Abstract
Background
Prostate cancer progresses slowly when present in low risk forms but can be lethal when it progresses to metastatic disease. A non-invasive test that can detect significant prostate cancer is needed to guide patient management.
Methods
Capillary electrophoresis/mass spectrometry has been employed to identify urinary peptides that may accurately detect significant prostate cancer. Urine samples from 823 patients with PSA (<15 ng/ml) were collected prior to biopsy. A case–control comparison was performed in a training set of 543 patients (nSig = 98; nnon-Sig = 445) and a validation set of 280 patients (nSig = 48, nnon-Sig = 232). Totally, 19 significant peptides were subsequently combined by a support vector machine algorithm.
Results
Independent validation of the 19-biomarker model in 280 patients resulted in a 90% sensitivity and 59% specificity, with an AUC of 0.81, outperforming PSA (AUC = 0.58) and the ERSPC-3/4 risk calculator (AUC = 0.69) in the validation set.
Conclusions
This multi-parametric model holds promise to improve the current diagnosis of significant prostate cancer. This test as a guide to biopsy could help to decrease the number of biopsies and guide intervention. Nevertheless, further prospective validation in an external clinical cohort is required to assess the exact performance characteristics.
Subject terms: Prostate cancer, Diagnostic markers, Proteomic analysis
Background
Prostate cancer (PCa) is ranked as the second most frequently diagnosed cancer in men,1 and the most frequent non-skin cancer in developed countries.2 Although PCa is diagnosed in 15–20% of men, the lifetime risk of death due to PCa is very low (3%),3 mainly because low-risk forms progress slowly and the disease is well treatable in early stages. PCa diagnosis is currently mostly based on serum prostate-specific antigen (PSA) testing, digital rectal examination (DRE) and confirmed by a multi-core prostatic biopsy.4 Multiple factors not related to prostate malignancy may affect the level of blood PSA [inflammation, infection or presence of benign prostate hyperplasia (BPH)]. Therefore, PSA lacks specificity particularly in the intermediate range, with only 22–27% of those patients with PSA between 4–10 ng/ml to be positively confirmed with PCa after biopsy.5 In addition, PSA screening and multicore biopsy have increased the detection rate of small, localised, well-differentiated PCa,6 resulting in over-diagnosis and over-treatment.6–9 For those patients presenting with an indolent or clinically non-significant cancer (Gleason score (GS) < 7),10 immediate treatment may not be beneficial and ideal management may be a conservative approach, such as active surveillance (AS).11 Management of patients with non-significant PCa currently relies on repeated biopsies, series of PSA measurements and DRE, while the uncertainty to properly assess PCa imposes a significant social and economic burden on patients and health insurances because of the side effects and treatment costs.12 For these reasons, better stratification of the risk for significant PCa (Sig PCa) appears beneficial to guide patient management.
Aimed at improving on the current discrimination of Sig PCa by non-invasive means, capillary-electrophoresis coupled to mass spectrometry (CE–MS) was employed to identify peptides specific for PCa in urine samples from patients with clinically significant and non-significant PCa. Urine was selected, as it presents several advantages over blood or tissue, among others: easy, non-invasive repeated sampling, effortless availability and high stability of the proteome. Although several candidate biomarkers have been described,13–15 the currently available single biomarkers lack diagnostic accuracy for routine clinical application. At the same time, the high biological variability of PCa suggests that a combination of clearly defined, -omics derived biomarkers, rather than a single biomarker, may provide higher accuracy to detect cancer.16–18 In this study, we aimed to establish a biomarker model to detect Sig PCa.
Methods
Study population and design
A case–control study was performed on patients who underwent a transrectal ultrasound (TRUS)-guided prostate biopsy from January 2013 to July 2015 in the Urology department, Reina Sofia Hospital, Cordoba, Spain, as part of the ONCOVER project. Ethical approval was obtained by the Reina Sofia Hospital Research Ethics Committee and informed consent was obtained from all participants for the project. ONCOVER cohort included patients who attended the urology clinic of Reina Sofia Hospital with a recommendation for a prostate biopsy according to clinical practice.19 Patients provided a urine sample and underwent blood testing just before undergoing a prostate biopsy. Recommendations for biopsy indication were: suspicious findings on DRE, PSA > 10 ng/mL, or PSA 3–10 ng/mL if free PSA ratio was low (usually, <25–30%), and in patients with previous biopsies, a persistently suspicious indication of PCa (persistently elevated PSA, suspicious DRE, etc.). For transrectal prostate biopsy, 12 cores were obtained from patients undergoing the first biopsy procedure, and a minimum of 16 biopsy cores for those who had a previous biopsy. For this analysis, 823 PCa patients were included according to the following criteria: (a) PSA level <15 ng/mL on the day of the biopsy and (b) no previous diagnoses of PCa. For all 823 patients, complete records for all the main variables were available, including PSA, DRE, number of previous biopsies, 5-alpha-reductase inhibitor intake and pathology results. Information on prostate volume was additionally retrieved for 721 patients, based on the measurements that had been performed with TRUS during the biopsy. Because of missing data for 102 patients, and in order to avoid introducing any selection bias, for this analysis prostate volume was not included in the nomogram analysis, but only for comparison purposes (i.e., biomarkers compared to prostate volume). Patients treated with 5-alpha-reductase inhibitors for urinary symptoms were also included in the study, but excluded from the analysis for the comparison with PSA, as treatment with 5-alpha-reductase inhibitors is expected to affect PSA levels. All biopsy specimens were analysed by a urologic pathologist according to International Society of Urological Pathology 2005 modified criteria.20 Clinical and laboratory data, including among others: age, PSA level (on the day of biopsy), the results of DRE, number of previous biopsies, prior treatment with 5-alpha-reductase inhibitors, prostate volume by TRUS, urinary creatinine and pathology results were collected and presented in the Supplementary Table 1. A score based on the risk calculator of the European Randomised Study of Screening for Prostate Cancer (ERSPC) was calculated (http://www.prostatecancer-riskcalculator.com/seven-prostate-cancer-risk calculators). The formulas that were utilised in this study, were ERSPC- 3, for those patients during initial biopsy, and ERSPC- 4, for patients during repeated biopsy. For the above estimates, the variables that are considered are PSA and DRE and the result of previous biopsy for those patients who underwent (biopsy before (ERSPC-4). GS was used in this study to discriminate Sig PCa (GS ≥ 7) from non-Sig PCa.
MS analysis
CE–MS analysis was performed for the 823 urine samples, following the previously established protocols for samples preparation and data acquisition, previously described in detail.21 In brief, sample preparation was performed by diluting 700 µl urine aliquots from the urine collected from patients prior to the prostate biopsy, with two volumes (1.4 ml) alkaline buffer containing 2 M urea, 10 mM NH4OH and 0.02% sodium dodecyl sulphate (pH 10.5). Subsequently, the samples were filtered by Centrisart ultracentrifugation filters (Sartorius, Göttingen, Germany) to retain proteins/polypeptides below 20 kDa and were subsequently desalted over PD-10 columns (GE Healthcare, Munich, Germany). The peptide extracts were lyophilised and resuspended in high-performance liquid-chromatography (LC) grade water. CE–MS analysis and data processing were performed according to ISO13485 standards yielding quality controlled urinary data sets.21 Mass spectral ion peaks representing identical molecules at different charge states were deconvoluted into single masses using MosaiquesVisu software.22,23 The peak list characterises each peptide by its molecular mass (kDa), normalised migration time (min) and normalised signal intensity (AU).22,23 Normalisation of the CE–MS data were based on twenty nine collagen fragments that are generally not affected by disease and serve as internal standards.24 After normalisation, all proteomics datasets were deposited, matched, and annotated in a Microsoft SQL database and used as input in the presented study. Transformation of the data (log-transformation) was performed prior to the statistical analysis, as previously described.25
Peptide sequencing and matching
Matching of the amino acid sequences with the CE–MS acquired ion peaks was based on mass correlation between CE–MS and LC-tandem MS analysis. The amino acid sequence was determined by MS/MS analysis using either a PACE CE or a Dionex Ultimate 3000 RSLS nanoflow system (Dionex, Camberly UK) coupled to an Orbitrap Velos instrument (Thermo Scientific), as previously described.26 Protein matching and data analysis was based on Proteome Discoverer 1.2 (activation type: HCD; precursor mass tolerance: 5 ppm; fragment mass tolerance: 0.05 Da). No fixed modifications were selected, oxidation of methionine and proline were selected as variable modifications. The data were searched against the UniProt human database27 without enzyme specificity. Further validation of the obtained peptide identifications is based on the assessment of the peptide charge at the working pH of 2.2 and the CE-migration time results.28
Statistical analysis
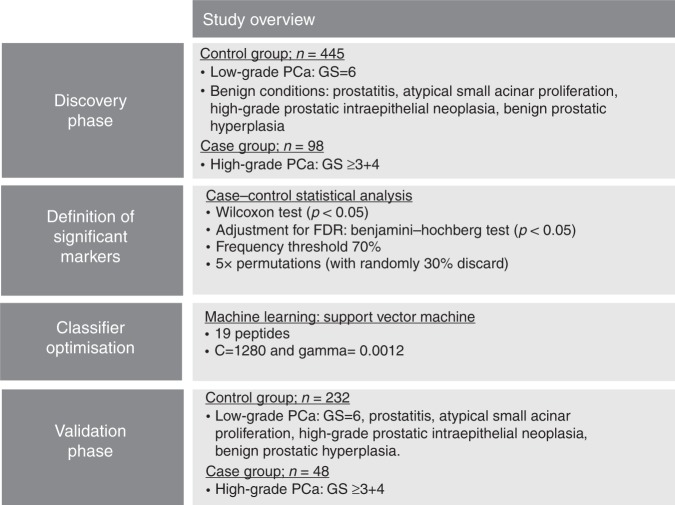
A case–control statistical comparison was performed to detect potentially Sig PCa biomarkers. The datasets were grouped into: (a) a case set of clinically Sig PCa (nSig = 146), including PCa patients with high-risk PCa (GS ≥ 7) and (b) a control set including clinically non-significant PCa (low-risk PCa; GS = 6) along with patients presenting with other aetiologies (n = 677). The groups were further divided into a discovery (nSig = 98 cases of Sig PCa; nnon-Sig = 445 controls) and validation set (nSig = 48 cases of Sig PCa, nnon-Sig = 232 controls), according to the ‘2/3–1/3 rule’, as previously described.16 Random sampling guarantees that each group/class is properly represented in all data subsets. Based on the literature,29 this commonly used strategy of allocating two-third of cases for training is close to optimal for large sized datasets (n ≥ 100) with strong signals (i.e., >85% full dataset accuracy).29
Further statistical analysis was performed to identify potential bias, considering the clinical data shown in Table 1. Mann–Whitney non-parametric test was used to investigate statistically significant differences between the two groups for continuous variables and chi-squared test for categorical variables, respectively. The urinary CE–MS profiles were compared for differences at the individual peptide excretion levels by applying the Wilcoxon rank sum test.25 A frequency threshold of 70% in at least one of the two groups was applied. To increase the validity of the statistical approach, permutation analysis was performed by randomly excluding 30% of the samples and repeated five times. Statistical correction of the estimated p values for multivariate testing was performed based on the Benjamini–Hochberg method.30 Only the peptides significant (p < 0.05) in all five permutation analyses were considered for further analysis.
Table 1.
Clinical and biochemical variables for the patients grouped into the discovery and validation set
| Baseline characteristics | Discovery phase (n = 543) | Validation phase (n = 280) | p Value discovery vs. validation | Group 1: Non-significant PCa/Controls (n = 677) | Group 2: Significant PCa (n = 146) | p Value group 1 vs. group 2 |
|---|---|---|---|---|---|---|
| Median age (IQR; y) | 64.0 (11.0) | 63.5 (12.0) | 0.2947a | 63.0 (11.5) | 68.0 (10.3) | <0.0001a |
| PSA median (IQR; ng/ml) | 5.4 (3.6) | 5.0 (3.1) | 0.2060 a | 5.1 (3.3) | 6.1 (4.1) | 0.0013a |
| Digital rectal examination (Pos/Neg) | 104/439 | 40/240 | 0.0576b | 94/583 | 50/96 | 0.1453b |
| Previous biopsies (Y/N) | 139/404 | 69/ 211 | 0.9895b | 187/480 | 21/125 | 0.0007b |
| Median number of previous biopsies (IQR) | 1 (1) | 1 (0) | 0.6366a | 1 (1) | 1 (0) | 0.007a |
| Prostate volume (IQR; ml) | 35.0 (25.0; n = 481) | 36.0 (18.4; n = 240) | 0.6701a | 37.4 (23; n = 594) | 28.0 (18; n = 127) | <0.0001a |
| PSA density (IQR; ng/ml2) | 0.14 (0.11; n = 481) | 0.14 (0.10; n = 240) | 0.3156a | 0.20 (0.09; n = 594) | 0.13 (0.15; n = 127) | <0.0001a |
| 5α-reductase treatment (Y/N) | 18/ 524 | 6/ 274 | 0.4640b | 22/655 | 2/144 | 0.2219b |
| Median urinary creatinine (IQR; mmol/L) | 7.6 (4.7) | 8.3 (4.9) | 0.0875a | 7.9 (4.7) | 7.8 (4.7) | 0.5319a |
| Significant PCa | 98 (18.0%) | 48 (17.1%) | 0.9345b | |||
| Non-significant PCa | 445 (82.0%) | 232 (82.9%) | 0.8532b | |||
| Disease pathology—significant PCa | ||||||
| Gleason 3 + 4/4 + 3 | 63 (64.3%)/20 (20.4%) | 32 (66.6%)/9 (18.8%) | 0.9290b/0.9945b | |||
| Gleason 8 | 9 (9.2%) | 5 (10.4%) | 0.9459b | |||
| Gleason≥9 | 6 (6.1%) | 2 (4.2%) | 0.9308b | |||
| Disease pathology—non-significant PCa | ||||||
| Gleason 6 | 99 (22.1%) | 32 (13.8%) | 0.0126b | |||
| Benign—non-PCa aetiologies | ||||||
| Benign prostatic hyperplasia; BPH | 307 (69.1%) | 175 (75.4%) | 0.1033b | |||
| Prostatic intraepithelial neoplasia; PIN | 22 (5.0%) | 12 (5.2%) | 0.9425b | |||
| Atypical small acinar proliferation; ASAP | 17 (3.8%) | 13 (5.6%) | 0.3766b | |||
aMann–Whitney test
bChi-squared test
IQR interquartile range, N not received, Neg negative, PCa prostate cancer, Pos positive, Y received
Optimisation of the SVM-based biomarker model
The urinary peptide-based classifier was developed in the training set, using MosaCluster (version 1.7.0), a support vector machine (SVM)-based software. The classifier was optimised based on the shortlisted PCa specific biomarkers with each biomarker representing one dimension in the n-dimensional parameter space.17 In additiony, the cut-off was established based on the discovery set. In the independent validation, the sensitivity and specificity estimates for the SVM-based peptide marker pattern were calculated based on the number of correctly classified samples. The receiver operating characteristic (ROC) plots and the respective confidence intervals (95% CI) were based on exact binomial calculations and were calculated in MedCalc 12.7.5.0 (Mariakerke, Belgium). Area under the curve (AUC) values were then compared using DeLong tests. Statistical comparisons of the classification scores in the validation cohort were performed by the Kruskal–Wallis rank sum test using MedCalc. To address the potential clinical utility of the models, we performed decision curve analysis, as proposed by Vickers and Elkin.31 This method has the advantage of not requiring the specification of the relative cost for false-positives and false-negatives, defining a net benefit as a function of the decision threshold at which one would consider obtaining a biopsy. For the analysis MedCalc 12.7.5.0 (Mariakerke, Belgium) and R version 3.2.3 were used.
Results
Study cohort for patients with clinically significant and non-significant PCa
Proteomics profiling data were acquired from 823 patients suspicious for PCa. Out of those, 677 (82.3%) presented with non-significant PCa (GS = 6), benign or atypical conditions (control group) and 146 (17.7%) were included in the case group due to presence of Sig PCa. Men with Sig PCa were significantly older [median age = 68; interquartile range (IQR) = 10.3] compared to men from control group (median age = 63; IQR = 11.5; p < 0.0001). In addition, patients from the control group had significantly lower PSA levels (median = 5.1 ng/ml; IQR = 3.3) compared to those from case group [median = 6.1 ng/ml; IQR = 4.1; p = 0.0013]. Within the control group, 480 (70.9%) did not undergo any previous negative biopsy, while for patients with Sig PCa, the respective proportion was 85.6% (n = 125); (p = 0.0007). The clinical characteristics along with the sample distribution are presented in the Table 1.
Development of a biomarker model based on CE–MS urinary peptide profiling
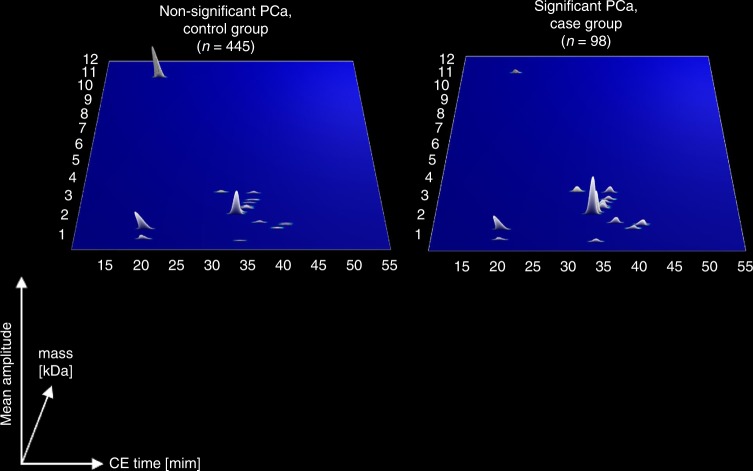
For the identification of CE–MS specific biomarkers, a case–control comparison was performed in the discovery set of 543 patients, schematically depicted in Fig. 1. The comparison enabled the identification of 19 peptides displaying statistically significant differences in their distribution between patients with Sig PCa compared to the control group (Supplementary Table 2). The graphical depiction of the compiled urinary profiling signatures is comparatively presented in Fig. 2. Using the 19 statistically altered peptide markers an SVM machine learning algorithm was adopted and optimise to develop a classifier (Fig. 1).
Fig. 1.
Schematic representation of the study design and the analytical workflow for the development of urine CE–MS-based biomarker panel
Fig. 2.
Compiled average urinary profiling signatures of the patients with significant and non-significant PCa. The molecular mass (0.1–12 kDa) is shown on a logarithmic scale and is plotted against normalised migration time (15–55 min). Signal intensity is encoded by peak height and colour
Independent validation of the SVM-based biomarker model
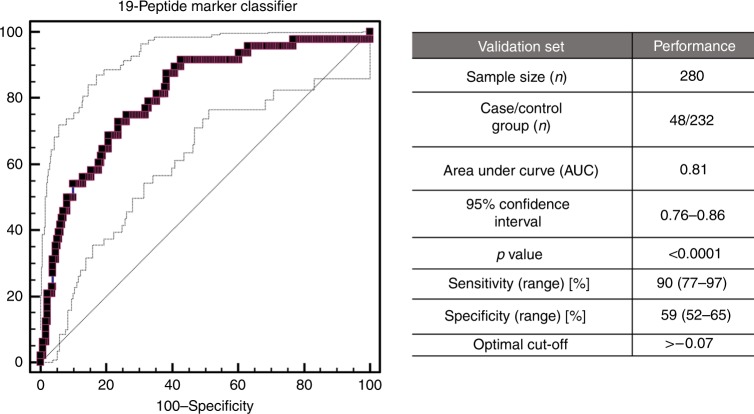
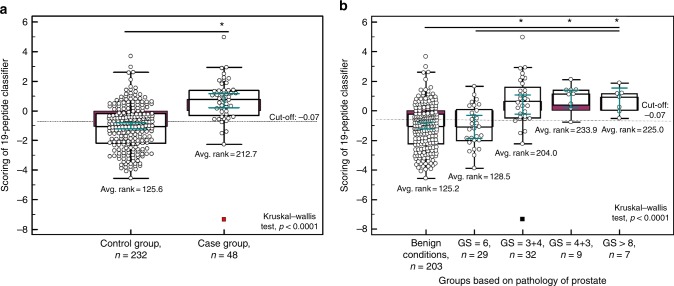
Validation of the 19-biomarker model in the independent set (n = 280), in line with the recommendations for biomarker identification and reporting in clinical proteomics,25 resulted in an overall AUC value of 0.81 ranged from 0.76 to 0.86 (95% CI: p < 0.0001). Fig. 3 presents the ROC curve, which at the pre-defined cut-off of −0.07 resulted in sensitivity levels of 90% (77–97; 95% CI) and specificity of 59% (52–65; 95% CI), respectively. Additional statistical analysis was performed, by application of a post hoc rank sum test to compare the scores between the case and control groups. As depicted in Fig. 4a, the classification of each group differs at the significance level of p < 0.0001. Moreover, as shown in Fig. 4b, there is a gradual increase in the 19-biomarker model score, as GS increases, while a significant difference is observed between the 19-biomarker model scores of GS 6 tumours and GS ≥ 7 (p < 0.0001).
Fig. 3.
Receiver operating characteristics (ROC) analysis performed in the independent validation cohort, displaying the performance of the 19-biomarker panel for discriminating the case group (nSig = 48) from the control group (nnon-Sig = 232). ROC characteristics, such as area under the curve (AUC), 95% confidence intervals (CI), and p value are provided for the classification of Sig PCa patients
Fig. 4.
a Classification scores, presented in Box-and-Whisker plots grouped according to the case group (nSig = 48) and control group (nnon-Sig = 232). b Classification scores displaying the level of discrimination across the different Gleason score. A post hoc rank-test was performed using Kruskal–Wallis test. *p < 0.05
Comparative analysis of the 19-biomarker model with clinical parameters
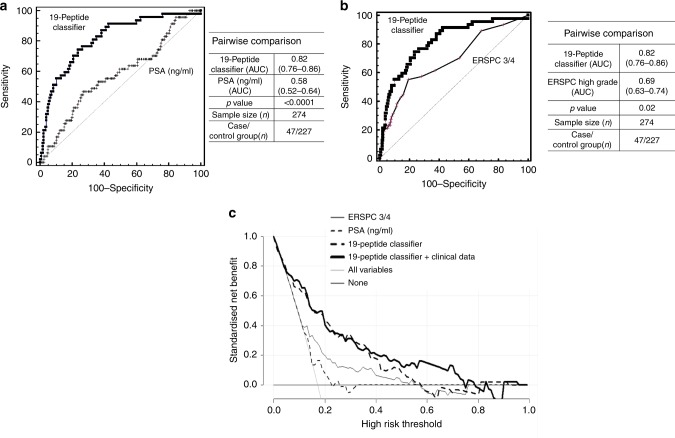
A direct comparison of the 19-biomarker model with PSA was performed in the validation set. Of note, out of 280 patients, 6 patients had received previous treatment with 5-alpha-reductase inhibitors, therefore for the comparative analysis only 274 patients were considered. As depicted in Fig. 5a, the multi-peptide model significantly outperformed the PSA testing with the AUC values at 0.82 and 0.58, respectively (p < 0.0001). For those patients where clinical records on prostate volume were available (n = 240), an additional comparison between the 19-biomarker model and the prostate volume was performed, indicating a significantly better accuracy for the 19-biomarker model (AUC of 0.81) compared to prostate volume (AUC of 0.64; p = 0.0103). Moreover, logistic regression analysis was performed for the available clinical variables to assess the potential significant predictive value of each of those in the discrimination of Sig PCa. The included clinical parameters were: (a) the result of DRE, (b) presence of previous biopsy, (c) the number of previous biopsies, (d) prostate volume and (e) age. Based on the statistical comparison significant contribution to the outcome is revealed for age (odds ratio of 1.1, p = 0.0366), PSA (odds ratio of 1.2, p = 0.0162) and the 19-biomarker model (odds ratio of 2.2, p < 0.0001), while the presence and number of previous biopsies, prostate volume and the result of DRE were not significant predictors of Sig PCa. Combination of the significant variables (19-biomarker model, PSA and age) into a nomogram through the regression equation, resulted in an improved AUC value of 0.83, although not statistically significant (p = 0.4344) compared to the 19-biomarker model alone. In order to investigate if the 19-peptide classifier can present an added value over the current state-of-the-art, the SVM-based score from the 19-biomarker model was further compared with the estimates of the ERSPC risk calculator for detecting high risk PCa (ERSPC—3/4), as presented in Fig. 5b. The 19-peptide classifier showed significantly better performance (AUC = 0.82; p = 0.02) compared to the ERSPC estimates (AUC = 0.69). To assess the clinical benefit of the 19-biomarker model, a decision curve analysis was additionally performed. Based on the net plotting against the threshold probabilities for the comparisons between the 19-biomarker model alone and with clinical variables (PSA, age), PSA and ERSPC estimates, there is a clear benefit of the biomarker model, particularly in the lower range of the risk thresholds (Fig. 5c).
Fig. 5.
a Comparative analysis depicted by receiver operating characteristics (ROC) curves for the 19-biomarker panel and the PSA levels (n = 274). b Added value of the 19-biomarker panel over ERSPC -3/4 for high risk (Gleason ≥ 7) (n = 274; six patients from the validation set were excluded as previously treated with 5 alpha reductase inhibitors). c Results of the decision curve analysis. The net benefit for the prediction of Sig PCa on biopsy is shown, by using the different models as a function of the risk threshold, compared to the benefits of strategies for treating all patients (grey thin line) and treating none (grey thick line)
Sequencing of peptide biomarkers
Among the 19 peptide biomarkers, sequences could be obtained for 17, while 2 peptides could not be sequenced. The majority (14/17) were originated from various collagens. Peptide fragments originating from alpha-1 collagen of types (I), (XI), (XVII), (XXI) and alpha-2 type (I), (V), (IX), were most prominent and fragments of collagen type (VIII) chain were also identified. All the collagen peptide fragments are of increased abundance in the Sig PCa cases, apart from collagen alpha-1 (XVII) chain and collagen alpha-1(XXI), which are presented with decreased abundance. Interestingly, among the collagen fragments, a unique motif (pGP) is very prominent. The three remaining peptide markers were a fragment of protein phosphatase 1 regulatory subunit 3A, which was identified with decreased abundance and fractalkine or chemokine (C-X3-C motif) ligand 1 and Semaphorin-7A, both upregulated in the group of patients with Sig PCa.
Discussion
Insignificant PCa are slowly progressing forms that may be better managed conservatively without immediate treatment. Nevertheless, as insignificant forms can progress to significant cancer, frequent monitoring is required to timely and accurately detect the progression. Currently, routine monitoring is based on either PSA, although associated low accuracy, or invasive biopsies.32 More accurate non-invasive biomarkers are required to improve on the discrimination of Sig PCa. In this study, a biomarker model based on urinary peptides was established and validated in 823 patients suspicious for presence of PCa. This peptide panel enables the discrimination of non-significant PCa from clinically significant forms with high sensitivity and moderate specificity. The lower specificity is mostly attributed to the misclassification of clinically non-significant PCa (mainly GS of 6) as clinically significant forms. The clinical consequence of this observation can be weighted as acceptable, since patients with a positive score based on the 19-biomarker model would further undergo biopsy to rule out the presence of significant cancer.
The 19-biomarker model performs significantly better, when compared to PSA levels and also, when compared to the ERSPC risk calculator, demonstrating an added value of the biomarkers. Comparison with other clinical variables was also performed indicating a significant improvement of the 19-biomarker model, although particularly for prostate volume, missing data for 34 patients from the validation set do compromise the statistical power. An additional decision curve analysis was performed to assess the clinical benefit of the 19-biomarker model, in comparison with the current clinical standards, PSA and ERSPC calculator, demonstrating an improved net benefit of the 19-biomarker model, particularly in the low range of risk threshold.
Nowadays, several biomarkers have been tested in order to discriminate Sig PCa, such as 4K score test, PHI, PCA3, SelectMDx).33 A direct comparison with those markers, was unfortunately not possible in the context of the presented study, as paired data were not available (as different cohorts and approximations were performed). However, the initial results shown in this study with an AUC higher than 0.80, is within the range of 0.74–0.90 which is shown by other biomarkers34,35 and clearly justify implementation of this approach in a future investigative setting. In line with this and in order to facilitate comparisons, an additional prospective validation study design is planned, similar to other studies, such as the step approximation of the STHLM3 study, which was able to identify up to 21% of Sig PCa in patients with a PSA between 1 and 3 ng/ml.35 In the prospective evaluation, inclusion of multiparametric magnetic resonance imaging is planned, as it has demonstrated an added value in the diagnostic approximation for Sig PCa with a high NPV, improving the detection of Sig PCa.36,37
Regarding the biomarker identity, sequences could be obtained from 17 of the 19 peptide markers, most of them derived from collagen origin and being in increased abundance. Collagen fragments represent the majority of urinary peptides, even in healthy individuals.18 The increase in specific collagen fragments may depict extracellular matrix rearrangements, associated with tumour invasion and resulting in proteolytic products, which are subsequently excreted in urine. Previous studies,18,38 reporting on CE–MS based biomarkers for detection of PCa (for discrimination of PCa patients from those without malignancy), also identified collagen fragments as being increased in abundance in cancer patients.18,38 In the present study, a slightly different clinical design was followed, as the aim was to discriminate in patients that had PCa, those presenting with significant cancer from those with non-significant cancer. In the study by Theodorescu et al.,18 four sequences out of twelve could be obtained, with one biomarker common in both studies: a fragment of Collagen alpha-1(I) chain. The other three biomarkers described by Theodorescu et al.18 were not identified as significantly altered in this study, while an enrichment was observed for other sequences belonging to collagen alpha-1 and collagen alpha-2 chains, protein phosphatase 1 regulatory subunit 3A and fractalkine. The observed differences are attributed in part to the advancements of the technology enabling a better sequence coverage, but also the different clinical context, which in this study was the identification of differentially abundant cancer biomarkers between two cancer forms. A pGP motif was present in most of the collagen sequences. The pGP motif is a chemoattractant derived from proteolytic cleavage of collagen by matrix metalloproteinases. pGP motif binds to (C–X–C motif) receptors and is thus associated with neutrophil attraction in inflamed tissues.39 In addition, protein phosphatase 1 regulatory subunit 3A, which is considered as a tumour suppressing molecule was identified with decreased abundance.40 Overall, the observations at the urinary peptides of the patients with Sig PCa, depict features of cancer progression and tumour related inflammation.
The specific clinical impact of the non-invasive biomarker model would primarily be to guide patient management and reduce the number of invasive biopsies. As such, high sensitivity is required, for correct detection of significant PCa. In view of a positive test, the treating physician is alerted to perform a more thorough investigation, improving the overall accuracy in detection of Sig PCa. Lower specificity would result in more misclassifications of non-significant PCa as potentially significant, and as a consequence prostate biopsy to rule out significant cancer. Therefore, a false positive result will be clarified upon biopsy.
These encouraging results should be interpreted considering the limitations of the study: Firstly, although the use of TRUS biopsy for PCa diagnosis suffers from random error and false negative results in comparison with trans-perineal template biopsy,37 which might have affected the results (underestimate the specificity and overestimate the sensitivity) of the present study, it should be noted that TRUS biopsy is the accepted standard method in the current clinical practice and mostly used in biomarkers studies. Secondly, comparison with prostate biopsy pathology and not prostatectomy specimens is a similar limitation, possibly affecting the results in the same way, but prostate biopsy is the first approximation to diagnose and to stablish the risk category of the patients, so that it might represent more clearly the clinical practice. Thirdly, urine was collected with no prostate stimulation which could diminish the number of peptides specifically derived from the prostate secretion. Moreover, this study was performed retrospectively, however, on samples that were prospectively collected. Furthermore, the exact potential benefit for patients has to be assessed in a prospective trial. However, based on the data presented, implementation of this approach in an investigative setting appears highly justified.
The data presented in this study could demonstrate the utility of a multiple-marker approach for improved non-invasive detection of Sig PCa. Taking into consideration the increased variability which is caused by the high intra-tumour heterogeneity, an intrinsic characteristic of cancer, a single biomarker is not expected to enable the discrimination of Sig PCa from non-significant with high accuracy. Therefore, a combination of biomarkers appears to be the currently best option to guide biopsies and AS. Effective discrimination between clinically significant and non-significant PCa is expected to have a positive impact on reducing biopsies, improving patient compliance and also guide a more thorough examination in case of a positive result. The benefit for the management of patients under AS is also evident, as discrimination of the Sig PCa will result in improved guidance for initiation of definite treatment. Overall, improved non-invasive patient stratification is expected to present a positive impact on PCa patient management, by improving patient compliance and reducing over-treatment and the associated costs. The results of this study, although highly significant, will be assessed in a prospective trial to also determine the exact value in the context of patient management.
Supplementary information
Acknowledgements
E. Gómez-Gómez thanks The Carlos III Health Institute (ISCIII) and the European Social Funds (FSE) which funds his Rio Hortega research grant contract (CM16/00180). The biological samples repository node (Córdoba, Spain) is gratefully acknowledged for coordination tasks in the selection of urine samples. Ipek Guler from the methodology department of the Maimonides Institute of Biomedical Research of Cordoba is also gratefully acknowledged for collaboration in the statistical analysis.
Author contributions
M.F., E.G.G, H.M., and J.C.V. carried out the conception and design of the study; M.F., E.G.G., A.B.P., J.V.R. and J.C.V. contributed to the data acquisition; M.F., E.G.G., A.L., H.M. and J.C.V. carried out the analysis and interpretation of data; M.F. and E.G.G. drafted the paper; A.B.P., J.V.R., A.L., Z.C., A.S.M., R.M.L., M.J.R.T., H.M. and J.C.V. carried out a critical revision of the paper for important intellectual content; M.F. and A.L. performed the statistical analysis; R.M.L., M.J.R.T., H.M. and J.C.V. supervised the work.
Competing interests
H.M. is the founder and co-owner of Mosaiques Diagnostics. M.F. and A.L. are employed by Mosaiques Diagnostics.
Funding
This work was supported by The Spanish Ministerio de Economía y Competitividad (MINECO) and FEDER programme which are gratefully acknowledged for the financial support (Projects “Development of methods for early cancer detection; ONCOVER—detection system of volatile compounds for early diagnosis of lung, colon and prostate cancer”, CCB.030PM). This research was also supported in part by the BioGuidePCa (E! 11023, Eurostars) funded by BMBF (Germany) and PCaProTreat (H2020-MSCA-IF-2017-800048).
Ethics approval and consent to participate
This study was performed as part of the ONCOVER project. Ethical approval was obtained by the Reina Sofia Hospital Research Ethics Committee in accordance with the Declaration of Helsinki and informed consent was obtained from all participants for the project.
Data availability
All data generated or analysed during this study are included in this published article.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Maria Frantzi, Enrique Gomez Gomez
These authors contributed equally: Harald Mischak, Julia Carrasco Valiente
Supplementary information
Supplementary information is available for this paper at 10.1038/s41416-019-0472-z.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, et al. Cancer statistics, 2006. CA: Cancer J. Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 4.Mottet N, Bellmunt J, Bolla M, Briers E, Cumberbatch MG, De Santis M, et al. EAU-ESTRO-SIOG guidelines on prostate cancer. Part 1: screening, diagnosis, and local treatment with curative intent. Eur. Urol. 2017;71:618–629. doi: 10.1016/j.eururo.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Gretzer MB, Partin AW. PSA levels and the probability of prostate cancer on biopsy. Eur. Urol. Suppl. 2002;1:21–27. doi: 10.1016/S1569-9056(02)00053-2. [DOI] [Google Scholar]
- 6.Roobol MJ, Kranse R, Bangma CH, van Leenders AG, Blijenberg BG, van Schaik RH, et al. Screening for prostate cancer: results of the Rotterdam section of the European randomized study of screening for prostate cancer. Eur. Urol. 2013;64:530–539. doi: 10.1016/j.eururo.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 7.Arnold M, Karim-Kos HE, Coebergh JW, Byrnes G, Antilla A, Ferlay J, et al. Recent trends in incidence of five common cancers in 26 European countries since 1988: Analysis of the European Cancer Observatory. Eur. J. cancer. 2015;51:1164–1187. doi: 10.1016/j.ejca.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Center MM, Jemal A, Lortet-Tieulent J, Ward E, Ferlay J, Brawley O, et al. International variation in prostate cancer incidence and mortality rates. Eur. Urol. 2012;61:1079–1092. doi: 10.1016/j.eururo.2012.02.054. [DOI] [PubMed] [Google Scholar]
- 9.Haas GP, Delongchamps N, Brawley OW, Wang CY, de la Roza G. The worldwide epidemiology of prostate cancer: perspectives from autopsy studies. Can. J. Urol. 2008;15:3866–3871. [PMC free article] [PubMed] [Google Scholar]
- 10.Godtman RA, Holmberg E, Khatami A, Stranne J, Hugosson J. Outcome following active surveillance of men with screen-detected prostate cancer. Results from the Goteborg randomised population-based prostate cancer screening trial. Eur. Urol. 2013;63:101–107. doi: 10.1016/j.eururo.2012.08.066. [DOI] [PubMed] [Google Scholar]
- 11.Hayes JH, Ollendorf DA, Pearson SD, Barry MJ, Kantoff PW, Lee PA, et al. Observation versus initial treatment for men with localized, low-risk prostate cancer: a cost-effectiveness analysis. Ann. Intern. Med. 2013;158:853–860. doi: 10.7326/0003-4819-158-12-201306180-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lotan Y. Controlling health care costs for prostate cancer. Eur. Urol. 2013;64:17–18. doi: 10.1016/j.eururo.2012.09.026. [DOI] [PubMed] [Google Scholar]
- 13.van den Bergh RC, Ahmed HU, Bangma CH, Cooperberg MR, Villers A, Parker CC. Novel tools to improve patient selection and monitoring on active surveillance for low-risk prostate cancer: a systematic review. Eur. Urol. 2014;65:1023–1031. doi: 10.1016/j.eururo.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 14.Tosoian JJ, Ross AE, Sokoll LJ, Partin AW, Pavlovich CP. Urinary biomarkers for prostate cancer. Urol. Clin. North Am. 2016;43:17–38. doi: 10.1016/j.ucl.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Hormaechea-Agulla D, Gomez-Gomez E, Ibanez-Costa A, Carrasco-Valiente J, Rivero-Cortes E, LL F, et al. Ghrelin O-acyltransferase (GOAT) enzyme is overexpressed in prostate cancer, and its levels are associated with patient’s metabolic status: Potential value as a non-invasive biomarker. Cancer Lett. 2016;383:125–134. doi: 10.1016/j.canlet.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 16.Frantzi M, van Kessel KE, Zwarthoff EC, Marquez M, Rava M, Malats N, et al. Development and Validation of Urine-based Peptide Biomarker Panels for Detecting Bladder Cancer in a Multi-center Study. Clin. Cancer Res. 2016;22:4077–4086. doi: 10.1158/1078-0432.CCR-15-2715. [DOI] [PubMed] [Google Scholar]
- 17.Frantzi M, Metzger J, Banks RE, Husi H, Klein J, Dakna M, et al. Discovery and validation of urinary biomarkers for detection of renal cell carcinoma. J. Proteom. 2014;98:44–58. doi: 10.1016/j.jprot.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 18.Theodorescu D, Schiffer E, Bauer HW, Douwes F, Eichhorn F, Polley R, et al. Discovery and validation of urinary biomarkers for prostate cancer. Proteom. Clin. Appl. 2008;2:556–570. doi: 10.1002/prca.200780082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez-Gomez E, Carrasco-Valiente J, Blanca-Pedregosa A, Barco-Sanchez B, Fernandez-Rueda JL, Molina-Abril H, et al. European randomized study of screening for prostate cancer risk calculator: external validation, variability, and clinical significance. Urology. 2017;102:85–91. doi: 10.1016/j.urology.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Epstein JI, Allsbrook WC, Jr., Amin MB, Egevad LL, Committee IG. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am. J. Surg. Pathol. 2005;29:1228–1242. doi: 10.1097/01.pas.0000173646.99337.b1. [DOI] [PubMed] [Google Scholar]
- 21.Mischak H, Vlahou A, Ioannidis JP. Technical aspects and inter-laboratory variability in native peptide profiling: the CE-MS experience. Clin. Biochem. 2013;46:432–443. doi: 10.1016/j.clinbiochem.2012.09.025. [DOI] [PubMed] [Google Scholar]
- 22.Wittke S, Fliser D, Haubitz M, Bartel S, Krebs R, Hausadel F, et al. Determination of peptides and proteins in human urine with capillary electrophoresis-mass spectrometry, a suitable tool for the establishment of new diagnostic markers. J. Chromatogr. A. 2003;1013:173–181. doi: 10.1016/S0021-9673(03)00713-1. [DOI] [PubMed] [Google Scholar]
- 23.Kaiser T, Hermann A, Kielstein JT, Wittke S, Bartel S, Krebs R, et al. Capillary electrophoresis coupled to mass spectrometry to establish polypeptide patterns in dialysis fluids. J. Chromatogr. A. 2003;1013:157–171. doi: 10.1016/S0021-9673(03)00712-X. [DOI] [PubMed] [Google Scholar]
- 24.Siwy J, Mullen W, Golovko I, Franke J, Zurbig P. Human urinary peptide database for multiple disease biomarker discovery. Proteom. Clin. Appl. 2011;5:367–374. doi: 10.1002/prca.201000155. [DOI] [PubMed] [Google Scholar]
- 25.Dakna M, Harris K, Kalousis A, Carpentier S, Kolch W, Schanstra JP, et al. Addressing the challenge of defining valid proteomic biomarkers and classifiers. BMC Bioinforma. 2010;11:594. doi: 10.1186/1471-2105-11-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein J, Papadopoulos T, Mischak H, Mullen W. Comparison of CE-MS/MS and LC-MS/MS sequencing demonstrates significant complementarity in natural peptide identification in human urine. Electrophoresis. 2014;35:1060–1064. doi: 10.1002/elps.201300327. [DOI] [PubMed] [Google Scholar]
- 27.UniProt C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45(D1). [DOI] [PMC free article] [PubMed]
- 28.Zurbig P, Renfrow MB, Schiffer E, Novak J, Walden M, Wittke S, et al. Biomarker discovery by CE-MS enables sequence analysis via MS/MS with platform-independent separation. Electrophoresis. 2006;27:2111–2125. doi: 10.1002/elps.200500827. [DOI] [PubMed] [Google Scholar]
- 29.Dobbin KK, Simon RM. Optimally splitting cases for training and testing high dimensional classifiers. BMC Med. Genom. 2011;4:31. doi: 10.1186/1755-8794-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- 31.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med. Decis. Mak. 2006;26:565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Briganti A, Fossati N, Catto JWF, Cornford P, Montorsi F, Mottet N, et al. Active surveillance for low-risk prostate cancer: The European Association of Urology Position in 2018. Eur. Urol. 2018;74:357–368. doi: 10.1016/j.eururo.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 33.Alford AV, Brito JM, Yadav KK, Yadav SS, Tewari AK, Renzulli J. The use of biomarkers in prostate cancer screening and treatment. Rev. Urol. 2017;19:221–234. doi: 10.3909/riu0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Neste L, Hendriks RJ, Dijkstra S, Trooskens G, Cornel EB, Jannink SA, et al. Detection of high-grade prostate cancer using a urinary molecular biomarker-based risk score. Eur. Urol. 2016;70:740–748. doi: 10.1016/j.eururo.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 35.Gronberg H, Adolfsson J, Aly M, Nordstrom T, Wiklund P, Brandberg Y, et al. Prostate cancer screening in men aged 50-69 years (STHLM3): a prospective population-based diagnostic study. Lancet Oncol. 2015;16:1667–1676. doi: 10.1016/S1470-2045(15)00361-7. [DOI] [PubMed] [Google Scholar]
- 36.Kasivisvanathan V, Rannikko AS, Borghi M, Panebianco V, Mynderse LA, Vaarala MH, et al. MRI-targeted or standard biopsy for prostate-cancer diagnosis. New Engl. J. Med. 2018;378:1767–1777. doi: 10.1056/NEJMoa1801993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmed HU, El-Shater Bosaily A, Brown LC, Gabe R, Kaplan R, Parmar MK, et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet. 2017;389:815–822. doi: 10.1016/S0140-6736(16)32401-1. [DOI] [PubMed] [Google Scholar]
- 38.Schiffer E, Bick C, Grizelj B, Pietzker S, Schofer W. Urinary proteome analysis for prostate cancer diagnosis: cost-effective application in routine clinical practice in Germany. Int. J. Urol. 2012;19:118–125. doi: 10.1111/j.1442-2042.2011.02901.x. [DOI] [PubMed] [Google Scholar]
- 39.Gaggar A, Jackson PL, Noerager BD, O’Reilly PJ, McQuaid DB, Rowe SM, et al. A novel proteolytic cascade generates an extracellular matrix-derived chemoattractant in chronic neutrophilic inflammation. J. Immunol. 2008;180:5662–5669. doi: 10.4049/jimmunol.180.8.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takakura S, Kohno T, Shimizu K, Ohwada S, Okamoto A, Yokota J. Somatic mutations and genetic polymorphisms of the PPP1R3 gene in patients with several types of cancers. Oncogene. 2000;19:836–840. doi: 10.1038/sj.onc.1203388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article.