Abstract
Macrophages (Mϕ) are an important component of the innate immune system; they play critical roles in the first line of defense to pathogen invasion and modulate adaptive immunity. MicroRNAs (miRNAs) are a newly recognized, important level of gene expression regulation. However, their roles in the regulation of Mϕ and the immune system are still not fully understood. In this report, we provide evidence that the conserved miR-183/96/182 cluster (miR-183/96/182) modulates Mϕ function in their production of reactive nitrogen (RNS) and oxygen species (ROS) and their inflammatory response to Pseudomonas aeruginosa (PA) infection and/or lipopolysaccharide (LPS) treatment. We show that knockdown of miR-183/96/182 results in decreased production of multiple proinflammatory cytokines in response to PA or LPS treatment in Mϕ-like Raw264.7 cells. Consistently, peritoneal Mϕ from miR-183/96/182-knockout versus wild-type mice are less responsive to PA or LPS, although their basal levels of proinflammatory cytokines are increased. In addition, overexpression of miR-183/96/182 results in decreased production of nitrite and ROS in Raw264.7 cells. We also provide evidence that DAP12 and Nox2 are downstream target genes of miR-183/96/182. These data suggest that miR-183/96/182 imposes global regulation on various aspects of Mϕ function through different downstream target genes.
Key Words: MicroRNA, miR-183/96/182 cluster, Macrophages, Pseudomonas aeruginosa, Lipopolysaccharides, DNAX-activating protein 12 kDa, DAP12, NADPH oxidase 2, Nox2, Inflammation, Reactive oxygen species, Reactive nitrogen species
Introduction
Myeloid phagocytes, including macrophages (Mϕ), neutrophils, and dendritic cells, are important components of the innate immune system and provide a first line of host defense against pathogen invasion. The cells are capable of ingestion and destruction of invading organisms, producing cytokines to evoke inflammatory responses, processing and presenting microbial antigens on their surface, and eliciting and modulating adaptive immune responses [1]. However, an uncontrolled inflammatory response, including persistence of neutrophils, also contributes to the collateral damage to host tissues during the host response to infection [2]. A deeper understanding of the molecular mechanisms underlying these processes is of great importance to develop new strategies to enhance innate immunity, eliminate the invading pathogen, coordinate adaptive immunity, and avoid bystander tissue damage.
In this regard, microRNAs (miRNAs) are a newly recognized, important mechanism of gene expression regulation at a posttranscriptional level [3]. They are proven to play important roles in all organ systems under physiological as well as pathological conditions [4] and are viable therapeutic targets for the treatment of human diseases [5]. miRNAs target their downstream messenger RNAs (mRNAs) by base pairing to their target sites with sequence complementarity, mainly in the 3′ untranslated region, and induce the breakdown and/or inhibition of translation of the targeted mRNAs [3]. At least 30% of the protein-coding genes are estimated to be regulated by miRNAs. One miRNA can target up to hundreds of downstream target mRNAs, while 1 mRNA can be targeted by multiple miRNAs, forming an intricate regulatory network. miRNAs impose quantitative regulation of their downstream target genes, making them an ideal mechanism to fine tune the functions of both innate and adaptive immune systems [6].
The miR-183/96/182 cluster (miR-183/96/182) is a conserved polycistronic paralogous miRNA cluster, including miR-183, -96 and -182 (in the sequence of transcription), which are clustered within 5 kb in the genomes of both mice and humans [7]. It was originally identified as a sensory organ-specific miRNA cluster [7, 8]. We and others have shown that miR-183/96/182 is required for the normal function of all major sensory organs, including vision [8], hearing and balance [8, 9, 10], olfaction [11], and pain [12]. Recently, mounting evidence from our group and others demonstrated that member(s) of miR-183/96/182 are also expressed and play important roles in both innate [13, 14, 15, 16] and adaptive immune systems [17, 18, 19], suggesting that miR-183/96/182 provides one of the first shared genetic links between the immune and traditional sensory systems, supporting the recently emerging concept of the immune system as a sensory system [20].
In our recent study on miR-183/96/182 in a mouse model of bacterial keratitis [13], we hypothesized that, since miR-183/96/182 is highly expressed in all sensory neurons, and the cornea is one of the most such innervated tissues, miR-183/96/182 may play a significant role in its response to Pseudomonas aeruginosa (PA) through its modulation of neuroinflammation in the cornea. PA is a gram-negative bacterium and an important human pathogen. It is also the most commonly recovered causative organism in contact lens-related keratitis in developed countries and induces rapid corneal destruction and blindness [21]. Our data provided evidence that inactivation of miR-183/96/182 in mice resulted in a significantly decreased inflammatory response with decreased severity of keratitis [13]. Furthermore, we showed that miR-183/ 96/182 is expressed in myeloid cells, Mϕ and neutrophils in both mice and humans. In both species, miR-182 and miR-183 predominated among the 3 components of miR-183/96/182 [13]. More importantly, we showed that miR-183/96/182 limits phagocytosis and intracellular bacterial killing capacity in both Mϕ and neutrophils and that knockdown or knockout (ko) of miR-183/96/182 in Mϕ and neutrophils reversed those functions [13].
Based on these data, we hypothesize that miR-183/ 96/182 plays important roles in myeloid phagocyte functions and regulates their functions in inflammatory responses to bacterial infection, for example. To test this hypothesis, we investigated the role of miR-183/96/182 in Mϕ with regard to the ability of the cluster to modulate the production of inflammatory cytokines and reactive nitrogen (RNS) and oxygen species (ROS) in response to PA and/or lipopolysaccharide (LPS) stimulation. We provide evidence that miR-183/96/182 regulates the production of proinflammatory cytokines, RNS, and ROS in Mϕ. Mechanistically, we provide evidence that DNAX-activating protein 12 kDa (DAP12) (also known as TYRO protein tyrosine kinase binding protein [TYROBP]), and NADPH oxidase 2 (Nox2) (also known as cytochrome b-245 β polypeptide [Cybb]), which play important roles in cytokine and RNS/ROS production in Mϕ, are potential downstream target genes of miR-183/96/182.
Materials and Methods
Mice
All animals were handled in accordance with NIH guidelines and the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. All animal protocols were approved by the Institutional Animal Care and Use Committee at the Wayne State University. miR-183/96/182 ko mice, miR-183CGT/GT, are on a 129S2/BL6-mixed background [8] and were originally derived from a gene trap (GT) embryonic stem cell clone [22]. Male and female young adult (8-week-old) ko mice and their wild-type (wt) littermates (miR-183C+/+) were used in this study.
In vitro Transfection of miRNA Mimics and Anti-miRs in Mouse Raw264.7 Cells and Stimulation with PA and LPS
Raw264.7 Mϕ-like cells, a cell line derived from the mouse, were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) (referred to as Raw cells from here on). 4 × 105 Raw cells were plated in a 48-well plate and transfected with anti-miR-183/anti-miR-182(/anti-miR-96) (10 nM for each) or negative control anti-miR oligonucleotides with scrambled sequences (10 nM; Exiqon, Vedbaek, Denmark), or miRNA mimics of miR-183/miR-182(/miR-96) (10 nM for each), or negative control mimics (10 nM; ThermoFisher) using RNAiMax Lipofectamine (ThermoFisher) as described before [23]. All experiments were conducted using 3–4 replicate wells. Forty-eight hours after transfection, PA (ATCC strain 19660; 1 × 107 colony forming units/well; multiplicity of infection [MOI] 25) or LPS (100 ng/mL) (from PA serotype 10; Sigma-Aldrich) was added. Six hours after PA or LPS treatment, cells were harvested for RNA preparation, and the supernatant was harvested for ELISA.
Isolation of Mouse Elicited Peritoneal Mϕ and Stimulation with PA or LPS
Mouse peritoneal Mϕ were isolated as described previously [13]. Briefly, Mϕ were elicited into the peritoneal cavity by intraperitoneal injection of 1 mL of 3% Brewer's thioglycollate medium (BD Biosciences, Sparks, MD, USA) 5 days before harvesting. Cells were collected by peritoneal lavage with DMEM containing 10% heat-inactivated fetal bovine serum (FBS; Invitrogen Life Technologies, Carlsbad, CA, USA), penicillin (100 U/mL; Invitrogen), and streptomycin (100 μg/mL; Invitrogen). Cells were stained with 0.4% trypan blue, and viable cells (> 95%) were counted with a hemocytometer. Peritoneal Mϕ were seeded in 48-well plates at a density of 1 × 105 cells/well (n = 4 wells/group) (in 250 µL/well) and incubated at 37°C overnight. The following day, Mϕ were washed once with growth medium (DMEM containing 1% FBS) and stimulated with LPS (100 ng/mL) for 6 h at 37°C. In a separate similar experiment, Mϕ were challenged with PA (ATCC strain 19660, MOI 25) for 6 h. Mϕ were mock treated with growth medium and incubated for 6 h at 37°C to serve as the negative controls.
RNA Preparation and Real-Time RT-PCR
Total RNA was prepared using the miRVana miRNA isolation kit (Life Technologies, Foster City, CA, USA) for miRNA or the RNeasy (Qiagen, Frederick, MD, USA) for mRNA studies as described before [13]. Quantitative (q) RT-PCR for miRNAs was performed using TaqMan miRNA primers and the RT-PCR kit (Life Technologies) on a CFX Connect Real-time System (Bio-Rad, Hercules, CA, USA) with snRNA U6 as an endogenous control; qRT-PCR assays of protein-coding genes were performed using the QuantiFast SYBR Green RT-PCR kit and QuantiTect primers (Qiagen) with 18s rRNA as endogenous controls as described before [13].
Western Blot Analysis
Western blot was performed following a standard protocol as we described previously [24]. Briefly, the protein lysate from Raw cells was homogenized using a pellet pestle motor (Fisher Scientific, Chicago, IL, USA) in RIPA buffer with a protease inhibitor cocktail, including 0.5 μM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF); 0.4 μM aprotinin, 10 μM leupeptin, 20 μM bestatin, 7.5 μM pepstatin A, and 7.0 μM E-64 (Sigma). Samples were sonicated followed by centrifugation at 12,000 g for 20 min. Total protein was quantified by the Micro BCA protein assay kit (ThermoFisher). Total protein samples (up to 40 µg) were run on a 16% SDS-PAGE in Tris-glycine-SDS buffer and then electroblotted onto nitrocellulose membranes (Bio-Rad). After blocking with 3% BSA TBST (TBS containing 0.05% Tween 20 and 3% BSA) for 1 h, the membranes were probed with primary antibodies for DAP12 (1: 500; Santa Cruz Biotechnology, Santa Cruz, CA, USA) in 3% BSA TBST overnight at 4°C. After 3 washes with TBST, the membranes were incubated with HRP secondary antibody (1: 1,000) diluted in TBST with 3% BSA at room temperature for 2 h. Bands were developed with SuperSignal West Femto chemiluminescent substrate (ThermoScientific) and visualized using a Kodak image station, 4000R Pro molecular imaging system (Carestream Health Inc., Rochester, NY, USA). Band intensity was quantified by using AlphaView software.
ELISA
Protein levels for IL-1β, Cxcl2, Ccl2, TNFα, and DAP12 were tested using ELISA kits (R&D Systems, Minneapolis, MN, USA [for IL-1β, Cxcl2, Ccl2, and TNFα]; MyBioSource.com, San Diego, CA, USA [for DAP12]) as described before [13]. Briefly, 50 µL of the supernatant of the culture medium (for TNFα, IL-1β, Ccl2, and Cxcl2) or cell lysate (for DAP12) were used for ELISA according to the manufacturer's instruction. Sensitivities of ELISAs were 2.31 pg/mL for IL-1β, 1.5 pg/mL for Cxcl2, 2 pg/mL for Ccl2, 1.88 pg/mL for TNFα, and 0.163 ng/mL for DAP12.
Griess Assay
Raw cells (1 × 105) were plated in a 48-well plate and transfected with miRNA mimics of miR-183, -182, and -96 (10 nM for each) or negative control mimics (10 nM; ThermoFisher) using RNAiMax Lipofectamine (ThermoFisher) as described above. Forty-eight hours after transfection, PA (ATCC strain 19660, MOI 25) was added. Three hours after PA treatment, the supernatant was collected to perform the Griess assay for nitrite determination according to the manufacturer's instructions (ThermoFisher Scientific). Briefly, in a microplate, for each well, 20 µL of Griess reagent, 150 µL of samples, and 130 µL of deionized water were added and incubated for 30 min at room temperature. After incubation, absorbance was measured using a Spectra Max plus 384 microplate reader (Molecular Devices) at 548 nm. All experiments were conducted with 4 replicate wells.
ROS Assay
Raw cells were plated (1 × 105) in a 96-well plate and transfected with anti-miR-183/-182/-96 (10 nM for each) or negative control anti-miR oligonucleotides with scrambled sequences (10 nM; Exiqon, Vedbaek, Denmark), using RNAiMax Lipofectamine (ThermoFisher) as described before [13]. Forty-eight hours after transfection, an ROS assay was performed using the CM-H2DCFDA general oxidative stress indicator kit (ThermoFisher Scientific) according to the manufacturer's instructions. Briefly, a working solution of 1 µM CM-H2DCFDA was loaded to the cells and incubated for 30 min at 37°C. After incubation, the cells were washed twice with PBS; fresh media (DMEM + 1% FBS) were added followed by challenge with PA (strain 19660, ATCC, MOI 25). Three hours after treatment, fluorescence was measured with approximate fluorescence excitation and emission at 492–495/517–527 nm using a Spectra Max Gemini EM plate reader (Molecular Devices). All experiments were conducted with 4 replicate wells.
Statistical Analysis
A two-tailed Student's t test was used to determine significance. p < 0.05 was considered significant. When the comparison was made among more than 2 conditions, one-way ANOVA with Bonferroni's multiple comparison test was employed. Each experiment was repeated at least twice to ensure reproducibility, and data from a representative experiment are shown. All quantitative data are expressed as the mean ± standard error of the mean (SEM).
Results
PA and LPS Induce the Expression of miR-183 and miR-182 in Raw Cells
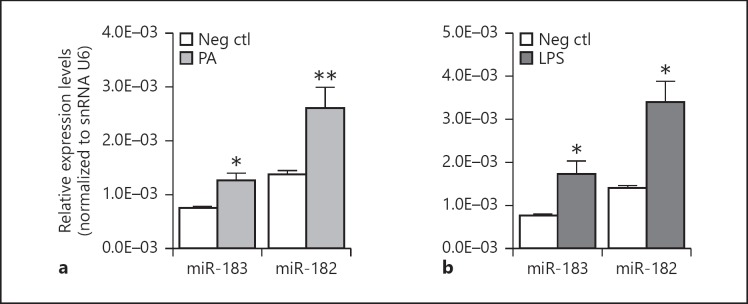
To test the response of miR-183/96/182 in Mϕ to PA and to its major outer membrane component, LPS, we treated Raw cells with PA (ATCC19660, MOI 25) or LPS (100 ng/mL). Six hours later, we harvested the cells and performed qRT-PCR analysis on the processed RNA. Our data showed that both PA and LPS treatment significantly induced the expression of miR-182 and miR-183 (Fig. 1), suggesting that miR-183/96/182 is involved in the Mϕ response to PA and LPS stimulation. miR-96, which is barely expressed in Mϕ and neutrophils [13], was not significantly changed (data not shown), suggesting cell type- and miRNA-specific maturation mechanisms in the posttranscriptional maturation of miR-183, -182, and -96, given that all 3 miRNAs are cotranscribed from the same gene [7, 8].
Fig. 1.
PA (ATCC strain 19660, MOI 25) (a) and LPS (100 ng/mL) treatment (b) for 6 h induced expression of miR-183 and miR-182 in Raw cells. * p < 0.05; ** p < 0.01 vs. mock-treated negative control (Neg ctl). n = 4/group. The error bars represent SEM in all figures.
miR-183/96/182 Regulates Cytokine Production in Response to PA or LPS
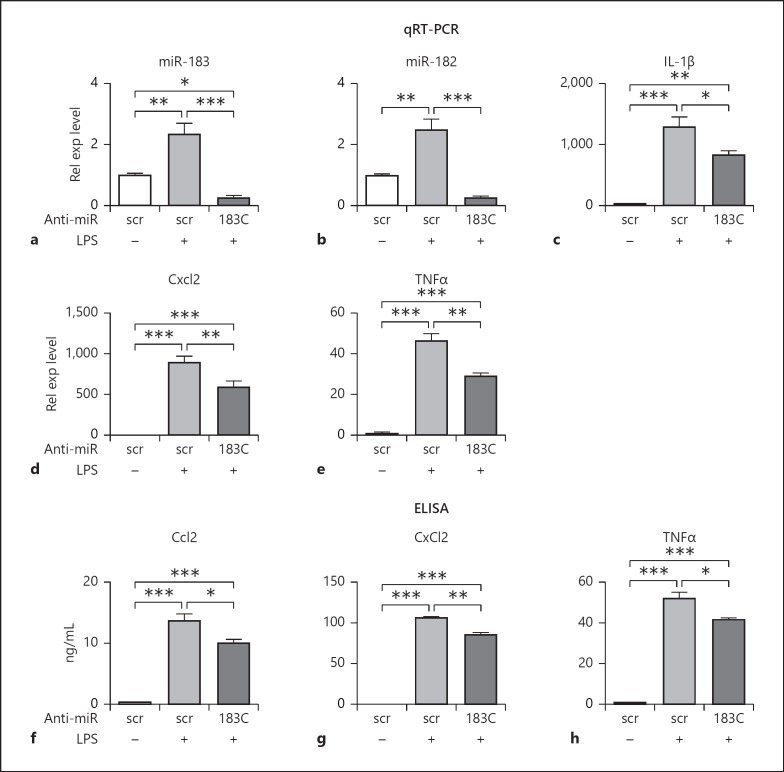
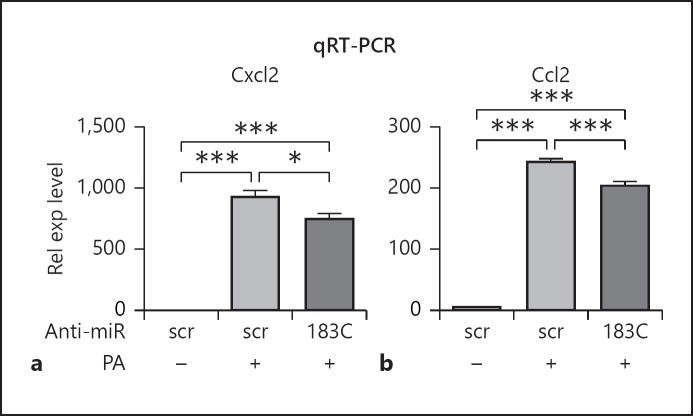
To begin to understand the roles of miR-183/96/182 in the inflammatory response in Mϕ, we knocked down the function of the cluster in Raw cells by transfection of anti-miR-183 and -182 (anti-miR-183/-182; 10 nM each), the functional components of miR-183/96/182 in Mϕ (Fig. 2) [13]. Forty-eight hours after transfection, cells were treated with PA (ATCC strain 19660, MOI 25) or LPS (100 ng/mL) for 6 h. qRT-PCR results showed that miR-183 and -182 were down-regulated (Fig. 2a, b) when compared to the negative controls, while the expression of miR-96 was unaffected (data not shown). Furthermore, qRT-PCR analysis showed that knockdown of miR-183/96/182 resulted in significant reduction in LPS-induced expression of IL-1β, Cxcl2, and TNFα at mRNA levels (Fig. 2c–e); ELISA further demonstrated that knockdown of miR-183/96/182 function significantly inhibited LPS-induced production of Ccl2, Cxcl2, and TNFα at the protein level in the supernatant (Fig. 2f–h). Knockdown of miR-183/96/182 also inhibited PA-induced production of Ccl2 and Cxcl2 at the mRNA level (Fig. 3).
Fig. 2.
Knockdown of miR-183/96/182 function in Raw cells results in a decreased inflammatory response to LPS treatment. a, b qRT-PCR analysis of miR-182 and miR-183 in Raw cells. c–e qRT-PCR analysis of IL-1β (c), Cxcl2 (d), and TNFα (e). Relative expression (Rel exp) levels were normalized to the negative control with scrambled sequence (scr) without treatment. f–h ELISA of Ccl2, Cxcl2, and TNFα. 183C, anti-miR-183 cluster. * p < 0.05. ** p < 0.01; *** p < 0.001. n = 4/condition.
Fig. 3.
Knockdown of miR-183/96/182 function in Raw cells results in a decreased inflammatory response to PA treatment. qRT-PCR analysis of Cxcl2 (a) and Ccl2 (b). Relative expression (Rel exp) levels were normalized to the negative control with scrambled sequence (scr) without treatment. 183C, anti-miR-183 cluster. * p < 0.05; *** p < 0.001. n = 4/condition.
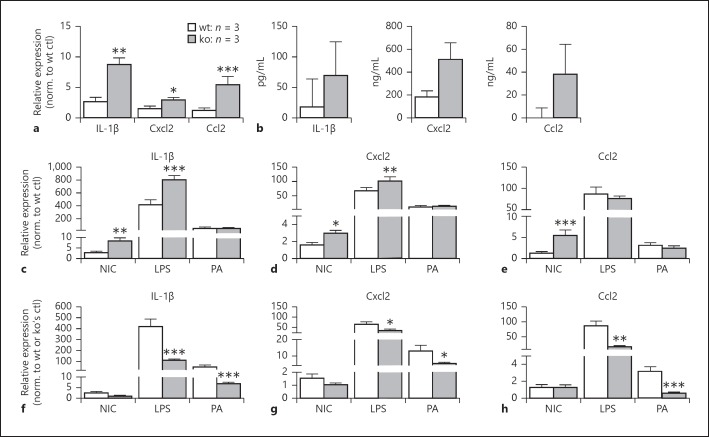
Inactivation of miR-183/96/182 Increases Basal Levels of Proinflammatory Cytokine Expression in Mϕ but Their Responsiveness to PA and LPS Stimulation Decreases
To study the in vivo functions of miR-183/96/182 in Mϕ, we isolated elicited peritoneal Mϕ from miR-183/ 96/182 ko and wt mice and stimulated them with PA (ATCC strain 19660, MOI 25) or LPS (100 ng/mL). Six hours after stimulation, we harvested cellular RNA for qRT-PCR and supernatant for ELISA. Our qRT-PCR results showed that the unstimulated elicited peritoneal Mϕ from the ko mice have increased basal levels of multiple proinflammatory cytokines/chemokines, including IL-1β, Cxcl2, and Ccl2, when compared to the ones from wt control mice (Fig. 4a). ELISA confirmed this trend at the protein level (Fig. 4b). Furthermore, after PA and LPS treatment, although the expression of these cytokines/chemokines was increased in the Mϕ from both ko and wt mice (Fig. 4c–e), the peritoneal Mϕ from ko mice showed significantly decreased induction when normalized to their own nontreated controls (Fig. 4f–h), suggesting decreased responsiveness to LPS and PA challenge.
Fig. 4.
Inactivation of miR-183/96/182 increases basal levels of proinflammatory cytokine expression in Mϕ from knockout (ko) mice, but reduces their responsiveness to PA and LPS stimulation. a, b qRT-PCR (a) and ELISA (b) of nontreated Mϕ of wild-type (wt) and ko mice. Relative expression levels were normalized to nontreated wt control (wt ctl) in a. c–h qRT-PCR in Mϕ challenged with LPS or PA. Relative expression levels were normalized to wt noninfected control (NIC) (c–e) or normalized to the wt or ko own NIC (f–h). * p < 0.05; ** p < 0.01; *** p < 0.001.
miR-183/96/182 Modulates Mϕ ROS and RNS Production
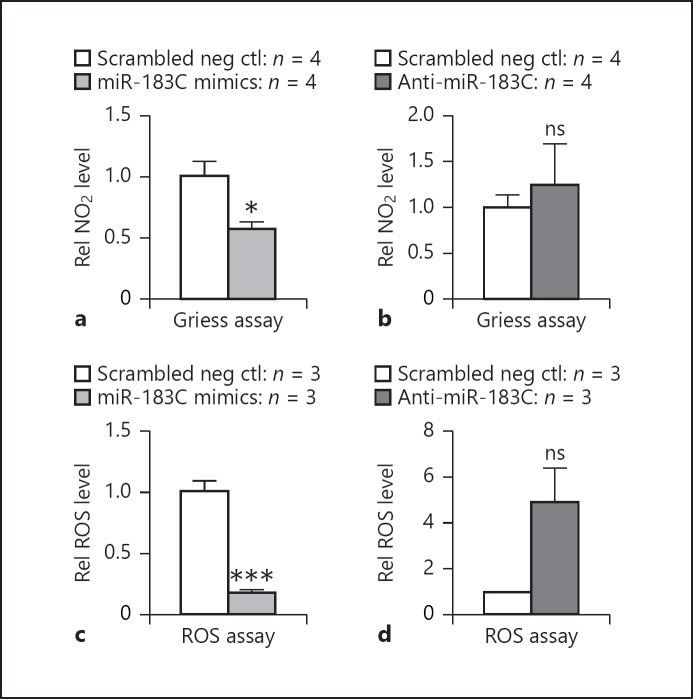
Previously, we showed that knockdown of miR-183/ 96/182 in Raw cells increased their phagocytosis and intracellular killing capacity, while overexpression of miR-183/96/182 led to decreased phagocytosis and intracellular killing of PA [13]. Consistently, peritoneal neutrophils from miR-183/96/182 ko mice showed an increased phagocytosis and intracellular killing of PA compared to cells from wt control mice [13], suggesting miR-183/ 96/182 limits the phagocytosis and intracellular killing capacity of innate phagocytes. It is well known that phagocyte-derived ROS and RNS play a key role in intracellular killing and host defense to microbial pathogens [25]. Therefore, we hypothesized that miR-183/96/182 limits the phagocytosis and intracellular killing capacity of innate phagocytes through ROS and/or RNS production regulation. To test this hypothesis, we used a Griess assay to detect nitrite (NO2−) in Raw cells. Our results showed that overexpression of miR-183/96/182 led to a decreased level of nitrite in Raw cells (Fig. 5a). To test the effect of miR-183/96/182 on ROS production, we performed an ROS assay using CM-H2DCFDA as an indicator of intracellular ROS. Overexpression of miR-183/96/182 resulted in decreased ROS production (Fig. 5c), while knockdown of miR-183/96/182 led to an increased trend of ROS production in Raw cells challenged by PA (Fig. 5c). These findings suggest that miR-183/96/182 directly regulates ROS and RNS production in phagocytes, which contributes to its modulation of their ability to kill invading bacteria intracellularly.
Fig. 5.
miR-183/96/182 modulates the production of reactive nitrogen species (RNS) and reactive oxygen species (ROS) in Raw cells. Griess (a, b) and ROS assays (c, d) in miR-183/96/182 cluster (miR-183C) mimic-transfected (a, c) and anti-miR-183C-transfected (b, d) Raw cells. * p < 0.05; *** p < 0.001.
DAP12 and Nox2 Are Potential Target Genes of miR-183/96/182 Mediating Its Functions in Mϕ
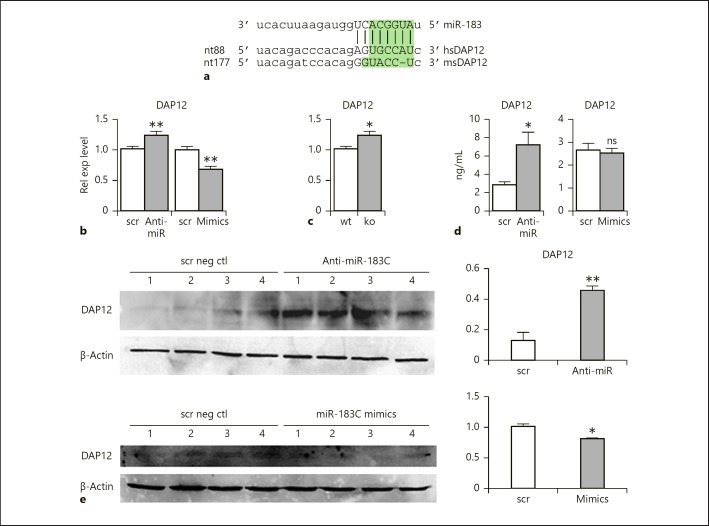
To uncover the molecular mechanisms underlying the regulatory roles of miR-183/96/182 in Mϕ, we performed target prediction (by miRanda algorithms. www.microrna.org) and functional annotation studies as we described before [8]. Among predicted target genes, DAP12 emerged as an appealing candidate as a functional mediator of miR-183/96/182 regulation of Mϕ. DAP12 is an important signaling adaptor protein that pairs with receptors on myeloid cells and natural killer (NK) cells [26]. It facilitates cell surface localization of its associated receptors to activate downstream signaling pathways [27]. In Mϕ, DAP12 inhibits TLR-induced proinflammatory cytokine production [28]. DAP12-deficient Mϕ exhibited increased production of proinflammatory cytokines in response to different pathogens and/or their products, including LPS [28]. Therefore, we hypothesized that if miR-183/96/182 targets DAP12 in Mϕ, knockdown or inactivation of miR-183/96/182 would increase the expression of DAP12, which may contribute to the inhibition of PA- or LPS-induced inflammatory responses.
Previously, Donatelli et al. [29] experimentally confirmed that miR-183 targeted DAP12 in human primary NK cells, a myeloid cell line, U-937, and in Hela cells. However, careful examination of the sequences of the predicted target sites of human and mouse DAP12 transcripts showed that, although the transcript of human DAP12 carries a conserved target site for miR-183, similar to what Donatelli et al. reported [29] (Fig. 6a), the target site is not conserved at the seed sequences in mouse DAP12 (Fig. 6a). In spite of this, considering that (1) the human and mouse DAP12 transcripts shared high homology in this region (Fig. 6a); (2) that other sequences and structural features beyond the seed sequence also contribute to the miRNA target recognition [30, 31]; and (3) that an important function tends to be conserved between mice and humans, we went on to test whether miR-183/96/182 modulates the expression of DAP12 in Mϕ. Our results showed that knockdown of miR-183/96/182 in Raw cells resulted in significant upregulation of DAP12 expression, while overexpression of miR-183/96/182 led to significantly decreased expression of DAP12 mRNA levels (Fig. 6b). Consistently, DAP12 is significantly increased in peritoneal Mϕ of miR-183/96/182 ko mice compared to wt controls (Fig. 6c). ELISA and Western blot analysis further confirmed that knockdown of miR-183/96/182 resulted in increased expression of DAP12 at protein levels (Fig. 6d, e). These data suggest that miR-183/96/182 targets DAP12 in mouse Mϕ in spite of lacking a canonical target site with perfect complementarity to the seed sequence, and that DAP12 may mediate the functions of miR-183/96/182 in Mϕ modulating their inflammatory responses to PA challenge.
Fig. 6.
DAP12 is a direct target of miR-183/96/182. a Sequence alignment of miR-183 with a canonical target site in human (hs) DAP12 (www.microrna.org). Lack of a canonical target site with perfect complementarity to the seed sequence (shaded in green rectangle) is seen in mouse (ms) DAP12. b Knockdown and overexpression of miR-183/96/182 in Raw cells results in significant up- and down-regulation of DAP12, respectively, by qRT-PCR analysis. Relative expression (Rel exp) levels were normalized to negative controls with scrambled sequence (scr). Anti-miR, cells transfected with anti-miR-183 cluster; mimics, cells transfected with mimics of miR-183 cluster. n = 4/condition. c DAP12 is significantly up-regulated in the elicited peritoneal Mϕ of miR-183/96/182 knockout (ko; n = 4) mice compared to wild-type controls (wt; n = 4). d ELISA of DAP12 in Raw cells treated with anti-miR-183 cluster (anti-miR) or negative control anti-miR with scrambled sequences (scr). n = 4/genotype. e Western blot analysis of DAP12 expression. * p < 0.05; ** p < 0.01. ns, nonsignificant.
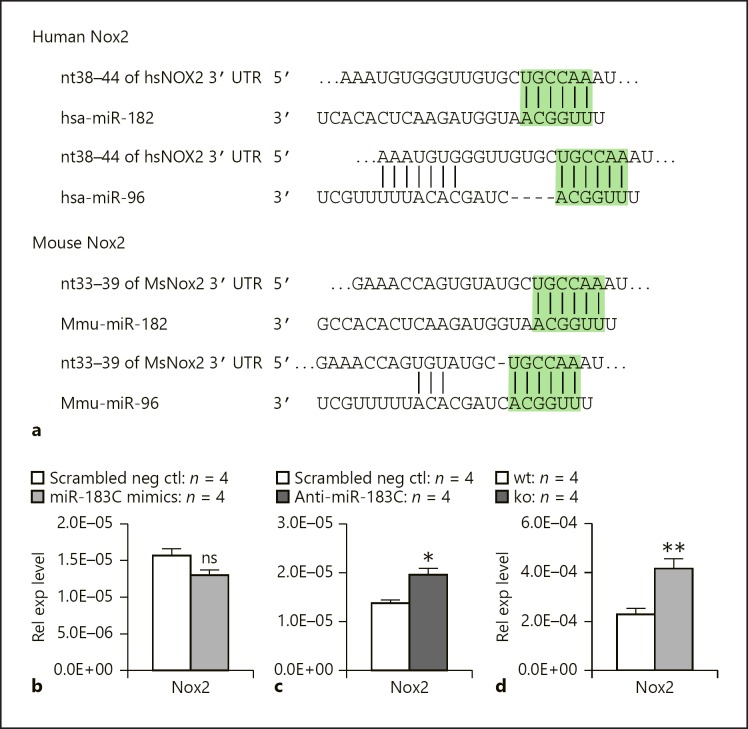
Nox2 is the NOX family isoform expressed in professional phagocytes, including Mϕ and neutrophils [32, 33]. Nox2 is the key enzyme required to generate superoxide (O2−) from NAPDH and oxygen, and other microbicidal ROS and RNS [32, 33]. Target prediction using the TargetScan algorithm (targetscan.org) suggested that Nox2 is a target of miR-182 and miR-96 in both humans and mice (Fig. 7a). Our results showed that overexpression of miR-183/96/182 in Raw cells resulted in decreased expression of Nox2 (Fig. 7b), while knockdown of miR-183/96/182 led to significantly increased Nox2 expression (Fig. 7c). Consistently, Nox2 was significantly upregulated in peritoneal Mϕ of miR-183/96/182 ko mice compared to wt controls (Fig. 7d), suggesting that miR-183/96/182 targets endogenous Nox2 and regulates its expression in Mϕ. This is consistent with our observation that overexpression of miR-183/96/182 decreased nitrite and ROS production (Fig. 5a, c), while knockdown of miR-183/96/182 resulted in an increased trend of RNS and ROS production in PA-challenged Raw cells (Fig. 5b, d), suggesting that miR-183/96/182 limits the intracellular killing capacity of PA infection [13] through its modulation of Nox2.
Fig. 7.
Nox2 is a direct target of miR-183/96/182. a Sequence alignment of miR-182 and miR-96 with their predicted target sites in the 3′ untranslated region of hu man (hs) and mouse (Ms) Nox2 transcripts (based on data from targetscan.org). The sequences in the green rectangle are alignments with seed sequences of miRNAs. b Overexpression of miR-183/96/182 in Raw cells results in a decreased expression of Nox2 by qRT-PCR analysis. c Knockdown of miR-183/96/182 in Raw cells results in an increased expression of Nox2 by qRT-PCR analysis. d Nox2 is upregulated in peritoneal Mϕ of miR-183/96/182 ko mice by qRT-PCR analysis. Relative expression levels were normalized to 18s rRNA.* p < 0.05; ** p < 0.01. ns, nonsignificant.
Discussion
Herein, we demonstrated that stimulation of Raw cells with PA and LPS resulted in significant upregulation of miR-183 and -182 (Fig. 1), the functional components of miR-183/96/182 in Mϕ [13], suggesting that miR-183/ 96/182 is involved in the inflammatory response of Mϕ to PA infection. Knockdown of the cluster in Raw cells in vitro resulted in decreased production of multiple proinflammatory factors (Fig. 2, 3), suggesting that physiologically miR-183/96/182 promotes the inflammatory response to PA infection in Mϕ, and that the induction of miR-183/96/182 upon PA and LPS treatment (Fig. 1) may contribute to the inflammatory response. This is consistent with our previous observation of a decreased inflammatory response in the cornea of miR-183/96/182 ko mice compared to wt control mice upon PA infection of the cornea [13]. The mammalian cornea contains resident Mϕ in the stroma [34], which are thought to play important roles in the corneal response to PA infection [34]. Since the corneal resident Mϕ share the same myeloid origin as the peritoneal Mϕ, postulating from our result, we predict that miR-183/96/182 may have a similar role in the inflammatory response of corneal-resident Mϕ to PA infection, contributing to the overall corneal response to PA infection and development of PA keratitis.
It is intriguing that, in the peritoneal elicited Mϕ from miR-183/96/182 ko mice, the basal level of multiple proinflammatory cytokines was increased when compared to their wt controls (Fig. 4a, b). This seemed contradictory to the result of acute knockdown of miR-183/-182 in vitro in Raw cells (Fig. 2, 3). However, when looking into the fold change in induction of these cytokines upon PA and LPS stimulation, there is a significantly reduced fold increase in these cytokines (Fig. 4f–h), suggesting a decreased responsiveness of Mϕ from ko mice to PA infection or LPS stimulation. We speculate that, in the ko mice, Mϕ may have adapted to the loss of miR-183/96/182 to keep homeostasis in vivo through yet unknown mechanisms, and this resulted in increased basal levels of inflammatory cytokines; however, upon PA infection or LPS stimulation, loss of miR-183/96/182 resulted in decreased responsiveness, which is in line with our observation in Raw cells that knockdown of miR-183/96/182 function resulted in decreased production of proinflammatory factors (Fig. 2, 3).
The observation of increased basal levels of proinflammatory cytokines in the Mϕ of the ko mice also may suggest a potential role of miR-183/96/182 in the functional differentiation and polarity of Mϕ. Supporting this hypothesis, our preliminary gene expression analysis on multiple M1 and M2 Mϕ markers showed that in primary peritoneal Mϕ of miR-183/96/182 ko mice, multiple M1 markers, including IL-6 and prostaglandin-endoperoxide synthase 2 (PSTG2) (online suppl. Fig. 1; for all online suppl. material, see www.karger.com/doi/10.1159/ 000495472) in addition to IL-1β, Cxcl2, and Ccl2 (Fig. 4a), as well as a M2 marker, IL-10, a powerful anti-inflammatory cytokine (in the keratitis model) [35], are simultaneously upregulated, when compared to wt controls (online suppl. Fig. 1), while another M2 marker, TGFβ, is significantly decreased (online suppl. Fig. 1), suggesting potential roles of miR-183/96/182 in Mϕ differentiation or polarity, and, possibly, the development of myeloid-derived suppressor cells, which are immature myeloid cells that often express both M1 and M2 markers [36]. Wurm et al. [14] showed that miR-182 regulated granulopoiesis through its mutually inhibitory interaction with C/EBPα, a master regulator of myelopoiesis [14]. Forced miR-182 expression inhibited the expression of C/EBPα and impaired granulopoiesis [14]. Other reports suggested that both miR-182 and -183 promoted osteoclastogenesis [15, 16]. These reports substantiated the hypothesis that miR-183/96/182 may have a regulatory role in myeloid cell differentiation. Further investigation on the role of miR-183/96/182 in the differentiation and function of Mϕ, including corneal resident Mϕ, appears warranted.
Previously, we showed that, in addition to its regulation of inflammatory responses to PA infection, overexpression of miR-183/96/182 in Raw cells resulted in decreased phagocytosis and intracellular killing of PA, while knockdown of the cluster in Raw cells led to enhanced phagocytosis and intracellular killing [13]. Consistently, elicited peritoneal neutrophils from miR-183/96/182 ko mice showed increased phagocytosis and intracellular killing, suggesting that miR-183/96/182 limits the ability of phagocytosis and intracellular bacterial killing in myeloid phagocytes. To begin to understand the underlying mechanism, we performed Griess and ROS assays on Raw cells and showed that overexpression of miR-183/96/182 resulted in significantly decreased production of nitrite and ROS upon PA challenge (Fig. 5a, c). These results suggest that miR-183/96/182 regulates the phagocytosis and intracellular killing capacity of myeloid phagocytes through its modulation of their ROS and RNS production.
DAP12 is a type I membrane protein containing a single immunoreceptor tyrosine-based activation motif (ITAM) and an important signaling adaptor protein [26, 27]. It facilitates the cell surface localization of associated receptors on myeloid cells, including Mϕ, dendritic cells, osteoclasts, microglia, and NK cells to activate downstream signaling pathways, playing an important role in the activation of myeloid and NK cells [26, 27]. In NK cells, DAP12 is the common adaptor for many NK cell-activating receptors. Upon ligand binding, DAP12 is tyrosine phosphorylated by Src kinases and proceeds through the phosphatidylinositide 3-kinase/extracellular signal-regulated kinase activation pathway to mobilize lytic granules that ultimately kill target cells [29]. Inactivation of DAP12 in mice impaired the function of the activating Ly49D receptors in NK cells and Ly49D-mediated killing of target cells [37]. Previously, Donatelli et al. [29] experimentally confirmed that miR-183 targeted human DAP12 in NK cells and U937 cells (monocytic cell line). They showed that in the tumor environment, miR-183/96/182 was induced by TGFβ and prevented NK cells from killing tumor cells by inhibiting DAP12 expression. Therefore, miR-183/96/182 is recognized as a NK cell immunosuppressor.
To test whether DAP12 is targeted by miR-183/96/182 in mouse myeloid cells, we transfected Raw cells with miR-183/96/182 mimics or anti-miRs and showed that overexpression of miR-183/96/182 decreased, while knockdown of miR-183/96/182 increased the expression levels of endogenous DAP12 at both RNA and protein levels (Fig. 6). In addition, expression of DAP12 at the mRNA level was significantly increased in peritoneal Mϕ of miR-183/96/182 ko mice compared to wt controls, suggesting that DAP12 is targeted by miR-183 in mice, in spite of the fact that the transcript of mouse DAP12 lacks a conserved canonical target site for miR-183 with a perfect complementarity to the seed sequences (Fig. 6a). This suggests that other features around the seed sequences may have contributed to the determination of the target site. Further studies are required to determine the exact mechanism for miR-183 targeting DAP12 in the mouse.
DAP12, with its associated receptor, triggering receptor expressed on myeloid cells-2 (TREM-2), has been shown to be a negative regulator of TLR-induced proinflammatory cytokine production in Mϕ [28, 38]. DAP12-deficient Mϕ exhibited increased production of proinflammatory cytokines in response to different TLR stimuli, including LPS-TLR4, synthetic bacterial lipopeptide-TLR2/1, CpG DNA-TLR9, zymosan-TLR2, poly(I:C)-TLR3, and peptidoglycan-TLR2, as well as to whole bacteria, e.g., Escherichia coli and Staphylococcus aureus [28], suggesting that DAP12 normally limits a TLR-induced inflammatory response. Our data showed that inactivation or knockdown of miR-183/96/182 increased the expression of DAP12 in Mϕ or Raw cells (Fig. 6), while LPS- and PA-induced production of proinflammatory cytokines was decreased (Fig. 2, 3, 4), suggesting that DAP12 mediated the miR-183/96/182 function to promote proinflammatory cytokine production. This is consistent with the observation that TREM-2 also limits the production of proinflammatory cytokines in Mϕ and promotes host resistance against PA keratitis [39], similar to what we observed in miR-183/96/182 ko mice [13], suggesting that the DAP12-TREM-2 pathway contributes to miR-183/96/182 modulation on the proinflammatory responses of Mϕ and overall corneal responses to PA infection.
In addition to DAP12, we also provided evidence that Nox2 – an isoform of NOX family genes expressed in professional phagocytes including Mϕ and neutrophils [32], is targeted by miR-183/96/182 (Fig. 7). Nox2, also known as Cybb or gp91phox (91-kDa glycoprotein of phagocyte oxidase), is a transmembrane protein and the largest subunit of the NADPH oxidase in phagocytes. The complete NADPH oxidase is composed of multiple subunits. At resting conditions, Nox2/gp91phox and p22phox form a heterodimer, cytochrome b558. Upon microbial infection, Nox2 is rapidly activated when cytosolic subunits, p40phox, p47phox, and p67phox, and Rac are translocated to the membrane-bound cytochrome b558 to form the active NADPH oxidase enzyme complex. The active enzyme complex transports electrons from cytoplasmic NAPDH to extracellular or phagosomal oxygen to generate superoxide (O2−), which is a ROS and also serves as a precursor to form other microbicidal ROS and RNS [32]. Therefore, Nox2 is a crucial enzyme in antimicrobial host defense and in the regulation of inflammation [32]. That Nox2 is targeted by miR-183/96/182 provides a potential molecular mechanism underlying its regulation of ROS and NOS production upon PA infection (Fig. 5) and intracellular killing of PA that we observed previously [13].
miRNAs are quantitative regulators of gene expression. One miRNA often targets multiple protein-coding genes in the same pathway or functional network, and imposes modest regulation on each of these genes. In concert, the modest regulation of multiple genes maintains the homeostasis of the signaling pathway or network under physiological conditions. Defects in a miRNA can result in simultaneous dysregulation of multiple genes, leading to significant functional consequences when the composite impact passes a threshold. Here, we present evidence that miR-183/96/182 targets at least 2 genes in myeloid phagocytes, DAP12 and Nox2, each of which plays an important role in different yet overlapping aspects of the Mϕ and neutrophil functions, including phagocytosis, bacterial killing, and inflammatory responses to bacterial infection. When miR-183/96/182 is knocked down or inactivated, deregulation on each of these genes has contributed to the phenotypes observed at both cellular (Fig. 2, 3, 4, 5) and organismic levels [13]. Target prediction analyses and experimental data from other organ systems have shown that miR-183/96/182 regulates hundreds of downstream target genes [7, 8, 18]; therefore, we predict that DAP12 and Nox2 are not the only functional targets of miR-183/96/182 in Mϕ and neutrophils to mediate its functions. We further predict that miR-183/96/182 may modulate other aspects of Mϕ and neutrophil functions, e.g., migration and chemotaxis, through targeting multiple genes involved in cytoskeletal reorganization [unpubl. data]. Through this mechanism, miR-183/96/182 imposes a global regulation on Mϕ and neutrophil functions.
In summary, we provide the first direct evidence that miR-183/96/182 is induced in Mϕ upon PA infection. It modulates the inflammatory response and ROS and RNS production of Mϕ in response to PA infection in vitro. This regulation is mediated, at least in part, through targeting both DAP12 and Nox2, suggesting that miR-183/96/182 plays an important role in innate immunity. Manipulation of miR-183/96/182 levels in innate immune cells may modulate various aspects of their function and, therefore, have therapeutic potential in the treatment of related diseases. Knockdown or inactivation of miR-183/96/182 in Mϕ increased ROS and RNS production, which may contribute to their increased bactericidal capacity, suggesting the potential to combat antibiotic-resistant bacterial infection by enhancing innate immunity.
Disclosure Statement
The authors disclose no potential conflicts of interest.
Supplementary Material
Supplementary data
Acknowledgments
This work was supported by grants R01 EY02605902 (S.X.), R01EY016058 (L.D.H.), and P30EY004068 (L.D.H.) from the National Eye Institute, National Institutes of Health, and by an unrestricted RPB (Research to Prevent Blindness) grant to the Department of Ophthalmology, Kresge Eye Institute. L.D.H. is a recipient of an Alcon Research Institute Award. We apologize to the many colleagues whose work was not cited because of space limitation.
References
- 1.Rivera A, Siracusa MC, Yap GS, Gause WC. Innate cell communication kick-starts pathogen-specific immunity. Nat Immunol. 2016 Apr;17((4)):356–63. doi: 10.1038/ni.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Segal AW. How neutrophils kill microbes. Annu Rev Immunol. 2005;23((1)):197–223. doi: 10.1146/annurev.immunol.23.021704.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004 Jan;116((2)):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet. 2007;8((1)):215–39. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- 5.van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007 Sep;117((9)):2369–76. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol. 2016 Apr;16((5)):279–94. doi: 10.1038/nri.2016.40. [DOI] [PubMed] [Google Scholar]
- 7.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007 Aug;282((34)):25053–66. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 8.Lumayag S, Haldin CE, Corbett NJ, Wahlin KJ, Cowan C, Turturro S, et al. Inactivation of the microRNA-183/96/182 cluster results in syndromic retinal degeneration. Proc Natl Acad Sci USA. 2013 Feb;110((6)):E507–16. doi: 10.1073/pnas.1212655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, Langford C, et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009 May;41((5)):614–8. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mencía A, Modamio-Høybjør S, Redshaw N, Morín M, Mayo-Merino F, Olavarrieta L, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009 May;41((5)):609–13. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 11.Fan J, Jia L, Li Y, Ebrahim S, May-Simera H, Wood A, et al. Maturation arrest in early postnatal sensory receptors by deletion of the miR-183/96/182 cluster in mouse. Proc Natl Acad Sci USA. 2017 May;114((21)):E4271–80. doi: 10.1073/pnas.1619442114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng C, Li L, Zhang MD, Bengtsson Gonzales C, Parisien M, Belfer I, et al. miR-183 cluster scales mechanical pain sensitivity by regulating basal and neuropathic pain genes. Science. 2017 Jun;356((6343)):1168–71. doi: 10.1126/science.aam7671. [DOI] [PubMed] [Google Scholar]
- 13.Muraleedharan CK, McClellan SA, Barrett RP, Li C, Montenegro D, Carion T, et al. Inactivation of the miR-183/96/182 Cluster Decreases the Severity of Pseudomonas aeruginosa-Induced Keratitis. Invest Ophthalmol Vis Sci. 2016 Apr;57((4)):1506–17. doi: 10.1167/iovs.16-19134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wurm AA, Zjablovskaja P, Kardosova M, Gerloff D, Bräuer-Hartmann D, Katzerke C, et al. Disruption of the C/EBPα-miR-182 balance impairs granulocytic differentiation. Nat Commun. 2017 Jun;8((1)):46. doi: 10.1038/s41467-017-00032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller CH, Smith SM, Elguindy M, Zhang T, Xiang JZ, Hu X, et al. RBP-J-Regulated miR-182 Promotes TNF-α-Induced Osteoclastogenesis. J Immunol. 2016 Jun;196((12)):4977–86. doi: 10.4049/jimmunol.1502044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ke K, Sul OJ, Rajasekaran M, Choi HS. MicroRNA-183 increases osteoclastogenesis by repressing heme oxygenase-1. Bone. 2015 Dec;81:237–46. doi: 10.1016/j.bone.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 17.Stittrich AB, Haftmann C, Sgouroudis E, Kühl AA, Hegazy AN, Panse I, et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. 2010 Nov;11((11)):1057–62. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- 18.Ichiyama K, Gonzalez-Martin A, Kim BS, Jin HY, Jin W, Xu W, et al. The MicroRNA-183-96-182 Cluster Promotes T Helper 17 Cell Pathogenicity by Negatively Regulating Transcription Factor Foxo1 Expression. Immunity. 2016 Jun;44((6)):1284–98. doi: 10.1016/j.immuni.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelada S, Sethupathy P, Okoye IS, Kistasis E, Czieso S, White SD, et al. miR-182 and miR-10a are key regulators of Treg specialisation and stability during Schistosome and Leishmania-associated inflammation. PLoS Pathog. 2013;9((6)):e1003451. doi: 10.1371/journal.ppat.1003451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veiga-Fernandes H, Freitas AA. The S(c)ensory Immune System Theory. Trends Immunol. 2017 Oct;38((10)):777–88. doi: 10.1016/j.it.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Hazlett LD. Corneal response to Pseudomonas aeruginosa infection. Prog Retin Eye Res. 2004 Jan;23((1)):1–30. doi: 10.1016/j.preteyeres.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Hansen J, Floss T, Van Sloun P, Füchtbauer EM, Vauti F, Arnold HH, et al. A large-scale, gene-driven mutagenesis approach for the functional analysis of the mouse genome. Proc Natl Acad Sci USA. 2003 Aug;100((17)):9918–22. doi: 10.1073/pnas.1633296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cowan C, Muraleedharan CK, O'Donnell JJ, 3rd, Singh PK, Lum H, Kumar A, et al. MicroRNA-146 inhibits thrombin-induced NF-κB activation and subsequent inflammatory responses in human retinal endothelial cells. Invest Ophthalmol Vis Sci. 2014 Jul;55((8)):4944–51. doi: 10.1167/iovs.13-13631. [DOI] [PubMed] [Google Scholar]
- 24.Zhuang P, Muraleedharan CK, Xu S. Intraocular Delivery of miR-146 Inhibits Diabetes-Induced Retinal Functional Defects in Diabetic Rat Model. Invest Ophthalmol Vis Sci. 2017 Mar;58((3)):1646–55. doi: 10.1167/iovs.16-21223. [DOI] [PubMed] [Google Scholar]
- 25.Fang FC. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat Rev Microbiol. 2004 Oct;2((10)):820–32. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 26.Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature. 1998 Feb;391((6668)):703–7. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- 27.Takaki R, Watson SR, Lanier LL. DAP12: an adapter protein with dual functionality. Immunol Rev. 2006 Dec;214((1)):118–29. doi: 10.1111/j.1600-065X.2006.00466.x. [DOI] [PubMed] [Google Scholar]
- 28.Hamerman JA, Tchao NK, Lowell CA, Lanier LL. Enhanced Toll-like receptor responses in the absence of signaling adaptor DAP12. Nat Immunol. 2005 Jun;6((6)):579–86. doi: 10.1038/ni1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donatelli SS, Zhou JM, Gilvary DL, Eksioglu EA, Chen X, Cress WD, et al. TGF-β-inducible microRNA-183 silences tumor-associated natural killer cells. Proc Natl Acad Sci USA. 2014 Mar;111((11)):4203–8. doi: 10.1073/pnas.1319269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007 Jul;27((1)):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robins H, Li Y, Padgett RW. Incorporating structure to predict microRNA targets. Proc Natl Acad Sci USA. 2005 Mar;102((11)):4006–9. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singel KL, Segal BH. NOX2-dependent regulation of inflammation. Clin Sci (Lond) 2016 Apr;130((7)):479–90. doi: 10.1042/CS20150660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Oliveira-Junior EB, Bustamante J, Newburger PE, Condino-Neto A. The human NADPH oxidase: primary and secondary defects impairing the respiratory burst function and the microbicidal ability of phagocytes. Scand J Immunol. 2011 May;73((5)):420–7. doi: 10.1111/j.1365-3083.2010.02501.x. [DOI] [PubMed] [Google Scholar]
- 34.Brissette-Storkus CS, Reynolds SM, Lepisto AJ, Hendricks RL. Identification of a novel macrophage population in the normal mouse corneal stroma. Invest Ophthalmol Vis Sci. 2002 Jul;43((7)):2264–71. [PMC free article] [PubMed] [Google Scholar]
- 35.Hazlett LD, Jiang X, McClellan SA. IL-10 function, regulation, and in bacterial keratitis. J Ocul Pharmacol Ther. 2014 Jun;30((5)):373–80. doi: 10.1089/jop.2014.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Condamine T, Gabrilovich DI. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. 2011 Jan;32((1)):19–25. doi: 10.1016/j.it.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bakker AB, Hoek RM, Cerwenka A, Blom B, Lucian L, McNeil T, et al. DAP12-deficient mice fail to develop autoimmunity due to impaired antigen priming. Immunity. 2000 Sep;13((3)):345–53. doi: 10.1016/s1074-7613(00)00034-0. [DOI] [PubMed] [Google Scholar]
- 38.Hamerman JA, Jarjoura JR, Humphrey MB, Nakamura MC, Seaman WE, Lanier LL. Cutting edge: inhibition of TLR and FcR responses in macrophages by triggering receptor expressed on myeloid cells (TREM)-2 and DAP12. J Immunol. 2006 Aug;177((4)):2051–5. doi: 10.4049/jimmunol.177.4.2051. [DOI] [PubMed] [Google Scholar]
- 39.Sun M, Zhu M, Chen K, Nie X, Deng Q, Hazlett LD, et al. TREM-2 promotes host resistance against Pseudomonas aeruginosa infection by suppressing corneal inflammation via a PI3K/Akt signaling pathway. Invest Ophthalmol Vis Sci. 2013 May;54((5)):3451–62. doi: 10.1167/iovs.12-10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data