancer-inducing mechanism. Annu Rev Pharmacol Toxicol. 2005;45:629–656. doi: 10.1146/annurev.pharmtox.45.120403.095832. [DOI] [PubMed] [Google Scholar]
- [39].Sawan C, Herceg Z. Histone modifications and cancer. Adv Genet. 2010;70:57–85. doi: 10.1016/B978-0-12-380866-0.60003-4. [DOI] [PubMed] [Google Scholar]
- [40].Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- [41].Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9(3):219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- [44].Kowalski MP, Krude T. Functional roles of non-coding Y RNAs. Int J Biochem Cell Biol. 2015;66:20–29. doi: 10.1016/j.biocel.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Nakano M, Nakajima M. Current knowledge of microRNA-mediated regulation of drug metabolism in humans. Exp Opin Drug Metab Toxicol. 2018;14(5):493–504. doi: 10.1080/17425255.2018.1472237. [DOI] [PubMed] [Google Scholar]
- [46].Lodato NJ, Melia T, Rampersaud A, Waxman DJ. Sex-differential responses of tumor promotion-associated genes and dysregulation of novel long noncoding RNAs in constitutive androstane receptor-activated mouse liver. Toxicol Sci. 2017; 159(1):25–41. doi: 10.1093/toxsci/kfx114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Li P, Ruan X, Yang L, et al. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015;21(3):455–467. doi: 10.1016/j.cmet.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lan X, Yan J, Ren J, et al. A novel long noncoding RNA Lnc-HC binds hnRNPA2B1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism. Hepatology. 2016;64(1):58–72. doi: 10.1002/hep.28391. [DOI] [PubMed] [Google Scholar]
- [49].Chen L, Bao Y, Piekos SC, Zhu K, Zhang L, Zhong XB. A transcriptional regulatory network containing nuclear receptors and long noncoding RNAs controls basal and drug-induced expression of cytochrome P450s in HepaRG cells. Mol Pharmacol. 2018;94(1):749–759. doi: 10.1124/mol.118.112235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ning B, Su Z, Mei N, et al. Toxicogenomics and cancer susceptibility: advances with next-generation sequencing. J Environ Sci Health C: Environ Carcinog Ecotoxicol Rev. 2014;32(2):121–158. doi: 10.1080/10590501.2014.907460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Akhtar MM, Micolucci L, Islam MS, Olivieri F, Procopio AD. Bioinformatic tools for microRNA dissection. Nucleic Acids Res. 2016;44(1):24–44. doi: 10.1093/nar/gkv1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2007;36(Database):D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9(11):831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12(2):99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- [56].Yang Z, Wang L. Regulation of microRNA expression and function by nuclear receptor signaling. Cell Biosci. 2011;1(1):31.doi: 10.1186/2045-3701-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in Cancer. Curr Genomics. 2010;11(7):537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Bolleyn J, De Kock J, Rodrigues RM, Vinken M, Rogiers V, Vanhaecke T. MicroRNAs as key regulators of xenobiotic biotransformation and drug response. Arch Toxicol. 2015;89(9):1523–1541. doi: 10.1007/s00204-014-1314-7. [DOI] [PubMed] [Google Scholar]
- [59].Osada H, Takahashi T. MicroRNAs in biological processes and carcinogenesis. Carcinogenesis. 2007;28(1):2–12. doi: 10.1093/carcin/bgl185. [DOI] [PubMed] [Google Scholar]
- [60].Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6(6):590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kalinina TS, Kononchuk VV, Gulyaeva LF. Expression of hormonal carcinogenesis genes and related regulatory microRNAs in uterus and ovaries of DDT-treated female rats. Biochem Moscow. 2017;82(10):1118–1128. doi: 10.1134/S0006297917100042. [DOI] [PubMed] [Google Scholar]
- [62].Sun Z, Zhang Z, Ji M, et al. BDE47 induces rat CYP3A1 by targeting the transcriptional regulation of miR-23b. Sci Rep. 2016;6:31958.doi: 10.1038/srep31958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wang Y, Zhang Z, Wang H, et al. miR-138–1 regulates aflatoxin B1-induced malignant transformation of BEAS-2B cells by targeting PDK1. Arch Toxicol. 2016; 90(5):1239–1249. doi: 10.1007/s00204-015-1551-4. [DOI] [PubMed] [Google Scholar]
- [64].Yu D, Green B, Marrone A, et al. Suppression of CYP2C9 by microRNA hsa-miR-128–3p in human liver cells and association with hepatocellular carcinoma. Sci Rep. 2015;5(1):8534. doi: 10.1038/srep08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Chuturgoon AA, Phulukdaree A, Moodley D. Fumonisin B(1) modulates expression of human cytochrome P450 1b1 in human hepatoma (Hepg2) cells by repressing Mir-27b. Toxicol Lett. 2014;227(1):50–55. doi: 10.1016/j.toxlet.2014.02.026. [DOI] [PubMed] [Google Scholar]
- [66].Kalscheuer S, Zhang X, Zeng Y, Upadhyaya P. Differential expression of microRNAs in early-stage neoplastic transformation in the lungs of F344 rats chronically treated with the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis. 2008;29(12):2394–2399. doi: 10.1093/carcin/bgn209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Choi YM, An S, Lee EM, et al. CYP1A1 is a target of miR-892a-mediated post-transcriptional repression. Int J Oncol. 2012;41(1):331–336. doi: 10.3892/ijo.2012.1418. [DOI] [PubMed] [Google Scholar]
- [68].Patel SA, Bhambra U, Charalambous MP, et al. Interleukin-6 mediated upregulation of CYP1B1 and CYP2E1 in colorectal cancer involves DNA methylation, miR27b and STAT3. Br J Cancer. 2014;111(12):2287–2296. doi: 10.1038/bjc.2014.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hou L, Wang D, Baccarelli A. Environmental chemicals and microRNAs. Mutat Res. 2011;714(1–2):105–112. doi: 10.1016/j.mrfmmm.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Skinner MK, Guerrero-Bosagna C. Environmental signals and transgenerational epigenetics. Epigenomics. 2009;1(1):111–117. doi: 10.2217/epi.09.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Yu HW, Cho WC. The role of microRNAs in toxicology. Arch Toxicol. 2015;89(3): 319–325. doi: 10.1007/s00204-014-1440-2. [DOI] [PubMed] [Google Scholar]
- [72].Marrone AK, Beland FA, Pogribny IP. Noncoding RNA response to xenobiotic exposure: an indicator of toxicity and carcinogenicity. Expert Opin Drug Metab Toxicol. 2014;10(10):1409–1422. doi: 10.1517/17425255.2014.954312. [DOI] [PubMed] [Google Scholar]
- [73].Thai P, Statt S, Chen CH, Liang E, Campbell C, Wu R. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol. 2013;49(2):204–211. doi: 10.1165/rcmb.2013-0159RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Izzotti A, Calin GA, Steele VE, Croce CM, De Flora S. Relationships of microRNA expression in mouse lung with age and exposure to cigarette smoke and light. FASEB J. 2009;23(9):3243–3250. doi: 10.1096/fj.09-135251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ngalame NN, Tokar EJ, Person RJ, Xu Y, Waalkes MP. Aberrant microRNA expression likely controls RAS oncogene activation during malignant transformation of human prostate epithelial and stem cells by arsenic. Toxicol Sci. 2014; 138(2):268–277. doi: 10.1093/toxsci/kfu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Izzotti A, Pulliero A. The effects of environmental chemical carcinogens on the microRNA machinery. Int J Hyg Environ Health. 2014;217(6):601–627. doi: 10.1016/j.ijheh.2014.01.001. [DOI] [PubMed] [Google Scholar]
- [77].Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- [78].Humans IWGotEoCRt. Tobacco smoke and involuntary smoking. IARC Monogr Eval Carcinog Risks Hum. 2004;83:1–1438. [PMC free article] [PubMed] [Google Scholar]
- [79].Akopyan G, Bonavida B. Understanding tobacco smoke carcinogen NNK and lung tumorigenesis. Int J Oncol. 2006;29(4):745–752. [PubMed] [Google Scholar]
- [80].Escriva L, Font G, Manyes L. In vivo toxicity studies of fusarium mycotoxins in the last decade: a review. Food Chem Toxicol. 2015;78:185–206. doi: 10.1016/j.fct.2015.02.005. [DOI] [PubMed] [Google Scholar]
- [81].Chen X, Cheung ST, So S, et al. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13(6):1929–1939. doi: 10.1091/mbc.02-02-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kim JW, Ye Q, Forgues M, et al. Cancer-associated molecular signature in the tissue samples of patients with cirrhosis. Hepatology. 2004;39(2):518–527. doi: 10.1002/hep.20053. [DOI] [PubMed] [Google Scholar]
- [83].Smith MW, Yue ZN, Geiss GK, et al. Identification of novel tumor markers in hepatitis C virus-associated hepatocellular carcinoma. Cancer Res. 2003;63(4): 859–864. [PubMed] [Google Scholar]
- [84].Maihofner C, Charalambous MP, Bhambra U, et al. Expression of cyclooxygenase-2 parallels expression of interleukin-1beta, interleukin-6 and NF-kappaB in human colorectal cancer. Carcinogenesis. 2003;24(4):665–671. doi: 10.1093/carcin/bgg006. [DOI] [PubMed] [Google Scholar]
- [85].Murray GI, Taylor MC, McFadyen MC, et al. Tumor-specific expression of cyto-chrome P450 CYP1B1. Cancer Res. 1997;57(14):3026–3031. [PubMed] [Google Scholar]
- [86].Frolkis A, Dieleman LA, Barkema HW, et al. Environment and the inflammatory bowel diseases. Can J Gastroenterol. 2013;27(3):e18–24. doi: 10.1155/2013/102859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Molodecky NA, Kaplan GG. Environmental risk factors for inflammatory bowel disease. Gastroenterol Hepatol (NY). 2010;6(5):339–346. [PMC free article] [PubMed] [Google Scholar]
- [88].Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Gill P, Bhattacharyya S, McCullough S, et al. MicroRNA regulation of CYP 1A2, CYP3A4 and CYP2E1 expression in acetaminophen toxicity. Sci Rep. 2017;7(1): 12331.doi: 10.1038/s41598-017-11811-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Chen Y, Zeng L, Wang Y, et al. The expression, induction and pharmacological activity of CYP1A2 are post-transcriptionally regulated by microRNA hsa-miR-132–5p. Biochem Pharmacol. 2017;145:178–191. doi: 10.1016/j.bcp.2017.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Wei JH, Luo QQ, Tang YJ, et al. Upregulation of microRNA-320 decreases the risk of developing steroid-induced avascular necrosis of femoral head by inhibiting CYP1A2 both in vivo and in vitro. Gene. 2018;660:136–144. doi: 10.1016/j.gene.2018.03.045. [DOI] [PubMed] [Google Scholar]
- [92].Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66(18):9090–9098. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- [93].Mao M, Wu Z, Chen J. MicroRNA-187–5p suppresses cancer cell progression in non-small cell lung cancer (NSCLC) through down-regulation of CYP1B1. Biochem Biophys Res Commun. 2016;478(2):649–655. doi: 10.1016/j.bbrc.2016.08.001. [DOI] [PubMed] [Google Scholar]
- [94].Chang I, Mitsui Y, Fukuhara S, et al. Loss of miR-200c up-regulates CYP1B1 and confers docetaxel resistance in renal cell carcinoma. Oncotarget. 2015;6(10): 7774–7787. doi: 10.18632/oncotarget.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Nakano M, Fukushima Y, Yokota S, et al. CYP2A7 pseudogene transcript affects CYP2A6 expression in human liver by acting as a decoy for miR-126. Drug Metab Dispos. 2015;43(5):703–712. doi: 10.1124/dmd.115.063255. [DOI] [PubMed] [Google Scholar]
- [96].Jin Y, Yu D, Tolleson WH, et al. MicroRNA hsa-miR-25–3p suppresses the expression and drug induction of CYP2B6 in human hepatocytes. Biochem Pharmacol. 2016;113:88–96. doi: 10.1016/j.bcp.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Zhang SY, Surapureddi S, Coulter S, Ferguson SS, Goldstein JA. Human CYP2C8 is post-transcriptionally regulated by microRNAs 103 and 107 in human liver. Mol Pharmacol. 2012;82(3):529–540. doi: 10.1124/mol.112.078386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Rieger JK, Reutter S, Hofmann U, Schwab M, Zanger UM. Inflammation-associated microRNA-130b down-regulates cytochrome P450 activities and directly targets CYP2C9. Drug Metab Dispos. 2015;43(6):884–888. doi: 10.1124/dmd.114.062844. [DOI] [PubMed] [Google Scholar]
- [99].Yu D, Green B, Tolleson WH, et al. MicroRNA hsa-miR-29a-3p modulates CYP2C19 in human liver cells. Biochem Pharmacol. 2015;98(1):215–223. doi: 10.1016/j.bcp.2015.08.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Li J, Xie M, Wang X, et al. Sex hormones regulate cerebral drug metabolism via brain miRNAs: down-regulation of brain CYP2D by androgens reduces the analgesic effects of tramadol. Br J Pharmacol. 2015;172(19):4639–4654. doi: 10.1111/bph.13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Zeng L, Chen Y, Wang Y, et al. MicroRNA hsa-miR-370–3p suppresses the expression and induction of CYP2D6 by facilitating mRNA degradation. Biochem Pharmacol. 2017;140:139–149. doi: 10.1016/j.bcp.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Wang Y, Yu D, Tolleson WH, et al. A systematic evaluation of microRNAs in regulating human hepatic CYP2E1. Biochem Pharmacol. 2017;138:174–184. doi: 10.1016/j.bcp.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Mohri T, Nakajima M, Fukami T, Takamiya M, Aoki Y, Yokoi T. Human CYP2E1 is regulated by miR-378. Biochem Pharmacol. 2010;79(7):1045–1052. doi: 10.1016/j.bcp.2009.11.015. [DOI] [PubMed] [Google Scholar]
- [104].Miao L, Yao H, Li C, et al. A dual inhibition: microRNA-552 suppresses both transcription and translation of cytochrome P450 2E1. Biochim Biophys Acta. 2016; 1859(4):650–662. doi: 10.1016/j.bbagrm.2016.02.016. [DOI] [PubMed] [Google Scholar]
- [105].Nakano M, Mohri T, Fukami T, et al. Single-nucleotide polymorphisms in cyto-chrome P450 2E1 (CYP2E1) 3′-untranslated region affect the regulation of CYP2E1 by miR-570. Drug Metab Dispos. 2015;43(10):1450–1457. doi: 10.1124/dmd.115.065664. [DOI] [PubMed] [Google Scholar]
- [106].Wei Z, Jiang S, Zhang Y, et al. The effect of microRNAs in the regulation of human CYP3A4: a systematic study using a mathematical model. Sci Rep. 2015; 4(1):4283. doi: 10.1038/srep04283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Shi Y, Liu Y, Wei Z, et al. Hsa-miR-27a is involved in the regulation of CYP3A4 expression in human livers from Chinese Han population. Pharmacogenomics. 2015;16(12):1379–1386. doi: 10.2217/pgs.15.82. [DOI] [PubMed] [Google Scholar]
- [108].Pan YZ, Gao W, Yu AM. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos. 2009;37(10):2112–2117. doi: 10.1124/dmd.109.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Liu JE, Ren B, Tang L, et al. The independent contribution of miRNAs to the missing heritability in CYP3A4/5 functionality and the metabolism of atorvastatin. Sci Rep. 2016;6(1):26544. doi: 10.1038/srep26544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Yu D, Wu L, Gill P, et al. Multiple microRNAs function as self-protective modules in acetaminophen-induced hepatotoxicity in humans. Arch Toxicol. 2018;92(2): 845–858. doi: 10.1007/s00204-017-2090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Yan L, Liu J, Zhao Y, et al. Suppression of miR-628–3p and miR-641 is involved in rifampin-mediated CYP3A4 induction in HepaRG cells. Pharmacogenomics. 2017; 18(1):57–64. doi: 10.2217/pgs-2016-0088. [DOI] [PubMed] [Google Scholar]
- [112].Wang B, Zheng L, Chou J, et al. CYP4Z1 3’UTR represses migration of human breast cancer cells. Biochem Biophys Res Commun. 2016;478(2):900–907. doi: 10.1016/j.bbrc.2016.08.048. [DOI] [PubMed] [Google Scholar]
- [113].Xi XP, Zhuang J, Teng MJ, et al. MicroRNA-17 induces epithelial-mesenchymal transition consistent with the cancer stem cell phenotype by regulating CYP7B1 expression in colon cancer. Int J Mol Med. 2016;38(2):499–506. doi: 10.3892/ijmm.2016.2624. [DOI] [PubMed] [Google Scholar]
- [114].Kumar P, Luo Y, Tudela C, Alexander JM, Mendelson CR. The c-Myc-regulated microRNA-17 ~ 92 (miR-17 ~ 92) and miR-106a~363 clusters target hCYP19A1 and hGCM1 to inhibit human trophoblast differentiation. Mol Cell Biol. 2013; 33(9):1782–1796. doi: 10.1128/MCB.01228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Wang L, Gao Z, Wang L, Gao Y. Loss of miR-125b contributes to upregulation of CYP24 in uraemic rats. Nephrology (Carlton). 2016;21(12):1063–1068. doi: 10.1111/nep.12714. [DOI] [PubMed] [Google Scholar]
- [116].Xin J, Zheng LM, Sun DK, Li XF, Xu P, Tian LQ. miR-204 functions as a tumor suppressor gene, at least partly by suppressing CYP27A1 in glioblastoma. Oncol Lett. 2018;16(2):1439–1448. doi: 10.3892/ol.2018.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Singh R, Yadav V, Kumar S, Saini N. MicroRNA-195 inhibits proliferation, invasion and metastasis in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1. Sci Rep. 2015;5(1):17454. doi: 10.1038/srep17454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Kurzawski M, Dziedziejko V, Post M, et al. Expression of genes involved in xenobiotic metabolism and transport in end-stage liver disease: up-regulation of ABCC4 and CYP1B1. Pharmacol Rep. 2012;64(4):927–939. doi: 10.1016/S1734-1140(12)70888-5. [DOI] [PubMed] [Google Scholar]
- [119].Pan X, Kent R, Won KJ, Jeong H. Cholic acid feeding leads to increased CYP2D6 expression in CYP2D6-humanized mice. Drug Metab Dispos. 2017;45(4):346–352. doi: 10.1124/dmd.116.074013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Sharma D, Turkistani AA, Chang W, Hu C, Xu Z, Chang T. Negative regulation of human Pregnane X receptor by MicroRNA-18a-5p: evidence for suppression of MicroRNA-18a-5p expression by rifampin and rilpivirine. Mol Pharmacol. 2017; 92(1):48–56. doi: 10.1124/mol.116.107003. [DOI] [PubMed] [Google Scholar]
- [121].Vachirayonstien T, Yan B. MicroRNA-30c-1–3p is a silencer of the pregnane X receptor by targeting the 3’-untranslated region and alters the expression of its target gene cytochrome P450 3A4. Biochim Biophys Acta. 2016;1859(9):1238–1244. doi: 10.1016/j.bbagrm.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Oda Y, Nakajima M, Tsuneyama K, et al. Retinoid X receptor α in human liver is regulated by miR-34a. Biochem Pharmacol. 2014;90(2):179–187. doi: 10.1016/j.bcp.2014.05.002. [DOI] [PubMed] [Google Scholar]
- [123].Takagi S, Nakajima M, Mohri T, Yokoi T. Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J Biol Chem. 2008;283(15):9674–9680. doi: 10.1074/jbc.M709382200. [DOI] [PubMed] [Google Scholar]
- [124].Zhang CL, Wang H, Yan CY, Gao XF, Ling XJ. Deregulation of RUNX2 by miR-320a deficiency impairs steroidogenesis in cumulus granulosa cells from polycystic ovary syndrome (PCOS) patients. Biochem Biophys Res Commun. 2017;482(4): 1469–1476. doi: 10.1016/j.bbrc.2016.12.059. [DOI] [PubMed] [Google Scholar]
- [125].Wang M, Liu M, Sun J, et al. MicroRNA-27a-3p affects estradiol and androgen imbalance by targeting Creb1 in the granulosa cells in mouse polycytic ovary syndrome model. Reprod Biol. 2017;17(4):295–304. doi: 10.1016/j.repbio.2017.09.005. [DOI] [PubMed] [Google Scholar]
- [126].Miao N, Wang X, Hou Y, Feng Y, Gong Y. Identification of male-biased microRNA-107 as a direct regulator for nuclear receptor subfamily 5 group A member 1 based on sexually dimorphic microRNA expression profiling from chicken embryonic gonads. Mol Cell Endocrinol. 2016;429:29–40. doi: 10.1016/j.mce.2016.03.033. [DOI] [PubMed] [Google Scholar]
- [127].Dai A, Sun H, Fang T, et al. MicroRNA-133b stimulates ovarian estradiol synthesis by targeting Foxl2. FEBS Lett. 2013;587(15):2474–2482. doi: 10.1016/j.febslet.2013.06.023. [DOI] [PubMed] [Google Scholar]
- [128].Wang L, Li C, Li R, et al. MicroRNA-764–3p regulates 17beta-estradiol synthesis of mouse ovarian granulosa cells by targeting steroidogenic factor-1. In Vitro Cell Dev Biol Anim. 2016;52(3):365–373. doi: 10.1007/s11626-015-9977-9. [DOI] [PubMed] [Google Scholar]
- [129].Liu J, Li X, Yao Y, Li Q, Pan Z, Li Q. miR-1275 controls granulosa cell apoptosis and estradiol synthesis by impairing LRH-1/CYP19A1 axis. Biochim Biophys Acta Gene Regul Mech. 2018;1861(3):246–257. doi: 10.1016/j.bbagrm.2018.01.009. [DOI] [PubMed] [Google Scholar]
- [130].He J, Guo X, Liu ZQ, Yang PC, Yang S. Micro RNA-550a interferes with vitamin D metabolism in peripheral B cells of patients with diabetes. Cell Biochem Funct. 2016;34(8):640–646. doi: 10.1002/cbf.3240. [DOI] [PubMed] [Google Scholar]
- [131].de Wildt SN, Kearns GL, Leeder JS, van den Anker JN. Cytochrome P450 3A: ontogeny and drug disposition. Clin Pharmacokinet. 1999;37(6):485–505. doi: 10.2165/00003088-199937060-00004. [DOI] [PubMed] [Google Scholar]
- [132].Zhou SF. Drugs behave as substrates, inhibitors and inducers of human cyto-chrome P450 3A4. Curr Drug Metab. 2008;9(4):310–322. [DOI] [PubMed] [Google Scholar]
- [133].Kamiyama Y, Matsubara T, Yoshinari K, Nagata K, Kamimura H, Yamazoe Y. Role of human hepatocyte nuclear factor 4 alpha in the expression of drug-metabolizing enzymes and transporters in human Hepatocytes assessed by use of small interfering RNA. Drug Metab Pharmacokinet. 2007;22(4):287–298. doi: 10.2133/dmpk.22.287. [DOI] [PubMed] [Google Scholar]
- [134].Pavek P, Dvorak Z. Xenobiotic-induced transcriptional regulation of xenobiotic metabolizing enzymes of the cytochrome P450 superfamily in human extrahepatic tissues. Curr Drug Metab. 2008;9(2):129–143. doi: 10.2174/138920008783571774. [DOI] [PubMed] [Google Scholar]
- [135].Monostory K, Dvorak Z. Steroid regulation of drug-metabolizing cytochromes P450. Curr Drug Metab. 2011;12(2):154–172. [DOI] [PubMed] [Google Scholar]
- [136].Juřica J, Dovrtĕlová G, Nosková K, Zendulka O. Bile acids, nuclear receptors and cytochrome P450. Physiol Res. 2016;65(Suppl 4):S427–S440. [DOI] [PubMed] [Google Scholar]
- [137].Lu Y, Cederbaum AI. CYP2E1 and oxidative liver injury by alcohol. Free Radic Biol Med. 2008;44(5):723–738. doi: 10.1016/j.freeradbiomed.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Bieche I, Narjoz C, Asselah T. Reverse transcriptase-PCR quantification of mRNA levels from cytochrome (CYP)1, CYP2 and CYP3 families in 22 different human tissues. Pharmacogenet Genomics. 2007;17(9):731–742. doi: 10.1097/FPC.0b013e32810f2e58. [DOI] [PubMed] [Google Scholar]
- [139].Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther. 1994;270(1):414–423. [PubMed] [Google Scholar]
- [140].Prescott LF. Paracetamol, alcohol and the liver. Br J Clin Pharmacol. 2000;49(4): 291–301. doi: 10.1046/j.1365-2125.2000.00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Seitz HK, Becker P. Alcohol metabolism and cancer risk. Alcohol Res Health. 2007; 30(1):38–41, 44–37. [PMC free article] [PubMed] [Google Scholar]
- [142].Zhao Y, Li H, Fang S, et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016;44(D1):D203–208. doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- [144].Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res. 2015;116(4):737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- [145].Wu GC, Pan HF, Leng RX, et al. Emerging role of long noncoding RNAs in auto-immune diseases. Autoimmun Rev. 2015;14(9):798–805. doi: 10.1016/j.autrev.2015.05.004. [DOI] [PubMed] [Google Scholar]
- [146].Luo Q, Chen Y. Long noncoding RNAs and Alzheimer’s disease. Clin Interv Aging. 2016;11:867–872. doi: 10.2147/CIA.S107037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- [148].Zhao XY, Lin JD. Long noncoding RNAs: a new regulatory code in metabolic control. Trends Biochem Sci. 2015;40(10):586–596. doi: 10.1016/j.tibs.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Huarte M The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11): 1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- [150].Dempsey JL, Cui JY. Long non-coding RNAs: a novel paradigm for toxicology. Toxicol Sci. 2017;155(1):3–21. doi: 10.1093/toxsci/kfw203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Karlsson O, Baccarelli AA. Environmental health and long non-coding RNAs. Curr Environ Health Rep. 2016;3(3):178–187. doi: 10.1007/s40572-016-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10(6):925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Semin Cell Dev Biol. 2011;22(4):366–376. doi: 10.1016/j.semcdb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- [156].Wang YT, Yan L, Liu JY, et al. The HNF1 alpha-regulated LncRNA HNF1 alpha-AS1 is involved in the regulation of cytochrome P450 expression in human liver tissues and Huh7 cells. J Pharmacol Exp Ther. 2019;368(3):353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Biomarkers Definitions Working G Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69(3):89–95. [DOI] [PubMed] [Google Scholar]
- [158].Wild CP. Environmental exposure measurement in cancer epidemiology. Mutagenesis. 2008;24(2):117–125. doi: 10.1093/mutage/gen061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Mayeux R Biomarkers: potential uses and limitations. Neurorx. 2004;1(2):182–188. doi: 10.1602/neurorx.1.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42(8):1273–1281. doi: 10.1016/j.biocel.2009.12.014. [DOI] [PubMed] [Google Scholar]
- [161].Vrijens K, Bollati V, Nawrot TS. MicroRNAs as potential signatures of environmental exposure or effect: a systematic review. Environ Health Perspect. 2015; 123(5):399–411. doi: 10.1289/ehp.1408459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Bolha L, Ravnik-Glavac M, Glavac D. Long noncoding RNAs as biomarkers in Cancer. Dis Markers. 2017;2017:7243968.doi: 10.1155/2017/7243968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Etheridge A, Lee I, Hood L, Galas D, Wang K. Extracellular microRNA: a new source of biomarkers. Mutat Res. 2011;717(1–2):85–90. doi: 10.1016/j.mrfmmm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005;14(8):1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- [165].Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr. 2009;21(2):243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Valencia-Quintana R, Sanchez-Alarcon J, Tenorio-Arvide MG, et al. The microRNAs as potential biomarkers for predicting the onset of aflatoxin exposure in human beings: a review. Front Microbiol. 2014;5:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Baccarelli A, Ghosh S. Environmental exposures, epigenetics and cardiovascular disease. Curr Opin Clin Nutr Metab Care. 2012;15(4):323–329. doi: 10.1097/MCO.0b013e328354bf5c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Zhang Y, Jia Y, Zheng R, et al. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem. 2010;56(12):1830–1838. doi: 10.1373/clinchem.2010.147850. [DOI] [PubMed] [Google Scholar]
- [169].Antoine DJ, Dear JW, Lewis PS, et al. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology. 2013;58(2):777–787. doi: 10.1002/hep.26294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Yamaura Y, Nakajima M, Takagi S, Fukami T, Tsuneyama K, Yokoi T. Plasma microRNA profiles in rat models of hepatocellular injury, cholestasis, and steatosis. PLoS One. 2012;7(2):e30250. doi: 10.1371/journal.pone.0030250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem. 2009;55(11):1944–1949. doi: 10.1373/clinchem.2009.125310. [DOI] [PubMed] [Google Scholar]
- [172].Nishimura Y, Kondo C, Morikawa Y, et al. Plasma miR-208 as a useful biomarker for drug-induced cardiotoxicity in rats. J Appl Toxicol. 2015;35(2):173–180. doi: 10.1002/jat.3044. [DOI] [PubMed] [Google Scholar]
- [173].Marrone AK, Beland FA, Pogribny IP. The role for microRNAs in drug toxicity and in safety assessment. Expert Opin Drug Metab Toxicol. 2015;11(4):601–611. doi: 10.1517/17425255.2015.1021687. [DOI] [PubMed] [Google Scholar]
- [174].Harrill AH, McCullough SD, Wood CE, Kahle JJ, Chorley BN. MicroRNA bio-markers of toxicity in biological matrices. Toxicol Sci. 2016;152(2):264–272. doi: 10.1093/toxsci/kfw090. [DOI] [PubMed] [Google Scholar]
- [175].Fuchs TC, Hewitt P. Biomarkers for drug-induced renal damage and nephrotoxicity-an overview for applied toxicology. AAPS J. 2011;13(4):615–631. doi: 10.1208/s12248-011-9301-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Koturbash I, Tolleson WH, Guo L, et al. microRNAs as pharmacogenomic bio-markers for drug efficacy and drug safety assessment. Biomarkers Med. 2015;9(11): 1153–1176. doi: 10.2217/bmm.15.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Schwarzenbach H Clinical relevance of circulating, cell-free and exosomal microRNAs in plasma and serum of breast cancer patients. Oncol Res Treat. 2017; 40(7–8):423–429. doi: 10.1159/000478019. [DOI] [PubMed] [Google Scholar]
- [178].Nakamura K, Sawada K, Yoshimura A, Kinose Y, Nakatsuka E, Kimura T. Clinical relevance of circulating cell-free microRNAs in ovarian cancer. Mol Cancer. 2016; 15(1):48. doi: 10.1186/s12943-016-0536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11(3):145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- [180].Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101(10): 2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- [182].Brase JC, Wuttig D, Kuner R, Sultmann H. Serum microRNAs as non-invasive bio-markers for cancer. Mol Cancer. 2010;9:306.doi: 10.1186/1476-4598-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Robinson WH, Lindstrom TM, Cheung RK, Sokolove J. Mechanistic biomarkers for clinical decision making in rheumatic diseases. Nat Rev Rheumatol. 2013;9(5): 267–276. doi: 10.1038/nrrheum.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]