Abstract
The NLRP1 inflammasome attenuates inflammatory bowel disease (IBD) progression and colitis-associated tumorigenesis. A possible mechanism postulates that the lack of the NLRP1 inflammasome creates permissive niches in the gut for pathogenic bacteria to flourish, causing dysbiosis and increased IBD susceptibility. To evaluate this hypothesis, we characterized the gut microbiome of wild-type, Nlrp1b−/−, and Asc−/− mice under naïve conditions by sequencing the V3 region of the 16s rRNA gene. For both genetically modified mouse lines, the microbiome composition reflected overrepresentation of bacteria associated with dysbiosis relative to wild-type animals. Measurement of short- and medium-chain fatty acids by mass spectrometry further revealed significant differences between genotypes. However, prior to concluding that the NLRP1 inflammasome plays a role in regulating the composition of the microbiome, we evaluated two additional strategies for cohousing wild-type and Nlrp1b−/− mice: breeding homozygous parents and cohousing at weaning, and breeding from heterozygous parents and cohousing littermates. We found that maternal influence was the greater predictor of microbiome composition rather than genotype. With the rise in microbiome research across disciplines, our study should be viewed as a cautionary example that illustrates the importance of careful breeding and housing strategies when evaluating host-microbiome interactions.
Keywords: Nod-like receptor, Microbiome, Inflammatory bowel disease, Experimental colitis, Colitis-associated cancer, Short-chain fatty acid, ASC, Dysbiosis
Introduction
Nearly 4 million people worldwide suffer from inflammatory bowel disease (IBD). IBD is characterized by chronic and relapsing inflammation in the gastrointestinal (GI) tract and has two primary clinical manifestations, either ulcerative colitis or Crohn's disease. Beyond the direct role of dysregulated inflammation on IBD progression, the loss of immune system homeostasis in the GI tract during IBD is also associated with an increased risk of developing a unique form of colorectal cancer, termed colitis-associated cancer, in patient populations [1]. Although the direct cause of IBD remains unclear, it has increasingly become evident that the microbiome plays a role in the development, progression, and/or risk of disease. Indeed, several studies have noted that patients with IBD possess dysbiotic microbiomes relative to healthy controls [2, 3, 4, 5, 6, 7]. During Crohn's disease and ulcerative colitis, the protective epithelium of the GI tract becomes damaged and uncontrolled host microbiome translocation from the lumen drives inflammation. Thus, the composition of the host microbiome can dramatically influence the resultant immune response and disease progression.
Nucleotide-binding domain and leucine-rich repeat-containing (NLR) proteins are a group of intracellular pattern recognition receptors (PRRs) responsible for sensing a variety of pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs). In the GI tract, NLR family members are essential mediators of inflammation during IBD and maintain immune system homeostasis [8]. There are 22 distinct NLR and NLR-like proteins identified in humans and several murine paralogs, many of which have yet to be fully characterized. One subgroup of NLRs has been found to form multiprotein complexes, defined as inflammasomes, which function to activate IL-1β and IL-18. Inflammasome formation is also associated with a unique form of inflammatory cell death, termed pyroptosis. Upon stimulation with a specific PAMP or DAMP, a given NLR senses a specific pattern and oligomerizes, activating apoptotic speck protein containing a CARD (Pycard; ASC), which results in the cleavage and activation of pro-caspase-1 to its mature form. Activated caspase-1 cleaves pro-IL-1β and pro-IL-18 into their respective active cytokines that are subsequently released from the cell.
Several NLR family members possess structural motifs that allow them to function as intracellular sensors and participate in inflammasome formation. In the context of IBD, NLRP1, NLRP3, NLRC4, and NLRP6 are well-characterized inflammasome-forming NLRs that significantly modulate the progression of experimental colitis and/or colitis-associated tumorigenesis in mouse models [9, 10, 11, 12, 13, 14]. Likewise, the inflammasome adaptor protein ASC and the relevant caspases, caspase-1 and caspase-11, also modulate inflammation in the gut during IBD and cancer [9, 10, 11, 15, 16]. In general, the majority of studies evaluating NLR inflammasomes have found that loss of any specific NLR or inflammasome component results in increased GI inflammation, epithelial cell barrier disruption, and inflammation-driven tumorigenesis. Inflammasome formation has been suggested to maintain immune system homeostasis in the gut and attenuate IBD progression through a variety of mechanisms, including promoting epithelial cell regeneration and repair, facilitating T-cell differentiation, and controlling cell death [8, 17]. This evidence has strongly suggested that a function of NLR family members in the gut is likely associated with their recognition of pathogenic and commensal members of the host microbiome and their modulation of a balanced host immune response following loss of epithelial cell barrier integrity [18, 19].
Our research team previously demonstrated that NLRP1 attenuates IBD and colitis-associated tumorigenesis progression [9]. Notably, we also saw evidence of increased expression of NLRP1 in human subjects with active ulcerative colitis relative to healthy controls [9]. Beyond IBD, polymorphisms in the NLRP1 gene have been associated with several autoimmune disorders, including vitiligo, celiac disease, rheumatoid arthritis, systemic lupus erythematosus, and type I diabetes [8]. It is clear that NLRP1 plays an important role in modulating inflammatory diseases, but the direct mechanism/s has not yet been defined. Using Nlrp1b−/− and Asc−/− mice in the dextran sulfate sodium (DSS) model, we found that animals lacking components of the NLRP1 inflammasome demonstrated significantly increased morbidity, colon inflammation, and tumorigenesis compared to wild-type littermate control animals. These observations were correlated with reduced levels of IL-1β and IL-18 [9]. Of specific relevance to the current work, antibiotic administration and cohousing studies between Nlrp1b−/−, Asc−/−, and wild-type animals suggested that sensitivity to DSS was strongly associated with each animal's microbiome [9]. However, beyond these broad antibiotic and cohousing observations, the compositions of the microbiome in these animals have not been characterized. It was our belief that NLRP1 might be responsible for sensing a specific bacteria or bacterial component that is associated with the microbiome, and that by identifying the baseline bacterial constituents we might be able to conclude that the NLRP1 inflammasome attenuates IBD and colitis-associated tumorigenesis by limiting the growth of this, yet to be identified, bacteria or PAMP.
Our initial hypothesis predicted that the loss of any individual NLR inflammasome would establish permissive niches in the gut and lead to significant dysbiosis in the host microbiome that drives dysregulated inflammation in the GI tract. Here, we evaluate this hypothesis in animals lacking components of the NLRP1 inflammasome. Our data indicate that the microbiome profiles for singly housed Nlrp1b−/−, Asc−/−, and wild-type mice are distinct prior to any exacerbation via chemically induced colitis and are highly dysbiotic. Microbiome dysbiosis in the Nlrp1b−/− and Asc−/− mice are associated with significant changes in the metabolic profile, including short-chain fatty acid (SCFA) and medium-chain fatty acid (MCFA) metabolism measured via mass spectrometry (GC/MS), thus suggesting a functional link between the dysbiotic host microbiome and susceptibility to IBD in the context of NLRP1 inflammasome-deficient mice.
However, before accepting this conclusion, we further evaluated the impact of different breeding and housing strategies on our findings. These additional studies were based on recent reports associated with NLRP6, which is an inflammasome-forming NLR that is widely accepted as having a direct influence on inflammation in the gut by regulating the intestinal microbial content [12, 15, 18, 20]. Two individual teams concluded that NLRP6 does not influence the microbiome, but instead observed that maternal influence was the greater predictor of gut microbiome content [21, 22]. In both cases, cohousing littermates from heterozygous parents ameliorated any significant differences in microbiome bacteria composition. Armed with this information, we evaluated two different methods of cohousing Nlrp1b−/− and wild-type mice to determine if these findings held true for NLRP1. Similar to the previous findings with NLRP6, our data demonstrate that maternal influence and housing, rather than genetic factors, are responsible for the differences observed in the microbiome composition.
Materials and Methods
Experimental Animals
The characterization and generation of Nlrp1b−/− and Asc−/− mice has previously been described [23, 24]. All experiments were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals and following the protocol approved by the Virginia Tech IACUC. Wild-type, Nlrp1b−/−, and Asc−/− mice were housed under standard specific pathogen-free (SPF) conditions in vivarium space at the Virginia Maryland College of Veterinary Medicine. These facilities are American Association for Laboratory Animal Care (AALAC) accredited and the SPF status of our colony is routinely verified using standard best practices. The list of excluded and included agents in our vivarium is described in online supplementary Table S1 (for all online suppl. material, see www.karger.com/doi/10.1159/000495850). The singly housed animals used in these studies were maintained as independent homozygous colonies, while the littermates were derived from heterozygous breeder pairs generated by crossing our independent homozygous mice within our vivarium and housed in the same physical location (room and rack). All experiments were conducted with 6- to 17-week-old mice that were maintained on the C57Bl/6J background.
For cohousing studies, pups born from singly housed wild-type and singly housed Nlrp1b−/− breeder pairs (matched-cohoused) and pups born from heterozygote Nlrp1b+/− breeder pairs (littermate-cohoused) were weaned and cohoused at 1: 1 ratios for 4 weeks prior to assessments. Cohoused wild-type and knockout mice were given autoclaved water with ampicillin (1 mg/mL), streptomycin (5 mg/mL), and vancomycin (0.25 mg/mL) daily for 2 weeks to normalize their respective microbiomes. At 12 weeks of age (matched-cohoused) or 7 weeks of age (littermate-cohoused), the animals were sacrificed and colonic contents were collected. All studies were controlled with gender- and age-matched wild-type animals that were maintained under SPF conditions and received 2018 chow (LabDiet) and water ad libitum.
Fecal Harvest
Tools were washed and autoclaved prior to harvest. Mice were euthanized, and separate sets of tools were designated and used for each genotype for necropsy in a biological safety cabinet. Colonic contents were collected and samples were immediately placed on dry ice. Fecal bacterial DNA was isolated using the QIAamp DNA Stool Mini Kit following the manufacturer's instructions for isolation of DNA from stool for pathogen detection (Qiagen, Germany).
16S rRNA Gene Sequencing and Data Analysis
To amplify the V3 region of the bacterial 16S rRNA gene for sequencing, two rounds of PCR reactions were used. Primers used in the first round PCR have the overhang sequences with Illumina adapters. Forward primer: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCA GACTCCTACGGGAGGCAG; reverse primer: GTCTCGTGGGCTCGGAGATGTGTATA AGAGACAGCGTATTACCGCGGCTGCTG. The PCR conditions were 98°C for 3 min; 15 cycles of 98°C for 30 s; and 62°C for 30 s, 72°C for 30 s. The second round of PCR was used to add the index to the amplicons for sequencing. PCR conditions were 95°C for 3 min; 8 cycles of 95°C for 30 s; and 55°C for 30 s, 72°C for 30 s. Sequencing was performed on the MiSeq platform in multiplex. Although no single hypervariable region can distinguish every individual bacteria, the V3 region was chosen because previous literature has found that it contains maximum nucleotide heterogeneity and discriminatory power [25]. Analysis of sequencing data was conducted using Mothur [26]. Briefly, Mothur was used to group or assign 16S rRNA sequences into OTUs using 97% similarity. Classifications were determined by comparing sequences to the Greengene database (gg_13_8_99). Classified OTUs were used to determine the relative abundance of bacterial phyla and family in each sample. To adjust for differences in sequencing depth, all samples were normalized to the same number in the following analysis. Principal coordinate analysis was used to assess community similarity among all samples and Bray-Curtis distances between the communities were displayed in a two-dimensional space [27]. Linear discriminant analysis effect size (LEfSe; https://huttenhower.sph.harvard.edu/galaxy/) was used to compare the differential bacterial abundance with default settings [28]. Bacterial DNA sequencing data were uploaded to the NCBI as PRJNA506222.
SCFA and MCFA Detection
Fecal samples were harvested as described above. Samples were weighed, and sterile molecular-grade water (Sigma-Aldrich) was added to each. Samples were placed on a tilt table for 4 h at room temperature, vortexed once every hour to break up remaining stool pellets, and allowed to settle. For analysis, 900 μL of liquid was removed and 10 μL of 85% phosphoric acid was added to each vial. Volatile fatty acid (VFA) analysis was carried out using an Agilent 6890 gas chromatograph (Agilent, Wilmington, DE, USA) equipped with a split injector, a flame-ionization detector, and Chemstation software for data analysis. VFA separation was performed using a NukolTM fused silica 15 m × 0.53 mm capillary column with 0.5-μm film thickness. Helium was used as the carrier gas at a flow rate of 15 mL/min with a split ratio of 2: 1. The oven temperature was maintained at 80°C for 3 min and then increased to 140°C at a rate of 6°C per minute and held for 1 min. The injector temperature was 200°C and the detector temperature was 250°C. VFAs are expressed as mg/L of individual species (C2-C7 fatty acids), and then divided by weight of total feces collected in grams to a final measurement of mg/L/g feces.
Statistical Analysis
Data were analyzed using GraphPad Prism version 6 (GraphPad, San Diego, CA, USA). We utilized the Student two-tailed t test for comparisons between two experimental groups. Multiple comparisons were conducted using one-way and two-way ANOVA where appropriate followed by Mann-Whitney or Tukey post-test for multiple pairwise examinations. Correlation was also computed using GraphPad Prism. Changes were identified as statistically significant with p < 0.05. Mean values were reported together with the standard error of the mean or standard deviation, as appropriate. Statistical analyses for α-diversity and β-diversity were compared by nonparametric Mann-Whitney U tests and nonparametric multidimensional ANOVA. Distance-based redundancy analysis determined the contribution of different variables to microbiota profile variations.
Data Availability
The datasets used and/or analyzed during the current study have been made public on NCBI's GenBank comprehensive database.
Results
Singly Housed Mice Lacking the Pan-Inflammasome Adaptor ASC Have a Distinct Gut Microbiome Population
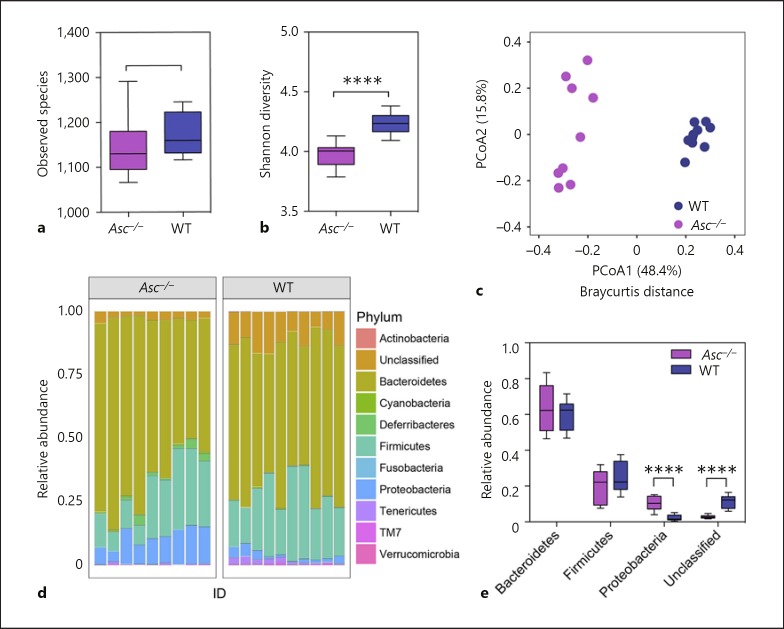
Mice lacking the pan-inflammasome adaptor protein ASC are highly sensitive to experimental colitis and demonstrate significantly increased inflammation-driven colon tumorigenesis [9, 10, 11, 15, 29]. Surprisingly, there are few comprehensive studies evaluating the microbiome composition in the Asc−/− mice under naïve conditions and, of those that have been conducted, there is currently conflicting data regarding the role of ASC in shaping the commensal gut microbiota composition [18, 21]. Here, we sought to evaluate the contribution of ASC and NLR inflammasome signaling on the composition of the gut microbiome in our vivarium, under nonpathological and SPF housing conditions. Mice were necropsied and stool was collected directly from the colons of 9 Asc−/− and 10 wild-type animals, without scraping but by removing each pellet with sterile forceps. Thus, it is likely that mucosal-adherent bacteria were underrepresented in the samples. Through 16S sequencing, we observed significant, genotype-associated changes in microbial ecology between these separately housed knockout animals and wild-type mice (Fig. 1a-e). Although we did not investigate the difference in observed species between the genotypes, the Shannon's diversity index of the gut microbiome was significantly decreased in Asc−/− mice relative to wild-type mice (Fig. 1a, b). We next determined the β-diversity to assess the differences in diversity among fecal specimens from the Asc−/− and wild-type mice (Fig. 1b). Principal coordinate analysis (PCoA) of Bray-Curtis distances illustrated that, although cage and housing was a relevant factor, the main separation of the fecal communities was determined by mouse genotype (Fig. 1c). Phylum level analysis indicated that the fecal microbial community composition is dominated by Bacteroidetes, Firmicutes, Proteobacteria, and unclassified bacteria (Fig. 1d; online suppl. Table S2). Despite inter-individual differences, relative abundance was significantly higher for Proteobacteria and lower for unclassified bacteria in Asc−/− mice (Fig. 1e). Together, these findings show that our Asc−/− colony has a different gut microbiome composition compared to our wild-type colony in the absence of an applied stressor.
Fig. 1.
Inflammasome-deficient mice have a unique microbiome relative to wild-type animals. a The observed species number between Asc−/− (purple) and wild-type (blue) animals. b The Shannon diversity index shows Asc−/− mice have significantly less diverse populations than wild-type mice. c PCoA of 16s rRNA sequences using Bray-Curtis separation reveals distinct separation and clustering based on genotype (ANOSIM R = 0.9827, p = 0.001, 999 permutations). d Relative abundance of bacteria in Asc−/− and wild-type animals by phylum. e Relative abundance of observed phyla differences between wild-type and Asc−/− mice revealed that Asc−/− mice have a significantly increased abundance of Proteobacteria and significantly decreased abundance of unclassified bac-teria compared to wild-type animals. Wild-type (blue), n = 10; Asc−/− (purple), n = 9.
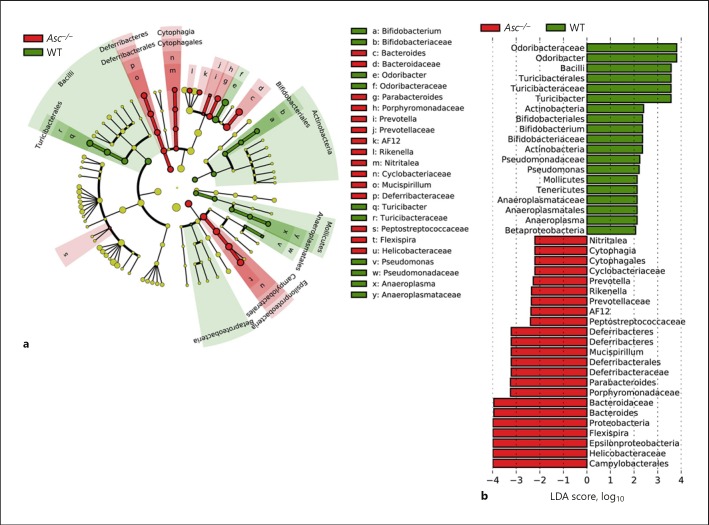
We next sought to evaluate the specific taxonomic variations in the gut microbiomes from Asc−/− and wild-type mice. We used LEfSe to classify specific bacteria that were differentially represented in the Asc−/− and wild-type microbiomes [28] (Fig. 2a). The wild-type mice had a higher abundance of bacteria from Odoribacter, Turicibacter, Bifidobacterium, Pseudomonas, and Anaeroplasma genera, with respective LDA scores (log10) greater than 2 under all-against-all strategy (Fig. 2a, b). Conversely, Asc−/− mice had a higher abundance of Bacteroides, Parabacteroides, Rikenella, AF12, Helicobacter, Prevotella, Mucispirillum, and Flexispira genera, with respective LDA scores greater than 2 (Fig. 2a, b). These findings are relatively consistent with those originally reported by Elinav et al. [18], including the findings of increased Prevotella. This is not surprising, as the experimental conditions for this assessment most closely match those of Elinav et al. [18], rather than assessments of ex-germ-free animals evaluated by Mamantopoulos et al. [21]. Combined, these data suggest that in mice lacking the pan-inflammasome adaptor protein ASC, there is an outgrowth of bacteria that could be implicated in a dysbiotic microbiome. This interpretation would be consistent with the previously reported increased sensitivity of these mice to models of experimental colitis and colitis-associated cancer [18, 29].
Fig. 2.
The absence of inflammasome signaling results in dysbiosis and an overabundance of specific bacterial genera. a Cladogram displaying differential bacterial abundance in Asc−/− mice (red) and wild-type mice (green). b LEfSe differential analysis displaying bacterial abundance in Asc−/− mice and wild-type mice reveals that significant differences exist between the microbial populations. The graph was generated using the LEfSe program. Wild-type (green), n = 10; Asc−/− (red), n = 9.
The NLRP1 Inflammasome Appears to Be Critical for Maintaining a Healthy Colonic Microbial Ecosystem
As with other PRRs, the inflammasome-forming NLRs are highly conserved and evolutionarily designed to sense specific PAMPs associated with bacteria from the same order, family, and/or genus. Thus, we postulated that loss of inflammasome signaling would result in the expansion of very specific bacteria populations in the gut microbiome. In essence, bacteria typically sensed by a specific NLR would be able to more readily expand into permissive niches in the absence of inflammasome signaling that would normally drive the host innate immune system to limit the expansion of commensal or pathogenic elements of the gut microbiota. Our data associated with microbiome changes in the Asc−/− mice support this hypothesis and suggest that NLR inflammasomes may be important in maintaining GI homeostasis via modulation of the composition of the microbiome.
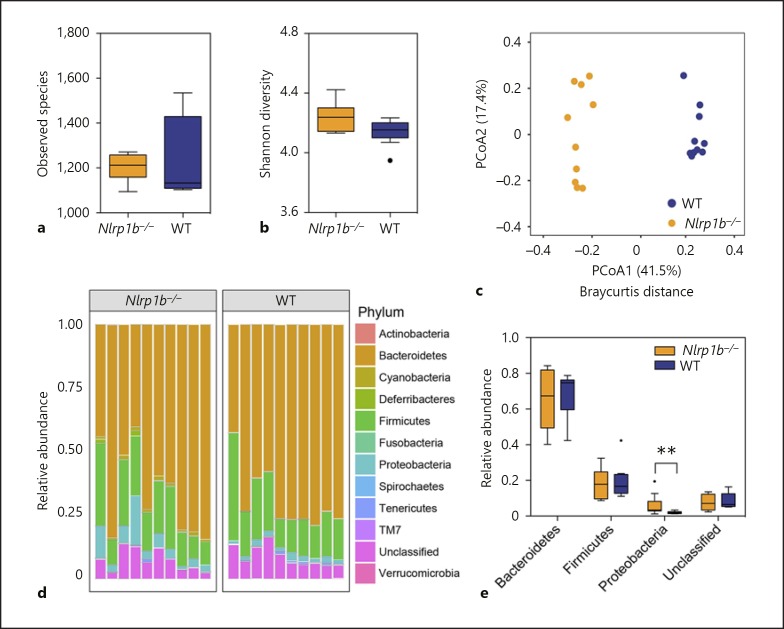
It has been postulated that the NLRP3 and NLRP6 inflammasomes regulate immune system homeostasis in the gut, in part through modulating specific bacteria populations in the host microbiome. While the mechanisms associated with the function of these NLRs in modulating the host microbiome is currently an area of intense research, it is reasonable to conclude that these microbiome differences are associated, either directly or indirectly, with the increased pathogenesis of experimental colitis in the Nlrp3−/− and Nlrp6−/− mice [11, 15, 18, 19, 20]. As previously stated, similar to NLRP3 and NLRP6, mice lacking NLRP1 also demonstrate significantly increased progression of experimental colitis and colitis-associated cancer [9]. However, virtually nothing is known about the microbiome composition of Nlrp1b−/− animals. Thus, based on the similarity of the phenotypes observed in animals lacking a functional NLRP1 inflammasome with Nlrp3−/− and Nlrp6−/− animals, we next sought to evaluate the microbiome composition in the Nlrp1b−/− mice. As described above for the Asc−/− studies, stool was collected directly from the colons of 10 Nlrp1b−/− and 10 wild-type mice, and the V3 region of 16s rRNA was sequenced. Due to the fact that two different cohorts of wild-type mice were used for the Asc−/− and Nlrp1b−/− versus wild-type comparisons, we compared the two groups (online suppl. Fig. S1; online suppl. Table S4). Despite variations in PCoA between the four groups, the wild-type group compared with Asc−/− and the wild-type group compared with Nlrp1b−/− clustered together, indicating that genotype contributed to the composition of the microbiome for the two wild-type groups (online suppl. Fig. S1B). We did not observe a significant difference in alpha diversity between wild-type and Nlrp1b−/− mice (Fig. 3a, b). However, PCoA of Bray-Curtis distances revealed significant separation of microbial communities based on genotype (Fig. 3c). Likewise, phylum level analysis indicated that the relative abundance of Proteobacteria was significantly higher in Nlrp1b−/− mice (Fig. 3d-e; online suppl. Table S3). Together, these data suggest that our Nlrp1b−/− colony has a different gut microbiome composition compared to our wild-type colony.
Fig. 3.
NLRP1 appears to be essential for maintaining the composition of the host microbiome. a The observed species number between Nlrp1b−/− (orange) and wild-type (blue) animals. b The Shannon diversity index shows Nlrp1b−/− mice have no significant difference in population diversity compared to wild-type mice. c PCoA of 16s rRNA sequences using Bray-Curtis separation revealed distinct separation and clustering based on genotype (ANOSIM R = 0.9851, p = 0.001, 999 permutations). d Relative abundance of bacteria in Nlrp1b−/− and wild-type animals by phylum. e Relative abundance of observed phyla differences between wild-type and Nlrp1b−/− mice revealed that Nlrp1b−/− mice have significantly increased abundance of Proteobacteria compared to wild-type animals. Wild-type (blue), n = 10; Nlrp1b−/− (orange), n = 10.
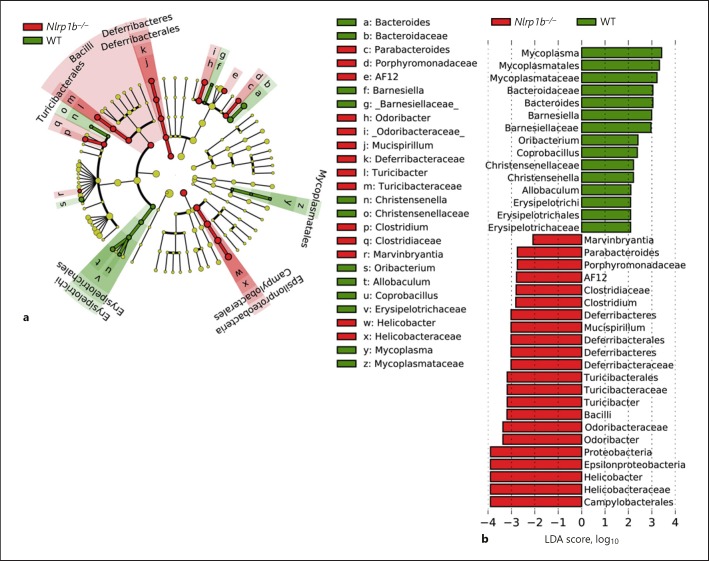
While NLRP1 was one of the first NLRs identified, it is one of the least characterized. For example, it is still unclear what microbial-associated molecular patterns or signals are recognized by NLRP1, especially in the context of GI health and disease. To date, NLRP1 has only been shown to recognize anthrax lethal toxin (LeTx) and Toxoplasma gondii [30, 31], neither of which are likely associated with either experimental colitis or cancer pathogenesis in the Nlrp1b−/− mice. Thus, it is unclear what specific microbial families and genera may flourish in the gut microbiome in the absence of NLRP1. To evaluate the microbial ecosystem in greater detail, we utilized LEfSe to identify and classify specific bacteria in the microbiomes from singly housed and separated Nlrp1b−/− and wild-type mice (Fig. 4a, b). In wild-type mice, a higher abundance of bacteria from the Bacteroides, Oribacterium, Allobaculum, and Barnesiella genera were observed, while in the Nlrp1b−/− mice we observed increased representation from bacteria associated with Helicobacter, Parabacteroides, Clostridium, Odoribacter, Turicibacter, and Mucispirillum genera with respective LDA scores greater than 2 (Fig. 4a, b). Indeed, many of these bacteria genera were also consistently increased in the Asc−/− mice (Fig. 2). Interestingly, the abundance of several of the genera in the microbiomes in the Nlrp1b−/− mice are associated with more commensal and pathogenic species known to be associated with inflammation and cancer in the GI tract, including H. pylori [32], H. hepaticus [33], M. schaedleri [34], and C. difficile [35]. It is tempting to speculate that the previously reported increased sensitivity of these knockout animals in models of experimental colitis and colitis-associated tumorigenesis could be due to changes in their microbiome composition [9].
Fig. 4.
Loss of NLRP1 results in excessive expansion of specific bacteria genera associated with IBD and cancer. a Cladogram display of differential bacterial abundance in in Nlrp1b−/− mice (red) and wild-type mice (green). b LEfSe differential analysis displaying bacterial abundance in Nlrp1b−/− mice and wild-type mice revealed that significant differences exist between the microbial populations. The graph was generated using the LEfSe program. Wild-type (green), n = 10; Nlrp1b−/− (red), n = 10.
Alterations in the GI Microbiome in NLRP1 Inflammasome-Deficient Mice Results in Significant Shifts in the Metabolic Profile
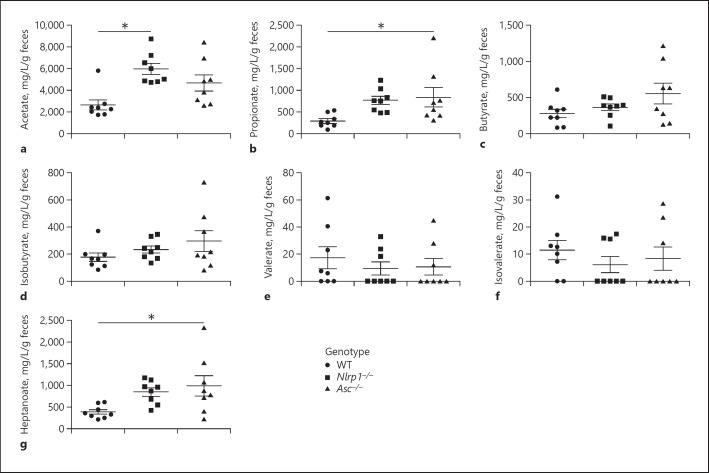
Changes in the composition of the gut microbiome can have dramatic effects on biochemistry and metabolism in the GI tract. Thus, we next sought to evaluate the functional consequences of the changes in the microbial ecosystems in the Asc−/− and Nlrp1b−/− mice. To assess metabolic changes associated with bacteria metabolism in the gut, we quantified fecal levels of SCFA and MCFA using mass spectrometry (GC/MS; Fig. 5). SCFAs are a necessary waste product, required to balance redox equivalent production in the anaerobic environment of the gut [36]. SCFAs are saturated aliphatic organic acids that consist of one to five carbons, of which acetate (C2), propionate (C3), and butyrate (C4) are the most abundant (≥90–95%) in the gut and are associated with carbohydrate fermentation [37]. We found significant differences in SCFA levels in the feces from Nlrp1b−/− and Asc−/− mice compared to the wild-type animals (Fig. 5a, b, g). Specifically, Nlrp1b−/− mice had significantly increased levels of acetate (5,978.6 mg/L/g feces) compared to the wild-type animals (2,642.9 mg/L/g feces; Fig. 5a). Similarly, Asc−/− mice had significantly increased levels of propionate (837.4 mg/L/g feces) compared to the wild-type animals (292.4 mg/L/g feces; Fig. 5b). While not attaining statistical significance, we also observed trending increases in propionate in the Nlrp1b−/− mice and acetate and butyrate in the Asc−/− animals compared to the wild-type mice (Fig. 5a-c). In addition to these common SCFA, we also evaluated isobutyrate (iC4), valerate (C5), and isovalerate (iC5), which are typically much less abundant (approx. 5–10%) and are associated with protein fermentation. Unlike the SCFAs associated with carbohydrate metabolism, we did not observe any significant differences in levels of isobutyrate, valerate, or isovalerate (Fig. 5d-f). Finally, we also evaluated the MCFA heptanoate (C7; Fig. 5g). MCFAs have been identified as the most discriminatory metabolites between healthy controls and patients with IBD [38]. Interestingly, we observed a significant increase in heptanoate in the Asc−/− mice (997.3 mg/L/g feces) compared to the wild-type animals (387.8 mg/L/g feces; Fig. 5g). While not reaching statistical significance, we also observed a trend towards increased heptanoate in the Nlrp1b−/− mice (826.5 mg/L/g feces) compared to wild-type animals.
Fig. 5.
SCFA and MCFA levels significantly differ based on genotype. Abundance of the acetate (C2; a), propionate (C3; b), butyrate (C4; c), isobutyrate (iC4; d), valerate (C5; e), isovalerate (iC5; f), and heptanoate (C7; g) in mg/L/g of feces of wild-type, Nlrp1b−/−, and Asc−/− mice. n = 8 mice/genotype. * p < 0.05.
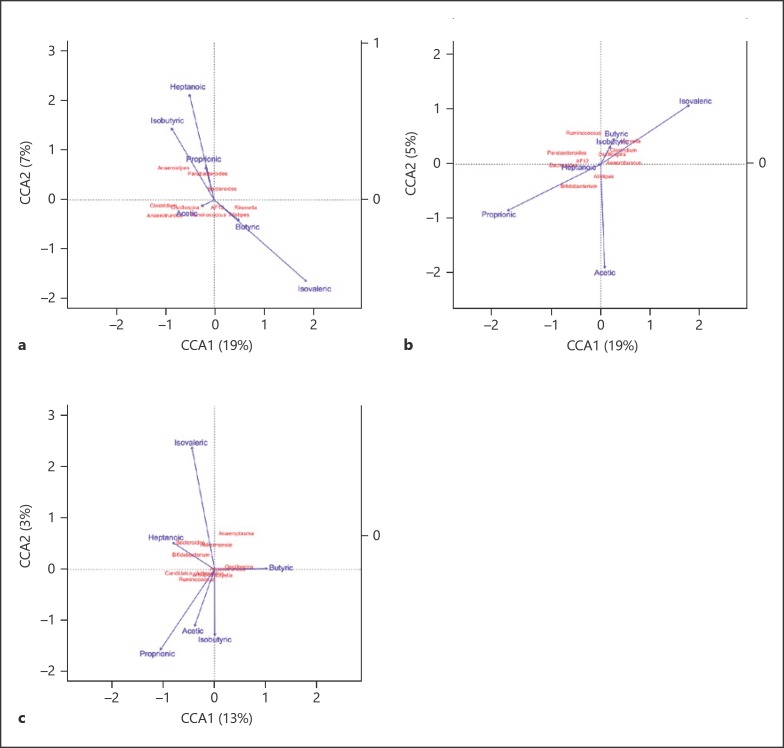
To better determine the influence of any specific bacteria within the microbiomes on the SCFA and MCFA abundance, we conducted canonical correspondence analysis between the fatty acids and the top ten most abundant genera from the microbiota profiles of each genotype (Fig. 6). In Asc−/− mice, Anaerostipes was ordered in relation to isobutyrate (Fig. 6a). Bifidobacterium was ordered in relation to propionate in the Nlrp1b−/− group (Fig. 6b). In the wild-type case, Oscillospira and Anaerotruncus were ordered in relation to butyrate (Fig. 6c). Together, these data suggest that the changes in the composition of the microbiome in the Nlrp1b−/− and Asc−/− mice are sufficient to alter levels of metabolites that reflect an imbalance in GI homeostasis.
Fig. 6.
Canonical correspondence analysis between fatty acids and microbial profiles revealed insight into metabolic differences in microbiomes. Ordination diagrams from canonical correspondence analysis (CCA) of genera abundances and SCFAs and MCFAs from Asc−/− (a), Nlrp1b−/− (b), and wild-type (c) mice. Blue arrows indicate the direction and magnitude of SCFAs associated with bacterial community structures. Bacterial communities are represented in red.
Maternal Influence, Not Genotype, Is the Biggest Factor on Microbiome Composition between Cohoused Wild-Type and Nlrp1b−/− Mice
Some of the earliest data suggesting a role for the microbiome in the sensitivity of Nlrp3−/− and Nlrp6−/− mice utilized cohousing studies with wild-type animals [11, 14]. These cohousing studies demonstrated that experimental colitis progression could be significantly attenuated in mice lacking NLRP3 or NLRP6 inflammasomes by cohousing with wild-type animals [11, 14]. Conversely, the wild-type mice demonstrated increased disease progression during cohousing with either Nlrp3−/− or Nlrp6−/− mice. Similar to these prior studies, we have also shown that the NLRP1 inflammasome-deficient phenotype is partially transmissible between Nlrp1b−/− mice to wild-type animals, when weaned and cohoused together and evaluated in models of experimental colitis [9]. Wild-type mice cohoused with Nlrp1b−/− animals displayed increased weight loss patterns compared to non-cohoused wild-type mice. However, this increased weight loss did not reach the same level as singly housed Nlrp1b−/− animals [9]. Notably, Nlrp1b−/− mice housed with wild-type animals were not protected from the phenotype during the experimental colitis model, which illustrates that pathogenicity can be transferred, but not protection [9]. To better define this previous observation, we sought to characterize the microbiome in both Nlrp1b−/− and wild-type mice following cohousing (Fig. 7). We were interested in two different strategies for cohousing knockout mice with wild-type mice, as both have been used to support previous claims in the field. We postulated that the cohoused wild-type mice would develop a microbiome population more consistent with the mice lacking the NLRP1 inflammasome, whereas the microbiome in the cohoused Nlrp1b−/− mice would remain relatively unchanged, and that this would remain consistent regardless of cohousing strategy.
Fig. 7.
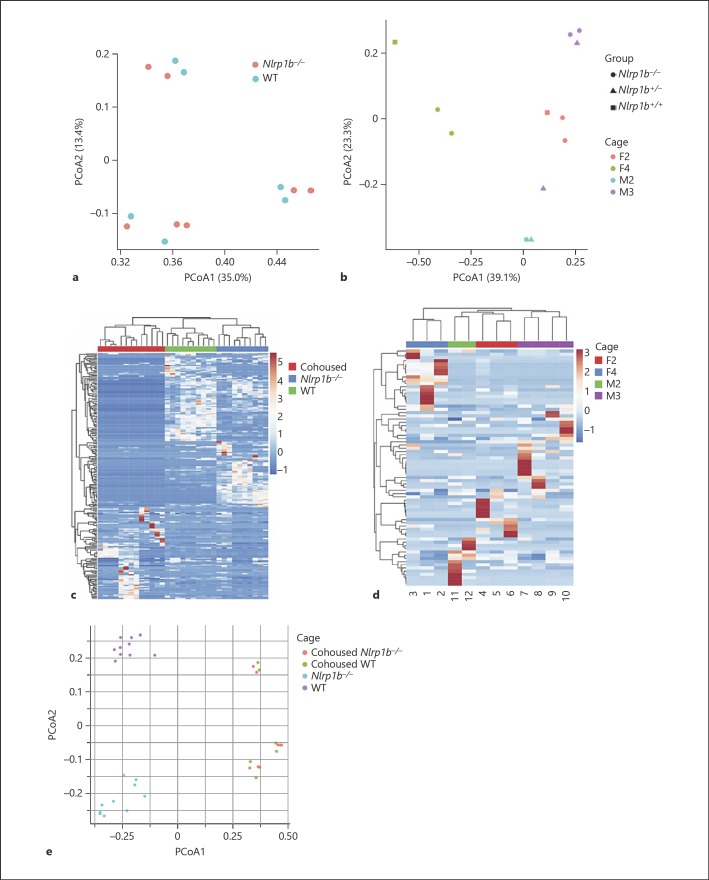
Cohousing strategy revealed that maternal influence is the driving factor in distinct microbiome populations in Nlrp1b−/− and wild-type animals. a PCoA of 16s rRNA sequences using Bray-Curtis separation identified three separate clusters with wild-type (blue) and Nlrp1b−/− (peach) cohoused with wild-type. These clusters correspond to individual housing for each group of mice. b PCoA of 16s rRNA sequences using Bray-Curtis separation of littermate-cohoused wild-type, heterozygote, and Nlrp1b−/− mice showed that cage is the predominant influence on separation (ANOSIM R = 0.7687, p = 0.001, 999 permutations). c The heatmap of > 1,000 OTUs revealed differences between the wild-type, Nlrp1b−/−, and matched-cohoused animals. d The heatmap of > 1,000 OTUs revealed that the major separations between the littermate-cohoused animals is dependent on cage. e PCoA of 16s rRNA sequences using Bray-Curtis separation identified separate clusters based on cages between wild-type individually housed (purple), Nlrpb1−/− individually housed (teal), wild-type (green) cohoused with Nlrp1b−/−, or Nlrp1b−/− (peach) cohoused with wild-type. Wild-type, n = 10 singly housed; Nlrp1b−/−, n = 10 singly housed; matched-cohoused, n = 13; littermate-cohoused, n = 12.
We employed two distinct cohousing strategies. In the first, age- and gender-matched Nlrp1b−/− and wild-type mice were weaned together (1: 1; matched-cohoused) while in the second Nlrp1b−/− and wild-type littermates from the offspring of Nlrp1b+/− mice were weaned together (littermate-cohoused). In both cases, the animals were treated with a standard antibiotic cocktail in their drinking water to ablate the existing bacterial components of their respective microbiomes. After a 2-week course of antibiotic treatment, the animals were transitioned back to normal drinking water. At 12 weeks of age (matched-cohoused) and 7 weeks of age (littermate-cohoused), fecal samples were collected following necropsy. Through 16s sequencing, we found that both matched-cohoused (Fig. 7a) and littermate-cohoused (Fig. 7b) animals clustered together mainly based on cage. When comparing the relative abundance at the genus level, we picked the most abundant genera (more than average 0.5% in all samples) and observed significant increases in bacteria from the Bacteroides and Clostridium genera in matched-cohoused mice (online suppl. Table S5), and no reliable significant differences in littermate-cohoused mice (online suppl. Table S6).
We originally predicted that genotype would be the main contributor of microbiome composition, and therefore anticipated the Nlrp1b−/− mice and wild-type ani mals would have significantly different microbiome populations following either cohousing strategy, where the Nlrp1b−/− microbiome would remain stable and the wild-type microbiome would migrate towards the Nlrp1b−/− animal's composition. These observations would be consistent with our prior observations following cohousing in the experimental colitis models, which revealed that the Nlrp1b−/− phenotype was unchanged whereas the wild-type mice became more sensitive to models of experimental colitis [9]. We grouped the microbiomes of matched-cohoused Nlrp1b−/− and wild-type mice and compared against singly housed Nlrp1b−/− and singly housed wild-type mice (Fig. 7c), and observed what appeared to be separation between the three groups. PCoA analysis between the three groups indicated that the microbiome of matched-cohoused Nlrp1b−/− and wild-type mice are different and unique following cohousing, suggesting that even partial transfer of the Nlrp1b−/− microbiome is sufficient to skew the GI ecosystem of wild-type mice and potentially enhance susceptibility to experimental colitis (Fig. 7e). However, clustering analysis based on Bray-Curtis distance showed that this separation was dependent on cage rather than genotype (online suppl. Fig. S2). Interestingly, the littermate-cohoused cohort separated predominantly based on cage (Fig. 7d) and not on genotype. This suggests that a matched knock out and wild-type cohousing strategy is not sufficient to determine genetic influences on the microbiome. Taken together, it is evident that the maternal influence in the Nlrp1b−/− mice, and not genetic makeup, is the biggest indicator of microbiome composition.
Discussion
Within the NLR field, we are not alone in drawing the conclusion that the microbiome likely plays a role in the phenotype observed in NLR knockout animals. Beyond NLRP1, NLRP3, NLRC4, and NLRP6 have been consistently shown to significantly modulate the progression of experimental colitis and/or colitis-associated tumorigenesis in mouse models [9, 10, 11, 12, 13, 14]. Likewise, the inflammasome adaptor protein ASC and the relevant caspases, caspase-1 and caspase-11, have also been shown to dramatically modulate inflammation in the gut during IBD and cancer [9, 10, 11, 15, 16]. In general, the majority of studies evaluating NLR inflammasomes have found that loss of any specific NLR or inflammasome component results in increased GI inflammation, epithelial cell barrier disruption, and inflammation-driven tumorigenesis. It makes logical sense that the next direction in the field is to investigate the microbiome of these animals, especially because cohousing NLR knockout animals with wild-type mice is sufficient to, at least partially, transfer susceptibility to DSS [9, 10, 11, 12, 13, 14]. Notably, there has been significant controversy in the PRR field, including both TLRs and NLRs, associated with conflicting results in models of experimental colitis and/or cancer. Despite the consensus data that demonstrates the loss of any specific inflammasome-forming NLR results in significantly increased susceptibility in DSS-induced experimental colitis models [9, 10, 11, 12, 16, 18], there are also counter observations reported that reveal either no differences in phenotype between the NLR-deficient animals and wild-type mice, or even opposite results that suggest attenuating NLR inflammasomes actually protect against disease pathogenesis [29]. Unlike the other NLRs, NLRP6 seemed to be exempt from conflicting results in colitis studies and, until recently, it has been widely accepted that the main role of this NLR was in regulating the intestinal microbiome composition [21]. However, this paradigm has recently also been questioned with results that conclude that NLRP6 and ASC do not actually play a role in influencing the gut microbiome composition [21, 22]. These studies demonstrated that maternal effects and caging have a stronger influence than genotype when different forms of cohousing were employed [21, 22]. These data led us to further investigate our initial conclusions that the NLRP1 inflammasome functions to regulate the intestinal microbiome, and provided the rationale for our cohousing comparisons. In the present study, we report our initial findings that the microbiome composition of singly housed and homozygous bred Nlrp1b−/− and Asc−/− mice were populated with bacteria that appeared to suggest that the NLRP1 inflammasome regulates microbial ecology, consistent with data on the role of NLRP6 [15, 18, 20]. We investigated and found differences not only in the genera of bacteria of our individually bred and housed knockout animals, but also in the abundance of SCFA and MCFA.
The preliminary motivation for the data generated in the present report was to identify families and genera of bacteria that flourish in the absence of a functional NLRP1 inflammasome in an attempt to define the driving factor of the increased sensitivity to colitis in knockout animals. The NLRP1 inflammasome senses anthrax LeTx and T. gondii in mice and muramyl dipeptide in humans [30, 31]. Consistent with the previous in vitro data, both Nlrp1−/− mouse strains fail to properly process pro-IL-1β and pro-IL-18 following in vivo exposure with these agents [23]. While these data provide significant mechanistic insight into NLRP1 inflammasome function, recognition of anthrax LeTx and T. gondii do not explain its protective effects during experimental colitis and colitis-associated cancer. The results from our initial microbiome analysis of our separately and individually bred and housed knockout mice identified several bacteria genera that contain species associated with inflammation and cancer in the GI tract, including H. pylori, H. hepaticus, M. schaedleri, and C. difficile, all of which drive colitis and/or tumorigenesis through diverse mechanisms, including: degrading the protective mucus layer in the gut, producing barrier-degrading toxins and metabolites, restricting the growth of probiotic bacteria, and directly inducing acute inflammation [32, 33, 34, 35]. This point is particularly compelling in the initial hypothesis that the composition of the microbiome of these animals is the culprit for the sensitivity to colitis in the absence of NLRP1. For example, we found strong relationships between loss of NLRP1 signaling and the increased abundance of Bacil li, Deferribacteres, and Epsilonproteobacteria classes (Fig. 4). Thus, these data would support future studies evaluating species that are representative of these classes in an effort to identify specific bacteria species associated with NLRP1 inflammasome formation and function in the gut. Moreover, this concept is reinforced by the finding that Bacilli species are overabundant in the absence of NLRP1 inflammasome signaling, which is consistent with the already defined role of NLRP1 sensing B. anthracis (a member of the Bacilli class). Thus, it is tempting to speculate that changes in the host microbiome that are directly associated with loss of NLRP1 inflammasome function may underlie many of these disorders.
However, prior to concluding that the NLRP1 inflammasome regulates colonic homeostasis, we compared our work to that of the evolving story and debate surrounding the role of the NLRP6 inflammasome in the regulation of the gut microbiome. Similar to NLRP1, NLRP6 inflammasome-deficient mice were also found to be more sensitive to intestinal inflammation and tumorigenesis in the chemically induced colitis and colitis-associated tumorigenesis models [12, 18]. While it is clear that altered processing of pro-IL-18 in the colonic epithelial cells contributes to disease pathogenesis in these animals, these studies and subsequent confirmatory data concluded that altered fecal microbiota during colitis also contributed to intestinal hyperplasia, inflammatory cell recruitment, and exacerbation [18]. Pertinently, antibiotic treatment attenuated colitis progression in the Nlrp6−/− mice and cohousing studies demonstrated that the colitogenic activity of the microbiota is transferable to wild-type animals [18]. The loss of NLRP6 inflammasome signaling was found to lead to significant expansion of the bacterial phyla Bacteroidetes (Prevotellaceae) and reductions in members of the Lactobacillus genus and in the Firmicutes phylum [18]. More detailed mechanistic studies reported that the expansion of Prevotellaceae was facilitated by attenuated goblet cell mucin production in the Nlrp6−/− animals, which established a permissive niche in the GI tract [15]. Our initial data most closely resembles that of Elinav et al. [18] in terms of the homozygous bred and singly housed Asc−/−, and even some of the changes observed in the Nlrp1b−/− mice. This is expected as our matched cohoused experimental design most closely reflected those utilized by these studies and the data show that separately bred and housed mice lacking NLR inflammasome components have significantly different microbiome compositions under naïve conditions. ASC is a critical adaptor protein for inflammasome formation by functioning as a bridge between NLRs containing PYD domains and caspase-1, as well as enhancing the activity of NLRs containing CARD domains. Due to its role as a pan-inflammasome adaptor protein, the role of ASC in IBD and colitis-associated cancer has been well studied. In models based on chemically induced colitis, loss of ASC results in a highly severe phenotype compared to other individual NLR-deficient genotypes [reviewed in 8]. The consensus data agree that mice lacking functional ASC fail to generate mature IL-1β and IL-18 in the colon during models of experimental colitis, which results in immune system and epithelial barrier defects [11]. The combination of these defects was suggested to result in enhanced permeability and bacteria translocation in the Asc−/− mice [9].
However, subsequent studies utilizing littermate-controlled Nlrp6−/− mice and ex-germ-free littermate-controlled Asc−/− mice revealed that regulation of the gut microbiota was independent of genotype [21]. Here, the authors clearly show that housing and maternal effects contribute to the commensal gut microbiota composition, rather than components of the NLRP6 inflammasome [21]. Indeed, these findings are consistent with our results associated with Nlrp1b−/− mice, using a similar experimental design. Thus, similar to NLRP6, the absence of the NLRP1 inflammasome does not appear to significantly influence the microbiome of cohoused homozygous littermates bred from heterozygous parents. The most deterministic factor on the microbial composition in our animals are maternal and housing factors, and not genetic. Because our previous studies evaluating NLRP1 in experimental colitis and colitis-associated cancer utilized littermate control animals, we can thus conclude that microbiome differences are likely not a significant component of the phenotypes observed in the Nlrp1b−/− mice in these models [9].
The microbiome is consistently cited as a potential factor influencing disease progression in not only IBD and colitis-associated cancer, but the gamut of seemingly non-GI-related diseases, from Alzheimer's disease to cardiovascular and metabolic disorders [39, 40]. Indeed, in the vast majority of studies that produce data that differ from the consensus in any given field, changes in the host microbiome is routinely cited as a possible explanation, but is rarely evaluated. Based on the data presented here, it is certainly possible that differences in the microbiome underlie many of the conflicting results in the NLR field, albeit not in the way typically portrayed. Taken together with other reports on NLRP6, our data suggest that the NLR inflammasomes likely play less of a role in directly regulating the host microbiome then initially proposed. The consensus data that are building, which is supported by our findings here, indicate that housing and maternal contributions are stronger variables than genotype in driving microbiome differences. Our data clearly suggest that future studies evaluating NLRs in the context of GI inflammation and microbiome interactions should always utilize littermate control animals, and proper housing should be a major consideration in experimental design and data analysis.
Statement of Ethics
All experiments were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals and following the protocol approved by the Virginia Tech IACUC.
Disclosure Statement
The authors declare that they have no conflicts of interest.
Funding Sources
Grants were awarded from the US National Institutes of Health (I.C.A.; R03 DK105975 and K01 DK092355); the Via College of Osteopathic Medicine (VCOM) One Health Center Seed Funding (I.C.A.); Virginia Maryland College of Veterinary Medicine Internal Research Competition (I.C.A.), and the Virginia Tech Institute for Critical Technology and Applied Sciences (I.C.A.). V.M.R.-S. was supported through the American Association of Immunologists Careers in Immunology Fellowship Program. This work was supported, in part, by the Intramural Research Program of the National Institute of Environmental Health Sciences (ES101965 to P.A.W.).
Author Contributions
Design and execution of the experiments were conducted by V.M.R.-S., Y.Q., C.A.T., K.E.H., D.K.M., and K.E. V.M.R.-S. and C.A.T. were responsible for mouse breeding, genotyping, and general husbandry. Experimental data interpretation was conducted by V.M.R.-S., Y.Q., C.A.T., K.E.H., D.K.M., K.E., P.A.W., and I.C.A. All authors contributed to the writing and revising of the manuscript.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Acknowledgements
The authors would like to thank Dr. Beverly H. Koller (The University of North Carolina at Chapel Hill) for generously providing the Nlrp1b−/− mice used in this study. We also wish to acknowledge the assistance of the Teaching and Research Animal Care Support Service (TRACSS) at the Virginia Maryland College of Veterinary Medicine, as well as our technician, graduate students, and undergraduate research students, including Dr. Sheryl Coutermarsh-Ott, Daniel Rothschild, and Bettina Heid. We would also like to especially acknowledge Jody Smiley from the Virginia Tech Sludge Lab for assistance with fecal metabolite analysis. We gratefully acknowledge outstanding technical support in 16s sequencing from the Epigenomics Core Facility at NIEHS.
References
- 1.Francescone R, Hou V, Grivennikov SI, Cytokines IB. Cytokines, IBD, and colitis-associated cancer. Inflamm Bowel Dis. 2015 Feb;21((2)):409–18. doi: 10.1097/MIB.0000000000000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alipour M, Zaidi D, Valcheva R, Jovel J, Martínez I, Sergi C, et al. Mucosal Barrier Depletion and Loss of Bacterial Diversity are Primary Abnormalities in Paediatric Ulcerative Colitis. J Crohn's Colitis. 2016 Apr;10((4)):462–71. doi: 10.1093/ecco-jcc/jjv223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casén C, Vebø HC, Sekelja M, Hegge FT, Karlsson MK, Ciemniejewska E, et al. Deviations in human gut microbiota: a novel diagnostic test for determining dysbiosis in patients with IBS or IBD. Aliment Pharmacol Ther. 2015 Jul;42((1)):71–83. doi: 10.1111/apt.13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hedin CR, van der Gast CJ, Stagg AJ, Lindsay JO, Whelan K. The gut microbiota of siblings offers insights into microbial pathogenesis of inflammatory bowel disease. Gut Microbes. 2017 Jul;8((4)):359–65. doi: 10.1080/19490976.2017.1284733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Q, Wang C, Tang C, He Q, Li N, Li J. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn's disease. J Clin Gastroenterol. 2014 Jul;48((6)):513–23. doi: 10.1097/MCG.0000000000000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mottawea W, Chiang CK, Mühlbauer M, Starr AE, Butcher J, Abujamel T, et al. Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn's disease. Nat Commun. 2016 Nov;7((1)):13419. doi: 10.1038/ncomms13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Putignani L, Del Chierico F, Vernocchi P, Cicala M, Cucchiara S, Dallapiccola B, Dysbiotrack Study Group Gut Microbiota Dysbiosis as Risk and Premorbid Factors of IBD and IBS Along the Childhood-Adulthood Transition. Inflamm Bowel Dis. 2016 Feb;22((2)):487–504. doi: 10.1097/MIB.0000000000000602. [DOI] [PubMed] [Google Scholar]
- 8.Ringel-Scaia VM, McDaniel DK, Allen IC. The Goldilocks Conundrum: NLR Inflammasome Modulation of Gastrointestinal Inflammation during Inflammatory Bowel Disease. Crit Rev Immunol. 2016;36((4)):283–314. doi: 10.1615/CritRevImmunol.2017019158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams TM, Leeth RA, Rothschild DE, Coutermarsh-Ott SL, McDaniel DK, Simmons AE, et al. The NLRP1 inflammasome attenuates colitis and colitis-associated tumorigenesis. J Immunol. 2015 Apr;194((7)):3369–80. doi: 10.4049/jimmunol.1402098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti TD. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010 Mar;32((3)):379–91. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen IC, TeKippe EM, Woodford RM, Uronis JM, Holl EK, Rogers AB, et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med. 2010 May;207((5)):1045–56. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen GY, Liu M, Wang F, Bertin J, Núñez G. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J Immunol. 2011 Jun;186((12)):7187–94. doi: 10.4049/jimmunol.1100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu B, Elinav E, Huber S, Booth CJ, Strowig T, Jin C, et al. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Proc Natl Acad Sci USA. 2010 Dec;107((50)):21635–40. doi: 10.1073/pnas.1016814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu B, Elinav E, Huber S, Strowig T, Hao L, Hafemann A, et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc Natl Acad Sci USA. 2013 Jun;110((24)):9862–7. doi: 10.1073/pnas.1307575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang JP, Brown EM, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014 Feb;156((5)):1045–59. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupaul-Chicoine J, Yeretssian G, Doiron K, Bergstrom KS, McIntire CR, LeBlanc PM, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010 Mar;32((3)):367–78. doi: 10.1016/j.immuni.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 17.Hebel K, Rudolph M, Kosak B, Chang HD, Butzmann J, Brunner-Weinzierl MC. IL-1β and TGF-β act antagonistically in induction and differentially in propagation of human proinflammatory precursor CD4+ T cells. J Immunol. 2011 Dec;187((11)):5627–35. doi: 10.4049/jimmunol.1003998. [DOI] [PubMed] [Google Scholar]
- 18.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011 May;145((5)):745–57. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaki MH, Lamkanfi M, Kanneganti TD. The Nlrp3 inflammasome: contributions to intestinal homeostasis. Trends Immunol. 2011 Apr;32((4)):171–9. doi: 10.1016/j.it.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levy M, Thaiss CA, Zeevi D, Dohnalová L, Zilberman-Schapira G, Mahdi JA, et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell. 2015 Dec;163((6)):1428–43. doi: 10.1016/j.cell.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mamantopoulos M., et al. Nlrp6- and ASC-Dependent Inflammasomes Do Not Shape the Commensal Gut Microbiota Composition. Immunity. 2017;47((2)):p. 339 e4–348 e4. doi: 10.1016/j.immuni.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Lemire P, Robertson SJ, Maughan H, Tattoli I, Streutker CJ, Platnich JM, et al. The NLR Protein NLRP6 Does Not Impact Gut Microbiota Composition. Cell Reports. 2017 Dec;21((13)):3653–61. doi: 10.1016/j.celrep.2017.12.026. [DOI] [PubMed] [Google Scholar]
- 23.Kovarova M, Hesker PR, Jania L, Nguyen M, Snouwaert JN, Xiang Z, et al. NLRP1-dependent pyroptosis leads to acute lung injury and morbidity in mice. J Immunol. 2012 Aug;189((4)):2006–16. doi: 10.4049/jimmunol.1201065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004 Jul;430((6996)):213–8. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 25.Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 2007 May;69((2)):330–9. doi: 10.1016/j.mimet.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schloss PD. A high-throughput DNA sequence aligner for microbial ecology studies. PLoS One. 2009 Dec;4((12)):e8230. doi: 10.1371/journal.pone.0008230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011 Feb;5((2)):169–72. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011 Jun;12((6)):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bauer C, Duewell P, Mayer C, Lehr HA, Fitzgerald KA, Dauer M, et al. Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut. 2010 Sep;59((9)):1192–9. doi: 10.1136/gut.2009.197822. [DOI] [PubMed] [Google Scholar]
- 30.Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006 Feb;38((2)):240–4. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 31.Ewald SE, Chavarria-Smith J, Boothroyd JC. NLRP1 is an inflammasome sensor for Toxoplasma gondii. Infect Immun. 2014 Jan;82((1)):460–8. doi: 10.1128/IAI.01170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JY, Park HW, Choi JY, Lee JS, Koo JE, Chung EJ, et al. Helicobacter pylori Infection with Atrophic Gastritis Is an Independent Risk Factor for Advanced Colonic Neoplasm. Gut Liver. 2016 Nov;10((6)):902–9. doi: 10.5009/gnl15340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daniel SG, Ball CL, Besselsen DG, Doetschman T, Hurwitz BL. Functional Changes in the Gut Microbiome Contribute to Transforming Growth Factor β-Deficient Colon Cancer. mSystems. 2017 Sep;2((5)):e00065-17. doi: 10.1128/mSystems.00065-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robertson BR, O'Rourke JL, Neilan BA, Vandamme P, On SL, Fox JG, et al. Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol. 2005 May;55((Pt 3)):1199–204. doi: 10.1099/ijs.0.63472-0. [DOI] [PubMed] [Google Scholar]
- 35.Cojocariu C, Stanciu C, Stoica O, Singeap AM, Sfarti C, Girleanu I, et al. Clostridium difficile infection and inflammatory bowel disease. Turk J Gastroenterol. 2014 Dec;25((6)):603–10. doi: 10.5152/tjg.2014.14054. [DOI] [PubMed] [Google Scholar]
- 36.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013 Sep;54((9)):2325–40. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cook SI, Sellin JH. Review article: short chain fatty acids in health and disease. Aliment Pharmacol Ther. 1998 Jun;12((6)):499–507. doi: 10.1046/j.1365-2036.1998.00337.x. [DOI] [PubMed] [Google Scholar]
- 38.De Preter V, Machiels K, Joossens M, Arijs I, Matthys C, Vermeire S, et al. Faecal metabolite profiling identifies medium-chain fatty acids as discriminating compounds in IBD. Gut. 2015 Mar;64((3)):447–58. doi: 10.1136/gutjnl-2013-306423. [DOI] [PubMed] [Google Scholar]
- 39.de J.R.D.-P.V., A.S. Forlenza, and O.V. Forlenza, Relevance of gut microbiota in cognition, behaviour and Alzheimer's disease. Pharmacol Res. 2018 doi: 10.1016/j.phrs.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Brial F, Le Lay A, Dumas ME, Gauguier D. Implication of gut microbiota metabolites in cardiovascular and metabolic diseases. Cell Mol Life Sci. 2018 Nov;75((21)):3977–90. doi: 10.1007/s00018-018-2901-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Data Availability Statement
The datasets used and/or analyzed during the current study have been made public on NCBI's GenBank comprehensive database.