Abstract
Background
Intratumoural T-cell infiltrate intensity cortes wrelaith clinical outcome in stage II/III colorectal cancer (CRC). We aimed to determine whether this association varies across this heterogeneous group.
Methods
We performed a pooled analysis of 1804 CRCs from the QUASAR2 and VICTOR trials. Intratumoural CD8+ and CD3+ densities were quantified by immunohistochemistry in tissue microarray (TMA) cores, and their association with clinical outcome analysed by Cox regression. We validated our results using publicly available gene expression data in a pooled analysis of 1375 CRCs from seven independent series.
Results
In QUASAR2, intratumoural CD8+ was a stronger predictor of CRC recurrence than CD3+ and showed similar discriminative ability to both markers in combination. Pooled multivariable analysis of both trials showed increasing CD8+ density was associated with reduced recurrence risk independent of confounders including DNA mismatch repair deficiency, POLE mutation and chromosomal instability (multivariable hazard ratio [HR] for each two-fold increase = 0.92, 95%CI = 0.87–0.97, P = 3.6 × 10−3). This association was not uniform across risk strata defined by tumour and nodal stage: absent in low-risk (pT3,N0) cases (HR = 1.03, 95%CI = 0.87–1.21, P = 0.75), modest in intermediate-risk (pT4,N0 or pT1-3,N1-2) cases (HR = 0.92, 95%CI = 0.86–1.0, P = 0.046) and strong in high-risk (pT4,N1-2) cases (HR = 0.87, 95%CI = 0.79–0.97, P = 9.4 × 10−3); PINTERACTION = 0.090. Analysis of tumour CD8A expression in the independent validation cohort revealed similar variation in prognostic value across risk strata (PINTERACTION = 0.048).
Conclusions
The prognostic value of intratumoural CD8+ cell infiltration in stage II/III CRC varies across tumour and nodal risk strata.
Subject terms: Colorectal cancer, Tumour biomarkers, Tumour immunology
Background
Colorectal cancer (CRC) is a substantial cause of morbidity and mortality worldwide. More than half of cases are diagnosed at stage II/III, for which management is typically curative-intent resection followed by adjuvant chemotherapy depending on recurrence risk. Unfortunately, current risk stratification—based on factors such as lymph node involvement, pT4 primary or absence of DNA mismatch repair deficiency (MMR-D)1,2—is imprecise, leading to considerable under- and over-treatment.3,4 Of the efforts to improve this, perhaps the most promising involves the quantification of the intratumoural T-cell infiltrate, high density of which is associated with improved clinical outcome in CRC.5–12 Importantly, in several studies, this relationship has been shown to persist after adjustment for MMR-D.9–15 However, these studies have not adjusted for other potential confounders such as chromosomal instability (CIN)—present in more than two thirds of CRCs16 and associated with decreased T-cell infiltrate and poor prognosis17,18—or POLE mutation, which correlates with enhanced immunogenicity and excellent outcome.19 In most cases, they have also not addressed the clinically important question of whether the prognostic value of intratumoural T-cell infiltrate in stage II/III CRC is uniform across pT/N stage-based risk strata, or indeed by molecular factors such as KRAS and BRAF mutation, and finally, they are limited by their use of non-trial, observational series, which are well recognised to suffer a greater risk of bias than meticulously curated clinical trial samples.20
In this study, we sought to address these shortcomings by pooled analysis of two large clinical trials and a validation cohort of seven independent series.
Methods
Patient selection for biomarker study
Details of the QUASAR2 and VICTOR trials have been reported previously.21,22 Briefly, QUASAR2 was an open-label, randomised, controlled trial investigating the addition of bevacizumab to capecitabine in the adjuvant treatment of stage II/III CRC. The final intention to treat population comprised 1941 patients of whom 1715 (88.3%) had colonic tumours and 226 (11.6%) had rectal tumours. The VICTOR trial investigated the efficacy of rofecoxib (a selective cyclooxygenase-2 [COX-2] inhibitor) following completion of standard therapy (surgery ± adjuvant chemotherapy) for stage II–III CRC. The intention to treat analysis included 2434 cases of whom 1592 (65.4%) had colonic tumours and 842 (34.6%) had tumours of the rectum/rectosigmoid junction. Cases from the QUASAR2 and VICTOR trials were identified for inclusion in this biomarker study based on the availability of tumour tissue microarrays (TMAs) and clinical outcome data. Cases treated with preoperative radiotherapy were excluded. All tumours were either stage II or III and had undergone confirmed R0 resection. Data on molecular covariables were not mandated for study inclusion but were available in most cases (see Statistical Analysis). Details of the seven series which formed our pooled validation cohort have either been previously published,23–27 or are freely available from the NCBI Gene Expression Omnibus (GEO).28 These were all non-experimental datasets, selected for their documentation of primary tumour and nodal stage, CD8A expression and clinical outcome.
Tumour molecular analysis and immunohistochemistry
Tissue microarrays were constructed using punches taken from the centre of the tumour in formalin-fixed paraffin-embedded blocks following identification by the study pathologists; the tumour invasive margin was not sampled. Molecular analyses and immunohistochemistry (IHC) for CD8 and CD3 were performed as reported previously (See Supplementary Methods for full details).16,19,21,29,30 Marker positive cells were quantified by computerised analyses using ImmunoPath 1.3.9.0 (Room4, Crowborough, UK), and expressed as the proportion of CD8+ or CD3+ cells in the total number of cell nuclei across all TMA cores for each case. In addition to the analysis of TMA cores, a subset of 51 cases from the QUASAR2 trial also underwent similar analysis of full face tissue sections to permit comparison between of measurements between the two methods, and comparison of marker densities between the tumour centre and its invasive margin. In view of their similar characteristics,19 POLE-mutant and MMR-D tumours were combined for all analyses. Molecular analyses in the validation series have been previously reported.23–26 Expression of CD8A, which encodes the CD8 receptor, was performed by either RNAseq23 or expression arrays.24–26 Gene expression data were log2 transformed, if not already done, and scaled to permit pooling of series.
Statistical analysis
Full details of the statistical methods used in this biomarker study are provided in Supplementary Methods. Analyses were performed and reported in accordance with the REMARK guidelines,31 and are detailed in Table S1. Survival curves were plotted using the Kaplan–Meier method and compared by the log-rank test. Our primary and secondary objectives were the association of CD8+ density, analysed as a continuous variable, with time to CRC recurrence (TTR) (defined as the time from randomisation to CRC relapse, with censoring at last contact or death in case of no recurrence), and overall survival (OS), respectively, in the pooled QUASAR2 and VICTOR cohorts. Exploratory objectives were the association of CD8+ cell density with clinical outcome after dichotomisation, and according to tumour and nodal stage, and other clinically relevant risk factors. These objectives were evaluated by pooled univariable and multivariable Cox proportional hazards models, stratified by trial. CD8+ cell density was log2 transformed prior to inclusion in regression models (see Supplementary Statistical Methods). Missing covariables data were uncommon (maximum 11.6% missing data for any covariable) and were imputed by multiple imputation by chained equations prior to regression (see Statistical Methods for details). For the final multivariable models, we prespecified the inclusion of variables of clinical importance or known prognostic value and those that demonstrated statistically significant association with CD8+ cell density in our prior analysis. The remaining variables were subjected to stepwise backward elimination to remove those which did not contribute to model fit. Details of model diagnostics are provided in the Supplementary Methods. Statistical tests were two-sided, and hypothesis testing was performed at the 5% significance level.
Results
QUASAR2 and VICTOR trial patient details
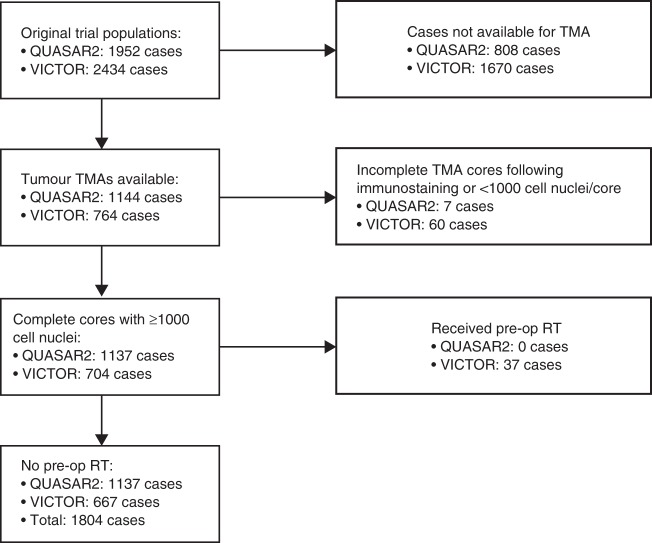
The CONSORT diagram for this biomarker study is shown in Fig. 1, and the baseline demographic characteristics and clinicopathological and molecular variables of the QUASAR2 and VICTOR cases analysed are presented in Table S2. The cases informative for biomarker analyses were similar to the total trial populations in respect of age, sex, disease stage, use of systemic therapy, disease recurrence and death to the total in both cases (Table S3). Given their similarity, we combined both studies for most subsequent analyses.
Fig. 1.
CONSORT diagram for biomarker-evaluable patients. TMA, tissue microarray; Pre-op RT, pre-operative radiotherapy
T-cell infiltration in the QUASAR2 study
Analysis of intratumoural TMA cores (taken from the tumour centre) from the QUASAR2 cohort revealed similar numbers of infiltrating CD8+ and CD3+ cells per mm2 to those reported in previous reports15 (Table S4), and a highly statistically significant correlation between the densities (defined as the proportion of positive cells to total cell nuclei) of both markers (Spearman rho 0.65; P = 2.2 × 10−16). Complementary analysis of whole tissue sections in a subset of 51 cases revealed similar positive correlations between estimates of CD8+ infiltrate in the tumour centre with those from TMA cores (Spearman rho 0.64; P = 3.1 × 10−5), and between CD8+ cell density in the tumour centre and invasive margin (Spearman rho 0.73, P = 1.3 × 10−9). In light of these findings, we proceeded to use the results obtained from analysis of TMA cores to examine the association of CD8+ and CD3+ cell density, alone and in combination, with time to CRC recurrence (TTR) in this trial population, given the known prognostic value of these markers in CRC.7,14 This revealed that intratumoural CD8, but not CD3, was significantly associated with TTR in univariable analysis, and in a model that included both markers (Table S4). As the CD8-only model also had similar discrimination ability and smaller Akaike information criterion (AIC—an estimator of model quality, which balances goodness of fit and parsimony), compared with the bivariate model (Table S5), we focused our subsequent analyses on CD8 infiltrate alone.
CD8+ cell density in the combined QUASAR2 and VICTOR cohorts
Unadjusted analyses of the combined trials demonstrated that CD8+ cell density was significantly associated with stage II versus III disease, tumour right-sidedness, MMR-D or POLE mutation, lack of chromosomal instability (CIN), BRAF mutation and absence of disease recurrence and death, but not age, sex, pT stage or KRAS mutation (Table S6). Similar associations were evident in the individual trials with the exception of BRAF mutation and CIN status, which were not statistically significant in VICTOR perhaps owing to its smaller size.
CD8+ cell density and clinical outcome in the pooled QUASAR2 and VICTOR cohorts
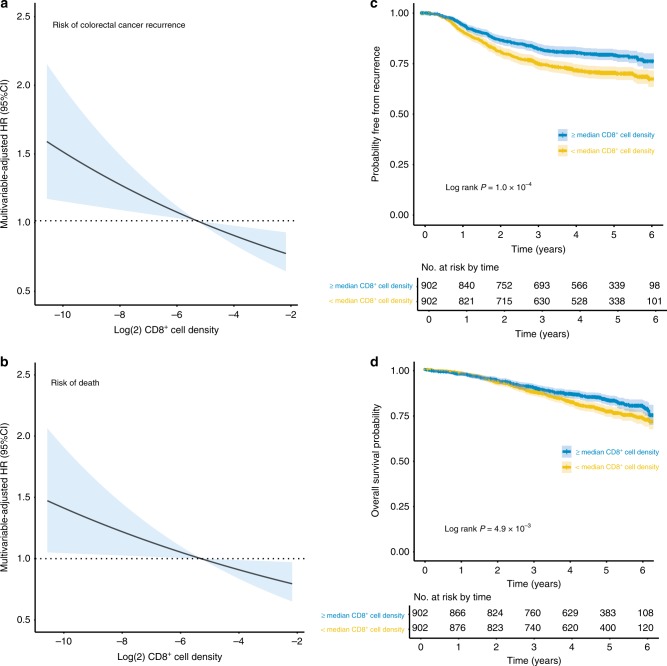
We next determined the relationship between tumour CD8+ density (analysed as a log2 transformed, continuous variable) and CRC recurrence in the pooled trials. Increasing CD8+ density was associated with significantly longer TTR in univariable analysis (HR = 0.90 for each two-fold increase; 95% CI = 0.85–0.95, P = 1.7 × 10−4) (Table 1). While the prognostic factors MMR-D/POLE mutation and CIN were also significantly associated with TTR in univariable analyses, only CD8+ density remained prognostic in multivariable analyses (HR = 0.92; 95% CI = 0.87–0.97, P = 3.6 × 10−3) (Table 2, S7, Fig. 2a). Corresponding analysis of overall survival (OS) revealed that CD8+ density, but not MMR-D/POLE mutation or CIN, was associated with significantly reduced mortality (multivariable-adjusted HR = 0·93; 95% CI = 0·87–0·99, P = 0·024) (Table 1, Fig. 2b). For comparison with previous reports, we divided cases into CD8+-high and CD8+-low categories at the sample median, noting that this resulted in comparable proportions to studies that have used a cut point defined by its relationship with clinical outcome.8,13 Multivariable analysis of TTR and OS confirmed better outcome for CD8+-high tumours (HR = 0.71, 95% CI = 0.59–0.87, P = 7.2 × 10−4, and HR = 0.71; 95% CI = 0.57–0.88, P = 1.6 × 10−3) (Fig. 2c, d). These effect sizes were similar to those reported for a similar recent analysis of 600 cases from the North Central Cancer Treatment Group (NCCTG) N1047 trial32
Table 1.
Univariable and multivariable analyses of time to colorectal cancer recurrence and overall survival in pooled VICTOR and QUASAR2 trial population
| No. | TTR events | OS events | Univariable analysis | Multivariable analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Time to recurrence | Overall survival | Time to recurrence | Overall survival | ||||||||
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | ||||
| Age (continuous) | 1804 | 435 | 350 | 1.01 (1.00–1.02) | 0.10 | 1.03 (1.02–1.04) | 6.2 × 10–6 | 1.00 (0.99–1.01) | 0.43 | 1.02 (1.01–1.04 | 1.0 × 10−4 |
| Sex | |||||||||||
| Male | 1083 | 281 | 222 | 1.0 (ref) | – | 1.0 (ref) | – | 1.0 (ref) | – | 1.0 (ref) | – |
| Female | 721 | 154 | 128 | 0.83 (0.68–1.01) | 0.063 | 0.89 (0.72–1.11) | 0.31 | 0.83 (0.68–1.02) | 0.075 | 0.89 (0.71–1.11) | 0.30 |
| Location | |||||||||||
| Left | 972 | 167 | 172 | 1.0 (ref) | – | 1.0 (ref) | – | 1.0 (ref) | – | 1.0 (ref) | – |
| Right | 730 | 167 | 158 | 1.08(0.93–1.31) | 0.45 | 0.77 (0.62–0.96) | 0.019 | 1.05 (0.85–1.31) | 0.63 | 0.83 (0.65–1.06) | 0.13 |
| Stage | |||||||||||
| II | 708 | 115 | 91 | 1.0 (ref) | – | 1.0 (ref) | – | 1.0 (ref) | – | 1.0 (ref) | – |
| III | 1096 | 320 | 259 | 1.89 (1.52–2.40) | 6.4 × 10−9 | 1.85 (1.45–2.35) | 5.6 × 10−7 | 1.96 (1.57–2.44) | 1.34 × 10−9 | 2.03 (1.59–2.60) | 1.3 × 10−8 |
| Primary tumour | |||||||||||
| pT1-3 | 1239 | 248 | 186 | 1.0 (ref) | – | 1.0 (ref) | – | 1.0 (ref) | – | 1.0 (ref) | – |
| pT4 | 552 | 191 | 158 | 1.96 (1.61–2.37) | 1.0 × 10−11 | 2.12 (1.70–2.61) | 1.1 × 10−11 | 2.14 (1.76–2.60) | 3.0 × 10−14 | 2.20 (1.77–2.74) | 1.3 × 10−12 |
| BRAF mutation | |||||||||||
| Wild-type | 1412 | 325 | 250 | 1.0 (ref) | |||||||
| Mutant | 194 | 55 | 52 | 1.33 (1.00–1.78) | 0.048 | 1.68 (1.25–2.27) | 6.5 × 10−4 | 1.59 (1.18 - 2.17) | 2.7 × 10−3 | 1.55 (1.12–2.16) | 8.0 × 10−3 |
| MMR & POLE status | |||||||||||
| MMR-P & POLE wild-type | 1412 | 357 | 272 | 1.0 (ref) | – | 1.0 (ref) | – | 1.0 (ref) | – | 1.0 (ref) | – |
| MMR-D or POLE-mutant | 230 | 40 | 42 | 0.68 (0.49–0.94) | 0.022 | 1.00 (0.73–1.39) | 0.98 | 0.72 (0.50–1.05) | 0.090 | 0.96 (0.65–1.41) | 0.85 |
| Chromosomal instability | |||||||||||
| CIN low | 550 | 110 | 88 | 1.0 (ref) | – | 1.0 (ref) | – | 1.0 (ref) | – | 1.0 (ref) | – |
| CIN high | 1049 | 278 | 214 | 1.37 (1.10–1.71) | 0.0051 | 1.27 (0.99–1.62) | 0.063 | 1.17 (0.93–1.49) | 0.18 | 1.21 (0.94–1.58) | 0.14 |
| Bevacizumab treatment | |||||||||||
| No | 1222 | 279 | 223 | 1.0 (ref) | – | 1.0 (ref) | – | 1.0 (ref) | – | 1.0 (ref) | – |
| Yes | 582 | 156 | 127 | 1.31 (1.03–1.67) | 0.027 | 1.25 (0.96–1.63) | 0.095 | 1.28 (1.00–1.62) | 0.047 | 1.23 (0.95–1.60) | 0.12 |
| CD8+ cell density (continuous, log2 transformed) | 1804 | 435 | 350 | 0.90 (0.85–0.95) | 1.7 × 10−4 | 0.91 (0.86–0.98) | 3.2 × 10−3 | 0.92 (0.87–0.97) | 3.6 × 10−3 | 0.93 (0.87–0.99) | 0.024 |
Univariable hazard ratios are derived from complete case analyses. Multivariable-adjusted hazard ratios are adjusted for all other covariables listed, and represent estimates derived from ‘final’ Cox models following stepwise backward elimination of candidate variables that did not contribute to model fit using the likelihood ratio test (NB: forced entry variables and variables significantly associated with CD8+ cell density were not subjected to variable selection). Results from ‘full’ Cox models including all candidate predictors (age, sex, tumour location, disease stage, primary tumour stage, BRAF mutation, KRAS mutation, MMR-D/POLE mutation, CIN, adjuvant chemotherapy, adjuvant bevacizumab and adjuvant rofecoxib), both before and after the addition of CD8+ cell density are provided in Table S5. The addition of CD8+ cell density to the model containing all other covariables was associated with an improvement in model fit in both the ‘final’ Cox model above (Akaike Information Criterion [AIC] = 5634.0 vs. AIC 5640.6; Likelihood ratio test for comparison of nested models: P = 3.6 × 10−3), and the initial, ‘full’ Cox model (Table S3). TTR time to colorectal cancer recurrence, OS overall survival, HR hazard ratio, 95% CI 95% confidence interval, pT pathological tumour (T) stage, MMR DNA mismatch repair, MMR-P mismatch repair proficient, MMR-D mismatch repair deficient, POLE-mutant pathogenic POLE exonuclease domain mutation
Table 2.
Colorectal cancer recurrence and overall survival according to tumour risk strata and CD8+ cell density in pooled VICTOR and QUASAR2 trial population
| No. | No. events | Predicted proportion event free at 3 years | Univariable analysis | Multivariable analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 25th centile CD8+ cell density (95% CI) | 75th centile CD8+ cell density (95% CI) | HR (95% CI) | P | P INTERACTION | HR (95% CI) | P | P INTERACTION | |||
| Time to recurrence | ||||||||||
| Low risk (pT3, N0) | 453 | 60 | 0.90 (0.88 –0.93) | 0.90 (0.88–0.93) | 1.02 (0.86–1.20) | 0.85 | 0.072 | 1.03 (0.87–1.21) | 0.75 | 0.090 |
| Intermediate risk (pT4, N0 or pT1-3, N1/2) | 1035 | 242 | 0.79 (0.76–0.82) | 0.82 (0.80–0.85) | 0.91 (0.85–0.98) | 0.016 | 0.92 (0.86–1.0) | 0.046 | ||
| High risk (pT4, N1/2) | 303 | 132 | 0.58 (0.53–0.64) | 0.69 (0.63–0.75) | 0.86 (0.78–0.96) | 4.7 × 10−3 | 0.87 (0.79–0.97) | 9.4 × 10−3 | ||
| Overall survival | ||||||||||
| Low risk (pT3, N0) | 453 | 52 | 0.95 (0.94–0.97) | 0.95 (0.93–0.96) | 1.03 (0.87–1.23) | 0.72 | 0.051 | 1.04 (0.87–1.24) | 0.69 | 0.056 |
| Intermediate risk (pT4, N0 or pT1-3, N1/2) | 1035 | 177 | 0.89 (0.87–0.91) | 0.90 (0.88–0.92) | 0.94 (0.86–1.03) | 0.20 | 0.94 (0.86–1.03) | 0.22 | ||
| High risk (pT4, N1/2) | 303 | 120 | 0.75 (0.71–0.80) | 0.82 (0.78–0.86) | 0.88 (0.79–0.97) | 0.017 | 0.88 (0.79–0.98) | 0.022 | ||
Point estimates of probability of colorectal cancer recurrence and overall survival are derived from univariable Cox regression of CD8+ cell density as a continuous variable (corresponding estimates by the Kaplan–Meier estimator for cases dichotomised at the median CD8+ cell density are shown in Table S6). Both point estimates and univariable hazard ratios are derived from complete case analyses. Multivariable-adjusted hazard ratios are adjusted for age, sex, tumour location, BRAF mutation, MMR-D/POLE mutation, CIN and adjuvant bevacizumab. Tests for interaction are from the cross product term of tumour risk stratum and log2 CD8+ cell density in bi-variable and multivariable models
HR hazard ratio, 95% CI 95% confidence interval, pT pathological tumour (T) stage
Fig. 2.
Relationship between tumour CD8+ cell density, colorectal cancer recurrence, and overall survival in pooled QUASAR2 and VICTOR studies. Multivariable-adjusted hazard ratios for a colorectal cancer recurrence and b overall survival according to tumour (log2 transformed) CD8+ cell density (i.e. each unit change represents a two-fold increase or decrease in CD8+ cell density). Hazard ratio is shown relative to the recurrence risk of tumours with CD8+ cell density at the median of the pooled study populations. Corresponding Kaplan–Meier plots for colorectal cancer recurrence (c) and overall survival (d) for tumours with high and low CD8+ cell density using a cut point at the sample median. Shaded area in panels indicates 95% confidence interval (95% CI) for estimate of hazard ratio/survival function. P values in c, d were obtained by the Log-rank test
Prognostic value of intratumoural CD8+ cell density by tumour and molecular risk strata
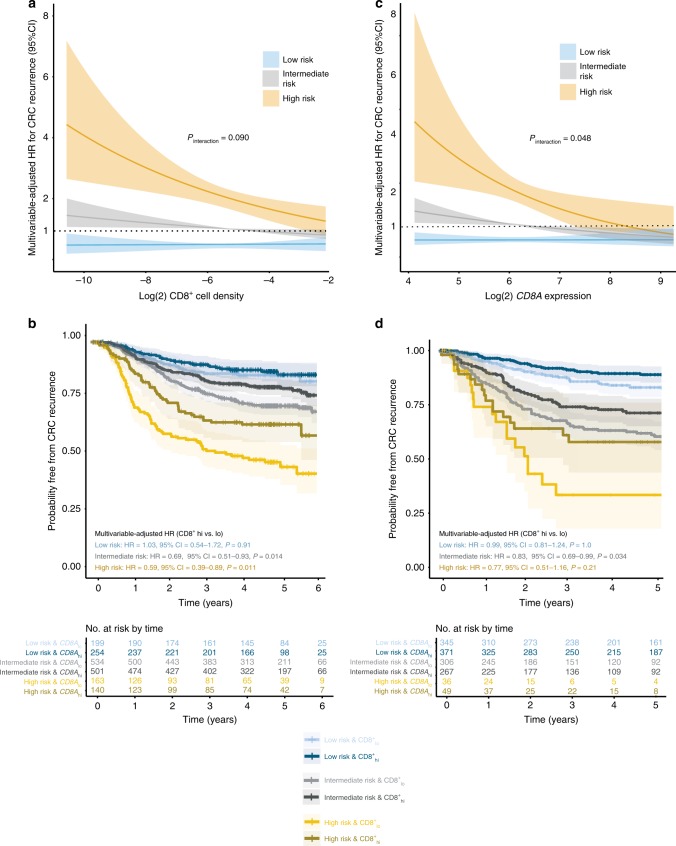
We next explored whether the prognostic value of CD8+ cell infiltrate varied by clinically relevant clinicopathological and molecular factors in our trial cohorts. CD8+ cell density was significantly associated with recurrence irrespective of patient age (<70 vs. ≥70 years), sex, tumour sidedness, KRAS and BRAF mutation, and while the correlation was not statistically significant in MMR-D/POLE-mutant or CIN-low subgroups, the hazard ratios were similar to those in MMR-P/POLE-wild-type and CIN-high subgroups, respectively. In contrast, corresponding univariable and confirmatory multivariable analysis suggested this association (between CD8+ cell infiltrate and TTR) differed between pT1-3 vs. pT4 tumours (multivariable-adjusted HR = 0.96, 95% CI = 0.89–1.04, P = 0.35 vs. HR = 0.87, 95% CI = 0.80–0.95, P = 1.9 × 10−3), and to a lesser extent, between node negative and node positive tumours (HR = 0.94, 95% CI = 0.84–1.06, P = 0.31; and HR = 0.91, 95% CI = 0.85–0.97, P = 5.0 × 10−3, respectively). As pT4 primary and node positivity portended similarly reduced TTR in multivariable analysis (Table 1), and pT4,N0 and pT1-3,N1/2 tumours had very similar outcome in the pooled trials (Fig. S1A), we grouped tumours into low (pT1-3, N0), intermediate (pT4, N0 or pT1-3, N1 or N2) and high (pT4, N2) risk groups and examined the correlation of CD8+ cell density with TTR across these strata. This suggested an apparent variation in its association, from essentially absent in low-risk cases (HR = 1.03, 95% CI = 0.87–1.21, P = 0.75), to modest in intermediate-risk cases (HR = 0.92, 95% CI = 0.86–1.00, P = 0.046) and strong in high-risk cases (HR = 0.87, 95% CI = 0.79–0.97, P = 9.4 × 10−3), although formal testing for an interaction was not statistically significant (PINTERACTION = 0.090) (Table 2, Fig. 3a). The apparent discordance translated into even greater variation between strata in the absolute risk of recurrence of tumours with sparse and dense CD8+ cell infiltrate. For example, in the low-risk group, 3 year recurrence-free probabilities for tumours with CD8+ cell density at the 25th and 75th centiles were similar at 90.2% (95% CI 87.8–92.7%) and 90.1% (95% CI 87.7–92.71%), respectively, while the corresponding proportions in the high-risk group were 58.3% (95% CI = 52.8–64.4%) and 68.6% (95% CI = 63.1–74.6%) (Table 2, Fig. S2). The variation between risk strata was also evident, albeit less obvious, when tumours were classified into CD8+ high and low groups based on the sample median, as defined above (Fig. 3b, Table S8). Analysis of overall survival revealed a similar tendency to differences in outcome by CD8+ cell density between strata (PINTERACTION = 0.056), (Table 2).
Fig. 3.
Relationship between tumour CD8+ cell density and colorectal cancer recurrence according to primary tumour and nodal status in the pooled QUASAR2 and VICTOR studies, and in the validation cohort. a Multivariable-adjusted estimates of the risk of colorectal cancer recurrence in pooled QUASAR2 and VICTOR studies according to tumour (log2 transformed) CD8+ cell density for low-risk (pT1-3, N0), intermediate-risk (pT4, N0 and pT1-3, N1/2) and high-risk (pT4, N1/2) tumours (i.e. each unit change represents a two-fold increase or decrease in CD8+ cell density). b Kaplan–Meier curves showing probability of colorectal cancer recurrence in the QUASAR2 and VICTOR studies for CD8+ high and CD8+ low tumours (divided at the pooled cohort median) across risk strata. c Multivariable-adjusted estimates of the risk of colorectal cancer recurrence according to tumour CD8A expression in the pooled validation series for low-risk (pT1-3, N0), intermediate-risk (pT4, N0 and pT1-3, N1/2) and high-risk (pT4, N1/2) tumours. CD8A expression values within series were transformed to have mean of zero and unit standard deviation before pooling, and were re-scaled to the mean and standard deviation within the largest series (GSE39582) for ease of interpretation. d Kaplan–Meier curves showing probability of colorectal cancer recurrence in the pooled validation series between CD8A high and CD8A low tumours (divided at the pooled cohort median) across risk strata. P value in a, c indicates results of test for interaction between tumour risk stratum and log2 CD8A expression in multivariable Cox model. Shaded areas in a, b, c, d indicate 95% confidence intervals
External validation of results
As the analysis across risk strata was not prespecified, we sought to validate our findings in an independent cohort. Although no publicly available datasets have quantified CD8 infiltrate by IHC, we identified five series with gene expression data including CD8A (which encodes the CD8 alpha chain) in addition to The Cancer Genome Atlas (TCGA) series and a further set of 264 tumours with CD8A expression data. In total, our validation set included 1375 stage II/III tumours with details of tumour and nodal staging, MMR and CIN status and clinical outcome data (Fig. S1B). In keeping with previous studies, tumour CD8A expression was significantly associated with TTR in multivariable analysis (HR for each 2-fold increase = 0.86, 95% CI = 0.76–0.97, P = 0.018). Interestingly, and concordant with our previous results, the strength of this association varied between low, intermediate and high-risk pT/N strata (HR = 0.99, 95% CI = 0.81–1.24, P = 1.0 vs. HR = 0.83, 95% CI = 0.69–0.99, P = 0.034 vs. HR = 0.77, 95% CI = 0.51–1.16, P = 0.21, respectively; PINTERACTION = 0.048) (Fig. 3c, d, Tables S9, S10).
Discussion
In this analysis of 1804 stage II/III CRCs from two clinical trials with external validation, we have shown that while the association of intratumoural CD8+ cell density with recurrence is independent of MMR-D, POLE mutation and CIN, it appears to vary by primary tumour and nodal status, from absent or minimal in pT3,N0 disease, to strong in pT4,N1/N2 disease. This variation could not easily be explained by fewer events in the low-risk groups, as the hazard ratios in these approximated unities in both the primary and the validation cohorts. Our study both strengthens the evidence for the prognostic value of tumour CD8+ infiltrate in CRC, and suggests that the clinical implementation of this novel marker will require careful consideration. Indeed, among stage pT3, N0 tumours—where MMR status often guides decisions on adjuvant chemotherapy1—we found no evidence that CD8+ cell density was independently prognostic, suggesting that further work is required to identify immune biomarkers of clinical value in this subgroup. In contrast, patients with pT4,N1/2, cancers with low sparse intratumoural CD8+ infiltrate had dismal prognosis, raising the possibility of trials testing intensified or novel adjuvant therapies in this subgroup. Importantly, the prognostic value of CD8+ cell density did not vary by other clinically relevant factors, including sidedness, MMR-D, KRAS and BRAF mutation, suggesting that it may complement these in risk stratification.
Arguably the best-known test for evaluation of the anti-tumour immune response is the “Immunoscore”, developed by Galon et al.15,33 While the immune markers and cut points used to define categories have evolved during its development, it currently classifies tumours based on CD8+ and CD3+ cell density in the tumour centre and its invasive margin. The strength of correlation between these markers, and the superior prognostication provided by CD8+ density in our preliminary analysis caused us to focus on this marker alone. Though the different methodologies preclude direct comparison, it is notable that our dichotomised analysis revealed a hazard ratio for recurrence of stage III disease broadly similar to that recently reported for Immunoscore in cases from the NCCTG N0147 trial.32 However, avoiding classification of continuous variables has several potential advantages in this setting, as may provide more refined prognostication, and avoids the pitfall of assigning substantially different prognoses to tumours with marker values falling narrowly either side of a cut point.34 Interestingly, the variation in the prognostic value of tumour CD8 infiltrate by tumour risk strata was much less obvious when analysed as a dichotomiaed variable.
Our study has limitations. For logistical reasons, our analysis used tumour cores rather than whole sections, and focusesd predominantly on CD8+cells, as these were more strongly prognostic than CD3+cells in our initial analysis. It will therefore be important to determine the impact of evaluation of a larger area, including the tumour invasive margin, as well as the impact of adding additional immune markers, including and beyond CD3, on prognostication; particularly given the recent results reported for Immunoscore in this setting.15 Emerging multispectral platforms are of particular relevance to the latter, though these are not yet ready for clinical implementation. Other priorities will be to define the value of intraepithelial and intrastromal CD8+ cell density, which our software did not discriminate between, and the impact of intratumoural heterogeneity on prognosis. Finally, because most patients in these trials were treated with adjuvant cytotoxic chemotherapy, though not oxaliplatin, we were not able to determine whether intratumoural CD8+ density predicts recurrence risk in the absence of such treatment, or under oxaliplatin-based therapy.
In summary, in our large study with independent validation, we have confirmed that intratumoural CD8+ cell density is independent prognostic factor in stage II/III CRC, and shown that this association appears to vary by tumour and nodal risk strata. Confirmation of this finding, and defining the underlying mechanisms, will be important questions for future studies, as will be the investigation of whether CD8+ density as can be used to identify patients who could benefit from adjuvant immunotherapy.
Supplementary information
Author contributions
M.G., I.T. and D.N.C. designed the study. M.G., E.D., A.S., J.B., A.N., M.N., K.L., D.O., W.K., H.D., R.K., D.K. and R.A.L. collected the data. M.G., A.S., I.T., R.A.L. and D.N.C. interpreted the data. M.G., G.N. and D.N.C. did the data analysis. M.G., I.T. and D.N.C. wrote the paper.
Competing interests
M.N. is a director of Room 4 who own the rights to immunopath, an image analysis program used in this study to assess T-cell counts. D.K. is an employee of, and owns stock in Oxford Cancer Biomarkers, and has received research grants from Roche, and Merck and personal fees from Bayer, Incyte, Medscape, Oxford Cancer Biomarkers, OxOnc, and ASCO. The remaining authors declared no competing interests.
Ethics approval and consent to participate
Ethical approval for patient recruitment and sample collection in the VICTOR and QUASAR2 trials was approved centrally and at all recruiting centres. Ethical approval for anonymised tumour molecular analysis was granted by Oxfordshire Research Ethics Committee B (Approval No 05/Q1605/66). Approval from the Regional South Eastern Health Authorities of Norway for including molecular and clinical data of the Oslo series in international collaborative studies (1.2005.1629; 2010/1805)
Funding
Funding for this study was provided by: Cancer Research UK (C6199/A10417 and C399/A2291), the European Union Seventh Framework Programme (FP7/207- 2013) grant 258236 collaborative project SYSCOL, European Research Council project EVOCAN, the Oxford Cancer Centre, the Oxford NIHR Comprehensive Biomedical Research Centre (BRC), and core funding to the Wellcome Trust Centre for Human Genetics from the Wellcome Trust (090532/Z/09/Z). M.G. is funded by a Wellcome Trust Clinical Training Fellowship. D.N.C. is funded by a Health Foundation/Academy of Medical Sciences Clinician Scientist Fellowship. A.S. is funded by a Norwegian Cancer Society Scientist Fellowship (no. 6824048-2016). R.A.L. is supported by the Research Council of Norway (no. 250993) and the Norwegian Cancer Society (no. 182759-2016). M.N. and D.O. were supported by Cancer Research UK and the National Institute for Health Research through the UCL Experimental Cancer Medicine Centre (to D.O.) and UCL Hospitals Biomedical Research Centre (to M.N.). The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, the Department of Health or the Wellcome Trust. The costs of open access publishing were funded by The Wellcome Trust (090532/Z/09/Z). The study funders had no role in the design, analysis, interpretation, or manuscript preparation for this biomarker study. D.N.C. had full access to all study data and had final responsibility for submission of this report.
Data availability
The datasets pertaining to VICTOR and QUASAR2 used during the current study are available from the corresponding author on reasonable request. The datasets used to validate our findings are available from the GEO database and The Cancer Genome Atlas (TCGA) (https://www.ncbi.nlm.nih.gov/geo/; http://firebrowse.org/).
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Mark A. Glaire, Enric Domingo
Change history
9/23/2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41416-019-0540-4.
References
- 1.Labianca R, Nordlinger B, Beretta GD, Mosconi S, Mandala M, Cervantes A, et al. Early colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2013;24(Suppl 6):vi64–vi72. doi: 10.1093/annonc/mdt354. [DOI] [PubMed] [Google Scholar]
- 2.Dienstmann R, Mason MJ, Sinicrope FA, Phipps AI, Tejpar S, Nesbakken A, et al. Prediction of overall survival in stage II and III colon cancer beyond TNM system: a retrospective, pooled biomarker study. Ann. Oncol. 2017;28:1023–1031. doi: 10.1093/annonc/mdx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Land SR, Kopec JA, Cecchini RS, Ganz PA, Wieand HS, Colangelo LH, et al. Neurotoxicity from oxaliplatin combined with weekly bolus fluorouracil and leucovorin as surgical adjuvant chemotherapy for stage II and III colon cancer: NSABP C-07. J. Clin. Oncol. 2007;25:2205–2211. doi: 10.1200/JCO.2006.08.6652. [DOI] [PubMed] [Google Scholar]
- 4.Rosmarin Dan, Palles Claire, Church David, Domingo Enric, Jones Angela, Johnstone Elaine, Wang Haitao, Love Sharon, Julier Patrick, Scudder Claire, Nicholson George, Gonzalez-Neira Anna, Martin Miguel, Sargent Daniel, Green Erin, McLeod Howard, Zanger Ulrich M., Schwab Matthias, Braun Michael, Seymour Matthew, Thompson Lindsay, Lacas Benjamin, Boige Valérie, Ribelles Nuria, Afzal Shoaib, Enghusen Henrik, Jensen Søren Astrup, Etienne-Grimaldi Marie-Christine, Milano Gérard, Wadelius Mia, Glimelius Bengt, Garmo Hans, Gusella Milena, Lecomte Thierry, Laurent-Puig Pierre, Martinez-Balibrea Eva, Sharma Rohini, Garcia-Foncillas Jesus, Kleibl Zdenek, Morel Alain, Pignon Jean-Pierre, Midgley Rachel, Kerr David, Tomlinson Ian. Genetic Markers of Toxicity From Capecitabine and Other Fluorouracil-Based Regimens: Investigation in the QUASAR2 Study, Systematic Review, and Meta-Analysis. Journal of Clinical Oncology. 2014;32(10):1031–1039. doi: 10.1200/JCO.2013.51.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(September):1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 6.Pagès F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N. Engl. J. Med. 2005;353:2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 7.Marisa Laetitia, Svrcek Magali, Collura Ada, Becht Etienne, Cervera Pascale, Wanherdrick Kristell, Buhard Olivier, Goloudina Anastasia, Jonchère Vincent, Selves Janick, Milano Gerard, Guenot Dominique, Cohen Romain, Colas Chrystelle, Laurent-Puig Pierre, Olschwang Sylviane, Lefèvre Jérémie H, Parc Yann, Boige Valérie, Lepage Côme, André Thierry, Fléjou Jean-François, Dérangère Valentin, Ghiringhelli François, de Reynies Aurélien, Duval Alex. The Balance Between Cytotoxic T-cell Lymphocytes and Immune Checkpoint Expression in the Prognosis of Colon Tumors. JNCI: Journal of the National Cancer Institute. 2017;110(1):68–77. doi: 10.1093/jnci/djx136. [DOI] [PubMed] [Google Scholar]
- 8.Galon J, Mlecnik B, Marliot F, Ou F-S, Bifulco CB, Lugli A, et al. Validation of the Immunoscore (IM) as a prognostic marker in stage I/II/III colon cancer: Results of a worldwide consortium-based analysis of 1,336 patients. J. Clin. Oncol. 2016;34:3500–3500. doi: 10.1200/JCO.2016.34.15_suppl.3500. [DOI] [Google Scholar]
- 9.Eriksen AC, Sorensen FB, Lindebjerg J, Hager H, dePont Christensen R, Kjaer-Frifeldt S, et al. The Prognostic value of tumor-infiltrating lymphocytes in stage II colon cancer. A nationwide population-based study. Transl. Oncol. 2018;11:979–987. doi: 10.1016/j.tranon.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berntsson J, Svensson MC, Leandersson K, Nodin B, Micke P, Larsson AH, et al. The clinical impact of tumour-infiltrating lymphocytes in colorectal cancer differs by anatomical subsite: a cohort study. Int. J. Cancer. 2017;141:1654–1666. doi: 10.1002/ijc.30869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daster S, Eppenberger-Castori S, Hirt C, Soysal SD, Delko T, Nebiker CA, et al. Absence of myeloperoxidase and CD8 positive cells in colorectal cancer infiltrates identifies patients with severe prognosis. Oncoimmunology. 2015;4:e1050574. doi: 10.1080/2162402X.2015.1050574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deschoolmeester V, Baay M, Van Marck E, Weyler J, Vermeulen P, Lardon F, et al. Tumor infiltrating lymphocytes: an intriguing player in the survival of colorectal cancer patients. BMC Immunol. 2010;11:19. doi: 10.1186/1471-2172-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity. 2016;44:698–711. doi: 10.1016/j.immuni.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 14.Wirta EV, Seppala T, Friman M, Vayrynen J, Ahtiainen M, Kautiainen H, et al. Immunoscore in mismatch repair-proficient and -deficient colon cancer. J. Pathol. Clin. Res. 2017;3:203–213. doi: 10.1002/cjp2.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pages F, Mlecnik B, Marliot F, Bindea G, Ou FS, Bifulco C, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391:2128–2139. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 16.Mouradov D, Domingo E, Gibbs P, Jorissen RN, Li S, Soo PY, et al. Survival in stage II/III colorectal cancer is independently predicted by chromosomal and microsatellite instability, but not by specific driver mutations. Am. J. Gastroenterol. 2013;108:1785–1793. doi: 10.1038/ajg.2013.292. [DOI] [PubMed] [Google Scholar]
- 17.Hveem TS, Merok MA, Pretorius ME, Novelli M, Baevre MS, Sjo OH, et al. Prognostic impact of genomic instability in colorectal cancer. Br. J. Cancer. 2014;110:2159–2164. doi: 10.1038/bjc.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davoli Teresa, Uno Hajime, Wooten Eric C., Elledge Stephen J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355(6322):eaaf8399. doi: 10.1126/science.aaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domingo E, Freeman-Mills L, Rayner E, Glaire M, Briggs S, Vermeulen L, et al. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: a retrospective, pooled biomarker study. Lancet Gastroenterol. Hepatol. 2016;1:207–216. doi: 10.1016/S2468-1253(16)30014-0. [DOI] [PubMed] [Google Scholar]
- 20.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J. Natl Cancer Inst. 2009;101:1446–1452. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerr RS, Love S, Segelov E, Johnstone E, Falcon B, Hewett P, et al. Adjuvant capecitabine plus bevacizumab versus capecitabine alone in patients with colorectal cancer (QUASAR 2): an open-label, randomised phase 3 trial. Lancet Oncol. 2016;17:1543–1557. doi: 10.1016/S1470-2045(16)30172-3. [DOI] [PubMed] [Google Scholar]
- 22.Midgley RS, McConkey CC, Johnstone EC, Dunn JA, Smith JL, Grumett SA, et al. Phase III randomized trial assessing rofecoxib in the adjuvant setting of colorectal cancer: final results of the VICTOR trial. J. Clin. Oncol. 2010;28:4575–4580. doi: 10.1200/JCO.2010.29.6244. [DOI] [PubMed] [Google Scholar]
- 23.Network TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staub Eike, Groene Joern, Heinze Maya, Mennerich Detlev, Roepcke Stefan, Klaman Irina, Hinzmann Bernd, Castanos-Velez Esmeralda, Pilarsky Christian, Mann Benno, Brümmendorf Thomas, Weber Birgit, Buhr Heinz-Johannes, Rosenthal André. An expression module of WIPF1-coexpressed genes identifies patients with favorable prognosis in three tumor types. Journal of Molecular Medicine. 2009;87(6):633–644. doi: 10.1007/s00109-009-0467-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, Barany F, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc. Natl Acad. Sci. USA. 2009;106:7131–7136. doi: 10.1073/pnas.0902232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sveen A, Bruun J, Eide PW, Eilertsen IA, Ramirez L, Murumagi A, et al. Colorectal cancer consensus molecular subtypes translated to preclinical models uncover potentially targetable cancer cell dependencies. Clin. Cancer Res. 2018;24:794–806. doi: 10.1158/1078-0432.CCR-17-1234. [DOI] [PubMed] [Google Scholar]
- 28.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Domingo E, Church DN, Sieber O, Ramamoorthy R, Yanagisawa Y, Johnstone E, et al. Evaluation of PIK3CA mutation as a predictor of benefit from nonsteroidal anti-inflammatory drug therapy in colorectal cancer. J. Clin. Oncol. 2013;31:4297–4305. doi: 10.1200/JCO.2013.50.0322. [DOI] [PubMed] [Google Scholar]
- 30.Domingo E, Ramamoorthy R, Oukrif D, Rosmarin D, Presz M, Wang H, et al. Use of multivariate analysis to suggest a new molecular classification of colorectal cancer. J. Pathol. 2013;229:441–448. doi: 10.1002/path.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumour MARKer prognostic studies (REMARK) Br. J. Cancer. 2005;93:387–391. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinicrope FA, Shi Q, Hermitte F, Heying EN, Benson AB, Gill S, et al. Association of immune markers and Immunoscore with survival of stage III colon carcinoma (CC) patients (pts) treated with adjuvant FOLFOX: NCCTG N0147 (Alliance) J. Clin. Oncol. 2017;35:3579–3579. doi: 10.1200/JCO.2017.35.15_suppl.3579. [DOI] [Google Scholar]
- 33.Galon J, Mlecnik B, Bindea G, Angell HK, Berger A, Lagorce C, et al. Towards the introduction of the ‘Immunoscore’ in the classification of malignant tumours. J. Pathol. 2014;232:199–209. doi: 10.1002/path.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altman DG, Royston P. The cost of dichotomising continuous variables. BMJ. 2006;332:1080. doi: 10.1136/bmj.332.7549.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets pertaining to VICTOR and QUASAR2 used during the current study are available from the corresponding author on reasonable request. The datasets used to validate our findings are available from the GEO database and The Cancer Genome Atlas (TCGA) (https://www.ncbi.nlm.nih.gov/geo/; http://firebrowse.org/).