Abstract
Bacterial biofilms constitute a critical problem in hospitals, especially in resuscitation units or for immunocompromised patients, since bacteria embedded in their own matrix are not only protected against antibiotics but also develop resistant variant strains. In the last decade, an original approach to prevent biofilm formation has consisted of studying the antibacterial potential of host communication molecules. Thus, some of these compounds have been identified for their ability to modify the biofilm formation of both Gram-negative and Gram-positive bacteria. In addition to their effect on biofilm production, a detailed study of the mechanism of action of these human hormones on bacterial physiology has allowed the identification of new bacterial pathways involved in biofilm formation. In this review, we focus on the impact of neuropeptidic hormones on bacteria, address some future therapeutic issues, and provide a new view of inter-kingdom communication.
Keywords: Hormones, Peptides, Biofilm, Virulence, Mechanism of action, Bacterial sensor, Pseudomonas aeruginosa, Staphylococci
Introduction
Biofilms are defined as complex microbial communities of cells that are attached to a substratum (biotic or abiotic), an interface, or to each other and embedded into a matrix of self-produced extracellular polymeric compounds [1]. Biofilm formation endows bacteria with a high tolerance to antimicrobials and host immune defense mechanisms, thus enabling the pathogens to survive in hostile environments and disperse and colonize new niches. In addition, a small percentage of persister cells developing within the biofilm is known to be highly tolerant to antibiotics and has typically been involved in causing the relapse of infections [2]. The medical consequence for the infected host is the persistency of chronic infections in organs in which the biofilm is established [3]. While the molecular mechanisms that allow Gram-negative bacteria to resist to antibiotics within biofilms are well documented, this is not yet the case for Gram-positive bacteria [4, 5]. Based on naturally occurring antimicrobial and host defense peptides, the design of small synthetic peptides with broad-spectrum antibiofilm activity has provided a glimmer of hope in the fight against pan-antibiotic-resistant bacteria, the so-called superbugs [6, 7], providing a really promising start for the development of new antibacterial agents [8]. These peptides have been tested either alone [9] or in association with antibiotics since they could act as biofilm-dispersive agents to improve the action of antibiotics [10]. Some of these peptides, whether endogenous or synthetic (i.e., LL-37 and peptide 1018), do not require a specific binding to a bacterial target such as a sensor protein [9]. However, as for antibiotics, some cationic antimicrobial peptides (AMP) such as LL-37 or magainin II induce the emergence of bacterial resistance often through the activation of 2-component systems (TCS) [11, 12, 13].
In the search for new strategies or bacterial targets to counteract bacterial infection and/or biofilm formation, the potential as antimicrobials of eukaryotic communication factors released constantly by host cells (hormones and cytokines) during a surgery (norepinephrine) [14] or during infections (natriuretic peptides, substance P [SP], or neuropeptide Y) [15, 16, 17] has emerged. This concept originated in 1930, when it was suspected that epinephrine could favor bacterial infections [18]. This hypothesis was validated by the studies of Lyte and Ernst [19], which showed that epinephrine and more generally catecholamines enhance bacterial virulence. This work opened a new research field named “microbial endocrinology” [20, 21] that refers to bacterial sensitivity to host hormones. The discovery that the bacterial response to epinephrine and norepinephrine was not only restricted to Escherichia coli [22] suggested that this phenomenon was widely encountered in host-bacteria relationships. In addition, since in humans these catecholamines act through specific eukaryotic receptors, it was tempting to explore the possibility that the bacterial sensitivity to these hormones was relayed via specific bacterial sensors. In this context, a crucial step in the comprehension of this mechanism was achieved by the studies of Sperandio et al. [23] and Clarke et al. [24], since they identified QseC as the bacterial sensor for catecholamines. In addition, this sensor further appeared to be widespread since homologs of E. coli QseC have been referenced in 24 bacterial species [25]. Interestingly, it was observed that bacteria expressed several subtypes of catecholamine sensors [26], as in human cells where numerous receptor subtypes exist. In parallel to the characterization of catecholamine effects on bacteria, several studies have focused on the impact of eukaryotic communication compounds (i.e., neurotransmitters, cytokines, and hormones) and a clear consensus emerged that numerous eukaryotic molecules have a physiological effect on a large variety of bacteria [27, 28] and that these activities are quite often dependent on binding to a bacterial sensor. For instance, it was shown that interferon-γ led to the induction of the Pseudomonas aeruginosa lectin PA-I via activation of the quorum-sensing network that is required for the production of virulence factors [29]. Noticeably, this effect was shown to be triggered by direct binding of interferon-γ on OprF [29], which is a multifunctional outer membrane porin [30] that is required for P. aeruginosa virulence regulation [31] and involved in regulation of biofilm formation [32]. It has also been observed that dynorphin A, an endogenous opioid, modifies P. aeruginosa physiology [33]. It is interesting to mention that opioid receptor agonists are able to mimic the effect of dynorphin A on P. aeruginosa, especially by enhancing bacterial adhesion properties [33], suggesting that the effect of this human peptide is relayed by a bacterial sensor showing a pharmacological profile similar to that of its eukaryotic counterpart [33]. In the same vein, it was shown that morphine can trigger P. aeruginosa virulence [34]. This seemingly exotic idea that bacteria would express sensor proteins presenting pharmacological activities similar to that of the eukaryote receptors was already suggested for a human hormone by Yamashita et al. [35]. In this case, somatostatin, which binds to receptors localized on the eukaryotic cell membrane that are coupled to adenylate or guanylate cyclases [36], is able to modify Helicobacter pylori physiology by enhancing the cAMP (cyclic adenosine monophosphate) and/or cGMP (cyclic guanosine monophosphate) intrabacterial concentration [35]. This concept, in which human hormones may have an impact on bacteria through bacterial cyclases, was also verified in Pseudomonas species. Indeed, natriuretic peptides were shown to modify the virulence of P. aeruginosa [37] and P. fluorescens [38] after modulation of cAMP or cGMP intrabacterial concentration.
The data presented above show that the activation of bacterial sensors by host communication compounds results in the modulation of bacterial virulence, adhesion, and/or biofilm formation. Next to these physiological observations, identification of the putative bacterial sensors for these human hormones and the deciphering of the mechanisms of action of these hormones on bacteria could serve the scientific community to identify new bacterial targets to counteract bacterial virulence and biofilm formation. The originality of this review, which is focused on the neuropeptide hormone family, lies in the fact that it describes not only the impact of these compounds on biofilm formation and on virulence regulation but also the corresponding bacterial sensors and the mechanisms of action triggered by the binding of neuropeptide hormones on their target(s). This review will begin with the sensitivity of Gram-negative bacteria to host signal compounds and it will continue with the response of Gram-positive bacteria.
Effects of Peptidic Hormones on Gram-Negative Bacteria
Dynorphins and P. aeruginosa
Hormone Presentation
Dynorphins are involved among others in pain and stress responses after binding to the κ-opioid receptor family. Dynorphin A (1–17), dynorphin B (1–13) (Table 1), and α-neoendorphin are generated from the common precursor prodynorphin by proteolytic cleavage and share a highly conserved N-terminal sequence and charge distribution [39]. Since opioids have been shown to accumulate during inflammation [40], this highlights that bacteria can be exposed to dynorphins during infection.
Table 1.
List of the eukaryotic peptide hormones that have an effect on bacteria
| Peptides | Sequence and structure | UniProt/PubChem (CID)a accession numbers | Organs and cells that produce the peptide hormone | Bacteria encountered in the producing organ | Bacteria susceptible to the peptide (molecular target) | Key references |
|---|---|---|---|---|---|---|
|
|
P01213/13133805/25081093 | Cerebellumb Lymphoid nodeb Skeletal muscleb Gall bladderb Pancreasb Oral mucosab Stomachb Intestineb Kidneyb Testisb Placentab |
Bacteria from the oral microbiota | P. aeruginosa (MvfR or ParS) | 33, 39, 46 |
|
|
P61278/16129681/16133849 | Hypothalamusb Cerebral cortexb Adrenal cellsb Pancreasb Stomachb Duodenumb Small intestineb Colonb Rectumb |
Bacteria from the intestinal microbiota | H. pylori (SSTR-2-like protein) | 35, 51 |
|
|
P01160 (human)/ 16166338 (rat) | Heartb Skin | Bacteria involved in endocarditis Bacteria from the skin microbiota |
S. epidermidis (molecular target not yet identified) S. aureus (molecular target not yet identified) |
110, 117–119 |
|
|
P16860/71308561 | Heart Skin | Bacteria involved in endocarditis | P. aeruginosa (molecular target not yet identified) | 37, 117,118 |
|
|
P23582/16179407 | Prostate (RNA) Retina (RNA) Testis (RNA) Brain Bone Testis Granulosa cells Thyroid Thymus Skin Endothelial cells |
P. aeruginosa S. aureus Bacteria from the skin microbiota |
P. aeruginosa (AmiC) S. epidermidis (molecular target not yet identified) S. aureus (molecular target not yet identified) |
37, 68, 69, 77, 110, 117, 118, 120–123 |
|
|
P20366/36511 | Cerebral cortexb Thyroid glandb Testisb Adrenal glandb Spleenb Bronchusb Nasopharynxb Pancreasb Salivary glandb Stomachb Intestineb Skinb |
Bacteria from the skin microbiota |
B. cereus (EfTu) S. epidermidis (EfTu) S. aureus (EfTu) P. fluorescens (molecular target not yet identified) |
83, 88, 97, 99, 124 |
|
|
P06881/56841902 | Thyroid glandb Skin |
Bacteria from the skin microbiota | S. epidermidis (DnaK) | 83, 106, 109, 124 |
The amino sequence and the 2-D structure of each peptide, the UniProt accession number, and the PubChem accession number of the corresponding peptide, the organs and cells that produce the peptide (protein form or RNA), the family of bacteria encountered in the organs producing the corresponding peptide hormone, and the bacteria sensitive to the corresponding peptide and the bacterial molecular targets are shown.
Data retrieved from the National Center for Biotechnology Information PubChem Compound Database (https://pubchem.ncbi.nlm.nih.gov, accessed September 3, 2018) [125].
Data retrieved from the Human Protein Atlas (https://www.proteinatlas.org/, accessed September 3, 2018) [126].
Impact of Dynorphins on P. aeruginosa
It has been shown that P. aeruginosa virulence increases after exposure to opioid agonists or dynorphins [33]. Indeed, the contact of P. aeruginosa with these compounds results in activation of the Pseudomonas quinolone signal (PQS) quorum-sensing system, pyocyanin (PCN) production, and consequently activation of bacterial virulence. More interestingly, exposure of P. aeruginosa to various concentrations of dynorphin A (100–400 µM) resulted in a dose-dependent effect on PCN production [33]. The biological relevance of this finding was further highlighted using a Caenorhabditis elegans nematode infection model, which showed enhanced virulence when P. aeruginosa cells were treated with dynorphins. Since the 3 quorum-sensing-related signal compounds PQS, 4-hydroxy-2-heptylquinoline (HHQ), and 4-hydroxy-2-heptylquinoline- N-oxide (HQNO) were overproduced, the authors investigated the levels of PQS-regulated PA-I lectin mRNA [41, 42]. Since the authors observed that the synthetic ĸ-agonist U-50,488 mimicked the dynorphin effects on P. aeruginosa PCN and quorum-sensing quinolone signaling molecule production and on expression of the pqsABCDE operon, it has been speculated that, like U-50,488, dynorphins would increase the expression of lectin PA-I in P. aeruginosa by increasing the PQS level. This is underlined by the facts that: (1) the productions of lectin PA-I and PCN were described to be very similarly regulated, (2) PQS was previously shown to enhance the attachment of P. aeruginosa to stainless steel coupons [42], and (3) lectin PA-I is involved in attachment to surfaces and subsequent biofilm formation [43]. The effect of dynorphins was also assayed on preformed biofilm, but neither induction of rhamnolipids nor induction of the PQS system could be observed in this condition [44].
Hormone Bacterial Targets and Mechanism of Action
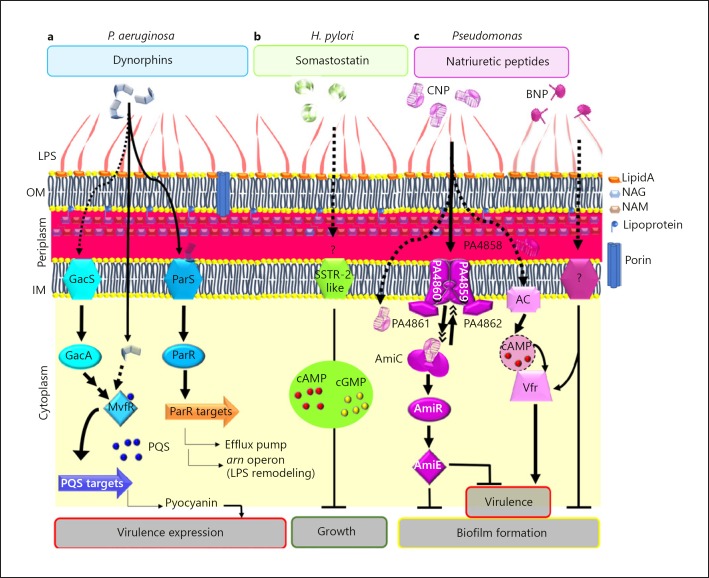
The virulence and PQS production regulator MvfR was first suggested to be the bacterial sensor for dynorphins [33] (Fig. 1). This hypothesis is backed by the observation that MvfR is associated with the cytoplasmic membrane during the exponential phase of growth before being released into the cytoplasm by proteolytic degradation [45]. Recently, the effect of dynorphins on P. aeruginosa was suggested to be triggered by the TCS ParRS rather than by MvfR, independently of the quorum-sensing system [46]. Interestingly, ParRS TCS has been involved in AMP resistance [47] since ParRS was shown to be required for induction of the lipopolysaccharide (LPS) remodeling arn operon by the cationic cyclolipopeptide polymyxin B [47]. Two other mechanisms of multidrug resistance are attributed to the ParRS system: overproduction of the MexXY efflux pump and downregulation of the porin oprD gene, resulting in an active efflux of antibiotics and a decreased membrane permeability to imipenem, respectively [48] (Fig. 1). Since dynorphin was shown to bind directly the inner membrane (IM) sensor ParS leading to activation of the cytoplasmic regulator ParR, these data suggest that P. aeruginosa could perceive this host factor as a classical AMP and therefore adapt its physiological answer.
Fig. 1.
Host peptidic hormones and Gram-negative bacteria. Model of the regulatory pathways involved in the effects of dynorphins on P. aeruginosa (a), somatostatin on H. pylori (b) and natriuretic peptides on P. aeruginosa (c). Arrows represent positive regulation and T-bar-finishing lines indicate negative regulation. OM, outer membrane; AC, adenylate cyclase; NAG, N-acetylglucosamine; NAM, N-acetylmuramic acid.
Although the dynorphin bacterial target and the associated mechanism of action of this peptide on P. aeruginosa remain unclear, it appears that bacteria are able to sense the opioid dynorphin peptides, resulting in a complex bacterial response.
Therapeutics Future
Opioid compounds are widely used in human health and affect the immune host defense [49]. A growing concern is related to the use of these compounds in humans as, like in animals, opioid use is associated with an increased risk of bacterial infections like pneumococcal diseases [50]. A better understanding of how endogenous opioids as well as synthetic ones interact with bacterial cells may help in the identification of potential bacterial targets and the development of dual-therapy in high-risk patients. Knowing the structure-activity relationships between the bacterial sensor and opioids would lead to the development of compounds that might not be recognized by bacteria but that would keep their helpful effect on pain release in humans.
Somatostatin and H. pylori
Hormone Presentation
Somatostatins belong to a neuropeptide family that has numerous functions, with effects on nearly every tissue or organ of the human body, with a special mention of the gastrointestinal tract. Somatostatins are produced after proteolytic processing of a pre-pro-somatostatin precursor. This process generates 2 cyclic peptides named according to their length, i.e., somatostatin-14 (SST-14) and somatostatin-28 (SST-28), that are characterized by a 12-amino acid loop [51] (Table 1). Somatostatins exert their biological effects after binding 1 of the 5 somatostatin receptor (SSTR) subtypes (SSTR-1 to SSTR-5) expressed in humans [52, 53], which are coupled to an adenylate cyclase through a G-protein.
Impact of Somatostatin on H. pylori
H. pylori is a bacterium that colonizes stomach and intestine mucosa where peptidic hormones such as somatostatin and gastrin are released. Gastrin peptides exist in 2 active forms (gastrin-17 and gastrin-34). A study conducted by Yamashita et al. [35] showed that the SST-14 somatostatin exerted a dose-dependent (1 µM to 10 pM) inhibition of H. pylori growth. In the same study, the authors concluded that the somatostatin effect on H. pylori was highly specific since gastrin peptides had no activity against H. pylori while somatostatin did not affect E. coli growth [35]. Nevertheless, this specificity should be confirmed since another study showed that gastrin could bind to a non-defined H. pylori sensor, increasing the bacterial growth rate [54]. Since somatostatin effects on human cells are relayed mainly by adenylate cyclases and occasionally by guanylate-cyclases modifying cAMP and cGMP intracellular concentrations, respectively, the impact of 2 cell-permeable analogs of cAMP and cGMP on H. pylori has been evaluated. Surprisingly, it has been observed that 8-bromo-cGMP fully mimicked the somatostatin inhibition of H. pylori growth while di-butyryl cAMP stimulated H. pylori growth, whereas cAMP and cGMP intrabacterial concentrations were both enhanced when H. pylori was exposed to somatostatin (10 pM; 48 h) [35]. These discrepancies suggested nonetheless that somatostatin acts on H. pylori through a specific sensor target.
Hormonal Bacterial Targets and Mechanism of Action
With the aim of characterizing the H. pylori somatostatin sensor, binding studies using radiolabeled SST-14 were performed. This enabled the identification of a saturable site presenting a dissociation constant (KD) of 0.31 nmol/L [35], confirming the presence of a specific target on H. pylori for SST-14. In the same vein, using immunoglobulins directed against the SSTR subtypes, it has been observed that IgG against SSTR-2 are able to block somatostatin effects on H. pylori whereas IgG against SSTR-1 have no impact [35]. Although these data remain fragmentary since the bacterial target was not identified at that time, they suggest the presence of a bacterial somatostatin sensor presenting functional similarities with its human counterpart SSTR-2 (Fig. 1).
Therapeutics Future
H. pylori biofilm establishment in the stomach is a source of gastritis and peptic ulcers. In the search for natural compounds able to block H. pylori biofilm formation, curcumin has shown interesting potential [55]. However, it is important to keep in mind that curcumin is able to reduce both somatostatin and gastrin release [55]. Regarding the impact of these hormones on H. pylori described above, it is important to mention that all of these parameters must be taken into account in the race to discover natural compounds able to prevent stomach disease.
Natriuretic Peptides and P. aeruginosa
Hormone Presentation
Natriuretic peptides constitute a family of hormones first identified for their cardiovascular and osmoregulation activities [56]. In addition to these well characterized functions, numerous activities have been attributed to these peptides [57]. The 3 main members of this family are: atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP), the C-type natriuretic peptide (CNP), which are mainly released in human blood by cardiomyocytes (ANP and BNP) and endothelial cells (CNP). This family of peptides is completed by urodilatin, uroguanylin, and guanylin, which are released by epithelial cells in the intestine or in the kidney lumen, and dendroaspis natriuretic peptide, which is a component of snake venom [58]. These peptides are synthesized as pre-pro-hormones that are cleaved in the cytoplasm first as pro-hormones and finally as the biologically active form by the action of a membrane enzyme during the releasing process [57, 59, 60, 61]. The length of these peptides is between 22 and 38 amino acids [57] (Table 1), and their structure is characterized by a disulfide bridge leading to a 17 amino-acid loop conferring an omega form. This structure is encountered in some cationic AMP such as rhesus θ-defensin-1 from the macaque or tyrocidin from Bacillus [62, 63, 64, 65]. These data added to the clinical observations showing that N-terminal-pro-BNP release is enhanced in the presence of E. coli or LPS in blood [15], and in vitro studies demonstrating that LPS added to cultured endothelial cells stimulated CNP release by these cells [66] have prompted some laboratories to study in more detail the impact of natriuretic peptides on the physiology of Gram-negative bacteria.
Impact of Natriuretic Peptides on P. aeruginosa
The first work dedicated to the impact of natriuretic peptides on bacteria was performed by Krause et al. [67]. In that study, it was shown that, among the natriuretic peptides tested, human BNP (hBNP) possesses antimicrobial activity against both Gram-positive and Gram-negative bacteria and that hANP and urodilatin are active as well, but with a weaker activity, while hCNP does not show any direct activity against bacteria [67]. This work constituted a starting point to evaluate the antimicrobial potential of natriuretic peptides, even if the minimum inhibitory concentration measured for hBNP was around 5 µM against Gram-positive bacteria and around 30–35 µM against Gram-negative bacteria [67], which are concentrations still quite far above their physiological concentration in human blood [56].
In this context, study of the impact of lower concentrations of natriuretic peptides on bacterial physiology was required to evaluate their real potential. It was observed that, whereas hBNP and hCNP have no impact at 1 and 0.1 µM on P. aeruginosa growth [37, 68], bacterial exposure to these peptides at 1 µM stimulates the virulence of P. aeruginosa and P. fluorescens [37, 38]. In addition, both hBNP and hCNP at 0.1 µM strongly prevented P. aeruginosa biofilm formation [69]. The most active peptide was hCNP, which was able to impair biofilm formation in a dose-dependent manner, and which was still able at 10 nM to reduce by a half the biofilm formation by P. aeruginosa [68].
Hormone Bacterial Targets and Mechanism of Action
In P. aeruginosa, production of virulence factors and biofilm establishment are inversely regulated at least partly by intracellular cyclic nucleotides [70]. The provirulent effect of CNP on P. aeruginosa was shown to result from the increase in the cAMP concentration [37], which activates the virulence factor regulator Vfr [71] (Fig. 1). In addition, in human cells, CNP was shown to bind both on the natriuretic peptide receptor (NPR) subtypes B (NPR-B) and C (NPR-C) [72], regulating cGMP and cAMP intracellular concentrations, respectively [72, 73]. Altogether, these data suggested that a putative bacterial CNP sensor could be present in P. aeruginosa. This hypothesis was reinforced by a study showing that isatin, an antagonist of NPR-A and NPR-C receptors in rats, totally blocked the antibiofilm activity of CNP [69]. In this context, it was clearly suspected that the P. aeruginosa sensor for CNP could present some similarities with the eukaryotic NPR. An in silico search for protein structure similarities between the 3 types of human NPR (NPR-A, NPR-B, and NPR-C) and the P. aeruginosa whole proteome (www.pseudomonas.com) [74] was then performed. The best score was obtained for the acetamide sensor AmiC [75]. Comparisons of human NPR-C and P. aeruginosa AmiC in terms of amino acid distribution and 3-D model structures based on molecular docking approaches allowed the proposal of AmiC as a putative bacterial sensor for CNP [69]. This hypothesis was validated by microscale thermophoresis assays using purified AmiC, in which CNP was shown to directly bind AmiC with a KD of 2 µM [69]. It is interesting to note that this binding was highly specific since: (1) BNP displayed no affinity for AmiC, suggesting that the antibiofilm effects of BNP are mediated by another pathway in bacteria (Fig. 1), and (2) NPR-C but not NPR-A agonists can bind AmiC [69]. Altogether these data proved that a bacterial sensor for a hormone and its eukaryotic receptor counterpart could have similar pharmacological profiles.
The amiC gene belongs to the ami operon which contains 4 or 5 genes depending on the P. aeruginosa strain (www.pseudomonas.com) [74]. In the absence of acetamide, AmiC binds and sequesters the anti-terminator factor regulator AmiR [76]. When acetamide [76] or CNP [69] binds to AmiC, AmiR is released and the whole operon is overexpressed. It is tempting to propose that the aliphatic amidase AmiE, which is the final product of the ami operon, could be responsible for the antibiofilm effect of CNP on P. aeruginosa. This hypothesis was recently reinforced since it was demonstrated that overproduction of AmiE impaired P. aeruginosa biofilm formation [77] (Fig. 1). One outstanding issue remains: how and where does CNP bind AmiC in vivo? Up to now, AmiC was classified as being a cytoplasmic protein (www.pseudomonas.com) [74]. However, this protein possesses a periplasmic signature in its sequence, suggesting that AmiC could move to the IM. It has therefore been suggested that during CNP exposure P. aeruginosa would address a pool of AmiC close to or within the bacterial IM [69] (Fig. 1). In addition, it has been shown that the presence of a protein named PA4858, which presents an amino acid sequence closely related to AmiC, is essential to seeing the inhibitory effect of CNP on biofilm [69]. In this context, AmiC could interact with CNP through the putative CNP periplasmic binding protein PA4858 [78]. It is speculated that the protein PA4858 could finally facilitate binding between AmiC and CNP [69], probably through a multiprotein complex including PA4859 to PA4862 (Fig. 1).
Therapeutics Future
It was recently shown that the CNP concentration used to affect P. aeruginosa physiology is important, since CNP at 1 µM induced both specific effects through the AmiC sensor and nonspecific effects likely through its interaction with the bacterial membranes, triggering virulence and/or antibiofilm activities [68] (Fig. 1). Conversely, CNP concentrations of 0.1 µM or less inhibited biofilm formation via binding to AmiC without an apparent virulence increase [68]. These data suggest that CNP or CNP derivatives could become interesting drugs for inhibition of P. aeruginosa biofilm formation either alone or in association with antibiotics, provided that a low hormone concentration is used. Among CNP derivatives, the osteocrin peptide, which displayed an affinity for AmiC in the nanomolar range, could be an interesting candidate [69]. In any case, the enhancement of the bacterial AmiE concentration induced after binding of CNP to AmiC must also be taken into account since it was shown that AmiE overproduction converts P. aeruginosa into a nonvirulent bacterium using an acute lung infection mouse model [77]. Finally, we can speculate that the development of such drugs could be facilitated by the fact that natriuretic peptides are already used as therapeutic drugs. Indeed, nesiritide (BNP) has been used since 2001 in patients with acute heart failure [79] and an agonist for natriuretic peptide receptors is actually in clinical trials (phase IIa) for future use as a bronchodilatory drug [80] and this could serve as a launch pad for delivery of natriuretic peptides in lung of patients suffering from P. aeruginosa chronic infections.
Effects of Peptidic Hormones on Gram-Positive Bacteria
The interest shown in the investigations on the effects of neuropeptides on Gram-positive bacteria was driven by the development of cosmetic sciences and the 1223UE regulation which required, since 2009, a complete scientific demonstration of all claims of the absence of adverse effects (regulation 1223/2009). In this regard, the pressure from consumers for preservative-free cosmetics or cosmetic containing a minimum of preservatives was of major concern since, in the absence of preservatives, any substance could modulate the development, virulence, and/or biofilm formation activity of skin-associated bacteria. The skin microbiota, which is the second most important one in the human body (i.e., 1012 microorganisms including more than 80% of bacteria) [81], was initially regarded as a potential issue due to the presence of numerous opportunistic pathogens. At this time, the whole skin microbiota is considered as a potential new target of cosmetic compounds. Cutaneous bacteria are continuously exposed to sweat and a mean of 25% are localized deep in the skin through hair follicles and sebaceous or sweat glands [82]. Human skin represents the largest neuroendocrine organ of the human body [83], and cutaneous bacteria are continuously exposed to skin neuropeptides diffusing in sweat and though the tissue matrix [84, 85].
SP and Bacteria
Hormone Presentation
SP was the first skin neuropeptide whose activity towards cutaneous bacteria was investigated. SP is an undecapeptide of the tachykinin family (Table 1), which is released in the skin by sensory C-type fibers [83]. It was the first skin neuropeptide to be identified and it is also the most abundant. Its role is not limited to neurotransmission and it is well known as a causative factor of neurogenic/antidromic inflammation, characterized by local plasma extravasion and edema in the absence of any exogenous signal [86]. In eukaryotes, SP is recognized by NK-1 receptors, although 2 other types of NK receptors with a lower affinity also exist [87].
Impact of SP on Bacteria
The first study of the effect of SP on bacteria was performed using a cutaneous strain of Bacillus cereus as a model [88]. This species was not selected because of it abundance in skin, although strains of B. cereus are part of the commensal cutaneous flora [89], but because of its potential virulence. Exposure of Bacillus cereus to SP was associated with a rapid (5 min) and important (400 %) increase in virulence that was associated with a significant rise of biofilm thickness (+63% after 5 h) [88]. The sensitivity of B. cereus to SP was considered high, since these values were obtained at micromolar concentrations and with a nanomolar threshold. The biofilm structure itself was also affected by the exposure to the neuropeptide with the formation of mushroom-like structures and a decrease in biofilm stability. The reversed sequence peptide of SP and neurokinin A, the natural ligand of human NK-2 receptors, did not show any activity, demonstrating the specificity of the action of SP peptides, suggesting the presence of a specific SP sensor in B. cereus.
Hormone Bacterial Targets and Mechanism of Action
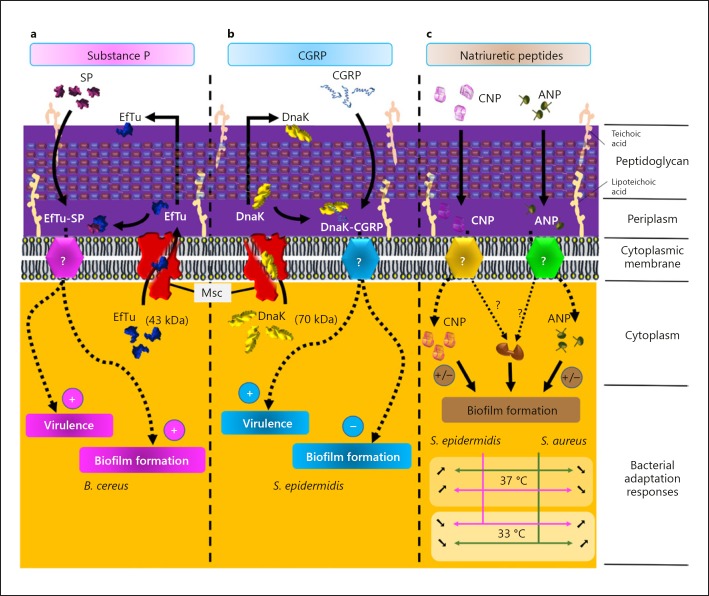
The ligand of SP in B. cereus was identified by immunoprecipitation, Western blot and MALDI-TOF/TOF analysis as the thermo unstable ribosomal elongation factor (EfTu) (Fig. 2). EfTu is produced in excess (4- to 14-fold) with regard to its stoichiometric association with ribosomes [90]. It is a member of a family of molecules identified as “moonlighting” proteins, i.e., proteins which possess alternative functions alongside their principal function. These different functions must be exercised by the same polypeptide chain and they must be realized into 2 different compartments of the cell. In this way, numerous enzymes or chaperones could be secreted outside of the cell or inserted into the membrane where they acquire generally the role of environmental sensor [91, 92, 93]. EfTu fits perfectly with this definition since this elongation factor is not only intracellular but it was also clearly found on the cell membrane [94, 95, 96], suggesting that in addition to its role in translation EfTu has a variety of other functions. SP was found to display similar effects on biofilm formation in the case of Staphylococcus epidermidis and S. aureus, with an increase in thickness after exposure to SP (1 µM) [97]. Like for B. cereus, EfTu was identified as the SP sensor in Staphylococcus [97]. EfTu is a highly conserved protein which is expressed in all bacteria as well as in archaea and eukaryotes (in mitochondria) [98]. It was thus tempting to hypothesize that SP is also capable of acting on other bacterial species, including Gram-negative ones. So far, the only Gram-negative bacterium on which the effect of SP has been investigated is P. fluorescens and it was observed that, as in Gram-positive species, SP stimulated biofilm formation [99]. The target of SP in P. fluorescens was not identified but it is interesting to note that in Pseudomonas, including P. aeruginosa, EfTu was also identified as a sensor for the neurotransmitter γ-aminobutyric acid (GABA) [100, 101].
Fig. 2.
Host peptidic hormones and Gram-positive bacteria. Model of the regulatory pathways involved in the effects of SP on B. cereus (a), CGRP on S. epidermidis (b), and natriuretic peptides on staphylococci (c).
Therapeutics Future
Different complementary studies revealed that the effect of SP on biofilm formation can be inhibited by high-ionic-force thermal water, probably through chelation, and by cosmetic products able to block the access of SP to the membrane [88]. The concentration of SP in skin and/or sweat can be increased during inflammation, stress, or even a nervous breakdown [84, 85] (these events are known to be frequently associated with skin disorders). Cutaneous bacteria, whose virulence and biofilm formation activities appear to be closely regulated by SP, are likely to take part in skin alterations. This concept was at the origin of the development of new cosmetic products, as well as new drugs with dermatological clinical applications [102]. Indeed, it was shown that, in reconstructed skin, SP-treated S. epidermidis promotes overexpression of the keratinocytes integrin-α-5 and chemokine ligand 10, which are both typical markers of psoriasis, suggesting that SP, in addition to its direct activity on skin cells [103], should be somehow involved in the development of psoriasis through its action on skin bacteria [97]. In addition, it has been observed that S. aureus biofilm formation and its persistence are associated with numerous skin disorders such as folliculitis decalvans [104], psoriasis, acne, and rosacea [105], suggesting that the impact of SP on bacteria biofilm should be evaluated in relation to these skin disorders.
Calcitonin Gene-Related Peptide and Bacteria
Hormone Presentation
Calcitonin gene-related peptide (CGRP) is a 37 amino-acid peptide (Table 1) cosecreted with SP by skin nerve terminals and it presents functions quite similar to those of SP in skin [83]. Considering its concomitant secretion with SP, CGRP was logically the second neuropeptide whose potential action on cutaneous bacteria was investigated.
Impact of CGRP on Bacteria
Until now, the effect of CGRP had only been investigated in S. aureus and S. epidermidis, but the 2 Staphylococci behaved differently. Whereas S. aureus was not sensitive to CGRP with no modification of bacterial virulence regulation and biofilm formation even at the highest concentration tested (1 µM), S. epidermidis showed an extreme sensitivity to CGRP with a nanomolar threshold [106]. In S. epidermidis, CGRP induced an increase in virulence of the same range as that observed with SP, which was followed by increased surface hydrophobicity and a decrease in biofilm formation at least in dynamic conditions [106] (Fig. 2).
Hormone Bacterial Targets and Mechanism of Action
Unexpectedly, the effects of SP and CGRP on S. epidermidis were not additive but antagonistic, suggesting that both peptides were acting on a common and easily saturable bacterial target. Surprisingly, this common step in the mechanism of action of SP and CGRP was not the recognition site, as CGRP was found to act through another moonlighting protein, i.e., the chaperone DnaK [106] (Fig. 2). However, the SP sensor EfTu and the CGRP sensor DnaK share the fact that they do not have export sequence signals and that, as shown in E. coli, these proteins should be exported through the cytoplasmic membrane by mechanosensitive channels (Msc) [107], presumably following transport by an MreB-dependent process [108]. In line with the involvement of Msc channels, blocking them using the specific inhibitor gadolinium chloride inhibited both EfTu and DnaK export and markedly inhibited the effects of SP and CGRP on S. epidermidis [106]. The apparent absence of sensitivity of S. aureus to CGRP remains a puzzling question. It should be noted that DnaK is a much larger protein than EfTu (70 vs. 43 kDa) and that the pores formed by Msc channels in S. epidermidis and S. aureus are not identical in terms of width. Since the binding of SP and CGRP to their sensor should occur in the periplasmic space (Fig. 2), DnaK cannot go through Msc in S. aureus while EfTu can cross the pore because of it smaller size, explaining why the bacterium should be sensitive to SP (via EfTu) and not to CGRP as experimentally observed [109].
Therapeutics Future
Until now the effect of CGRP on S. epidermidis has not been a matter of cosmetic or clinical applications, presumably as these observations were recent, but these data confirmed that skin neuropeptides should play a central role in the control of cutaneous bacterial behavior.
Natriuretic Peptides and Gram-Positive Bacteria
We can consider that all known neuropeptides can be found in skin, although they should be only present in specific niches. This is the case of natriuretic peptides (see above), which are produced by endothelial cells of capillary vessels and can diffuse from blood at a close distance to capillaries. In this regard, natriuretic peptides should be particularly concentrated in a specific region of the hair follicle, i.e., the bulge, where capillary vessels are abundant. However, this region of the hair follicle is rich in sebum and it is colonized by bacteria such as S. aureus and Cutibacterium (Propionibacterium) acnes because of their lipophilic characteristic and tolerance to low oxygen local concentrations [81].
Impact of Natriuretic Peptides on Gram-Positive Bacteria
It was recently demonstrated that the effects of natriuretic peptides on S. aureus were highly dependent on the growth temperature. At 37°C, the biofilm formation activity of S. aureus was inhibited by ANP and CNP, whereas that of S. epidermidis was increased [110]. The opposite effect was observed at 33°C (Fig. 2). These results should be correlated to the skin microenvironment since, while in depth the temperature stabilizes at 37°C, the surface temperature is generally about 33°C. Very recent data suggest that the biofilm formation activity of C. acnes should be submitted to even more complex temperature regulation, or probably medium- and oxygen-dependent regulation by natriuretic peptides [111], as was observed for S. aureus.
Therapeutics Future
Natriuretic peptides, SP, and CGRP are known as key factors of neurogenic antidromic inflammation in skin. This process should be essential in atopic dermatitis [112], psoriasis [113], rosacea [114], and a large range of inflammation-related troubles [115]. The discovery of the effects of SP, CGRP, and natriuretic peptides on Gram-positive bacteria led to a new vision of neurogenic inflammation which involves the microbial skin flora as a central relay in the inflammation process but also as a potential new target for the treatment of these diseases [116].
Conclusion
Taken together, the original new field of research consisting of the study of the impact of eukaryotic communication signals on bacteria in general and biofilms especially suggests future applications mainly in cosmetology for Gram-positive bacteria and preferentially in the clinical field for Gram-negative bacteria. This review will encourage crossing of data obtained in these 2 application fields (i.e., cosmetic and clinical) in order to enlarge the spectrum of action of these peptides on bacteria. In addition, deciphering the mechanism of action of human hormones on bacteria will allow indirect discovery of new functions for bacterial proteins in term of virulence regulation or biofilm formation often far from the main functions for which they were primarily characterized, extending the concept of moonlighting proteins.
Disclosure Statement
The authors have no conflict of interests to declare.
Acknowledgement
T. Clamens and T. Rosay were recipients of a doctoral fellowship from the French Ministry of Research (MRE). This work was supported by grants from the Evreux Portes de Normandie, the Conseil Général de l'Eure, the European Union (FEDER), the Normandy region, and the French Association “Vaincre la Mucoviscidose.”
References
- 1.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002 Apr;15((2)):167–93. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciofu O, Rojo-Molinero E, Macià MD, Oliver A. Antibiotic treatment of biofilm infections. APMIS. 2017 Apr;125((4)):304–19. doi: 10.1111/apm.12673. [DOI] [PubMed] [Google Scholar]
- 3.Høiby N, Bjarnsholt T, Moser C, Bassi GL, Coenye T, Donelli G, et al. ESCMID Study Group for Biofilms and Consulting External Expert Werner Zimmerli ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect. 2015 May;21(Suppl 1):S1–25. doi: 10.1016/j.cmi.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 4.Felden B, Cattoir V. Bacterial Adaptation to Antibiotics through Regulatory RNAs. Antimicrob Agents Chemother. 2018 Apr;62((5)):62. doi: 10.1128/AAC.02503-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alav I, Sutton JM, Rahman KM. Role of bacterial efflux pumps in biofilm formation. J Antimicrob Chemother. 2018 Aug;73((8)):2003–20. doi: 10.1093/jac/dky042. [DOI] [PubMed] [Google Scholar]
- 6.Pletzer D, Coleman SR, Hancock RE. Anti-biofilm peptides as a new weapon in antimicrobial warfare. Curr Opin Microbiol. 2016 Oct;33:35–40. doi: 10.1016/j.mib.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pletzer D, Hancock RE. Antibiofilm Peptides: Potential as Broad-Spectrum Agents. J Bacteriol. 2016 Sep;198((19)):2572–8. doi: 10.1128/JB.00017-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mishra B, Reiling S, Zarena D, Wang G. Host defense antimicrobial peptides as antibiotics: design and application strategies. Curr Opin Chem Biol. 2017 Jun;38:87–96. doi: 10.1016/j.cbpa.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Fuente-Núñez C, Reffuveille F, Haney EF, Straus SK, Hancock RE. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 2014 May;10((5)):e1004152. doi: 10.1371/journal.ppat.1004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reffuveille F, de la Fuente-Núñez C, Mansour S, Hancock RE. A broad-spectrum antibiofilm peptide enhances antibiotic action against bacterial biofilms. Antimicrob Agents Chemother. 2014 Sep;58((9)):5363–71. doi: 10.1128/AAC.03163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McPhee JB, Lewenza S, Hancock RE. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol. 2003 Oct;50((1)):205–17. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- 12.Anaya-López JL, López-Meza JE, Ochoa-Zarzosa A. Bacterial resistance to cationic antimicrobial peptides. Crit Rev Microbiol. 2013 May;39((2)):180–95. doi: 10.3109/1040841X.2012.699025. [DOI] [PubMed] [Google Scholar]
- 13.Chen HD, Groisman EA. The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu Rev Microbiol. 2013;67((1)):83–112. doi: 10.1146/annurev-micro-092412-155751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alverdy J, Holbrook C, Rocha F, Seiden L, Wu RL, Musch M, et al. Gut-derived sepsis occurs when the right pathogen with the right virulence genes meets the right host: evidence for in vivo virulence expression in Pseudomonas aeruginosa. Ann Surg. 2000 Oct;232((4)):480–9. doi: 10.1097/00000658-200010000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vila G, Resl M, Stelzeneder D, Struck J, Maier C, Riedl M, et al. Plasma NT-proBNP increases in response to LPS administration in healthy men. J Appl Physiol (1985) 2008 Dec;105((6)):1741–5. doi: 10.1152/japplphysiol.90442.2008. [DOI] [PubMed] [Google Scholar]
- 16.Pascual DW. The role of tachykinins on bacterial infections. Front Biosci. 2004 Sep;9((1-3)):3209–17. doi: 10.2741/1473. [DOI] [PubMed] [Google Scholar]
- 17.Cani PD, Knauf C. How gut microbes talk to organs: the role of endocrine and nervous routes. Mol Metab. 2016 May;5((9)):743–52. doi: 10.1016/j.molmet.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renaud M, Miget A. Rôle favorisant des perturbations locales causées par l'adrénaline sur le développement des infections microbiennes. C R Seances Soc Biol Fil. 1930;1930((103)):1052–4. [Google Scholar]
- 19.Lyte M, Ernst S. Catecholamine induced growth of gram negative bacteria. Life Sci. 1992;50((3)):203–12. doi: 10.1016/0024-3205(92)90273-r. [DOI] [PubMed] [Google Scholar]
- 20.Lyte M, Ernst S. Alpha and beta adrenergic receptor involvement in catecholamine-induced growth of gram-negative bacteria. Biochem Biophys Res Commun. 1993 Jan;190((2)):447–52. doi: 10.1006/bbrc.1993.1068. [DOI] [PubMed] [Google Scholar]
- 21.Lyte M. Microbial endocrinology and infectious disease in the 21st century. Trends Microbiol. 2004 Jan;12((1)):14–20. doi: 10.1016/j.tim.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Freestone PP, Haigh RD, Williams PH, Lyte M. Stimulation of bacterial growth by heat-stable, norepinephrine-induced autoinducers. FEMS Microbiol Lett. 1999 Mar;172((1)):53–60. doi: 10.1111/j.1574-6968.1999.tb13449.x. [DOI] [PubMed] [Google Scholar]
- 23.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci USA. 2003 Jul;100((15)):8951–6. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci USA. 2006 Jul;103((27)):10420–5. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasko DA, Moreira CG, Li R, Reading NC, Ritchie JM, Waldor MK, et al. Targeting QseC signaling and virulence for antibiotic development. Science. 2008 Aug;321((5892)):1078–80. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reading NC, Rasko DA, Torres AG, Sperandio V. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc Natl Acad Sci USA. 2009 Apr;106((14)):5889–94. doi: 10.1073/pnas.0811409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lesouhaitier O, Veron W, Chapalain A, Madi A, Blier AS, Dagorn A, et al. Gram-negative bacterial sensors for eukaryotic signal molecules. Sensors (Basel) 2009;9((9)):6967–90. doi: 10.3390/s90906967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neuman H, Debelius JW, Knight R, Koren O. Microbial endocrinology: the interplay between the microbiota and the endocrine system. FEMS Microbiol Rev. 2015 Jul;39((4)):509–21. doi: 10.1093/femsre/fuu010. [DOI] [PubMed] [Google Scholar]
- 29.Wu L, Estrada O, Zaborina O, Bains M, Shen L, Kohler JE, et al. Recognition of host immune activation by Pseudomonas aeruginosa. Science. 2005 Jul;309((5735)):774–7. doi: 10.1126/science.1112422. [DOI] [PubMed] [Google Scholar]
- 30.Chevalier S, Bouffartigues E, Bodilis J, Maillot O, Lesouhaitier O, Feuilloley MG, et al. Structure, function and regulation of Pseudomonas aeruginosa porins. FEMS Microbiol Rev. 2017 Sep;41((5)):698–722. doi: 10.1093/femsre/fux020. [DOI] [PubMed] [Google Scholar]
- 31.Fito-Boncompte L, Chapalain A, Bouffartigues E, Chaker H, Lesouhaitier O, Gicquel G, et al. Full virulence of Pseudomonas aeruginosa requires OprF. Infect Immun. 2011 Mar;79((3)):1176–86. doi: 10.1128/IAI.00850-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouffartigues E, Moscoso JA, Duchesne R, Rosay T, Fito-Boncompte L, Gicquel G, et al. The absence of the Pseudomonas aeruginosa OprF protein leads to increased biofilm formation through variation in c-di-GMP level. Front Microbiol. 2015 Jun;6:630. doi: 10.3389/fmicb.2015.00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaborina O, Lepine F, Xiao G, Valuckaite V, Chen Y, Li T, et al. Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa. PLoS Pathog. 2007 Mar;3((3)):e35. doi: 10.1371/journal.ppat.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Babrowski T, Holbrook C, Moss J, Gottlieb L, Valuckaite V, Zaborin A, et al. Pseudomonas aeruginosa virulence expression is directly activated by morphine and is capable of causing lethal gut-derived sepsis in mice during chronic morphine administration. Ann Surg. 2012 Feb;255((2)):386–93. doi: 10.1097/SLA.0b013e3182331870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamashita K, Kaneko H, Yamamoto S, Konagaya T, Kusugami K, Mitsuma T. Inhibitory effect of somatostatin on Helicobacter pylori proliferation in vitro. Gastroenterology. 1998 Nov;115((5)):1123–30. doi: 10.1016/s0016-5085(98)70083-6. [DOI] [PubMed] [Google Scholar]
- 36.Reynaert H, Geerts A. Pharmacological rationale for the use of somatostatin and analogues in portal hypertension. Aliment Pharmacol Ther. 2003 Aug;18((4)):375–86. doi: 10.1046/j.1365-2036.2003.01657.x. [DOI] [PubMed] [Google Scholar]
- 37.Veron W, Lesouhaitier O, Pennanec X, Rehel K, Leroux P, Orange N, et al. Natriuretic peptides affect Pseudomonas aeruginosa and specifically modify lipopolysaccharide biosynthesis. FEBS J. 2007 Nov;274((22)):5852–64. doi: 10.1111/j.1742-4658.2007.06109.x. [DOI] [PubMed] [Google Scholar]
- 38.Veron W, Orange N, Feuilloley MG, Lesouhaitier O. Natriuretic peptides modify Pseudomonas fluorescens cytotoxicity by regulating cyclic nucleotides and modifying LPS structure. BMC Microbiol. 2008 Jul;8((1)):114. doi: 10.1186/1471-2180-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Connor C, White KL, Doncescu N, Didenko T, Roth BL, Czaplicki G, et al. NMR structure and dynamics of the agonist dynorphin peptide bound to the human kappa opioid receptor. Proc Natl Acad Sci USA. 2015 Sep;112((38)):11852–7. doi: 10.1073/pnas.1510117112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sobczak M, Sałaga M, Storr MA, Fichna J. Physiology, signaling, and pharmacology of opioid receptors and their ligands in the gastrointestinal tract: current concepts and future perspectives. J Gastroenterol. 2014 Jan;49((1)):24–45. doi: 10.1007/s00535-013-0753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winzer K, Falconer C, Garber NC, Diggle SP, Camara M, Williams P. The Pseudomonas aeruginosa lectins PA-IL and PA-IIL are controlled by quorum sensing and by RpoS. J Bacteriol. 2000 Nov;182((22)):6401–11. doi: 10.1128/jb.182.22.6401-6411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diggle SP, Winzer K, Chhabra SR, Worrall KE, Cámara M, Williams P. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol. 2003 Oct;50((1)):29–43. doi: 10.1046/j.1365-2958.2003.03672.x. [DOI] [PubMed] [Google Scholar]
- 43.Grishin AV, Krivozubov MS, Karyagina AS, Gintsburg AL. Pseudomonas Aeruginosa Lectins As Targets for Novel Antibacterials. Acta Naturae. 2015 Apr-Jun;7((2)):29–41. [PMC free article] [PubMed] [Google Scholar]
- 44.Alhede M, Bjarnsholt T, Jensen PO, Phipps RK, Moser C, Christophersen L, et al. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology. 2009 Nov;155((Pt 11)):3500–8. doi: 10.1099/mic.0.031443-0. [DOI] [PubMed] [Google Scholar]
- 45.Cao H, Krishnan G, Goumnerov B, Tsongalis J, Tompkins R, Rahme LG. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc Natl Acad Sci USA. 2001 Dec;98((25)):14613–8. doi: 10.1073/pnas.251465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright MH, Fetzer C, Sieber SA. Chemical Probes Unravel an Antimicrobial Defense Response Triggered by Binding of the Human Opioid Dynorphin to a Bacterial Sensor Kinase. J Am Chem Soc. 2017 May;139((17)):6152–9. doi: 10.1021/jacs.7b01072. [DOI] [PubMed] [Google Scholar]
- 47.Fernández L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock RE. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother. 2010 Aug;54((8)):3372–82. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muller C, Plésiat P, Jeannot K. A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and β-lactams in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2011 Mar;55((3)):1211–21. doi: 10.1128/AAC.01252-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang X, Liu R, Chen C, Ji F, Li T. Opioid System Modulates the Immune Function: A Review. Transl Perioper Pain Med. 2016;1((1)):5–13. [PMC free article] [PubMed] [Google Scholar]
- 50.Wiese AD, Griffin MR, Schaffner W, Stein CM, Greevy RA, Mitchel EF, Jr, et al. Opioid Analgesic Use and Risk for Invasive Pneumococcal Diseases: A Nested Case-Control Study. Ann Intern Med. 2018 Mar;168((6)):396–404. doi: 10.7326/M17-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reichlin S. Somatostatin. N Engl J Med. 1983 Dec;309((24)):1495–501. doi: 10.1056/NEJM198312153092406. [DOI] [PubMed] [Google Scholar]
- 52.Reisine T, Bell GI. Molecular biology of somatostatin receptors. Endocr Rev. 1995 Aug;16((4)):427–42. doi: 10.1210/edrv-16-4-427. [DOI] [PubMed] [Google Scholar]
- 53.Reisine T, Bell GI. Molecular properties of somatostatin receptors. Neuroscience. 1995 Aug;67((4)):777–90. doi: 10.1016/0306-4522(95)00072-q. [DOI] [PubMed] [Google Scholar]
- 54.Chowers MY, Keller N, Tal R, Barshack I, Lang R, Bar-Meir S, et al. Human gastrin: a Helicobacter pylori—specific growth factor. Gastroenterology. 1999 Nov;117((5)):1113–8. doi: 10.1016/s0016-5085(99)70396-3. [DOI] [PubMed] [Google Scholar]
- 55.Vetvicka V, Vetvickova J, Fernandez-Botran R. Effects of curcumin on Helicobacter pylori infection. Ann Transl Med. 2016 Dec;4((24)):479. doi: 10.21037/atm.2016.12.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Potter LR, Abbey-Hosch S, Dickey DM. Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocr Rev. 2006 Feb;27((1)):47–72. doi: 10.1210/er.2005-0014. [DOI] [PubMed] [Google Scholar]
- 57.Potter LR, Yoder AR, Flora DR, Antos LK, Dickey DM. Natriuretic peptides: their structures, receptors, physiologic functions and therapeutic applications. Handb Exp Pharmacol. 2009;191:341–66. doi: 10.1007/978-3-540-68964-5_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schweitz H, Vigne P, Moinier D, Frelin C, Lazdunski M. A new member of the natriuretic peptide family is present in the venom of the green mamba (Dendroaspis angusticeps) J Biol Chem. 1992 Jul;267((20)):13928–32. [PubMed] [Google Scholar]
- 59.Yan W, Wu F, Morser J, Wu Q. Corin, a transmembrane cardiac serine protease, acts as a pro-atrial natriuretic peptide-converting enzyme. Proc Natl Acad Sci USA. 2000 Jul;97((15)):8525–9. doi: 10.1073/pnas.150149097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Semenov AG, Tamm NN, Seferian KR, Postnikov AB, Karpova NS, Serebryanaya DV, et al. Processing of pro-B-type natriuretic peptide: furin and corin as candidate convertases. Clin Chem. 2010 Jul;56((7)):1166–76. doi: 10.1373/clinchem.2010.143883. [DOI] [PubMed] [Google Scholar]
- 61.Wu C, Wu F, Pan J, Morser J, Wu Q. Furin-mediated processing of Pro-C-type natriuretic peptide. J Biol Chem. 2003 Jul;278((28)):25847–52. doi: 10.1074/jbc.M301223200. [DOI] [PubMed] [Google Scholar]
- 62.Selsted ME. Theta-defensins: cyclic antimicrobial peptides produced by binary ligation of truncated alpha-defensins. Curr Protein Pept Sci. 2004 Oct;5((5)):365–71. doi: 10.2174/1389203043379459. [DOI] [PubMed] [Google Scholar]
- 63.Lee DW, Kim BS. Antimicrobial cyclic peptides for plant disease control. Plant Pathol J. 2015 Mar;31((1)):1–11. doi: 10.5423/PPJ.RW.08.2014.0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choi KY, Chow LN, Mookherjee N. Cationic host defence peptides: multifaceted role in immune modulation and inflammation. J Innate Immun. 2012;4((4)):361–70. doi: 10.1159/000336630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002 Jan;415((6870)):389–95. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 66.Suga S, Itoh H, Komatsu Y, Ogawa Y, Hama N, Yoshimasa T, et al. Cytokine-induced C-type natriuretic peptide (CNP) secretion from vascular endothelial cells—evidence for CNP as a novel autocrine/paracrine regulator from endothelial cells. Endocrinology. 1993 Dec;133((6)):3038–41. doi: 10.1210/endo.133.6.8243333. [DOI] [PubMed] [Google Scholar]
- 67.Krause A, Liepke C, Meyer M, Adermann K, Forssmann WG, Maronde E. Human natriuretic peptides exhibit antimicrobial activity. Eur J Med Res. 2001 May;6((5)):215–8. [PubMed] [Google Scholar]
- 68.Desriac F, Clamens T, Rosay T, Rodrigues S, Tahrioui A, Enault J, et al. Different Dose-Dependent Modes of Action of C-Type Natriuretic Peptide on Pseudomonas aeruginosa Biofilm Formation. Pathogens. 2018 Apr;7((2)):7. doi: 10.3390/pathogens7020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rosay T, Bazire A, Diaz S, Clamens T, Blier AS, Mijouin L, et al. Pseudomonas aeruginosa Expresses a Functional Human Natriuretic Peptide Receptor Ortholog: Involvement in Biofilm Formation. MBio. 2015 Aug;6((4)):6. doi: 10.1128/mBio.01033-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Almblad H, Harrison JJ, Rybtke M, Groizeleau J, Givskov M, Parsek MR, et al. The Cyclic AMP-Vfr Signaling Pathway in Pseudomonas aeruginosa Is Inhibited by Cyclic Di-GMP. J Bacteriol. 2015 Jul;197((13)):2190–200. doi: 10.1128/JB.00193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blier AS, Veron W, Bazire A, Gerault E, Taupin L, Vieillard J, et al. C-type natriuretic peptide modulates quorum sensing molecule and toxin production in Pseudomonas aeruginosa. Microbiology. 2011 Jul;157((Pt 7)):1929–44. doi: 10.1099/mic.0.046755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suga S, Nakao K, Hosoda K, Mukoyama M, Ogawa Y, Shirakami G, et al. Receptor selectivity of natriuretic peptide family, atrial natriuretic peptide, brain natriuretic peptide, and C-type natriuretic peptide. Endocrinology. 1992 Jan;130((1)):229–39. doi: 10.1210/endo.130.1.1309330. [DOI] [PubMed] [Google Scholar]
- 73.Špiranec K, Chen W, Werner F, Nikolaev VO, Naruke T, Koch F, et al. Endothelial C-Type Natriuretic Peptide Acts on Pericytes to Regulate Microcirculatory Flow and Blood Pressure. Circulation. 2018 Apr;138((5)):CIRCULATIONAHA.117.033383. doi: 10.1161/CIRCULATIONAHA.117.033383. [DOI] [PubMed] [Google Scholar]
- 74.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FS. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016 Jan;44(D1):D646–53. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilson SA, Wachira SJ, Drew RE, Jones D, Pearl LH. Antitermination of amidase expression in Pseudomonas aeruginosa is controlled by a novel cytoplasmic amide-binding protein. EMBO J. 1993 Sep;12((9)):3637–42. doi: 10.1002/j.1460-2075.1993.tb06037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilson S, Drew R. Cloning and DNA sequence of amiC, a new gene regulating expression of the Pseudomonas aeruginosa aliphatic amidase, and purification of the amiC product. J Bacteriol. 1991 Aug;173((16)):4914–21. doi: 10.1128/jb.173.16.4914-4921.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clamens T, Rosay T, Crépin A, Grandjean T, Kentache T, Hardouin J, et al. The aliphatic amidase AmiE is involved in regulation of Pseudomonas aeruginosa virulence. Sci Rep. 2017 Jan;7((1)):41178. doi: 10.1038/srep41178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011 Sep;8((10)):785–6. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 79.Colucci WS, Elkayam U, Horton DP, Abraham WT, Bourge RC, Johnson AD, et al. Nesiritide Study Group Intravenous nesiritide, a natriuretic peptide, in the treatment of decompensated congestive heart failure. N Engl J Med. 2000 Jul;343((4)):246–53. doi: 10.1056/NEJM200007273430403. [DOI] [PubMed] [Google Scholar]
- 80.Edelson JD, Makhlina M, Silvester KR, Vengurlekar SS, Chen X, Zhang J, et al. In vitro and in vivo pharmacological profile of PL-3994, a novel cyclic peptide (Hept-cyclo(Cys-His-Phe-d-Ala-Gly-Arg-d-Nle-Asp-Arg-Ile-Ser-Cys)-Tyr-[Arg mimetic]-NH(2)) natriuretic peptide receptor-A agonist that is resistant to neutral endopeptidase and acts as a bronchodilator. Pulm Pharmacol Ther. 2013 Apr;26((2)):229–38. doi: 10.1016/j.pupt.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Feuilloley M, Doléans-Jordheim A, Freney J. 3rd ed. Précis de Bactériologie Clinique; 2015. Chapitre 12: Flore microbienne de la peau saine; pp. pp. 1–15. [Google Scholar]
- 82.Lange-Asschenfeldt B, Marenbach D, Lang C, Patzelt A, Ulrich M, Maltusch A, et al. Distribution of bacteria in the epidermal layers and hair follicles of the human skin. Skin Pharmacol Physiol. 2011;24((6)):305–11. doi: 10.1159/000328728. [DOI] [PubMed] [Google Scholar]
- 83.Roosterman D, Goerge T, Schneider SW, Bunnett NW, Steinhoff M. Neuronal control of skin function: the skin as a neuroimmunoendocrine organ. Physiol Rev. 2006 Oct;86((4)):1309–79. doi: 10.1152/physrev.00026.2005. [DOI] [PubMed] [Google Scholar]
- 84.Severini C, Improta G, Falconieri-Erspamer G, Salvadori S, Erspamer V. The tachykinin peptide family. Pharmacol Rev. 2002 Jun;54((2)):285–322. doi: 10.1124/pr.54.2.285. [DOI] [PubMed] [Google Scholar]
- 85.Cizza G, Marques AH, Eskandari F, Christie IC, Torvik S, Silverman MN, et al. POWER Study Group Elevated neuroimmune biomarkers in sweat patches and plasma of premenopausal women with major depressive disorder in remission: the POWER study. Biol Psychiatry. 2008 Nov;64((10)):907–11. doi: 10.1016/j.biopsych.2008.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Steinhoff M, Ständer S, Seeliger S, Ansel JC, Schmelz M, Luger T. Modern aspects of cutaneous neurogenic inflammation. Arch Dermatol. 2003 Nov;139((11)):1479–88. doi: 10.1001/archderm.139.11.1479. [DOI] [PubMed] [Google Scholar]
- 87.Steinhoff MS, von Mentzer B, Geppetti P, Pothoulakis C, Bunnett NW. Tachykinins and their receptors: contributions to physiological control and the mechanisms of disease. Physiol Rev. 2014 Jan;94((1)):265–301. doi: 10.1152/physrev.00031.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mijouin L, Hillion M, Ramdani Y, Jaouen T, Duclairoir-Poc C, Follet-Gueye ML, et al. Effects of a skin neuropeptide (substance p) on cutaneous microflora. PLoS One. 2013 Nov;8((11)):e78773. doi: 10.1371/journal.pone.0078773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bottone EJ. Bacillus cereus, a volatile human pathogen. Clin Microbiol Rev. 2010 Apr;23((2)):382–98. doi: 10.1128/CMR.00073-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beck BD. Polymerization of the bacterial elongation factor for protein synthesis, EF-Tu. Eur J Biochem. 1979 Jul;97((2)):495–502. doi: 10.1111/j.1432-1033.1979.tb13137.x. [DOI] [PubMed] [Google Scholar]
- 91.Amblee V, Jeffery CJ. Physical Features of Intracellular Proteins that Moonlight on the Cell Surface. PLoS One. 2015 Jun;10((6)):e0130575. doi: 10.1371/journal.pone.0130575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Henderson B, Martin A. Bacterial moonlighting proteins and bacterial virulence. Curr Top Microbiol Immunol. 2013;358:155–213. doi: 10.1007/82_2011_188. [DOI] [PubMed] [Google Scholar]
- 93.Mani M, Chen C, Amblee V, Liu H, Mathur T, Zwicke G, et al. MoonProt: a database for proteins that are known to moonlight. Nucleic Acids Res. 2015 Jan;43((Database issue)):D277–82. doi: 10.1093/nar/gku954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Granato D, Bergonzelli GE, Pridmore RD, Marvin L, Rouvet M, Corthésy-Theulaz IE. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect Immun. 2004 Apr;72((4)):2160–9. doi: 10.1128/IAI.72.4.2160-2169.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006 May;125((4)):749–60. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 96.Dallo SF, Zhang B, Denno J, Hong S, Tsai A, Haskins W, et al. Association of Acinetobacter baumannii EF-Tu with cell surface, outer membrane vesicles, and fibronectin. ScientificWorldJournal. 2012;2012:128705. doi: 10.1100/2012/128705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.N'Diaye A, Mijouin L, Hillion M, Diaz S, Konto-Ghiorghi Y, Percoco G, et al. Effect of Substance P in Staphylococcus aureus and Staphylococcus epidermidis Virulence: Implication for Skin Homeostasis. Front Microbiol. 2016 Apr;7:506. doi: 10.3389/fmicb.2016.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baensch M, Frank R, Köhl J. Conservation of the amino-terminal epitope of elongation factor Tu in eubacteria and Archaea. Microbiology. 1998 Aug;144((Pt 8)):2241–6. doi: 10.1099/00221287-144-8-2241. [DOI] [PubMed] [Google Scholar]
- 99.Hillion M, Mijouin L, N'Diaye A, Percoco G, Ernault J, Follet-Gueye ML, et al. Pseudomonas fluorescens, a forgotten member of the human cutaneous microflora sensible to skin communication and defence peptides. Int J Curr Microbiol Appl Sci. 2014;3:910–25. [Google Scholar]
- 100.Dagorn A, Chapalain A, Mijouin L, Hillion M, Duclairoir-Poc C, Chevalier S, et al. Effect of GABA, a bacterial metabolite, on Pseudomonas fluorescens surface properties and cytotoxicity. Int J Mol Sci. 2013 Jun;14((6)):12186–204. doi: 10.3390/ijms140612186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dagorn A, Hillion M, Chapalain A, Lesouhaitier O, Duclairoir Poc C, Vieillard J, et al. Gamma-aminobutyric acid acts as a specific virulence regulator in Pseudomonas aeruginosa. Microbiology. 2013 Feb;159((Pt 2)):339–51. doi: 10.1099/mic.0.061267-0. [DOI] [PubMed] [Google Scholar]
- 102.Blanc C. Skin-O-Flor: les bactéries pacifiées. Pour la Science. 2015;453:3. [Google Scholar]
- 103.Amatya B, El-Nour H, Holst M, Theodorsson E, Nordlind K. Expression of tachykinins and their receptors in plaque psoriasis with pruritus. Br J Dermatol. 2011 May;164((5)):1023–9. doi: 10.1111/j.1365-2133.2011.10241.x. [DOI] [PubMed] [Google Scholar]
- 104.Matard B, Meylheuc T, Briandet R, Casin I, Assouly P, Cavelier-balloy B, et al. First evidence of bacterial biofilms in the anaerobe part of scalp hair follicles: a pilot comparative study in folliculitis decalvans. J Eur Acad Dermatol Venereol. 2013 Jul;27((7)):853–60. doi: 10.1111/j.1468-3083.2012.04591.x. [DOI] [PubMed] [Google Scholar]
- 105.Totté JE, van der Feltz WT, Bode LG, van Belkum A, van Zuuren EJ, Pasmans SG. A systematic review and meta-analysis on Staphylococcus aureus carriage in psoriasis, acne and rosacea. Eur J Clin Microbiol Infect Dis. 2016 Jul;35((7)):1069–77. doi: 10.1007/s10096-016-2647-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.N'Diaye AR, Leclerc C, Kentache T, Hardouin J, Poc CD, Konto-Ghiorghi Y, et al. Skin-bacteria communication: Involvement of the neurohormone Calcitonin Gene Related Peptide (CGRP) in the regulation of Staphylococcus epidermidis virulence. Sci Rep. 2016 Oct;6((1)):35379. doi: 10.1038/srep35379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Berrier C, Garrigues A, Richarme G, Ghazi A. Elongation factor Tu and DnaK are transferred from the cytoplasm to the periplasm of Escherichia coli during osmotic downshock presumably via the mechanosensitive channel mscL. J Bacteriol. 2000 Jan;182((1)):248–51. doi: 10.1128/jb.182.1.248-251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Defeu Soufo HJ, Reimold C, Linne U, Knust T, Gescher J, Graumann PL. Bacterial translation elongation factor EF-Tu interacts and colocalizes with actin-like MreB protein. Proc Natl Acad Sci USA. 2010 Feb;107((7)):3163–8. doi: 10.1073/pnas.0911979107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.N'Diaye A, Gannesen A, Borrel V, Maillot O, Enault J, Racine PJ, et al. Substance P and Calcitonin Gene-Related Peptide: Key Regulators of Cutaneous Microbiota Homeostasis. Front Endocrinol (Lausanne) 2017 Jan;8:15. doi: 10.3389/fendo.2017.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gannesen A, Lesouhaitier O, Netrusov AI, Plakunov V, Feuilloley M. The new role of human natriuretic peptides: regulation of monospecies and binary biofilms formation of skin Staphylococcus epidermidis and Staphylococcus aureus. Microbiology (Russia) 2018;87:597–609. doi: 10.3389/fmicb.2018.02912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gannesen A, Racine PJ, Robert M, Netrusov AI, Plakunov V, Feuilloley M. Skin-microbiote communication: Human natriuretic peptides as regulators of Propionibacterium acnes and Staphylococcus aureus biofilm formation. 9th International Conference on Skin Ageing and Challenges. Porto, Portugal February 26-27 2018. [Google Scholar]
- 112.Misery L. Atopic dermatitis and the nervous system. Clin Rev Allergy Immunol. 2011 Dec;41((3)):259–66. doi: 10.1007/s12016-010-8225-z. [DOI] [PubMed] [Google Scholar]
- 113.Raychaudhuri SP, Raychaudhuri SK. Role of NGF and neurogenic inflammation in the pathogenesis of psoriasis. Prog Brain Res. 2004;146:433–7. doi: 10.1016/S0079-6123(03)46027-5. [DOI] [PubMed] [Google Scholar]
- 114.Aubdool AA, Brain SD. Neurovascular aspects of skin neurogenic inflammation. J Investig Dermatol Symp Proc. 2011 Dec;15((1)):33–9. doi: 10.1038/jidsymp.2011.8. [DOI] [PubMed] [Google Scholar]
- 115.Meggs WJ. Neurogenic inflammation and sensitivity to environmental chemicals. Environ Health Perspect. 1993 Aug;101((3)):234–8. doi: 10.1289/ehp.93101234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Feuilloley MG. Antidromic neurogenic activity and cutaneous bacterial flora. Semin Immunopathol. 2018 May;40((3)):281–9. doi: 10.1007/s00281-018-0671-3. [DOI] [PubMed] [Google Scholar]
- 117.Edvinsson ML, Ahnstedt H, Edvinsson L, Andersson SE. Characterization of Relaxant Responses to Natriuretic Peptides in the Human Microcirculation In Vitro and In Vivo. Microcirculation. 2016 Aug;23((6)):438–46. doi: 10.1111/micc.12290. [DOI] [PubMed] [Google Scholar]
- 118.Drewett JG, Garbers DL. The family of guanylyl cyclase receptors and their ligands. Endocr Rev. 1994 Apr;15((2)):135–62. doi: 10.1210/edrv-15-2-135. [DOI] [PubMed] [Google Scholar]
- 119.Curry FE, Clark JF, Jiang Y, Kim MH, Adamson RH, Simon SI. The role of atrial natriuretic peptide to attenuate inflammation in a mouse skin wound and individually perfused rat mesenteric microvessels. Physiol Rep. 2016 Sep;4((18)):4. doi: 10.14814/phy2.12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sudoh T, Minamino N, Kangawa K, Matsuo H. C-type natriuretic peptide (CNP): a new member of natriuretic peptide family identified in porcine brain. Biochem Biophys Res Commun. 1990 Apr;168((2)):863–70. doi: 10.1016/0006-291x(90)92401-k. [DOI] [PubMed] [Google Scholar]
- 121.Xia W, Mruk DD, Cheng CY. C-type natriuretic peptide regulates blood-testis barrier dynamics in adult rat testes. Proc Natl Acad Sci USA. 2007 Mar;104((10)):3841–6. doi: 10.1073/pnas.0610100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sellitti DF, Koles N, Mendonça MC. Regulation of C-type natriuretic peptide expression. Peptides. 2011 Sep;32((9)):1964–71. doi: 10.1016/j.peptides.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 123.Zhang M, Su YQ, Sugiura K, Xia G, Eppig JJ. Granulosa cell ligand NPPC and its receptor NPR2 maintain meiotic arrest in mouse oocytes. Science. 2010 Oct;330((6002)):366–9. doi: 10.1126/science.1193573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Choi JE, Di Nardo A. Skin neurogenic inflammation. Semin Immunopathol. 2018 May;40((3)):249–59. doi: 10.1007/s00281-018-0675-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016 Jan;44(D1):D1202–13. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015 Jan;347((6220)):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]