Abstract
Bacterial challenge of keratinocytes with the abundant skin commensal Staphylococcus epidermidis induces distinct innate immune responses, but the underlying molecular mechanisms are still emerging. We report that the aryl hydrocarbon receptor (AhR) was activated in human primary keratinocytes infected with S. epidermidis, leading to induction of the AhR-responsive gene cytochrome P450 1A1 (CYP1A1). In addition, functional AhR was required for S. epidermidis-mediated induction of IL-1β expression in keratinocytes. AhR-dependent gene induction of IL-1β and CYP1A1 was mediated by factor(s) < 2 kDa secreted by S. epidermidis. Blockade of the AhR in a 3D organotypic skin equivalent infected with S. epidermidis attenuated the S. epidermidis-induced CYP1A1 and IL-1β expression. Moreover, S. epidermidis also induced expression of IL-1α and of the antimicrobial peptide human β-defensin-3 in an AhR-dependent manner in a 3D skin equivalent. An increased outgrowth of S. epidermidis on the surface of skin explants treated with a specific AhR inhibitor further indicate a pivotal role of the AhR in mediating an epidermal defense response. Taken together, our data expand the role of the AhR in innate immunity and support a previously unappreciated contribution for the AhR in cutaneous defense.
Keywords: Cutaneous innate defense, Aryl hydrocarbon receptor, Staphylococcus epidermidis, IL-1β
Introduction
The Gram-positive bacterium Staphylococcus epidermidis is an abundant member of the normal human skin microbiota [1]. Its presence on the skin is also associated with a risk of nosocomial S. epidermidis infections, especially infections associated with medical devices such as catheter-related infections [2]. However, there is increasing evidence that under normal conditions the presence of S. epidermidis on the skin surface seems to be favorable for the host [3] and strengthens cutaneous innate defense [4]. For example, S. epidermidis is able to dampen inflammatory processes after skin wounding [5] and it secretes antimicrobial factors, which restrict the growth of other competing microbes [3, 6]. In addition, S. epidermidis directly induces the expression of antimicrobial peptides in keratinocytes [7, 8, 9] or indirectly via S. epidermidis-specific T-cell responses [10]. Activation of dendritic cells and keratinocytes by S. epidermidis also induces distinct IL-1-mediated immune responses, indicating that IL-1 receptor targeting cytokines such as IL-1α and IL-1β are important mediators of host skin-commensal cross-talk [11, 12]. Since the molecular mechanisms underlying the S. epidermidis-mediated activation of IL-1 in keratinocytes remain elusive, the primary aim of this project was to gain more insight into the interaction of S. epidermidis and keratinocytes leading to IL-1β production. As a result, we report here a novel role of the aryl hydrocarbon receptor (AhR) in mediating a defense response in keratinocytes exposed to S. epidermidis.
The AhR was originally identified as a receptor to polycyclic aromatic hydrocarbons such as TCDD (2,3,7,8-tetrachlorodibenzo- p-dioxin) and other environmental pollutants and xenobiotics. It serves as a cytosolic ligand-activated transcription factor which, upon ligand binding, translocates into the nucleus, where it dimerizes with the AhR nuclear translocator (ARNT) followed by binding to specific sites in target gene promoters [13]. Much research in the last 15 years has revealed that the AhR is involved in various physiological aspects such as cell homeostasis, cell proliferation and differentiation, embryogenesis, carcinogenesis, inflammation, and host immunity [13, 14]. Several reports also indicate an important function of the AhR for maintaining the skin barrier [15, 16]. For example, functional AhR seems to be required for normal keratinocyte differentiation [17] and for mouse skin barrier integrity [18]. Moreover, activation of the AhR has been reported to dampen skin inflammation in psoriasis [19, 20] and to improve the skin barrier in atopic dermatitis [21]. Whereas these reports illustrate an important role of the AhR in cutaneous barrier homeostasis, the role of the AhR in cutaneous innate defense is less understood.
Here we provide evidence that S. epidermidis activates the AhR in human primary keratinocytes to induce an innate defense response. These data highlight a novel role of the AhR in cutaneous innate defense and show that the AhR is involved in sensing bacteria by keratinocytes.
Materials and Methods
Cell Culture and Stimulation
For stimulation experiments, pools of primary human keratinocytes derived from different donors (Promocell, Heidelberg, Germany) were used at passage 3–5 and cultured in 24-well plates in Keratinocyte Growth Medium 2 (KGM2) plus supplements (Promocell) without antibiotics at 37°C in a 5% CO2 atmosphere. S. epidermidis skin-derived clinical isolates (identity verified by MALDI-TOF mass spectrometry; MALDI Biotyper, Bruker, Billerica, MA, USA) and S. epidermidis ATCC 14990 were maintained on blood agar plates. For stimulation, bacteria were grown overnight at 37°C in tryptic soy broth, diluted and grown for an additional 3–4 h until the OD600 reached at least 0.2. Bacteria were harvested by centrifugation, washed twice with PBS and resuspended in cell culture medium KGM2 without supplements to the appropriate density. Confluent keratinocytes were stimulated with bacteria at a concentration of approximately 5 × 106 colony forming units (CFU) per well. Bacteria were centrifuged on cells at 350 g for 5 min followed by incubation at 37°C for the indicated time periods. For overnight stimulation, the culture medium was removed after 3 h and cells were washed twice with PBS to remove nonadherent bacteria. The cells were then incubated for an additional 17–19 h in culture medium supplemented with 200 μg/mL gentamicin. After incubation, cell supernatant was harvested for ELISA and cells were lysed for RNA isolation.
In some experiments the keratinocytes were stimulated for 20 h with culture supernatants of S. epidermidis. These supernatants were generated by incubating 8 mL (approx. 5 × 106 CFU/mL) of S. epidermidis in KGM2 in a petri dish at 37°C for 24 h. Subsequently, the bacteria were pelleted by centrifugation at 8.500 g and the remaining supernatant was sterile filtered (0.22 µm) and stored at −20°C until use. For some experiments, size fractionated supernatants were used. To this end, the supernatants were applied to 2- and 3-kDa ultrafiltration centrifugal units (Amicon/Merck, Darmstadt, Germany) and size-fractionated by centrifugation according to the supplier's protocol.
To evaluate the influence of the AhR, the specific AhR inhibitor CH223191 (10 µM, Cayman Chemical Company, Ann Arbor, MI, USA) was used. The inhibitor was applied to the medium 1 h before stimulation and remained during stimulation. DMSO (1: 6,000 diluted in medium) served as the vehicle control. To block the activity of NF-κB, we treated the cells with 5 µM of the specific NF-κB inhibitor BMS-345541 (Cayman Chemical Company) and DMSO (1: 4,000 diluted in medium) served as the vehicle control.
siRNA Experiments
Primary human keratinocytes were transfected with siRNA (5 nM) at a confluence of 40–60% using 1 µL of the transfection reagent HiPerFect (Qiagen, Hilden, Germany). After 16–20 h the medium was removed and the keratinocytes were further cultured for 3 days until stimulation with S. epidermidis. Two different AhR-specific “SilencerSelect” siRNAs (s1199 and s1200) and a nonsilencing control siRNA (4390844) were purchased from Life Technologies (Carlsbad, CA, USA).
Organotypic 3D Skin Equivalent
The organotypic 3D skin equivalent was constructed as recently described [22]. The skin equivalent was stimulated with S. epidermidis by application of approximately 2.5 × 107 CFU/mL in 20 µL of KGM2 without supplements for approximately 24 h. During stimulation the culture medium in the external well was treated with DMSO (1: 6,000 as vehicle control) or with the specific AhR inhibitor CH223191 (10 µM).
Real-Time PCR Analysis
Total RNA of the keratinocytes was isolated using the reagent Crystal RNAmagic according to the manufacturer's protocol (Biolabproducts, Gödenstorf, Germany). In total, 0.5 µg of the isolated RNA was reverse transcribed to cDNA using an oligo dT primer and 12.5 units of reverse transcriptase (PrimeScript RT Reagent Kit, TaKaRa Bio, Saint-Germain-en-Laye, France). cDNA corresponding to 10 ng total RNA served as the template in a real-time PCR. Real-time PCR was performed in a StepOne Real-Time PCR System (Applied Biosystem, Carlsbad, CA, USA) using SYBR Premix Ex Taq II (TaKaRa Bio) as previously described [23]. The following intron-spanning primers were used: IL-1β: 5′-AAG CCC TTG CTG TAG TGG TG-3′ (forward primer) and 5′-GAA GCT GAT GGC CCT AAA CA-3′ (reverse primer); CYP1A1: 5′-CAC CAT CCC CCA CAG CAC-3′ (forward primer) and 5′-ACA AAG ACA CAA CGC CCC TT-3′ (reverse primer); CYP1B1: 5′-TAT CAC TGA CAT CTT CGG CG-3′ (forward primer) and 5′-CTG CAC TCG AGT CTG CAC AT-3′ (reverse primer); IL-1α: 5′-TGT GAC TGC CCA AGA TGA AG-3′ (forward primer) and 5′-AAG TTT GGA TGG GCA ACT GA-3′ (reverse primer); hBD-3: 5′-TGT TTG CTT TGC TCT TCC TGT-3´ (forward primer) and 5′-CGC CTC TGA CTC TGC AAT AA-3′ (reverse primer); AhR: 5′-TCA GTT CTT AGG CTC AGC GTC-3′ (forward primer) and 5′-AGT TAT CCT GGC CTC CGTTT-3′ (reverse primer). Standard curves were generated for each primer set with serial dilutions of cDNA. All quantifications were normalized to the housekeeping gene RPL38 (ribosomal protein L38) using the primer pair: 5′-TCA AGG ACT TCC TGC TCA CA-3′ (forward primer) and 5′-AAA GGT ATC TGC TGC ATC GAA-3′ (reverse primer). Relative expression is given as a ratio between expression of the specific gene and RPL38 gene expression.
AhR and NF-κB Luciferase Gene Reporter Assay
To determine activation of the AhR, we used the firefly luciferase reporter plasmid pGUDLUC6.1 (a generous gift from Dr. M. Denison, U.C. Davis). The pGUDLUC6.1 plasmid is derived from the pGUDLUC1.1 plasmid [24] and contains four AhR-responsive elements (dioxin-responsive elements, DREs) and no other known regulatory elements [25]. Human primary keratinocytes were cotransfected using 1 μL of the transfection reagent Fugene HD (Promega, Madison, WI, USA) with 300 ng of pGUDLUC6.1 and 30 ng of a renilla luciferase expression plasmid (pGL4.74[hRluc/TK]; Promega), which served as an internal control. After 24 h the cells were stimulated with S. epidermidis as described above. Subsequently, keratinocytes were lysed with 150 µL of passive lysis buffer (Promega) and firefly and renilla luciferase activities were determined using the Dual Luciferase assay system (Promega) on a TD-20/20 luminometer (Promega). Luciferase activity was calculated by the amount of firefly luciferase activity normalized to the amount of renilla luciferase activity. For determination of NF-κB activity, we performed the same experiment as described above using an NF-κB firefly luciferase reporter plasmid instead of pGUDLUC6.1 [23].
ELISA
Secreted protein levels in the cell culture supernatants were measured by specific ELISA for IL-1β and pro-IL-1β (catalog No. DY-201 and DLBP00; R&D Systems, Minneapolis, MN, USA) and for IL-1α (catalog No. 445804; BioLegend, San Diego, CA, USA). ELISA was performed according to the manufacturer's protocol. The detection limit of the IL-1β, pro-IL-1β, and IL-1α ELISA were at 3.4–7.8 pg/mL, 11.6–22.2 pg/mL, and 3.4–7.8 pg/mL, respectively.
Immunostaining
For immunostaining, the organotypic 3D skin equivalent was fixed by formalin and embedded in paraffin. Staining was performed with rabbit anti-IL-1β antibody (1: 200, Bioss, Woburn, MA, USA) followed by biotinylated pig anti-rabbit IgG (1: 300; Dako Cytomation, Glostrup, Denmark) and avidin/biotinylated enzyme complex (Elite ABC complex, Vector Laboratories, Peterborough, UK) and a horseradish peroxidase substrate (HRP; NovaRed, Vector laboratorie, Burlingame, CA, USA). Slides were counterstained with hematoxylin and mounted with Eukitt (poly[butyl methacrylate-co-methyl methacrylate]; O. Kindler, Freiburg, Germany).
iGLuc Reporter System to Measure Proteolytic Cleavage of Pro-IL-1β
To analyze whether the AhR may influence proteolytic processing of pro-IL-1β, we transfected the keratinocytes with a luciferase-based inflammasome and protease activity reporter termed iGLuc [26]. This reporter, which contains Gaussia luciferase fused to a part of pro-IL-1β, releases luciferase activity in the supernatant of the keratinocytes upon proteolytic cleavage of pro-IL-1β. This plasmid (300 ng/well) was transfected in the keratinocytes 24 h before stimulation using 1 µL of Fugene HD (Promega). After stimulation, the cell culture supernatant was harvested and luciferase activity was determined on a TD-20/20 luminometer (Promega) by mixing a 4.4-µM coelenterazine (Carl Roth, Karlsruhe, Germany) solution with the cell culture supernatant at the ratio of 1: 1 (35 µL).
Ex vivo Skin Explant Model for Studying the Influence of the AhR on S. epidermidis Growth
Human skin explants were derived from abdomen or breast reduction surgeries after written informed consent was obtained. The use of skin material for this study was approved by the local ethics committee of the Medical Faculty, University of Kiel, Germany (D 414/09; D 442/16). Skin samples were washed with PBS, subcutaneous fat was carefully removed and the samples were cut in defined pieces (0.25 cm2). For stimulation a 1.5-mL microtube was filled with 400 µL of KGM2 without supplements and the specific AhR inhibitor CH223191 (10 µM) or DMSO as vehicle. Subsequently, the skin samples were placed on the medium with the epidermis facing up. Only the dermis was submerged by medium, allowing the epidermis to be exposed to an air interface similar to the in vivo situation. S. epidermidis was prepared for stimulation as described above and 20 µL of the bacteria solution (OD600 approx. 0.2) was added centrally onto the epidermis. After 5 h of incubation at 37°C in a 5% CO2 atmosphere, 600 µL of PBS was added, mixed by vortexing, and the skin was lysed by sonification on ice. Several dilutions were prepared in PBS and 50 µL were plated on tryptic soy broth agar plates. After overnight incubation at 37°C, the CFUs were counted.
Results
S. epidermidis Activates an AhR Reporter in Human Primary Keratinocytes
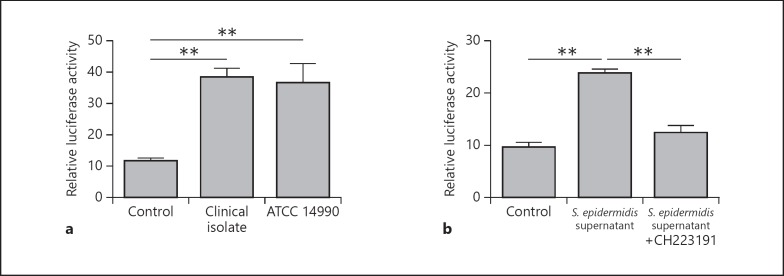
To determine whether S. epidermidis is able to activate the AhR in keratinocytes, we transfected keratinocytes with an AhR luciferase reporter plasmid. Stimulation of these cells with S. epidermidis activated the AhR as measured by induction of luciferase activity (Fig. 1a). A clinical isolate as well as an ATCC strain of S. epidermidis activated the AhR reporter, suggesting that S . epidermidis in general has the capacity to activate the AhR (Fig. 1a). Culture supernatants of S. epidermidis also induced the AhR reporter in keratinocytes, indicating that S. epidermidis secretes factor(s) activating the AhR. This activation was inhibited by the specific AhR inhibitor CH223191 (Fig. 1b).
Fig. 1.
S. epidermidis activates an AhR reporter in human keratinocytes. To investigate whether S. epidermidis is able to activate the AhR, we transfected human primary keratinocytes with an AhR firefly luciferase reporter plasmid and a renilla luciferase control plasmid. The keratinocytes were stimulated with living S. epidermidis (clinical isolate) compared to the S. epidermidis ATCC strain 14990 (a) and with culture supernatants of S. epidermidis (clinical isolate) in the presence or absence of the specific AhR inhibitor CH223191 (b). AhR activation was determined by analyzing luciferase activity which was determined as the ratio between firefly and renilla luciferase activities in each sample. Data are the means ± SEM of 3 stimulations (** p < 0.01, Student t test).
S. epidermidis Activates the AhR-Responsive Genes CYP1A1 and CYP1B1 in Human Primary Keratinocytes
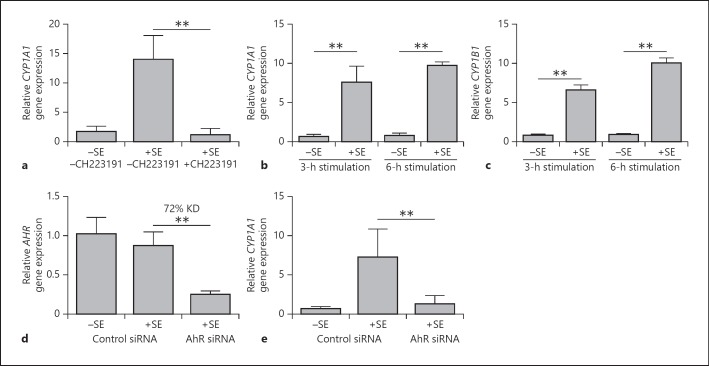
Stimulation of keratinocytes with S. epidermidis induced gene expression of the AhR-responsive gene CYP1A1 (Fig. 2a). The S. epidermidis-mediated gene induction of CYP1A1 occurred already after 3 h (Fig. 2b). Similarly, another AhR-responsive gene, CYP1B1, was also induced by S. epidermidis already after 3 h (Fig. 2c). The use of the AhR inhibitor CH223191 completely abolished the S. epidermidis-mediated CYP1A1 induction, confirming the dependency on the AhR (Fig. 2a). Accordingly, S. epidermidis-mediated induction of CYP1A1 gene expression was almost completely blocked in keratinocytes treated with an AhR-specific siRNA (Fig. 2d, e). The use of the S. epidermidis strain ATCC 14990 and another AhR-specific siRNA revealed similar results (see online suppl. Fig. 1a, b; for all online suppl. material, see www.karger.com/doi/10.1159/000492162).
Fig. 2.
S. epidermidis (SE) induces the AhR-responsive gene CYP1A1 in human keratinocytes. a Human primary keratinocytes were stimulated with a clinical isolate of SE in the presence or absence of the specific AhR inhibitor CH223191, and gene expression of CYP1A1 was determined by real-time PCR. b, c Human primary keratinocytes were stimulated for the indicated time periods with a clinical isolate of SE. Gene expression of CYP1A1 (b) and CYP1B1 (c) were determined by real-time PCR. d, e Human primary keratinocytes were transfected with a control siRNA or an AhR-specific siRNA (s1199) and stimulated with a clinical isolate of SE. Knockdown efficiency (KD) of AhR gene expression (d) and CYP1A1 gene expression (e) were analyzed by real-time PCR. Data are the means ± SEM of 6 (a, d, e) or 3 (b, c) stimulations (** p < 0.01, Student t test).
Supernatants of three different skin-derived S. epidermidis isolates induced CYP1A1 expression in primary keratinocytes, indicating that S. epidermidis in general has the capacity to activate AhR signaling in human keratinocytes (see online suppl. Fig. 2a). In addition, supernatants of S. warneri and Corynebacterium amycolatum induced CYP1A1 gene expression in primary keratinocytes, demonstrating that AhR activation is not restricted to S. epidermidis (see online suppl. Fig. 2b).
S. epidermidis Induces AhR-Dependent IL-1β Expression in Human Primary Keratinocytes
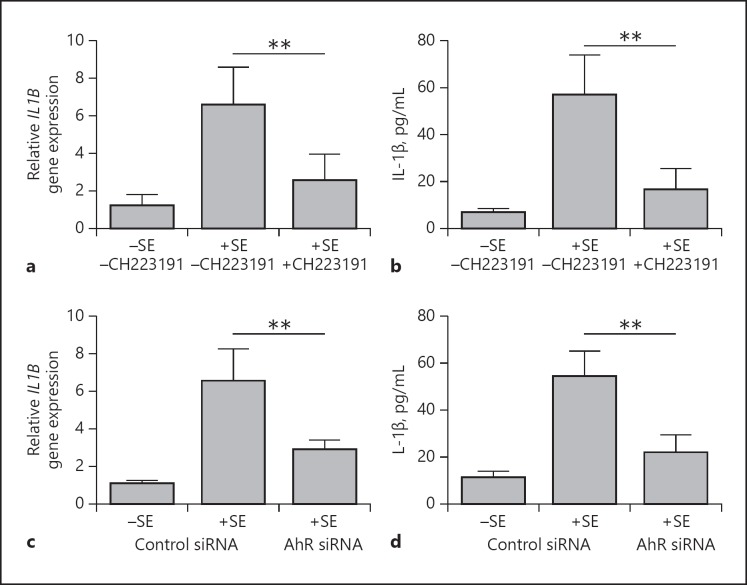
Stimulation of keratinocytes with S. epidermidis resulted in an increased induction of IL-1β gene expression and protein secretion. This induction was significantly inhibited by cotreatment with the specific AhR inhibitor CH223191 (Fig. 3a, b). Similarly, the induction of IL-1β gene expression and protein secretion were significantly inhibited after siRNA-mediated AhR knockdown indicating the participation of the AhR in the S. epidermidis-induced IL-1β expression (Fig. 3c, d, and 2d for knockdown efficiency). Similar results were obtained with another AhR-specific siRNA and S. epidermidis strain ATCC 14990 (see online suppl. Fig. 3a, b, and online suppl. Fig. 1a for knockdown efficiency). Since the IL-1β ELISA detects 10% of unprocessed pro-IL-1β, we also used an ELISA specific for pro-IL-1β. This revealed that the majority of released IL-1β in keratinocytes stimulated with S. epidermidis represents the processed mature IL-1β form (see online suppl. Fig. 4).
Fig. 3.
The AhR mediates S. epidermidis (SE)-induced IL-1β expression in human primary keratinocytes. a, b Human primary keratinocytes were stimulated with a clinical isolate of SE in the presence or absence of the specific AhR inhibitor CH223191. IL-1β gene expression and protein secretion were determined by real-time PCR (a) and ELISA (b), respective ly. c, d Human primary keratinocytes were transfected with a control siRNA and an AhR-specific siRNA (s1199) and stimulated with a clinical isolate of SE. IL-1β gene expression and protein secretion was determined by real-time PCR (c) and ELISA (d), respectively. Bars are the means ± SEM of 6 stimulations (** p < 0.01; Student t test).
Activation of the AhR by S. epidermidis Does Not Influence Processing of IL-1β or NF-κB Activity
To assess whether the AhR may directly influence proteolytic cleavage of pro-IL-1β, we transfected primary keratinocytes with a luciferase-based inflammasome and protease activity reporter plasmid (iGLuc). Treatment of these keratinocytes with living S. epidermidis induced luciferase activation, indicating proteolytic cleavage of IL-1β induced by S. epidermidis (see online suppl. Fig. 5a). This activation was not significantly influenced when the cells were cotreated with the specific AhR inhibitor CH223191 (see online suppl. Fig. 5a) although IL-1β secretion was significantly decreased by CH223191 (see online suppl. Fig. 5b). In addition, we also treated the keratinocytes with an AhR-specific siRNA, which resulted in 69% knockdown of AhR gene expression (see online suppl. Fig. 6a). Stimulation of these cells with living S. epidermidis induced less IL-1β release, whereas proteolytic cleavage of IL-1β monitored by the iGLuc reporter was not significantly affected by AhR-knockdown (see online suppl. Fig. 6b,c).
To investigate if the S. epidermidis-induced activation of the AhR influences NF-κB activity, we transfected human primary keratinocytes with an NF-κB luciferase reporter plasmid. Stimulation of these cells with living S. epidermidis induced activation of NF-κB. This activation was not influenced when the cells were cotreated with the specific AhR inhibitor CH223191 (see online suppl. Fig. 7). Since S. epidermidis induced the activation of NF-κB, we investigated whether NF-κB was involved in the S. epidermidis-mediated IL-1β induction. Blocking NF-κB diminished the induction of IL-1β gene expression in keratinocytes exposed to S. epidermidis (see online suppl. Fig. S8).
S. epidermidis Culture Supernatants Induce AhR-Dependent CYP1A1 and IL-1β Expression
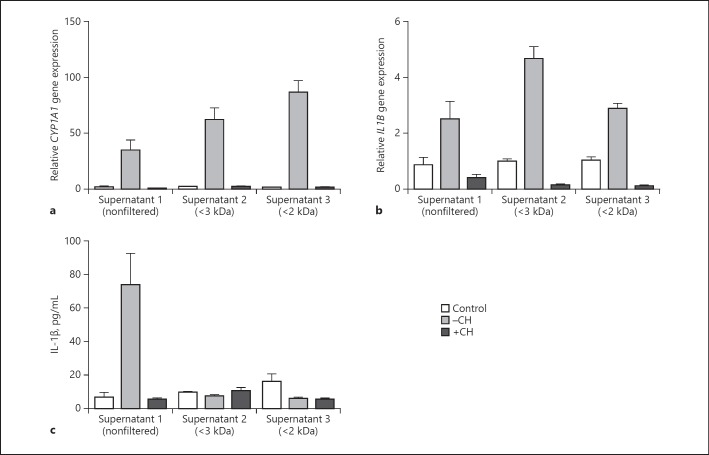
Keratinocytes were stimulated with culture supernatants of S. epidermidis in the presence of the specific AhR inhibitor CH223191 to explore whether S. epidermidis releases factors that induce CYP1A1 and IL-1β in an AhR-dependent manner. This revealed an induction of CYP1A1 gene expression that was blocked by CH223191 (Fig. 4a). Gene expression of CYP1A1 was increased after exposure to < 2- and < 3-kDa ultrafiltrates of S. epidermidis culture supernatants. This induction of CYP1A1 gene expression by the ultrafiltrates of S. epidermidis culture supernatants was blocked by CH223191 (Fig. 4a). IL-1β gene expression was also induced after exposure to nonfiltered as well as < 2- and < 3-kDa ultrafiltrates of S. epidermidis culture supernatants (Fig. 4b). This induction was blocked by the specific AhR inhibitor CH223191, indicating the involvement of the AhR in the induction of IL-1β gene expression by S. epidermidis culture supernatants. IL-1β protein release by the nonfiltered S. epidermidis culture supernatant was also increased and blocked by CH223191 (Fig. 4c). In contrast, the < 2- and < 3-kDa ultrafiltrates of S. epidermidis culture supernatants did not induce release of IL-1β (Fig. 4c).
Fig. 4.
S. epidermidis culture supernatants induce AhR-dependent CYP1A1 and IL-1β expression. Human primary keratinocytes were stimulated without (control) or with different culture supernatants of S. epidermidis, either nonfiltered supernatant (supernatant 1) or supernatant with a molecular weight < 3 kDa (supernatant 2) or < 2 kDa (supernatant 3). Stimulations were done in the absence (–CH) or presence (+CH) of the specific AhR inhibitor CH223191. a CYP1A1 gene expression was determined by real-time PCR. b, c IL-1β gene expression and protein secretion was determined by real-time PCR (b) and ELISA (c), respectively. Bars are the means ± SEM of 3 stimulations.
S. epidermidis Induces AhR-Dependent CYP1A1 and IL-1β Expression in an Organotypic Skin Equivalent
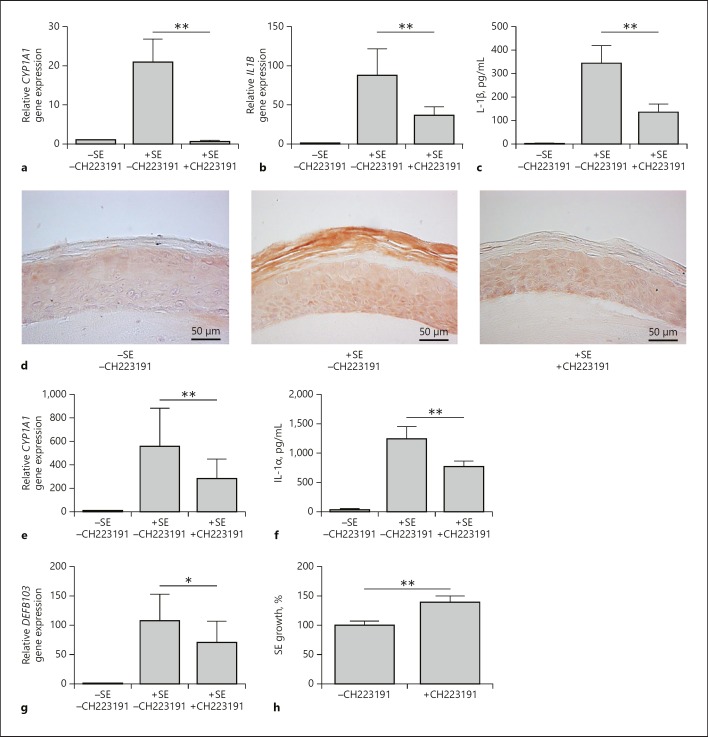
Stimulation of an organotypic 3D skin equivalent with S. epidermidis in the presence of CH223191 revealed an AhR-dependent induction of CYP1A1 and IL-1β (Fig. 5a–c). In concordance, IL-1β immunostaining of the 3D skin equivalent exposed to S. epidermidis showed an intensified IL-1β immunoreactivity, which was reduced by cotreatment with CH223191 (Fig. 5d).
Fig. 5.
The AhR mediates activation of skin innate defense by S. epidermidis (SE). An organotypic 3D skin equivalent was stimulated with living SE for 24 h in the presence or absence of the AhR inhibitor CH223191 (10 µM). Gene expression of CYP1A1 (a) and IL-1β (b) was analyzed by real-time PCR. c Secretion of IL-1β into the medium was determined by ELISA. d Protein expression of IL-1β was analyzed by immunohistochemistry. e Gene expression of IL-1α was analyzed by real-time PCR. f Secretion of IL-1α into the medium was investigated by ELISA. g Gene expression of hBD-3 was analyzed by real-time PCR. Data are the means ± SEM of 7 different skin models (* p < 0.05; ** p < 0.01; Student ratio paired t test). h To assess the functional relevance of the AhR to control the growth of SE on the human skin surface, skin explants were exposed to living SE for 5 h in the presence or absence of the specific AhR inhibitor CH223191. The amounts of SE in the presence of the AhR inhibitor CH223191 were compared to the amounts of SE on the skin without treatment, which were set as 100%. Bars are the means ± SEM of 9 skin explants derived from 3 different donors (** p < 0.01; Student t test).
We also investigated whether other defense molecules might be induced by S. epidermidis in the 3D skin equivalent in an AhR-dependent manner. We found that the S. epidermidis-mediated induction of IL-1α (Fig. 5e, f) and of the antimicrobial peptide hBD-3 (Fig. 5g) were partially blocked after inhibition of the AhR.
The AhR Contributes to Controlling the Growth of S. epidermidis on Human Skin
To assess the functional role of the AhR on the interaction between S. epidermidis and skin, we investigated if inactivation of the AhR may influence the outgrowth of S. epidermidis on the skin surface of human skin explants. To this end, we used an ex vivo model where we applied living S. epidermidis for 5 h on the surface of human skin explants. Inactivation of the AhR by CH223191 led to a significant outgrowth of S. epidermidis as compared to skin treated with vehicle only (Fig. 5h). These results indicate a functional relevance of the AhR to controlling the growth of S. epidermidis on human skin.
Discussion
S. epidermidis is an abundant member of the human cutaneous microbiota and has been shown to play an important role in skin defense through its activation of cutaneous defense mechanisms [10, 12]. It has been reported that induction of cutaneous defense can be mediated by S. epidermidis-induced IL-1 signaling, which promotes protective effector T-cell responses [12]. However, the direct S. epidermidis-mediated induction of IL-1 in keratinocytes is less explored. Our results provide evidence that, besides NF-κB, the AhR is crucial for the S. epidermidis-mediated gene induction of IL-1β in keratinocytes.
The role of the AhR in skin innate immunity is not well understood. It has been shown that Malassezia yeasts are able to secrete various indole derivatives as putative AhR ligands that induced the AhR-responsive genes CYP1A1 and CYP1B1 in HaCaT keratinocytes [27]. Nonetheless, it has not been reported whether the AhR plays a role in human keratinocytes in the interaction between keratinocytes and bacteria. Recent research demonstrated that the AhR is able to sense pigmented virulence factors produced by the pulmonary pathogens Pseudomonas aeruginosa and Mycobacterium tuberculosis, thus highlighting a novel role of the AhR as a pattern recognition receptor [28]. Our data indicate that S. epidermidis releases small (< 2 kDa) factor(s) that activate the AhR in keratinocytes. Since the known AhR activators are typically derived from small aromatic hydrocarbons, it is likely that S. epidermidis releases similar compounds. In addition, it remains to be shown whether the factor(s) released by S. epidermidis directly bind to the AhR or increase the amount of endogenous AhR ligands, such as 6-formylindolo[3, 2-b]carbazole (FICZ) through inhibition of CYP450 enzymes [29, 30, 31].
There are contradictory studies reporting either a pro- or an anti-inflammatory role of the AhR. Our study demonstrates that antagonizing the AhR leads to a reduced expression of IL-1β in keratinocytes exposed to S. epidermidis. In addition, it has been reported that transgenic mice expressing the constitutive active form of the AhR in keratinocytes showed an increased expression of various cytokines, including IL-1β [32]. Iyer et al. [33] reported that AhR activation in intestinal epithelial cells by oxazole compounds induced inflammation. In line with a proinflammatory role of the AhR, the induction of IL-1β in leukocytes derived from bronchoalveolar lavage fluid of P. aeruginosa-infected mice was reduced in AhR-deficient cells [28]. On the other hand, other studies reported that AhR-deficient cells showed an exacerbated expression of cytokines in reaction to inflammatory stimuli. For example, mouse macrophages treated with AhR-siRNA expressed more IL-1β mRNA upon LPS stimulation [34]. Moreover, in mouse skin, the lack of AhR caused hyperinflammation, and AhR signaling exerted an anti-inflammatory effect in keratinocytes [19]. These contrasting outcomes of AhR activation may be related to the fact that AhR ligands may serve as activators or suppressors of subsequent signal transduction pathways depending on the nature and concentration of the ligands and the cell-specific environment [13, 35]. In this context, it has been reported that different tryptophan-derived metabolites may act as AhR agonists and antagonists [36]. Furthermore, isoflavones have been identified as AhR agonists and antagonists depending on the species and cell-specific context [37]. The recent demonstration that indole and 3-methyl indole are potent ligands and activators of the human AhR, but only weak activators of the mouse AhR, highlights the importance of also considering potential differences between activation of the AhR in humans and mice [38].
Recently, it has been reported that the inflammasome activation in mouse macrophages by LPS in combination with nigericin, ATP, or alum was inhibited by activation of the AhR through the dioxin-derivative TCDD [34]. Therefore, we sought to determine whether the inflammasome is affected by AhR in our experimental system. To this end, we transfected the keratinocytes with a luciferase-based inflammasome and protease activity reporter (iGLuc). This reporter indicates proteolytic processing of IL-1β [26]. Blocking AhR activity by the use of the specific AhR inhibitor CH223191 as well as by AhR-specific siRNA did not significantly affect S. epidermidis-induced iGLuc activity. These data indicate that the AhR has no direct influence on proteolytic processing of pro-IL-1β in the keratinocytes stimulated with S. epidermidis.
As S. epidermidis is able to activate the transcription factor NF-κB and because a cross-talk of the AhR and NF-κB has been described [39], we investigated whether the S. epidermidis-mediated AhR activation may influence NF-κB. Although we observed that AhR activation in keratinocytes by S. epidermidis did not directly influence NF-κB activation, we found that activation of both transcription factors, NF-κB and AhR, were crucial for the S. epidermidis-mediated induction of IL-1β in keratinocytes. This is in concordance with the necessity of NF-κB and AhR for IL-1β expression in TLR- and M. tuberculosis-activated dendritic cells and macrophages [40, 41].
In addition to IL-1β, we also found that other S. epidermidis-induced defense molecules are regulated via the AhR. In particular, the S. epidermidis-induced expression of IL-1α and the antimicrobial peptide hBD-3 in the 3D organotypic skin depended on the AhR. These data indicate that the AhR mediates the expression of diverse innate defense molecules in keratinocytes, thus highlighting an important role of the AhR in innate cutaneous defense. This is supported by the observed outgrowth of S. epidermidis on the surface of skin explants treated with an AhR inhibitor.
In conclusion, our study provides novel insight into the molecular interaction between the skin commensal S. epidermidis and keratinocytes, and further strengthens the hypothesis that the AhR evolved not only to sense environmental toxins, but also to participate in host-microbe interactions. Understanding how commensals modulate host defense may provide innovative strategies for the treatment or prophylaxis of infectious and inflammatory diseases.
Disclosure Statement
The authors declare no conflicts of interest.
Supplementary Material
Supplementary data
Acknowledgments
We would like to thank Heilwig Hinrichs and Cornelia Wilgus for excellent technical assistance. We thank Dr. M. S. Denison (University of California, Davis, CA, USA) for his generous gift of the pGUDLUC6.1 vector, and Prof. Dr. Veit Hornung for providing the iGLuc plasmid. We thank Prof. R. Podschun (Institute for Infection Medicine, Kiel, Germany) for his help to verify the identity of the bacteria by MS-analyses. We thank Doris Scharinger for editorial help with the manuscript. This study was supported by grants from the German Research Foundation given to J. Harder (HA 3386/5-1/-2) and in parts by funding of the medical faculty of the University of Kiel.
References
- 1.Kloos WE, Musselwhite MS. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl Microbiol. 1975;30:381–385. doi: 10.1128/am.30.3.381-395.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Otto M. Molecular basis of Staphylococcus epidermidis infections. Semin Immunopathol. 2012;34:201–214. doi: 10.1007/s00281-011-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen GJ, Bruggemann H. Bacterial skin commensals and their role as host guardians. Benef Microbes. 2014;5:201–215. doi: 10.3920/BM2012.0062. [DOI] [PubMed] [Google Scholar]
- 4.Gallo RL, Nakatsuji T. Microbial symbiosis with the innate immune defense system of the skin. J Invest Dermatol. 2011;131:1974–1980. doi: 10.1038/jid.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai Y, Di Nardo A, Nakatsuji T, Leichtle A, Yang Y, Cogen AL, Wu ZR, Hooper LV, Schmidt RR, von Aulock S, Radek KA, Huang CM, Ryan AF, Gallo RL. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med. 2009;15:1377–1382. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, Shafiq F, Kotol PF, Bouslimani A, Melnik AV, Latif H, Kim JN, Lockhart A, Artis K, David G, Taylor P, Streib J, Dorrestein PC, Grier A, Gill SR, Zengler K, Hata TR, Leung DY, Gallo RL. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. 2017;9:4680. doi: 10.1126/scitranslmed.aah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li D, Lei H, Li Z, Li H, Wang Y, Lai Y. A novel lipopeptide from skin commensal activates TLR2/CD36-p38 MAPK signaling to increase antibacterial defense against bacterial infection. PLoS One. 2013;8:e58288. doi: 10.1371/journal.pone.0058288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ommori R, Ouji N, Mizuno F, Kita E, Ikada Y, Asada H. Selective induction of antimicrobial peptides from keratinocytes by staphylococcal bacteria. Microb Pathog. 2013;56:35–39. doi: 10.1016/j.micpath.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Wanke I, Steffen H, Christ C, Krismer B, Gotz F, Peschel A, Schaller M, Schittek B. Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J Invest Dermatol. 2011;131:382–390. doi: 10.1038/jid.2010.328. [DOI] [PubMed] [Google Scholar]
- 10.Naik S, Bouladoux N, Linehan JL, Han SJ, Harrison OJ, Wilhelm C, Conlan S, Himmelfarb S, Byrd AL, Deming C, Quinones M, Brenchley JM, Kong HH, Tussiwand R, Murphy KM, Merad M, Segre JA, Belkaid Y. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520:104–108. doi: 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chng KR, Tay AS, Li C, Ng AH, Wang J, Suri BK, Matta SA, McGovern N, Janela B, Wong XF, Sio YY, Au BV, Wilm A, De Sessions PF, Lim TC, Tang MB, Ginhoux F, Connolly JE, Lane EB, Chew FT, Common JE, Nagarajan N. Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare. Nat Microbiol. 2016;1:16106. doi: 10.1038/nmicrobiol.2016.106. [DOI] [PubMed] [Google Scholar]
- 12.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, Spencer S, Hall JA, Dzutsev A, Kong H, Campbell DJ, Trinchieri G, Segre JA, Belkaid Y. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol. 2014;32:403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 14.Noakes R. The aryl hydrocarbon receptor: a review of its role in the physiology and pathology of the integument and its relationship to the tryptophan metabolism. Int J Tryptophan Res. 2015;8:7–18. doi: 10.4137/IJTR.S19985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esser C, Rannug A. The aryl hydrocarbon receptor in barrier organ physiology, immunology, and toxicology. Pharmacol Rev. 2015;67:259–279. doi: 10.1124/pr.114.009001. [DOI] [PubMed] [Google Scholar]
- 16.Furue M, Takahara M, Nakahara T, Uchi H. Role of AhR/ARNT system in skin homeostasis. Arch Dermatol Res. 2014;306:769–779. doi: 10.1007/s00403-014-1481-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Bogaard EH, Podolsky MA, Smits JP, Cui X, John C, Gowda K, Desai D, Amin SG, Schalkwijk J, Perdew GH, Glick AB. Genetic and pharmacological analysis identifies a physiological role for the AHR in epidermal differentiation. J Invest Dermatol. 2015;135:1320–1328. doi: 10.1038/jid.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haas K, Weighardt H, Deenen R, Kohrer K, Clausen B, Zahner S, Boukamp P, Bloch W, Krutmann J, Esser C. Aryl hydrocarbon receptor in keratinocytes is essential for murine skin barrier integrity. J Invest Dermatol. 2016;136:2260–2269. doi: 10.1016/j.jid.2016.06.627. [DOI] [PubMed] [Google Scholar]
- 19.Di Meglio P, Duarte JH, Ahlfors H, Owens ND, Li Y, Villanova F, Tosi I, Hirota K, Nestle FO, Mrowietz U, Gilchrist MJ, Stockinger B. Activation of the aryl hydrocarbon receptor dampens the severity of inflammatory skin conditions. Immunity. 2014;40:989–1001. doi: 10.1016/j.immuni.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith SH, Jayawickreme C, Rickard DJ, Nicodeme E, Bui T, Simmons C, Coquery CM, Neil J, Pryor WM, Mayhew D, Rajpal DK, Creech K, Furst S, Lee J, Wu D, Rastinejad F, Willson TM, Viviani F, Morris DC, Moore JT, Cote-Sierra J. Tapinarof is a natural AhR agonist that resolves skin inflammation in mice and humans. J Invest Dermatol. 2017;137:2110–2119. doi: 10.1016/j.jid.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 21.van den Bogaard EH, Bergboer JG, Vonk-Bergers M, van Vlijmen-Willems IM, Hato SV, van der Valk PG, Schroder JM, Joosten I, Zeeuwen PL, Schalkwijk J. Coal tar induces AHR-dependent skin barrier repair in atopic dermatitis. J Clin Invest. 2013;123:917–927. doi: 10.1172/JCI65642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rademacher F, Simanski M, Schröder L, Mildner M, Harder J. The role of RNase 7 in innate cutaneous defense against Pseudomonas aeruginosa. Exp Dermatol. 2017;26:227–233. doi: 10.1111/exd.13166. [DOI] [PubMed] [Google Scholar]
- 23.Roth SA, Simanski M, Rademacher F, Schröder L, Harder J. The pattern recognition receptor NOD2 mediates Staphylococcus aureus-induced IL-17C expression in keratinocytes. J Invest Dermatol. 2014;134:374–380. doi: 10.1038/jid.2013.313. [DOI] [PubMed] [Google Scholar]
- 24.Garrison PM, Tullis K, Aarts JM, Brouwer A, Giesy JP, Denison MS. Species-specific recombinant cell lines as bioassay systems for the detection of 2,3,7,8-tetrachlorodibenzo-p-dioxin-like chemicals. Fundam Appl Toxicol. 1996;30:194–203. doi: 10.1006/faat.1996.0056. [DOI] [PubMed] [Google Scholar]
- 25.Long WP, Pray-Grant M, Tsai JC, Perdew GH. Protein kinase C activity is required for aryl hydrocarbon receptor pathway-mediated signal transduction. Mol Pharmacol. 1998;53:691–700. doi: 10.1124/mol.53.4.691. [DOI] [PubMed] [Google Scholar]
- 26.Bartok E, Bauernfeind F, Khaminets MG, Jakobs C, Monks B, Fitzgerald KA, Latz E, Hornung V. iGLuc: a luciferase-based inflammasome and protease activity reporter. Nat Methods. 2013;10:147–154. doi: 10.1038/nmeth.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magiatis P, Pappas P, Gaitanis G, Mexia N, Melliou E, Galanou M, Vlachos C, Stathopoulou K, Skaltsounis AL, Marselos M, Velegra ki A, Denison MS, Bassukas ID. Malassezia yeasts produce a collection of exceptionally potent activators of the Ah (dioxin) receptor detected in diseased human skin. J Invest Dermatol. 2013;133:2023–2030. doi: 10.1038/jid.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moura-Alves P, Fae K, Houthuys E, Dorhoi A, Kreuchwig A, Furkert J, Barison N, Diehl A, Munder A, Constant P, Skrahina T, Guhlich-Bornhof U, Klemm M, Koehler AB, Bandermann S, Goosmann C, Mollenkopf HJ, Hurwitz R, Brinkmann V, Fillatreau S, Daffe M, Tummler B, Kolbe M, Oschkinat H, Krause G, Kaufmann SH. AhR sensing of bacterial pigments regulates antibacterial defence. Nature. 2014;512:387–392. doi: 10.1038/nature13684. [DOI] [PubMed] [Google Scholar]
- 29.Schiering C, Wincent E, Metidji A, Iseppon A, Li Y, Potocnik AJ, Omenetti S, Henderson CJ, Wolf CR, Nebert DW, Stockinger B. Feedback control of AHR signalling regulates intestinal immunity. Nature. 2017;542:242–245. doi: 10.1038/nature21080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smirnova A, Wincent E, Vikstrom Bergander L, Alsberg T, Bergman J, Rannug A, Rannug U. Evidence for new light-independent pathways for generation of the endogenous aryl hydrocarbon receptor agonist FICZ. Chem Res Toxicol. 2016;29:75–86. doi: 10.1021/acs.chemrestox.5b00416. [DOI] [PubMed] [Google Scholar]
- 31.Wincent E, Bengtsson J, Mohammadi Bardbori A, Alsberg T, Luecke S, Rannug U, Rannug A. Inhibition of cytochrome P4501-dependent clearance of the endogenous agonist FICZ as a mechanism for activation of the aryl hydrocarbon receptor. Proc Natl Acad Sci USA. 2012;109:4479–4484. doi: 10.1073/pnas.1118467109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tauchi M, Hida A, Negishi T, Katsuoka F, Noda S, Mimura J, Hosoya T, Yanaka A, Aburatani H, Fujii-Kuriyama Y, Motohashi H, Yamamoto M. Constitutive expression of aryl hydrocarbon receptor in keratinocytes causes inflammatory skin lesions. Mol Cell Biol. 2005;25:9360–9368. doi: 10.1128/MCB.25.21.9360-9368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iyer SS, Gensollen T, Gandhi A, Oh SF, Neves JF, Collin F, Lavin R, Serra C, Glickman J, de Silva PSA, Sartor RB, Besra G, Hauser R, Maxwell A, Llebaria A, Blumberg RS. Dietary and microbial oxazoles induce intestinal inflammation by modulating aryl hydrocarbon receptor responses. Cell. 2018;173:1123–1134. doi: 10.1016/j.cell.2018.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huai W, Zhao R, Song H, Zhao J, Zhang L, Zhang L, Gao C, Han L, Zhao W. Aryl hydrocarbon receptor negatively regulates NLRP3 inflammasome activity by inhibiting NLRP3 transcription. Nat Commun. 2014;5:4738. doi: 10.1038/ncomms5738. [DOI] [PubMed] [Google Scholar]
- 35.Julliard W, Fechner JH, Mezrich JD. The aryl hydrocarbon receptor meets immunology: friend or foe? A little of both. Front Immunol. 2014;5:458. doi: 10.3389/fimmu.2014.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin UH, Lee SO, Sridharan G, Lee K, Davidson LA, Jayaraman A, Chapkin RS, Alaniz R, Safe S. Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol Pharmacol. 2014;85:777–788. doi: 10.1124/mol.113.091165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wall RJ, He G, Denison MS, Congiu C, Onnis V, Fernandes A, Bell DR, Rose M, Rowlands JC, Balboni G, Mellor IR. Novel 2-amino-isoflavones exhibit aryl hydrocarbon receptor agonist or antagonist activity in a species/cell-specific context. Toxicology. 2012;297:26–33. doi: 10.1016/j.tox.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hubbard TD, Murray IA, Bisson WH, Lahoti TS, Gowda K, Amin SG, Patterson AD, Perdew GH. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci Rep. 2015;5:12689. doi: 10.1038/srep12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogel CF, Sciullo E, Li W, Wong P, Lazennec G, Matsumura F. RelB, a new partner of aryl hydrocarbon receptor-mediated transcription. Mol Endocrinol. 2007;21:2941–2955. doi: 10.1210/me.2007-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kado S, Chang WL, Chi AN, Wolny M, Shepherd DM, Vogel CF. Aryl hydrocarbon receptor signaling modifies Toll-like receptor-regulated responses in human dendritic cells. Arch Toxicol. 2017;91:2209–2221. doi: 10.1007/s00204-016-1880-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Memari B, Bouttier M, Dimitrov V, Ouellette M, Behr MA, Fritz JH, White JH. Engagement of the aryl hydrocarbon receptor in Mycobacterium tuberculosis-infected macrophages has pleiotropic effects on innate immune signaling. J Immunol. 2015;195:4479–4491. doi: 10.4049/jimmunol.1501141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data