Abstract
Background
Long non-coding RNAs (lncRTNAs) are a new focus in cancer research. Although lncRNAs have no protein coding capacity, they are important in epigenetics as well as in regulating gene expression, playing an important role in various cancers. In the current study, we investigated the roles of lncRNA-D16366 in hepatocellular carcinoma (HCC) and expected to find a new biomarker for early detection and prognosis of the disease.
Material/Methods
Quantitative real-time polymerase chain reaction (qRT-PCR) was used to detect the expression of lncRNA-D16366 in tissue and serum samples. The relationship between lncRNA-D16366 expression and clinicopathologic characteristics of patients with HCC was analyzed to estimate whether it was involved in malignancy development. Then, potential diagnostic and prognostic values were evaluated via receiver operating characteristic (ROC) curve, Kaplan-Meier and Cox regression analysis, respectively.
Results
lncRNA-D16366 was proved to be decreased in the tissues and serum among patients with HCC compared with the corresponding controls. Its expression was influenced by tumor size, HbsAg, portal vein tumor thrombus, Child-Pugh score, therapies, and neoplasm metastasis. It had high diagnostic value, with an AUC of 0.752, accompanied by a sensitivity of 65.5% and a specificity of 84.6%. In addition, it was related to the prognosis of HCC.
Conclusions
lncRNA-D16366 was decreased in HCC, and might be an independent diagnostic and prognostic indicator in the disease.
MeSH Keywords: Diagnosis, Liver Neoplasms, Prognosis, RNA Caps
Background
Hepatocellular carcinoma (HCC) is one of the most common malignant cancers and has a high mortality rate [1,2]. The disease has steadily increasing incidence and mortality rates, especially in Asia and Africa [3]. Surgical resection is the most effective therapeutic method for HCC treatment, but high recurrence rate, early distant metastasis, frequent resistance to chemo and radiation therapies, and lack of symptoms in early stages lead to poor prognosis [4–6]. Therefore, exploring effective bio-markers is of great importance for early detection and improvement of prognosis in HCC.
Long non-coding RNAs (lncRNAs) are located in the cell nucleus or cytoplasm and have a length of more than 200 nucleotides [7]. Although lncRNAs have no protein coding capacity, they are important in epigenetics as well as in regulating gene expression [8,9]. It has been confirmed that lncRNAs take part in brain development, embryonic development, histological differentiation, and organogenesis [10,11]. Recently, studies demonstrated that lncRNAs are also involved in many complex cell processes in cancers [10–15]. For example, lncRNA-D16366 was confirmed to be decreased in HCC [16]. However, its role in HCC diagnosis and prognosis remains unclear.
Here, we assessed the expression of lncRNA-D16366 and its correlation with clinicopathologic characteristics of patients with HCC. The diagnostic and prognostic values of lncRNA-D16366 were also estimated through ROC curve, Kaplan-Meier analysis, and Cox regression analysis.
Material and Methods
Patients and samples
This study was conducted in Xianyang Central Hospital and was approved by the Research Ethics Committee of Xianyang Central Hospital. All patients had never received any therapies before sampling and signed written informed content in advance.
A total of 107 patients (62 males and 45 females) who were diagnosed with HCC via histopathological examination were recruited from Xianyang Central Hospital from September 2011 to July 2013. Paired tumor tissues and adjacent ones as well as serum samples were preserved at the same time. Serum samples from 58 patients, including 28 with hepatitis B virus (HBV), 18 with alcoholic liver disease (ALD), and 12 with fatty liver disease, were also obtained and taken as benign controls. Serum from 85 healthy people with matched age were taken as healthy controls. Tissues samples were frozen in liquid nitrogen while serum samples were immediately put into blood collection tubes containing EDTA. All samples were stored at −80°C for RNA extraction. The expression of lncRNA-D16366 was detected through qRT-PCR analysis.
RNA extraction and qRT-PCR analysis
Total RNA was isolated from tumor tissues, adjacent tissues, and serum samples. Reverse transcription was conducted with a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) to synthesize the first chain of cDNA. Then, RT-PCR reaction was performed with β-actin as internal control in the Applied Biosystems 7900 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). The expression of lncRNA-D16366 was evaluated using the comparative cycle threshold (CT).
Follow-up
A 5-year follow-up was conducted for 107 patients with HCC. Overall survival time referred to the duration between the day of diagnosis to the end of follow-up. Clinical information, including age, sex, tumor size, tumor site, HbsAg, AFP, portal vein tumor thrombus, Child-Pugh, therapies, and neoplasm metastasis were recorded in a database. The information of follow-up was obtained through telephone call or letter, and updated every 3 months. Patients who died from unexpected events or other diseases were excluded from our study.
Statistical analysis
Statistical analysis was conducted adopting SPSS version 13.0 for Windows (SPSS, Inc, IL, USA). All data are presented as mean ±SD. Differences between 2 groups were analyzed by t test, while those among 3 groups were assessed via one-way ANOVA. The relationship between lncRNA-D16366 expression and clinicopathological characteristics of patients with HCC was evaluated via chi-square test. Diagnostic value of lncRNA-D16366 was estimated through building ROC curves. Kaplan-Meier and Cox regression analyses were used to estimate the prognostic value of lncRNA-D16366 in HCC. P<0.05 was considered to be statistically significant.
Results
Expression of lncRNA-D16366 in patients with HCC
The expression level of lncRNA-D16366 in tumor tissues and adjacent ones was detected through qRT-PCR assay. As shown in Figure 1, lncRNA-D16366 expression in tumor tissues (0.212±0.001) was significantly lower than that in adjacent ones (1.772±0.076) (P<0.05).
Figure 1.
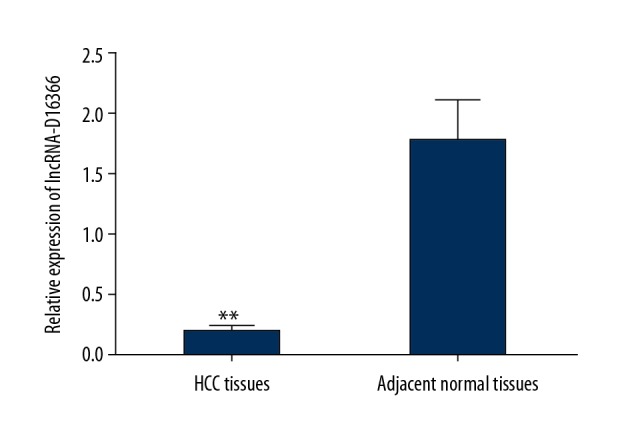
The expression of lncRNA-D16366 in tissue samples. lncRNA-D163663 was downregulated in tumor tissues compared with adjacent ones. ** P<0.01 represents significant difference between 2 groups.
Serum lncRNA-D16366 expression in patients with HCC, in benign controls, and in healthy controls
QRT-PCR analysis was also used to detect serum lncRNA-D16366 expression in each group. The results demonstrated that lncRNA-D16366 expression in preoperative serum was lower than that in postoperative serum among 107 patients with HCC (Figure 2, P<0.01). Serum lncRNA-D16366 expression was decreased in patients with HCC compared with benign controls and healthy controls (Figure 3, P<0.01). We also found that serum lncRNA-D16366 level in benign controls was lower than that in healthy controls (P<0.05). The results indicated that serum lncRNA-D16366 expression could distinguish HCC patients from other cases with benign liver diseases.
Figure 2.
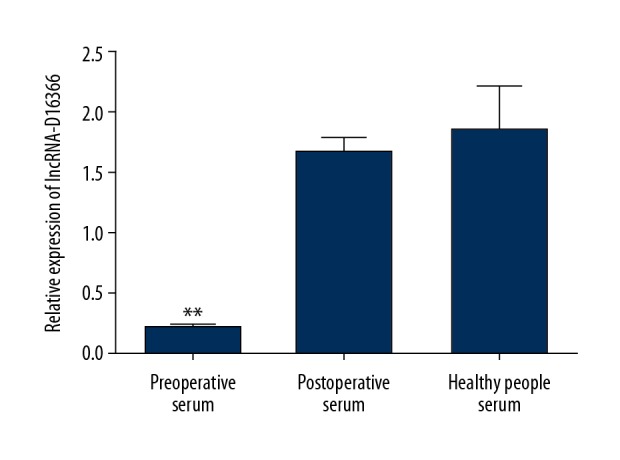
Serum expression of lncRNA-D16366 before and after surgery. lncRNA-D16366 expression in preoperative serum was significantly lower than that in postoperative serum and that in serum from healthy people. ** P<0.01 or * P<0.05 represents significant difference between 2 groups.
Figure 3.
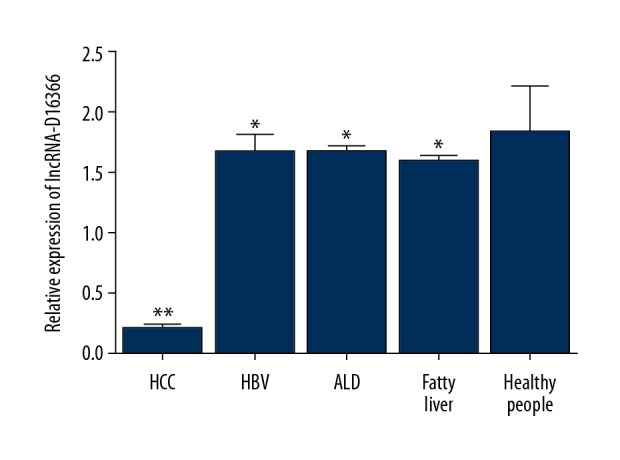
Serum lncRNA-D16366 expression in patients with HCC, patients with benign liver diseases, and healthy people. It was obviously lower in patients with HCC than in the other 2 groups (P<0.05). ** P<0.01 or * P<0.05 represents significant difference between 2 groups.
The relationship between lncRNA-D16366 expression and clinicopathological characteristics
In this study, 107 patients with HCC were divided into high- and low-expression groups according to their mean expression levels of lncRNA-d16366 (0.212) in HCC tissues. Fifty-one patient were in the high lncRNA D16366 group, while the others were in the low lncRNA-D16366 group. Then, we analyzed the relationship between lncRNA-D16366 and clinicopathological characteristics. The results showed that tumor size (P<0.001), HbsAg (P=0.005), portal vein tumor thrombus (P=0.001), Child-Pugh score (P=0.033), therapies (P=0.003), and neoplasm metastasis (P<0.001) significantly influenced the expression of lncRNA-D16366 (Table 1).
Table 1.
The relationship between expression of lncRNA-D16366 and clinicopathological characteristics.
| Clinicopathological characteristics | n | lncRNA-D16366 expression | P | |
|---|---|---|---|---|
| High | Low | |||
| Age | 0.762 | |||
| 40–50 | 25 | 12 | 13 | |
| 51–60 | 47 | 24 | 23 | |
| 61–70 | 35 | 15 | 20 | |
| Sex | 0.164 | |||
| Female | 45 | 25 | 20 | |
| Male | 62 | 26 | 36 | |
| Tumor size | 0.000 | |||
| <3 cm | 43 | 34 | 9 | |
| 3–5 cm | 33 | 13 | 20 | |
| 5–10 cm | 24 | 2 | 22 | |
| >10 cm | 7 | 2 | 5 | |
| Tumor site | 0.282 | |||
| Left | 49 | 27 | 22 | |
| Right | 43 | 19 | 24 | |
| Both | 15 | 5 | 10 | |
| HbsAg | 0.005 | |||
| No | 50 | 31 | 19 | |
| Yes | 57 | 20 | 37 | |
| AFP | 0.783 | |||
| <20 ng/mL | 41 | 20 | 21 | |
| 20–400 ng/mL | 34 | 17 | 17 | |
| >400–1000 ng/mL | 21 | 8 | 13 | |
| >1000 ng/mL | 11 | 6 | 5 | |
| Portal vein tumor thrombus | 0.001 | |||
| No | 42 | 12 | 30 | |
| Yes | 65 | 39 | 26 | |
| Child-Pugh | 0.033 | |||
| A | 9 | 8 | 1 | |
| B | 62 | 28 | 34 | |
| C | 36 | 15 | 21 | |
| Therapies | 0.003 | |||
| TACE | 35 | 9 | 26 | |
| Conservative treatment | 32 | 16 | 16 | |
| Combined therapy | 40 | 26 | 14 | |
| Neoplasm metastasis | 0.000 | |||
| No | 40 | 28 | 12 | |
| Yes | 67 | 23 | 44 | |
Diagnostic value of lncRNA-D16366 in HCC
To explore diagnostic value of lncRNA-D16366, an ROC curve was established. The outcome showed an AUC of 0.752 with a sensitivity of 65.5% and a specificity of 84.6%. The ideal cut-off value was 0.196 (Figure 4).
Figure 4.
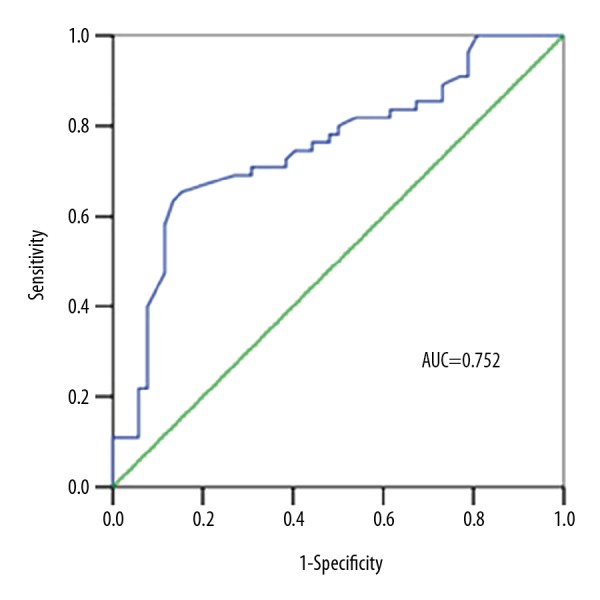
ROC curve for patients with HCC based on the expression of lncRNA-D16366.
Association between the expression of lncRNA-D16366 and overall survival
To analyze prognostic value of lncRNA-D16366 in HCC, Kaplan-Meier and Cox regression analyses were performed. Kaplan-Meier analysis demonstrated that overall survival of patients in the low lncRNA-D16366 expression group was shorter than in the high lncRNA-D16366 expression group (Figure 5). Log-rank testing revealed the result possessed statistical significance (P<0.001). Then, multivariate analysis was conducted through Cox regression analysis. Portal vein tumor thrombus (HR=3.104, 95CI%=1.197–8.048, P=0.020) and low expression of lncRNA-D16366 (HR=6.472, 95CI%=2.046–20.470, P=0.001) appear to be related to the prognosis of HCC (Table 2). Low expression level of lncRNA-D16366 was an unfavorable factor for the prognosis in HCC.
Figure 5.
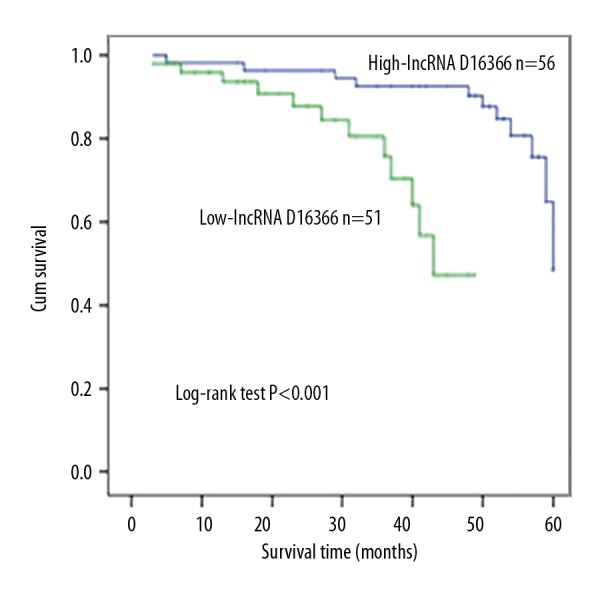
Kaplan-Meier analysis for patients with HCC based on the expression of lncRNA-D16366.
Table 2.
Cox regression analysis for the prognostic value of lncRNA-D16366 in HCC.
Discussion
HCC is the sixth most common cancer and the third leading cause of cancer-related deaths, and accounts for 70% to 85% of total liver cancers worldwide [1,17,18]. Rapid infiltrating growth, early metastasis, high-grade malignancy, and poor prognosis are important characteristics of HCC [19]. Accurate bio-markers are useful in predicting the prognosis of different diseases, including HCC. Studies have proposed many bio-markers for HCC prognosis, such as miRNAs, specific expression gene, and lncRNAs. Among them, lncRNAs offer a new direction and have attracted much attention.
In previous studies, many virtual lncRNAs have been studied in HCC. For instance, H19 was the first non-coding RNA reported to show abnormal expression in HCC [20]. Li et al. found that in HCC, maternally expressed gene 3(MEG3) was downregulated and played an inhibitory role in the malignancy growth [21]. Wu et al. found that the over-expression of lncRNA HOTAIR enhanced carcinogenic activity of HCC cells, and predicted unfavorable clinical outcomes [22]. Metastasis-associated lung and adenocarcinoma transcript 1(MALAT1) was reported to promote the viability, mobilization force, and invasiveness of HCC cells through sponging miR-204 and releasing SIRT1 [23]. lncRNA-HULC and lncRNA-HANR were both upregulated in patients with HCC [24,25]. Zhang et al. found that the dysregulation of lncRNAs influences the pathogenesis of HBV-HCC [26]. These results show that lncRNAs play important roles in the development and progression of HCC.
In our study, we detected the expression of lncRNA-D16366 in both tissue and serum samples, showing its downregulation in patients with HCC, which was consistent with the view of Liu et al. [16]. To further explore whether it is involved in the development of HCC, we explored the association of lncRNA-D16366 with clinicopathological characteristics, and found that tumor size, HbsAg, portal vein tumor thrombus, Child-Pugh scores, therapies received, and neoplasm metastasis were significantly associated with the expression of lncRNA-D16366 in HCC.
We also investigated the clinical significance of lncRNA-D16366. Our results suggest that its abnormal expression is related to the diagnosis or prognosis of HCC. The high diagnostic value of lncRNA-D16366 was demonstrated by an ROC curve with an AUC of 0.752, a sensitivity of 65.5%, and a specificity of 84.6%. In addition, Kaplan-Meier analysis indicated lncRNA-D16366 expression is correlated with the prognosis of HCC. Therefore, we conducted a multivariate analysis with Cox regression analysis. The outcome verified that lncRNA-D16366 predicted prognosis of HCC.
Conclusions
lncRNA-D16366 is downregulated in patients with HCC. Its abnormal expression could be used as an independent diagnostic and prognostic marker for this lethal cancer. However, its mechanisms and other effects remain unknown. Moreover, this study is limited by its small sample. Hence, further studies are needed.
Footnotes
Source of support: The Project of Shaanxi Science and Technology Committee (2017SF-232); Hospital Project of Xianyang Central Hospital (2017G05, 2017Z01)
Conflict of interests
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Choi WT, Kakar S. Immunohistochemistry in the diagnosis of hepatocellular carcinoma. Gastroenterol Clin North Am. 2017;46:311–25. doi: 10.1016/j.gtc.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol. 2017;34:153–59. doi: 10.1053/j.semdp.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 6.Zhang S, Yue M, Shu R, et al. Recent advances in the management of hepatocellular carcinoma. J BUON. 2016;21:307–11. [PubMed] [Google Scholar]
- 7.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 8.Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformatics. 2017;15:177–86. doi: 10.1016/j.gpb.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: Spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. 2016;17:756–70. doi: 10.1038/nrm.2016.126. [DOI] [PubMed] [Google Scholar]
- 10.Dey BK, Mueller AC, Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5:e944014. doi: 10.4161/21541272.2014.944014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akhade VS, Pal D, Kanduri C. Long Noncoding RNA: Genome organization and mechanism of action. Adv Exp Med Biol. 2017;1008:47–74. doi: 10.1007/978-981-10-5203-3_2. [DOI] [PubMed] [Google Scholar]
- 12.Weidle UH, Birzele F, Kollmorgen G, Ruger R. Long non-coding RNAs and their role in metastasis. Cancer Genomics Proteomics. 2017;14:143–60. doi: 10.21873/cgp.20027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun W, Yang Y, Xu C, Guo J. Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genet. 2017;216–217:105–10. doi: 10.1016/j.cancergen.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–81. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Ye C, Xiong H, et al. Dysregulation of long non-coding RNA in breast cancer: An overview of mechanism and clinical implication. Oncotarget. 2017;8:5508–22. doi: 10.18632/oncotarget.12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu WT, Lu X, Tang GH, et al. LncRNAs expression signatures of hepatocellular carcinoma revealed by microarray. World J Gastroenterol. 2014;20:6314–21. doi: 10.3748/wjg.v20.i20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dimitroulis D, Damaskos C, Valsami S, et al. From diagnosis to treatment of hepatocellular carcinoma: An epidemic problem for both developed and developing world. World J Gastroenterol. 2017;23:5282–94. doi: 10.3748/wjg.v23.i29.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlachterman A, Craft WW, Jr, Hilgenfeldt E, et al. Current and future treatments for hepatocellular carcinoma. World J Gastroenterol. 2015;21:8478–91. doi: 10.3748/wjg.v21.i28.8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chedid MF, Kruel CRP, Pinto MA, et al. Hepatocellular carcinoma: Diagnosis and operative management. Arq Bras Cir Dig. 2017;30:272–78. doi: 10.1590/0102-6720201700040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pope C, Mishra S, Russell J, et al. Targeting H19, an imprinted long non-coding RNA, in hepatic functions and liver diseases. Diseases. 2017;5(1) doi: 10.3390/diseases5010011. pii: E11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Ren M, Zhao Y, et al. MicroRNA-26a inhibits proliferation and metastasis of human hepatocellular carcinoma by regulating DNMT3B-MEG3 axis. Oncol Rep. 2017;37:3527–35. doi: 10.3892/or.2017.5579. [DOI] [PubMed] [Google Scholar]
- 22.Wu L, Zhang L, Zheng S. Role of the long non-coding RNA HOTAIR in hepatocellular carcinoma. Oncol Lett. 2017;14:1233–39. doi: 10.3892/ol.2017.6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou Z, Xu X, Zhou L, et al. The long non-coding RNA MALAT1 promotes the migration and invasion of hepatocellular carcinoma by sponging miR-204 and releasing SIRT1. Tumour Biol. 2017;39:1010428317718135. doi: 10.1177/1010428317718135. [DOI] [PubMed] [Google Scholar]
- 24.Li SP, Xu HX, Yu Y, et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7:42431–46. doi: 10.18632/oncotarget.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao J, Lv Y, Jin F, et al. LncRNA HANR promotes tumorigenesis and increase of chemoresistance in hepatocellular carcinoma. Cell Physiol Biochem. 2017;43:1926–38. doi: 10.1159/000484116. [DOI] [PubMed] [Google Scholar]
- 26.Zhang B, Han S, Feng B, et al. Hepatitis B virus X protein-mediated non-coding RNA aberrations in the development of human hepatocellular carcinoma. Exp Mol Med. 2017;49:e293. doi: 10.1038/emm.2016.177. [DOI] [PMC free article] [PubMed] [Google Scholar]


