Abstract
Antimicrobial peptides (AMPs) are a diverse family of peptides that defend the mucosal surfaces of the oral cavity and other locations. Many AMPs have multiple functions and properties that influence aspects of innate defense and colonization by microorganisms. The human oral cavity is home to the second-most diverse microbiome, and the health of the mouth is influenced by the presence of these bacteria as well as by extrinsic factors such as periodontitis and smoking. This study hypothesized that the AMP profile is different in the presence of extrinsic factors and that this would also be reflected in the bacteria present. The AMP profile was analyzed by quantitative selected-reaction-monitoring mass spectrometry analysis and 40 bacterial species were quantified by DNA-DNA hybridization in saliva donated by 41 individuals. Periodontal status was assessed through dental examination and smoking status through medical charting. Periodontal health (in nonsmokers) was associated with a higher abundance of ribonuclease 7, protachykinin 1, β-defensin 128, lipocalin 1, bactericidal permeability-increasing protein fold-containing family B member 3, and bone-marrow proteoglycan. Nonsmoking periodontal disease was associated with an abundance of neutrophil defensin 1 and cathelicidin. However, 7 AMPs were overabundant in periodontal disease in smokers: adrenomedullin, eosinophil peroxidase, 3 different histones, myeloperoxidase, and neutrophil defensin 1. There were no differentially abundant AMPs in smokers versus nonsmokers with periodontal health. Correlation network inference of healthy nonsmokers, healthy smokers, nonsmoking periodontitis, or smoking periodontitis donors demonstrated very different networks growing in complexity with increasing numbers of stressors. The study highlights the importance of the interaction between the oral cavity and its resident microbiota and how this may be influenced by periodontal disease and smoking.
Key Words: Antimicrobial peptides, Periodontitis, Inflammation, Saliva
Introduction
Salivary antimicrobial peptides (AMPs) are the first line of innate defense in the oral and gastrointestinal system. The antimicrobial capacity of saliva permits colonization of the oral cavity but prevents extensive microbial colonization of the oral and oropharyngeal tissues. A sudden absence of salivary flow, e.g., in intensive care patients [1], causes increased inflammation in the oral gingiva and mucosa, probably due to the increased oropharyngeal colonization of potential pathogenic microorganisms [1]. Long-term attenuation of salivary flow, as in xersostomia, is also associated with an increased incidence of caries lesion as well as oral fungal infections [2]. The reason why xerostomia causes oral diseases and permits microbiological overgrowth in the oropharyngeal tissues is due to the physical capacity of saliva to wash away, and thereby inhibit the growth of, microbiological biofilms, but also because of the rich variety of salivary AMPs.
The salivary AMPs range from the relatively large agglutinating mucins to the smaller membrane-perforating cathelicidins and defensins [3]. With respect to the large variety of smaller peptides, their individual antimicrobial capacity and their relevance in oral disease have been the focus of many studies. For example, a lack of cathelicidin LL-37 has been suggested to be the biological mechanism responsible for the aggressive periodontitis in subjects with Kostmann syndrome [4]. Interestingly, many AMPs also possess proapoptotic and proinflammatory properties. Our group, as well as others, have reported that cathelicidin LL-37 has antimicrobial and LPS-neutralizing properties at low concentrations but proapoptotic at higher levels [5, 6]. Similarly, the chemoattractant CCL28 displays chemotactic activity as well as broad-spectrum antimicrobial activity [7].
In other tissues, such as in the lung and the intestinal crypts, AMPs reach levels of bactericidal activity, keeping microbial activity at very low levels [8]. The oral cavity is constantly being exposed to bacterial, viral, and fungal activity, and is dependent on a symbiosis between a strong resident microbiome and the innate immune responses for the maintenance of health. In the oral cavity, the AMPs do not reach the same levels as in the lung, allowing microorganisms to inhabit the oral cavity. The oral AMPs do, however, reach the levels at which they can neutralize bacterial products such as LPS [5, 9], thereby disarming the bacteria and changing the ability of some microorganisms to compete to inhabit the oral cavity. However, the influence of AMPs on the composition of oral microbial communities is, by and large, unknown.
Certain conditions create an oral milieu that is more favorable for oral dysbiosis, such as environmental factors like smoking and systemic factors like diabetes. Smoking has been widely investigated in its role in periodontitis risk and increases this risk approximately 6-fold [10]. To our knowledge, no studies have investigated the impact of smoking and periodontal disease on the salivary AMP profile.
The oral microbiota is the second-most complex microbial community in the human body and, like the gut, interacts with the immune system to induce tolerance of commensal organisms. Pathogenic organisms can proliferate in diseases of the oral cavity such as periodontitis, and they cause inflammatory and immune responses that are equivalent to the microorganisms themselves (if not more important) in the destruction of soft tissues. Initial characterization of the oral flora is part of the history of microscopy [11], but techniques used in the past 2 decades, such as DNA-DNA hybridization, established the importance of the complexes defined by Socransky et al. [12] for periodontal disease. More recently, next-generation sequencing has elaborated on the complexities of the oral microbiome. Socransky et al. [12] used mathematical modeling to define complexes, denoted by colors, that were associated with clinical-measures periodontal disease (red complex), whereas as others were associated with bacteria found to bridge health and disease (orange complex), or in periodontally healthy donors (yellow complex). These classifications have been used greatly in research into periodontal disease.
The hypothesis of this study was that saliva constitutes an AMP profile with specific AMPs or clusters of AMPs that may be correlated with certain bacteria or clusters of bacteria. We also hypothesize that the AMP profile may be different under certain circumstances, e.g., in smokers and subjects with periodontal disease. Knowledge of which AMPs cluster with which microbes in healthy and dysbiotic conditions will allow for future studies to investigate if the AMP profile can be an instrument by which healthy oral microbiota can be sculptured. The aim of this study was to map the human salivary AMP profile and investigate which AMPs correlate with certain bacterial clusters. A second aim was to investigate which AMPs are up- or downregulated in subjects with periodontitis, both smokers and nonsmokers.
Methods
Cohort
One thousand subjects, representative of the population of Skåne County in Sweden, were selected from the Swedish Government's Person Address Register, SPAR. Individuals between the ages of 20 and 89 years were selected. Of these 1,000 subjects, 451 underwent a dental clinical examination, had their medical history recorded, and had saliva collected. Of these 451 subjects, 41 diabetes-free individuals were selected based on their periodontal status and smoking habits. Periodontal disease was defined as having bone loss (BL) of ≥1/3 of the root length and/or a pocket depth of > 5 mm as well as bleeding on probing (BoP) > 20%. Among subjects with the most severe periodontitis, 10 nonsmokers (defined as never or former smokers) and 9 current smokers with the highest number of pockets with a probing depth of 4–5 mm were selected. Periodontal health was defined as not having any teeth with a periodontal probing depth > 5 mm and no bone loss > 1/3 of the root length. Among the periodontally healthy, 11 nonsmokers and 11 current smokers were frequency-matched by age based on the corresponding periodontitis/smoking status group.
Clinical Examination
The dental clinical examination has been reported previously [13]. It included radiographs, i.e., bitewing radiographs in the premolar and molar areas as well as a panoramic radiograph. Periodontal pockets exceeding 3 mm were recorded at 4 sites of each tooth using an Hu-Friedy 1-mm-graded periodontal probe. The presence or absence of plaque and bleeding on probing were also recorded. Bone loss on panoramic radiographs corresponding to > 1/3 of the root length was noted as well.
Medical History
Medical history charting included questions on blood pressure, cardiovascular disease (CVD), diabetes, rheumatic diseases, and other diseases. Any medication prescribed by a doctor was also recorded. The medical history also included questions on present or past usage of tobacco, Swedish snuff, and smoking. Smoking status was recorded from the medical charting and participants were defined as never/former smokers or current smokers.
Sampling and Saliva Handling
Saliva was harvested by having the subject chew on a sterile piece of paraffin for 5 min and expectorate into a tube. The sample was immediately frozen at −20°C and later at −80°C. At a later time point, the saliva was thawed and spun down to allow the formation of a pellet and a supernatant. The pellet was analyzed by checkerboard DNA-DNA hybridization with respect to 40 different oral species, and the supernatant was used for proteomics.
Checkerboard DNA-DNA Hybridization
The salivary pellet was sent to the Forsyth Institute in Boston on dry ice for further handling. At the Forsyth, the pellet was resuspended in sterile Tris-EDTA buffer and each sample was individually analyzed using checkerboard DNA-DNA hybridization with respect to 40 different species representing commensal oral microflora as well as bacteria associated with caries and periodontitis: A. naeslundii, S. constellatus, E. nodatum, P. gingivalis, A. actinomycetemcomitans, F. nucleatum subsp. vincentii, C. rectus, T. socranskii, E. saburreum, P. micra, V. parvula, A. oris, S. anginosus, S. sanguinis, H. pylori, S. oralis, C. ochracea, A. israelli, S. intermedia, T. denticola, S. mutans, P. nigrescens, A. odontolyticus, F. nucleatum subsp. polymorphum, C. showae, F. periodonticum, N. mucosa, F. nucleatum subsp. nucleatum, C. gingivalis, S. gordonii, T. forsythia, S. noxia, S. salivarius, P. melaninogenica, S. mitis, E. corrodens, G. morbillorum, C. sputigena, L. bucallis, C. gracilis, and P. intermedia.
Preparation of Saliva
The total protein concentration of saliva supernatant was assessed using the Bradford assay: 10 µg of salivary protein was diluted with HPLC-grade water up to 20 µL and denatured with 10 M urea and 100 mM ammonium bicarbonate (Sigma-Aldrich). Subsequently, the samples were submitted to in-solution digestion. First, the protein samples were reduced with 5 mM Tris-2-carboxyethyl phosphine (TCEP, Sigma-Aldrich) at 37°C for 60 min. 10 mM of 2-Iodoacetamide (IAA) was then added to the samples and they were incubated in the dark for 30 min. Samples were diluted 5× in 0.1 M ammonium bicarbonate before trypsin digestion (2.5 ng/µL) was performed overnight at 37°C. The next morning, 10% formic acid (FA) was added to terminate the reaction until the pH reached 2–3. For all dilutions, HPLC-grade water was used (Sigma-Aldrich). The C18 peptide clean-up protocol was performed afterwards using Harvard Apparatus C18 Macro SpinColumns (#744101), according to the manufacturer's protocol. Samples were reconstituted in 2% acetonitrile (ACN) and 0.1 % FA in water.
Selected-Reaction-Monitoring Mass Spectrometry
To construct a selected-reaction-monitoring mass spectrometry (SRM-MS) method capable of quantifying AMPs in saliva, we selected 117 proteins from previous MS experiments, microarray data [14, 15], and the literature [3], and synthesized 340 peptides corresponding to AMPs within the 117 proteins. The 20 most intense fragments of each peptide were estimated from previously acquired shotgun MS data, and the resulting 6,800 transitions were subsequently tested on the triple quadrupole mass spectrometer. These efforts generated SRM-MS assays for all the 117 proteins covered by at least 1 peptide per protein and at least 6 transitions per peptide. At the same time, we recorded the retention time off the chromatography column to enabled scheduled SRM-MS analysis, and then established the reference library needed to automatically score the results generated from the saliva samples with our software analysis tools [16]. The produced data were processed through an in-house-developed software pipeline [17]. The SRM-MS measurements were performed on a TSQ Vantage triple quadrupole mass spectrometer (QqQ) or a TSQ Quantiva QqQ (both from Thermo Scientific). Chromatographic separations of peptides were performed with Easy-nLC II systems (Thermo Scientific). Peptides were loaded with a constant pressure rate of 280 bars onto the analytical columns, either PicoChip (PCH7515-105H354-FS25, New Objective) or EasySpray (ES802, Thermo Scientific) with 10 µL of 97% solvent A (0.1% FA in water) and 3% solvent B (0.1% FA in ACN). The peptides were eluted with a linear gradient from 92% solvent A and 8% solvent B to 35% solvent B over 10 or 34 min with a constant flow of 300 nL/min. The raw data were acquired using Xcalibur software (Thermo Scientific) and then processed and analyzed with SRM-MS analysis software Skyline38 with manual validation and inspection of the results. This included retention times, similarity between library spectra and acquire data (dotp), and the similarity of relative fragment ion intensity ratios between heavy and light peptide signals (rdotp).
Statistics and Data Analysis
Data were analyzed using SPSS v23. The correlation between peptide and bacterial quantities was calculated using the Spearman correlation. Fold-change of AMPs and bacteria were calculated based on the raw data values. Due to data normality distribution, the raw data were log10-transformed to allow for parametric tests. ANCOVA analysis was used to compare between group fold-change in AMP quantity. The ANCOVA analysis was adjusted for age. p < 0.05 were considered statistically significant.
Heat maps were made with R-based ClustVis [18] using log10-transformed peptide quantities or untransformed bacterial quantities. Rows and columns were clustered using Manhattan distance and average linkage. Peptides present in < 75% of all samples were excluded from the heat maps. Network graphs were generated using Gephi [19] using statistically significant correlations with r > 0.75. Specifically, the nodes represent the AMPs and bacteria investigated here. These nodes are connected by lines (edges) showing the positive (red) and negative (green) correlations between the analytes. Correlations within the AMPs or bacterial groups alone have not been investigated and are not shown here. The size of each node shows the proportional size compared to the overall average detected. The width of the edges shows the strength of the correlation with greater width equating to stronger correlation. Where correlations between analytes are less than r = 0.75 and p = 0.05, edges are not shown. Unconnected nodes were removed for clarity of viewing.
Results
Table 1 describes the demographic characteristics, tobacco usage, and periodontal parameters of all parti-cipants. From the cohort of 41 subjects, there were 10 nonsmoking and 9 smoking subjects with periodontal disease. The periodontally healthy smokers were significantly younger than smokers with periodontal disease. Participants with periodontitis (vs. periodontally healthy) had lower tooth counts, and an increased bleeding index, prevalence, and proportion of deep periodontal pockets (all p < 0.05).
Table 1.
Demographic and dental status of the whole cohort, subdivided into subjects with periodontal disease, periodontally healthy, smokers, non-smokers, diabetics and non-diabetics
| All | Perio nonsmoker | Healthy nonsmoker | Perio smoker | Healthy smoker | |
|---|---|---|---|---|---|
| Subjects,n | 41 | 10 | 11 | 9 | 11 |
| Age, years | 50.1±16.3 | 51.3±17.8§ | 44.8±12.3 | 51.3±15.0§ | 33.2±11.2 |
| Female gender | 20 (48.8) | 5 (50) | 6 (54.5) | 4 (44) | 5 (45.5) |
| Current smoker | 20 (48.8) | 0 (0) | 0 (0) | 9 (100) | 11 (100) |
| Never-smoker | 13 (31.7) | 4 (40) | 9 (81.8) | 0 (0) | 0 (0) |
| Former smoker | 8 (19.5) | 6 (60) | 2 (18.2) | 0 (0) | 0 (0) |
| Swedish snuff user | 4 (9.8) | 1 (10) | 1 (9.1) | 0 (0) | 2 (18.2) |
| Tooth count | 25.0±4.0 | 24.5±4.1§ | 27.1±1.0 | 22.3±3.8§ | 26.8±1.5 |
| BoP, % | 37.1±27.4 | 69.1±23.9§ | 5.9±9.4 | 47.8±18.2§ | 26.2±21.6 |
| Prevalence, PPD ≥6 mm | 2.8±4.8 | 9.7±6.3§ | 0 | 5.8±4.0§ | 0 |
| No/mild periodontitis | 22 (53.7) | 0 (0) | 11 (100) | 0 (0) | 11 (100) |
| Moderate/severe periodontitis | 19 (46.3) | 10 (100) | 0 (0) | 9 (100) | 0 (0) |
Values express n (%) or mean ± SD, unless otherwise indicated. BoP, bleeding on probing; PPD, probing pocket depth; Perio, with periodontal disease.
Significantly different compared to the corresponding nonexposed group (perio nonsmoker vs. healthy nonsmoker and perio smoker versus healthy smoker).
AMP Profile
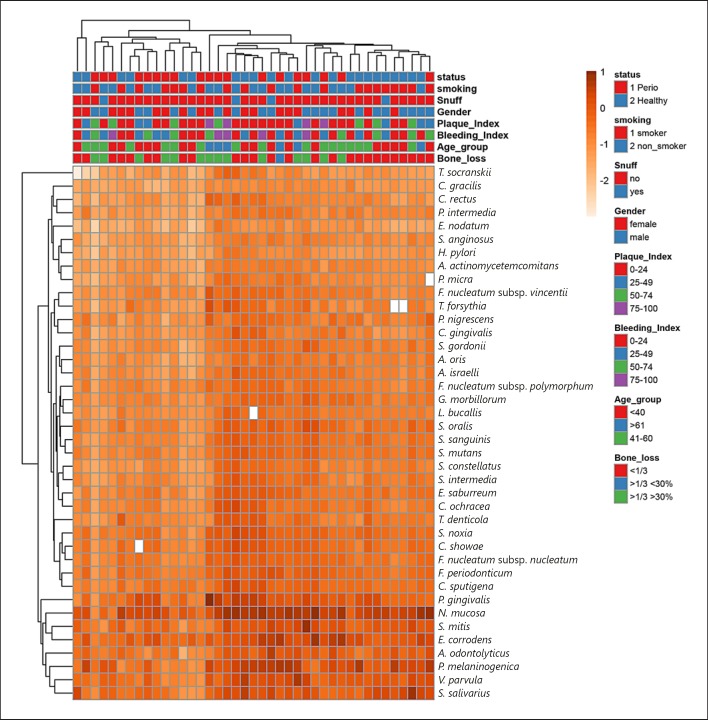
A total of 63 antimicrobial proteins and peptides were detected in saliva (online suppl. Table 1; for all online suppl. material, see www.karger.com/doi/10.1159/000494146) by SRM-MS and are visualized in Figure 1 with the associated demographic data. Online supplementary Figure 1 includes graphical representation of the abundance of all peptides based on the 4 groups (periodontitis, healthy, periodontitis in smokers, and healthy smokers). The highest quantity of peptide was found for neutrophil-derived neutrophil defensin 1.
Fig. 1.
Visualization of all peptide signatures for all samples. Peptide data were log10-transformed, as shown on the scale bar for colors. Imputation is used for missing value estimation. Missing data squares are represented in white. Both rows and columns are clustered using Manhattan distance and average linkage (63 rows and 41 columns).
In total, 5 peptides were significantly more abundant in periodontal healthy donors versus diseased subjects after age adjustment, namely ribonuclease 7 (RNase 7), protachykinin 1, β-defensin 128, bactericidal permeability-increasing fold-containing family B member 3 (BPIB3) and bone-marrow proteoglycan. Lipocalin 1 was only significantly abundant in healthy in a t test comparison, but not after adjustment for age in an ANCOVA analysis of covariance (Table 2). As demonstrated in Table 2, neutrophil defensin 1 and cathelicidin were overabundant in periodontal disease in nonsmokers. The relative overabundance of AMPs in smokers with periodontal disease compared to healthy smokers was 2- to 2.6-fold; neutrophil defensin 1 and cathelicidin were overabundant in nonsmokers with periodontitis and healthy nonsmokers by 3.3-fold and 2.2-fold, respectively. An age-adjusted ANCOVA analysis resulted in no statistically significantly abundant AMPs in smokers with periodontal disease and healthy smokers, but in nonsmokers the AMPs stayed significant.
Table 2.
Differential abundance of AMPs among nonsmoking participants who were periodontally healthy and those with periodontal disease
| Peptide | Group | Fold change | p on log valuet test | p on log value ANCOVA (age-adjusted) |
|---|---|---|---|---|
| Ribonuclease 7 | nPnS/PnS | 19.3 | 0.001 | 0.001 |
| Protachykinin 1 | nPnS/PnS | 3.7 | 0.01 | 0.006 |
| β-Defensin128 | nPnS/PnS | 3.5 | 0.002 | 0.0005 |
| Lipocalin 1 | nPnS/PnS | 2.9 | 0.049 | 0.065 |
| BPIB3 | nPnS/PnS | 2.3 | 0.025 | 0.009 |
| Bone-marrow proteoglycan | nPnS/PnS | 2.1 | 0.015 | 0.02 |
| Neutrophil defensin 1 | nPnS/PnS | 0.3 | 0.034 | 0.048 |
| Cathelicidin | nPnS/PnS | 0.45 | 0.006 | 0.007 |
p values are based on Student’s t test and ANCOVA adjusted for age for independent samples on log10-transformed levels of AMPs. BPIB3, bactericidal permeability-increasing fold-containing family B member 3; nPnS, nonsmokers with no periodontal disease; PnS, nonsmokers with periodontal disease.
The relative abundance of AMPs in subjects with periodontal disease and periodontally healthy subjects was statistically significant for 7 peptides in a t test comparison, but in none when adjusting for age in the ANCOVA analysis. In smokers, adrenomedullin, eosinophil peroxidase (EPX), histone (H)2A type 2A, H2B type 2E, H2B type F-S, myeloperoxidase (MPO) and neutrophil defensin 1 were all overabundant in periodontal disease compared to healthy subjects. There was no difference in the abundance of specific AMPs in periodontally healthy smokers versus nonsmokers (Table 3).
Table 3.
Differential abundance of AMPs in periodontally healthy smokers versus smokers with periodontal disease
| Peptide | Group | Fold change | p value1 | p value2 |
|---|---|---|---|---|
| Adrenomedullin | PS/nPS | 2.3 | 0.04 | 0.602 |
| Eosinophil peroxidase | PS/nPS | 2.2 | 0.025 | 0.153 |
| Histone H2A type 2A | PS/nPS | 2.6 | 0.028 | 0.183 |
| Histone H2B type 2E | PS/nPS | 2.2 | 0.046 | 0.310 |
| Histone H2B type F-S | PS/nPS | 2.0 | 0.042 | 0.245 |
| Myeloperoxidase | PS/nPS | 2.3 | 0.014 | 0.101 |
| Neutrophil defensin 1 | PS/nPS | 2.3 | 0.021 | 0.082 |
p values are based on Student’st test and ANCOVA adjusted for age for independent samples on log10-transformed levels of AMPs:
unadjusted
adjusted for age. nPS, smokers with no periodontal disease; PS, smokers with periodontal disease.
Bacterial Profile
Checkerboard DNA-DNA hybridization was used to target 40 different oral bacterial species representing commensal oral microflora as well as bacteria associated with caries and periodontitis. All the bacteria were detected in at least 52% of the saliva samples, with a median detection rate of 95%. The data are visualized in Figure 2.
Fig. 2.
Visualization of all bacterial signatures for all samples. Bacterial data were log10-transformed, as shown on the scale bar for colors. Imputation is used for missing value estimation. Missing data squares are represented in white. Both rows and columns are clustered using Manhattan distance and average linkage (40 rows and 41 columns).
Correlation between AMPs and Bacteria
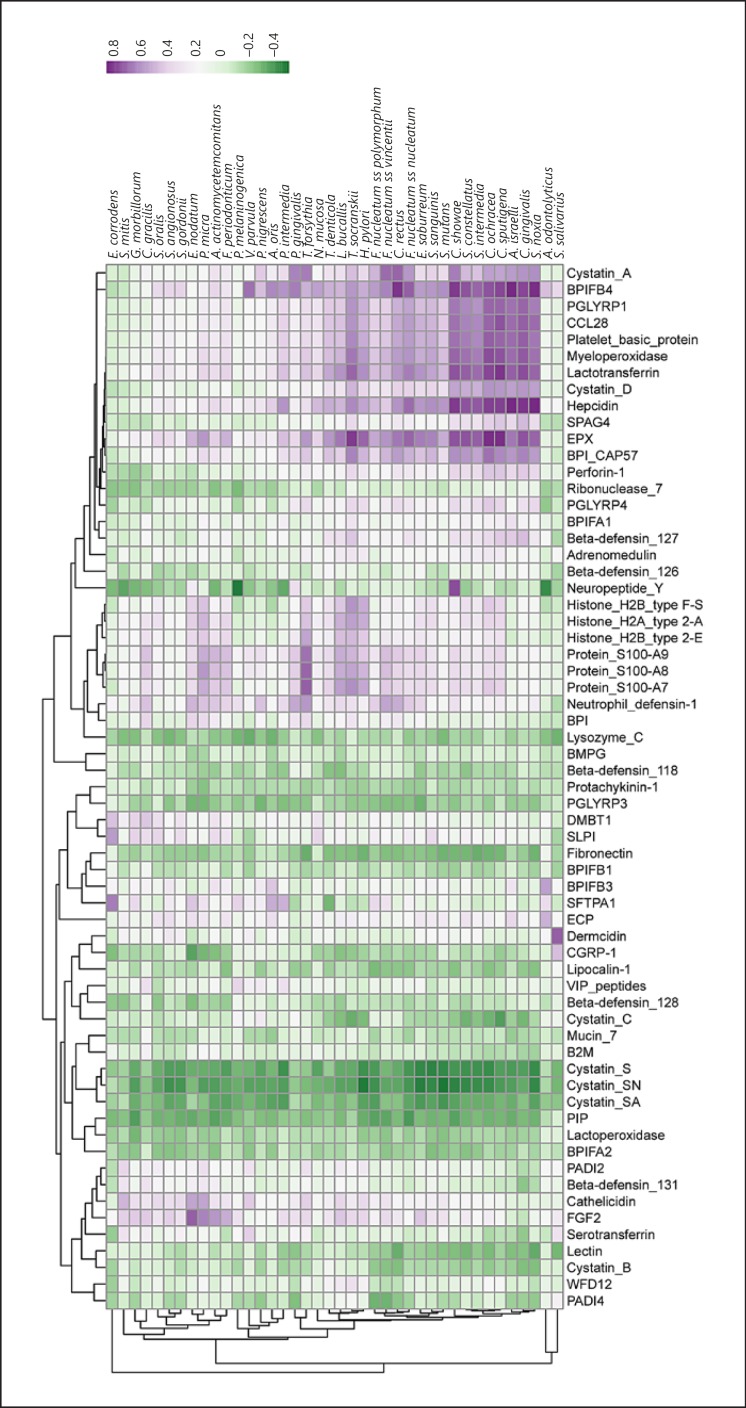
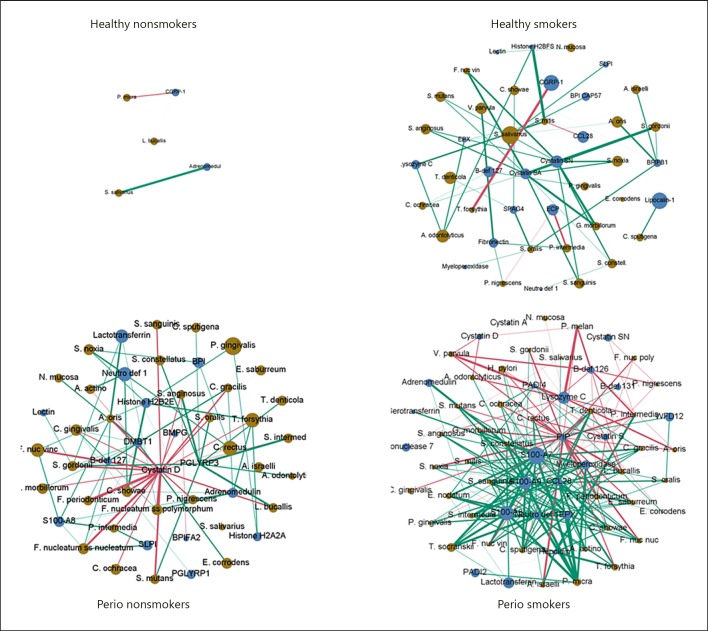
A correlation matrix comparing AMP and bacterial relationships in the whole cohort (Fig. 3) was constructed to illustrate the pattern of AMP-bacteria correlation across all samples. It is apparent that there are 3 groups of peptides in these correlations: those predominantly positively correlated (in purple); those predominantly negatively correlated (in green); and some that showed a mixed response across the bacteria and peptides. The more positively correlated peptides and bacteria appear to have peptides that could be derived from an immune response, i.e., MPO, EPX, histones, and S100 proteins. Therefore, an exploration of only the most highly correlated (r > 0.75) and significantly (p < 0.05) correlated peptides and bacteria were examined for groups of patients. Figure 4 shows the network graphs generated by this analysis. It is clear to see that there are few significant and highly correlated nodes in healthy nonsmoking participants; however, the number of nodes and connections (edges) increases with both smoking and periodontal disease. In healthy smokers, higher quantities of salivary peptides such as cystatins as well as CGRP and yellow complex bacteria (e.g., S. mutans and S. salivarius) appear (Fig. 4). In periodontitis patients, a greater quantity of the red complex bacterium P. gingivalis becomes apparent alongside more neutrophil-derived peptides such as neutrophil defensin 1. In periodontal smokers, there is a much more complex and connected network with high quantities of S100 proteins potentially derived from lysing neutrophils. Accordingly, the average degree increases across the networks (healthy nonsmokers 1.2, healthy smokers 2.3, periodontitis nonsmokers 3.4, and periodontitis smokers 7.4), with concomitant increases in the number of nodes and edges.
Fig. 3.
Clustering of AMP abundance based on correlation to bacterial abundance. Purple represents positive correlation and green represents negative correlation.
Fig. 4.
Network analysis of significantly correlated (Spearman correlation: r > 0.75, p < 0.05) peptides and bacteria. Network graphs drawn with Gephi using Fruchterman-Reingold layout and no overlap. Size of node is proportional to the average quantity detected in the condition relative to overall average. Node colors denote bacterium (brown) or peptide (blue). Color of edges represents positive (red) or negative (green) correlation. Perio, periodontitis.
Discussion
This study has mapped the relative abundance of 63 AMPs and the presence of 40 bacterial species known to be important in the oral cavity. It has investigated correlations between specific AMPs and clusters of AMPs with bacterial species, and clusters of species, and determined which salivary AMPs are more/less abundant in periodontal health and disease in the presence or absence of the most important risk factor of periodontal disease, namely smoking.
Of the 63 AMPs, there was a wide variation between individual donors; however, higher quantities of various AMPs could be associated with either health or disease, or with or without smoking. When examined for different roles, it could be seen that clusters of diverse salivary AMPs were found together rather than clustering of AMPs with similar functions. This type of clustering has been observed previously by Choi et al. [20], who demonstrated that P. gingivalis LPS-binding proteins in whole saliva could be detected alongside high frequencies of diverse AMPs, including cystatins, lipocalin 1, and perforin 1.
Some AMPs were of particular interest as they have either rarely been investigated or appear to be new to investigations into periodontitis. In our data, RNase 7 was 19 times overabundant in periodontal health versus disease in nonsmokers and correlated inversely with most bacteria. RNase 7 is strongly cationic and specifically expressed in the epithelial cells of the skin, and the respiratory and genitourinary tracts, and is induced by inflammatory agents and bacterial infection [21]. Interestingly, RNAse 7 is found more abundantly in skin samples from travelers unaffected by Staphylococcus aureus-positive skin infection than in travelers with S. aureus-positive skin infection, suggesting that it plays a role in the maintenance of healthy, noncolonised health states [22]. RNase 7 has, to our knowledge, not been studied in the context of periodontal disease previously.
Protachykinin is also more abundant in periodontal health and has not previously been reported in its own right. However, it is a precursor of substance P and substance K. Substance P is a well-known neurotransmitter and neuromodulator with antimicrobial capacity [23]. Substance P is reportedly upregulated in inflammation, but a recent paper from Haririan et al. [24] confirmed higher salivary levels of substance P in healthy subjects than in subjects with periodontal disease.
When examining how AMP abundance correlated with bacterial species in all the samples, a cluster of highly correlated AMPs and bacteria could be seen (Fig. 3). These AMPs included cystatins A and D, BPIFB4, CCL28, MPO, and EPX, lactotransferrin, platelet basic protein, and PGLYRP1. Many of these peptides are of leukocyte, particularly neutrophil, origin. The bacterial species vary, being Gram-positive or Gram-negative, and they belong to the range of complexes in Socransky et al. [12]. Notable absences though, are the bacteria most associated with periodontal disease, the red complex bacteria. Periodontal lesions have an abundance of neutrophils and, even in health, these leukocytes are travelling through the periodontium to appraise the microbe-rich environment of the oral cavity. It is possible that the abundance of these peptides and these bacteria represent an induction of a robust immune response by species that are easily recognized by the body.
Notable inverse correlations were observed between cystatins SN, S, and SA, and many bacterial species (Fig. 3). These proteins are part of the cystatin superfamily: 2 of the families are represented in family 1, the stefins (cystatins A-D), and in family 2 cystatins (SA, S, and SN). Family 1 members lack disulfide bonds whereas family 2 members have 2 internal disulfide bonds; it seems that family 2 has a much lower inhibitory activity [25]. Perhaps the role of family 2 cystatins differs from the more efficient role played by family 1 in cysteine protease inhibition. Cystatins S, SA, and SN are generally expressed extracellularly, in saliva, tear fluid, and blood [26]. Previous studies have shown that cystatin SA can inhibit the growth of both P. gingivalis and A. actinomycetemcomitans, potentially through a surface-binding mechanism rather than by inhibiting protease activity [27, 28].
When examining the data as a whole by correlation network analysis (Fig. 4), healthy nonsmoking individuals showed a very sparsely connected network, suggesting a capacity to adapt to changes, but perturbation of the healthy condition by either smoking or periodontitis alters this to create correlated networks that have many more connections and potential interdependencies. In the periodontitis nonsmoking samples, cystatin D was positively correlated with 16 bacteria. This saliva peptide inhibits the cathepsin cysteine proteinases and has the highest preference for cathepsin S [29]. Cathepsin S is produced by many cell types, including leukocytes, suggesting that the correlation with bacteria may indicate an increased leukocyte response which the salivary glands are trying to contain. In the periodontitis smoker samples there was a negative correlation between the S100 peptides (A7-A9) and many bacteria, forming a dense cluster of interactions. S100A8 and A9 form the heterodimer, calprotectin, which has broad-spectrum antimicrobial activity [30] reflected by the numerous bacteria in these donors. In the periodontitis smoker samples, there were also negative correlations with prolactin-inducible protein (PIP) and lysozyme C. PIP is produced by the salivary glands and has multiple functions including the proteolysis of fibronectin [31] and the agglutination of bacteria, including Streptococci [32]. Lysozyme C is a glycosyl hydrolase with well-known bacteriolytic functions produced widely, including by the salivary glands. The negative correlation of these peptides with various species of bacteria may indicate differential routes for the manipulation of the oral microbiome.
When designing this study, we used a broad approach for the definition of AMPs; however, there may be peptides with antimicrobial features reported in the saliva that we were not able to detect with the design that we deployed, and we may also have included peptides with questionable antibacterial activity. Online supplementary Table 1 presents a detailed description of all AMPs by full name and UniProt code. Some of the AMPs quantified here are precursors of the form that exerts antibacterial activity, but they are still considered relevant in this setting (e.g., cathelicidin is quantified here but not its active form LL-37). In the case of BPI, we were able to detect both BPI and its precursor BPI CAP57, which are presented separately.
In order to capture both bacteria and AMPs, we chose the model of using salivary supernatant for peptides and pellet for bacteria. The model may be criticized, as cationic peptides can bind to larger molecules that may end up in the pellet. Also, free 16S rRNA is lost using this design. On the other hand, cationic peptides, such as cathelicidin and defensins are highly abundant in this study, and as the study focused on AMPs, it was beneficial to exclude abundant free 16S rRNA from devitalized bacteria. One benefit of using salivary supernatant and pellet is that less saliva is needed, allowing the inclusion of subjects with a low salivary flow. An alternative approach would have been to use the site-specific gingival crevicular fluid (GCF) which flows into the saliva. GCF is regularly used in biomarker analysis in periodontal disease and it is rich in AMPs. However, in this study, the interest lay in the analysis of the fluid in the whole mouth. Focusing on GCF may have provided even greater insight into AMP regulation of the oral microbiota, but no GCF samples were available.
The ideal design of this study would be to include all 451 subjects of the cohort, but this was not feasible due to the expense and the length of time associated with SRM-MS analysis. When selecting subjects from a larger cohort, there is the question of prioritizing the exposure variable or the proper matching of confounding variables. Here, we prioritized the exposure variable “periodontitis”, as the variable “smoking” is included based on its impact on periodontitis. The periodontal health diagnosis here focused on pocket depth and bone loss, which unfortunately allowed for the inclusion of subjects with gingivitis.
Another selection of participants, to more accurately elucidate the impact of smoking, would be to sort smokers based on pack years. That would, however, compromise the main objective of the study, i.e., to elucidate the association of periodontitis and AMPs. Adjustment for age was in the form of 2 steps: (i) including younger periodontitis subjects and older healthy subjects, and (ii) analyzing covariance using ANCOVA, allowing for adjustment.
The cells, tissues, and fluids of the oral cavity allow bacterial colonization, but it is the innate immune response and especially the salivary AMPs that dictate the milieu that will promote the colonization of certain bacterial species. This study was an attempt to capture that interaction. Our data also show the shift in AMP profile from health to periodontal disease, and in smokers versus nonsmokers with and without periodontal disease, suggesting yet another mechanism by which smoking impacts the pathogenesis of periodontal disease. The data from this study will result in future oral AMP-related research focusing on the reported AMPs.
Statement of Ethics
The study was approved by The Regional Ethical Review Board, Lund, Sweden.
Disclosure Statement
All authors declare no conflicts of interest.
Supplementary Material
Supplementary data
Supplementary data
Acknowledgements
This paper was made possible through grants from the Swedish Research Council, and the Crafoord, Magnus Bergvall, and Lars Hierta Memorial Foundations.
References
- 1.Dennesen P, van der Ven A, Vlasveld M, Lokker L, Ramsay G, Kessels A, et al. Inadequate salivary flow and poor oral mucosal status in intubated intensive care unit patients. Crit Care Med. 2003 Mar;31((3)):781–6. doi: 10.1097/01.CCM.0000053646.04085.29. [DOI] [PubMed] [Google Scholar]
- 2.Mandel ID. The role of saliva in maintaining oral homeostasis. J Am Dent Assoc. 1989 Aug;119((2)):298–304. doi: 10.14219/jada.archive.1989.0211. [DOI] [PubMed] [Google Scholar]
- 3.Gorr SU, Abdolhosseini M. Antimicrobial peptides and periodontal disease. J Clin Periodontol. 2011 Mar;38(Suppl 11):126–41. doi: 10.1111/j.1600-051X.2010.01664.x. [DOI] [PubMed] [Google Scholar]
- 4.Pütsep K, Carlsson G, Boman HG, Andersson M. Deficiency of antibacterial peptides in patients with morbus Kostmann: an observation study. Lancet. 2002 Oct;360((9340)):1144–9. doi: 10.1016/S0140-6736(02)11201-3. [DOI] [PubMed] [Google Scholar]
- 5.Jönsson D, Nilsson BO. The antimicrobial peptide LL-37 is anti-inflammatory and proapoptotic in human periodontal ligament cells. J Periodontal Res. 2012 Jun;47((3)):330–5. doi: 10.1111/j.1600-0765.2011.01436.x. [DOI] [PubMed] [Google Scholar]
- 6.Säll J, Carlsson M, Gidlöf O, Holm A, Humlén J, Ohman J, et al. The antimicrobial peptide LL-37 alters human osteoblast Ca2+ handling and induces Ca2+-independent apoptosis. J Innate Immun. 2013;5((3)):290–300. doi: 10.1159/000346587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hieshima K, Ohtani H, Shibano M, Izawa D, Nakayama T, Kawasaki Y, et al. CCL28 has dual roles in mucosal immunity as a chemokine with broad-spectrum antimicrobial activity. J Immunol. 2003 Feb;170((3)):1452–61. doi: 10.4049/jimmunol.170.3.1452. [DOI] [PubMed] [Google Scholar]
- 8.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003 Sep;3((9)):710–20. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 9.Bociek K, Ferluga S, Mardirossian M, Benincasa M, Tossi A, Gennaro R, et al. Lipopolysaccharide Phosphorylation by the WaaY Kinase Affects the Susceptibility of Escherichia coli to the Human Antimicrobial Peptide LL-37. J Biol Chem. 2015 Aug;290((32)):19933–41. doi: 10.1074/jbc.M114.634758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergstrom J, Preber H. Tobacco use as a risk factor. J Periodontol. 1994;65(5s):545–50. doi: 10.1902/jop.1994.65.5.545. [DOI] [PubMed] [Google Scholar]
- 11.Lane N. The unseen world: reflections on Leeuwenhoek (1677) 'Concerning little animals’. Philos Trans R Soc Lond B Biol Sci. 2015 Apr;370((1666)):370. doi: 10.1098/rstb.2014.0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998 Feb;25((2)):134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 13.Lundegren N, Axtelius B, Akerman S. Oral health in the adult population of Skane, Sweden: a clinical study. Acta Odontol Scand. 2011 doi: 10.3109/00016357.2011.640279. [DOI] [PubMed] [Google Scholar]
- 14.Demmer RT, Behle JH, Wolf DL, Handfield M, Kebschull M, Celenti R, et al. Transcriptomes in healthy and diseased gingival tissues. J Periodontol. 2008 Nov;79((11)):2112–24. doi: 10.1902/jop.2008.080139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jönsson D, Ramberg P, Demmer RT, Kebschull M, Dahlén G, Papapanou PN. Gingival tissue transcriptomes in experimental gingivitis. J Clin Periodontol. 2011 Jul;38((7)):599–611. doi: 10.1111/j.1600-051X.2011.01719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malmström L, Malmström J, Selevsek N, Rosenberger G, Aebersold R. Automated workflow for large-scale selected reaction monitoring experiments. J Proteome Res. 2012 Mar;11((3)):1644–53. doi: 10.1021/pr200844d. [DOI] [PubMed] [Google Scholar]
- 17.Malmström L, Marko-Varga G, Westergren-Thorsson G, Laurell T, Malmström J. 2DDB - a bioinformatics solution for analysis of quantitative proteomics data. BMC Bioinformatics. 2006 Mar;7((1)):158. doi: 10.1186/1471-2105-7-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015 Jul;43(W1):W566-70. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacomy M, Venturini T, Heymann S, Bastian M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS One. 2014 Jun;9((6)):e98679. doi: 10.1371/journal.pone.0098679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi S, Baik JE, Jeon JH, Cho K, Seo DG, Kum KY, et al. Identification of Porphyromonas gingivalis lipopolysaccharide-binding proteins in human saliva. Mol Immunol. 2011 Sep;48((15-16)):2207–13. doi: 10.1016/j.molimm.2011.06.434. [DOI] [PubMed] [Google Scholar]
- 21.Harder J, Schroder JM. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J Biol Chem. 2002 Nov;277((48)):46779–84. doi: 10.1074/jbc.M207587200. [DOI] [PubMed] [Google Scholar]
- 22.Zanger P, Holzer J, Schleucher R, Steffen H, Schittek B, Gabrysch S. Constitutive expression of the antimicrobial peptide RNase 7 is associated with Staphylococcus aureus infection of the skin. J Infect Dis. 2009 Dec;200((12)):1907–15. doi: 10.1086/648408. [DOI] [PubMed] [Google Scholar]
- 23.Brogden KA, Guthmiller JM, Salzet M, Zasloff M. The nervous system and innate immunity: the neuropeptide connection. Nat Immunol. 2005 Jun;6((6)):558–64. doi: 10.1038/ni1209. [DOI] [PubMed] [Google Scholar]
- 24.Haririan H, Andrukhov O, Böttcher M, Pablik E, Wimmer G, Moritz A, et al. Salivary Neuropeptides, Stress and Periodontitis. J Periodontol. 2017 Sep;•••:1–15. doi: 10.1902/jop.2017.170249. [DOI] [PubMed] [Google Scholar]
- 25.Nandy SK, Seal A. Structural Dynamics Investigation of Human Family 1 & 2 Cystatin-Cathepsin L1 Interaction: A Comparison of Binding Modes. PLoS One. 2016 Oct;11((10)):e0164970. doi: 10.1371/journal.pone.0164970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abrahamson M, Alvarez-Fernandez M, Nathanson CM. Cystatins. Biochem Soc Symp. 2003:179–199. doi: 10.1042/bss0700179. [DOI] [PubMed] [Google Scholar]
- 27.Ganeshnarayan K, Velliyagounder K, Furgang D, Fine DH. Human salivary cystatin SA exhibits antimicrobial effect against Aggregatibacter actinomycetemcomitans. J Periodontal Res. 2012 Oct;47((5)):661–73. doi: 10.1111/j.1600-0765.2012.01481.x. [DOI] [PubMed] [Google Scholar]
- 28.Blankenvoorde MF, van't Hof W, Walgreen-Weterings E, van Steenbergen TJ, Brand HS, Veerman EC, et al. Cystatin and cystatin-derived peptides have antibacterial activity against the pathogen Porphyromonas gingivalis. Biol Chem. 1998 Nov;379((11)):1371–5. [PubMed] [Google Scholar]
- 29.Chapman HA, Riese RJ, Shi GP. Emerging roles for cysteine proteases in human biology. Annu Rev Physiol. 1997;59((1)):63–88. doi: 10.1146/annurev.physiol.59.1.63. [DOI] [PubMed] [Google Scholar]
- 30.Damo SM, Kehl-Fie TE, Sugitani N, Holt ME, Rathi S, Murphy WJ, et al. Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc Natl Acad Sci USA. 2013 Mar;110((10)):3841–6. doi: 10.1073/pnas.1220341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naderi A, Vanneste M. Prolactin-induced protein is required for cell cycle progression in breast cancer. Neoplasia. 2014;16:329–342. doi: 10.1016/j.neo.2014.04.001. e321-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nistor A, Bowden G, Blanchard A, Myal Y. Influence of mouse prolactin-inducible protein in saliva on the aggregation of oral bacteria. Oral Microbiol Immunol. 2009 Dec;24((6)):510–3. doi: 10.1111/j.1399-302X.2009.00543.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data