ABSTRACT
Highly conserved DM domain-containing transcription factors (Doublesex/MAB-3/DMRT1) are responsible for generating sexually dimorphic features. In the Drosophila central nervous system, a set of Doublesex (Dsx)-expressing neuroblasts undergo apoptosis in females whereas their male counterparts proliferate and give rise to serotonergic neurons crucial for adult mating behaviour. Our study demonstrates that the female-specific isoform of Dsx collaborates with Hox gene Abdominal-B (Abd-B) to bring about this apoptosis. Biochemical results suggest that proteins AbdB and Dsx interact through their highly conserved homeodomain and DM domain, respectively. This interaction is translated into a cooperative binding of the two proteins on the apoptotic enhancer in the case of females but not in the case of males, resulting in female-specific activation of apoptotic genes. The capacity of AbdB to use the sex-specific isoform of Dsx as a cofactor underlines the possibility that these two classes of protein are capable of cooperating in selection and regulation of target genes in a tissue- and sex-specific manner. We propose that this interaction could be a common theme in generating sexual dimorphism in different tissues across different species.
KEY WORDS: Abdominal-B, Doublesex, Drosophila, Neuroblast, Apoptosis, Sexual dimorphism
Highlighted Article: Drosophila DoublesexF collaborates with Abdominal-B to generate a sexually dimorphic central nervous system.
INTRODUCTION
Sexual reproduction is central to animal existence and propagation. Irrespective of the different upstream molecular events, generation of sexually dimorphic traits converge down to a highly conserved DM domain containing non-classical zinc-finger proteins such as Doublesex (Dsx) in Drosophila, Male abnormal 3 (MAB-3) in Caenorhabditis elegans and Doublesex/Male-abnormal-3 Related Transcription factor 1 (Dmrt1) in vertebrates (Matson and Zarkower, 2012). The DM domain is an intertwined zinc finger-containing DNA binding module (Erdman and Burtis, 1993; Zhu et al., 2000) associated with sexual development and reproduction across different species (Baker and Ridge, 1980; Burtis and Baker, 1989; Shen and Hodgkin, 1988; Raymond et al., 2000; Chong et al., 2013; Matson and Zarkower, 2012; Raymond et al., 1999; Ross et al., 2005).
Apoptosis also plays an important role in generating sexual dimorphism in neural and non-neural tissues of Drosophila (DeFalco et al., 2003; Billeter et al., 2006b; Billeter et al., 2006a; Wang et al., 2011; Birkholz et al., 2013a; Kimura et al., 2005; Ren et al., 2016; Truman et al., 1993), C. elegans (Peden et al., 2007; Conradt and Horvitz, 1999) and vertebrates (Davis et al., 1996; Price et al., 1977; Roberts et al., 1999). Drosophila is a useful system for studying sexual dimorphism, wherein Dsx is an executive transcription factor (TF) of the sex determination hierarchy. Male and female isoforms of Dsx have a common region containing DNA-binding and dimerization domains followed by a sex-specific C-terminal region responsible for generating sex-specific features (Burtis and Baker, 1989; Erdman and Burtis, 1993). The Hox gene Abdominal-B (Abd-B) and its homologues are also known for their role in generating sexual dimorphism across different species (Kalis et al., 2010; Satokata et al., 1995; Dolle et al., 1991; DeFalco et al., 2004; Wang and Yoder, 2012; Wang et al., 2011; Jeong et al., 2006; Williams et al., 2008; Kopp et al., 2000; Tanaka et al., 2011; Foronda et al., 2012). Proteins Dsx and AbdB are known to function together in cuticle pigmentation as well as in growth and differentiation of Drosophila genital discs (Sanchez et al., 2001; Kopp et al., 2000; Williams et al., 2008). However, the precise details of Dsx and AbdB interaction remains unclear. Similarly, although the role of Dsx in generating sexual dimorphism in the central nervous system (CNS) has been well investigated (Lee et al., 2002; Baker and Ridge, 1980; Taylor and Truman, 1992; Villella and Hall, 1996; Billeter et al., 2006b; Waterbury et al., 1999; Zhou et al., 2014; Rideout et al., 2010), its possible interaction with a spatial determinant such as AbdB has not been tested.
In Drosophila, neural stem cells (neuroblasts, NBs) in abdominal segments (A3-A7) of the larval ventral nerve cord (VNC) undergo Hox gene abdominal-A (abd-A)-mediated apoptosis in the late larval stage (Bello et al., 2003). This apoptosis is mediated by the RHG family genes grim and reaper, activated through an enhancer lying within a 22 kb genomic region called the neuroblast regulatory region (NBRR) (Arya et al., 2015; Tan et al., 2011). It has been subsequently shown that AbdA, along with another TF Grainyhead (Grh), transcriptionally activates apoptotic genes through a 1 kb enhancer located within the NBRR (Khandelwal et al., 2017).
In terminal segments (A8-A10) of the embryonic VNC, which expresses Abd-B, a subpopulation of 62 NBs (Birkholz et al., 2013a) undergo apoptosis while remaining cells enter quiescence (Monedero Cobeta et al., 2017). Only 12 of these 62 NBs exit quiescence in early larval stages and are referred to as terminal NBs (tNBs) (Taylor and Truman, 1992; Truman and Bate, 1988). Four of these 12 tNBs express the doublesex (dsx) gene and are referred to hereafter as Dsx+tNBs. In females, these cells undergo apoptosis mediated by DsxF, whereas DsxM mediates their continued proliferation in males until the late third instar larval stage (L3) (Fig. 1A) (Taylor and Truman, 1992; Truman and Bate, 1988; Birkholz et al., 2013a), resulting in additional serotonergic neurons that are crucial for male mating behaviour in adults (Billeter et al., 2006a,b). Interestingly, although Dsx-mediated sex-specific neuronal apoptosis occurs across different regions of the developing CNS (Billeter et al., 2006b; Kimura et al., 2008; Sanders and Arbeitman, 2008), Dsx-mediated larval NB apoptosis has so far been reported specifically in females and only in the AbdB-expressing region of the CNS (Birkholz et al., 2013a; Taylor and Truman, 1992; Truman and Bate, 1988). This offers a unique opportunity to understand if (and how) spatial patterning genes could collaborate with the sex determination hierarchy to pattern the CNS.
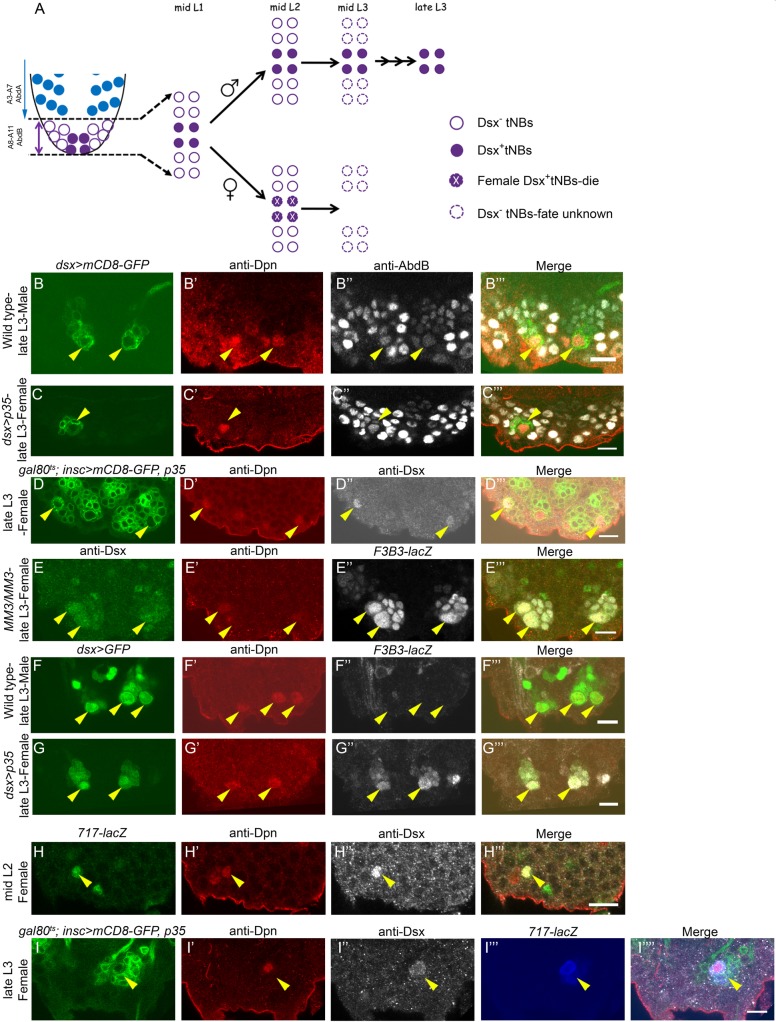
Fig. 1.
Sexually dimorphic expression of abdominal apoptotic enhancer in Dsx+tNBs. (A) Schematic of terminal segments (A8-A11) of larval VNC. (B,C) Dsx+tNBs are marked by dsx-GAL4 and express AbdB in males (n=10) (B″) and females (n=11) (C″). In females, cell death was blocked by p35. (D) Blocking cell death from mid-L2 rescues Dsx+tNBs in female VNCs (see Fig. S2B for TS protocol, n=10). (E) F3B3-lacZ expresses in Dsx+tNBs of MM3 mutant female VNCs (n=11) until late L3. (F,G) F3B3-lacZ shows sex-specific expression in Dsx+tNBs of female VNCs (n=10) (G″) but not in male VNCs (n=11) (F″). (H) 717-lacZ is expressed in mid-L2, prior to apoptosis in Dsx+tNBs of female VNCs (n=12). (I) 717-lacZ sustains its expression in Dsx+tNBs in female VNCs until late L3 when apoptosis is blocked (Fig. S2A for TS protocol, n=8). n number of VNCs analysed. Yellow arrowheads indicate Dsx+tNBs. Scale bars: 10 µm. All images are single confocal sections.
In the current body of work, we investigate the molecular mechanism of female-specific apoptosis of Dsx+tNBs in the larval CNS. We found that Hox gene Abd-B uses DsxF as a cofactor to cause sex-specific transcriptional activation of the RHG family of apoptotic genes, to generate sexual dimorphism in the structure of the developing CNS. Biochemical results suggest that AbdB and Dsx are capable of physically interacting with each other through their highly conserved homeodomain (HD) and DM domain, respectively. This interaction translates into cooperative binding of the two proteins on the DNA, which in vivo translates into sex-specific activation of the apoptotic enhancer in females. Congruent to this, mutagenesis of cooperative binding motifs is sufficient to abrogate female-specific apoptotic enhancer activity in vivo.
Collectively, our insights suggest that spatial determinants such as AbdB can utilize the sex-specific isoform of Dsx as a cofactor to select and activate the respective target genes in a tissue- and sex-specific manner. Considering the wide-ranging role of the dsx gene, we suggest that its capacity to collaborate with region-specific factors such as the Hox genes (in this case, AbdB) or other HD proteins could be a common theme for generating sexual dimorphism during development.
RESULTS
Sexually dimorphic expression of abdominal apoptotic enhancer in Dsx+tNBs
Dsx-expressing embryonic NBs express AbdB (Birkholz et al., 2013a), therefore we started out by testing whether Dsx and AbdB are also co-expressed in larval NBs. We found that NBs (marked by Deadpan, Dpn) in all of the four dsx-GAL4-marked tNB lineages (dsx-GAL4>mCD8-GFP) expressed Hox gene Abd-B in both males (Fig. 1B-B‴) and females (Fig. 1C-C‴). This was in contrast to abdominal NBs, which are Hox− and die on becoming Hox+ (or AbdA+) (Bello et al., 2003). To establish the precise time of apoptosis of Dsx+tNBs in female VNC, we expressed cell death blocker p35 in larval stages using a temporally inducible GAL4 system [tub-GAL80ts; insc-GAL4 used in temperature shift (TS) experiments]. We recovered all four Dsx+tNBs (4.0±0.89, n=12, N=3 where n is the number of VNCs analysed and N the number of experiments) for an early first instar larva stage (L1) TS (for TS experimental protocol, see Fig. S2A), an average of three of these cells (2.8±0.92, n=10, N=3) for mid-L2 TS (Fig. S2B; Fig. 1D-D‴) and none in early L3 TS (Fig. S2C), suggesting that these cells undergo apoptosis in approximately mid- to late L2.
Larval abdominal NBs apoptosis in A3-A7 segments requires grim and reaper expression, through an apoptotic enhancer lying within a 22 kb NBRR. A 54 kb genomic deletion (MM3) deletes the NBRR (Tan et al., 2011), whereas a smaller deletion called M22 deletes a 14.5 kb region within the NBRR. We found that female larval VNCs of genotype M22/MM3 show a block of Dsx+tNBs apoptosis (Fig. S1A-A″), suggesting that the apoptotic enhancer crucial for their death also lies within the 14.5 kb region uncovered by M22 deletion. A 1 kb region (referred to as F3B3) within M22 was identified as the abdominal NB apoptotic enhancer (Khandelwal et al., 2017). We tested and found that the 1 kb region (F3B3-lacZ) and its 717 bp subfragment (717-lacZ) were also expressed in Dsx+tNBs at the mid-L2 stage in female VNCs prior to cell death; thereby establishing precise temporal expression of the enhancer (1.5±0.52 Dsx+lacZ+tNBs; n=12 VNCs, N=3) (Fig. 1H-H‴). A hallmark of an apoptotic enhancer is its capacity to sustain expression until the target cells undergo apoptosis. Congruent to this, we observed that F3B3-lacZ and 717-lacZ were expressed and sustained until late L3 in all four Dsx+tNBs when cell death was blocked in female VNCs either by MM3 deletion (Fig. 1E-E‴) or by p35 expression (Fig. 1I-I″″) (Fig. S2A for TS protocol; 4.0±0.5 Dsx+lacZ+tNBs, n=8 VNCs, N=4).
Dsx+tNB apoptosis is female-specific (Birkholz et al., 2013a) and, as expected, we observed that F3B3-lacZ and 717-lacZ were specifically expressed in Dsx+tNBs of female VNCs (Fig. 1G″ and Fig. 1I‴), but not in male VNCs (Fig. 1F″ and Fig. S1B).
Collectively, these results suggested that AbdB and Dsx express in larval tNBs and that the abdominal apoptotic enhancer is activated in a sexually dimorphic fashion to cause death of Dsx+tNBs from the mid-L2 stage in females.
Dsx+tNB apoptosis in females requires AbdB but not Exd, Hth, Grh and Notch
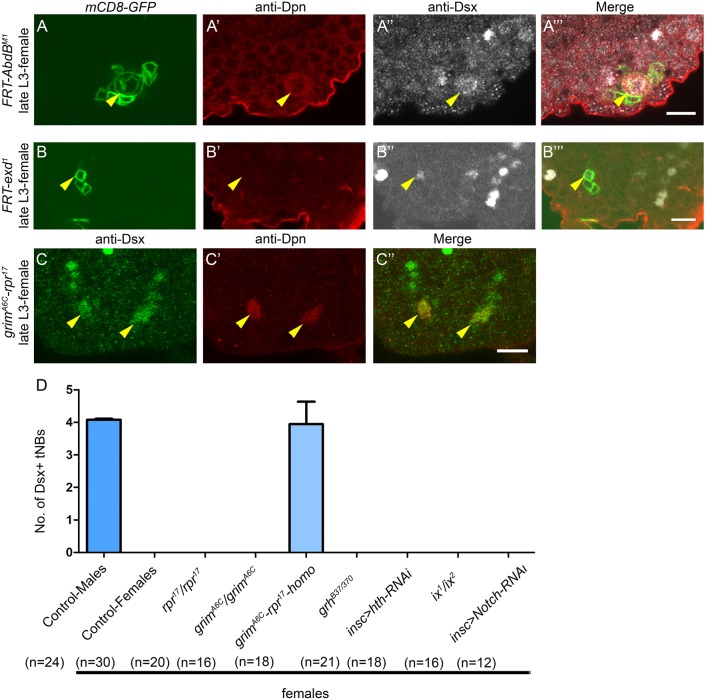
Because Dsx-mediated NB cell death is reported only in the AbdB-expressing region of the female larval CNS, we tested the role of AbdB in sex-specific apoptosis. To this end, MARCM clones were made for the loss-of-function AbdBM1 allele. We could successfully recover a Dsx-expressing NB clone mutant for the AbdBM1 allele in the terminal region of the female larval CNS (Fig. 2A-A‴) (9 MARCM clones analysed from 16 VNCs) showing that AbdB is crucial for apoptosis. Next, we tested known Hox cofactors Extradenticle (Exd) and Homothorax (Hth) for their expression (Fig. S4) and role in this apoptosis. In the case of exd1 mutant MARCM clones analysed in late L3, we could recover remnants of GFP-marked lineages that stained for Dsx, but none of these lineages contained any surviving NBs (12 lineages from 26 VNCs) (Fig. 2B-B‴). However, we recovered abdominal mutant clones with surviving NBs, showing that the mutant line was working (Fig. S5). This indicated that Dsx+tNBs divided before undergoing apoptosis and that Exd does not play a role in this apoptosis. Next, we tested whether Hth could substitute for Exd as a Hox cofactor. This was done by making hthP2 MARCM clones. Here again, we did not recover any Dsx+ lineages in female VNCs. RNA interference (RNAi)-mediated knockdown of Hth protein from early L1 (Fig. S2A for TS protocol) did not yield any Dsx+ lineages (n=18 VNCs, N=3) in female VNCs, although there was a potent knockdown of Hth protein in larval thoracic NBs (Khandelwal et al., 2017).
Fig. 2.
Dsx+tNB apoptosis in females requires AbdB and occurs through RHG genes grim and reaper. (A) Dsx+tNB mutants for AbdBM1 do not undergo apoptosis in female VNCs (n′=9). (B) exd1 mutant Dsx+tNBs die normally (n′=12). GFP-marked exd1 mutant lineage expressing Dsx protein and lacking the NB is shown. (C) grimA6C-rpr17 double-mutant homozygous female VNCs (n=18) exhibit a block of Dsx+tNB apoptosis. (D) Quantification of the Dsx+tNBs across various genotypes in the female VNC. n number of VNCs, n′ number of MARCM clones analysed. Yellow arrowheads indicate Dsx+tNBs. Scale bars: 10 µm. All images are single confocal sections. Graphs show mean±s.d.
Because Grh and Notch have been shown to work with AbdA in abdominal NBs apoptosis (Khandelwal et al., 2017), we assessed their role in Dsx+tNB apoptosis in females. Both Grh and Notch were expressed in Dsx+tNBs in mid-L2 (Fig. S4). Neither a CNS-specific null allelic combination of grh (grh370/B37) (Fig. 2D; n=21, N=2) nor knockdown of Notch using RNAi (from early L1; Fig. S2A for TS protocol) resulted in any ectopic Dsx+tNB apoptosis in female VNCs in late L3 (Fig. 2D; n=12, N=3). However, we did recover abdominal NBs in both the cases (Fig. S5).
Subsequently, homozygous deletions for RHG genes grim, reaper and grim-reaper double mutants were tested. We recovered four Dsx+tNBs only for the homozygous double deletion (grim-reaper) (Fig. 2C-C″ and Fig. 2D; 3.94±0.64, n=18 VNCs, N=3), suggesting that both grim and reaper are required for Dsx+tNB apoptosis in females (as in the case of abdominal NB cell death).
Collectively, these results show that Dsx+tNBs seem to rely only on Hox (AbdB) and DsxF for sex-specific activation of apoptotic genes grim and reaper in female VNCs, unlike apoptosis in abdominal NBs that depends on Hox (AbdA), Exd, Grh and Notch.
AbdB regulates Dsx in tNBs
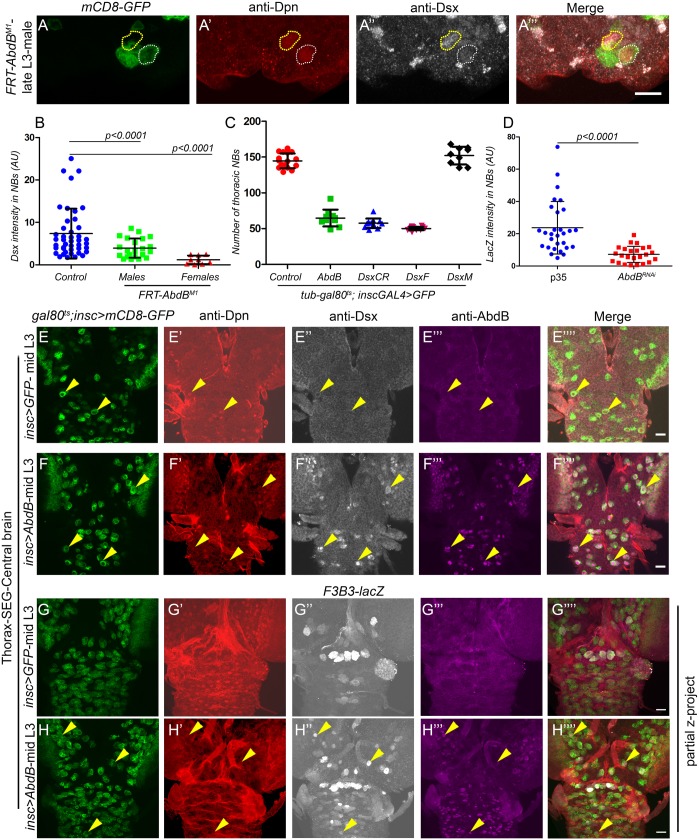
AbdB is known to regulate Dsx expression in non-neural tissues (Kopp et al., 2000; Wang et al., 2011; Wang and Yoder, 2012; Tanaka et al., 2011; Foronda et al., 2012); therefore, we tested regulation of Dsx expression in Dsx+tNBs by checking AbdBM1 MARCM clones in both females and males. We observed a decrease in Dsx staining in AbdBM1 mutant NBs (marked by mCD8-GFP) in both female (Fig. 2A″) and male VNCs (Fig. 3A″). Quantitative comparison showed that mutant NBs of both males (3.97±2.271, 22 MARCM clones analysed in 21 VNCs, N=4) and females (1.18±1.01, 9 clones in 16 VNCs, N=4) demonstrated a significant decrease in Dsx levels compared with control NBs (7.35±5.87, 20 clones in 43 male VNCs, N=4) (Fig. 3B). Next, ectopic expression of AbdB was tested for its capacity to induce Dsx in NBs of the anterior CNS (thorax, subesophageal ganglia and central brain region), where Dsx is not normally expressed (in NBs). We observed that induction of AbdB (Fig. S2E for TS protocol) in the anterior regions of the CNS (Fig. 3F‴) ectopically induced Dsx expression in NBs in both males and females compared with controls (Fig. 3E″ versus 3F″; n=12 VNCs, N=2).
Fig. 3.
AbdB regulates Dsx in tNBs. (A) Dsx+tNB mutant for AbdBM1 in male VNCs (n′=22) (white dotted line) show a decrease in levels of Dsx compared with adjacent wild-type control cells (yellow dotted line). (B) Graph comparing Dsx intensity in control NBs (n′=43) (in male VNCs) versus the NB mutant for AbdBM1 in male (n′=22) and female VNCs (n′=9). (C) Graph comparing the total number of thoracic NBs found in control VNC (n=17) versus AbdB (n=10), DsxCR (n=12), DsxF (n=14) and DsxM (n=9) overexpressing male VNCs at late L3 (Fig. S2E for TS protocol for AbdB and Fig. S2B for others). (D) Graph comparing F3B3-lacZ intensity in Dsx+tNBs in female VNCs as control (blocked for apoptosis using p35, n=9) versus AbdB knockdown (n=9) (Fig. S2A for TS protocol). (E-H) Ectopic expression of AbdB in anterior region of CNS induces Dsx (n=12) (F″) and F3B3-lacZ (n=8) (H″) in NBs compared with wild-type controls (n=11 for E″, n=10 for G″) (Fig. S2E for TS protocol). Both male and female VNCs were analysed. Representative data and images shown here are from males, except in B where both male and female data are shown and in D where only female data is shown. n number of VNCs, n′ number of MARCM clones analysed. Yellow arrowheads indicate NBs. Scale bars: 10 µm for A; 20 µm for E-H. All images are single confocal sections except panels G and H. Graphs show mean±s.d. Significance (P-value) is from two-tailed Student's unpaired t-test.
Ectopic expression of AbdB in the anterior region of the CNS is known to cause apoptosis of thoracic NBs (Bello et al., 2003). Congruent to this we also found that AbdB could induce apoptotic enhancer F3B3-lacZ in anterior regions of the CNS both in males (Fig. 3H″, n=8 VNCs, N=2) and females, as scored by an increase in intensity of lacZ and number of lacZ-expressing cells (Fig. 3G″ versus 3H″). The expression of F3B3-lacZ seen in control CNS (Fig. 3G″) was background leaky expression of the enhancer-lacZ. Comparison of the number of surviving GFP-marked thoracic NB lineages for control (144.6±2.5 NBs, n=17 VNCs, N=3) versus AbdB overexpression in males (64.80±3.7 NBs, n=10 VNCs, N=3; Fig. 3C) and females (69.8±8.8, n=12 VNC, N=3), showed a marked decrease in the latter two cases. Comparison of F3B3-lacZ expression in Dsx+tNBs for AbdB knockdown by RNAi versus p35-expressing controls was carried out for female VNCs in late L3 (Fig. S2A for TS protocol). Analysis showed that AbdB-RNAi could consistently downregulate F3B3-lacZ in NBs (7.19±5.0; measured from 24 NBs, n=9 VNCs, N=3) compared with p35-expressing controls (23.6±16.2; measured from 28 NBs, n=9 VNCs, N=3; Fig. 3D). A similar experiment could not be done for Dsx because none of the RNAi lines tested were potent enough to significantly knock down Dsx expression in NBs.
These results show that AbdB regulates Dsx expression in tNBs of both the male and female CNS. The ectopic induction and knockdown experiments suggest that AbdB can transcriptionally regulate the apoptotic enhancer in Dsx+tNBs of female VNCs.
Dsx common region is enough to cause NB apoptosis
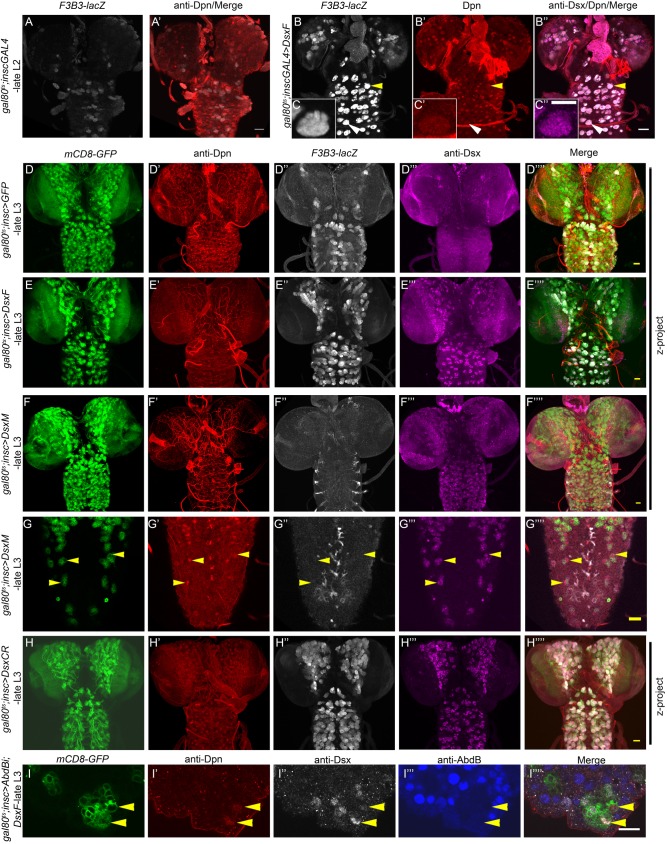
Because DsxF promotes apoptosis, whereas DsxM blocks apoptosis of Dsx+tNBs (Birkholz et al., 2013a), we tested whether DsxF could induce F3B3-lacZ in the anterior CNS. We observed that ectopic expression of DsxF in mid-L2 (Fig. S2F for TS protocol) induced F3B3-lacZ expression in NBs of the anterior CNS 7 h later (Fig. 4A versus 4B, n=14 VNCs, N=3; Fig. 4C-C″, shown as inset to Fig. 4B-B″). Expectedly, many of the thoracic NBs expressing DsxF eventually underwent apoptosis in late L3 (Fig. S2B for TS protocol; Fig. 4D versus 4E), as judged by comparison of the number of GFP-marked surviving thoracic lineages in L3 (50.14±0.6 NBs, n=14 VNCs, N=2; Fig. 3C) compared with controls (144.6±2.5 NBs n=17, N=2; Fig. 3C). In contrast, ectopic expression of DsxM did not result in any NB apoptosis in the thoracic region (152.0±4.1 NBs, n=9 VNCs, N=2; Fig. 3C, Fig. 4D versus 4F). In fact, DsxM repressed the background expression of F3B3-lacZ in the anterior CNS (Fig. 4F″). DsxM also repressed developmental apoptosis as well as F3B3-lacZ expression in larval abdominal NBs (yellow arrowhead in Fig. 4G-G″″; n=12 VNCs, N=3). To test the role of the female-specific C-terminal region in apoptosis, we ectopically expressed a Dsx form that lacked the entire female-specific region and contained only the Dsx common region (hereafter referred to as DsxCR; Fig. 5A). Interestingly, we observed that DsxCR expression was capable of causing thoracic NB apoptosis in late L3 (57.50±1.9 NBs, n=12 VNCs; Fig. 3C, Fig. 4D versus 4H) with an efficiency almost comparable to that of DsxF (50.14±0.6 NBs, n=14; Fig. 3C). We also observed that DsxCR was capable of causing the death of all four Dsx+tNBs in the male VNC (n=18 VNC; Fig. S2A for TS protocol) and ectopically inducing F3B3-lacZ in the anterior CNS (Fig. 4D″ versus 4H″).
Fig. 4.
Role of sex-specific region of Dsx in apoptosis. (A,B) Compared with controls (n=10) (A,A′), induction of DsxF in mid-L2 (B-B″) activates F3B3-lacZ in NBs of the anterior CNS in late L2 (n=14) (Fig. S2F for TS protocol). (C) Induction of F3B3-lacZ in thoracic NBs in response to ectopic expression of DsxF. (D,E,H) Compared with controls, misexpression of both DsxF (n=17) and DsxCR (n=12) cause induction of F3B3-lacZ (D″ vs E″; D″ vs H″) as well as NB apoptosis in the anterior CNS at late L3 (D vs E; D vs H) (Fig. S2B for TS protocol). (F) Ectopic expression of DsxM (n=11) in the anterior CNS (F‴) represses the leaky expression of F3B3-lacZ in thoracic segments (F″) and does not cause apoptosis of thoracic NBs (F). (G) DsxM also represses F3B3-lacZ (n=12) (G″) and blocks apoptosis of abdominal NBs (G). (I) Simultaneous overexpression of DsxF and knockdown of AbdB in the female VNCs (n=8) blocks apoptosis of tNBs (Fig. S2A for TS protocol), suggesting that both AbdB and DsxF are required for NB apoptosis. Both male and female VNCs were analysed. Representative data and images are shown here are from males, except in F,G,I where only female data is shown. n number of VNCs analysed. Yellow arrowheads indicate NBs. Scale bars: 20 µm for A,B,D-H; 10 µm for C,I. All images are single confocal sections except panels D,E,F,H.
Fig. 5.
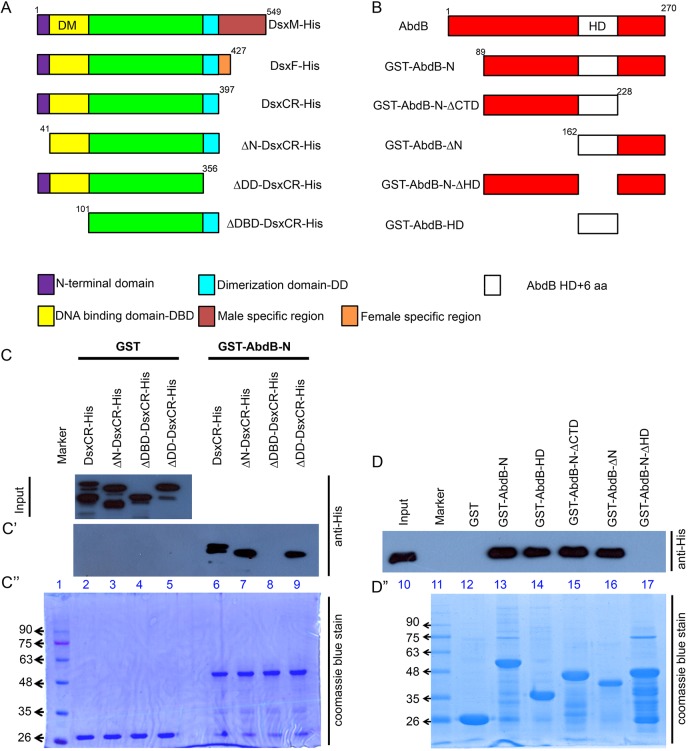
AbdB and Dsx interact physically in vitro. (A,B) Schematic of full-length and domain-deleted proteins used in the case of Dsx (A) and AbdB-r (B). (C,D) Western blot of the GST pulldown for bacterially expressed His-tagged input cell lysates of DsxCR and various domain deletions of Dsx. Samples were tested with GST alone, GST-AbdB-N (C′) and various GST-tagged domain deletions of AbdB-N (D). Pulldown shows that AbdB-N and DsxCR interact through their HD (lanes 13-17) and DM domain (lanes 6-9), respectively. (C″-D″) SDS-PAGE stained with Coomassie Blue depicts almost equal loading of the GST-tagged protein samples.
These results indicated that the common region of Dsx protein is sufficient to bring about NB apoptosis, and that the female-specific region complements this capacity whereas the male-specific region seems to inhibit it.
AbdB and DsxF are necessary for apoptosis
Our results suggest that both AbdB and DsxF are capable of inducing tNB apoptosis. Because AbdB seems to positively regulate Dsx in NBs, we wanted to know whether both AbdB and DsxF are necessarily required for Dsx+tNB apoptosis or whether AbdB executes apoptosis solely by regulating DsxF expression in tNBs. To test this, we induced a simultaneous RNAi-mediated knockdown of AbdB and overexpression of DsxF from early L1 and examined its effect in late L3 (Fig. S2A for TS protocol). We could successfully recover as many as 12 Dsx-expressing NBs (Fig. 4I-I″″) in the terminal region of female VNCs in late L3 (n=8 VNCs, N=2) compared with controls (expressing just DsxF), which had none. This implied that even though DsxF was expressed in NBs, knockdown of AbdB was sufficient to block their cell death and both AbdB and DsxF are needed for NB apoptosis in the terminal region of female VNCs.
AbdB and DsxCR interact in vitro
Next, we decided to test the physical interaction between AbdB and Dsx. We started out by using the Dsx common region (DsxCR) and tested its interaction with AbdB (Fig. 5A). Both morphogenetic (AbdB-m) and regulatory (AbdB-r) isoforms of AbdB are expressed in the CNS (Birkholz et al., 2013b). Because Dsx+tNBs reside in PS13, which mainly expresses the AbdB-r isoform, as well as for ease of handling, we chose a truncated version of AbdB-r for our biochemical experiments that lacks the first 89 amino acids (hereafter referred to as AbdB-N; Fig. 5B).
In a GST-pulldown assay, we observed that although GST alone did not pull down His-DsxCR (lane 2, Fig. 5C′ and lane 12, Fig. 5D), GST-AbdB-N could successfully pull down His-DsxCR (approximately 55 kDa; lane-6, Fig. 5C′ and lane 13, Fig. 5D). This suggested that Dsx and AbdB interact with each other in vitro and that the interaction is mediated through a domain lying outside the sex-specific region of the protein. Next, we made a series of truncations for both Dsx and AbdB to map the respective interaction domains (detailed in Fig. 5A,B and Fig. S6B,C). We found that although individual deletions of the N-terminal domain (ΔN-DsxCR) or the dimerization domain (ΔDD-DsxCR) of Dsx did not alter its binding with AbdB (lanes 7 and 9, Fig. 5C′), deletion of the DM domain or the DNA-binding domain (ΔDBD-DsxCR, 41-101 amino acids) completely abolished the interaction between the two proteins (lane 8, Fig. 5C′). Reciprocal mapping of the domain within AbdB responsible for its interaction with DsxCR was carried out using four constructs centred around the AbdB HD region (60 amino acids of the HD and, additionally, two amino acids N-terminal to the HD and four C-terminal to the HD; Fig. 5B and Fig. S6C). For the ease of representation, we chose to call the entire stretch of 66 amino acids AbdB-HD (Fig. 5B). We found that AbdB-HD retained its ability to interact with DsxCR when the regions C-terminal and N-terminal to HD were deleted (lanes 15 and 16, Fig. 5D). However, deletion of AbdB-HD (AbdB-N-ΔHD) abolished its interaction with DsxCR completely (lane 17, Fig. 5D). Expectedly, AbdB-HD alone was sufficient to pull down His-DsxCR (lane 14, Fig. 5D). These results show that AbdB and Dsx interact with each other in vitro through the DM domain (DBD) of Dsx and the HD of AbdB.
DsxCR and AbdB show cooperative binding on the apoptotic enhancer
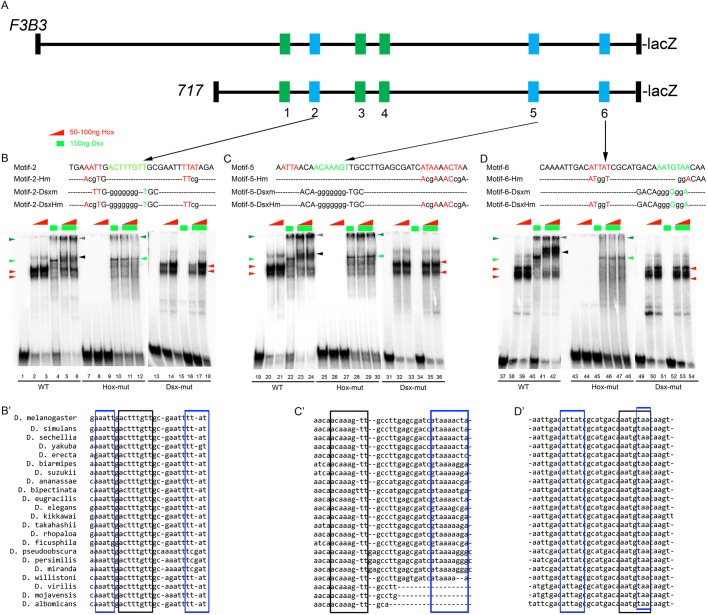
To test whether the interaction between DsxCR and AbdB also took place on the DNA, we analysed the 1 kb enhancer for potential Dsx and AbdB binding sites. We identified 11 potential Dsx sites conforming to the variation of the consensus sequence (RNNACWAWGTNNY) (Luo et al., 2011; Clough et al., 2014). All these sites had AT-rich sequences (potential Hox binding sites) within 20 bp proximity (Sorge et al., 2012). Six out of eleven motifs showed DsxCR binding by EMSA (Fig. 6 and Fig. S3). These six Dsx-binding motifs were concentrated in a highly conserved 717 bp subfragment of the 1 kb apoptotic enhancer (motifs 1-6, Fig. 6A). Three of the six binding motifs (motifs 2, 5 and 6) were found to be highly conserved (Fig. 6B′-D′) and were analysed in detail.
Fig. 6.
DsxCR and AbdB cooperatively bind on the sex-specific apoptotic enhancer. (A) Schematic of F3B3-lacZ and 717-lacZ depicting six motifs containing Dsx and AbdB binding sites. Blue motifs exhibit cooperative binding of DsxCR and AbdB whereas green motifs do not (refer to Fig. S3). (B-D) EMSA for AbdB, DsxCR and AbdB-DsxCR binding on wild-type (WT), AbdB mutant and Dsx mutant oligos for motifs 2, 5 and 6. WT motif sequences are colour-coded for Hox (red) and Dsx (green) binding sites. Mutant variants are shown in lower case. Red arrowheads indicate Hox–DNA complex; green and dark green arrowheads indicate Dsx monomer and dimer complexes with DNA, respectively. Black arrowhead indicates Hox–Dsx–DNA cooperative complex. Grey arrowhead indicates cooperative complex with two molecules of Dsx. (B′-D′) Sequence comparison across multiple species for three motifs analysed. Dsx and Hox binding sites are highlighted by black and blue boxes, respectively.
We used two different concentrations of Hox protein AbdB (50 and 100 ng) and a fixed concentration of DsxCR (150 ng) and found that these proteins bound individually to motif 2 (lanes 2-4, Fig. 6B), motif 5 (lanes 20-22, Fig. 6C) and motif 6 (lanes 38-40, Fig. 6D). Dsx protein is known to have a dimerization domain and therefore Dsx binds on DNA both as a monomer and dimer. The specificity of the binding for AbdB and Dsx was established using oligos mutant for potential AbdB and Dsx binding sites. We found that an oligo mutant for the Hox binding site resulted in loss of AbdB binding for all three binding motifs (lanes 8-9, 26-27 and 44-45; Fig. 6B-D), leaving the Dsx binding either intact in the case of motif 5 or partly affected in the case of motifs 2 and 6 (lanes10, 28 and 46, Fig. 6B-D). Reciprocally we found that an oligo mutant for potential Dsx binding sites showed a complete loss of DsxCR binding (lanes 16, 34 and 52, Fig. 6B-D) leaving AbdB binding mostly unaltered (lanes 14-15, 32-33 and 50-51, Fig. 6B-D) in all three motifs. Interestingly, we found that AbdB protein seems to form a cooperative complex of slower mobility in the presence of DsxCR, as indicated by the black arrowhead in Fig. 6B-D and by an increase in binding intensity of the DsxCR dimer band (grey arrowhead), which was observed for all three motifs (lanes 5-6, 23-24, 41-42, Fig. 6B-D). This cooperative synergism of the two proteins is lost in the case of oligos with mutation for either AbdB (lanes 11-12, 29-30 and 47-48, Fig. 6B-D) or Dsx binding sites (lanes 17-18, 35-36 and 53-54, Fig. 6B-D).
These results show that AbdB and DsxCR cooperatively interact with each other on three binding motifs (motifs 2, 5 and 6) found on the sex-specific apoptotic enhancer and their interaction on DNA is probably mediated through the HD of AbdB and the DM domain of Dsx.
DsxF helps AbdB to select and activate the apoptotic enhancer
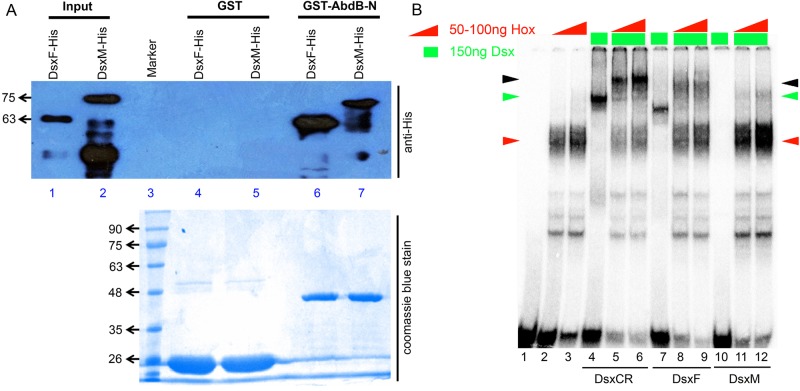
Because DsxM represses apoptosis of abdominal NBs (Fig. 4G′), we decided to test the isoform-specific interaction of Dsx with AbdB. For this, AbdB was tested for its interaction with full-length male- and female-specific isoforms of Dsx in a GST pulldown assay. Here, we observed that although GST alone did not pull down either of the isoforms (lanes 4 and 5, Fig. 7A), GST-tagged AbdB-N could successfully pull down both DsxF and DsxM (approximately running at 63 and 75 kDa, respectively; lanes 6 and 7, Fig. 7A). We observed that AbdB interacted with DsxF much more strongly than with DsxM. Because Dsx+tNB apoptosis and enhancer expression is female-specific, we tested AbdB and DsxF for cooperative interaction on motif 5. We observed that although AbdB (lanes 2 and 3, Fig. 7B), DsxCR (lane 4, Fig. 7B) and DsxF (lane 7, Fig. 7B) bound individually to DNA, there was no significant binding of DsxM (lane 10, Fig. 7B) on the DNA. In the presence of AbdB, we observed a lower mobility cooperative complex for both DsxF and DsxCR (lanes 5-6 and 8-9, Fig. 7B), but we did not observe a significant band of similar or lower mobility for DsxM, considering its larger size (lanes 11 and 12, Fig. 7B), although we did observe an increase in AbdB band intensity. These results suggest that although DsxCR and DsxF cooperatively interact with AbdB to bind on DNA, DsxM does not (Fig. 7B). The results also suggest that the DsxCR region is capable of forming a complex with AbdB on the apoptotic motifs of the enhancer; furthermore, the female-specific region facilitates complex formation but the male-specific region seems to interfere with the same.
Fig. 7.
Sex-specific interaction of Dsx with AbdB. (A) Western blot of the GST pulldown shows that DsxF interaction with AbdB is stronger than that of DsxM. Bacterially expressed His-tagged input cell lysates of DsxF and DsxM (lanes 1 and 2) are pulled down by GST-AbdB (lanes 6 and 7) but not by GST alone (lanes 4 and 5). SDS-PAGE stained with Coomassie Blue depicts almost equal loading of the GST-tagged protein samples. (B) EMSA for DsxCR, DsxF and DsxM with AbdB on motif 5 shows that DsxCR and DsxF bind cooperatively with AbdB and DsxM does not. Red arrowheads indicate Hox–DNA complex, green arrowheads indicate Dsx–DNA complex, black arrowhead indicates Hox–Dsx–DNA cooperative complex.
These results support the apoptosis-inducing potential of the Dsx isoforms tested earlier. DsxF (and DsxCR), which formed a cooperative complex with AbdB on the apoptotic enhancer, were capable of inducing apoptosis of Dsx+tNBs in females whereas DsxM, which does not show cooperation with AbdB on the apoptotic enhancer, repressed the death of Dsx+tNBs in females.
This also suggests that Dsx isoforms can act as a sex-specific cofactor for Hox gene Abd-B and can help it to select its target genes.
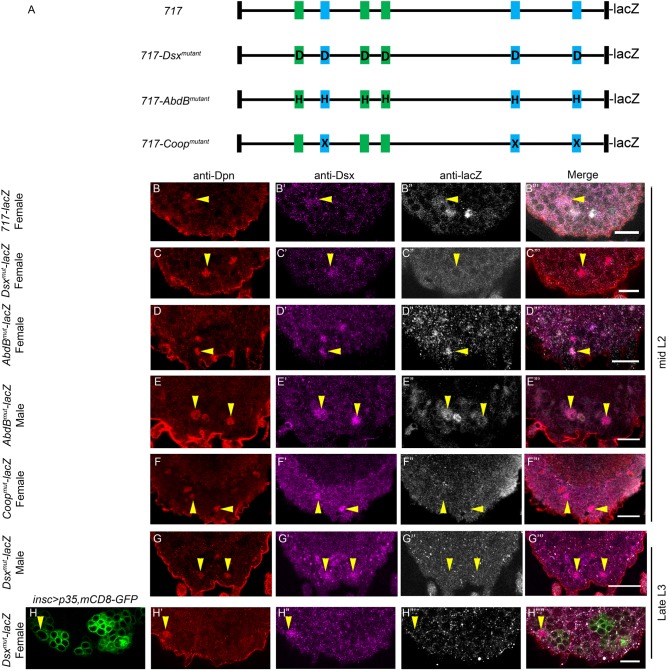
DsxF is required for activity of the apoptotic enhancer
To test the in vivo relevance of the binding sites, we mutagenized six Dsx binding sites that bound DsxCR (717-Dsxmutant-lacZ; mutations as shown in Fig. 6B-D, and supplementary Materials and Methods). We tested the activity of the mutagenized enhancer in mid-L2 in Dsx+tNBs of the female VNC. Although we observed the expression of 717-lacZ in Dsx+tNBs of the female VNC (Fig. 8B-B‴; n=21 VNCs, N=4), we could not observe any lacZ expression in the case of 717-Dsxmutant-lacZ at the same time in these cells (Fig. 8C-C‴). To rule out any delay in enhancer initiation, we tested 717-Dsxmutant-lacZ for its expression in late L3 by temporally blocking Dsx+tNB apoptosis using p35 (Fig. S2A for TS protocol). We found that 717-Dsxmutant-lacZ did not show any expression in the Dsx+tNBs of female VNCs in late L3 (Fig. 8H-H″″). In contrast to what is known for other Dsx target genes, we observed that 717-Dsxmutant-lacZ did not show any ectopic expression of lacZ in Dsx+tNBs of the male VNC (Fig. 8G-G‴). This suggests that DsxM-mediated repression of apoptosis and enhancer-lacZ (Fig. 4G) is through a mechanism other than simple DsxM binding on the enhancer and silencing. This also suggests that the Dsx binding motif in the enhancer is crucial for its activation in the female VNC, implying a role for DsxF in sex-specific enhancer initiation.
Fig. 8.
Enhancer mutagenesis analysis in vivo. (A) Schematic of 717-lacZ, 717-Dsxmutant-lacZ, 717-AbdBmutant-lacZ and 717-Coopmutant-lacZ. (B-F) Shows the enhancer-lacZ expression for wild-type and various mutant versions of the enhancer in Dsx+tNBs at mid-L2. 717-lacZ expresses normally (n=21) (B″) but Dsxmutant-lacZ (n=14) (C″) and Coopmutant-lacZ (n=12) (F″) fail to express in Dsx+tNBs of female VNCs in mid-L2. AbdBmutant-lacZ expression is unaffected in Dsx+tNBs of females (n=9) (D″), but male VNCs show ectopic expression in Dsx+tNBs where it is normally not expressed (n=11) (E″). In contrast, Dsxmutant-lacZ expression is not observed in either male (n=15) (G-G‴) or cell death-blocked female VNCs (n=8) (H-H″″) in late L3 (Fig. S2A for TS protocol). n indicates the number of VNCs analysed. Yellow arrowheads indicate Dsx+tNBs. Scale bars: 10 µm. All the images are single confocal sections.
AbdB acts as a repressor of the apoptotic enhancer
Next, AbdB binding sites were mutagenized across the six motifs, leaving neighbouring Dsx binding sites intact (717-AbdBmutant-lacZ; mutations as shown in Fig. 6B-D, see supplementary Materials and Methods for details). We found that reporter lacZ expression in the case of 717-AbdBmutant-lacZ in the female VNC at mid-L2 was unaffected in Dsx+tNBs (Fig. 8D-D‴). Interestingly, in the case of male VNCs, we found that reporter lacZ was activated in Dsx+tNBs (Fig. 8E-E‴). This suggested that AbdB binding on the enhancer normally keeps it in a repressed state.
Because DsxCR and DsxF cooperate with AbdB on three motifs (motifs 2, 5 and 6) in the enhancer and DsxM does not, we hypothesized that these motifs are central to sex-specific activation of the apoptotic enhancer and, therefore, cell death in female VNCs. To test this, we mutagenized both Dsx and AbdB binding sites in these three cooperative motifs (717-Coopmutant-lacZ; mutations as shown in Fig. 6B-D). Mutagenesis of AbdB-Dsx binding sites found in these motifs was sufficient to abrogate enhancer activity in Dsx+tNB in the female VNC (Fig. 8F-F‴). This underlined the importance of the three motifs and the cooperative interaction of AbdB-Dsx on these motifs for sex-specific activation of the enhancer.
These results show that AbdB normally functions as a repressor of the apoptotic enhancer. Perhaps its capacity to form a cooperative complex with DsxF converts it into an activator in the complex, and it therefore plays a central role in sex-specific activation of the apoptotic enhancer.
Dsx is the trigger for apoptosis
An enhancer mutant for Dsx binding sites failed to initiate the expression of the reporter gene, and AbdB alone functions as a repressor of the wild-type enhancer. Therefore, we explored whether DsxF can act as a trigger for activation of apoptosis in females by quantifying Dsx levels using antibody staining from late L1 to mid-L2 (temporal window prior to apoptosis). Because it is difficult to sex the larvae at L1, quantification was carried out for a mixed population of male and female larvae. We observed a steady increase in levels of Dsx across these stages (with almost no expression in late L1), although the size of the cells remained almost constant (Fig. 9). A simultaneous increase in AbdB expression levels was also observed in the same time window. This led us to propose that levels of Dsx increase in both sexes, but this acts as a trigger for apoptosis of Dsx+tNBs only in females.
Fig. 9.
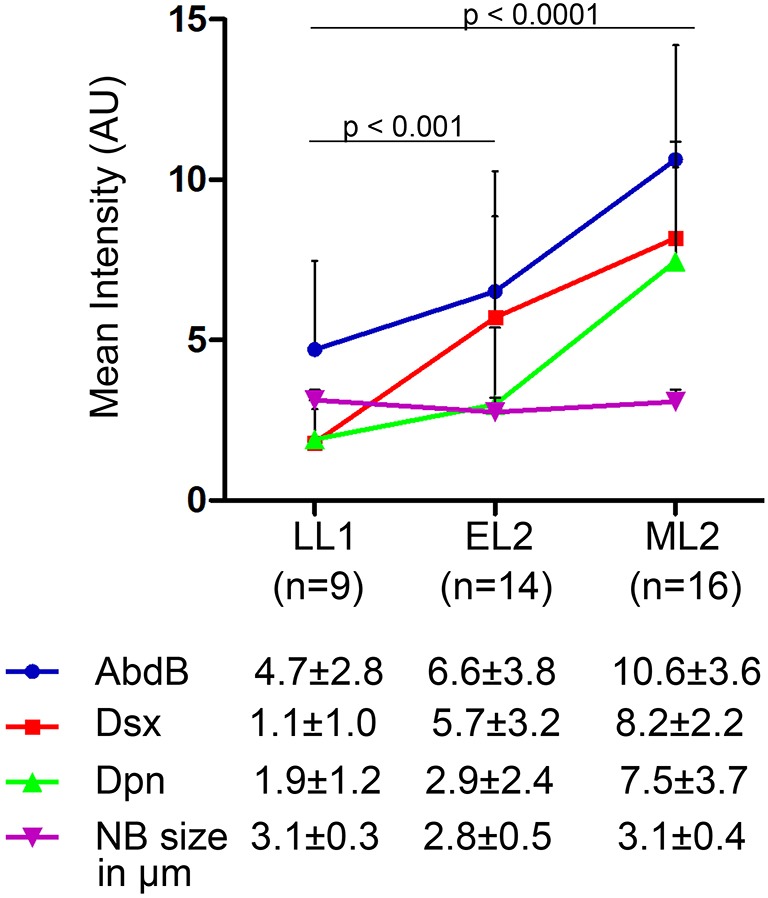
DsxF act as the trigger for Dsx+tNBs apoptosis in female VNCs. Graph showing increasing levels of Dsx, AbdB and Dpn staining along with NB size, across different stages. n number of VNCs analysed. Significance (P-value) was calculated using the two-tailed Student's paired t-test for Dsx levels calculated across three stages.
DISCUSSION
Multicellular organisms are known to be a mosaic of cells and tissues, which are on a “need to know basis” as far as sex is concerned (Hempel and Oliver, 2007; Lee et al., 2002; Rideout et al., 2010; Robinett et al., 2010). These cells and tissues have to tightly regulate the expression of sex-specific genes and their dimorphic targets such as Lgr3 (Meissner et al., 2016), dsx (Zhou et al., 2014), bab1 (Williams et al., 2008), Yp genes (An and Wensink, 1995; Burtis et al., 1991; Coschigano and Wensink, 1993) and fmo-2 (Luo and Baker, 2015); more so because their expression might be required in one tissue (or sex) and detrimental in another (Clough et al., 2014). Sexually dimorphic enhancers are thought to function as genetic switches (Tanaka et al., 2011; Williams and Carroll, 2009) and are central to such tight regulation. They are expected to have binding sites for both sex-specific and tissue- or region-specific TFs. Enhancers for the yolk protein (Yp) genes (An and Wensink, 1995; Burtis et al., 1991; Coschigano and Wensink, 1993), bab1 (Kopp et al., 2000; Williams et al., 2008) and the RHG genes (required for female-specific Dsx+tNB apoptosis) are the only known examples of this kind. In Dsx+tNBs, we observed that AbdB and Dsx work in hierarchical as well as collaborative fashion, as reported earlier for non-neural tissues (Wang et al., 2011; Wang and Yoder, 2012; Kopp et al., 2000; Tanaka et al., 2011; Williams et al., 2008). We found that these two proteins physically interact through the highly conserved HD (of AbdB) and DM domain (of Dsx) (Fig. 5). AbdB and Dsx also show cooperative interaction on the sexually dimorphic apoptotic enhancer (Fig. 6), which has not been reported earlier (Williams et al., 2008). This identifies an unexpected novel functional link between the two kinds of TFs and underlines their capacity to use each other as partner or cofactor to influence specific outcomes in male and female bodies. Because Dsx+tNBs apoptosis is specific to terminal segments of the female VNC, it is suggested that context and interaction of AbdB and DsxF on the enhancer ensures that RHG genes are activated only in females (and not in males, or in NBs of other regions of the developing CNS).
Our results also suggest that although the Dsx common region is crucial for DNA binding and its interaction with AbdB (for target gene selection), the sex-specific regions have minimal impact on the same. In fact, the latter seem to play an important role in determining the regulatory output of the AbdB–Dsx complex, depending on the context (target gene, cell type and sex). Therefore, it is equally plausible to expect that DsxM and AbdB also cooperate in selection and regulation, albeit on a different set of target genes. This is in agreement with the finding that both DsxF and DsxM have a similar genomic occupancy pattern and therefore have many common target genes, yet many of them have tissue- and sex-specific expression (Clough et al., 2014). We propose that this could be a consequence of their interaction with spatial TFs such as AbdB or other HD-containing factors (including other Hox genes). Insights from mutagenesis studies on the Yp gene fat body enhancer element (o-r) support this idea (Coschigano and Wensink, 1993), wherein Dsx has been suggested to work with spatial TFs (such as AEF-1 and C/EBP) to localize Yp gene expression to the female fat body. In the fat body, DsxF was suggested to synergize with a C/EBP class of TFs to activate Yp genes, whereas the C-terminal region of DsxM was thought to interfere with this complex formation and hence its activation (Coschigano and Wensink, 1993).
Enhancer mutagenesis and the Dsx expression profile prior to apoptosis imply a role of DsxF in female-specific activation of RHG genes in Dsx+tNBs. However, a relative delay in the expression of Dsx compared with AbdB suggests a role for additional regulation. The temporal series TF Castor (Isshiki et al., 2001) has been shown to play a role in abdominal NBs apoptosis through regulation of the grh gene (Maurange et al., 2008). However, we did not find Grh protein to be crucial for Dsx+tNB apoptosis, raising the possibility of Dsx being a direct target of Castor.
Considering the literature, we expected that DsxM binding on the enhancer would actively silence apoptosis in males (Luo and Baker, 2015; Coschigano and Wensink, 1993). Interestingly, it was the mutagenesis of AbdB binding sites that activated the enhancer in males. This implies that AbdB (and not DsxM) acts as a repressor for the wild-type enhancer. It also suggests that the capacity of AbdB to form a cooperative complex with DsxF perhaps converts the two into an activator complex on the death enhancer (exclusively in females). In thoracic NBs that lack Dsx expression, misexpression of Hox genes causes NB apoptosis (Bello et al., 2003); we believe that Grh plays an important role in this apoptosis (Khandelwal et al., 2017).
The cellular role of Hox genes (like any other TF) is expected to be highly context dependent. For instance, Hox genes have been shown to directly regulate RHG genes to function both as promoters and repressors of apoptosis in different contexts (Bello et al., 2003; Miguel-Aliaga and Thor, 2004; Lohmann et al., 2002). More specifically, the role of AbdB in the survival of dMP2 and dMP1 neurons in embryonic VNC (Miguel-Aliaga and Thor, 2004) is most likely executed by direct binding and repression of a common neuronal enhancer for grim and reaper genes. Similarly, female-specific motor neurons expressing FS-Ilp7 could also be using AbdB for their selective survival, or for their male-specific death. It is of interest to check the former cell types for Dsx expression and their possible cooperative role with AbdB to repress RHG genes in both sexes; however, the sex-specific survival of motor neurons expressing FS-Ilp7 is more intriguing, wherein the detailed mechanism of male-specific apoptosis that occurs in the absence of Dsx expression is yet to be elucidated (Garner et al., 2018).
Even though abdominal NBs and Dsx+tNBs employ the same enhancer for apoptosis, they use different molecular strategies. In abdominal NBs, a pulse of AbdA in mid-L3 triggers apoptosis (Bello et al., 2003) and requires Grh, Exd and Notch signalling (Khandelwal et al., 2017). Dsx+tNBs, however, utilize increasing levels of DsxF as a trigger to recruit otherwise repressive AbdB into a cooperative activator complex specifically in females but not in males (Fig. S7). This is a clever modification of the abdominal apoptotic strategy by replacing Grh, Notch and Exd with Dsx to engineer the cell death of a desired cell type in females at an earlier time point. Because there is a considerable overlap between Dsx and DMRT1 target genes (Murphy et al., 2010), utilization of HD-containing TFs (as cofactors) can be a general strategy used by both Dsx and DMRT1 to carry out their tissue-, cell type- and sex-specific gene regulations.
MATERIALS AND METHODS
Fly stocks and fly husbandry
Fly stocks used were F3B3-lacZ, 717-lacZ, M22/TM6Tb, FRT-exd1 (Khandelwal et al., 2017); dsx-GAL4 (S. F. Goodwin, University of Oxford, UK) (Rideout et al., 2010); MM3/TM6Tb and grimA6C/TM6Tb (Kristin White, Harvard Medical School, USA) (Tan et al., 2011); FRT-AbdBM1 (E. Sanchéz-Herrero, CSIC-UAM, Spain); grhB37/CyO and grh370/CyO (Sarah Bray, University of Cambridge, UK) (Almeida and Bray, 2005); hsflp, FRT19A, tub-GAL80; tub-GAL4, UAS-mCD8-GFP/CyO-GFP (H. Reichert, University of Basel, Switzerland); UAS-dcr2; inscGAL4 UAS-mCD8-GFP; tub-GAL80ts (J. A. Knoblich, IMBA, Austria) (Neumüller et al., 2011); UAS-DsxM, UAS-DsxF [G. Lee, University of Tennessee, USA, and Bloomington Drosophila Stock Center (BDSC), 44224 and 44223] (Lee et al., 2002); Canton-S (BDSC, 64349); UAS-DsxCR-DPiM (K. VijayRaghavan, NCBS, India) (Guruharsha et al., 2011); ix1 (BDSC, 205); ix2 (BDSC, 372); UAS-p35 [Drosophila Genomics Resource Center (DGRC), 108019]; UAS-AbdB (DGRC, 106120); elav[C155]-GAL4, UAS-mCD8-GFP, hsflp1, w (BDSC, 5146); yw; FRT82B tub-GAL80-LL3 (BDSC, 5135); Notch-RNAi (BDSC 28981); AbdB-RNAi [Vienna Drosophila Resource Center (VDRC), 11024]; and hth-RNAi (NIG-17117-R4 and R2).
The following transgenic lines were used: 717-Dsxmutant-lacZ, 717-AbdBmutant-lacZ and 717-Coop-mutant-lacZ, generated by site-specific insertion at attP40-25C6. The rpr17 deletion line was generated by mobilization of the P-element, inserted 891 bp from the 5′-region of rpr transcript (BDSC-14326) [the deletion line was confirmed through genomic PCR mapping (Fig. S1)]. A recombinant strain of grimA6C-rpr17/TM6B,Tb was obtained after screening 20 independent recombinant lines for block of abdominal and terminal pNB apoptosis.
All the fly stocks and crosses were maintained at 25°C unless otherwise stated. For fly crosses, 4 h egg collections were carried out and the larvae reared at 25°C. All larvae were dissected at the desired larval stage.
Immunohistochemistry and image acquisition
Larvae of the desired sex, genotype and age were dissected as described earlier (Khandelwal et al., 2017) with the following variations: fixation was done for 30 min at room temperature and immunostaining with primary antibodies overnight at 4°C. The primary antibodies used were rabbit anti-Dpn (1:5000) (Khandelwal et al., 2017), mouse anti-Grh (1:2000) (Khandelwal et al., 2017), mouse Abd-B (1:20; 1A2E9, DSHB), rat anti-Dsx (1:3000; Fig. S6F), mouse anti-Exd (1:20; B11M, DSHB), mouse anti-NICD (1:50; C17.9C6, DSHB), guinea pig anti-Hth (1:100; GP52, Richard Mann), chicken anti-β-gal (1:2000; ab9361, Abcam). For the anti-Dsx antibody, N-terminal His-tagged DsxCR fusion protein [amino acid (aa) residues 1-397] (Fig. 5A) was used to raise the antibody at the in-house animal facility.
All brain samples were mounted in 70% glycerol and fluorescent images acquired using a Zeiss LSM 700 inverted confocal microscope and analysed using ZEN 2012 software. The pNBs were counted by scanning the entire scans in the region of interest while taking care that no cell was counted twice. The images were processed using ImageJ and Adobe Photoshop CS2 software. For all quantifications, the Student's unpaired t-test was used to assess significance, except for Fig. 9A where the Student's paired t-test was used (GraphPad Prism software).
Temperature shift experiments
Fly crosses were set up at 18°C between virgin females of the genotype UAS-dcr2; inscGAL4-UASmCD8-GFP; tub-GAL80ts with males of genotypes UAS-p35; UAS-AbdB-RNAi; UAS-Notch-RNAi; UAS-Hth-RNAi; UAS-DsxF; UAS-DsxM; F3B3-lacZ, UAS-DsxF; F3B3-lacZ, UAS-DsxM; UAS-DsxCR-DPiM; F3B3-lacZ, UAS-DsxCR-DpiM; UAS-AbdB; UAS-AbdB, F3B3-lacZ; and UAS-AbdB-RNAi, UAS-DsxF with suitable controls, as detailed in the genotype section. Egg collections were carried out at 4 h and the larvae reared at 18°C until the desired stage (time) and were then shifted to 30°C. The larvae were sexed, separated at specific times and subsequently dissected according to the requirement of each individual experiment. Six different TS protocols were employed, as described in Fig. S2A-F. The specific protocol used in each experiment is indicated by the figure number. The lac-Z levels were quantified for Dsx+tNBs as described previously (Khandelwal et al., 2017).
Mosaic Analysis of Cell repressible marker (MARCM)
MARCM clones were generated as described previously (Lee and Luo, 2001) by crossing flies of relevant genotypes (detailed below). All the eggs collected for a duration of 4 h at 25°C were subjected to periodic heat shock of 1 h at 37°C after every 12 h, starting from the time of egg laying. Larvae were dissected at the late third instar larval stage. Dsx intensity was calculated as described above.
AbdBM1 clones
Females of the genotype elav[C155]-GAL4, UASmCD8-GFP, hsflp1, w; FRT82B tub-GAL80-LL3 were crossed with males of genotype FRT-AbdBM1/TM6B,Tb.
exd1 clones
Males of the genotype hsflp, FRT19A, tubGAL80; tub-GAL4, UAS-mCD8-GFP/CyO-GFP were crossed with females of FRT- exd1/FM7.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays (EMSAs) were performed as described previously (Khandelwal et al., 2017). The following protein constructs were used for EMSA studies: N-terminal GST-tagged AbdB-N (aa residues 89-270) (Figs 6 and 7), N-terminal His-tagged DsxCR (aa residues 1-397) (Fig. 6), C-terminal His-tagged DsxF (aa residues 1-427), DsxM (aa residues 1-549) (Fig. 7). Dsx protein was purified and dialyzed in standard buffers containing 10 µM Zn2+SO4. All the binding reactions were set up in a 20 μl volume and incubated at room temperature for 40 min. EMSA images for Fig. 6B-D are a combination of multiple gels, as described in the legend. Each set of six lanes for wild-type, Hox mutant and Dsx mutant oligos are from the same gel. EMSA images in Fig. 7B, which compare the relative binding efficiencies of DsxCR, DsxF and DsxM, are from a single gel.
GST pulldown experiments
The following constructs were used for the affinity pulldown assay: N-terminal His-tagged fusion proteins of DsxCR (aa residues 1-397), DsxΔN (aa residues 41-397), DsxΔDBD (aa residues 101-397) and DsxΔDD (aa residues 1-356); C-terminal His-tagged fusion proteins of DsxF (aa residues 1-427) and DsxM (aa residues 1-549); N-terminal GST-tagged fusion proteins were used for AbdB-N (aa residues 89-270), AbdB-HD (aa residues 162-228), AbdB-NΔCTD (aa residues 89-228), AbdB-ΔN (aa residues 162-270) and AbdB-NΔHD (aa residues 89-161 were deleted). Bacterial cultures were induced at OD600 0.5 with 0.5 mM IPTG at 18°C for 2 h. The affinity pulldown experiments were performed as described in a previous report (Khandelwal et al., 2017). Representative blots from three repetition of the experiments are shown in Fig. 5 and Fig. 7.
Genotypes analysed
Fig. 1B-B‴: w/Y; UAS-mCD8-GFP/CyO; dsx-GAL4/dsx-GAL4
Fig. 1C-C‴: w/w; UAS-mCD8-GFP/+; dsx-GAL4/UAS-p35
Fig. 1D-D‴: UAS-Dcr2/w; inscGAL4, UAS-mCD8-GFP/+; tub-GAL80ts/UAS-p35
Fig. 1E-E‴: w/w; F3B3-lacZ/CyO; MM3/MM3
Fig. 1F-F‴: w/Y; UAS-nls-GFP/F3B3-lacZ; dsx-GAL4/+
Fig. 1G-G‴: w/w; UAS-nls-GFP/F3B3-lacZ; dsx-GAL4/UAS-p35
Fig. 1H-H‴: w/w; 717-lacZ/717-lacZ
Fig. 1I-I″″: UAS-Dcr2/w; inscGAL4, UAS-mCD8-GFP/717-lacZ; tub-GAL80ts/UAS-p35
Fig. 2A-A‴: elavGAL4, UAS-mCD8-GFP, hsFLP w/w; FRT82B tub-GAL80/ FRT82B-AbdBM1
Fig. 2B-B‴: hsflp, FRT19A, tub-GAL80/FRT19A-exd1; tubGAL4,UAS-mCD8-GFP/+
Fig. 2C-C″:
w/w; grimA6C-rpr17/grimA6C-rpr17 and Canton S
w/w; +/+; grimA6C/grimA6C
w/w; +/+; rpr17/rpr17
Fig. 2D:
Canton S
w/w; +/+; rpr17/rpr17
w/w; +/+; grimA6C/grimA6C
w/w; +/+; grimA6C-rpr17/grimA6C-rpr17
w/w; grhB37/370
UAS-dcr2/w; inscGAL4, UAS-mCD8-GFP/+; tub-GAL80ts/UAS-Hth-RNAi
w/w; ix1/2
UAS-dcr2/w; inscGAL4, UAS-mCD8-GFP/+; tub-GAL80ts/UAS-Notch-RNAi
Fig. 3A-A‴:
elavGAL4, UAS-mCD8-GFP, hsFLP w/Y; FRT82B tub-GAL80/ FRT82B-AbdBM1 was the genotype of larvae dissected. White dotted cell is FRT82B-AbdBM1/FRT82B-AbdBM1 and yellow dotted cell is FRT82-AbdBM1/FRT82B- tub-GAL80 for 3rd chromosome.
Fig. 3B:
elavGAL4, UAS-mCD8-GFP, hsFLP w/Y; FRT82B tub-GAL80/FRT82B-AbdBM1
elavGAL4, UAS-mCD8-GFP, hsFLP w/Y; FRT82B tub-GAL80/FRT82B-AbdBM1
elavGAL4, UASmCD8-GFP, hsFLP w/w; FRT82B tub-GAL80/ FRT82B-AbdBM1
Fig. 3C:
UAS-Dcr2/Y; inscGAL4, UAS-mCD8-GFP; tub-GAL80ts/tub-GAL80ts
UAS-Dcr2/Y; inscGAL4, UAS-mCD8-GFP/UAS-AbdB; tub-GAL80ts/+
UAS-Dcr2/Y; inscGAL4, UAS-mCD8-GFP/+; tub-GAL80ts/UAS-DsxCR
UAS-Dcr2/Y; inscGAL4, UAS-mCD8-GFP/+; tub-GAL80ts/UAS-DsxF
UAS-Dcr2/Y; inscGAL4, UAS-mCD8-GFP/UAS-DsxM; tub-GAL80ts/+
Fig. 3D:
UAS-Dcr2/w; inscGAL4, UAS-mCD8-GFP/F3B3-lacZ; tub-GAL80ts/UAS-p35
UAS-Dcr2/w; inscGAL4, UAS-mCD8-GFP/F3B3-lacZ; tub-GAL80ts/UAS-AbdB-RNAi
Fig. 3E-E″″: UAS-Dcr2/Y; inscGAL4, UAS-mCD8-GFP; tub-GAL80ts/tub-GAL80ts
Fig. 3F-F″″: UAS-Dcr2/Y; inscGAL4, UAS-mCD8-GFP/UAS-AbdB; tub-GAL80ts/+
Fig. 3G-G″″: UAS-Dcr2/Y; inscGAL4, UAS-mCD8-GFP/+; tub-GAL80ts/F3B3-lacZ
Fig. 3H-H″″: UAS-Dcr2/Y; inscGAL4, UAS-mCD8-GFP/UAS-AbdB; tub-GAL80ts/F3B3-lacZ
Comparison of surviving thoracic NBs in AbdB-overexpressing males and females was carried out for the following two genotypes:
Males: UAS-Dcr2/Y; inscGAL4, UAS-mCD8-GFP/UAS-AbdB; tub-GAL80ts/+;
Females: UAS-Dcr2/w; inscGAL4, UAS-mCD8-GFP/UAS-AbdB; tub-GAL80ts/+.
Fig. 4A-A′: UAS-Dcr2/Y; inscGAL4, UAS-mCD8-GFP/F3B3-lacZ; tub-GAL80ts/+
Fig. 4B-B″ and C-C″: UAS-Dcr2/Y; inscGAL4, UAS-mCD8-GFP/F3B3-lacZ; tub-GAL80ts/UAS-DsxF
Fig. 4D-D″″: UAS-Dcr2/Y; inscGAL4, UAS-mCD8-GFP/F3B3-lacZ; tub-GAL80ts/+
Fig. 4E-E″″: UAS-Dcr2/Y; inscGAL4, UAS-mCD8-GFP/F3B3-lacZ; tub-GAL80 ts/UAS-DsxF
Fig. 4F-G″″: UAS-Dcr2/Y; inscGAL4, UAS-mCD8-GFP/UAS-DsxM; tub-GAL80ts/F3B3-lacZ
Fig. 4H-H″″: UAS-Dcr2/Y; inscGAL4, UAS-mCD8-GFP/F3B3-lacZ; tub-GAL80ts/UAS-DsxCR
Fig. 4I-I″″: UAS-Dcr2/w; inscGAL4, UAS-mCD8-GFP/+; tub-GAL80ts/UAS-DsxF, UAS-AbdB-RNAi
Fig. 8B-B‴: w/w; 717-lacZ/717-lacZ; +/+
Fig. 8C-C‴: w/w; 717-Dsxmutant-lacZ/717-Dsxmutant-lacZ; +/+
Fig. 8D-D‴: w/w; 717-AbdBmutant-lacZ/ 717-AbdBmutant-lacZ; +/+
Fig. 8E-E‴: w/Y; 717-AbdBmutant-lacZ/717-AbdBmutant-lacZ; +/+
Fig. 8F-F‴: w/w; 717-Coopmutant-lacZ/717-Coopmutant-lacZ;+/+
Fig. 8G-G‴: w/Y; 717-Dsxmutant-lacZ/717-Dsxmutant-lacZ; +/+
Fig. 8H-H″″: UAS-Dcr2/w; inscGAL4, UAS-mCD8-GFP/Dsxmutant-lacZ; tub-GAL80ts/UAS-p35
Fig. 9A: w/w; UAS-mCD8-GFP/CyO; dsx-GAL4/ dsx-GAL4 and w/Y; UAS-mCD8-GFP/CyO; dsx-GAL4/dsx-GAL4
Supplementary Material
Acknowledgements
We thank B. Baker, E. Sanchéz-Herrero, S. Goodwin, G. Lee, Kristine White, R. Mann, J. Knoblich, G. Hasan, K. VijayRaghavan, L. S. Shashidhara, R. Mishra and G. Ratnaparkhi for various reagents and advice with experiments; the BDSC, VDRC, NIG-Fly and DGRC-Japan stock centres for fly lines; and the CDFD animal facility, Bioklone Biotech (Chennai), the Developmental Studies Hybridoma Bank at The University of Iowa for antibodies and TFF at NCBS-CCAMP (Bangalore) for transgenic flies. We acknowledge B. Swain, P. Kalyani, C. S. Singh for their assistance in various phases of the project. R.J. thanks all the members of Laboratory of Drosophila Neural Development (LDND) and his colleagues for their support in setting up LDND at its new location.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: R.J.; Methodology: N.G., A.B., R.J.; Formal analysis: N.G., A.B., R.J.; Investigation: N.G., A.B., R.K., S.G.R., R.J.; Data curation: N.G., A.B.; Writing - original draft: N.G., A.B., R.J.; Writing - review & editing: N.G., A.B., R.J.; Visualization: R.J.; Supervision: R.J.; Project administration: R.J.; Funding acquisition: R.J.
Funding
This study was funded by the Wellcome Trust DBT India Alliance Intermediate Fellowship to R.J. (500171/Z/09/Z); the Centre for DNA Fingerprinting and Diagnostics core funds; the Department of Biotechnology, India [BT/PR26385/MED/122/110/2017 and BT/PR27455/BRB/10/1647/2018]; the Department of Science and Technology India [EMR/2016/003775]; the Indian Council of Medical Research [3/1/3/JRF-2011/HRD-59 (42219) Fellowship to N.G.]; and the University Grants Commission India [22/06/2014(i)EU-V, 2061430472 Fellowship to A.B.]. Deposited in PMC for immediate release.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.175158.supplemental
References
- Almeida M. S. and Bray S. J. (2005). Regulation of post-embryonic neuroblasts by Drosophila Grainyhead. Mech. Dev. 122, 1282-1293. 10.1016/j.mod.2005.08.004 [DOI] [PubMed] [Google Scholar]
- An W. and Wensink P. C. (1995). Integrating sex- and tissue-specific regulation within a single Drosophila enhancer. Genes Dev. 9, 256-266. 10.1101/gad.9.2.256 [DOI] [PubMed] [Google Scholar]
- Arya R., Sarkissian T., Tan Y. and White K. (2015). Neural stem cell progeny regulate stem cell death in a Notch and Hox dependent manner. Cell Death Differ. 22, 1378-1387. 10.1038/cdd.2014.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B. S. and Ridge K. A. (1980). Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics 94, 383-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello B. C., Hirth F. and Gould A. P. (2003). A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron 37, 209-219. 10.1016/S0896-6273(02)01181-9 [DOI] [PubMed] [Google Scholar]
- Billeter J.-C., Rideout E. J., Dornan A. J. and Goodwin S. F. (2006a). Control of male sexual behavior in Drosophila by the sex determination pathway. Curr. Biol. 16, R766-R776. 10.1016/j.cub.2006.08.025 [DOI] [PubMed] [Google Scholar]
- Billeter J.-C., Villella A., Allendorfer J. B., Dornan A. J., Richardson M., Gailey D. A. and Goodwin S. F. (2006b). Isoform-specific control of male neuronal differentiation and behavior in Drosophila by the fruitless gene. Curr. Biol. 16, 1063-1076. 10.1016/j.cub.2006.04.039 [DOI] [PubMed] [Google Scholar]
- Birkholz O., Rickert C., Berger C., Urbach R. and Technau G. M. (2013a). Neuroblast pattern and identity in the Drosophila tail region and role of doublesex in the survival of sex-specific precursors. Development 140, 1830-1842. 10.1242/dev.090043 [DOI] [PubMed] [Google Scholar]
- Birkholz O., Vef O., Rogulja-Ortmann A., Berger C. and Technau G. M. (2013b). Abdominal-B and caudal inhibit the formation of specific neuroblasts in the Drosophila tail region. Development 140, 3552-3564. 10.1242/dev.096099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtis K. C. and Baker B. S. (1989). Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56, 997-1010. 10.1016/0092-8674(89)90633-8 [DOI] [PubMed] [Google Scholar]
- Burtis K. C., Coschigano K. T., Baker B. S. and Wensink P. C. (1991). The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J. 10, 2577-2582. 10.1002/j.1460-2075.1991.tb07798.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong T., Collins J. J. III, Brubacher J. L., Zarkower D. and Newmark P. A. (2013). A sex-specific transcription factor controls male identity in a simultaneous hermaphrodite. Nat. Commun. 4, 1814 10.1038/ncomms2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough E., Jimenez E., Kim Y. A., Whitworth C., Neville M. C., Hempel L. U., Pavlou H. J., Chen Z. X., Sturgill D., Dale R. K. et al. (2014). Sex- and tissue-specific functions of Drosophila doublesex transcription factor target genes. Dev. Cell 31, 761-773. 10.1016/j.devcel.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt B. and Horvitz H. R. (1999). The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell 98, 317-327. 10.1016/S0092-8674(00)81961-3 [DOI] [PubMed] [Google Scholar]
- Coschigano K. T. and Wensink P. C. (1993). Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes Dev. 7, 42-54. 10.1101/gad.7.1.42 [DOI] [PubMed] [Google Scholar]
- Davis E. C., Popper P. and Gorski R. A. (1996). The role of apoptosis in sexual differentiation of the rat sexually dimorphic nucleus of the preoptic area. Brain Res. 734, 10-18. 10.1016/0006-8993(96)00298-3 [DOI] [PubMed] [Google Scholar]
- Defalco T. J., Verney G., Jenkins A. B., Mccaffery J. M., Russell S. and VAN Doren M. (2003). Sex-specific apoptosis regulates sexual dimorphism in the Drosophila embryonic gonad. Dev. Cell 5, 205-216. 10.1016/S1534-5807(03)00204-1 [DOI] [PubMed] [Google Scholar]
- Defalco T., LE Bras S. and van Doren M. (2004). Abdominal-B is essential for proper sexually dimorphic development of the Drosophila gonad. Mech. Dev. 121, 1323-1333. 10.1016/j.mod.2004.07.001 [DOI] [PubMed] [Google Scholar]
- Dolle P., Izpisua-Belmonte J. C., Brown J. M., Tickle C. and Duboule D. (1991). HOX-4 genes and the morphogenesis of mammalian genitalia. Genes Dev. 5, 1767-1767. 10.1101/gad.5.10.1767 [DOI] [PubMed] [Google Scholar]
- Erdman S. E. and Burtis K. C. (1993). The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. EMBO J. 12, 527-535. 10.1002/j.1460-2075.1993.tb05684.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foronda D., Martín P. and Sánchez-Herrero E. (2012). Drosophila Hox and sex-determination genes control segment elimination through EGFR and extramacrochetae activity. PLoS Genet. 8, e1002874 10.1371/journal.pgen.1002874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner S. R. C., Castellanos M. C., Baillie K. E., Lian T. and Allan D. W. (2018). Drosophila female-specific Ilp7 motoneurons are generated by Fruitless-dependent cell death in males and by a double-assurance survival role for Transformer in females. Development 145, dev150821 10.1242/dev.150821 [DOI] [PubMed] [Google Scholar]
- Guruharsha K. G., Rual J.-F., Zhai B., Mintseris J., Vaidya P., Vaidya N., Beekman C., Wong C., Rhee D. Y., Cenaj O. et al. (2011). A protein complex network of Drosophila melanogaster. Cell 147, 690-703. 10.1016/j.cell.2011.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel L. U. and Oliver B. (2007). Sex-specific DoublesexM expression in subsets of Drosophila somatic gonad cells. BMC Dev. Biol. 7, 113 10.1186/1471-213X-7-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T., Pearson B., Holbrook S. and Doe C. Q. (2001). Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106, 511-521. 10.1016/S0092-8674(01)00465-2 [DOI] [PubMed] [Google Scholar]
- Jeong S., Rokas A. and Carroll S. B. (2006). Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell 125, 1387-1399. 10.1016/j.cell.2006.04.043 [DOI] [PubMed] [Google Scholar]
- Kalis A. K., Murphy M. W. and Zarkower D. (2010). EGL-5/ABD-B plays an instructive role in male cell fate determination in the C. elegans somatic gonad. Dev. Biol. 344, 827-835. 10.1016/j.ydbio.2010.05.516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal R., Sipani R., Rajan S. G., Kumar R. and Joshi R. (2017). Combinatorial action of Grainyhead, Extradenticle and Notch in regulating Hox mediated apoptosis in Drosophila larval CNS. PLoS Genet. 13, e1007043 10.1371/journal.pgen.1007043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K.-I., Ote M., Tazawa T. and Yamamoto D. (2005). Fruitless specifies sexually dimorphic neural circuitry in the Drosophila brain. Nature 438, 229-233. 10.1038/nature04229 [DOI] [PubMed] [Google Scholar]
- Kimura K.-I., Hachiya T., Koganezawa M., Tazawa T. and Yamamoto D. (2008). Fruitless and doublesex coordinate to generate male-specific neurons that can initiate courtship. Neuron 59, 759-769. 10.1016/j.neuron.2008.06.007 [DOI] [PubMed] [Google Scholar]
- Kopp A., Duncan I. and Carroll S. B. (2000). Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature 408, 553-559. 10.1038/35046017 [DOI] [PubMed] [Google Scholar]
- Lee T. and Luo L. (2001). Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24, 251-254. 10.1016/S0166-2236(00)01791-4 [DOI] [PubMed] [Google Scholar]
- Lee G., Hall J. C. and Park J. H. (2002). Doublesex gene expression in the central nervous system of Drosophila melanogaster. J. Neurogenet. 16, 229-248. 10.1080/01677060216292 [DOI] [PubMed] [Google Scholar]
- Lohmann I., Mcginnis N., Bodmer M. and Mcginnis W. (2002). The Drosophila Hox gene deformed sculpts head morphology via direct regulation of the apoptosis activator reaper. Cell 110, 457-466. 10.1016/S0092-8674(02)00871-1 [DOI] [PubMed] [Google Scholar]
- Luo S. D. and Baker B. S. (2015). Constraints on the evolution of a doublesex target gene arising from doublesex's pleiotropic deployment. Proc. Natl. Acad. Sci. USA 112, E852-E861. 10.1073/pnas.1501192112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S. D., Shi G. W. and Baker B. S. (2011). Direct targets of the D. melanogaster DSXF protein and the evolution of sexual development. Development 138, 2761-2771. 10.1242/dev.065227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson C. K. and Zarkower D. (2012). Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 13, 163-174. 10.1038/nrg3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurange C., Cheng L. and Gould A. P. (2008). Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell 133, 891-902. 10.1016/j.cell.2008.03.034 [DOI] [PubMed] [Google Scholar]
- Meissner G. W., Luo S. D., Dias B. G., Texada M. J. and Baker B. S. (2016). Sex-specific regulation of Lgr3 in Drosophila neurons. Proc. Natl. Acad. Sci. USA 113, E1256-E1265. 10.1073/pnas.1600241113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel-Aliaga I. and Thor S. (2004). Segment-specific prevention of pioneer neuron apoptosis by cell-autonomous, postmitotic Hox gene activity. Development 131, 6093-6105. 10.1242/dev.01521 [DOI] [PubMed] [Google Scholar]
- Monedero Cobeta I., Salmani B. Y. and Thor S. (2017). Anterior-posterior gradient in neural stem and daughter cell proliferation governed by spatial and temporal Hox control. Curr. Biol. 27, 1161-1172. 10.1016/j.cub.2017.03.023 [DOI] [PubMed] [Google Scholar]
- Murphy M. W., Sarver A. L., Rice D., Hatzi K., Ye K., Melnick A., Heckert L. L., Zarkower D. and Bardwell V. J. (2010). Genome-wide analysis of DNA binding and transcriptional regulation by the mammalian Doublesex homolog DMRT1 in the juvenile testis. Proc. Natl. Acad. Sci. USA 107, 13360-13365. 10.1073/pnas.1006243107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumüller R. A., Richter C., Fischer A., Novatchkova M., Neumüller K. G. and Knoblich J. A. (2011). Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell 8, 580-593. 10.1016/j.stem.2011.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden E., Kimberly E., Gengyo-Ando K., Mitani S. and Xue D. (2007). Control of sex-specific apoptosis in C. elegans by the BarH homeodomain protein CEH-30 and the transcriptional repressor UNC-37/Groucho. Genes Dev. 21, 3195-3207. 10.1101/gad.1607807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J. M., Donahoe P. K., Ito Y. and Hendren W. H. III (1977). Programmed cell death in the Müllerian duct induced by Müllerian inhibiting substance. Am. J. Anat. 149, 353-375. 10.1002/aja.1001490304 [DOI] [PubMed] [Google Scholar]
- Raymond C. S., Kettlewell J. R., Hirsch B., Bardwell V. J. and Zarkower D. (1999). Expression of Dmrt1 in the genital ridge of mouse and chicken embryos suggests a role in vertebrate sexual development. Dev. Biol. 215, 208-220. 10.1006/dbio.1999.9461 [DOI] [PubMed] [Google Scholar]
- Raymond C. S., Murphy M. W., O'sullivan M. G., Bardwell V. J. and Zarkower D. (2000). Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 14, 2587-2595. 10.1101/gad.834100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q., Awasaki T., Huang Y.-F., Liu Z. and Lee T. (2016). Cell Class-Lineage Analysis Reveals Sexually Dimorphic Lineage Compositions in the Drosophila Brain. Curr. Biol. 26, 2583-2593. 10.1016/j.cub.2016.07.086 [DOI] [PubMed] [Google Scholar]
- Rideout E. J., Dornan A. J., Neville M. C., Eadie S. and Goodwin S. F. (2010). Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat. Neurosci. 13, 458-466. 10.1038/nn.2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts L. M., Hirokawa Y., Nachtigal M. W. and Ingraham H. A. (1999). Paracrine-mediated apoptosis in reproductive tract development. Dev. Biol. 208, 110-122. 10.1006/dbio.1998.9190 [DOI] [PubMed] [Google Scholar]
- Robinett C. C., Vaughan A. G., Knapp J.-M. and Baker B. S. (2010). Sex and the single cell. II. There is a time and place for sex. PLoS Biol. 8, e1000365 10.1371/journal.pbio.1000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. M., Kalis A. K., Murphy M. W. and Zarkower D. (2005). The DM domain protein MAB-3 promotes sex-specific neurogenesis in C. elegans by regulating bHLH proteins. Dev. Cell 8, 881-892. 10.1016/j.devcel.2005.03.017 [DOI] [PubMed] [Google Scholar]
- Sanchez L., Gorfinkiel N. and Guerrero I. (2001). Sex determination genes control the development of the Drosophila genital disc, modulating the response to Hedgehog, Wingless and Decapentaplegic signals. Development 128, 1033-1043. [DOI] [PubMed] [Google Scholar]
- Sanders L. E. and Arbeitman M. N. (2008). Doublesex establishes sexual dimorphism in the Drosophila central nervous system in an isoform-dependent manner by directing cell number. Dev. Biol. 320, 378-390. 10.1016/j.ydbio.2008.05.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satokata I., Benson G. and Maas R. (1995). Sexually dimorphic sterility phenotypes in Hoxa10-deficient mice. Nature 374, 460-463. 10.1038/374460a0 [DOI] [PubMed] [Google Scholar]
- Shen M. M. and Hodgkin J. (1988). mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans. Cell 54, 1019-1031. 10.1016/0092-8674(88)90117-1 [DOI] [PubMed] [Google Scholar]
- Sorge S., Ha N., Polychronidou M., Friedrich J., Bezdan D., Kaspar P., Schaefer M. H., Ossowski S., Henz S. R., Mundorf J. et al. (2012). The cis-regulatory code of Hox function in Drosophila. EMBO J. 31, 3323-3333. 10.1038/emboj.2012.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Yamada-Mabuchi M., Arya R., ST Pierre S., Tang W., Tosa M., Brachmann C. and White K. (2011). Coordinated expression of cell death genes regulates neuroblast apoptosis. Development 138, 2197-2206. 10.1242/dev.058826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Barmina O., Sanders L. E., Arbeitman M. N. and Kopp A. (2011). Evolution of sex-specific traits through changes in HOX-dependent doublesex expression. PLoS Biol. 9, e1001131 10.1371/journal.pbio.1001131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B. J. and Truman J. W. (1992). Commitment of abdominal neuroblasts in Drosophila to a male or female fate is dependent on genes of the sex-determining hierarchy. Development 114, 625-642. [DOI] [PubMed] [Google Scholar]
- Truman J. W. and Bate M. (1988). Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev. Biol. 125, 145-157. 10.1016/0012-1606(88)90067-X [DOI] [PubMed] [Google Scholar]
- Truman J. W., Taylor B. J. and Awad T. A. (1993). Formation of the adult nervous system. In The Development of Drosophila melanogaster (ed. Bate M. A.), pp.1245-1275. CSHL Press. [Google Scholar]
- Villella A. and Hall J. C. (1996). Courtship anomalies caused by doublesex mutations in Drosophila melanogaster. Genetics 143, 331-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. and Yoder J. H. (2012). Hox-mediated regulation of doublesex sculpts sex-specific abdomen morphology in Drosophila. Dev. Dyn. 241, 1076-1090. 10.1002/dvdy.23791 [DOI] [PubMed] [Google Scholar]
- Wang W., Kidd B. J., Carroll S. B. and Yoder J. H. (2011). Sexually dimorphic regulation of the Wingless morphogen controls sex-specific segment number in Drosophila. Proc. Natl. Acad. Sci. USA 108, 11139-11144. 10.1073/pnas.1108431108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterbury J. A., Jackson L. L. and Schedl P. (1999). Analysis of the doublesex female protein in Drosophila melanogaster: role on sexual differentiation and behavior and dependence on intersex. Genetics 152, 1653-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T. M. and Carroll S. B. (2009). Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat. Rev. Genet. 10, 797-804. 10.1038/nrg2687 [DOI] [PubMed] [Google Scholar]
- Williams T. M., Selegue J. E., Werner T., Gompel N., Kopp A. and Carroll S. B. (2008). The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell 134, 610-623. 10.1016/j.cell.2008.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Pan Y., Robinett C. C., Meissner G. W. and Baker B. S. (2014). Central brain neurons expressing doublesex regulate female receptivity in Drosophila. Neuron 83, 149-163. 10.1016/j.neuron.2014.05.038 [DOI] [PubMed] [Google Scholar]
- Zhu L., Wilken J., Phillips N. B., Narendra U., Chan G., Stratton S. M., Kent S. B. and Weiss M. A. (2000). Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers. Genes Dev. 14, 1750-1764. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.