ABSTRACT
Some DMRT family genes including arthropod dsx, nematode mab-3, and vertebrate dmrt1 are involved in sex determination and/or differentiation in bilaterian animals. Although there have been some reports about evolutionary analyses of the family by using its phylogenetic trees, it is still undecided as to whether these three sex determination-related genes share orthologous relationships or not. To clarify this question, we analyzed evolutional relationships among the family members in various bilaterians by using not only phylogenetic tree analysis, but also synteny analysis. We found that only four genes, dmrt2a/2b, dmrt3, dmrt4/5 and dmrt93B were commonly present in invertebrate bilateria. The syntenies of dmrt2a/2b-dmrt3 and dmrt4/5-dmrt93B are conserved before and after two rounds of whole genome duplication in the ancestral vertebrate. Importantly, this indicates that dmrt1 must have appeared in the common vertebrate ancestor. In addition, dmrt1, dsx, or mab-3 formed each different cluster at a distance in our phylogenetic tree. From these findings, we concluded that the three sex determination-related genes, dmrt1, dsx, and mab-3 have no orthologous relationships, and suggested independent evolution for sex determination and differentiation in the DMRT gene family. Our results may supply clues about why sex-determining systems have diverged during animal evolution.
KEY WORDS: DMRT, DM domain, Sex determination, Sex differentiation, Gene duplication
Summary: Three DMRT family genes, vertebrate dmrt1, arthropod dsx and nematode mab-3, involved in sex determination and primary sex differentiation have no orthologous relationships, indicating independent evolution in bilaterian animals.
INTRODUCTION
The doublesex and mab-3 related transcription factor (DMRT) family is well-conserved in bilaterian animals and is characterized by a DNA-binding region called the DM domain (Matson and Zarkower, 2012). The domain was named from Drosophila melanogaster Dsx and Caenorhabditis elegans Mab-3 proteins, both of which play important roles in sex differentiation (Matson and Zarkower, 2012). Most animals have multiple DMRT genes. In mammals, there are eight DMRT genes, DMRT1-DMRT8 (Veith et al., 2006; Bellefroid et al., 2013). Veith et al. previously suggested that DMRT7 and DMRT8 are mammalian-specific DMRT genes (Veith et al., 2006). Most of the DMRT proteins play roles in various developmental processes including myogenesis, somitogenesis, neurogenesis and gametogenesis (O'Day, 2010; Bellefroid et al., 2013; Zhang et al., 2014a; Yu et al., 2015).
Some DMRT genes are well-studied in sexual determination and differentiation in somatic cells of the gonads. DMRT1 is a regulator of testicular formation and/or male determination in gonadal somatic cells in various vertebrate species (Yoshimoto and Ito, 2011; Zhao et al., 2015). Dmrt1 is necessary for somatic-cell masculinization in mice (Raymond et al., 2000). In chickens, the Z-linked dmrt1 induces male sex determination by its gene dosage (Smith et al., 2009). The dmrt1 paralogs, the Y-linked dmy/dmrt1by in teleost fish (Oryzias latipes) and the W-linked dmw in the African clawed frog (Xenopus laevis) are sex-determining genes (Matsuda et al., 2002; Nanda et al., 2002; Yoshimoto et al., 2008; Mawaribuchi et al., 2012). On the other hand, in germ cells of the gonads, Dmrt1, Dmrt6, Dmrt7 and Dmrt11E are involved in spermatogenesis in some bilaterian species (Kawamata and Nishimori, 2006; Matson et al., 2010; Zhang et al., 2014a,b; Yu et al., 2015). Dmrt1 and Dmrt4 play roles in oogenesis and folliculogenesis, respectively, in female mice (Balciuniene et al., 2006; Krentz et al., 2011). Moreover, many DMRT genes are implicated in non-gonadal development. Dmrt2 participates in myogenesis and somitogenesis in some vertebrates (Meng et al., 1999; Seo et al., 2006; Sato et al., 2010). Dmrt3, Dmrt4, Dmrt5, Dmd-5 and Dmrt93B engage in neurogenesis in some bilaterian species (Huang et al., 2005; O'Day, 2010; Andersson et al., 2012; Saulnier et al., 2013; De Clercq et al., 2016; Oren-Suissa et al., 2016). The molecular function of DMRT8 is not yet known. Importantly, only the three types of the DMRT family genes – that is, dmrt1 homologs, dsx and mab-3 – are known to function in sex determination and/or somatic sex differentiation to date.
Some researchers suggested that DMRT1 may be a vertebrate equivalent of dsx, that is, dsx ortholog (Ottolenghi et al., 2002; Kato et al., 2011; Clough et al., 2014). Other researchers discussed that they could not yet conclude that dmrt1 is the mammalian ortholog of dsx and mab-3 from their sequence comparisons (Raymond et al., 2000). Moreover, the phylogenetic trees of DMRT family proteins showed that two clusters consisting of dmrt1 and dsx do or do not form a sister group (Toyota et al., 2013; Wexler et al., 2014). In addition, the synteny analysis of DMRT family genes has not been reported in invertebrates. Collectively, it is still an undecided question as to whether these sex determination-related genes, dmrt1, dsx, and mab-3 are orthologous or not. Interestingly, our recent report indicated that the ancestral gene of vertebrate dmrt1 might have emerged not for sex determination but for germ-cell development (Mawaribuchi et al., 2017a), suggesting that dmrt1 might not be a functional ortholog of dsx and mab-3.
The divergence of the DMRT family genes for gonadal and non-gonadal functions remains unclear. In addition, the synteny analysis of these genes in invertebrates is rarely performed. In this study, the evolutionary relationships of the DMRT genes in bilateria were examined by not only phylogenetic tree, but also synteny analysis. We found that four DMRT genes, dmrt2a/2b, dmrt3, dmrt4/5 and dmrt93B were commonly present in invertebrate bilateria. The syntenies of dmrt2a/2b-dmrt3 and dmrt4/5-dmrt93B are conserved before and after 2R-WGD in the ancestral vertebrate. As for the sex determination-related DMRT genes, the evolutionary analyses revealed that dmrt1 might have appeared in the common vertebrate ancestor, and that there are independent and different clusters for dmrt1, dsx and mab-3 in our phylogenetic tree. These results suggested that the three sex determination-related genes, dmrt1, dsx and mab-3 might emerge independently in each taxon and obtain new functions for sex determination and/or primary sex differentiation.
RESULTS AND DISCUSSION
A common ancestor of bilateria must have possessed three DMRT family genes, dmrt2a/2b, dmrt4/5 and dmrt93B
The syntenic relationships of the DMRT family genes in invertebrates have not been investigated in detail. To perform the synteny analyses of the family genes between invertebrate and vertebrate bilateria, we obtained the sequences from the GenBank or various genomes by BLAST search (Table S1). In mammals, there are eight DMRT genes, DMRT1-DMRT8 (Veith et al., 2006; Bellefroid et al., 2013). Johnsen and Andersen reported that dmrt2 (dmrt2a) and dmrt2b or dmrt4 and dmrt5 might emerge from their ancestral genes, respectively, through the two rounds of whole genome duplication (2R-WGD) (Johnsen and Andersen, 2012). This supports the finding that Dmrt4 and Dmrt5 may possess redundant roles during neurogenesis (Parlier et al., 2013). Interestingly, we could obtain no dmrt2b sequences in tetrapod genome databases (Table S1). In contrast, Kato et al. indicated the close relationship between dmrt11E and dmrt2 or dmrt99B and dmrt4/5 from their sequence similarity (Kato et al., 2008). Based on these findings, the DMRT gene families could be classified into eight major subsets, DMRT1, DMRT2a/2b (dmrt2, dmrt2a and dmrt2b), DMRT3, DMRT4/5 (dmrt4, dmrt5 and dmrt99B), DMRT6, DMRT7, DMRT8 and DMRT93B. The dmrt93B orthologs belonging to the eighth DMRT family were newly identified in not only invertebrates, but also some vertebrate species (Table S1, see details below). Importantly, the BLAST search revealed that only four genes, dmrt2a/2b, dmrt3, dmrt4/5 and dmrt93B were commonly present in invertebrate bilateria (Table S1).
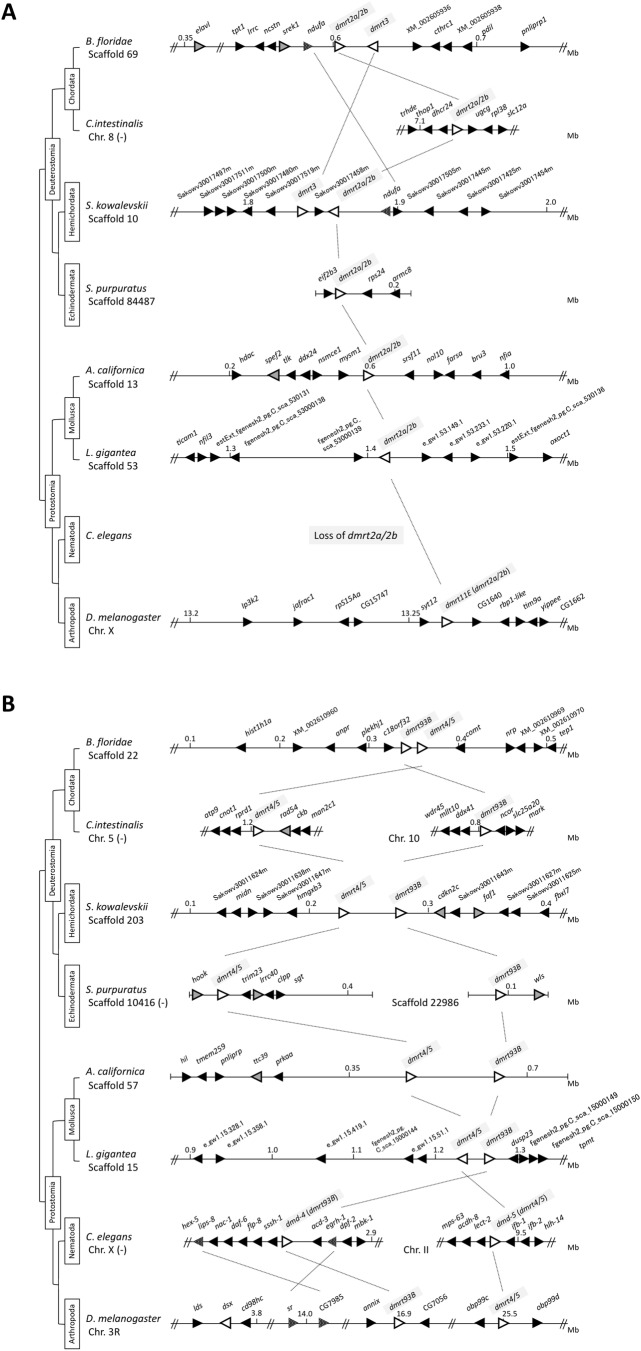
Then we performed synteny analyses of the DMRT genes in invertebrate bilateria including four deuterostome species (Chordata, Cephalochordata, Branchiostoma floridae; Chordata, Urochordata, Ciona intestinalis; Hemichordata, Saccoglossus kowalevskii; Echinodermata, Strongylocentrotus purpuratus) and four protostome species (Mollusca, Aplysia californica; Mollusca, Lottia gigantea; Nematoda, C. elegans; Arthropoda, D. melanogaster) (Fig. 1). A synteny analysis of dmrt2a/2b in these eight species indicated the presence of the dmrt3 gene in close proximity to the dmrt2a/2b locus in deuterostomia (Fig. 1A). In deuterostomia, three genes, dmrt2a/2b, dmrt3 and ndufa were found to be syntenic between B. floridae and S. kowalevskii, suggesting that dmrt3 could have emerged through a gene duplication event of dmrt2a/2b during deuterostome evolution (Fig. 1A). In addition, we found that the dmrt4/5 (dmrt99B) and dmrt93B genes were located adjacent to each other in two deuterostomes (B. floridae and S. kowalevskii) and two protostomes (A. californica and L. gigantea) (Fig. 1B). These two genes were present on the same chromosome in D. melanogaster (Fig. 1B). CG7985 (lips-8) and sr (egrh-1) appeared to be located near dmrt93B (dmd-4) in D. melanogaster and C. elegans (Fig. 1B). Taken together, these results suggested that a common ancestor before the divergence of deuterostomes and protostomes possessed three subsets of the DMRT gene family, dmrt2a/2b, dmrt4/5 and dmrt93B.
Fig. 1.
Synteny analysis of DMRT genes in invertebrates. (A) Synteny of dmrt2a/b (dmrt11E) and dmrt3. (B) Synteny of dmrt4/5 (dmrt99B) and dmrt93B. The synteny analysis was performed in eight species of invertebrate bilateria (B. floridae; C. intestinalis; S. kowalevskii; S. purpuratus; A. californica; L. gigantea; C. elegans; D. melanogaster). Triangles indicate genes and their tips correspond to their 3′-ends. White and black triangles represent DMRT genes and surrounding genes, respectively. Gray triangles represent genes that have been found in the areas surrounding DMRT genes in both vertebrates and invertebrates (Fig. 2). Spotted triangles represent genes showing synteny between invertebrates. Chr., chromosome; (−), reverse relationship.
The syntenies of dmrt2a/2b-dmrt3 and dmrt4/5-dmrt93B are conserved before and after two rounds of whole genome duplication (2R-WGD) in a common ancestor of vertebrates
From the above results, we found that dmrt2a/2b, dmrt3, dmrt4/5 and dmrt93B might be present in a common vertebrate ancestor. It is believed that 2R-WGD occurred in the common ancestor of vertebrates. Based on this premise, we next examined synteny relationships of the DMRT family genes between invertebrates and vertebrates (Fig. 2). In the spotted gar Lepisosteus oculatus, which belongs to the Holostei infraclass in the Actinopterygii class, the two dmrt clusters dmrt1-dmrt3-dmrt2a and dmrt6-dmr2b were localized to the region encompassing the hook, pde4, dock, foxd, pgm, kank and glis family members in linkage groups (LG) 2 and 10, respectively (Fig. 2A). In addition, dmrt4 or dmrt5 was localized to the region encompassing the ttc39, elavl and cdkn2 family members in LG4 or LG10, respectively. Two sets of faf, foxd, glis, keap1, smarca and pgm, three sets of cdkn2, dock and hook, or four sets of elavl, kank and pde paralogues were observed in the linkage groups in close proximity to the dmrt family members, indicating the presence of traces of 2R-WGD (Fig. 2A). A gene corresponding to the ancestral gene of the elavl family was localized in the vicinity of dmrt2a/2b and dmrt3 on scaffold 69 in B. floridae (Fig. 1A). The ancestral cdkn2- and faf-related genes or ancestral ttc39-related gene were found near dmrt4/5 or dmrt93B on scaffold 203 in S. kowalevskii and scaffold 57 in A. californica, respectively (Fig. 1B). The hook-related gene was found in the vicinity of dmrt4/5 on scaffold 10416 in S. purpuratus. Moreover, srek1 on scaffold 69 and spef2 on scaffold 13 in B. floridae and A. californica, respectively, near dmrt2a/2b corresponded to the regions around dmrt1-dmrt3-dmrt2a cluster on LG 2 in L. oculatus (Figs 1A and 2A). rad54 near dmrt4/5 on chromosome 5 in C. intestinalis, lrrc40 near dmrt4/5 on scaffold 10416 in S. purpuratus, and wls near dmrt93B on scaffold 22986 in S. purpuratus corresponded to the regions on LG 10 in L. oculatus (Figs 1B and 2A). Interestingly, no dmrt genes were identified on LG6 or LG19 (Fig. 2A).
Fig. 2.
Synteny analysis of DMRT genes in vertebrates. (A) Synteny of DMRT family genes in L. oculatus. This synteny shows a trace of 2R-WGD. (B) Synteny of DMRT1, DMRT2, DMRT3, DMRT4 and dmrt93B. (C) Synteny of dmrt2b, DMRT5 and DMRT6. The synteny analyses were performed in six species of vertebrates (H. sapiens, X. laevis, L. chalumnae, O. latipes, L. oculatus and C. milii). Triangles indicate genes and their tips correspond to their 3′-ends. White and black triangles represent DMRT genes and the surrounding genes, respectively. Chr., chromosome; LG, linkage group; (−), reverse relationship.
We next performed synteny analysis of DMRT family genes using six species of vertebrates, mammalian Homo sapiens, amphibian X. laevis, sarcopterygian Latimeria chalumnae, actinopterygian O. latipes and L. oculatus, and chondrichthyan Callorhinchus milii (Fig. 2B,C). The synteny of the DMRT1-DMRT3-DMRT2 cluster was well-conserved in all of the vertebrate species examined (Fig. 2B). However, the dmrt6-dmr2b cluster was only conserved in the spotted gar (L. oculatus) and coelacanth (L. chalumnae) (Fig. 2C). The tandem arrays of dmrt4 and dmrt93B were conserved in the elephant shark (C. milii) and coelacanth (L. chalumnae) (Fig. 2B) and in some invertebrate species (Fig. 1B). Namely, the syntenies of dmrt2a/2b-dmrt3 and dmrt4/5-dmrt93B are conserved before and after the 2R-WGD. dmrt2a and dmrt2b or dmrt4 and dmrt5 must have evolved from dmrt2a/2b or dmrt4/5, respectively, through the 2R-WGD.
DMRT1 and DMRT6 are vertebrate-specific genes
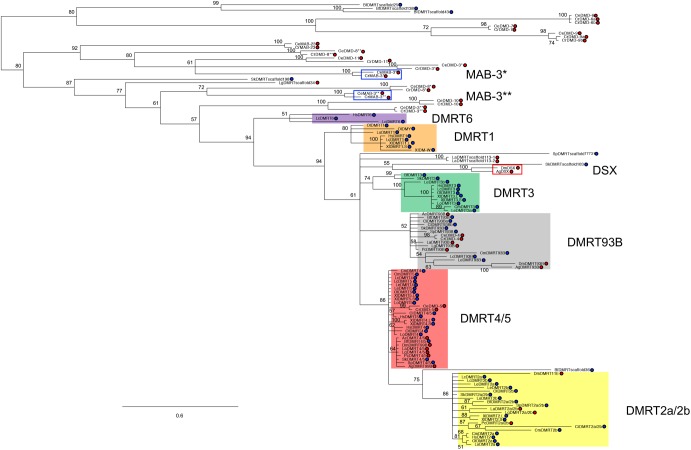
DMRT2a/2b, DMRT3, DMRT4/5 and DMRT93B were commonly present in invertebrate bilateria (Figs 1, 2; Table S1). The synteny analyses also indicated that DMRT7 or DMRT8 are specific in mammalian and reptilian or mammalian species, respectively (Fig. S1). Our recent study reported that lamprey dmrt1 is primarily expressed in germ cells, suggesting that the ancestral vertebrate dmrt1 might have emerged for germ-cell development (Mawaribuchi et al., 2017a). We also found that dmrt6 was pseudogenized in chondrichthyes, Leucoraja erinacea (Table S1). To clarify when dmrt1 and dmrt6 emerged, we constructed Bayesian and maximum likelihood phylogenetic trees of DMRT family members without mammalian- and reptilian-specific DMRT7 and mammalian-specific DMRT8. We analyzed the members in 19 species representing eight different phyla in bilaterians; Brachiopoda, Mollusca, Priapulida, Nematoda, Arthropoda, Hemichordata, Echinodermata and Chordata (Fig. 3; Fig. S2, and Table S1). Chordata included nine species from various taxa including Urochordata, Cephalochordata Chondrichthyes, Actinopterygii, Sarcopterygii, Amphibia and Mammalia. The DM domain regions, which are the only conserved regions among the family members in bilaterian animals, were used for the phylogenetic constructions. The DMRT1 cluster contained Dmrt1 orthologues and the paralogues encoded by the O. latipes and X. laevis sex-determining genes dmy/dmrt1by and dmw, respectively (Fig. 3; Fig. S2) (Matsuda et al., 2002; Nanda et al., 2002; Yoshimoto et al., 2008). As expected, there were no invertebrate genes in the DMRT1 cluster (Fig. 3). In addition, the phylogenetic trees indicate the following viewpoints. The DMRT2a/2b cluster included vertebrate Dmrt2 (Dmrt2a) and Dmrt2b, and invertebrate bilateria Dmrt2a/2b and arthropoda Dmrt11E. The DMRT3 cluster consisted of DMRT3 orthologues in deuterostomes. The DMRT4/5 cluster consisted of the vertebrata Dmrt4 and Dmrt5, invertebrate bilateria Dmrt4/5, arthropoda Dmrt99B and nematoda Dmd-5. The DMRT93B cluster consisted of Dmrt93B from most invertebrate bilateria, nematoda Dmd-4, and Dmrt93B from some fishes, suggesting that dmrt93B may have been lost during tetrapoda evolution. The DMRT6 cluster was comprised of only vertebrate DMRT6 orthologues. The Dsx and Mab-3 clusters consisted of arthropods and nematodes, respectively. These results indicated that DMRT1 and DMRT6 are vertebrate-specific genes. Accordingly, dmrt1 and dmrt6 genes might emerge through gene duplication during vertebrate evolution.
Fig. 3.
Bayesian tree of bilaterian DMRT family genes. The tree was constructed by MrBayes5D using the protein sequences of the DM domains from 19 species representing eight different phyla in bilateria (see Fig. S3). Brachiopoda, Lingula anatina (La); Mollusca, Aplysia californica (Ac); Mollusca, L. gigantea (Lg); Priapulida, Priapulus caudatus (Pc); Nematoda, C. elegans (Ce); Nematoda, Caenorhabditis remanei (Cr); Arthropoda, Anopheles gambiae (Ag); Arthropoda, D. melanogaster (Dm); Hemichordata, Saccoglossus kowalevskii (Sk); Echinodermata, Strongylocentrotus purpuratus (Sp); Chordata, Urochordata, Ciona intestinalis (Ci); Chordata, Cephalochordata, Branchiostoma floridae (Bf); Chordata, Vertebrata, Chondrichthyes, C. milii (Cm); Chordata, Vertebrata, Chondrichthyes, L. erinacea (Le); Chordata, Vertebrata, Actinopterygii, L. oculatus (Lo); Chordata, Vertebrata, Actinopterygii, O. latipes (Ol); Chordata, Vertebrata, Sarcopterygii, L. chalumnae (Lc); Chordata, Vertebrata, Amphibia, X. laevis (Xl); Chordata, Vertebrata, Mammalia, H. sapiens (Hs). Model test was performed by Aminosan (rtREV+F_Gamma). Blue and red circles represent Deuterostomia and Protostomia, respectively. * and ** indicate DM domain regions on 5′ and 3′ sides, respectively. The numbers indicate posterior probability. The values less than 50% were collapsed.
Interestingly, some dmrt genes including arthropod Dsx and nematode Mab-3 did not belong to the eight major subsets of DMRT in bilaterians. These diverged genes mediated through gene duplication might have evolved for taxa diversity. Especially, C. elegans and C. remanei possess many DMRT family members, which might have been derived from the high rate of spontaneous gene duplication in nematodes (Lipinski et al., 2011).
DMRT1 is a homolog but not an ortholog of arthropod dsx and nematode mab-3
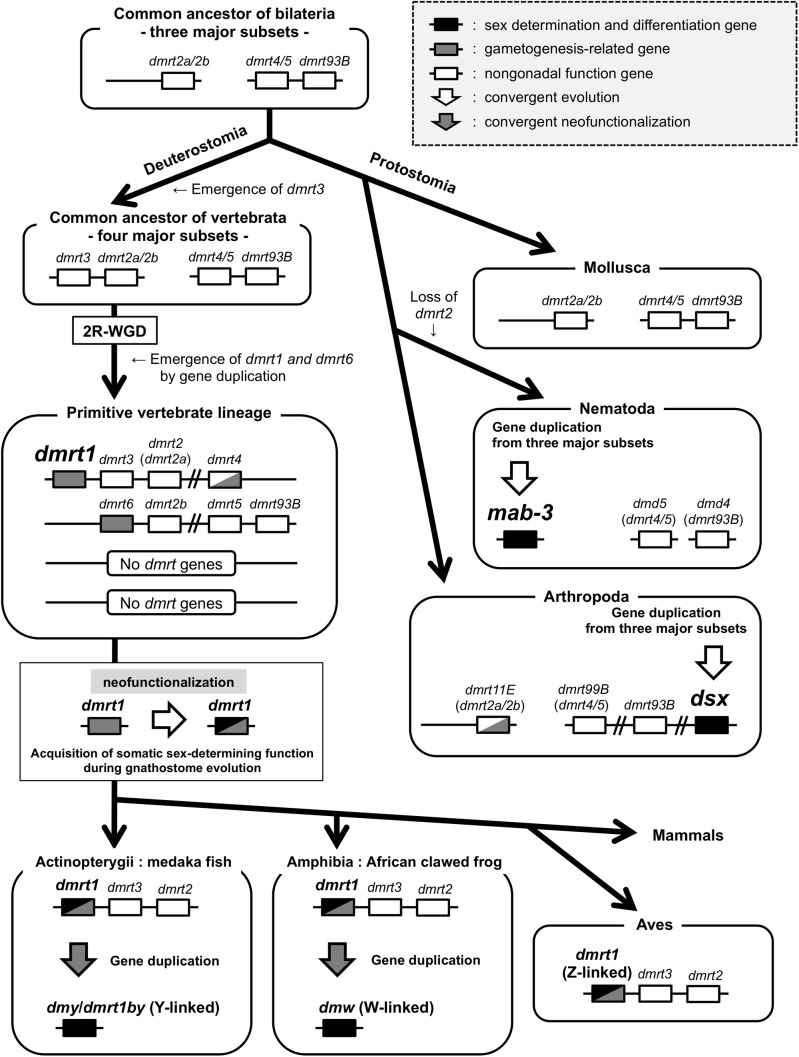
As mentioned in the Introduction section, there is no clear conclusion to the question whether dmrt1, dsx and mab-3 are orthologous to one another or not. However, our recent report indicated that the ancestral vertebrate dmrt1 gene might have emerged not for sex determination but for germ-cell development (Mawaribuchi et al., 2017a). Our syntenic and phylogenetic analyses in this study showed that a common ancestor of vertebrata must have possessed only four DMRT family genes, dmrt2a/2b, dmrt3, dmrt4/5 and dmrt93B (Figs 1, 2, 3; Fig. S2). The syntenies of dmrt2a/2b-dmrt3 and dmrt4/5-dmrt93B are conserved before and after 2R-WGD in a common ancestor of vertebrates (Figs 1 and 2). In addition, dmrt1 and dmrt6 might emerge in the primitive vertebrate lineage (Fig. 3; Fig. S2). Importantly, the dsx and mab-3 genes have been found only in the subphylum Hexapoda among Arthropoda and the phylum Nematoda, respectively (Fig. 3; Fig. S2, and Table S1) (Price et al., 2015). These findings suggested that DMRT1 is not orthologous to arthropod dsx and nematode mab-3. We then summarized molecular evolution of the DMRT gene family in bilateria (Fig. 4). dsx, mab-3 and Dmrt1 play important roles in sex determination and/or sex differentiation (Raymond et al., 2000; Smith et al., 2009; Matson and Zarkower, 2012). Oryzias latipes and X. laevis sex-determining genes, dmy/dmrt1by and dmw, independently evolved from duplication of dmrt1 during the species diversity in each taxon (Matsuda et al., 2002; Nanda et al., 2002; Kondo et al., 2004; Bewick et al., 2011; Mawaribuchi et al., 2017b). Other DMRT genes have not been known to be involved in sex determination and sex differentiation to date. Then, we propose the independent evolution of dmrt1 homologs, dsx and mab-3 for sex determination and primary sex differentiation in the DMRT gene family.
Fig. 4.
Evolutionary history for the DMRT family genes in bilateria. This figure was constructed based on figures in this study. The common ancestor of bilaterian animals possessed three ancestral genes, dmrt2a/2b, dmrt4/5 and dmrt93B. In protostomia, sex-determining and/or primary sex-differentiating genes, arthropod dsx and nematode mab-3 independently arose in each taxon. dmrt3 could have emerged during deuterostome evolution. A common ancestor of vertebrata must have possessed four DMRT family genes, dmrt2a/2b, dmrt3, dmrt4/5 and dmrt93B. The syntenies of dmrt2a/2b-dmrt3 and dmrt4/5-dmrt93B are conserved before and after two rounds of whole genome duplication in the ancestral vertebrate. dmrt1 gene might have emerged for germ-cell development in the primitive vertebrate lineage, and then acquired sex-determining function during gnathostome evolution. (Mawaribuchi et al., 2017a). Moreover, two sex-determining genes, the medaka fish dmy/dmrt1by and African clawed frog dmw, evolved independently through dmrt1 duplication by convergent neofunctionalization. Other DMRT genes are not known to be involved in somatic sex determination and differentiation to date.
MATERIALS AND METHODS
Sequence analysis
The DMRT gene sequences were obtained from the GenBank or various databases and genomes by BLAST (Table S1). Synteny analyses were also performed by BLAST using the obtained sequences and various genome sequences (Table S1). The protein sequences were aligned using MUSCLE (https://www.megasoftware.net), and gaps (insertions/deletions) were removed (Fig. S3). A best-fit protein substitution model was selected by Aminosan (https://www.fifthdimension.jp). Maximum likelihood and Bayesian phylogenetic analyses were performed using MEGA7 and MrBayes5D, respectively, with an rtREV+F+G model (https://www.megasoftware.net, https://www.fifthdimension.jp).
Supplementary Material
Acknowledgements
We thank Dr Haramoto at the National Institute of AIST and Prof. Oota at the University of Tokyo for their comments on the work.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.M., M.I.; Validation: S.M., M.I.; Formal analysis: S.M.; Investigation: S.M.; Data curation: S.M., Y.I.; Writing - original draft: S.M., M.I.; Writing - review & editing: S.M., M.I.; Visualization: S.M.; Supervision: S.M., M.I.; Project administration: M.I.; Funding acquisition: M.I.
Funding
This study was supported in part by Grant-in-Aid for Takahashi Industrial and Economic Research Foundation (M.I.) and Scientific Research from the Ministry of Education Culture, Sports, Science and Technology (M.I).
Data availability
IDs of sequences used in this study are shown in Table S1.
Supplementary information
Supplementary information available online at http://bio.biologists.org/lookup/doi/10.1242/bio.041962.supplemental
References
- Andersson L. S., Larhammar M., Memic F., Wootz H., Schwochow D., Rubin C.-J., Patra K., Arnason T., Wellbring L., Hjälm G. et al. (2012). Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature 488, 642-646. 10.1038/nature11399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciuniene J., Bardwell V. J. and Zarkower D. (2006). Mice mutant in the DM domain gene Dmrt4 are viable and fertile but have polyovular follicles. Mol. Cell. Biol. 26, 8984-8991. 10.1128/MCB.00959-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellefroid E. J., Leclère L., Saulnier A., Keruzore M., Sirakov M., Vervoort M. and De Clercq S. (2013). Expanding roles for the evolutionarily conserved Dmrt sex transcriptional regulators during embryogenesis. Cell. Mol. Life Sci. 70, 3829-3845. 10.1007/s00018-013-1288-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick A. J., Anderson D. W. and Evans B. J. (2011). Evolution of the closely related, sex-related genes DM-W and DMRT1 in African clawed frogs (Xenopus). Evolution 65, 698-712. 10.1111/j.1558-5646.2010.01163.x [DOI] [PubMed] [Google Scholar]
- Clough E., Jimenez E., Kim Y.-A., Whitworth C., Neville M. C., Hempel L. U., Pavlou H. J., Chen Z.-X., Sturgill D., Dale R. K. et al. (2014). Sex- and tissue-specific functions of Drosophila doublesex transcription factor target genes. Dev. Cell 31, 761-773. 10.1016/j.devcel.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq S., Keruzore M., Desmaris E., Pollart C., Assimacopoulos S., Preillon J., Ascenzo S., Matson C. K., Lee M., Nan X. et al. (2016). DMRT5 together with DMRT3 directly controls hippocampus development and neocortical area map formation. Cereb. Cortex 28, 493-509. 10.1093/cercor/bhw384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Hong C. S., O'Donnell M. and Saint-Jeannet J. P. (2005). The doublesex-related gene, XDmrt4, is required for neurogenesis in the olfactory system. Proc. Natl. Acad. Sci. USA 102, 11349-11354. 10.1073/pnas.0505106102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen H. and Andersen Ø. (2012). Sex dimorphic expression of five dmrt genes identified in the Atlantic cod genome. The fish-specific dmrt2b diverged from dmrt2a before the fish whole-genome duplication. Gene 505, 221-232. 10.1016/j.gene.2012.06.021 [DOI] [PubMed] [Google Scholar]
- Kato Y., Kobayashi K., Oda S., Colbourn J. K., Tatarazako N., Watanabe H. and Iguchi T. (2008). Molecular cloning and sexually dimorphic expression of DM-domain genes in Daphnia magna. Genomics 91, 94-101. 10.1016/j.ygeno.2007.09.002 [DOI] [PubMed] [Google Scholar]
- Kato Y., Kobayashi K., Watanabe H. and Iguchi T. (2011). Environmental sex determination in the branchiopod crustacean Daphnia magna: deep conservation of a Doublesex gene in the sex-determining pathway. PLoS Genet. 7, e1001345 10.1371/journal.pgen.1001345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata M. and Nishimori K. (2006). Mice deficient in Dmrt7 show infertility with spermatogenic arrest at pachytene stage. FEBS Lett. 580, 6442-6446. 10.1016/j.febslet.2006.10.066 [DOI] [PubMed] [Google Scholar]
- Kondo M., Nanda I., Hornung U., Schmid M. and Schartl M. (2004). Evolutionary origin of the medaka Y chromosome. Curr. Biol. 14, 1664-1669. 10.1016/j.cub.2004.09.026 [DOI] [PubMed] [Google Scholar]
- Krentz A. D., Murphy M. W., Sarver A. L., Griswold M. D., Bardwell V. J. and Zarkower D. (2011). DMRT1 promotes oogenesis by transcriptional activation of Stra8 in the mammalian fetal ovary. Dev. Biol. 356, 63-70. 10.1016/j.ydbio.2011.05.658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski K. J., Farslow J. C., Fitzpatrick K. A., Lynch M., Katju V. and Bergthorsson U. (2011). High spontaneous rate of gene duplication in Caenorhabditis elegans. Curr. Biol. 21, 306-310. 10.1016/j.cub.2011.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson C. K. and Zarkower D. (2012). Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 13, 163-174. 10.1038/nrg3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson C. K., Murphy M. W., Griswold M. D., Yoshida S., Bardwell V. J. and Zarkower D. (2010). The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev. Cell 19, 612-624. 10.1016/j.devcel.2010.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M., Nagahama Y., Shinomiya A., Sato T., Matsuda C., Kobayashi T., Morrey C. E., Shibata N., Asakawa S., Shimizu N. et al. (2002). DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417, 559-563. 10.1038/nature751 [DOI] [PubMed] [Google Scholar]
- Mawaribuchi S., Yoshimoto S., Ohashi S., Takamatsu N. and Ito M. (2012). Molecular evolution of vertebrate sex-determining genes. Chromosome Res. 20, 139-151. 10.1007/s10577-011-9265-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawaribuchi S., Musashijima M., Wada M., Izutsu Y., Kurakata E., Park M. K., Takamatsu N. and Ito M. (2017a). Molecular evolution of two distinct dmrt1 promoters for germ and somatic cells in vertebrate gonads. Mol. Biol. Evol. 34, 724-733. 10.1093/molbev/msw287 [DOI] [PubMed] [Google Scholar]
- Mawaribuchi S., Takahashi S., Wada M., Uno Y., Matsuda Y., Kondo M., Fukui A., Takamatsu N., Taira M. and Ito M. (2017b). Sex chromosome differentiation and the W- and Z-specific loci in Xenopus laevis. Dev. Biol. 426, 393-400. 10.1016/j.ydbio.2016.06.015 [DOI] [PubMed] [Google Scholar]
- Meng A., Moore B., Tang H., Yuan B. and Lin S. (1999). A Drosophila doublesex-related gene, terra, is involved in somitogenesis in vertebrates. Development 126, 1259-1268. [DOI] [PubMed] [Google Scholar]
- Nanda I., Kondo M., Hornung U., Asakawa S., Winkler C., Shimizu A., Shan Z., Haaf T., Shimizu N., Shima A. et al. (2002). A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. USA 99, 11778-11783. 10.1073/pnas.182314699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Day D. (2010). Genetic analysis of the function of Drosophila doublesex-related factor dmrt93B. UT GSBS Dissertations and Theses Paper 54. [Google Scholar]
- Oren-Suissa M., Bayer E. A. and Hobert O. (2016). Sex-specific pruning of neuronal synapses in Caenorhabditis elegans. Nature 533, 206-211. 10.1038/nature17977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottolenghi C., Fellous M., Barbieri M. and McElreavey K. (2002). Novel paralogy relations among human chromosomes support a link between the phylogeny of doublesex-related genes and the evolution of sex determination. Genomics 79, 333-343. 10.1006/geno.2002.6711 [DOI] [PubMed] [Google Scholar]
- Parlier D., Moers V., Van Campenhout C., Preillon J., Leclère L., Saulnier A., Sirakov M., Busengdal H., Kricha S., Marine J.-C. et al. (2013). The Xenopus doublesex-related gene Dmrt5 is required for olfactory placode neurogenesis. Dev. Biol. 373, 39-52. 10.1016/j.ydbio.2012.10.003 [DOI] [PubMed] [Google Scholar]
- Price D. C., Egizi A. and Fonseca D. M. (2015). The ubiquity and ancestry of insect doublesex. Sci. Rep. 5, 13068 10.1038/srep13068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond C. S., Murphy M. W., O'Sullivan M. G., Bardwell V. J. and Zarkower D. (2000). Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 14, 2587-2595. 10.1101/gad.834100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Rocancourt D., Marques L., Thorsteinsdóttir S. and Buckingham M. (2010). A Pax3/Dmrt2/Myf5 regulatory cascade functions at the onset of myogenesis. PLoS Genet. 6, e1000897 10.1371/journal.pgen.1000897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulnier A., Keruzore M., De Clercq S., Bar I., Moers V., Magnani D., Walcher T., Filippis C., Kricha S., Parlier D. et al. (2013). The doublesex homolog Dmrt5 is required for the development of the caudomedial cerebral cortex in mammals. Cereb. Cortex 23, 2552-2567. 10.1093/cercor/bhs234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo K. W., Wang Y., Kokubo H., Kettlewell J. R., Zarkower D. A. and Johnson R. L. (2006). Targeted disruption of the DM domain containing transcription factor Dmrt2 reveals an essential role in somite patterning. Dev. Biol. 290, 200-210. 10.1016/j.ydbio.2005.11.027 [DOI] [PubMed] [Google Scholar]
- Smith C. A., Roeszler K. N., Ohnesorg T., Cummins D. M., Farlie P. G., Doran T. J. and Sinclair A. H. (2009). The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461, 267-271. 10.1038/nature08298 [DOI] [PubMed] [Google Scholar]
- Toyota K., Kato Y., Sato M., Sugiura N., Miyagawa S., Miyakawa H., Watanabe H., Oda S., Ogino Y., Hiruta C. et al. (2013). Molecular cloning of doublesex genes of four cladocera (water flea) species. BMC Genomics 14, 239 10.1186/1471-2164-14-239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith A.-M., Klattig J., Dettai A., Schmidt C., Englert C. and Volff J.-N. (2006). Male-biased expression of X-chromosomal DM domain-less Dmrt8 genes in the mouse. Genomics 88, 185-195. 10.1016/j.ygeno.2006.01.003 [DOI] [PubMed] [Google Scholar]
- Wexler J. R., Plachetzki D. C. and Kopp A. (2014). Pan-metazoan phylogeny of the DMRT gene family: a framework for functional studies. Dev. Genes Evol. 224, 175-181. 10.1007/s00427-014-0473-0 [DOI] [PubMed] [Google Scholar]
- Yoshimoto S. and Ito M. (2011). A ZZ/ZW-type sex determination in Xenopus laevis. FEBS J. 278, 1020-1026. 10.1111/j.1742-4658.2011.08031.x [DOI] [PubMed] [Google Scholar]
- Yoshimoto S., Okada E., Umemoto H., Tamura K., Uno Y., Nishida-Umehara C., Matsuda Y., Takamatsu N., Shiba T. and Ito M. (2008). A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc. Natl. Acad. Sci. USA 105, 2469-2474. 10.1073/pnas.0712244105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Wu H., Wen Y., Liu Y., Zhou T., Ni B., Lin Y., Dong J., Zhou Z., Hu Z. et al. (2015). Identification of seven genes essential for male fertility through a genome-wide association study of non-obstructive azoospermia and RNA interference-mediated large-scale functional screening in Drosophila. Hum. Mol. Genet. 24, 1493-1503. 10.1093/hmg/ddu557 [DOI] [PubMed] [Google Scholar]
- Zhang T., Murphy M. W., Gearhart M. D., Bardwell V. J. and Zarkower D. (2014a). The mammalian Doublesex homolog DMRT6 coordinates the transition between mitotic and meiotic developmental programs during spermatogenesis. Development 141, 3662-3671. 10.1242/dev.113936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang H., Li M., Cheng Y., Jiang D., Sun L., Tao W., Zhou L., Wang Z. and Wang D. (2014b). Isolation of doublesex- and mab-3-related transcription factor 6 and its involvement in spermatogenesis in tilapia. Biol. Reprod. 91, 136 10.1095/biolreprod.114.121418 [DOI] [PubMed] [Google Scholar]
- Zhao L., Svingen T., Ng E. T. and Koopman P. (2015). Female-to-male sex reversal in mice caused by transgenic overexpression of Dmrt1. Development 142, 1083-1088. 10.1242/dev.122184 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.