Abstract
Mucor circinelloides is a pathogenic fungus and etiologic agent of mucormycosis. In 2013, cases of gastrointestinal illness after yogurt consumption were reported to the US FDA, and the producer found that its products were contaminated with Mucor. A previous study found that the Mucor strain isolated from an open contaminated yogurt exhibited virulence in a murine systemic infection model and showed that this strain is capable of surviving passage through the gastrointestinal tract of mice. In this study, we isolated another Mucor strain from an unopened yogurt that is closely related but distinct from the first Mucor strain and subsequently examined if Mucor alters the gut microbiota in a murine host model. DNA extracted from a ten-day course of stool samples was used to analyze the microbiota in the gastrointestinal tracts of mice exposed via ingestion of Mucor spores. The bacterial 16S rRNA gene and fungal ITS1 sequences obtained were used to identify taxa of each kingdom. Linear regressions revealed that there are changes in bacterial and fungal abundance in the gastrointestinal tracts of mice which ingested Mucor. Furthermore, we found an increased abundance of the bacterial genus Bacteroides and a decreased abundance of the bacteria Akkermansia muciniphila in the gastrointestinal tracts of exposed mice. Measurements of abundances show shifts in relative levels of multiple bacterial and fungal taxa between mouse groups. These findings suggest that exposure of the gastrointestinal tract to Mucor can alter the microbiota and, more importantly, illustrate an interaction between the intestinal mycobiota and bacteriota. In addition, Mucor was able to induce increased permeability in epithelial cell monolayers in vitro, which might be indicative of unstable intestinal barriers. Understanding how the gut microbiota is shaped is important to understand the basis of potential methods of treatment for gastrointestinal illness. How the gut microbiota changes in response to exposure, even by pathogens not considered to be causative agents of food-borne illness, may be important to how commercial food producers prevent and respond to contamination of products aimed at the public. This study provides evidence that the fungal microbiota, though understudied, may play an important role in diseases of the human gut.
Keywords: microbiota, mycobiota, mycobiome, Mucor, fungi in the guts
Introduction
Mucor circinelloides is a fungus that belongs to the Mucorales family in the early diverging Mucoromycota phylum (Spata-fora et al., 2016). Among other members of the Mucorales family, exposure to M. circinelloides causes the disease mucormycosis (Morin-Sardin et al., 2017). Mucormycosis is a disease that primarily affects immunocompromised individuals, such as those with neutropenia, as well as diabetes mellitus and burn patients (Chayakulkeeree et al., 2006). Despite the increased prevalence of mucormycosis that has resulted from an increase in immunocompromised patients, fatality rates reach as high as 90% in disseminated cases (Roden et al., 2005), and current treatment options remain limited.
M. circinelloides has also been implicated in food product spoilage and development of gastrointestinal mucormycosis and fungemia following consumption in both immunocompromised and immunocompetent individuals (Aboltins et al., 2006; Lazar et al., 2014), the cause of which has been identified as post-pasteurization with hyphal phase Mucor (Snyder et al., 2016). Additionally, M. circinelloides can produce the mycotoxin 3-nitropropionic acid (3NP) (Hollmann et al., 2008). 3NP is an irreversible inhibitor of succinate dehydrogenase (Alston et al., 1977), and ingestion of 3NP is known to result in neurodegenerative changes that have been studied in animal models of Huntington’s disease (Brouillet et al., 2005). 3NP has also been involved with food poisoning caused by contaminated sugarcane, in which the 3NP produced by Arthrinium species caused the onset of acute encephalopathy in humans during outbreaks of sugarcane mildew in China from 1972 to 1989 (He et al., 1995). Therefore, M. circinelloides serves as a potential health threat regarding food-borne illness in addition to its more traditional clinical role in mucormycosis.
In September 2013, a recall was issued for yogurt products produced by a company after complaints of spoiled product and gastrointestinal illness following consumption of the yogurt (USAToday, 2013). The causative agent of this spoilage was determined by both the U.S. Food and Drug Administration (FDA) and the company to be M. circinelloides (FDA, 2013). In our previous study, a M. circinelloides strain was isolated from a contaminated yogurt sample that a consumer had provided (Lee et al., 2014). This strain, named Mucho, was shown to be virulent in systemic exposure of mice in a manner comparable to that of strains isolated from clinical cases of mucormycosis. The study also noted that the colons of mice that ingested the strain tended to be shorter than those of mice that had not ingested it (Lee et al., 2014). This is of interest, as shortening of the colon is associated with the intestinal inflammation seen in ulcerative colitis, one of the inflammatory bowel diseases (IBDs) (Kim et al., 2012). It is then important to consider how ingestion of this fungus may affect the gut.
Recently, emphasis has been placed on the role of the microbiota in the pathology of various human health conditions. Studies examining the microbiota in IBDs, including Crohn’s disease, have also noted changes of composition in the disease state (Ott et al., 2008; Li et al., 2014; Hoarau et al., 2016; Liguori et al., 2016; Sokol et al., 2017; Miyoshi et al., 2018). These studies noted an overall decrease in beneficial bacteria, increase in the bacteria Serratia marcescens and Escherichia coli, decrease in the fungus Saccharomyces cerevisiae, and increase in the fungus Candida albicans. Inter-kingdom interactions were also observed through the positive correlation between Candida tropicalis and S. marcescens and E. coli (Hoarau et al., 2016; Sokol et al., 2017). Anorexia nervosa, characterized in part by severe malnourishment, has been associated with decreased gut diversity and alterations in microbial composition (Kleiman et al., 2015). Additionally, altered diversity was shown to be associated with psychiatric measures, such as an increased risk of depression (Kleiman et al., 2015). Obesity is associated with gastrointestinal illness and has also been associated with changes in the microbiota. Studies examining the bacteria associated with obesity have often found the condition to be negatively associated with relative abundance of Firmicutes members and positively associated with Bacteroides members (Devaraj et al., 2013). More recently, Mucor species abundance was shown to parallel the degree of weight loss in obese individuals, being more abundant in non-obese individuals (Rodriguez et al., 2015). This study also concluded that obese individuals could be distinguished from non-obese individuals through fungal composition, specifically utilizing the genera Aspergillus, Mucor, Penicillium, Saccharomyces, and Eupenicillium. From our current knowledge of the association between the microbiota and gastrointestinal illness, a reasonable hypothesis is that food-borne illness caused by M. circinelloides may also be associated with changes in the gut microbiota.
In this study, we isolated another M. circinelloides strain from a previously unopened yogurt that was within the company’s 2013 recall range. Interestingly, the newly isolated Mucor strain is closely related but distinct from Mucho, the first strain isolated in our previous study. Following ingestion of this newly isolated Mucor strain, the murine gut microbiota was characterized by analyzing the bacterial microbiota and mycobiota in stool samples over the course of ten days. Analysis of gut alpha diversity revealed changes in bacterial and fungal diversity after ingestion of Mucor, suggesting a shift in the gut environment. This finding was supported by taxa compositional analysis, which suggested changes in the abundance of specific bacterial species related to gut health. These results demonstrate an example of fungal-driven alteration of the gut microbiota, in which ingestion of Mucor led to a shift in the gut microbiota to a potentially unhealthy state. Going forward, more emphasis should be placed on the role of fungi in relation to food-borne illness, the role of fungi in the gut, and interactions between intestinal bacteria and fungi.
Materials and Methods
Strains and culture conditions
An unopened black cherry flavor yogurt within the recall range was obtained before being stored in a freezer for approximately one year (Supplementary data Text S1). Yogurt samples were transferred on PDAm (Potato Dextrose Agar media, containing 4 g of potato starch, 20 g of dextrose, and 15 g of agar per L) media containing ampicillin (100 μg/ml) and kanamycin (50 μg/ml) by using sterile wooden applicators. The plates were incubated at 30°C for three days, when mycelial growth was observed. Three mycelial colonies were then subject to two vegetative rounds of streak purification, and spores from each colony were collected as described previously (Lee et al., 2014). All three strains had identical DNA sequences at the ITS, LSU rDNA, and RPB1 loci (see below). One of them was selected and designated AzMucho for further study. The first M. circinelloides isolate, Mucho (Lee et al., 2014), and the human skin isolate 1006PhL (Findley et al., 2013) were also used for this study.
For spore production, the strains were grown on PDAm for 3 to 4 days in the light. Deionized distilled water (2 ml) was added to the plates, which were gently rubbed with a sterile spreader to release spores from the mycelia. The spore suspension was collected and transferred into sterile micro-centrifuge tubes. The number of spores was calculated by using a hemocytometer. The whole genome analysis was performed as previously described (Lee et al., 2014).
Phylogenetic analysis
The three loci including ITS, LSU rDNA, and RPB1 were used for analysis as previously described (Lee et al., 2014). Primers used for ITS are: ITS1, TCCGTAGGTGAACCTG CGG and ITS4, TCCTCCGCTTATTGATATGC; for LSU rRNA: D1/D2 LRDNA, GCATATCAATAAGCGGAGGAA AAG and LR3, GGTCCGTGTTTCAAGACGG; and for RPB1: RPB1-Ac, GARTGYCCDGGDCAYTTYGG and RPB1-Cr, CCNGCDATNTCRTTRTCCATRTA. The PCR amplicons were sequenced and analyzed. The whole genome analysis was performed as previously described for the first Mucho strain (Lee et al., 2014). The AzMucho assembly was submitted to NCBI under the project accession number PRJNA-431470.
Virulence tests and microbiota analysis after Mucor ingestion in a murine model
Four to six-week-old male BALB/C mice (18–20 g body weight, Charles River Laboratories) were used for the virulence test. The animals were randomly distributed into two cages before the spore challenge. Spores for exposures were washed and resuspended in sterile PBS. For virulence tests, PBS (200 ml) containing 1 × 106 spores of Mucor strains was administered via tail vein injection. Survival of the mice was examined twice daily and body weight was monitored daily. Animals that appeared moribund or in pain were euthanized appropriately. Survival curve was generated and analyzed by using Kaplan-Meier survival curves with PRISM (GraphPad ad Software, Inc.).
Two independent microbiota analyses were performed with mice housed at the animal facility at the Duke university or The University of Texas at San Antonio (UTSA). Four- to six-week old male BALB/C mice, purchased from Charles River Laboratories, were used. Upon arrival, the animals were randomly distributed into different cages (5 mice per cage) and established for 10 days in a regular room at each facility before Mucor spore challenge. After ingestion, the animals were placed in a BSL2 room and maintained under 12 h day-and-night cycles throughout the experiments. Cohorts of the animals were ingested, by oral gavage, either spores (1 × 106) of the AzMucho strain, spores (1 × 106) of the 1006PhL strain, or PBS alone at day 0 before the night cycle. Body weight of the exposed and control mice was monitored daily until termination of the experiment. Fresh stool from mice was collected daily before the night cycle for up to 10 days post infection (or 9 days post infection at UTSA). The collected stool samples were immediately frozen at −80°C for later analysis. Duplicate stool samples from the Mucor exposed mice at day 2 were placed on PDAm to ensure Mucor spores had reached the GI tract. Mucor colonies formed from the stool samples collected from all 5 exposed mice (data not shown).
All murine experiments were conducted at Duke University and UTSA in full compliance with all of the guidelines of the both Institutional Animal Care and Use Committees (IACUC) and in full compliance with the United States Animal Welfare Act (Public Law 98–198). The UTSA IACUC approved all the murine studies under protocol number MU-104–2/20, and the Duke University IACUC approved all of the murine studies under protocol number A061–12-03. The experiments were conducted in the Division of Laboratory Animal Resources (DLAR) facilities that are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC).
3-nitropropionic acid analysis
For 3-nitropropionic acid (3NP) analysis, commercially available yogurt of the same sort as the originally contaminated one were inoculated with 1 × 106 spores of AzMucho in triplicates. Spores were injected through the foil yogurt lid which was subsequently sealed with tape and the whole container stored at 4°C until bloating of the lids were visible (2 weeks). Uninfected control yogurts of the same variety were incubated simultaneously. Analysis of 3NP was performed as initially described by Muir and Majak (Muir and Majak, 1984). Briefly, each yogurt was homogenized and 2 ml of each sample was mixed with 1 ml of 0.6 N perchloric acid and incubated on ice for 30 min. From the uninfected yogurt containers, 2 ml were additionally spiked with 3NP (Sigma Aldrich) to a final concentration of 20 μM and processed as described above. All perchloric acid treated samples were centrifuged at 3,000 g for 15 min and 500 μl of supernatant was filtered through 0.2 μm polyvinylidene fluoride filters before high performance liquid chromatography (HPLC) photodiode array (PDA) analysis. The samples were separated on a ZORBAX Eclipse XDB-C18 column (Agilent, 4.6 mm by 150 mm with a 5 μm particle size) isocratically with 0.15% (v/v) orthophosphoric acid (pH 2) using a Flexar Binary Liquid Chromatography (LC) Pump (PerkinElmer) coupled to a Flexar LC Autosampler (Perkin Elmer) and a Flexar PDA Plus Detector (PerkinElmer). The PDA was set to 210 nm. Each sample was run for 15 min and after each sample the column was washed with 95% methanol for 5 min and equilibrated with 0.15% (v/v) orthophosphoric acid for 5 min before injection of the next sample. Identification of 3NP was performed using Chromera Manager (PerkinElmer) by comparison to an authentic standard (Sigma Aldrich).
Stool DNA isolation and sequencing for microbiota analysis
DNA was isolated from collected mouse stool samples by using the ZymoBiomics DNA Miniprep kit (Zymo Research Co.) with slight modifications made to the manufacturer’s protocol. Before bead beating, samples were incubated at 70°C for 5 min, then at 37°C for 30 min. Bead beating was broken up into 5 processes of 1 min each with 1 min in between each process. As described in Kleiman et al. (2017), bacterial sequences in the isolated DNA samples were identified by amplification of the V4 variable region of the 16S rRNA gene by PCR (forward primer 5´-GAGTGCCAGCMGCCGCGG TAA-3’, reverse primer 5’-ACGGACTACHVGGGTWTC TAAT-3’). Forward and reverse primers incorporated single nucleotide shifts in six different lengths. The first amplification used 200 ng of isolated DNA as a template, a mix of the forward and reverse 16S primers each at a concentration of 10 mM, and the KAPA2G Robust PCR Kit (KAPA Biosystems Inc.) (95°C for 3 min then 10 cycles of 95°C for 30 sec, 50°C for 30 sec, and 72°C for 30 sec, ending with 72°C for 5 min). A bacterial 16S V4 library for sequencing was generated via a second amplification using 5 ml of purified product from the first amplification as a template, 10 mM of forward and reverse primers containing Illumina MiSeq adapter sequences, 12 base pair barcodes (Caporaso et al., 2012) specific for each sample, and the KAPA HiFi HotStart ReadyMix (95°C for 3 min, then 22 cycles of 95°C for 30 sec, ending with 72°C for 5 min). Fungal sequences in the isolated DNA samples were identified by amplification of the ITS1 region (forward primer 5’-CTTGGTCATTTAGAGGAAGTAA-3’, reverse primer 5’-GCTGCGTTCTTCATCGATGC-3’). Forward and reverse primers incorporated single nucleotide shifts in six different lengths. First amplification used 200 ng of isolated DNA as a template, a mix of the forward and reverse ITS primers at a final concentration of 0.25 mM, and the TaKaRa Ex Taq PCR (Takara Bio USA Inc.) system (98°C for 10 sec, then 35 cycles of 98°C for 10 sec, 56.1°C for 30 sec, and 72°C for 2 min, ending with 72°C for 5 min). A fungal ITS1 library was constructed by a second amplification using 5 ml of purified product from the first amplification as a template, 10 mM of forward and reverse primers containing Illumina MiSeq adapter sequences, 12 base pair barcodes (Caporaso et al., 2012) specific for each sample, and the KAPA HiFi HotStart ReadyMix (95°C for 3 min, then 22 cycles of 95°C for 30 sec, ending with 72°C for 5 min). PCR products from each amplification were cleaned utilizing the HighPrep PCR clean-up kit. Final cleaned PCR products were then diluted to be made equal in concentrations, pooled, and sequenced by the High-Throughput Sequencing Facility at the University of North Carolina at Chapel Hill School of Medicine or by the Genome Core at The University of Texas at San Antonio.
Analysis of 16S and ITS1 sequences
Demultiplexing and classification of 16S rRNA gene sequence data were performed utilizing the Quantitative Insights into Microbial Ecology (QIIME) pipeline using default settings (Walters et al., 2010). Open reference operational taxonomic units (OTUs) were clustered utilizing the Uclust algorithm based on a 97% threshold for sequence similarity (Edgar, 2010). Taxonomic classification was determined using the Greengenes database. Classification of ITS1 data were performed utilizing the PIPITS pipeline (Gweon et al., 2015), which clusters OTUs at a 97% sequencing similarity threshold by utilizing the RDP classifier. Taxonomic classification was determined using the UNITE database (Kõljalg et al., 2005). The resulting OTU tables were filtered in QiIME to remove OTUs present in less than 25% of all samples and with an overall abundance less than 0.005% as a conservative cutoff (Bokulich et al., 2013) for alpha diversity analysis and a more relaxed cutoff of 0.05% for beta diversity and taxonomic composition analysis due to the small sample size used.
Statistical analysis
Bacterial and fungal species richness (alpha diversity) were characterized by the number of species and the Chao 1 estimator of diversity for each sample using QIIME (Chao, 1984). Differences in alpha diversity between exposed and unexposed mice were tested with a multiple linear regression, which accounted for the possible interaction between exposure status and time since exposure using R version 3.3.3 and R packages vegan version 2.4–3 and phyloseq 1.19.1 (RCore-Team, 2017). Factors considered to calculate overall model for alpha diversity included day, Mucor spore or PBS ingestion, and the interaction between day and ingestion. Beta diversity analysis was performed with principle coordinate analysis of Bray-Curtis distances and permutational analysis of variance using adonis implementation in R. Taxa abundance of bacterial and fungal groups were calculated at the phylum, class, order, family, and genus levels using QIIME, and graphs depicting these levels were generated using R. Statistical significance of groups was determined using the Kruskal-Wallis test together with false discovery rate (FDR) correction (Benjamini and Hochberg, 1995). Taxa analysis was confirmed and visualized using the Linear Discriminant Analysis (LDA) Effect Size (LEfSe) web application (Segata et al., 2011). An alpha level of 0.05 was used for all tests, but after FDR-correction, a level of 0.1 was used due to the small sample and exploratory nature of this study.
In vitro monolayer permeability assay after exposure of epithelial cells to Mucor
Caco-2 human colonic epithelial cell lines were cultured in MEM (Gibco) media supplemented with 10% heat-inactivated FBS (Gibco) in the presence of 1% penicillin/streptomycin (Gibco), 1% MEM Non-Essential Amino Acid (Gibco) and were grown at 37°C in a 5% CO2 humidified incubator. For epithelial monolayer development, 2.5 × 105 Caco-2 cells were seeded into 24-well trans-well plates with a pore size of 0.4 mm and were maintained under standard conditions. Monolayers were cultured until tight junctions formed between cells for three weeks. Caco-2 cell monolayers then were exposed to Mucor (AzMucho and 1006PhL). After 20 h of exposure, fluorescein isothiocyanate (FITC)-labeled dextran (Sigma) (50 ng/ml) was added to the upper chamber of the trans-wells, and monolayers were incubated for additional 4 h. Fluorescence was measured in the upper and lower chambers of the trans-well plates using a fluorometer at an excitation wavelength of 485 nm and an emission wavelength of 530 nm to determine the percent flux of FITC-dextran across the monolayer (Maharshak et al., 2015). Trypsin was used as a positive control for enhanced permeability. Three biological repeats were included for a statistic analysis (t-test with Welch’s correction) and the result was visualized by using PRISM (GraphPad).
Results
Isolation of a Mucor circinelloides strain from an unopened yogurt within the recall range
In 2013, a yogurt company voluntarily withdrew its yogurt products produced from a factory located in Twin Falls, ID after more than 300 consumers had reported gastrointestinal illnesses to the US FDA. In a previous study, we isolated a M. circinelloides strain from an opened yogurt container within the recall range and analyzed the virulence, whole genome sequences, and interactions of the strain with mammalian hosts (Lee et al., 2014).
For this study, a consumer in Phoenix, AZ, who was affected by the outbreak with associated illness, provided an unopened yogurt sample that was within the recall range and had been stored in a household freezer for about a year since the incident (Supplementary data Text S1). Subsequently, we isolated a M. circinelloides strain from this yogurt sample and named it AzMucho (see Materials and Methods). AzMucho was as virulent as the previous strain, Mucho (also isolated from a contaminated yogurt) in a murine systemic mucormycosis model (AzMucho vs Mucho, P = 0.8342) (Supplementary data Fig. S1). For phylogenic identification of AzMucho, a multi-locus sequence typing (MLST) analysis was performed with three of the genomic loci used for the fungal tree of life project (James et al., 2006). The loci include an intergenic spacer region (ITS) between two ribosomal sub-unit genes, a large ribosomal subunit gene (LSU rDNA), and an RNA polymerase subunit gene (RPB1). The sequences of the three loci amplified from the AzMucho genomic DNA were indistinguishable from those from Mucho (data not shown; Genbank accession numbers are Kj588204, Kj588205, and Kj588206, respectively) (Lee et al., 2014). Mucho and AzMucho were Mucor circinelloides f. circinelloides and not distinguishable based on this MLST analysis. Based on an analysis of the whole genome, the genome of AzMucho differed by 6899 variants compared to that of Mucho. Comparisons with the whole genome sequences of ATCC1207a (M. circinelloides f. griseocyanus), ATCC1207b (M. circinelloides f. griseocyanus), CBS277.49 (M. circinelloides f. lu-sitanicus), and 1006PhL (M. circinelloides f. circinelloides) showed 69433 shared variants separated Mucho and AzMucho from the rest of the sequenced population (Supplementary data Fig. S2). This suggests indicating that Mucho and AzMucho are closely related but distinct isolates and it is likely that at least two isolates, and possibly more, were responsible for the yogurt spoilage. Alternatively, both could be the same isolate after accumulation of different SNPs during vegetative growth.
Ingestion of AzMucho influences bacterial species richness (alpha diversity)
Our initial examination found that AzMucho did not produce the toxin 3NP (Supplementary data Fig. S3), which is known to be produced by some M. circinelloides isolates and other fungi (He et al., 1995; Hollmann et al., 2008). It is therefore possible that the GI discomfort that consumers experienced was associated with an alteration of the microbiota. We investigated the possible effect of ingestion of Mucor on the intestinal microbiota. Alpha diversity of gut microbiota after AzMucho ingestion by male BALB/C mice housed in two independent animal facilities, UTSA and Duke university, were analyzed (Supplementary data Fig. S4). In our previous study, Mucor can survive in the mouse GI tract at least for 10 days and our Mucor colony forming unit (CFU) analysis from the stool also supports it (Supplementary data Fig. S4) (Lee et al., 2014). The stool samples therefore were collected up to day 10 post infection from animals at Duke or day 9 from animals at UTSA. Mice housed at the UTSA facility also ingested heat killed AzMucho or another Mucor strain, 1006PhL.
Cohorts of five mice ingested, by oral gavage, either AzMucho spores (exposed group) or PBS alone (unexposed group) at the Duke facility. Mucor ingestion was not likely affecting overall health of exposed mice based on the body weight monitoring, in which only one mouse experienced weight loss and eventually recovered (Supplementary data Fig. S1B). Fresh stool was collected from the mice daily for ten days, and DNA was extracted from the samples. Bacterial and fungal sequences in the isolated DNA were identified as OTUs utilizing 16S V4 and ITS1 primers, respectively, and Illumina MiSeq sequencing (see Materials and Methods for details). Bacterial and fungal OTU diversity were estimated, through OTU counts and Chao1 indices, and were tested for outliers using Cook’s distance (Cook, 1977; Chao, 1984). The single sample outlier in bacteria and the two outliers in fungi with greatest Cook’s distances were identified for removal prior to alpha diversity analysis.
In analysis of the microbiota of the mice housed at Duke, the overall model for bacterial Chaol index (Fig. 1A left and Supplementary data Table Sl) was significant (F3,94 = 26.12, P < 0.05), exhibiting higher bacterial diversity after ingestion of Mucor and over the course of the 10 day-period. The overall model denotes a summary of the entire linear regression with all factors including day, strain ingested, and the interaction between day and strain. Both day and exposure status were significant predictors of the Chao1 index. However, there was no significant interaction between day and exposure status. The overall model for number of bacterial OTUs (Fig 1A right and Supplementary data Table S1) was significant (F3,94 = 19.76, P < 0.05). Both day and exposure status were significant predictors of number of OTUs, as seen in the Chao1 index. There was also a significant interaction between day and exposure status. These results show that exposed mice have a higher overall bacterial diversity than their unexposed counterparts. It is important to note that both groups of mice experienced an increase in diversity with time. This increase, even in unexposed mice, may be because the mice used were juveniles (four to six weeks old upon arrival at the Duke animal facility), and thus their microbiota may still have been developing. However, it appears that exposure to Mucor altered the natural growth of the developing microbiota by lowering the rate at which bacterial alpha diversity increased in these mice.
Fig. 1. Bacterial alpha diversity in stool samples from mice exposed to M. circinelloides through ingestion and unexposed mice during ten days of observation.
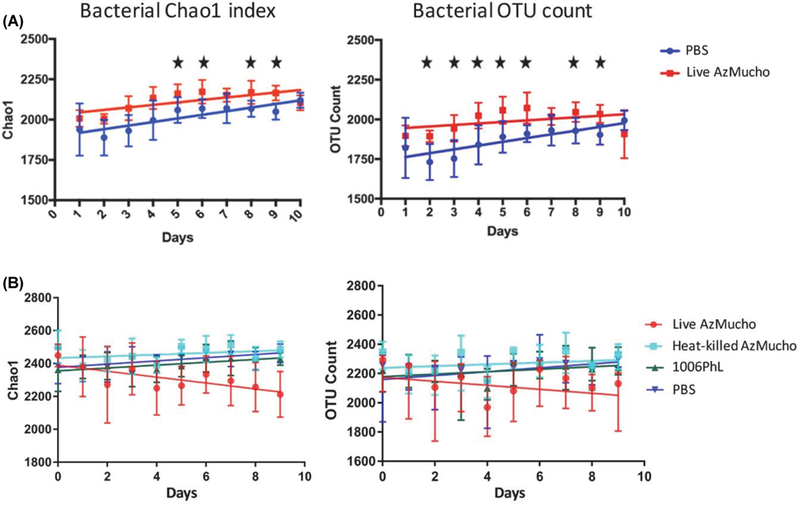
Alpha diversity was measured using Chao-1 index (left) and number of bacterial OTUs (right) present. The bacterial alpha diversity was measured from mice housed at the Duke facility (A) and the UTSA facility (B). Live AzMucho ingested mice showed a higher diversity than the unexposed mice at the Duke; on the other hand, live AzMucho ingested mice showed a lower diversity than the unexposed mice at the UTSA facility. Ingestion of heat-killed AzMucho and the 1006PhL strain did not alter alpha diversity compared to the unexposed mice. Standard error bars are shown. Significant days (identified in Supplementary data Table S1, P < 0.05) with the data from the mice at Duke are denoted with stars.
An independent experiment was performed in the UTSA facility. At day 0, mice in all cohorts exhibited similar alpha diversity, indicating that the changes presented here are reflective of the fungal spore ingestions (data not shown). Interestingly, the overall model for bacterial Chao1 index when comparing live AzMucho with the PBS control (Fig. 1B) was significant (F3,94 = 12.05, P < 0.05), exhibiting lower bacterial diversity after ingestion of Mucor and over the course of the 10 day-period. Both day and the interaction term between day and strain were significant predictors of the Chao 1 index. Similarly, the overall model for bacterial OTU count when comparing live AzMucho with the PBS control was significant (F3,94 = 7.801, P < 0.05), exhibiting lower bacterial diversity after ingestion of Mucor (Fig. 1B). Both day and the interaction term between day and strain were significant predictors of the number of OTUs. These results indicate that Mucor ingestion can cause a shift of bacterial abundance in either direction, dependent on the facility and/or animal cohorts. It is noteworthy that bacterial alpha diversity was increased in the microbiota of the mice housed at Duke but was decreased in mice housed at UTSA. Similar alpha diversity change variations have been observed in patients with Crohn’s disease, in which fungal alpha diversity was increased, decreased, or unaltered when compared to healthy control groups, depending on the cohorts in the studies (Ott et al., 2008; Li et al., 2014; Hoarau et al., 2016; Liguori et al., 2016; Sokol et al., 2017; Miyoshi et al., 2018). In this study, it is possible that the differences in diet (different food for animals) and environment may have affected the direction of bacterial alpha diversity change. However, this study at least shows that ingestion of AzMucho resulted in changes to bacterial alpha diversity.
We then examined if Mucor needs to be alive to cause bacterial abundance change. The overall model for bacterial Chaol index when comparing live AzMucho to heat killed AzMucho was significant (F3,95 = 16.14, P < 0.05), exhibiting lower bacterial diversity after ingestion of live Mucor than after ingestion of killed Mucor over the course of the 10-day period (Fig. 1B). The interaction term between day and strain was a significant predictor of the Chao1 index. Similarly, the overall model for bacterial OTU count when comparing live AzMucho with heat killed AzMucho was significant (F3,95 = 12.3, P < 0.05), exhibiting lower bacterial diversity after ingestion of live Mucor. The interaction term between day and strain was a significant predictor of the number of OTUs. Interestingly, the ingestion of heat killed AzMucho, although lower than live AzMucho, resulted in a change in alpha diversity compared to that of PBS, where the overall models for bacterial Chao1 and number of bacterial OTUs were significant (F9,95 = 4.775, P < 0.05 and F3,95 = 4.06, P < 0.05, respectively). This indicates that fungal spore component(s) can affect the bacterial microbiota (see Discussion).
Next, two different Mucor strains were compared. The overall model for bacterial Chao1 index when comparing live AzMucho to the 1006PhL strain was significant (F3,95 = 8.271, P < 0.05), exhibiting lower bacterial diversity after ingestion of AzMucho than after the alternative Mucor strain (Fig. 1B left). The interaction term between day and strain was a significant predictor of the Chao1 index. Similarly, the over-all model for bacterial OTU count when comparing live AzMucho with 1006PhL was significant (F3,95 = 7.19, P < 0.05), exhibiting lower bacterial diversity after ingestion of AzMucho. The interaction between day and strain was a significant predictor of the number of OTUs. Every one of the above described bacterial alpha diversity models remained significant after Bonferroni correction for multiple testing (data not shown). It appears that the bacterial alpha diversity changes after Mucor ingestion are likely strain-dependent. Considering all bacterial alpha diversity analyses together, ingestion of live AzMucho resulted in changes in bacterial abundance in two separate animal facilities and in a strain dependent manner. In addition, live Mucor is required to alter the bacterial abundance.
AzMucho ingestion effects on alpha diversity and abundance of fungal species
Considering animals housed at the Duke facility, the overall model for fungal Chao1 index (Fig. 2A) was non-significant (F3,94 = 2.55, P = 0.06), as was the overall model for number of fungal OTUs (Fig. 2B) (F3,94 = 1.32, P = 0.27). As with bacterial diversity, unexposed mice experienced an increase in fungal alpha diversity over time.
Fig. 2. Fungal alpha diversity in stool samples from mice exposed to M. circinelloides through ingestion and unexposed mice during ten days of observation.
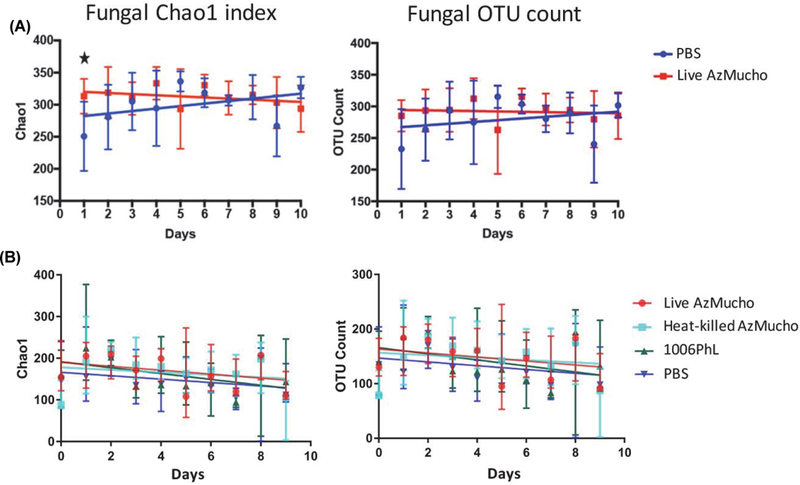
Alpha diversity was measured using Chao-1 index (left) and number of fungal OTUs (right) present. The fungal alpha diversity was measured from mice housed at the Duke facility (A) and the UTSA facility (B). The overall linear model was non-significant in mice at the Duke facility but significant in mice at the UTSA facility. Standard error bars are shown. A significant day (identified in Supplementary data Table S1, P < 0.05) with the data from the mice at Duke is denoted with stars.
However, in a test with animals housed at the UTSA facility, the overall model for fungal Chao1 index when comparing live AzMucho with the PBS control was significant (F3,94 = 3.698, P < 0.05), exhibiting higher fungal diversity after ingestion of Mucor spores over the course of the 10-day period (Fig. 2B). This remained significant after Bonferroni correction for multiple testing. Similarly, the overall model for fungal OTU count when comparing live AzMucho with the PBS control was significant (F3,94 = 2.964, P < 0.05), exhibiting higher fungal diversity (Fig. 2B) after ingestion of Mucor, although this did not remain significant after Bon-ferroni correction.
Interestingly, the overall model for fungal Chao1 index when comparing live AzMucho to heat killed AzMucho was not significant (F3,94 = 1.7, P = 0.1723). Similarly, the overall model for fungal OTU count when comparing live AzMucho with heat killed AzMucho was not significant (F3,95 = 1.213, P = 0.3082). These results indicate that dead AzMucho can also alter mycobiota, though to a lesser extent than live spores. It is therefore possible that cell wall associated fungal components) may be associated with the ability to induce fungal alpha diversity changes. Unlike bacterial alpha diversity, the heat killed AzMucho spores did not alter fungal alpha diversity, where the overall model for fungal Chao1 was non-significant (F3,95 = 2.162, P = 0.098), as was the overall model for fungal OTUs (F3,95 = 1.956, P = 0.126).
The overall model for fungal Chao1 index when comparing live AzMucho to the 1006PhL strain was significant (F3,95 = 3.257, P < 0.05), exhibiting higher fungal diversity after ingestion of AzMucho than after the alternative Mucor strain. Day was a significant predictor of the Chao1 index. This did not remain significant after Bonferroni correction. The over-all model for fungal OTU count when comparing live AzMucho with 1006PhL was marginally different (F3,95 = 2.67, P = 0.05196). This result further support that the shift in microbiota after Mucor ingestion can be strain-dependent. However, changes in fungal alpha diversity and abundance after AzMucho ingestion are seen to a lesser extent when compared to bacterial diversity. The patterns of these alterations in fungal alpha diversity in different Mucor spore conditions are congruent with the changes in bacterial alpha diversity, in which fungal alpha diversity changes are strain-dependent and require live Mucor spores.
Mucor ingestion alters beta diversity and abundance of certain taxa
Dissimilarity of intestinal microbiota of the animals housed at the Duke facility for all samples was visualized by principle coordinate analysis (PCoA) of Bray-Curtis distances (Bray and Curtis, 1957) and statistical significance was determined with the Adonis implementation of permutational analysis of variance (PERMANOVA) (Oksanen et al., 2012). Analysis of bacterial microbiota (Fig. 3A and Supplementary data Fig. S5), when accounting for both live AzMucho ingestion and day, revealed a significant effect of both factors, where bacteria beta diversity was increased upon exposure and through the course of 10 days (P = 0.016 for AzMucho ingestion and P = 0.001 for day). Analysis of fungal microbiota (Fig. 3B and Supplementary data Fig. S6), when accounting for both AzMucho ingestion and day, also revealed a significant effect of both factors, where similar to the bacterial beta diversity, the fungal beta diversity was also increased upon exposure and through the course of 10 days (P = 0.029 for AzMucho ingestion and P = 0.001 for day).
Fig. 3. Beta diversity between exposure groups at all time points.
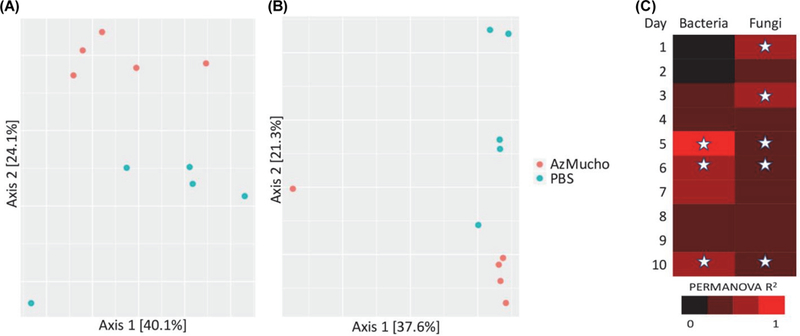
Bray-Curtis dissimilarity was visualized by PCoA for (A) bacterial and (B) fungi on day 6 with the data obtained from mice at Duke. (C) Effect size of Mucor exposure by live AzMucho for individual time points was calculated using PERMANOVA. Significant days (identified in Supplementary data Table S2, P < 0.05) are denoted with stars.
PERMANOVA analyses were then restricted to each individual day to explore on which specific days there was a significant effect of live AzMucho ingestion on beta diversity (Supplementary data Table S2 and Fig. 3C). There was a significant effect for bacterial samples on days 5, 6, and 10, and there was a significant effect for fungal samples on days 1, 3, 5, 6, and 10. These results have some overlap with the days of interest found during alpha diversity analysis and support the hypothesis that ingestion of AzMucho induced an alteration in the gastrointestinal microbiota of these mice. For ease of viewing, beta diversity plots were limited to individual days. Day 6 for both bacteria and fungi is shown for the purposes of this report (Fig. 3). Visualization of all days simultaneously and individually are available in supplemental data (Supplementary data Figs. S5 and S6).
In tests with animals housed at the UTSA facility, there was a significant effect of live AzMucho ingestion for the bacterial population on days 6 and 7, and on the fungal population on day 4 and 5 (Supplementary data Figs. S7 and S8). Interestingly, the ingestion of heat-killed AzMucho also showed a significant effect on bacterial population on days 3 and 4, and on fungal population on days 1, 4, and 7. Although heat-killed AzMucho did not significantly alter bacterial and fungal alpha diversities compared to live AzMucho, the dead Mucor spores can shift the bacterial and fungal populations (Supplementary data Fig. S7). This result further supports that fungal cell wall component(s) may contribute to the change in microbiota. Another strain 1006PhL, however, only had a significant effect on fungal population on day 1 (Supplementary data Fig. S7), which is in accordance with the bacterial and fungal alpha diversity analyses after AzMucho or 1006PhL ingestion. This further suggests that there is a Mucor strain dependent alteration of microbiota. For example, the 1006PhL strain may not encode gene(s) responsible for the microbiota change, or it may express the gene(s) at a lower level compared to AzMucho. Thus, the effect of 1006PhL ingestion resulted in subtle shift in microbiota.
Relative bacterial taxa composition of the intestinal microbiota of the animals housed at the Duke facility was compared between unexposed and exposed (live AzMucho compared to PBS ingestion) mice on day 8, based on the comparison of alpha diversity on individual days. Samples from days 5, 6, and 9, which also showed significant differences in alpha diversity, and all other time points were also subjected to compositional analysis (Supplementary data Table S3). By day 8, mice which ingested live AzMucho had significantly higher levels of the genus Bacteroides (P = 0.016) and significantly lower levels of the genus Akkermansia, particularly the species A. muciniphila (P = 0.016), than their unexposed counterparts (Fig. 4). Interestingly, Bacteroides species are known to be involved in inflammation in the gut (Wu et al., 1998; Saitoh et al., 2002; Bamias et al., 2007) and A. muciniphila is associated with healthy guts (Everard et al., 2013; Reunanen et al., 2015). Therefore, the shift in bacterial species suggests an unhealthy shift in the guts of exposed mice. In a separate experiment performed with animals housed in the UTSA facility, the genus Bacteriodes (P = 0.028) was also significantly higher by day 7 in the animals which ingested AzMucho. However, this change was not observed in animals which ingested heat-killed AzMucho or 1006PhL. The bacterial taxa changes in each group are listed in Supplementary data Tables S4, S5, and S6.
Fig. 4. Shift in bacterial microbiota composition in exposed mice on day 8 and in fungal microbiota compostion in exposed mice day 10.
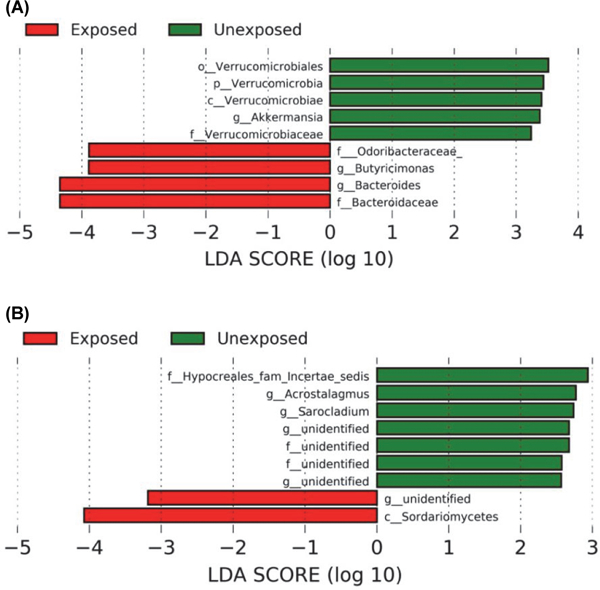
(A) LEfSe analysis was used to determine which bacterial taxonomic units showed a significant difference between groups before FDR-correction. The data were obtained from the mice housed at Duke. Further bacteria taxa analysis data are available in Supplementary data. Bacterial taxa enriched in the exposed group were indicated with a negative LDA score (red bars) and bacterial taxa abundant in unexposed group were indicated with a negative LDA score (green bars). (B) LEfSe analysis was used to determine which fungal taxonomic units showed a significant difference between groups before FDR-correction. The data were obtained from the mice housed at Duke. Further fungal taxa analysis data are available in Supplementary data. Fungal taxa enriched in the exposed group were indicated with a negative LDA score (red bars) and fungal taxa abundant in unexposed group were indicated with a negative LDA score (green bars). The LDA scores indicates the effect size of each differently enriched taxon. LDA stands for Linear Discriminant Analysis.
Because fungal alpha diversity differed only on day 1 in animals housed at the Duke facility, we compared the relative fungal taxa composition of the intestinal microbiota between unexposed and exposed (live AzMucho and PBS ingestion) mice on day 10, which served as the endpoint of data collection. Samples from day 1 and all other time points (Supplementary data Table S7) were also subjected to compositional analysis. By day 10, exposed mice showed significantly lower levels of the genera Acrostalagmus (P = 0.026) and Sarocladium (P = 0.047) than their unexposed counterparts (Fig. 5). The fungal taxa changes seen in the animals housed in the UTSA facility are listed in Supplementary data Tables S8, S9, and S10. Interestingly, these analyses did not reveal a significant difference in Mucor between the two groups. This may be due to Mucor spores having undergone lysis in the stomach, although some are capable of passage through the GI tract (Lee et al., 2014). Alternatively, Mucor spores are spread throughout the GI tract, so a single gavage of Mucor spores might not produce an amount of DNA sufficient to reach statistical significance in this taxonomic analysis.
Fig. 5. Exposure to AzMucho increases the permeability of epithelial cell monolayers.
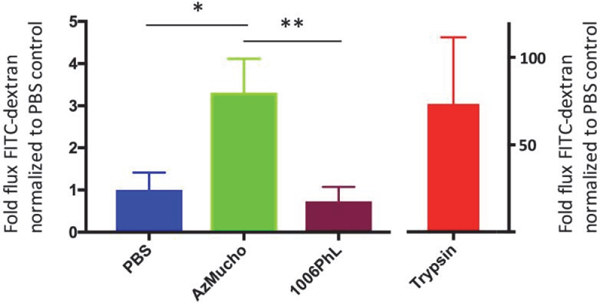
CaCo-2 cell monolayers were co-cultured with spores of AzMucho or 1006PhL or PBS. The flux of FITC-dextran across the monolayers was significantly higher when they were exposed to AzMucho compared to PBS (* P = 0.0217). However, when the monolayers were exposed to 1006PhL, the difference is not significant compared to PBS (P = 0.4369). AzMucho caused greater permeability compared to 1006PhL (** P = 0.0187). Trypsin treatment groups were served as a positive control. Each group was tested at least 3 times and two independent experiments exhibited the similar results. Data were visualized as mean ± SD. Statistical significance was measured using Welch’s t test.
Exposure to AzMucho increases monolayer permeability in epithelial cells
To investigate if AzMucho can permeabilize the epithelial monolayer barrier, CaCo-2 cell monolayers were exposed to AzMucho (105 spores) and 1006PhL (105 spores) along with PBS as a control (Fig. 6). Trypsin was also used as a positive cell permeabilizer. Upon exposure to AzMucho, the permeability of the CaCo-2 monolayers increased by approximately 3.3-fold compared to PBS control, which is significantly higher (P = 0.0217). These results suggest that AzMucho may have an ability to cause disruption of the enteric epithelial barrier. Interestingly, exposure to the 1006PhL strain did not result in an increase in the permeability at the given condition (1006PhL vs PBS, P = 0.4369; 1006PhL vs AzMucho, P = 0.0187). These results are congruent with the relatively lower shift in microbiota by ingestion of 1006PhL compared to that by ingestion of AzMucho.
Discussion
Our results demonstrate a significant change in bacterial alpha diversity in the intestinal microbiota of mice that ingested Mucor. Our results also show shifts in the composition of the microbiota, specifically through changes in the bacteria A. muciniphila and Bacteroides species. Interestingly, both bacterial taxa that differed between groups are known to be associated with gut health. Specifically, Bacteroides species have been implicated as a cause behind the inflammatory antibody response towards the gut microbiota seen in inflammatory bowel disease (IBD) (Saitoh et al., 2002). Bacteroides species have also been shown to be involved in the exacerbation of inflammation in the gut, which is associated with Crohn’s disease, as well as an increase in permeability of intestinal epithelial cells in patients with IBD (Wu et al., 1998; Bamias et al., 2007). Bacteroides species are present at higher levels in exposed mice, implying a shift in the microbial composition towards a potentially more pro-inflammatory environment. A. muciniphila, however, is known to be positively associated with gut health in that this species has been shown to control inflammation and strengthen epithelial barrier integrity within the gut (Everard et al., 2013; Reunanen et al., 2015). A. muciniphila is present at lower levels in exposed mice, thus bolstering the idea that the microbial composition has experienced a shift towards a potentially unhealthy environment. However, shifts in bacterial taxa were variably significant after FDR-correction, and no shift at any fungal taxa level maintained significance after FDR-correction (see Supplementary data Tables for FDR-corrections). This may be due in part to the small sample size (n = 5 per group), and thus limited statistical power, used. In addition, it is also possible that a single gavage of Mucor would not be sufficient to cause a serious dysbiosis, rather than the moderate alteration in the microbiota (bacteria and fungi) compositions seen in our experiments. The lack of significant alteration in abundance of Mucor species between exposed and unexposed animals indicates this as a possibility.
Interestingly, ingestion of heat killed AzMucho also resulted in changes in beta diversity. This may suggest that fungal cell wall associated factor(s) can affect the intestinal microbiota. The genome of Mucho also encodes gene clusters involved in production of potentially harmful toxins (Lee et al., 2014), and it is therefore possible that Mucor can produce cell wall associated toxin(s) that is detrimental to GI tract health. The overall lack of microbiota shift seen when using another Mucor strain, 1006PhL, can be explained by the possibility that the isolate may not produce the causal toxin(s), unlike Mucho or AzMucho. This observation is also in accordance with the findings that AzMucho may cause a leaky gut phenotype by testing the permeability assay using CaCo-2 cell tight junctions. On the other hand, the 1006PhL strain did not cause increased permeability (Fig. 6). Leaky gut or enhanced permeability of intestinal epithelial barrier is a common symptom in IBD and IBS (Odenwald and Turner, 2013). It is therefore possible that Mucor may play a role in the development of symptoms associated with GI discomfort. However, it is not clear that Mucor produces toxin(s) that would result in microbiota dysbiosis and/or disruption of enteric epithelial cell barrier in animals. Nevertheless, this strain dependent microbiota shift will provide a platform to identify how Mucor causes the microbiota shift.
Despite the small sample size used, overall, the examination of the composition of the gut microbiota of these mice supports the hypothesis that ingestion of AzMucho induces an alteration in the bacterial component of the gut in the days following ingestion. Our findings therefore suggest that consuming Mucor can alter the microbiota of the gut. It is possible that the alteration of the microbiota caused by Mucor might not be enough to be translated into a dysbiosis in the GI tracts of humans. Alternatively, Mucor might have affected processes in the stomach and/or small intestine that may not be apparent from the stool DNA analysis. Interestingly, in our separate study to analyze the bacterial and fungal microbiota in South Korean patients with Crohn’s disease (n = 35) compared to healthy individuals (n = 52), abundance of Mucor was found to be significantly higher in the patient group (Serrano, Mueller, Huh, Choi, and Lee, unpublished data). Interestingly however, Mucor was not found to be significantly increased in studies of patients with CD from the United Stated and European countries (Hoarau et al., 2016; Sokol et al., 2017).
The change in bacterial alpha diversity in our experiments is consistent with previous studies relating to diseases, such as Crohn’s disease, ulcerative colitis (UC), irritable bowel syndrome (IBS), and anorexia nervosa, in which the disease state is associated with altered alpha diversity and shifts in microbiota composition of the gut (Kleiman et al., 2015; Hoarau et al., 2016; Sokol et al., 2017). Specifically, these studies have demonstrated alterations in the prevalence of bacteria and fungi, such as Ruminococcaceae and Enterobacteriaceae genera, C. tropicalis, and Saccharomyces cerevisiae, which are linked to intestinal disorders (Kleiman et al., 2015; Hoarau et al., 2016; Sokol et al., 2017) (Mueller, Serrano, and Lee, unpublished data). This then suggests that ingestion of AzMucho alters the stability of the gut microbiota, possibly pushing towards a less healthy state by inducing an alteration of the resident bacteria. It may be, if this alteration is translatable to humans, that the gastrointestinal illness experienced by consumers of Mucor-contaminated yogurt might have been, in part, due to a disruption of their gut microbiota. This would be consistent with previously established associations between gut dysbiosis and gastrointestinal discomfort (Chichlowski and Rudolph, 2015; Botschuijver et al., 2017).
It is well known that induced alterations of the gut bacteria both affect gut health and often result in gastrointestinal discomfort, such as in the case of fungal overgrowth during the course of antibiotic use (Erdogan and Rao, 2015) and antibiotic-associated diarrhea caused by bacterial pathogens such as Clostridium difficile (Young and Schmidt, 2004). Intestinal fungi have the ability to cause inflammation in the gut. For example, improper recognition of fungi in the GI tract due to mutations in the gene encoding Dectin-1, a C-type lectin receptor, results in exacerbation of colitis (Iliev et al., 2012). A dysbiosis of the enteric mycobiota is linked to inflammation in Crohn’s disease (Li et al., 2014). In particular, modulation of purine metabolism by S. cerevisiae has been shown to be associated with worsening colitis and increased gut barrier permeability in a murine model (Chiaro et al., 2017). Candida colonization increases pro-inflammatory cytokine production that is associated with IBD (Kumamoto, 2011). Previous studies have further demonstrated an interaction between fungi and bacteria in the gut, and specifically, how that interaction relates to diseases of the gut (Hoarau et al., 2016; Sokol et al., 2017). Antifungal drug derived microbiota dysbiosis results in enhanced colitis and ex-acerbated allergy airway disease in a murine model (Wheeler et al., 2016; Leonardi et al., 2018). Our study, shown in Supplementary data Fig. S9, also supports fungal dysbiosis driven inflammation, in which after fluconazole and fluocytosine treatment the guts of mice secreted higher amounts of inflammatory cytokines, including IL-1b, IL-6, and INFg (see Supplementary data Fig. S9 and legend). Some fungi are also known, but to a lesser extent than bacteria, for their usefulness in promoting gut health. For example, Saccharomyces boulardii, a fungus which is commercially available as a probiotic supplement, is utilized in part to reduce the risk of development of antibiotic-associated diarrhea (Szajewska and Mrukowicz, 2005).
It is clear that both fungi and bacteria play important roles in the gut, and that maintaining a harmony between the two should be kept in mind when considering the prevention and treatment of gastrointestinal illness. Despite this, the role of fungi in the gut and in food-borne illness has been vastly understudied. It is evident then that more consideration should be given, and more studies conducted, regarding the role of the mycobiota in gut health.
Supplementary Material
Acknowledgements
We are indebted to Ian Carroll for insightful discussion, and to Anna Averette and Zanetta Chang for technical support. We thank John Rawls, Lawrence David, Ian Carroll, and Emily Bulik-Sullivan for critical reading the manuscript and providing constructive critiques. We thank Kathleen Dodge for her contribution of the yogurt analyzed in this study.
This work was funded by NIH/NIAID R03 AI119617, UTSA Start-up research fund, and UTSA GREAT (co-PI)_to S.C.L., by NIH grants GM100806 and AI080579 to Y.W., by NIH/NIAID R37 MERIT award AI39115-20 to J.H, by NIH/NIAID grant R01 AI065728-01 to N.P.K., and by Astellas Award CRES-15601 to S.C.L. and J.H.
References
- Aboltins CA, William ABP, and Solano TR 2006. Fungemia secondary to gastrointestinal Mucor indicus infection. Clin. Infect. Dis. 42, 154–155. [DOI] [PubMed] [Google Scholar]
- Alston TA, Mela L, and Bright HJ 1977. 3-Nitropropionate, the toxic substance of Indigofera, is a suicide inactivator of succinate dehydrogenase. Proc. Natl. Acad. Sci. USA 74, 3767–3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamias G, Okazawa A, Rivera-Nieves J, Arseneau KO, De La Rue SA, Pizarro TT, and Cominelli F 2007. Commensal bacteria exacerbate intestinal inflammation but are not essential for the development of murine ileitis. J. Immunol. 178, 1809–1818. [DOI] [PubMed] [Google Scholar]
- Benjamini Y and Hochberg Y 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300. [Google Scholar]
- Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, and Caporaso JG 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–U11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botschuijver S, Roeselers G, Levin E, Jonkers DM, Welting O, Heinsbroek SEM, de Weerd HH, Boekhout T, Fornai M, Masclee AA, et al. 2017. Intestinal fungal dysbiosis is associated with visceral hypersensitivity in patients with irritable bowel syndrome and rats. Gastroenterology 153, 1026–1039. [DOI] [PubMed] [Google Scholar]
- Bray JR and Curtis JT 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monograph. 27, 326–349. [Google Scholar]
- Brouillet E, Jacquard C, Bizat N, and Blum D 2005. 3-Nitropro-pionic acid: a mitochondrial toxin to uncover physiopathological mechanisms underlying striatal degeneration in Huntington’s disease. J. Neurochem. 95, 1521–1540. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A 1984. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11, 265–270. [Google Scholar]
- Chayakulkeeree M, Ghannoum MA, and Perfect JR 2006. Zygomycosis: the re-emerging fungal infection. Eur. J. Clin. Microbiol. Infect. Dis. 25, 215–229. [DOI] [PubMed] [Google Scholar]
- Chiaro TR, Soto R, Zac Stephens W, Kubinak JL, Petersen C, Gogokhia L, Bell R, Delgado JC, Cox J, Voth W, et al. 2017. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci. Transl. Med. 9, 9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichlowski M and Rudolph C 2015. Visceral pain and gastro-intestinal microbiome. J. Neurogastroenterol. Motil. 21, 172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook RD 1977. Detection of influential observation in linear regression. Technometrics 19, 15–18. [Google Scholar]
- Devaraj S, Hemarajata P, and Versalovic J 2013. The human gut microbiome and body metabolism: implications for obesity and diabetes. Clin. Chem. 59, 617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. [DOI] [PubMed] [Google Scholar]
- Erdogan A and Rao SSC 2015. Small intestinal fungal over-growth. Curr. Gastroenterol. Rep. 17, 16. [DOI] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Art C, Bindels LB, Guiot Y, Derrien MMN, Muccioli GG, Delzenne NM, et al. 2013. Crosstalk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 110, 9066–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA. 2013. Chobani, Inc. voluntarily recalls greek yogurt because of product concerns.
- Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Program NCS, et al. 2013. Human skin fungal diversity. Nature 498, 367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gweon HS, Oliver A, Taylor J, Booth T, Gibbs M, Read DS, Griffiths RI, Schonrogge K, and Bunce M 2015. PIPITS: an automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods Ecol. Evol. 6, 973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Zhang S, Qian F, and Zhang C 1995. Delayed dystonia with striatal CT lucencies induced by a mycotoxin (3-nitropro-pionic acid). Neurology 45, 2178–2183. [DOI] [PubMed] [Google Scholar]
- Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, Neut C, Vermeire S, Clemente J, Colombel JF, et al. 2016. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. mBIO 7, e0.1250–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmann M, Razzazi-Fazeli E, Grajewski J, Twaruzek M, Sulyok M, and Bohm J 2008. Detection of 3-nitropropionic acid and cytotoxicity in Mucor circinelloides. Mycotoxin Res. 24, 140–150. [DOI] [PubMed] [Google Scholar]
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. 2012. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336, 1314–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, et al. 2006. Reconstructing the early evolution of fungi using a six- gene phylogeny. Nature 443, 818–822. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Shajib MS, Manocha MM, and Khan WI 2012. Investigating intestinal inflammation in DSS-induced model of IBD. J. Vis. Exp. 60, 3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman SC, Bulik-Sullivan EC, Glenny EM, Zerwas SC, Huh EY, Tsilimigras MCB, Fodor AA, Bulik CM, and Carroll IM 2017. The gut-brain axis in healthy females: lack of significant association between microbial composition and diversity with psychiatric measures. PLoS One 12, e0170208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman SC, Watson HJ, Bulik-Sullivan EC, Huh EY, Tarantino LM, Bulik CM, and Carroll IM 2015. The intestinal microbiota in acute anorexia nervosa and during renourishment: relationship to depression, anxiety, and eating disorder psycho-pathology. Psychosom. Med. 77, 969–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koljalg U, Larsson KH, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, Erland S, Holland K, Kjoller R, Larsson E, et al. 2005. UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 166, 1063–1068. [DOI] [PubMed] [Google Scholar]
- Kumamoto CA 2011. Inflammation and gastrointestinal Candida colonization. Curr. Opin. Microbiol. 14, 386–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar SP, Lukaszewicz JM, Persad KA, and Reinhardt JF Rhinocerebral Mucor circinelloides infection in immunocompromised patient following yogurt ingestion. Del. Med. J. 86, 245. [PubMed] [Google Scholar]
- Lee SC, Billmyre RB, Li A, Carson S, Sykes SM, Huh EY, Mieczkowski P, Ko DC, Cuomo CA, and Heitman J 2014. Analysis of a food-borne fungal pathogen outbreak: virulence and genome of a Mucor circinelloides isolate from yogurt. mBIO 5, e01390–01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi I, Li X, Semon A, Li D, Doron I, Putzel G, Bar A, Prieto D, Rescigno M, McGovern Dermot PB, et al. 2018. CX3CR1+ mononuclear phagocytes control immunity to intestinal fungi. Science 359, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wang C, Tang C, He Q, Li N, and Li J 2014. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn’s disease. J. Clin. Gastroenterol. 48, 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguori G, Lamas B, Richard ML, Brandi G, da Costa G, Hoff-mann TW, Di Simone MP, Calabrese C, Poggioli G, Langella P, et al. 2016. Fungal dysbiosis in mucosa-associated microbiota of Crohn’s disease patients. J. Crohns Colitis 10, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharshak N, Huh EY, Paiboonrungruang C, Shanahan M, Thurlow L, Herzog J, Djukic Z, Orlando R, Pawlinski R, Ellermann M, et al. 2015. Enterococcus faecalis gelatinase mediates intestinal permeability via protease-activated receptor 2. Infect. Immun. 83, 2762–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi J, Sofia MA, and Pierre JF 2018. The evidence for fungus in Crohn’s disease pathogenesis. Clin. J. Gastroenterol. 11, 449–456. [DOI] [PubMed] [Google Scholar]
- Morin-Sardin S, Nodet P, Coton E, and Jany JL 2017. Mucor: A Janus-faced fungal genus with human health impact and industrial applications. Fungal Biol. Rev. 31, 12–32. [Google Scholar]
- Muir AD and Majak W 1984. Quantitative determination of 3-nitropropionic acid and 3-nitropropanol in plasma by HPLC. Toxicol. Lett. 20, 133–136. [DOI] [PubMed] [Google Scholar]
- Odenwald MA and Turner JR 2013. Intestinal permeability defects: Is it time to treat? Clin. Gastroenterol. Hepatol. 11, 1075–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin P, O’Hara R, Simpson G, Solymos P, Stevens M, and Wagner H 2012. Community ecology package R package version 2.0–2. R Development Core Team, http://www.r-project.org/. [Google Scholar]
- Ott SJ, Kühbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, Drews O, Weichert W, Timmis KN, and Schreiber S 2008. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scan. J. Gastroenterol. 43, 831–841. [DOI] [PubMed] [Google Scholar]
- RCoreTeam. 2017. R: a language and environment for statistical computing.
- Reunanen J, Kainulainen V, Huuskonen L, Ottman NA, Belzer C, Huhtinen H, de Vos WM, and Satokari RM 2015. Akker-mansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Appl. Environ. Microbiol. 81, 3655–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden MM, Zaoutis TE, Buchanan WL, Knudsen TA, Sarki- sova TA, Schaufele RL, Sein M, Sein T, Chiou CC, Chu JH, et al. 2005. Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clin. Infect. Dis. 41, 634–653. [DOI] [PubMed] [Google Scholar]
- Rodriguez MM, Perez D, Chaves FJ, Esteve E, Marin-Garcia P, Xifra G, Vendrell J, Jove M, Pamplona R, Ricart W, et al. Obesity changes the human gut mycobiome. Scientific Rep. 5, 14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S, Noda S, Aiba Y, Takagi A, Sakamoto M, Benno Y, and Koga Y 2002. Bacteroides ovatus as the predominant commensal intestinal microbe causing a systemic antibody response in inflammatory bowel disease. Clin. Diagn. Lab. Immunol. 9, 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, and Huttenhower C 2011. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder AB, Churey JJ, and Worobo RW 2016. Characterization and control of Mucor circinelloides spoilage in yogurt. Int. J. Food Microbiol. 228, 14–21. [DOI] [PubMed] [Google Scholar]
- Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, et al. 2017. Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora JW, Chang Y, Benny GL, Lazarus K, Smith ME, Berbee ML, Bonito G, Corradi N, Grigoriev I, Gryganskyi A, et al. 2016. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108, 1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajewska H and Mrukowicz J 2005. Meta-analysis: non-pathogenic yeast Saccharomyces boulardii in the prevention of antibiotic-associated diarrhoea. Aliment. Pharmacol. Ther. 22, 365–372. [DOI] [PubMed] [Google Scholar]
- USAToday. 2013. FDA receives dozens of reports of illness from yogurt.
- Walters WA, Pirrung M, Peña AG, Huttley GA, Zaneveld J, Kuczynski J, Knights D, Bittinger K, Costello EK, Turn-baugh PJ, et al. 2010. QIIME allows analysis of high-through-put community sequencing data. Nat. Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, Brown J, Funari VA, Wang HL, Crother TR, et al. 2016. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe 19, 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Lim KC, Huang J, Saidi RF, and Sears CL 1998. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc. Natl. Acad. Sci. USA 95, 14979–14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young VB and Schmidt TM 2004. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J. Clin. Microbiol. 42, 1203–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


