Abstract
Background
The present meta-analysis examined the diagnostic accuracy of T2 Candida for candidiasis.
Methods
The literature databases, such as PubMed, Embase, DVIO, Cochrane library, Web of Science, and CNKI, were searched on T2 Candida detection.
Results
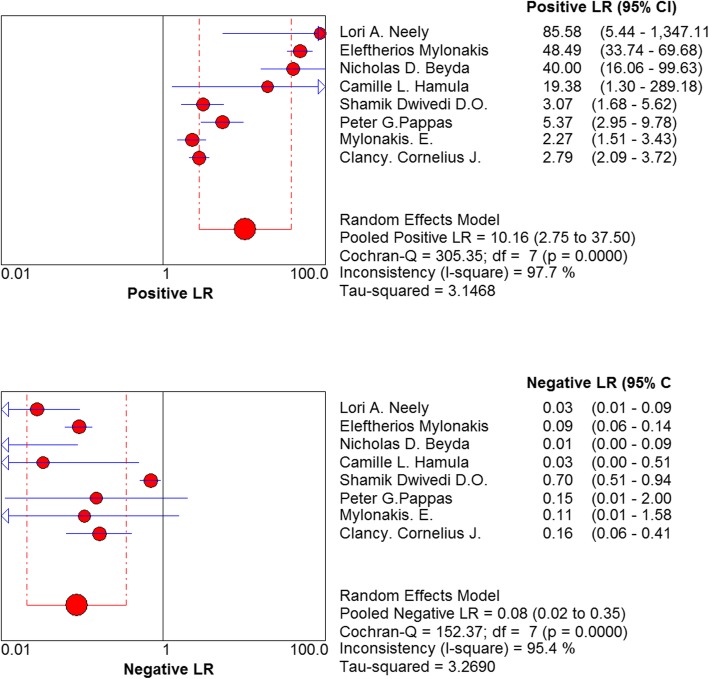
A total of 8 articles, comprising of 2717 research subjects, were included in the study. The pooled sensitivity and specificity were 0.91 (95% confidence interval (CI): 0.88–0.94) and 0.94 95% CI: 0.93–0.95), respectively. The pooled positive likelihood ratio and negative likelihood ratio was 10.16 (95% CI: 2.75–37.50) and 0.08 (95% CI: 0.02–0.35), respectively. The combined diagnostic odds ratio is 133.65 95% CI: 17.21–1037.73), and the AUC of SROC is 0.9702 [(SE = 0.0235), Q* = 0.9201(SE = 0.0381)].
Conclusions
The current evidence supported that T2 Candida has high accuracy and sensitivity and is of major clinical significance in the diagnosis of Candida infection.
Keywords: T2 Candida, Candida infection, Diagnosis, Meta-analysis
Background
Candidiasis is a major medical problem in the twenty-first century. The hospital-acquired candidiasis (candidemia and other forms of invasive candidiasis) is a life-threatening disease in patients with low immune function and severe illness [1, 2]. Presently, it is the fourth leading cause of nosocomial infections in the USA. Patients with candidiasis have about 50% mortality, and each incident would cause an extra healing cost of >$40,000 [3, 4]. In summary, despite the use of several antifungal agents, candidiasis is generally considered as a disease associated with medical advances and a leading cause of morbidity and mortality in the healthcare environment. This phenomenon might be attributed to the difficulty in diagnosing candidiasis and challenges in administering earlier and adequate antifungal therapy [5].
The delay in diagnosis and treatment of candidiasis is closely linked to the increase in mortality and healthcare costs, and hence, a rapid and effective detection method is an urgent requirement. Therefore, finding a rapid and sensitive diagnostic method that can directly test the entire clinical sample is essential. The technology known as T2 Candida investigated in the present study is expected to solve this problem.
The T2Candida Panel (T2 Biosystems) is an FDA-approved rapid diagnostic method that can detect candida pathogens in whole blood specifically and sensitively without the need for culture or nucleic acid extraction steps [6]. It uses T2MR technology [7]. The detecting density of T2MR is 1 colony forming unit (CFU)/mL of whole blood [6, 8]. Herein, we aimed to evaluate the diagnostic accuracy of T2 Candida by conducting a meta-analysis of the data extracted from relevant studies.
Methods
Database and literature search strategies
We searched the PubMed, Embase, OVID, Cochrane library, Web of Science, and China National Knowledge Infrastructure (CNKI) databases up to February 2018 and the official website of T2 Biosystems which developed T2 Candida, using the search phrase (T2 Magnetic Resonance OR T2MR OR T2 Candida OR T2Dx) AND (Candidiasis OR Candida OR Candidemia OR candidaemia OR fungemia). In addition, we scanned the references of all the studies included. The search results show that the latest research was updated in 2018 and the publication years ranged from 2013 to 2018.
Inclusion/exclusion conditions
In order to assess the diagnostic accuracy of T2 Candida, we conducted prospective or retrospective cohort and case-control studies and included those reporting true-positives (TP), false-positives (FP), false-negatives (FN), and true-negatives (TN) or the studies where these variables could be calculated from the other published data.
The studies that were duplicated publications or have no description of the available data and had no restrictions on the language, publication status, year of study, or participants’ ages were excluded. All records were independently selected by authors according to the inclusion criteria, and a consensus was found on each record.
Data extraction
Based on the inclusion conditions, two reviewers extracted the data from the studies separately. A third reviewer resolved the disagreements by discussion and in consultation.
Quality assessment
The risk of bias was assessed independently by two reviewers using the Review Manager 5.2 (Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2012) software according to the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2). There were four parts: patient selection, index test, reference standard, and flow and timing [9]. Each part was assessed in terms of the risk of bias; the first three parts were also rated based on the concerns about applicability. According to the “yes,” “no,” or “uncertain” responses of the relevant landmark questions included in each part, the “risk,” “high,” or “uncertainty” might be determined corresponding to the risk level of bias. The applicability sections are based primarily on the degree of matching with the appraisal problem. Concerns about the applicability are rated as “low,” “high,” or “unclear.” All qualified publications are independently rated by two review authors for quality. The disagreements were resolved by discussion and in consultation with a third reviewer.
Statistical analysis
Statistical analyses were performed using The Meta-disc software, version 1.4 [10]. We extracted the TP, FP, TN, and FN from each study, plotted the summary receiver operating characteristic (SROC) curve, and estimated the absolute diagnostic accuracy of each test. Spearman’s correlation coefficient was assessed to evaluate the heterogeneity of the included studies (threshold effects and non-threshold effects). The random-effect model was used if significant heterogeneity existed among the individual studies. In addition, the fixed-effect model was used to calculate the pooled sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, and the diagnostic odds ratio of the included studies. To assess the publication bias of the included studies, we used STATA 14.0 (StataCorp, College Station, Texas, USA) to perform Deeks’ regression test of funnel plot asymmetry and the Egger’s test.
Results
Summary of the included studies
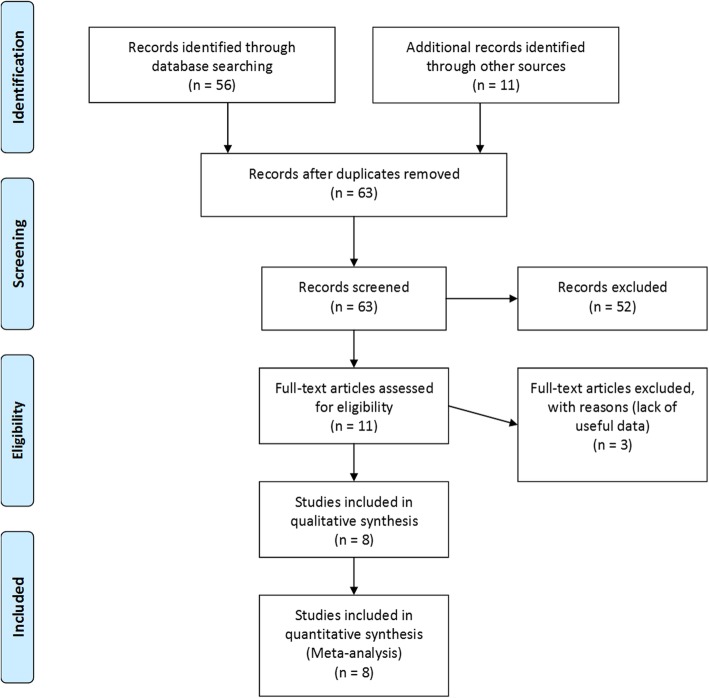
The following literature databases were searched: PubMed, Embase, OVID, Cochrane library, Web of Science, CNKI, and T2 Biosystems web pages, depending on the keywords. A total of 63 related documents were discovered. After reviewing the abstract, 52 studies that did not involve the detection efficiency of Candida were removed. The remaining articles examined the literature and excluded 3 studies due to the lack of experimental sensitivity and specificity. Finally, a total of 8 articles consisting of 2717 research subjects were included [6–8, 11–15] (Fig. 1). The relevant data in each literature were extracted according to the proposed effect-indicators (the study of C. albicans, C. tropicalis, C. parapsilosis, C. krusei, and C. glabrata) (Table 1).
Fig. 1.
Flow Chart of the Study Selection Process
Table 1.
Summary of the included studies
| Diagnostic Test | Author /year | Method | Size | Sample type | Country | TP | FP | FN | TN |
|---|---|---|---|---|---|---|---|---|---|
| T2Candida | Neely 2013 [6] | Culture | 133 | Whole blood | America | 88 | 0 | 2 | 43 |
| T2Candida | Mylonakis 2015 [8] | Culture | 1801 | Whole blood | America | 233 | 29 | 23 | 1516 |
| T2Candida | Beyda 2013 [13] | Culture | 270 | Whole blood | America | 90 | 4 | 0 | 176 |
| T2Candida | Hamula 2016 [14] | Culture | 24 | Whole blood | Athens | 15 | 0 | 0 | 9 |
| T2Candida | Dwivedi 2016 [12] | Culture | 200 | Whole blood | America | 11 | 22 | 17 | 150 |
| T2Candida | Pappas 2015 [11] | Culture | 91 | Whole blood | America & England | 3 | 14 | 0 | 74 |
| T2Candida | Mylonakis 2018 [15] | Culture | 46 | Whole blood | America | 7 | 16 | 0 | 23 |
| T2Candida | Clancy 2018 [7] | Culture | 152 | Whole blood | America | 32 | 37 | 4 | 79 |
Quality of the included study
Being dependent on the results of the QUADAS-2 (Fig. 2), the risk of bias in the study was low, and the clinical applicability was low.
Fig. 2.
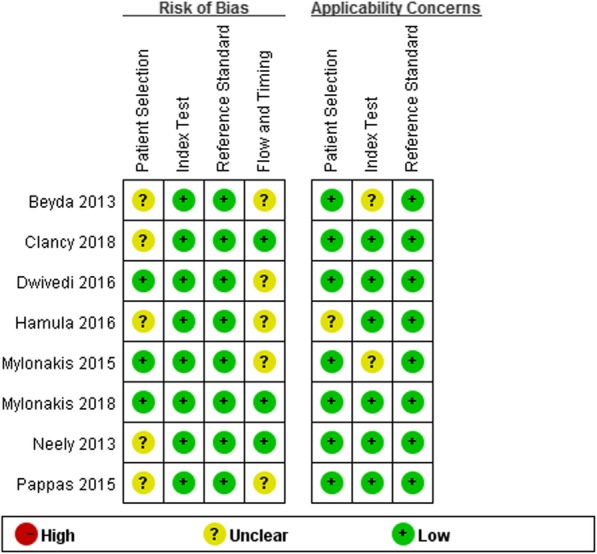
Quality evaluation plot of methodological
Risk analysis of publication bias
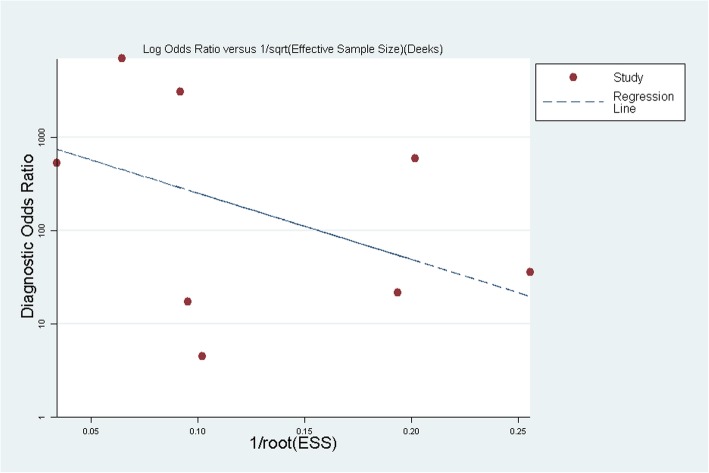
No publication bias was discovered in the funnel plot (Fig. 3). The Egger’s test showed that the publication bias of the study was small (P = 0.404).
Fig. 3.
Publication bias Plot for T2Candida
Threshold effects
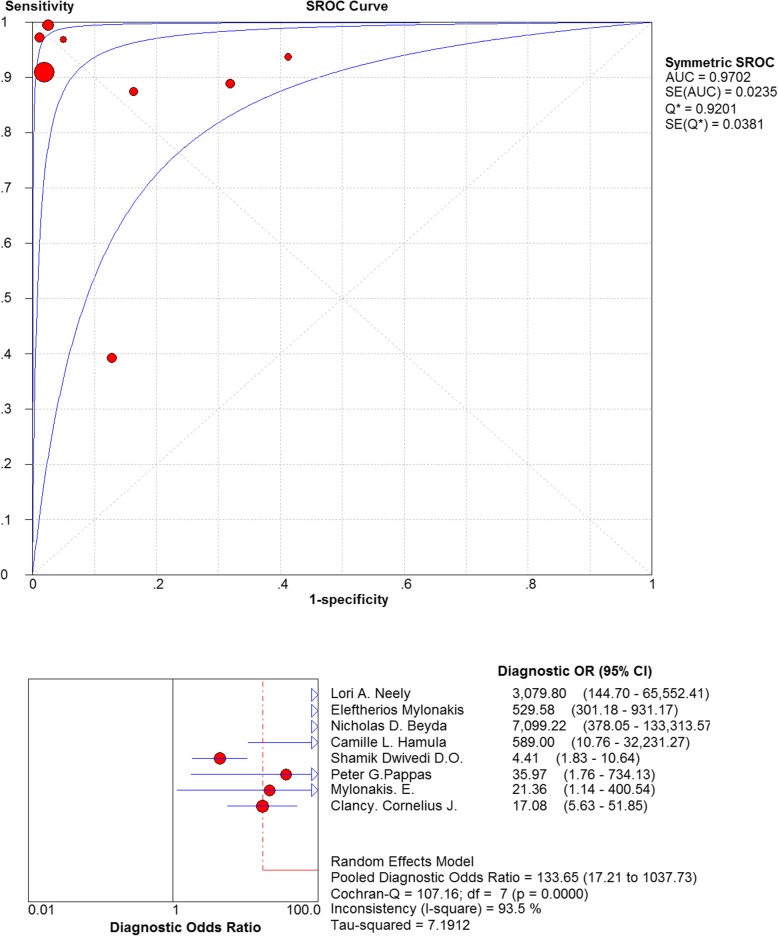
Firstly, the SROC curves were examined, and the scatter plots did not appear as “shoulder-arms” in the image formed on the SROC curve, indicating the absence of threshold effect (Fig. 4). Secondly, the Spearman’s correlation coefficient of the logarithm of sensitivity and specificity was = − 0.500, P-value = 0.207, which also indicated that there was no threshold effect in the included studies. Consequently, there is no threshold effect stated among the articles constituted in the present analysis. In terms of thresholds, each of the included studies was homogeneous.
Fig. 4.
Summary receiver operating curves and forest plot of diagnostic odds ratio for T2Candida
Non-threshold effects
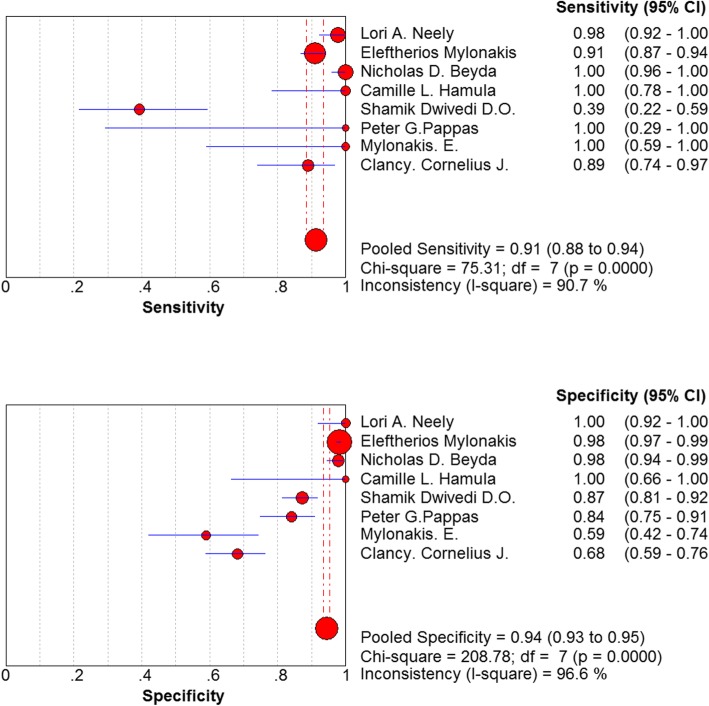
The non-threshold effect test was assessed using the χ2 test. The heterogeneity caused by non-threshold effects was evaluated using a diagnostic odds ratio (DOR). Cochran-Q = 107.16, P < 0.05 indicated a non-threshold effect heterogeneity among the included studies (Fig. 4). Another analysis indicated that the combined sensitivity was: χ2 = 75.31, I2 = 90.7%, the combined specificity was: χ2 = 208.78, I2 = 96.6%. These values confirmed the non-threshold effect of heterogeneity between the included studies (Fig. 5).
Fig. 5.
Forest plots of sensitivity and specificity for T2Candida
Diagnostic accuracy of T2 Candida for candidiasis
The pooled diagnostic ratio was 133.65 (95% confidence interval (CI): 17.21–1037.73) (Fig. 4), and the area under the SROC curve was: AUC = 0.9702 (SE = 0.0235), Q* = 0.9201 (SE = 0.0381). The pooled sensitivity of T2 Candida in the differential diagnosis of candidiasis was 0.91 (95% CI: 0.88–0.94), and the pooled specificity was 0.94 (95% CI: 0.93–0.95) (Fig. 5). The pooled positive likelihood ratio was as follows: 10.16 (95% CI: 2.75–37.50). The pooled negative likelihood ratio was as follows: 0.08 95% CI: 0.02–0.35) (Fig. 6).
Fig. 6.
Forest plots of positive likelihood ratio and negative likelihood ratio for T2Candida
Discussion
Blood culture has been considered as the diagnostic gold standard for invasive candidiasis. Nonculture diagnostic tests, such as an antigen, antibody, or β-D-glucan detection assays and polymerase chain reactions (PCR) are applied as the supplements to cultures in clinical practice.
The overall sensitivity of blood cultures for diagnosing invasive candidiasis is about 50% [16]. Blood cultures have a limitation; the slow turnaround time (median time to positivity of 2–3 days, ranging from 1 to ≥7 days) leads to a delay in diagnosis [17, 18]. Currently, Candida antigen has been detected by the anti-Candida antibody. The combined mannose/anti-mannose antibody assay is the optimal method using Candida antigen and anti-Candida antibody detection. The sensitivity/specificity of diagnosing invasive mannose candidiasis was 58%/93%, and the rate of combined determination was 83% according to a meta-analysis conducted on 14 studies [19]. In another technology, the anti-human IgG detection was 59%/83 and 86%, respectively [20]. However, antigen detection has a limitation as they can easily clear from the bloodstream [21]. The reliability of antibody detection in immunosuppressive hosts is poor, and hence, this assay is rarely used in the USA. The sensitivity and specificity for the diagnosis of invasive candidiasis were 75–80 and 80%, respectively based on a meta-analysis of β-D-glucan studies [22–24]. The true-positive results of β-D-glucan detection are not specific for invasive candidiasis, thereby indicating the possibility of an invasive fungal infection. Thus, the detection of β-D-glucan has poor specificity and sensitivity. In a recent meta-analysis, the sensitivity and specificity of PCR for suspecting invasive candidiasis were 95 and 92%, respectively [25], and among the putative invasive candidiasis, the sensitivities of PCR and blood culture were 85 and 38%, respectively. A major limitation of PCR studies is the lack of standardized methodologies and multicenter validation of the assay [5].
Candida sp. is a conditional pathogen that infects the body and can cause various diseases. It is a common infection in hospitals. According to the study from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997–2007, > 90% of the invasive diseases, such as candidiasis are triggered by the 5 most common pathogens: C. albicans, C. tropicalis, C. parapsilosis, C. krusei, and C. glabrata. [26]. In order to reduce the increased cost of treatment owing to the delay in diagnosis and reduce the poor prognosis of candidiasis caused by the delay in diagnosis and treatment, a rapid and accurate diagnosis is required urgently. Present, the emergence of new diagnostic technology, T2 Candida, can resolve this long-standing clinical problem. T2 Candida is the first FDA-approved rapid and automated molecular diagnostic test to detect Candida sp. directly from blood, being culture-free.
In the blood sample test, compared to the blood cultures (gold standard), T2 Candida can detect the equivalent proportion of Candida, and the test time decreased from an average of 2.6 days in blood culture to about 3–4 h [13]. Thus, this technology is significantly superior to the gold standard (BC) in the identification of candidiasis [27]. Compared to the other diagnostic tests, high combined sensitivity (91%) and specificity (94%) are noted. This can also lead to better antifungal stewardship (good thing to include) [28]. Nevertheless, according to the designer’s statement, T2 Candida can only detect 5 Candida species: C. albicans, C. tropicalis, C. parapsilosis, C. krusei, and C. glabrata. It’s the limitation of T2 Candida. But > 90% of the invasive diseases, such as candidiasis are triggered by these 5 pathogens [26]. So T2 Candida is a highly valued detecting tool.
By searching the relevant literature on technology and collecting the relevant data, we combined and analyzed the diagnostic test data of T2 Candida: pooled sensitivity: 0.91 (95% CI: 0.88–0.94), pooled specificity: 0.94 (95% CI: 0.93–0.95). Strikingly, the combined sensitivity and specificity are both high. Furthermore, the resulting SROC curve is distant from the middle diagonal and close to the upper left corner, and the AUC = 1.0, which indicates an improved accuracy of T2 Candida is better.
We were also concerned about the heterogeneity. This study has developed stringent criteria for the inclusion and exclusion of the studies, minimizing the sources of heterogeneity. I2 values of the pooled sensitivity and specificity were > 90%, indicating a large heterogeneity among the included studies. In terms of threshold effects, the included studies were homogenous as assessed by statistical analysis. However, in the non-threshold effect analysis, a non-threshold effect heterogeneity was detected between the included studies. Markedly, this meta-analysis could not perform subgroup analysis to explore its heterogeneity due to time constraint and the small number of studies. According to the included literature, heterogeneity caused by the non-threshold effects originated from the factors, such as disease severity and concomitant diseases and test conditions such as different technologies, tests, operators, standard tests, and age (adults or children). However, the sensitivity of T2 Candida obtained by the study of Shamik et al. was 39% [12], and the specificity of the study by Mylonakis et al. was 59% [8], which deviated significantly from the pooled prediction values. These characteristics were related to several factors, such as the performance of T2 Candida, predicting patients’ outcomes, and cost-efficiency in various settings.
In conclusion, T2 Candida, the novel detection technology, has high efficiency, high specificity and time efficiency.
Conclusions
In summary, the current meta-analysis suggested that T2 Candida can be considered as a novel detection technology with high sensitivity and specificity. The method had a rapid and accurate diagnostic ability, a potential to improve the prognosis of the disease, reduce unnecessary expenses, and shorten the detection period. Thus, the T2 Candida could be a significant improvement for the laboratory diagnosis of candidiasis.
Acknowledgements
I am deeply indebted to all the tutors and teachers in this study for their direct and indirect help to me.
Abbreviations
- AUC
Area under curve
- BC
Blood culture
- C
Candida
- CI
Confidence interval
- DOR
Diagnostic odds ratio
- Fig
Figure
- FN
False-negatives
- FP
False-positives
- ROC
Receiver operating characteristic
- SE
Standard error
- SROC
Summary receiver operating characteristic
- TN
True-negatives
- TP
True-positives
Authors’ contributions
Research design: XGG, YX. Relevant literature selection: CGZ, DLT, XC, XGG. Data extraction: CGZ, XC, XGG. Quality evaluation: CGZ, DLT, XGG. Data analyzation: XC, DLT, XGG. Writing – original draft: DLT, XC, XGG. Writing – review & revision: DLT, ZWL, YX, XGG. All authors have read and approved the final version of the manuscript.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Availability of data and materials
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Ethics approval and consent to participate
We searched and obtained data from literature databases and the original researches were ethically approved.
Consent for publication
Not applicable. There are no details on individuals reported within the manuscript.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Dong-Lan Tang, Xiao Chen and Chang-Guo Zhu contributed equally to this work.
Contributor Information
Dong-Lan Tang, Email: 2409688522@qq.com.
Xiao Chen, Email: 1329102698@qq.com.
Chang-Guo Zhu, Email: 1286632857@qq.com.
Zhong-wei Li, Email: 1397895814@qq.com.
Yong Xia, Email: gysyxy@gmail.com.
Xu-Guang Guo, Email: gysygxg@gmail.com.
References
- 1.Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis. 1999;29(2):239–244. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- 2.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39(3):309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 3.Zilberberg MD, Shorr AF, Kollef MH. Secular trends in candidemia-related hospitalization in the United States, 2000-2005. Infect Control Hosp Epidemiol. 2008;29(10):978–980. doi: 10.1086/591033. [DOI] [PubMed] [Google Scholar]
- 4.Zaoutis TE, Argon J, Chu J, Berlin JA, Walsh TJ, Feudtner C. The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin Infect Dis. 2005;41(9):1232–1239. doi: 10.1086/496922. [DOI] [PubMed] [Google Scholar]
- 5.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, et al. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis. 2016;62(4):e1–50. doi: 10.1093/cid/civ933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neely LA, Audeh M, Phung NA, Min M, Suchocki A, Plourde D, et al. T2 magnetic resonance enables nanoparticle-mediated rapid detection of candidemia in whole blood. Sci Transl Med. 2013;5(182):182ra154. doi: 10.1126/scitranslmed.3005377. [DOI] [PubMed] [Google Scholar]
- 7.Clancy Cornelius J, Pappas Peter G, Vazquez Jose, Judson Marc A, Kontoyiannis Dimitrios P, Thompson George R, Garey Kevin W, Reboli Annette, Greenberg Richard N, Apewokin Senu, Lyon G Marshall, Ostrosky-Zeichner Luis, Wu Alan H B, Tobin Ellis, Nguyen M Hong, Caliendo Angela M. Detecting Infections Rapidly and Easily for Candidemia Trial, Part 2 (DIRECT2): A Prospective, Multicenter Study of the T2Candida Panel. Clinical Infectious Diseases. 2018;66(11):1678–1686. doi: 10.1093/cid/cix1095. [DOI] [PubMed] [Google Scholar]
- 8.Mylonakis E, Clancy CJ, Ostrosky-Zeichner L, Garey KW, Alangaden GJ, Vazquez JA, et al. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: a clinical trial. Clin Infect Dis. 2015;60(6):892–899. doi: 10.1093/cid/ciu959. [DOI] [PubMed] [Google Scholar]
- 9.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 10.Zamora J, Abraira V, Muriel A, Khan K, Coomarasamy A. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med Res Methodol. 2006;6:31. doi: 10.1186/1471-2288-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pappas PG, Nguyen MH, Mylonakis E. T2Candida is more sensitive and rapid than blood culture for monitoring candidemia in patients with proven infection. Poster presentation. 2015. [Google Scholar]
- 12.Dwivedi S, Ordaya E, Kezlarian B, Kenny R, Tibbetts R. Novel T2 magnetic resonance assay compared to standard blood cultures for detection of candidemia. Open Forum Infect Dis. 2016. 10.1093/ofid/ofw172.1271.
- 13.Beyda ND, Alam MJ, Garey KW. Comparison of the T2Dx instrument with T2Candida assay and automated blood culture in the detection of Candida species using seeded blood samples. Diagn Microbiol Infect Dis. 2013;77(4):324–326. doi: 10.1016/j.diagmicrobio.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Hamula CL, Hughes K, Fisher BT, Zaoutis TE, Singh IR, Velegraki A. T2Candida provides rapid and accurate species identification in pediatric cases of candidemia. Am J Clin Pathol. 2016;145(6):858–861. doi: 10.1093/ajcp/aqw063. [DOI] [PubMed] [Google Scholar]
- 15.Mylonakis E, Zacharioudakis IM, Clancy CJ, Nguyen MH, Pappas PG. Efficacy of T2 magnetic resonance assay in monitoring candidemia after initiation of antifungal therapy: the serial therapeutic and antifungal monitoring protocol (STAMP) trial. J Clin Microbiol. 2018;56(4). 10.1128/JCM.01756-17 PMID: 29367293. [DOI] [PMC free article] [PubMed]
- 16.Clancy CJ, Nguyen MH. Finding the “missing 50%” of invasive candidiasis: how nonculture diagnostics will improve understanding of disease Spectrum and transform patient care. Clin Infect Dis. 2013;56(9):1284–1292. doi: 10.1093/cid/cit006. [DOI] [PubMed] [Google Scholar]
- 17.Pfeiffer CD, Samsa GP, Schell WA, Reller LB, Perfect JR, Alexander BD. Quantitation of Candida CFU in initial positive blood cultures. J Clin Microbiol. 2011;49(8):2879–2883. doi: 10.1128/JCM.00609-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ness MJ, Vaughan WP, Woods GL. Candida antigen latex test for detection of invasive candidiasis in immunocompromised patients. J Infect Dis. 1989;159(3):495–502. doi: 10.1093/infdis/159.3.495. [DOI] [PubMed] [Google Scholar]
- 19.Mikulska M, Calandra T, Sanguinetti M, Poulain D, Viscoli C, Infect TEC. The use of mannan antigen and anti-mannan antibodies in the diagnosis of invasive candidiasis: recommendations from the third European conference on infections in leukemia. Crit Care. 2010;14:6. doi: 10.1186/cc9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yera H, Sendid B, Francois N, Camus D, Poulain D. Contribution of serological tests and blood culture to the early diagnosis of systemic candidiasis. Eur J Clin Microbiol Infect Dis. 2001;20(12):864–870. doi: 10.1007/s100960100629. [DOI] [PubMed] [Google Scholar]
- 21.Ellepola AN, Morrison CJ. Laboratory diagnosis of invasive candidiasis. J Microbiol. 2005;43:65–84. [PubMed] [Google Scholar]
- 22.Karageorgopoulos DE, Vouloumanou EK, Ntziora F, Michalopoulos A, Rafailidis PI, Falagas ME. beta-D-glucan assay for the diagnosis of invasive fungal infections: a meta-analysis. Clin Infect Dis. 2011;52(6):750–770. doi: 10.1093/cid/ciq206. [DOI] [PubMed] [Google Scholar]
- 23.Lu Y, Chen Y-Q, Guo Y-L, Qin S-M, Wu C, Wang K. Diagnosis of invasive fungal disease using serum (1→3)-β-D-Glucan: a bivariate meta-analysis. Intern Med. 2011;50(22):2783–2791. doi: 10.2169/internalmedicine.50.6175. [DOI] [PubMed] [Google Scholar]
- 24.Onishi A, Sugiyama D, Kogata Y, Saegusa J, Sugimoto T, Kawano S, et al. Diagnostic accuracy of serum 1,3-beta-D-glucan for pneumocystis jiroveci pneumonia, invasive candidiasis, and invasive aspergillosis: systematic review and meta-analysis. J Clin Microbiol. 2012;50(1):7–15. doi: 10.1128/JCM.05267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avni T, Leibovici L, Paul M. PCR diagnosis of invasive candidiasis: systematic review and meta-analysis. J Clin Microbiol. 2011;49(2):665–670. doi: 10.1128/JCM.01602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Ellis D, Tullio V, et al. Results from the ARTEMIS DISK global antifungal surveillance study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and Voriconazole as determined by CLSI standardized disk diffusion. J Clin Microbiol. 2010;48(4):1366–1377. doi: 10.1128/Jcm.02117-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thaler M, Pastakia B, Shawker TH, O'Leary T, Pizzo PA. Hepatic candidiasis in cancer patients: the evolving picture of the syndrome. Ann Intern Med. 1988;108(1):88–100. doi: 10.7326/0003-4819-108-1-88. [DOI] [PubMed] [Google Scholar]
- 28.Pfaller MA, Wolk DM, Lowery TJ. T2MR and T2Candida: novel technology for the rapid diagnosis of candidemia and invasive candidiasis. Future Microbiol. 2016;11(1):103–117. doi: 10.2217/fmb.15.111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article [and its supplementary information files].