Abstract
Avian influenza vaccines exhibit poor immunogenicity in humans. We hypothesized that one factor underlying weak B cell responses was sequence divergence between avian and seasonal influenza hemagglutinin proteins limiting availability of adequate CD4 T cell help. To test this, a novel chimeric hemagglutinin protein (cH7/3) was derived, comprised of the stem domain from seasonal H3 HA and the head domain from avian H7. Immunological memory to seasonal influenza was established in mice, through strategies that included seasonal inactivated vaccines, Flumist and synthetic peptides derived from the H3 stalk domain. After establishment of memory, mice were vaccinated with H7 or cH7/3 protein. The cH7/3 antigen was able to recall H3-specific CD4 T cells, and this potentiated CD4 T cell response was associated with enhanced early germinal center response and rapid elicitation of antibodies H7, including antibodies specific for the H7 head domain. These results suggest that in pandemic situations, inclusion of CD4 T cell epitopes from seasonal viruses have the potential to overcome poor immunogenicity of avian vaccines, conferring greater subtype-specific reactivity to the antibody response.
Introduction
Avian-derived influenza A viruses pose a serious threat to human health, with increasing reports of direct avian-to-human transmission (1, 2). Because of their pandemic potential, there have been a number of efforts to assess responses to avian influenza vaccines (3, 4), which have revealed poor vaccine efficacy. Multiple and/or high dose(s) or use of adjuvants are typically required for seroprotection, suggesting that avian vaccines may be inherently poorly immunogenic. There have been several proposed explanations for why primary responses to avian influenza vaccines are so weak in humans. Because of the more highly conserved nature of the HA stem domain between seasonal (e.g. H3N2 and H1N1) and avian (e.g. H7N9, H5N1) viruses, it is possible that avian HA proteins preferentially boost stalk-reactive memory B cells leading to a low magnitude of neutralizing antibodies (3, 5). An alternative explanation for the poor antibody response to avian vaccines is a deficit in CD4 T cell help. While humans do have circulating, cross-reactive CD4 T cells to H5 and H7 HA (6-8), we have found that the abundance of these T cells is low relative to the abundance of CD4 T cell reactivity to circulating H3 and H1 epitopes (7). We found that in subjects previously primed with a H5N1 vaccine and boosted with an H5N1 vaccine years later, H5-specific antibodies were elicited (9). When the status of CD4 T cells in these subjects was examined (10), we found that prior to the second vaccination, they had accumulated circulating H5-specific memory CD4 T cells, which could be recruited into the subsequent response to vaccination. This enhanced HA-specific CD4 T cell response was correlated with H5-specific neutralizing antibody responses to the serologically distinct H5N1 vaccine. These data are consistent with the view that inadequate CD4 T cell help is a limiting factor in the production of high affinity neutralizing antibodies to novel avian vaccines, but that recruitment of CD4 T cell help established by previous vaccinations can enable robust antibody responses.
Here, based on the premise that suboptimal CD4 T cell memory for novel avian HA epitopes may limit the B cell responses to novel influenza vaccines, we have explored a novel approach to enhance antibody responses to avian HA. We have engineered and tested a unique chimeric vaccine construct, which consists of an H7 globular head and an H3-stem domain (“cH7/3”). We speculated that if CD4 T cell help is indeed a limiting factor in the antibody responses to avian viruses, then in the setting of host memory to seasonal influenza, drawing CD4 T cell help from the population of H3-specific, memory CD4 T cells would enhance responses. In this study, we find that in animals with CD4 T cell memory to seasonal influenza, cH7/3 vaccination elicits more influenza-specific helper CD4 T cells than elicited with full-length H7 vaccine, and that the enhanced CD4 T cell response is associated with a greater early germinal center responses and early H7-specific antibody response. These results suggest that in the case of an emerging pandemic, when time is limited or there are concerns about the use of adjuvants, such chimeric avian/seasonal vaccine constructs could be rapidly deployed in humans, allowing for more effective antibody response to be elicited in at risk populations.
Materials and Methods
cH7/3 protein antigens
Two different constructs were examined, shown in Supplemental Figure 1. Both proved to be comparable in efficacy. The first used a mammalian expression system (293 cells) with the H7 head derived from A/Anhui/1/2013 (H7N9) and HA2 domain was completely from derived from A/Perth/16/2009 (H3N2). The second construct, derived from A/Anhui/1/2013 (H7N9) with the stem from A/Hong Kong/2014 (H3N2). The head domain is exclusively H7, but the stalk domain contains an N-terminal extension of 52 amino acids (1–52) from H3 that extends into the stem domain of H3 when folded (see Supplemental Figure 1). A baculovirus expression system (11) was used. Both constructs were made in soluble form by eliminating the transmembrane and cytoplasmic domains and inserting foldon trimerization domain (12), allowing multimeric assembly and contained 6x histidine tag used for nickel affinity purification.
Animals, viruses, vaccinations and infections
Female A/JCr (H-2a), were obtained from Charles River Laboratories and maintained in a specific-pathogen-free facility at URMC, according to institutional guidelines (protocol # 2006–030), most recently approved Jan. 2018. Mice were anaesthetized with avertin (2,2,2-Tribromoethanol) via intraperitoneal injection and infected intranasally with 15μL LAIV (Flumist 2013–14) (13) or vaccinated subcutaneously in the footpad with licensed seasonal inactivated influenza vaccine (IIV) (Flulaval 2011) (14) or with synthetic peptides derived from H3 HA. For peptide priming, mice were given 5nmol each H3 469 (469 KQLRENAEDMGNGCFKI 485) and H3 475 (475 AEDMGNGCFKIYHKCDN 491) peptide, in the presence of IFA and 0.6μg/ml lipopolysaccharide (50% v/v). For IIV, mice were administered 15μg total HA in the presence of alum (50% v/v). Control or previously primed groups were vaccinated subcutaneously in base of the tail, otherwise noted in the Figure legend, using 1.5μg recombinant protein (H7 or cH7/3) in the presence of alum (50% v/v), diluted in PBS unless
ELISpot and ELISA assays
CD4 T cell ELISpot assays and serum antibody ELISAs were performed as previously described (15). For the ELISA assays, 0.5μg recombinant H7 protein (A/Anhui/1/2013) (BEI Resources) or intact H7 or stabilized H7 head only construct, prepared in baculovirus as described (11, 16), used as the source of coating antigen for the ELISA assays were gifts from Florian Krammer.
Flow cytometry
Single cell suspensions from the draining lymph node were blocked with rat anti-mouse CD16/32 Fc Block, (clone 2.4G2) for 10 min at 4°C. Next, antibodies comprised of CD19 (clone 1D3), CD138 (clone 281–1, Biolegend), Fas (clone Jo2), GL-7 (clone GL-7), B220 (clone RA3–6B2), CD4 (clone RAM4–5). All reagents were purchased from BD Biosciences unless otherwise indicated. Multimerized H7 B cell probe (A/Anhui/1/13) (12) was also directly added to samples to resolve antigen-specific germinal center B cells. Data was acquired as previously described (17).
Micro-neutralization
For microneutralization assays (MN), 50ul of Receptor Destroying Enzyme (RDE) treated mouse sera were serially diluted 2-fold in 1x MEM/BSA medium and each dilution is then mixed with100 TCID50 of H7N9 virus. The mixture was incubated at 37°C for 1hr. For positive control, virus was incubated with serum generated from H7N9 infection. Other controls included only virus (no serum) or only serum (no virus). To calculate the MN titers, infectivity of serum-virus mixture was determined by infecting the cells MDCK with neutralized virus and measuring CPE. CPE was monitored by HA assay and MN titer was determined by calculating the maximum dilution of mouse serum to inhibit CPE. The controls of virus medium mixture showed 100% HA positive and all cells were killed. The controls of serum-medium mixture showed no CPE and HA negative.
Results
Design of chimeric protein and identification of mouse vaccine model
The general strategy employed here was to create a hybrid HA protein, whose membrane distal “head” domain, containing most of the neutralizing antibody epitopes in HA (reviewed in (18)) was derived from H7, with the membrane proximal stalk domain derived from a seasonal influenza virus, which would have the potential to recruit memory CD4 T cells established by vaccination or infection. We chose H7 as the donor of the head domain and used its closest group 2 seasonal “relative” H3 (19) as the donor of the membrane proximal stalk domain, schematized in Supplemental Figure 1. We have found that many healthy humans have readily detectable CD4 T cells specific for H3 HA-derived epitopes (20, 21), which we speculated could be recruited into the response to vaccination with the chimeric construct. Two slightly different constructs were used, as described in Supplemental Data, both with the head domain derived from H7 and the stalk domain from H3. The main difference between the first and second being the source of the NH2 terminal extension that extends into the stem domain. However, the two constructs have behaved similarly in immunological and biological assays employed thus far and we indicate in the legend of each Figure which construct was used.
Mouse model of priming and vaccination
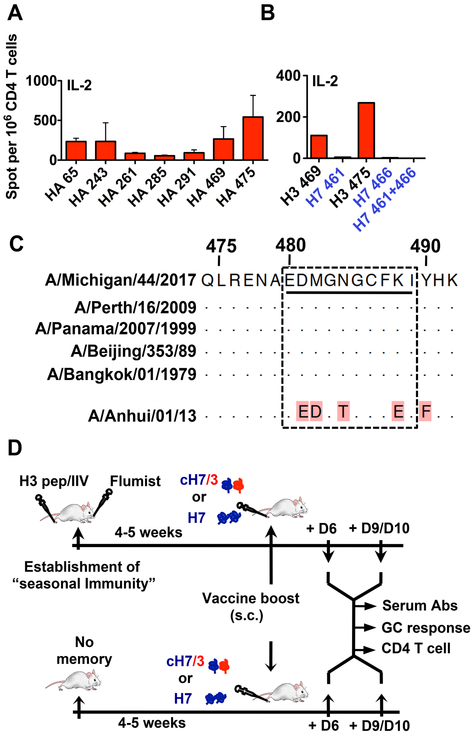
To test the hypothesis that recruitment of memory CD4 T cells would facilitate the H7-specific B cell response, we first identified a strain of mice that had highly conserved H3 derived epitopes within the HA2 domain. These epitopes also needed to be sufficiently divergent from the homologous segment of H7 to allow distinction of recruitment of memory H3-specific CD4 T cells from the primary response to H7. Several strains of mice were examined for CD4 T cell epitope specificity, using overlapping peptides, as we have previously described (22) in order to identify strains with H3-derived epitopes in the HA-2 domain. We found that CD4 T cells from A/J mice, expressing I-Ak and I-Ek, recognized several epitopes in H3, including major immunodominant epitopes in the stem region (Figure 1A). Peptides (469-KQLRENAEDMGNGCFKI-485) and (475-AEDMGNGCFKIYHKCDN-491) contain the shared epitope (475-AEDMGNGCFKI-484). The segment encoding this CD4 T cell epitope displayed no detectable cross reactive CD4 T cell recognition from the homologous segment in H7 (Figure 1B) and was highly conserved among many H3N2 viruses (Figure 1C) making this strain appropriate for the studies planned.
Figure 1. Definition and characterization of CD4 epitopes for study and experimental design.
A) IL-2 ELISpot assay used to identify the immunodominant CD4 T cell epitopes elicited to H3 stem domain. Initial screening revealed 7 candidate H3-derived peptides, which were used to stimulate antigen-specific CD4 T cells. B) IL-2 ELISpot to test any cross reactivity of H3 stem-specific CD4 T cells to the homologous H7 stem region. Results represent mean of two independent experiments with the range indicated. C) Sequence alignment shows homologous sequence in the H3 epitope regions from diverse H3N2 virus strains and lack of homology to the segment of H7. D) Schematic representation of the immunization protocol: Cold adapted vaccine Flumist and a trivalent inactivated influenza protein based vaccine, IIV (Flulaval) or H3-derived synthetic peptides were used to establish “seasonal CD4 T cell memory”. 4-5 weeks later, mice were vaccinated with the candidate H7 or cH7/3 vaccines. 6 or 10 days post-vaccination, sera from vaccinated mice were sampled to analyze H7-specific antibody responses. Draining LNs were sampled to analyze H3-specific CD4 T cells by ELISpot assay and for the identification of H7-specific GC B cells by flow-cytometry. Mice with no memory to seasonal H3 vaccinated with H7 or cH7/3 in parallel, served as controls.
cH7/3 vaccination elicits greater H7-specific B cell responses compared to full-length H7 protein when hosts have pre-existing CD4 T cell immunity to H3 MHC class II epitopes
The key question we sought to address here is whether the cH7/3 protein was able to recruit memory CD4 T cells established by contact with seasonal influenza A epitopes and if, by doing so, would the primary H7-specific antibody responses to vaccination be enhanced. To establish H3-specific CD4 T cell memory, three different priming regimens were used, illustrated in Figure 1D. Two licensed vaccines, the intranasally administered cold adapted vaccine Flumist and an inactivated influenza vaccine (IIV) were used in separate cohorts to establish complex immunological memory to seasonal influenza strains (H1N1, H3N2 and influenza B), mimicking human subjects. Vaccination with live, attenuated Flumist allowed recruitment of CD4 T cells from endogenously synthesized antigens, while vaccination with H3-derived synthetic peptides was used to allow assessment of exclusive CD4 T cell priming, with no concerns of priming the B cell repertoire. The synthetic peptides H3 (469–460 and 475–491) and IIV were introduced in the footpad with IFA and LPS, while the Flumist was introduced by intranasal vaccination. For all priming regimens, we were able to detect H3 stem-specific memory CD4 T cells in spleen and LN prior to boost (not shown).
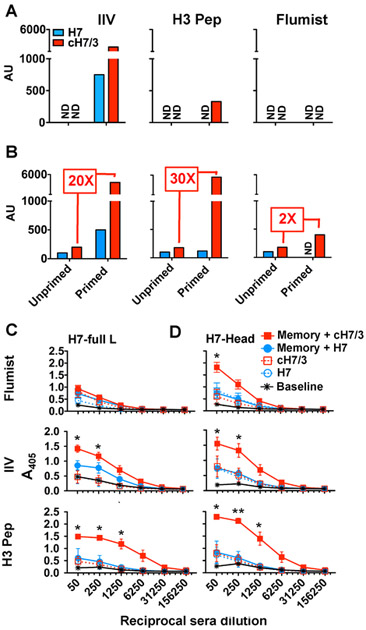
To compare the responses to the alternate vaccines, 30–35 days after priming, H7 WT or cH7/3 recombinant vaccines, adjuvanted in alum, were used for subcutaneous priming at the base of the tail. This mode of vaccination was chosen first, because in general, subcutaneous administration of vaccines are more robust than intramuscular delivery and used a site distal to the initial priming site so that the response would undergo a discreet immune response relative to the original priming for establishment of memory. Control mice with no previous exposure to influenza were vaccinated and compared in parallel. Six or 10 days after HA protein vaccination, sera from vaccinated mice were sampled, and pooled sera were tested for reactivity to a recombinant full-length H7 protein via ELISA assays, using titration assays. Figure 2A and 2B shows the results of these assays, represented as the serum dilution that achieves the same detectable reactivity on the linear portion of the antibody titration curve. At day 6, in the absence of memory, there were no detectable antibody responses to vaccination (“ND”). Strikingly, priming via all three modes to introduce H3-specific memory led to enhanced production of H7-specific antibodies after administration of the cH7/3 vaccine relative to that elicited by the full-length H7 vaccine. The enhanced antibody responses were evident, remarkably, as early as day 6 post-vaccination for the peptide-primed and IIV-primed mice. Serum antibody was most readily detected at day 10 post-vaccination in all three groups (Figure 2B). The degree of enhancement with immunological memory varied with the mode of priming, with IIV-primed host exhibiting 20-fold greater response than unprimed animals vaccinated with the cH7/3 vaccine, H3-derived peptide priming showing almost a 30-fold enhanced response to the cH7/3, while the Flumist primed mice exhibiting a 2-fold enhanced H7-specific antibody responses, comparing the titer elicited in the presence vs. absence of memory. These results, particularly compelling those involving peptide priming, indicate that elicitation of an antibody responses to avian HA from the naïve B cell repertoire can be dramatically enhanced by engaging CD4 T cells specific for seasonal HA epitopes.
Figure 2. H7-specific IgG serum antibodies are enhanced in cH7/3 vaccinated mice armed with immunological memory to H3.
Mice that had established CD4 T cell memory to seasonal H3 with one of 3 different vaccine formulations were vaccinated with full-length H7 or cH7/3a. At day 6 or 10 post-vaccination, sera were sampled and ELISA assays were performed to detect H7-reactive antibodies on pooled serum samples. Shown are IgG responses to full-length H7 (BEI resource, NR-45118) at A) D6 and B) D10 post-vaccination with full-length H7 (Blue bar) or chimera cH7/3 (Red bar) proteins. Data was plotted as arbitrary unit (AU) by converting the serum dilution within the treatment groups that achieved the same detectable reactivity on the linear portion of the antibody titration curve. Individual serum samples were also tested for reactivity to recombinant full-length H7 or H7-head proteins by ELISA assay at 10 days post-vaccination. Shown are IgG responses to C) full-length H7 and D) H7-head. Antibody responses from mice primed by Flumist-Upper panel, IIV- middle panel and H3 peptide-lower panel. Data represents the mean ± error for individual mice. Reactivity of sera from naïve mice are shown in orange lines and were never above background when tested. Note: Responses shown for IIV and H3 peptide are from mice receiving the cH7/3 chimera containing derived from the complete domain switch between H7 and H3, while responses shown for Flumist are from mice receiving the cH7/3b chimera. Student T-test (unpaired, non-parametric, two-tailed) was performed to calculate the significance among the groups (*p ≤ 0.05, **p ≤ 0.01), using GraphPad Prism v5.0a.
We next sought to examine the patterns among individually vaccinated mice, allowing statistical treatment of data. We also asked whether the serum antibodies elicited by the vaccines were reactive to the head domain of H7, containing the sites of HA that control binding and infection of susceptible cells. Single mouse serum samples were tested for reactivity with recombinant full-length H7 or an H7 head domain construct (23), using antibody titration and ELISA assays. Mice previously primed with H3 HA-2 derived peptides, Flumist or the IIV were sampled at day 10 post-vaccination. The results of these assays are shown in Figure 2C and 2D, where the dotted lines with open symbols represent the H7-specific antibodies elicited without memory, and the solid lines and symbols indicate the H7-specific antibodies elicited in the presence of memory. These experiments showed good agreement among individual mice with established CD4 T cell memory to H3 epitopes, via IIV, Flumist or with the H3-derived peptides. Relative to the response elicited by the full-length H7 protein vaccine in the presence seasonal memory the cH7/3 vaccine to promote greatly enhanced levels of circulating antibody to H7. Most importantly, in the mice vaccinated with the cH7/3 vaccine, there were readily detectable antibodies reactive with the head domain of the H7 HA (Figure 2D) indicating that these antibodies would likely provide protective immunity to infection. Interestingly, the serum antibodies elicited by the chimeric H7/H3 vaccine that was enhanced by CD4 memory to the H3 stalk appeared to be preferentially focused on the H7 head domain, indicating that enhanced CD4 T cell help can potentiate B cell reactivity to novel HA protein epitopes.
Early B cell responses to vaccination
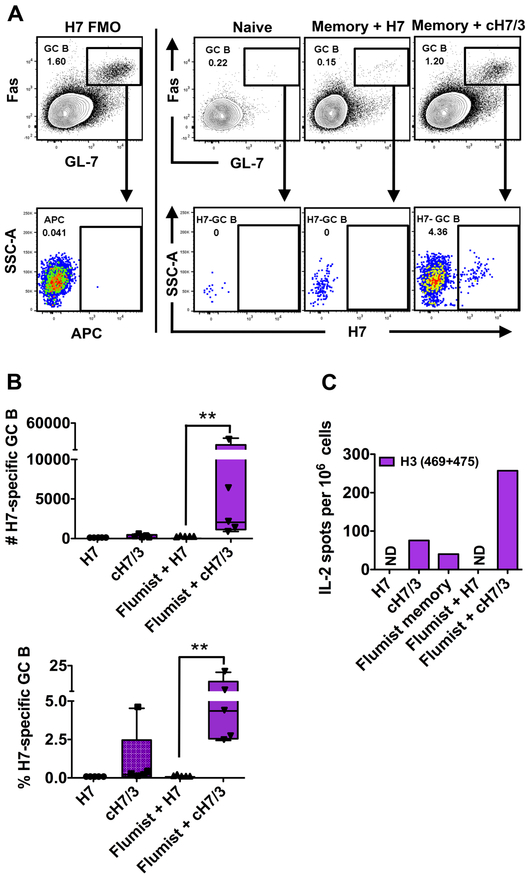
We next examined if the antibody response to vaccination was associated with a boost of CD4 T cells specific for H3 epitopes and an early germinal center (GC) B cell response. We also examined the specificity of the early B cell response, using an H7 protein probe to detect antigen-specific GC B cells. In this assay, a recombinant H7 protein that acts as a ligand for the B cell receptor (BcR) of virus antigen-specific B cells is coupled to a fluorochrome (12) and multimerized with streptavidin, allowing direct detection of H7-specific B cells. We first established the specificity of the H7 probe. In this study, shown in Figure 3, mice were vaccinated in parallel with either H7 protein or NP protein in alum. At day 14 post-vaccination, draining lymph node cells were examined for total germinal center responses and germinal center B cells that express immunoglobulin receptors specific for H7. As can be seen in Figure 3A, the H7 protein fluorescent probe has negligible reactivity with GC B cells from influenza nucleoprotein (NP) vaccinated mice (middle panel), but readily detects GC cells isolated from H7-protein vaccinated mice (right panel). Figure 3B also shows that GC B cells from H7-vaccinated mice do not react with an H1-derived probe, prepared in the same way as the H7 probe. Collectively, these controls verify the selectivity of these HA probe constructs in detecting B cells with immunoglobulin receptors specific for H7. In Figure 4, we show the results obtained from Flumist primed mice vaccinated with either H7 or the cH7/H3 vaccine. Controls for the detection of H7-specific B cells after priming memory-containing mice with the HA vaccines are shown in Figure 4A. The left-most single panels show the FMO controls. Lack of reactivity in B cells from lymph node cells from naïve mice is shown in the next panel. The two right-most panels are examples of the GC cells elicited in the mice vaccinated with H7 vaccine, and from mice receiving the cH7/3, respectively, as indicated above each panel. These data revealed a greatly enhanced GC response in lymph node cells isolated from the H7/H3-vaccinated mice, relative to the mice vaccinated with the full length H7 protein. The cH7/H3 vaccinated mice also showed readily detectable B cells bearing BcR for H7. A compilation of the results of this experiment is shown in Figure 4B, where both the total number (top) and percentage (bottom) of H7-specific B cells are shown. The values for individual mice are shown as small symbols within each panel. We found that 5/5 mice primed with Flumist mounted a detectable H7-specific GCB cell response after vaccination with cH7/3. Importantly, these H7-specific GC responses were much greater in frequency and number compared to the H7 vaccinated animals. All of these results suggest the importance of recruitment of memory CD4 T cells in enhancing the early B cell response to avian influenza vaccines. Figure 4C, representing the results of IL-2 cytokine EliSpot assays, confirms that the early GC response and enhanced serum antibody responses in the presence of seasonal CD4 T cell memory are associated with a boost in CD4 T cells reactive with the peptide epitopes derived from the stem of H3.
Figure 3. Fluorescent H7-probe specifically detects H7-speciic GC B cells, which are not detected by an H1 specific probe.
Mice were immunized subcutaneously in the footpad either with H7 protein or influenza nucleoprotein (NP) in alum. 14 days post-immunization, popliteal lymph nodes (pLN) were harvested and single cells analyzed by flow to detect H7-specific GC B cells. A) An APC-labeled H7- probe was used to identify H7-specific GC B cells. Representative staining is shown to illustrate the expression pattern of H7-specific GC B cells (CD4−CD19+B220hiCD138−GL−7+Fas+) in naïve mice, H7- immunized or NP-immunized mice as indicated in the top of the figures. H7 reactivity was only detected in the pLN from the H7 vaccinated mice. B) An APC-labeled H1-probe (A/California/04/09) was used in a separate aliquot of cells from the same cohorts of mice, showing the selectivity of binding.
Figure 4. Enhanced H7-specific GC B cell response correlates with the boost and mobilization of H3 memory CD4 T cells.
Mice were primed with Flumist by the intranasal route and were boosted 30 days later with the full length or cH7H3 vaccine subcutaneously in the footpad. Lymph node cells were isolated from the draining popliteal lymph node. A) Flow-cytometry for the identification of H7-specific GC B cells. An APC-labeled H7-probe was used to identify the HA-specific GC B cells, as described in Materials and Methods. Representative staining is shown to illustrate the expression pattern of H7-specific GC B cells (CD4−CD19+B220hiCD138−GL-7+Fas+) in H7 or cH7/3b immunized mice: Left panel: Control to establish gating for H7-specific GC B cells in the respective channel. From left to right on the remaining panels are examples of the analyses of GC and H7 reactivity are shown from lymph node cells isolated from naïve, H7-immunized and cH7/3b-immunized mice, respectively, as indicated above each panel. B) The top panel shows the number and the bottom panel shows the frequency of H7-reactive GC B cells 6 days post-immunization in mice vaccinated with H7 or cH7/3 chimera proteins in the presence or absence of memory to H3. Each symbol represents responses from a single animal. C) H3-specific memory CD4 T cells are boosted following cH7/3 vaccination. Mice were injected with Flumist to establish CD4 T cell memory to seasonal influenza and 4–5 weeks later were vaccinated subcutaneously with the candidate full-length or cH7/3 recombinant proteins. Unprimed mice with no prior memory to seasonal H3 influenza vaccinated in parallel were used as controls. Draining LNs (n = 4 per group) were sampled and pooled at 6 days post-immunization and IL-2 CD4 T cell ELISpot assays were performed. The peptides used for re-stimulation are indicated in the Figure. Data are represented as spots per million CD4 T cells. Student T-test (unpaired, non-parametric, two-tailed) was performed to calculate the significance among the groups (*p ≤ 0.05, **p ≤ 0.01), using GraphPad Prism v5.0a.
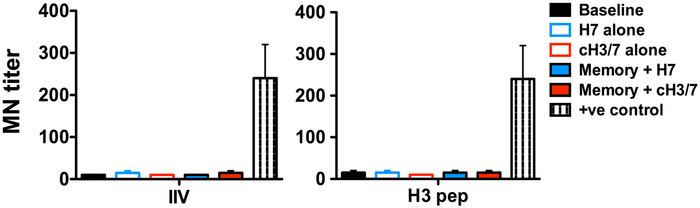
We have sought to test the ability of the day 10 sera from primed and vaccinated mice to neutralize H7N9 viruses, (Figure 5), but found these to be inadequate for neutralization for both IIV primed (left panel) and H3-peptide primed (right panel) groups after subsequent vaccination with either the full length H7 vaccine or the cH7H3 vaccine. We speculate that the difficulty in demonstrating the in vivo protective efficacy of the vaccine is due to the early days sera was sampled here (day 6 and day 10). At these time points, the primary B cell response to the H7 head, affinity maturation is still evolving in the host and secreted antibodies are still accumulating in sera. Typical assays testing for neutralization or in vivo protection is typically done after a prime/boost vaccination regimens and extended times post-vaccination prior to serum sampling (24-27). We cannot implement the prime boost strategy for the studies here, because most rigorous experimental design in this study relies on exclusive CD4 T cell priming in the host, which was provided by synthetic peptides derived from H3. Boosting with the cH7/H3 chimera would allow priming both the B cell repertoire and the CD4 T cell repertoire, thus confounding the interpretation of experiments to test this strategy. Nonetheless, the robust recognition of the avian head domain by the cH7H3 vaccine when tested by ELISA assays demonstrates that chimeric strategies enhances B cell targeting to the head, while allowing recruitment of CD4 T cells specific to highly conserved stalk domain that are established by seasonal vaccines and viruses.
Figure 5. MN titers for H7N9.
Cohorts of mice (10-12 in each group) that had CD4 T cell memory to seasonal H3 CD4 T cells, established with one of 2 different vaccine formulations (H3-peptide and IIV), were vaccinated with H7 or cH7/3 protein. 10 days post-vaccination, sera from vaccinated mice were sampled and micro-neutralization (MN) assay was performed to detect H7N9 neutralizing antibody titers. Shown here are MN titers to H7N9 at D10 post-vaccination with full-length H7 (Solid blue bar) or chimera cH7/H3 (solid red bar) proteins in the mice with memory to seasonal H3 established either by IIV (Left panel) or H3 peptide (right panel). Controls include MN titers with baseline serum (black solid bars), serum from mice vaccinated with H7 alone (open blue bar) or cH7/3 alone (open red bar). Serum from H7N9 infected mice was used as positive control. Data represents the mean ± range for individual mice.
Discussion
These studies revealed that selective recruitment of memory CD4 T cells specific for epitopes in the HA protein can potentiate humoral immunity to what is considered to be weakly immunogenic avian HA proteins. These memory CD4 T cells can be engaged through incorporation of CD4 T cell peptide epitopes from seasonal influenza HA-derived proteins into avian HA construct. Using several priming regimens to generate “seasonal” CD4 T cell immunity to influenza, we found that the presence of circulating H3-specific CD4 T cells prior to vaccination was associated with greatly enhanced early antibody responses to the chimeric cH7/3 vaccines, relative to that elicited by WT H7 protein vaccines. Importantly, the antibodies elicited by the cH7/3 vaccine were highly reactive with the H7 head domain, suggesting that most of these will be able to mediate sterilizing protection. Recent studies in both human and mouse vaccine recipients have provided evidence that ELISA assays and in particular those with HA “head-only” reagents often correlate better with protection from H7N9 than do conventional HAI or microneutalization assays (16, 24). The potentiated B cell response induced by the cH7/3 vaccine was associated with very early (day 6) serum antibody production and H7-specific GC cells and a boost in CD4 T cell reactivity for the epitopes derived from the H3 stem. Collectively, these results suggest that chimeric vaccines consisting of head domains from avian strains and stalk domains from seasonal strains of influenza will be able to recruit memory circulating and abundant CD4 T cells established by seasonal influenza (6-8). Overall, however, we are confident from the data presented here that in the context of normal memory established by seasonal influenza viruses or vaccines, early B cell and antibody responses directed to the head domain of the H7 will be enhanced after vaccination with the chimeric vaccine relative to a wild type H7 vaccine. Therefore, in the case of an emerging pandemic, this type of chimeric vaccine would be expected to provide better early protection from infection and seed the host with B cells specific for H7 HA
The encouraging results presented in this paper provide a proof of concept for derivation for influenza vaccines that can be rapidly deployed in the case of an emerging avian virus. Also here, in mice, there was only a single epitope in H3 from which memory CD4 T cells could be recruited. We have found in humans that the abundance of CD4 T cell reactivity to H3 is variable (28), and includes reactivity to the highly conserved stalk domain which more abundant than the cross-reactive memory to H7 (7). Therefore, the efficacy of this chimeric vaccine would likely depend on the specific characteristics of the human CD4 T cell repertoire. In general, we speculate that if used for human vaccination, chimeric HA vaccines described here will likely recruit many more memory CD4 T cells than that elicited by conventional avian vaccines, providing the needed CD4 T cell help for production of protective antibody responses to avian HA.
Supplementary Material
Key Points:
Human CD4 T cells have limited cross-reactive memory to avian H7 proteins.
CD4 T cell help can be provided to B cells with novel H7/H3 chimeric vaccines
Acknowledgements:
We would like to thank Florian Krammer for the gift of cH7/3 used for vaccination and for the head only and full length H7 protein constructs to test antibody specificity. We also thank Katherine Richards for excellent help in preparing this manuscript for publication. We acknowledged the valuable contribution of BEI resources (https://www.beiresources.org) for synthetic influenza peptides and influenza HA proteins.
This work was supported by NIH HHSN272201400005C to AJS, and Leading Advanced Projects for medical innovation (LEAP) from AMED (JP18am001007) and by the NIAID-funded Center for Research on Influenza Pathogenesis (CRIP, HHSN272201400008C) to YK. ATD was supported in part by 5T32AI007285.
References:
- 1.Trombetta C, Piccirella S, Perini D, Kistner O, and Montomoli E. 2015. Emerging Influenza Strains in the Last Two Decades: A Threat of a New Pandemic? Vaccines (Basel) 3: 172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morens DM, Taubenberger JK, and Fauci AS. 2013. H7N9 avian influenza A virus and the perpetual challenge of potential human pandemicity. MBio 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nichol KL, and Treanor JJ. 2006. Vaccines for seasonal and pandemic influenza. J Infect Dis 194 Suppl 2: S111–118. [DOI] [PubMed] [Google Scholar]
- 4.de Vries RD, Herfst S, and Richard M. 2018. Avian Influenza A Virus Pandemic Preparedness and Vaccine Development. Vaccines (Basel) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellebedy AH, Krammer F, Li GM, Miller MS, Chiu C, Wrammert J, Chang CY, Davis CW, McCausland M, Elbein R, Edupuganti S, Spearman P, Andrews SF, Wilson PC, Garcia-Sastre A, Mulligan MJ, Mehta AK, Palese P, and Ahmed R. 2014. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc Natl Acad Sci U S A 111: 13133–13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee LY, Ha do LA, Simmons C, de Jong MD, Chau NV, Schumacher R, Peng YC, McMichael AJ, Farrar JJ, Smith GL, Townsend AR, Askonas BA, Rowland-Jones S, and Dong T. 2008. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J Clin Invest 118: 3478–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richards KA, Nayak J, Chaves FA, DiPiazza A, Knowlden ZA, Alam S, Treanor JJ, and Sant AJ. 2015. Seasonal Influenza Can Poise Hosts for CD4 T-Cell Immunity to H7N9 Avian Influenza. J Infect Dis 212: 86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roti M, Yang J, Berger D, Huston L, James EA, and Kwok WW. 2008. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. J Immunol 180: 1758–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goji NA, Nolan C, Hill H, Wolff M, Noah DL, Williams TB, Rowe T, and Treanor JJ. 2008. Immune responses of healthy subjects to a single dose of intramuscular inactivated influenza A/Vietnam/1203/2004 (H5N1) vaccine after priming with an antigenic variant. J Infect Dis 198: 635–641. [DOI] [PubMed] [Google Scholar]
- 10.Nayak JL, Richards KA, Yang H, Treanor JJ, and Sant AJ. 2015. Effect of influenza A(H5N1) vaccine prepandemic priming on CD4+ T-cell responses. J Infect Dis 211: 1408–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hai R, Krammer F, Tan GS, Pica N, Eggink D, Maamary J, Margine I, Albrecht RA, and Palese P. 2012. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J Virol 86: 5774–5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whittle JR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS, Narpala SR, Yassine HM, Frank GM, Yewdell JW, Ledgerwood JE, Wei CJ, McDermott AB, Graham BS, Koup RA, and Nabel GJ. 2014. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J Virol 88: 4047–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carter NJ, and Curran MP. 2011. Live attenuated influenza vaccine (FluMist(R); Fluenz): a review of its use in the prevention of seasonal influenza in children and adults. Drugs 71: 1591–1622. [DOI] [PubMed] [Google Scholar]
- 14.Baras B, Bouveret N, Devaster JM, Fries L, Gillard P, Sanger R, and Hanon E. 2008. A vaccine manufacturer’s approach to address medical needs related to seasonal and pandemic influenza viruses. Influenza Other Respir Viruses 2: 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alam S, Knowlden ZA, Sangster MY, and Sant AJ. 2014. CD4 T cell help is limiting and selective during the primary B cell response to influenza virus infection. J Virol 88: 314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan GS, Leon PE, Albrecht RA, Margine I, Hirsh A, Bahl J, and Krammer F. 2016. Broadly-Reactive Neutralizing and Non-neutralizing Antibodies Directed against the H7 Influenza Virus Hemagglutinin Reveal Divergent Mechanisms of Protection. PLoS Pathog 12: e1005578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brewer MG, DiPiazza A, Acklin J, Feng C, Sant AJ, and Dewhurst S. 2017. Nanoparticles decorated with viral antigens are more immunogenic at low surface density. Vaccine 35: 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu NC, and Wilson IA. 2017. A Perspective on the Structural and Functional Constraints for Immune Evasion: Insights from Influenza Virus. J Mol Biol 429: 2694–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nabel GJ, and Fauci AS. 2010. Induction of unnatural immunity: prospects for a broadly protective universal influenza vaccine. Nat Med 16: 1389–1391. [DOI] [PubMed] [Google Scholar]
- 20.Richards KA, Treanor JJ, Nayak JL, and Sant AJ. 2018. Overarching Immunodominance Patterns and Substantial Diversity in Specificity and Functionality in the Circulating Human Influenza A and B CD4 T Cell Repertoire. J Infect Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richards KA, Topham D, Chaves FA, and Sant AJ. 2010. Cutting edge: CD4 T cells generated from encounter with seasonal influenza viruses and vaccines have broad protein specificity and can directly recognize naturally generated epitopes derived from the live pandemic H1N1 virus. J Immunol 185: 4998–5002. [DOI] [PubMed] [Google Scholar]
- 22.Richards KA, Chaves FA, and Sant AJ. 2009. Infection of HLA-DR1 transgenic mice with a human isolate of influenza a virus (H1N1) primes a diverse CD4 T-cell repertoire that includes CD4 T cells with heterosubtypic cross-reactivity to avian (H5N1) influenza virus. J Virol 83: 6566–6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krammer F, Margine I, Tan GS, Pica N, Krause JC, and Palese P. 2012. A carboxy-terminal trimerization domain stabilizes conformational epitopes on the stalk domain of soluble recombinant hemagglutinin substrates. PLoS One 7: e43603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamal RP, Blanchfield K, Belser JA, Music N, Tzeng WP, Holiday C, Burroughs A, Sun X, Maines TR, Levine MZ, and York IA. 2017. Inactivated H7 Influenza Virus Vaccines Protect Mice despite Inducing Only Low Levels of Neutralizing Antibodies. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pushko P, Pujanauski LM, Sun X, Pearce M, Hidajat R, Kort T, Schwartzman LM, Tretyakova I, Chunqing L, Taubenberger JK, and Tumpey TM. 2015. Recombinant H7 hemagglutinin forms subviral particles that protect mice and ferrets from challenge with H7N9 influenza virus. Vaccine 33: 4975–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Hilchey SP, DeDiego M, Perry S, Hyrien O, Nogales A, Garigen J, Amanat F, Huertas N, Krammer F, Martinez-Sobrido L, Topham DJ, Treanor JJ, Sangster MY, and Zand MS. 2018. Broad cross-reactive IgG responses elicited by adjuvanted vaccination with recombinant influenza hemagglutinin (rHA) in ferrets and mice. PLoS One 13: e0193680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crevar CJ, Carter DM, Lee KY, and Ross TM. 2015. Cocktail of H5N1 COBRA HA vaccines elicit protective antibodies against H5N1 viruses from multiple clades. Hum Vaccin Immunother 11: 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richards KA, Treanor JJ, Nayak J, Sant AJ 2018. Overarching immunodominance patterns and substantial diversity in specificity and functionality within the circulating human influenza A and B CD4 T cell repertoire. J. Infect. Disease. 218:1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.