Abstract
The human gut microbiota gained tremendous importance in the last decade as next-generation technologies of sequencing and multiomics analyses linked the role of the microbial communities to host physiology and pathophysiology. A growing number of human pathologies and diseases are linked to the gut microbiota. One of the main mechanisms by which the microbiota influences the host is through its interactions with the host immune system. These interactions with both innate and adaptive host intestinal and extraintestinal immunity, although usually commensalistic even mutualistic with the host, in some cases lead to serious health effects. In the case of allogenic hematopoietic stem cell transplantation (allo-HSCT), the disruption of the intestinal microbiota diversity is associated with acute graft-versus-host disease (GvHD). Causing inflammation of the liver, skin, lungs, and the intestine, GvHD occurs in 40–50% of patients undergoing allo-HSCT and results in significant posttransplantation mortality. In this review, we highlight the impact of the gut microbiota on the host immunity in GvHD and the potential of microbiota in alleviation or even prevention of GvHD.
Keywords: Gut microbiota, Stem cell transplantation, Immune system, Graft-versus-host disease, Microbiome
The human body is colonized by a multitude of microorganisms, commonly referred to as the microbiota, with their associated genomes being referred to as the microbiome. Although most of these organisms are bacteria, others include viruses, fungi, and archaea. The gastrointestinal tract maintains the largest microbial community with an estimated 100 trillion bacterial cells from 100–150 species [1]. The gut microbiota performs diverse functions for the host such as the development and maturation of the host immune system, the digestion of food, synthesis of essential amino acids, secondary metabolites, short-chain fatty acids (SCFAs) and vitamins, the metabolism of xenobiotics, modulation of the immune responses, and the resistance to pathogens. In a healthy person, the gut microbiota constitutes a balanced composition of different commensals, symbionts and pathobionts (see glossary in Table 1) which maintain functional homeostasis essential for human health. Being individual-specific, the composition of a healthy microbiota may be different for people according to their age, geographical location and genetics [2]. In addition, the gut microbiota may change over time as it is influenced by other external environmental factors such as major diet changes, antibiotics treatment, and important lifestyle changes. Sometimes, such changes alter microbiota composition with either a decrease in symbionts and commensals and/or an increase in pathobionts perturbing the symbiotic equilibrium. Perturbations in the gut microbiota have been associated with obesity, type 2 diabetes, metabolic syndrome, irritable bowel syndrome, autoimmune diseases, and neurodevelopmental disorders [3]. There are bidirectional interactions between the microbiota, immunity, and diet (Fig. 1). Changes in the gut microbiota can have profound effects on the host immune system. This could be seriously detrimental to the success of therapy for a variety of malignant and benign hematological diseases for which allogenic stem cell transplantation is the only curative treatment.
Table 1.
Glossary
| ARG | Antibiotic resistance gene, gene conferring resistance to antibiotics; the resistance can be intrinsic (naturally occurring, independent of previous exposure to antibiotics) or acquired (via horizontal gene transfer or by mutation) |
| Commensal | A nonharmful coexistence of an organism in a host |
| Bacteriotherapy | It is the administration of live bacteria (probiotics) or their products (bacteriocins) to restore health or cure disease |
| FMT | Fecal microbiota transplant; a process of transferring a fecal suspension containing the fecal microbiota, from a healthy individual into a recipient (patient) |
| Gut microbiome | It refers to the group of microbial genomes of the microbial communities of the gastrointestinal tract (also called the gut) |
| Gutmicrobiota | It is the ensemble of all microorganisms such as bacteria, archaea, fungi, and viruses inhabiting the gut |
| GvHD | Graft-versus-host disease; a complication after allogenic stem cell transplantation in which T cells from the donor (graft) immunologically attack the host’s tissue |
| Metagenomics | It is the study of the collective genetic material in a sample usually directly taken from an environment; it provides information on the taxonomic composition and functional potential of a microbial community |
| Metatranscriptomics | It is the study of the sets of transcripts of microorganisms from an environmental sample; it gives information on the functional profile of a microbial community within the given sample |
| Mutualistic | It is type of symbiotic relationship between two organisms in which each symbiont benefits from the activity of the other |
| Pathobiont | These are potentially pathological organisms which can cause diseases in predisposed hosts |
| Prebiotics | Nondigestible fiber compounds that are substrates for the microorganisms usually bacteria colonizing the gut having stimulatory effect on these bacteria |
| Probiotics | Live microorganisms which can confer health benefits to a host |
| SCFA | Short-chain fatty acids; produced by bacterial anaerobic fermentation in the gut and are main energy providers to colonocytes |
| Symbiont | An organism living in symbiosis with a dissimilar host in a mutualistic or parasitic (with potential to cause pathology) relationship |
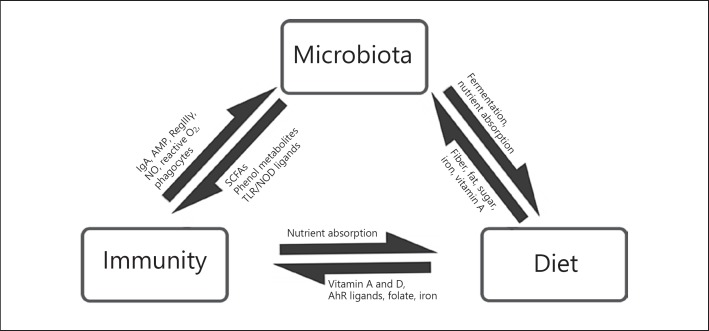
Fig. 1.
Bidirectional interactions between microbiota, immunity, and diet (adapted from Belkaid and Hand [4]). Dietary components such fiber, fat, sugars, and trace nutrients modulate microbiota composition and metabolic capacity. Microbiota help in digestion and nutrient uptake. Dietary nutrients and metabolites (such as AhR ligands) produced by microbiota modulate the immunity. The host immune system can affect the nutrient uptake. The immune system regulates microbiota by various mechanisms such as AMP, IgA, reactive oxygen species, and phagocytosis. The microbiota in turn produces metabolites that modulate the functioning of the immune system.
Interaction between the Gut Microbiota and the Host Immune System
Physical barriers in the gut such as the mucus layer produced by goblet cells, and the tight junctions regulate the relationship between the microbiota and the host [4]. Intestinal epithelial cells, dendritic cells, and macrophages express pattern recognition receptors such as Toll-like receptors and nucleotide-binding oligomerization domain-like receptors which recognize microbe-associated molecular patterns. Activation of these receptors triggers a range of effects from a proinflammatory cytokine response to the presentation of antigens to regulatory T cells (Tregs). Activation of Tregs conveys tolerance towards commensal bacteria from the initial colonization of the gut during early life [4]. Some gut bacteria produce SCFAs such as butyrate, propionate, and acetate. SCFAs play roles in both innate and adaptive immunity. Among other activities, SCFAs have histone deacetylase inhibitory activity through which they exert anti-inflammatory effects on the macrophages and the dendritic cells [5]. Butyrate is one of the most important energy sources for enterocytes and exerts anti-inflammatory activity by inhibiting NF-κB signaling and increasing IL-10 expression [6]. Moreover, it maintains the epithelial integrity by upregulation of the expression of tight junction proteins and the production of mucin by the goblet cells.
Whilst most bacteria occupy the gut lumen, segmented filamentous bacteria can penetrate the mucus layer and interact closely with the epithelial cells, inducing signalling events that lead to the differentiation of T helper (TH) 17 cells [7]. These are CD4+ effector T cells which are specialized in responses to extracellular bacteria and fungi by secretion of cytokines such as IL-17A, IL-17F, IL-21, and IL-22 [7]. Activation of TH17 cells is the host's protection against gastrointestinal pathogens. It is also associated with proinflammatory systemic effects [4]. The cytokines produced by TH17 cells induce secretion of antimicrobial peptides (AMPs) such as the α-defensins and RegIIIγ by the Paneth cells [4]. While strengthening the tight junctions, TH17 cells also promote the production of immunoglobulin A (IgA). T cell-dependent IgA secretion plays an important role in adaptive immunity. IgA maintains microbiota diversity and its compartmentalization. Loss of TH17 cells and their protective functions are associated with bacterial translocation [8]. B cells also secrete IgA that regulates interactions of the host with the commensals and prevent their adhesion to the epithelial surfaces [8]. In addition to protecting the intestinal epithelium from pathogenic bacteria, viruses and toxins, IgA can also downregulate proinflammatory responses and the expansion of Tregs cells.
Patients with hematological diseases requiring stem cell transplantation undergo extensive preconditioning chemotherapy as well as supportive measures such as antibiotic or antifungal treatments. These interventions result in the disruption of the gut microbiota and its equilibrium. Although antibiotic treatment in stem cell transplant patients is meant to avoid bacterial infections and is essential in many patients, recent studies link these to a higher risk of developing bacterial infections from opportune pathogens and subsequently graft-versus-host disease (GvHD). A significant number of patients receiving allogenic stem cell transplantation develop GvHD, which causes up to 30% mortality in these patients [9].
Allogenic Hematopoietic Stem Cell Transplantation and GvHD
Intensive treatment of hematological malignancies with radiation and chemotherapy targets the malignant cell clones but also results in severe damage to the gut epithelia. Compromised barrier allows microorganisms or bacterial products to enter the blood circulation [10]. Consequently, the immune response in the form of inflammatory cytokines leads to inflammation, bloodstream infections (BSI), i.e. bacteremia and fungemia, GvHD, and sepsis [10]. BSI are predominantly caused by Pseudomonas aeruginosa, Escherichia coli, and Candida albicans [10]. To prevent BSI in patients receiving al logenic hematopoietic stem cell transplantation (allo-HSCT), very often prophylactic antibiotics are used. There are conflicting reports about the use of antibiotics, with some studies reporting a remarkable decrease in the GvHD mortality and others associating it to increased morbidity and mortality. This is partly due to the indiscriminate use of antibiotics without considerations of patient-specific microbiota composition. Moreover, widespread and standard use of broad-spectrum antibiotics has resulted in an increasing number of antibiotic-resistant bacteria [11]. Both mortality as well as incidence of acute GvHD were higher in allo-HSCT recipients who were colonized by antibiotic-resistant bacteria [11, 12]. An integrated meta-omics analysis determined a higher number and expression of antibiotic-resistant genes (ARGs) in the gut microbiome of a patient after allo-HSCT who died due to GvHD [13]. Another retrospective study indicated that the treatment of neutropenic fever with antibiotics such as imipenem-cilastatin and piperacillin-tazobactam was linked to increased GvHD-related mortality within 5 years [14]. Other recent studies also link prophylactic use of antibiotics to higher transplant-related mortality [15, 16].
Studies focusing on the microbiota composition in patients before and after allo-HSCT, report a drastic loss of bacterial diversity after the treatment. This loss of diversity was more pronounced in patients who developed gut GvHD [17]. Loss of microbiota diversity is linked to an increased risk of infections and GvHD [18]. The decrease in diversity following allo-HSCT is often accompanied by the expansion of a single taxon. In most cases, at the onset of GvHD, an increase in the abundance of the members of the genus Enterococcus has been observed [17, 19]. This is frequently followed by BSI with vancomycin-resistant Enterococci and a decrease in the abundance of members of the order Clostridiales including Faecalibacterium spp. and Ruminococcus spp. [19, 20]. The majority of the members of the order Clostridiales are Gram-positive obligate anaerobes and are the most abundant commensal bacteria of the gut. Many of these are known butyrate producers [21]. Butyrate is involved in increased histone acetylation resulting in increased expression of antiapoptotic proteins such as JAM and occludin which are involved in barrier integrity [22]. Administration of 17 strains of butyrate-producing clostridia species improved gut epithelial integrity, reduced GvHD specific damage, and improved survival after allo-HSCT in a murine model [22, 23]. Butyrate produced by these commensal bacteria induces Treg cell expansion and differentiation [23, 24]. Recently, higher abundance of another Clostridiales member, the genus Blautia, was linked to reduced GvHD-related mortality [25].
In addition to producing immunomodulatory metabolites, the gut microbiota confers colonization resistance against pathogens [26] by preventing these from colonizing the gut. The commensals induce the production of AMPs by the Paneth cells. These AMPs keep the pathogens at bay. Severe gut GvHD correlates with a decrease in Paneth cells in biopsies [27]. The leaky epithelial barrier allows translocation of pathogens and/or antigens thereof into the systemic circulation causing BSI. Infiltration of these pathogenic antigens into other organs elicits local immune responses leading to organ damage as seen in GvHD.
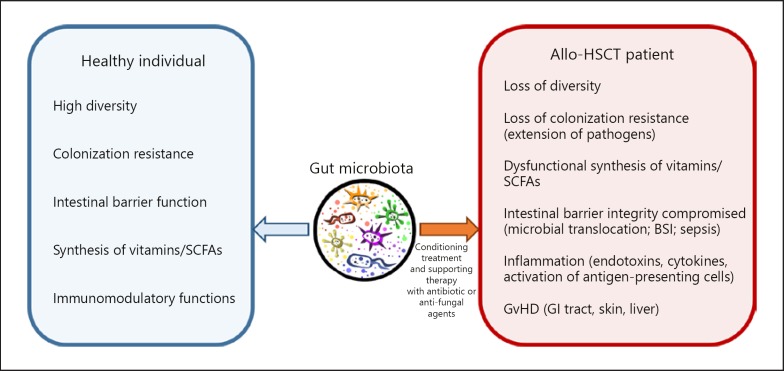
Summarizing, the host defense system comprises cellular immunity, intact intestinal barrier, and balanced microbiota. In patients undergoing allo-HSCT, very often these three defense mechanisms are compromised (Fig. 2). The therapy requires immunosuppression and cytotoxic chemotherapy resulting in severe neutropenia and massive intestinal barrier damage (mucositis). In addition, prophylactic antibiotics used to decontaminate the gut result in significant imbalance of the microbial community of the gut possibly favoring colonization by pathogens especially antibiotic-resistant strains. Ultimately, the microbial dissemination from the gut leads to sepsis and GvHD.
Fig. 2.
Gut microbiota in a healthy individual and in an allo-HSCT patient. Cancer conditioning treatment and supporting therapy with antibiotics or antifungal agents disrupt the microbiota and its functions (left panel) resulting in a range of side effects (right panel) that lead to GvHD causing significant mortality in allo-HSCT patients.
Potential of Microbiota as Therapeutic Intervention
Increasing understanding of the implication of the gut microbiota in GvHD is opening an opportunity towards evidence-based therapy for patients undergoing allo-HSCT. The modulation of gut microbiota according to the specific needs of patients will be an important part of the therapeutic strategy for allo-HSCT patients in the future. Moreover, dynamic monitoring of the microbiota may provide biomarker signatures for optimal therapeutic outcome and follow-up in the allo-HSCT patients.
Modulators of the Gut Microbiota
Recent research indicates that the modulation of gut microbiota may represent a potential therapeutic intervention to alleviate and even prevent GvHD in allo-HSCT patients. The gut microbiota can be temporarily manipulated by the choice of the antibiotics and the use of probiotics/prebiotics to repopulate the gut with commensals. The risk of invasive BSI is directly proportional to the microbial burden in the gut. It was shown that a 2-log reduction of bacterial and fungal colonization of the gut is enough to significantly reduce pathogenic microbial translocation from the gut. As such, antibiotic-independent approaches also hold promise as therapeutic options. Probiotics, fecal microbiota transplant (FMT), and prebiotics are all viable options. Other methods to modulate the gut immunity are strategies such as genetically engineered bacteria, CRISPR-Cas9 phagemids, bacterial ligands, and pathogen-specific antibodies.
Antibiotic Therapy
The use of broad-spectrum antibiotics clindamycin, piperacillin-tazobactam, and imipenem-cilastatin are linked to GvHD in patients [15]. In allo-HSCT patients, common pathogens are Gram-negative bacteria (Enterobacteriaceae) and Gram-positive bacteria such as coagulase-negative Staphylococci, Streptococcus viridans, Enterococcus, and Candida species. Identification and monitoring of these common known pathogens in patients could help in the choice of the antibiotics. Narrow-spectrum antibiotics, which target specific pathogens, while avoiding emergence of antibiotic resistant strains may prevent the complications caused by empiric antibiotics. About one-third of the pretransplant patients develop infection from Clostridium difficile, an opportune pathogen which colonizes the gut when the commensals especially the Clostridiales (anti-inflammatory clostridia or AIC) are depleted by broad-spectrum antibiotics [28]. The AIC reduction is also linked to expansion of Enterococcus spp. in patients who develop GvHD. As such, real-time monitoring of AIC with quantitative polymerase chain reaction (qPCR) or metabolomics [29] could be used to guide therapy in two ways; first in confirming that the antibiotics are not depleting the AIC below a critical range and secondly by providing in time decisions towards strategies to repopulate (probiotics) and/or sustain (antibiotics termination or prebiotics) the microbiota. Other gut-protective measures such as exogenous SCFAs or inducers of gut AMP can also be considered.
Antibiotic-independent approaches include the use of bacteriocins and bacteriophages. Bacteriocins are bacterial AMPs, and phages are viruses that infect and kill bacteria. These are natural components of the microbial communities and support competition, survival, and microbial diversity. Since these are very specific precision killers, they may be used to control specific pathogenic colonization of the gut. Although many studies with bacteriocins and bacteriophages are available and are ongoing for inflammatory bowel disease and Crohn's disease [30], to our knowledge there is limited or no information on their use in allo-HSCT patients.
Probiotics and Prebiotics
Specific strains of bacteria confer resistance to pathogens by either occupation of intestinal niches, competition for nutrients or even by induction of host immunity towards these pathogens. One such example is colonization with Barnesiella which was shown to protect against intestinal domination with vancomycin-resistant Enterococci and subsequent BSI [31].
Gut microbiota is directly influenced by diet. Several studies have demonstrated a protective effect of enteral feeding in GvHD [32]. On the other hand, total parenteral feeding enhances proinflammatory cytokines [33] and most likely aggravates the immune responses in allo-HSCT patients. The diet effects are due to the prebiotics (indigestible carbohydrates). Upon fermentation of these prebiotics by the microbiota, the SCFAs are produced. The SCFAs aliment and promote the intestinal enterocytes by providing energy and exerting an antiapoptotic effect on these. Exogenous butyrate was shown to restore barrier integrity, protect the enterocytes and alleviate GvHD. The SCFAs play a central role in anti-inflammatory pathways through the induction of Tregs which produce anti-inflammatory cytokines. This shift in balance towards anti-inflammatory cytokines in turn may directly modulate the incidence and the severity of GvHD.
Other bacterial metabolites that maintain the epithelial barrier integrity and functions are bile acids, polyamines, and aryl hydrocarbon receptor (AhR) ligands [34]. Bile acids have enteroprotective effects via the Farnesoid X receptor in the intestinal epithelium [34]. Polyamines such as spermine and spermidine have been shown to improve intestinal epithelial integrity by modulating E-cadherin expression via c-Myc. Spermine has been shown to modulate adaptive immunity by regulating macrophage activation and inhibiting proinflammatory cytokines. AhR ligands play an important role in the proper function and regeneration of the epithelial barrier in addition to secretion of AMPs by the Paneth cells. Lactobacilli produce indole-3-aldehyde from tryptophan which is a known AhR ligand. Other dietary components, especially those derived from the family Brassicaceae, are metabolized by the gut microbiota to indole-containing moieties and have protective effects on the intestinal mucosa. A study demonstrated reduced Enterococcus translocation and reduced mortality in HSCT patients who were administered fiber, glutamine, and oligosaccharides [35]. A clinical study investigating the effect of resistant starch as prebiotic to induce buty-rate production by the microbiota for the prevention of acute GvHD is ongoing (www.clinicaltrials.gov:NCT02763033). Another clinical study focusing on the effects of probiotics in both acute and chronic GvHD is underway (www.clinicaltrials.gov: NCT02805075, NCT02144701).
Fecal Microbiota Transplant
FMT is probably the most effective probiotic to restore the gut microbial diversity. FMT was used to successfully treat recurrent C. difficile infections (CDI) [36]. Some studies with small numbers of patients have demonstrated the beneficial effects of FMT, with 3 out 4 patients achieving complete remission of GvHD [37]. In the case of allogenic HSCT patients, repeated FMT in patients who developed GvHD resulted in alleviating the symptoms of the disease [38]. Yet another recent study demonstrated that FMT could reduce intestinal colonization by antibiotic-resistant strains in patients with hematological malignancies [39]. Patients with a history of colonization with antibiotic-resistant strains such as C. difficile may be prioritized to receive FMT before they undergo HSCT. In this direction, a clinical study is evaluating the role of autologous FMT in allo-HSCT patients to prevent posttransplant CDI (www.clinicaltrials.gov: NCT02269150). Another clinical study (www.clinical trials.gov: NCT02733744) is focusing on the benefits of FMT after allo-HSCT on the incidence of acute GvHD as well as 2-year survival in these patients.
Personalized Medicine
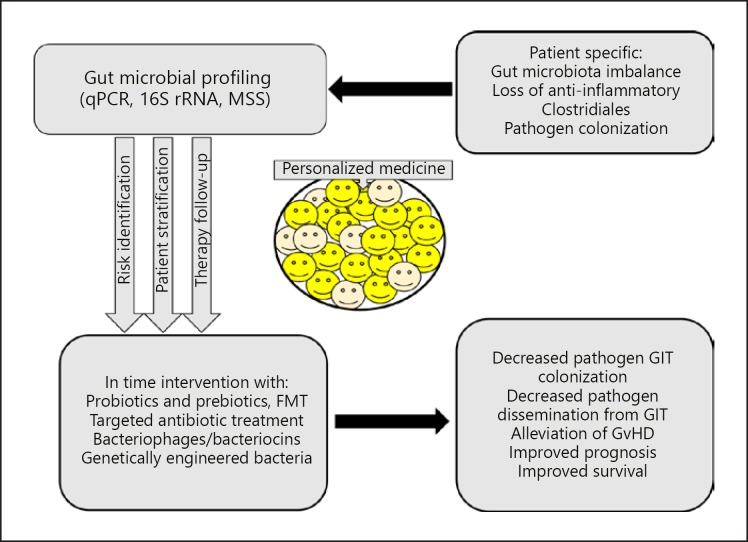
Some patients experience expansion of pathogenic strains and develop invasive infections and GvHD whereas others do not. This is partly because the gut microbiota and its interaction with the host is individual specific. As such, the microbial profiling provides an opportunity for personalized medicine (Fig. 3). To understand the causal links between microbiota and cytokines involved in GvHD, a study (www.clinicaltrials.gov: NCT02398708) will compare the fecal microbiota of patients who develop chronic GvDH with those who do not.
Fig. 3.
Gut microbiota profiling and personalized medicine. Changes in individual microbiota such as loss of diversity and anti-inflammatory Clostridiales or expansion of pathogen(s) could be investigated using qPCR, 16S rRNA, and/or metagenomics shotgun sequencing. The microbiota profiles will facilitate risk identification and stratification of patients, for example patients at risk for CDI infections. In this way, various intervention strategies could be used and followed up for effects. These individual specific therapeutic interventions could alleviate GvHD improving survival by decreasing pathogen burden and microbial dissemination. GIT, gastrointestinal tract.
Microbial Profiling
Frequently used techniques of microbial profiling are 16S rRNA sequencing and the unbiased high-resolution method of metagenomics shotgun sequencing (MSS). On the other hand, in situ hybridization and PCR have been used to identify and quantify bacteria.
The gene encoding the 16S ribosomal RNA subunit contains regions that are highly conserved between different bacterial species and nine hypervariable regions that are unique for specific bacterial species. Bacterial communities can be identified by sequencing these variable regions. This method allows comparison of identified bacterial taxa over time. It does not however provide the definitive identification of the bacterial strain. Sequences are clustered into phylotypes called operational taxonomic units based on their similarity. A reference database is needed for taxonomic identification. Open-source software such as Mothur and QIIME are used to analyze 16S rRNA sequencing data.
MSS provides an unbiased sequencing of all the genetic material present in a sample. The extracted gDNA undergoes massively parallel sequencing to provide a wide coverage of the microbial communities. MSS can detect very-low-abundance microorganisms due to high sequence coverage. The MSS data are used for taxonomic identification as well as functional analysis. Taxonomic classification is carried out using unique clade-specific marker genes or lowest common ancestor methods. At the same time, functionality could be assessed from the pathway mapping of the sequenced genes using, for example, Kyoto Encyclopedia of Genes or Genomes Orthology Database. MSS can also provide comprehensive information on the ARGs giving a snapshot of the resistome.
Microbial metagenomics sequencing and metabolomics profiling are important tools in understanding the correlation between taxon level changes and metabolic functions in the host. These techniques are expected to provide causal relationships and help in establishing functional correlates. For example, CDI is frequent in allo-HSCT patients, and some patients have predetermined higher risk for CDI before conditioning therapy. Stratifying this population of patients will help improve therapy success in these patients.
Prognosis/Therapy Follow-Up
Since the microbiota regulates host immunity and plays a potential role in the onset and severity of GvHD, the microbiota signatures could become very important therapy follow-up criteria. Increased gut microbial burden, e.g. increased abundance of E. coli is indicative of increased risk of bacteremia. Therefore, monitoring of gut Enterobacteriaceae strains (such as E. coli, Klebsiella spp. and Enterobacter spp.) in allo-HSCT patients could identify patients at risk and can be greatly helpful to minimize Enterobacteriaceae bacteremia. The interactions of the microbiota with host immunity can have long-term effects. These interactions could thus be used to identify patients at risk for disease relapse. A retrospective study on a large cohort linked the presence of SCFA-producing Gram-positive Eubacterium limosum with reduced risk of disease relapse [40].
Microbiota as Noninvasive Biomarker for GvHD
The main target organs of GvHD are skin, gastrointestinal tract, and the liver. The severity is determined based on the degree of involvement of the three principle target organs. The diagnosis is differential based on biopsy and histological examination. The therapy for GvHD is rarely started before a definitive diagnosis is made. Changes in the microbiota may provide noninvasive biomarker signatures to guide the therapy. For example, a qPCR for AIC species could be used to monitor the AIC concentration frequently. A critical depletion of the AIC could be indicative of high risk for GvHD, and appropriate therapy could be initiated. In addition, the same technique could allow monitoring the success of the therapy. The application of bacterial qPCR in routine clinical analysis for allo-HSCT patients warrants large studies including validation.
Urine metabolomics is another noninvasive way to identify bacterial metabolite biomarkers. Recently, indoxyl sulfate was shown to correlate with intestinal AIC abundance [30]. Low concentration of this metabolite was associated with poor outcome in GvHD patients.
Challenges in Widespread Use of Microbiota Modulation in HSCT Patients
Many of the challenges to the application of microbiota in the diagnosis, prognosis, and therapy of GvHD are of technical nature. Although culture-independent techniques of microbial profiling such as the 16S rRNA sequencing and the MSS sequencing have significantly advanced the microbiome research, they remain too costly, time consuming, and complex to be applied on a routine clinical basis. Another limitation of these techniques is that they give the relative abundance but not the absolute levels of the microbial communities. As such, these methods do not provide quantitative assessment of the total microbiota levels. Both the 16S rRNA and MSS techniques require refined computational methods and bioinformatics platforms. Validated databases are not only needed to draw taxonomic identification as in the case of 16S rRNA sequencing but also to find functional attributes by looking at the pathways and the involved genes in the case of MSS. As more scientists use these techniques, guidelines for the standardization and harmonization of sample processing and data acquisition will be required. This will be challenging as the microbiota is only partially known and more and more knowledge will be added in the coming years. Moreover, this means that there is a constant need to update the databases and the functional information. Bioinformatics tools will undergo continuous upgrading and development as huge datasets will be analyzed.
Species-specific qPCR could be a rapid method to obtain dynamic information on specific microbial communities. The technique is inexpensive and is clinically used in the screening of pathogens such as cytomegalovirus. In addition, qPCR has validated use in 16S rRNA sequencing for the quantification of bacterial groups and species. However, qPCR is limited by the fact that it will not provide an overall snapshot of the whole gut microbiota. The challenge in the routine clinical use of this method is to define specific and validated group(s) of bacteria that need to be continuously monitored in allo-HSCT patients. As more and more studies are published, it is expected that the scientific community could agree upon such a panel of specific microorganisms in the near future.
It is imperative to understand how the taxa level changes modulate the host homeostasis by influencing the metabolic functions. Including information from other omics levels especially from the metabolomics, may confirm the actual functional activity [41]. At the same time, although increasing information is becoming available on the bacteria, the involvement and impact of mycobiome and the virome in GvHD is poorly understood.
Currently, definite diagnosis of GvHD is difficult in the absence of noninvasive biomarkers. Real-time gut microbiota monitoring using qPCR could provide critical thresholds for specific microorganism that maintain epithelial barrier integrity and function. However, this method suffers from lack of clinical data and validated clinical efficacy.
Microbial metabolites could provide an early and precise indication of the gut microbiota health. Nevertheless, further investigations on the role of microbial metabolites in GvHD are required. Some examples include the IL-22 production in response to AhR ligands and Tregs differentiation, both of which have effects on the adaptive immune reactions.
FMT from healthy donors can restore microbiota diversity and function. The clinical efficacy of FMT in CDI infections is established. FMT has also been used to decolonize antibiotic-resistant strains. Although nowadays, several studies on the feasibility of FMT are planned while some are ongoing, there are concerns that FMT in immunosuppressed patients may provoke further infections. Recent work has demonstrated the use of encapsulated freeze-dried microorganisms may circumvent this risk [42]. Another limitation to the use of FMT is that donor and fecal screening is time consuming and may not accurately reflect the microbiota, for example the microorganisms living in the mucous layer may be underrepresented. Alternatively, bacteriotherapy using a defined mixture of fecal bacteria could be a solution. Several clinical studies on the investigation of FMT in the prevention and/or alleviation of GvHD are ongoing. These studies are expected to show the utility and efficacy of FMT as therapeutic strategy in allo-HSCT patients.
Perspectives for Therapeutic Strategies Involving the Gut Microbiota
The gut microbiota is associated with human health and disease. Current technology has facilitated the identification of the role of gut microbiota in disease processes and even established causality in some cases. Monitoring of changes in the gut microbiota could serve as noninvasive biomarker for GvHD. Profiling tools such as 16S rRNA, metagenomic shotgun sequencing (MSS) or qPCR provide valuable information on the relative abundance and dynamics of the microbiota. In a clinical setting, qPCR may be applied for real-time monitoring of specific microbial strains before, during, and after allo-HSCT, especially in high-risk patients.
Recent studies support the use of specific and preferably narrow spectrum antibiotics instead of broad-spectrum antibiotics which cause more collateral damage to intestinal mucosa and promote emergence of antibiotic-resistant microbial strains. The mechanistic understanding is also promoting new therapy options for such patients. One example of such targeted therapy is to conjugate a pathogen-specific antibody to an antibiotic [43]. Specific pathogens could also be targeted by using CRISPR-Cas9 phagemids [44]. On the colonization resistance front, genetically engineered bacteria may be used to compete with the pathogens [45]. Inducers of gut AMPs and of Toll-like receptors such as LPS [46] or pharmacological compounds [31, 47] could help reduce pathogenic microbial colonization of the gut and subsequently reduce the risk of GvHD.
High-throughput culture techniques, the so called culturomics, was applied to determine global microbial composition using MALDI-TOF mass spectrometry and identified more than 32,000 colonies in human stools [48]. This technique nevertheless misses the “noncultivable” microorganisms. Moreover, although a variety of culture conditions and microbial compositions can be tested, the interactions of the microbial communities with the human gut are not possible. Alternatively, a controlled microfluidics-based co-culture system such as HuMiX [49] allows growth of bacteria, intestinal epithelial cells, and human immune cells in different chambers separated by membranes. Other in vitro systems such as the gut-on-the-chip with stem cell-derived organoids supporting co-cultures with the microbiome could facilitate the study of the interactions of the microbial cultures with human gut cells [50]. Another area of major focus will be in situ noninvasive sampling from different parts of the gut to extend the current knowledge on the compartmentalization of the microbiota. A device allowing the isolation and identification of uncultivable microorganisms is the iChip, and its derivative iTip could provide further coverage of the human gut microbiota [50].
Conclusion
As more and more mechanistic insights are becoming available, gut microbiota will have a central role in the diagnostic and therapeutic strategies for patients undergoing allo-HSCT as well as in the alleviation or even prevention of GvHD. In the future, targeted manipulation of the gut immune system would be most likely applied as standard care for allo-HSCT patients to improve treatment success and to avoid complications such as the GvHD.
Disclosure Statement
There are no conflicts of interest whatsoever.
References
- 1.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, et al. The long-term stability of the human gut microbiota. Science. 2013 Jul;341((6141)):1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenhalgh K, Meyer KM, Aagaard KM, Wilmes P. The human gut microbiome in health: establishment and resilience of microbiota over a lifetime. Environ Microbiol. 2016 Jul;18((7)):2103–16. doi: 10.1111/1462-2920.13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palm NW, de Zoete MR, Flavell RA. Immune-microbiota interactions in health and disease. Clin Immunol. 2015 Aug;159((2)):122–7. doi: 10.1016/j.clim.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014 Mar;157((1)):121–41. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA. 2014 Feb;111((6)):2247–52. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vinolo MA, Rodrigues HG, Nachbar RT, Curi R. Regulation of inflammation by short chain fatty acids. Nutrients. 2011 Oct;3((10)):858–76. doi: 10.3390/nu3100858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 2008 Apr;28((4)):454–67. doi: 10.1016/j.immuni.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belkaid Y, Harrison OJ. Homeostatic Immunity and the Microbiota. Immunity. 2017 Apr;46((4)):562–76. doi: 10.1016/j.immuni.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrara JL, Smith CM, Sheets J, Reddy P, Serody JS. Altered homeostatic regulation of innate and adaptive immunity in lower gastrointestinal tract GVHD pathogenesis. J Clin Invest. 2017 Jun;127((7)):2441–51. doi: 10.1172/JCI90592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koh AY. The microbiome in hematopoietic stem cell transplant recipients and cancer patients: opportunities for clinical advances that reduce infection. PLoS Pathog. 2017 Jun;13((6)):e1006342. doi: 10.1371/journal.ppat.1006342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bilinski J, Robak K, Peric Z, Marchel H, Karakulska-Prystupiuk E, Halaburda K, et al. Impact of Gut Colonization by Antibiotic-Resistant Bacteria on the Outcomes of Allogeneic Hematopoietic Stem Cell Transplantation: A Retrospective, Single-Center Study. Biol Blood Marrow Transplant. 2016 Jun;22((6)):1087–93. doi: 10.1016/j.bbmt.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Scheich S, Reinheimer C, Brandt C, Wichelhaus TA, Hogardt M, Kempf VA, et al. Clinical impact of colonization with multidrug-resistant organisms on outcome after autologous stem cell transplantation: a retrospective single center study. Biol Blood Marrow Transplant. 2017 Sep;23((9)):1455–62. doi: 10.1016/j.bbmt.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 13.Kaysen A, Heintz-Buschart A, Muller EE, Narayanasamy S, Wampach L, Laczny CC, et al. Integrated meta-omic analyses of the gastrointestinal tract microbiome in patients undergoing allogeneic hematopoietic stem cell transplantation. Transl Res. 2017 Aug;186:79–94.e1. doi: 10.1016/j.trsl.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Shono Y, Docampo MD, Peled JU, Perobelli SM, Velardi E, Tsai JJ, et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci Transl Med. 2016 May;8((339)):339ra71. doi: 10.1126/scitranslmed.aaf2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber D, Jenq RR, Peled JU, Taur Y, Hiergeist A, Koestler J, et al. Microbiota disruption induced by early use of broad spectrum antibiotics is an independent risk factor of outcome after allogeneic stem cell transplantation. Biol Blood Marrow Transplant. 2017 May;23((5)):845–52. doi: 10.1016/j.bbmt.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Routy B, Letendre C, Enot D, Chénard-Poirier M, Mehraj V, Séguin NC, et al. The influence of gut-decontamination prophylactic antibiotics on acute graft-versus-host disease and survival following allogeneic hematopoietic stem cell transplantation. OncoImmunology. 2016 Dec;6((1)):e1258506. doi: 10.1080/2162402X.2016.1258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, et al. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant. 2014 May;20((5)):640–5. doi: 10.1016/j.bbmt.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taur Y, Jenq RR, Perales MA, Littmann ER, Morjaria S, Ling L, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014 Aug;124((7)):1174–82. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biagi E, Zama D, Nastasi C, Consolandi C, Fiori J, Rampelli S, et al. Gut microbiota trajectory in pediatric patients undergoing hematopoietic SCT. Bone Marrow Transplant. 2015 Jul;50((7)):992–8. doi: 10.1038/bmt.2015.16. [DOI] [PubMed] [Google Scholar]
- 20.Simms-Waldrip TR, Sunkersett G, Coughlin LA, Savani MR, Arana C, Kim J, et al. Antibiotic-induced depletion of Anti-Inflammatory Clostridia is associated with the development of GVHD in pediatric stem cell transplant patients. Biol Blood Marrow Transplant. 2017;23:820–9. doi: 10.1016/j.bbmt.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016 May;7((3)):189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathewson ND, Jenq R, Mathew AV, Koenigsknecht M, Hanash A, Toubai T, et al. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol. 2016 May;17((5)):505–13. doi: 10.1038/ni.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013 Aug;500((7461)):232–6. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 24.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013 Dec;504((7480)):446–50. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 25.Jenq RR, Taur Y, Devlin SM, Ponce DM, Goldberg JD, Ahr KF, et al. Intestinal Blautia Is Associated with Reduced Death from Graft-versus-Host Disease. Biol Blood Marrow Transplant. 2015 Aug;21((8)):1373–83. doi: 10.1016/j.bbmt.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013 Nov;13((11)):790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine JE, Huber E, Hammer ST, Harris AC, Greenson JK, Braun TM, et al. Low Paneth cell numbers at onset of gastrointestinal graft-versus-host disease identify patients at high risk for nonrelapse mortality. Blood. 2013 Aug;122((8)):1505–9. doi: 10.1182/blood-2013-02-485813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuster MG, Cleveland AA, Dubberke ER, Kauffman CA, Avery RK, Husain S, et al. Infections in Hematopoietic Cell Transplant Recipients: Results From the Organ Transplant Infection Project, a Multicenter, Prospective, Cohort Study. Open Forum Infect Dis. 2017 Mar;4((2)):ofx050. doi: 10.1093/ofid/ofx050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber D, Oefner PJ, Hiergeist A, Koestler J, Gessner A, Weber M, et al. Low urinary indoxyl sulfate levels early after transplantation reflect a disrupted microbiome and are associated with poor outcome. Blood. 2015 Oct;126((14)):1723–8. doi: 10.1182/blood-2015-04-638858. [DOI] [PubMed] [Google Scholar]
- 30.Mills S, Ross RP, Hill C. Bacteriocins and bacteriophage; a narrow-minded approach to food and gut microbiology. FEMS Microbiol Rev. 2017;41((Supp_1)):S129–S153. doi: 10.1093/femsre/fux022. [DOI] [PubMed] [Google Scholar]
- 31.Ubeda C, Bucci V, Caballero S, Djukovic A, Toussaint NC, Equinda M, et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun. 2013 Mar;81((3)):965–73. doi: 10.1128/IAI.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shallis RM, Terry CM, Lim SH. Changes in intestinal microbiota and their effects on allogeneic stem cell transplantation. Am J Hematol. 2018 Jan;93((1)):122–8. doi: 10.1002/ajh.24896. [DOI] [PubMed] [Google Scholar]
- 33.Sigalet DL, Mackenzie SL, Hameed SM. Enteral nutrition and mucosal immunity: implications for feeding strategies in surgery and trauma. Can J Surg. 2004 Apr;47((2)):109–16. [PMC free article] [PubMed] [Google Scholar]
- 34.Riwes M, Reddy P. Microbial metabolites and graft versus host disease. Am J Transplant. 2018 Jan;18((1)):23–9. doi: 10.1111/ajt.14443. [DOI] [PubMed] [Google Scholar]
- 35.Iyama S, Sato T, Tatsumi H, Hashimoto A, Tatekoshi A, Kamihara Y, et al. Efficacy of Enteral Supplementation Enriched with Glutamine, Fiber, and Oligosaccharide on Mucosal Injury following Hematopoietic Stem Cell Transplantation. Case Rep Oncol. 2014 Oct;7((3)):692–9. doi: 10.1159/000368714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013 Jan;368((5)):407–15. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 37.Kakihana K, Fujioka Y, Suda W, Najima Y, Kuwata G, Sasajima S, et al. Fecal microbiota transplantation for patients with steroid-resistant acute graft-versus-host disease of the gut. Blood. 2016 Oct;128((16)):2083–8. doi: 10.1182/blood-2016-05-717652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spindelboeck W, Schulz E, Uhl B, Kashofer K, Aigelsreiter A, Zinke-Cerwenka W, et al. Repeated fecal microbiota transplantations attenuate diarrhea and lead to sustained changes in the fecal microbiota in acute, refractory gastrointestinal graft-versus-host-disease. Haematologica. 2017 May;102((5)):e210–3. doi: 10.3324/haematol.2016.154351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bilinski J, Grzesiowski P, Sorensen N, Madry K, Muszynski J, Robak K, et al. Fecal Microbiota Transplantation in Patients With Blood Disorders Inhibits Gut Colonization With Antibiotic-Resistant Bacteria: Results of a Prospective, Single-Center Study. Clin Infect Dis. 2017 Aug;65((3)):364–70. doi: 10.1093/cid/cix252. [DOI] [PubMed] [Google Scholar]
- 40.Peled JU, Devlin SM, Staffas A, Lumish M, Khanin R, Littmann ER, et al. Intestinal Microbiota and Relapse After Hematopoietic-Cell Transplantation. J Clin Oncol. 2017 May;35((15)):1650–9. doi: 10.1200/JCO.2016.70.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muller EE, Pinel N, Laczny CC, Hoopmann MR, Narayanasamy S, Lebrun LA, et al. Community-integrated omics links dominance of a microbial generalist to fine-tuned resource usage. Nat Commun. 2014 Nov;5((1)):5603. doi: 10.1038/ncomms6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Staley C, Hamilton MJ, Vaughn BP, Graiziger CT, Newman KM, Kabage AJ, et al. Successful Resolution of Recurrent Clostridium difficile Infection using Freeze-Dried, Encapsulated Fecal Microbiota; Pragmatic Cohort Study. Am J Gastroenterol. 2017 Jun;112((6)):940–7. doi: 10.1038/ajg.2017.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehar SM, Pillow T, Xu M, Staben L, Kajihara KK, Vandlen R, et al. Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature. 2015 Nov;527((7578)):323–8. doi: 10.1038/nature16057. [DOI] [PubMed] [Google Scholar]
- 44.Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014 Nov;32((11)):1146–50. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P, et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015 Oct;526((7575)):719–22. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daguindau E, Gautier T, Chagué C, Pais de Barros JP, Deckert V, Lagrost L, et al. Is It Time to Reconsider the Lipopolysaccharide Paradigm in Acute Graft-Versus-Host Disease? Front Immunol. 2017 Aug;8:952. doi: 10.3389/fimmu.2017.00952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan D, Coughlin LA, Neubauer MM, Kim J, Kim MS, Zhan X, et al. Activation of HIF-1α and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat Med. 2015 Jul;21((7)):808–14. doi: 10.1038/nm.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012 Dec;18((12)):1185–93. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 49.Shah P, Fritz JV, Glaab E, Desai MS, Greenhalgh K, Frachet A, et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat Commun. 2016 May;7:11535. doi: 10.1038/ncomms11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arnold JW, Roach J, Azcarate-Peril MA. Emerging Technologies for Gut Microbiome Research. Trends Microbiol. 2016 Nov;24((11)):887–901. doi: 10.1016/j.tim.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]