Abstract
Staphylococcus aureus small-colony variants (SCVs) are associated with chronic, persistent, and relapsing courses of infection and are characterized by slow growth combined with other phenotypic and molecular traits. Although certain mechanisms have been described, the genetic basis of clinical SCVs remains often unknown. Hence, we adapted an episomal tool for rapid identification and investigation of putative SCV phenotype-associated genes via antisense gene silencing based on previously described Tnl0-encoded tet-regulatory elements. Targeting the SCV phenotype-inducing enoyl-acyl-carrier-protein reductase gene (fabI), plasmid pSN1-AS‘fabI’ was generated leading to antisense silencing, which was proven by pronounced growth retardation in liquid cultures, phenotype switch on solid medium, and 200-fold increase of antisense ‘fabI’ expression. A crucial role of TetR repression in effective regulation of the system was demonstrated. Based on the use of anhydrotetracycline as effector, an easy-to-handle one-plasmid setup was set that may be applicable to different S. aureus backgrounds and cell culture studies. However, selection of the appropriate antisense fragment of the target gene remains a critical factor for effectiveness of silencing. This inducible gene expression system may help to identify SCV phenotype-inducing genes, which is prerequisite for the development of new antistaphylococcal agents and future alternative strategies to improve treatment of therapy-refractory SCV-related infections by iatrogenically induced phenotypic switch. Moreover, it can be used as controllable phenotype switcher to examine important aspects of SCV biology in cell culture as well as in vivo.
Keywords: Staphylococcus aureus, small-colony variant (SCV), antisense silencing, phenotype switch, enoyl-acyl-carrier-protein reductase gene (fabI), tet-regulatory expression system
Introduction
The small-colony variant (SCV) phenotype represents a slow growing subpopulation of Staphylococcus aureus associated with chronic, persistent, and recurring infections, which are particularly difficult to diagnose and challenging in terms of treatment (Proctor et al., 2006; Vaudaux et al., 2006; Kahl et al., 2016). The colony morphology and physiological characteristics of SCVs differ widely from the WT, not only due to slower growth, pinpoint colonies, reduced or no pigmentation and hemolytic activity, respectively, but also in several biochemical traits such as altered expression of virulence factors, decreased respiration and coagulase activity as well as frequently by auxotrophism for hemin, menadione, thymidine, or fatty acids (Proctor et al., 2006; Seggewiß et al., 2006; Kriegeskorte et al., 2011, 2014; Proctor et al., 2014; Schleimer et al., 2018).
The identification of the genetic mechanisms leading to the phenotype switch are prerequisite to prevent SCV-related recurrence and chronic infection and to develop new antimicrobials not selecting for SCVs. Hitherto, however, only some phenotype switch-related genes and mechanisms were identified by in vitro generation of knockout mutants (including accC, accD, aroD, cspB, hemA, hemB, menA, menB, menD, plsX, sdhCAB, and thyA) and sequencing approaches (including accC, accD, aroB, aroC, aroD, ecfA, ecfT, fabF, fabI, hemA, hemB, hemC, hemD, hemE, hemG, hemH, menA, menB, menC, menE, menF, relA, stp, and thyA) (von Eiff et al., 1997; Bates et al., 2003; Schaaff et al., 2003; Chatterjee et al., 2008; Lannergård et al., 2008; Duval et al., 2010; Gao et al., 2010; Gaupp et al., 2010; Parsons et al., 2011, 2013, 2014; Köser et al., 2012; Wakeman et al., 2012; Hammer et al., 2013; Dean et al., 2014; Painter et al., 2015; Lin et al., 2016; Cao et al., 2017; Zhang et al., 2017; Bazaid et al., 2018; Giulieri et al., 2018; Schleimer et al., 2018; Vestergaard et al., 2018). Besides these SCVs triggered by mutational events restricted to one gene locus, SCVs being the consequence of combined mutations in two or more genes were also rarely described (Hammer et al., 2013; Bui and Kidd, 2015; James et al., 2019). Moreover, the differential expression of some key transcriptional regulators such as SigB and non-protein-coding RNAs (npcRNAs) were shown to contribute to the phenotype switch (Moisan et al., 2006; Abu-Qatouseh et al., 2010; Mitchell et al., 2013; Bui and Kidd, 2015; Tuchscherr et al., 2015). Nevertheless, the (potential) genetic mechanisms of several clinical SCVs remain unidentified and knockout mutant generation and sequencing approaches including whole genome sequencing (WGS) are time-consuming. Moreover, selective disruption of genes essential for growth is not feasible. Aggravating the analysis of clinical SCVs so far, indubitable identification of genes causative for the phenotype switch is only possible for stable SCVs that already underwent mutational adaption and by comparison with a revertant phenotype that spontaneously emerged from the SCV phenotype (Becker et al., 2006; Schleimer et al., 2018). Besides the rarely isolated stable clinical SCVs, in vitro selection for stable SCVs mandatory for WGS as well as for transcriptomics and proteomics can be performed by cultivation in the presence of sublethal concentrations of certain antibiotics (e.g., aminoglycosides) (Balwit et al., 1994) or by serially passaging bacteria in the presence of HeLa cells using modified gentamicin protection assays (McLean et al., 2019). By using these methods, however, the SCV-selective mutations only occur in the corresponding drug target genes or genes essential for antibiotic uptake (Schaaff et al., 2003; McLean et al., 2019). Further approaches selecting for a stable SCV phenotype include a chemostat to generate steady-state growth conditions with low nutrients and low growth rate for a prolonged time and with specific chemical stress (Bui and Kidd, 2015; Bui et al., 2015). Although this is an adequate method, it is also time-consuming and not applicable in every laboratory. Thus, there is an urgent need for simpler, faster, and more cost-effective methods to identify genes that trigger the phenotype switch upon mutational or transcriptional inactivation or downregulation.
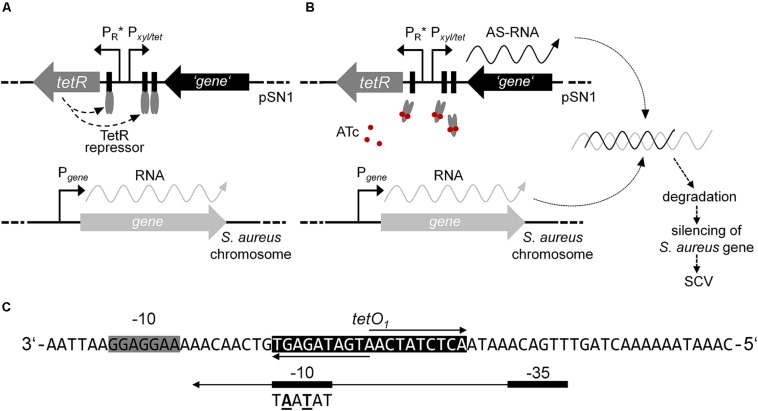
Inducible gene expression systems comprising repressor and operator regulatory elements have been reported for use in the genus Staphylococcus for selectively controlling the (over)expression of genes (Wieland et al., 1995; Ji et al., 1999; Jana et al., 2000; Helle et al., 2011). Amongst these, tetracycline- (Tc) or anhydrotetracycline (ATc)-inducible expression plasmids comprising improved elements originating from the transposable element Tn10 were used not only for overexpression, but also for regulated antisense silencing of distinct genes (Ji et al., 1999, 2004; Stary et al., 2010; Helle et al., 2011). This study focused on the adaption of the expression plasmid pRAB11 (Helle et al., 2011) for the identification of genes potentially triggering the phenotype switch from WT to SCV. Therefore, we developed an ATc-inducible antisense approach outlined in Figure 1. This tet-regulatory system is based on autoregulated expression of the repressor TetR, which is responsible for the adequate regulation of a target gene. TetR attaches to the cognate operator sequences within both its own promoter and in the divergently located promoter of the target gene, thus, inhibiting the transcription of the target gene (Meier et al., 1988). Upon binding of the effector, TetR undergoes a conformational change and the detachment from the operator proceeds (see Figure 1).
FIGURE 1.
Model of episomal ATc-inducible antisense silencing in S. aureus using plasmid pSN1. (A) In the absence of effector ATc, constitutively expressed homodimer TetR (gray ellipses) binds to the cognate operator sequences consequently inhibiting gene transcription via promoter Pxyl/tet and negatively regulating the rate of transcription of tetR. (B) Gene expression proceeds upon induction with ATc (red circles) that results in a conformational change of TetR followed by detachment from the cognate operator sequences. Hybridization of transcribed antisense (AS)-RNA to the complementary mRNA is presumptively followed by degradation and silencing of the respective gene maybe resulting in an SCV phenotype. (C) Sequence of promoter PR∗ according to Geissendörfer and Hillen (1990); extended Shine-Dalgarno sequence of tetR is indicated with a gray box and operator sequence tetO1 with a black box, respectively. The consensus sequence of the –10 site of promoter PtetR of plasmid pRAB11 is shown below the –10 sequence of PR∗ with the changed bases given in bold and underlined.
Using the further improved shuttle plasmid pSN1, silencing of the SCV phenotype-related gene fabI (encodes for enoyl-acyl-carrier-protein reductase) via production of antisense RNA was performed in a one-plasmid setup. Application of this type of episomal silencing system may provide an easy-to-use and fast tool for the identification of phenotype switch-related genes. The identified genes producing the SCV phenotype will not only allow the screening of clinical SCV isolates, in particular those with undefined auxotrophy, for mutations in these genes by simple Sanger sequencing. Furthermore, knockout mutant generation of the identified genes may also contribute to the identification of possible new auxotrophies and thus to the characterization of undefined SCV types, further simplifying the identification of SCVs. Another advantage of the pSN1 tool is that its inducer ATc is able to penetrate cell membranes. Thus, it may not only help to develop future alternative treatment options of therapy-refractory SCV-related infections, e.g., by iatrogenically induced phenotypic switch, but will also offer the opportunity to monitor the effects and consequences of the switching process on the host cell, organs, and the host organism. Finally, this tool may enable the fast suitability analysis of potential new drug target genes regarding their influence on an adverse phenotype switch to the SCV.
Materials and Methods
Bacterial Strains and Plasmids, Growth Conditions, and Antibiotics
All strains and plasmids used are listed in Table 1. Escherichia coli cells were cultivated and grown at 37°C in liquid or on solid Luria-Bertani medium (LB; Becton Dickinson, Franklin Lakes, NJ, United States). S. aureus strains were grown at 37°C in tryptic soy broth/agar (TSB/TSA; Becton Dickinson) or on Columbia blood agar (BBLTM Columbia agar with 5% sheep blood; Becton Dickinson). All liquid cultures were incubated with shaking at 160 rpm in 10 or 50 mL TSB in 100- or 500-mL glass baffled flasks, respectively. Ampicillin (Sigma-Aldrich, St. Louis, MO, United States) was used at a final concentration of 100 μg/mL for selection of E. coli. Chloramphenicol (AppliChem, Darmstadt, Germany) was used at a final concentration of 10 μg/mL for selection of S. aureus. ATc (IBA Lifesciences, Göttingen, Germany) was purchased in dry chemical form, prepared as 10 mM stock solution in 70% ethanol and used as effector agent at a final concentration of 0.4 μM.
TABLE 1.
Bacterial strains and plasmids with their precursors used in this study.
| Strain or plasmid | Description | Source or references |
| S. aureus strains | ||
| NCTC8325-4 | NCTC8325 derivative; agr+; 11-bp deletion in rsbU; cured of three prophages | Novick, 1967; Herbert et al., 2010 |
| RN4220 | NCTC8325-4 derivative; agr–; 11-bp deletion in rsbU; cured of three prophages; MNNG: r–m– | Iordanescu and Surdeanu, 1976; Herbert et al., 2010 |
| SA113 | NCTC8325 derivative; agr–; 11-bp deletion in rsbU; point mutation in tcaR; three prophages, Φ11, Φ12, and Φ13; MNNG: r–m– | Iordanescu and Surdeanu, 1976; Herbert et al., 2010 |
| SAS32/1 | SA113 pRAB11 | This study |
| SAS99/1 | SA113 pSN1 | This study |
| SAS118/2 | SA113 pSN1-AS‘fabI’ | This study |
| E. coli strain | ||
| One shotTM Top10 | E. coli K-12 derivative; F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 Δ lacX74 recA1 araD139 Δ(araleu)7697 galU galK rpsL (StrR) endA1 nupG | Invitrogen/Thermo Fisher Scientific |
| Plasmids | ||
| pCPP-31 | E. coli/B. subtilis shuttle plasmid; derived from plasmids pBR322, pUBll0 and pC194; neo | Band et al., 1983 |
| pWH3531 | E. coli/B. subtilis shuttle plasmid; pCPP-3 derivative carrying an 800-bp fragment comprising Tn10 derived elements tetR (encoding the TetR repressor) and PR∗ (mutant tetR promoter with poly-A block and improved −35 site), and the divergent Pxyl/tet fusion promoter from B. subtilis with 1x tetO; neo | Geissendörfer and Hillen, 1990 |
| pSK2361 | E. coli/Staphylococcus shuttle plasmid with pUC19 cloned into the HindIII site of pC194; bla cat | (unpublished data) Tinsley and Khan, 2007 |
| pALC20731 | E. coli/Staphylococcus shuttle plasmid; pSK236 derivative with 800-bp fragment from pWH353 comprising tetR, PR∗, and Pxyl/tet with 1x tetO (cloned into the PstI and SmaI sites); bla cat | Bateman et al., 2001 |
| pRMC21 | E. coli/Staphylococcus shuttle plasmid; pALC2073 derivative with mutated −10 sequence within the tetR promoter (5′-tag ag t-3′→ 5′-tat aa t-3′; PtetR) and extended MCS (KpnI, HpaI, BglII, SacI/BanII, and EcoRI); tetR, PtetR, and Pxyl/tet with1x tetO; bla cat | Corrigan and Foster, 2009 |
| pRAB112 | E. coli/Staphylococcus shuttle plasmid; pRMC2 derivative with tetR, PtetR, and Pxyl/tet with 2x tetO; bla cat | Helle et al., 2011 |
| pEX-A2 | E. coli standard plasmid; pUC ori; bla | Eurofins Genomics |
| pEX-A2-PR∗ | E. coli standard plasmid; pEX-A2 derivative with 90-bp synthetic fragment comprising PR∗ framed by XbaI and XhoI; bla | This study |
| pSN1 | E. coli/Staphylococcus shuttle plasmid; pRAB11 derivative with tetR, PR∗, and Pxyl/tet with 2x tetO; bla cat | This study |
| pSN1-AS‘fabI’ | E. coli/Staphylococcus shuttle plasmid; pSN1 with 382-bp fragment of fabI cloned in antisense orientation in the EcoRI and KpnI sites downstream of Pxyl/tet with 2x tetO; bla cat | This study |
1Precursor plasmids of pRAB11. 2Kindly provided by Ralph Bertram, Department of Microbial Genetics, University of Tübingen, Tübingen, Germany.
Genetic Manipulation in E. coli and S. aureus
One ShotTM Top10 chemically competent E. coli cells (Invitrogen/Thermo Fisher Scientific, Waltham, MA, United States) were used for propagation and cloning experiments. Transformation of E. coli was performed using standard techniques (Hanahan, 1983) and plasmid DNA cloned in E. coli was isolated with the Qiagen Plasmid Mini kit (Qiagen, Venlo, Netherlands) and used to transform restriction-deficient S. aureus RN4220 by electroporation (Augustin and Götz, 1990). Subsequently, plasmid DNA was introduced into S. aureus SA113 by electroporation. All constructed plasmids were verified after each cloning step via standard-PCR amplification using plasmid-specific oligonucleotides pSN1-F (5′-CTGGGCGAGTTTACGGGTTG-3′) and pSN1-R (5′-CAC ATGCAGCTCCCGGAGAC-3′) covering the tet-regulatory elements and, if present, the fabI fragment. The reaction conditions for standard-PCRs using Taq DNA polymerase (Segenetic, Borken, Germany) were 4 min initial denaturation at 95°C followed by 31 cycles of (i) denaturation at 95°C for 30 s, (ii) annealing at 58°C for 30 s, and (iii) extension at 72°C for 1 min. Final extension was performed at 72°C for 10 min. Analysis of PCR and restriction products was performed by agarose gel electrophoresis and purification using the QIAquick PCR Purification kit (Qiagen). This was followed by Sanger sequencing (Eurofins Genomics, Ebersberg, Germany). Restriction enzymes used for cloning were obtained from New England Biolabs (Frankfurt am Main, Germany). Genomic DNA of NCTC8325-4 was extracted after lysostaphin treatment (20 μg/mL, 1 h, 37°C) (Wak-Chemie Medical, Steinbach, Germany) using the QIAamp DNA Mini kit (Qiagen). Secondary sequence structure prediction was performed using the Mfold web server (Zuker, 2003) with default settings. The structure exhibiting the lowest ΔG was used for further analyses.
Construction of Plasmids for Episomal Silencing of Gene fabI
To introduce two base substitutions within the −10 site of promoter PtetR in pRAB11, a 90-bp sequence comprising promoter PR∗ (Geissendörfer and Hillen, 1990) framed by XbaI and XhoI sites was chemically synthesized and cloned into the multiple cloning site (MCS) of pEX-A2 (Eurofins Genomics) to yield pEX-A2-PR∗. Since restriction via XbaI and XhoI was not practicable, PR∗ was amplified with oligonucleotides PR∗-F (5′-GCGCTCTAGACATCATTAATTCCT-3′) and PR∗-R (5′-CGCGCTCGAGGGGATCCAAATAAA-3′) (restriction sites are written in bold) from pEX-A2-PR∗ via standard PCR followed by restriction with XbaI and XhoI. For the construction of pSN1, the XbaI-XhoI synthetic PR∗ sequence (see Figure 1C) was cloned into pRAB11 replacing PtetR and generating pSN1 that was transformed to S. aureus SA113 to generate SAS99/1.
For the construction of plasmid pSN1-AS‘fabI’, a 382-bp fragment of the N-terminal region of the fabI gene (SAOUHSC_00947, corresponding to nucleotides 18 to 399) was amplified from genomic DNA of NCTC8325-4 using oligonucleotides fabI-AS-F-EcoRI (5′-GCGCGAATTCCAAAACATATGTCATCATGGGAAT-3′) and fabI-AS-R-KpnI (5′-GCGCGGTACCTTTAGCTTCATGAG CCACAAT-3′) (restriction sites are written in bold) and the EcoRI/KpnI-digested PCR product was ligated in antisense orientation downstream of the Pxyl/tet promotor-operator fusion into the EcoRI and KpnI sites within the MCS of pSN1. The verified construct was subsequently transformed into SA113 yielding SAS118/2. Further fabI fragments with 75-bp, 100-bp, 150-bp, 200-bp, and 300-bp in size originating from different regions of fabI with and without the Shine-Dalgarno sequence were additionally amplified (used oligonucleotides and characteristics of each antisense “fabI” fragment are listed in Supplementary Tables S1, S2, respectively), cloned into plasmid pSN1, and transformed into SA113.
Growth Curve Analysis and tet-Regulated Antisense Silencing
For growth analysis applying gene silencing due to induction of antisense transcription of the fabI fragment, S. aureus SA113, SAS32/1, SAS99/1, and SAS118/2 were inoculated in 50 mL TSB (Becton Dickinson) containing chloramphenicol where appropriate. Concurrently, strains were inoculated as stated with and without ATc (IBA). The strains were cultured at 37°C on a rotary shaker at 160 rpm in 500-mL glass baffled flasks. Aliquots were taken every hour to determine the optical density (OD578nm).
Furthermore, TSA (Becton Dickinson) containing 0.2 μM and 0.4 μM ATc (IBA) was used for examination of the colony phenotype upon gene silencing. Therefore, samples were adjusted to McFarland 0.5 (in 0.9% NaCl), diluted (10–4) and 100 μl were streaked on agar that was then incubated overnight at 37°C. The different colony phenotypes, macroscopically divided into micro-SCVs, mini-SCVs, SCVs, and WTs, were counted and their percentage was calculated (mean of three independent measurements).
Gene Expression Analysis via Real-Time Quantitative PCR
Levels of fabI transcripts at several time points from three independent experiments were determined by real-time qPCR (run in triplicates). Cells were harvested at 2 h, 4 h, and 7 h during growth curve analysis. Harvesting, RNA isolation, DNase treatment, reverse transcription, and qPCR were performed as previously described (Ballhausen et al., 2014) with the exception of minor deviations in the qPCR protocol consisting of 40 cycles of 10 s at 95°C, 10 s at 55°C, and 30 s at 72°C. 16S rRNA (oligonucleotides, 16S-rRNA-1 and 16S-rRNA-2) (Ballhausen et al., 2014) and gyrB (gyrB-1, 5′-AATTGAAGCAGGCTATGTGT-3′ and gyrB-2, 5′-ATAGACCATTTTGGTGTTGG-3′) were used as references. Specific oligonucleotides used for determination of expression of the 382-bp fabI fragment were fabI-RT-1 (5′-AAATCAACCAGAAGCGCACT-3′) and fabI-RT-2 (5′-ACAAGAAGCCTTCACGTGAA-3′) both binding within the antisense fragment. The relative expression was calculated using the software CFX Manager v 3.1 (Bio-Rad Laboratories, Hercules, CA, United States) applying inter-run calibration. Expression values were normalized to strain SAS99/1 (2 h) harboring the empty pSN1 (control) to exclude chromosomal fabI expression.
Results and Discussion
Since SCV-caused infections are difficult to treat, it is of major interest to identify and characterize those genes, which are involved in the phenotypic switch from the WT to the SCV phenotype. However, the approaches used so far for this purpose are expensive, time-consuming, and/or difficult in handling. Moreover, clinically derived strain pairs consisting of both stable SCVs and revertant WTs are prerequisites. Therefore, this study focused on the generation of an easy-to-handle genetic tool for the rapid identification of SCV phenotype generation-related genes.
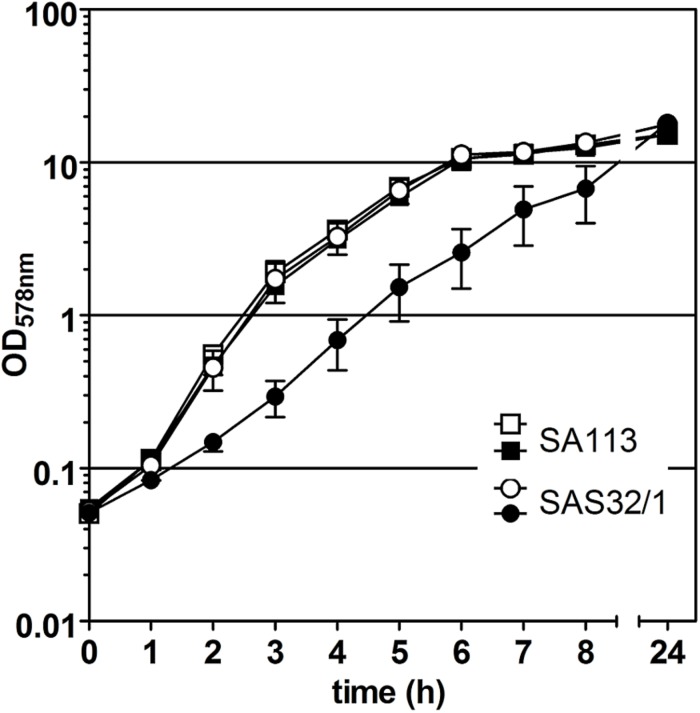
In our study, we adapted the Tn10-encoded Tc- or ATc-inducible tet-regulated repressor-operator expression system, which was first described in E. coli and then modified in several ways to meet the different requirements for Gram-positive bacteria (reviewed in Bertram and Hillen, 2008). For episomal antisense silencing of phenotype-related genes in S. aureus (see Figure 1), we employed plasmid pRAB11 (Helle et al., 2011) containing the tet-regulatory elements tetR (encodes the repressor), its enhanced autoregulated promoter PtetR (Geissendörfer and Hillen, 1990; Corrigan and Foster, 2009), and the divergently located promoter-operator fusion Pxyl/tet with two tetO sequences (Geissendörfer and Hillen, 1990; see Figure 1). Since for fatty acid metabolism-related gene fabI, mutations were already shown to be responsible for the phenotype switch to the SCV (Bazaid et al., 2018), we exemplarily used this gene in our experiments. Initial experiments to determine the suitability of pRAB11 unexpectedly revealed pronounced growth retardation for strain SAS32/1 carrying the empty pRAB11 upon induction with the effector ATc (Figure 2). ATc represents a derivative of Tc, but since growth of SA113 in ATc-containing TSB was not affected, its low antibiotic activity (Degenkolb et al., 1991) was not responsible for the slow growth phenomenon. During generation of plasmid pRAB11 for the use in Gram-positive bacteria, several modifications were performed (see Table 1; Geissendörfer and Hillen, 1990; Bateman et al., 2001; Corrigan and Foster, 2009; Helle et al., 2011). Amongst these, adaptation of the −10 sequence within the tetR-driving promoter PR∗ formally designed by Geissendörfer and Hillen (1990) resulted in the consensus sequence (5′-tag ag t-3′→ 5′-tat aa t-3′) (Corrigan and Foster, 2009), but unintentionally led to a disruption of its palindromic tetO1 sequence (see Figure 1C) and therefore, presumptively resulted in increased amounts of TetR due to loss of its negative regulation (Stary et al., 2010). Whilst increased amounts of TetR provide improved repression for Pxyl/tet promotor constructs harboring only one tetO (Corrigan and Foster, 2009) in the uninduced state, these high TetR concentrations may be toxic to the cells (Lutz and Bujard, 1997) as the unbound TetR homodimers are freely available in the induced state. Furthermore, since induction of the Pxyl/tet promoter may be negatively affected by overexpression of TetR (Ehrt et al., 2005), we adapted the system by restoring the tetO1 sequence within the TetR promoter generating plasmid pSN1. S. aureus cells harboring pSN1 (SAS99/1) showed no signs of growth retardation in presence of ATc (Figure 3) presumptively linked to the fact that, due to the restored tetO1, the negatively controlled transcription of tetR by TetR as in its original state (Yang et al., 1976; Beck et al., 1982; Wissmann et al., 1988) is reconstituted.
FIGURE 2.
Effect of pRAB11-derived TetR on growth of S. aureus strain SAS32/1. S. aureus wild type strain SA113 without pRAB11 was used as control. Filled symbols indicate the presence of 0.4 μM anhydrotetracycline (ATc).
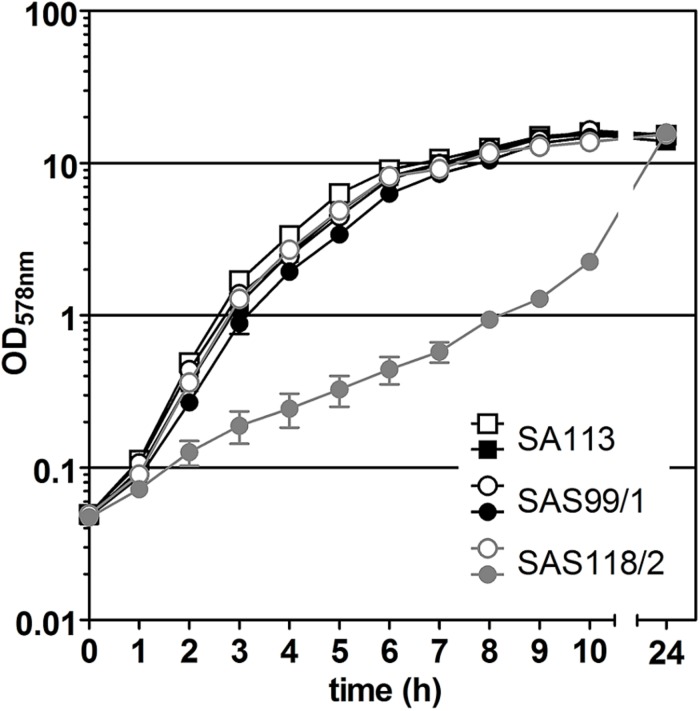
FIGURE 3.
Effect of episomal ‘fabI’ antisense expression controlled by pSN1-derived TetR in S. aureus SAS118/2 carrying pSN1-AS‘fabI’ (gray symbols). Black symbols indicate growth curves of S. aureus control strains SA113 and SAS99/1 (SA113 carrying empty pSN1). Filled symbols indicate the presence of 0.4 μM anhydrotetracycline (ATc).
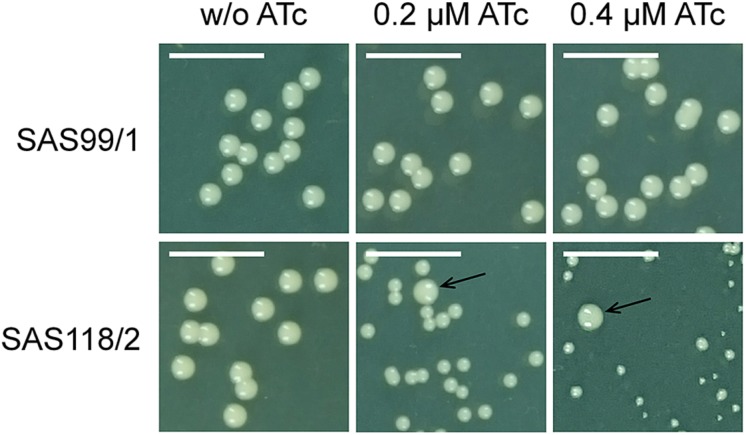
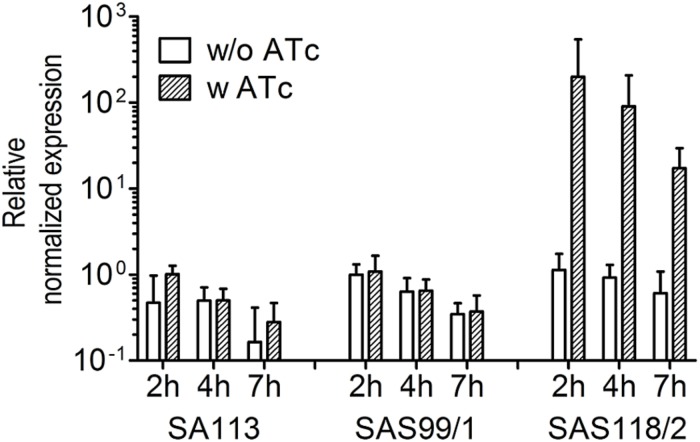
For episomal antisense silencing, a 382-bp fragment of the gene fabI mostly congruent with fragments used before (Ji et al., 2004; Stary et al., 2010) was cloned downstream of promoter Pxyl/tet in pSN1. Induction with ATc resulted in distinct retardation of growth (Figure 3) as shown before with chromosomally encoded tetR (Stary et al., 2010) as well as for episomal silencing with a plasmid harboring a leaky (Zhang et al., 2000) one-tetO-version that was induced with Tc (Ji et al., 2004). Phenotypic characterization of fabI antisense silencing on solid agar revealed colonies exhibiting the SCV phenotype only upon induction with ATc (Figure 4). The SCV phenotype was more pronounced on agar containing 0.4 μM ATc than on agar with 0.2 μM ATc. However, on agar with 0.4 μM ATc, SCVs were heterogeneous in size with 74.83% micro-SCVs, 19.14% mini-SCVs, and 3.73% SCVs, respectively. This differentiation into different SCV phenotypes despite the same treatment and environmental conditions is a phenomenon known for SCVs and implicates an adaption strategy during the infection and persistence process (Tuchscherr et al., 2011). It was therefore expected that the bacterial cells have the ability to react slightly different to the silencing of a particular gene and the environmental conditions. Unaffected WT cells remained for both ATc concentrations at a ratio of 1.17% for 0.2 μM and 2.30% for 0.4 μM ATc. This may indicate that some of the cells were able to escape induction with ATc, which needs to be further investigated. Since qPCR showed a considerably elevated expression of antisense ‘fabI’ only for strain SAS118/2 in the induced state (∼ 200-fold increase compared to induced control strain SAS99/1, both measured after 2 h of growth) (Figure 5), this indicates functional silencing due to pSN1-derived antisense expression.
FIGURE 4.
Phenotypic effect of episomal ‘fabI’ antisense silencing controlled by pSN1-derived TetR in S. aureus on TSA agar with and without anhydrotetracycline (ATc). The upper row shows control strain SAS99/1 harboring the empty plasmid pSN1 and the lower row shows ‘fabI’-carrying strain SAS118/2. Phenotype switch from wild type to SCV only occurs if ATc is contained in the medium; arrows indicate unaffected wild type cells. Scale bar indicates 5 mm.
FIGURE 5.
Relative patterns of ‘fabI’-antisense expression in S. aureus strains SA113, SAS99/1 (carrying pSN1), and SAS118/2 (carrying pSN1-AS‘fabI’) in response to 0.4 μM anhydrotetracycline (ATc). Dashed bars indicate the presence of 0.4 μM ATc, the effector of ‘fabI’-antisense expression in strains harboring pSN1-AS‘fabI’. Normalized expression is shown relative to control SAS99/1 carrying the empty pSN1 (2 h) to exclude chromosomal fabI expression.
Since not all antisense RNA fragments are effective in inhibiting gene expression in S. aureus (Ji et al., 2004), we analyzed several fabI fragments from different regions of fabI with and without Shine-Dalgarno sequence that exhibited 75- to 300-bp in size, a GC content of 29.3 to 36.7% and ΔG values of −45.6 to −11.8 kcal/mol (for comparison, the 382-bp fragment exhibited a GC content of 34.8% and a ΔG of −70.3 kcal/mol; see Supplementary Material). However, a silencing effect could only be observed for the 382-bp fabI fragment, not for any of the other fragments tested (data not shown) as it could be also not shown before for a fragment bigger than 500 bp (Ji et al., 2004). Therefore, contrary to expectations, the presence of the Shine-Dalgarno sequence does not seem to be relevant as it was described for gene silencing in E. coli (Stefan et al., 2008). Secondary sequence structure prediction for each fragment revealed different loop regions with unpaired bases that are complementary and with accessible conformation to hybridize with fabI mRNA (data not shown). Even though some of these fragments showed loop regions of a bigger size compared to the 382-bp fabI fragment, this seemed to have no impact on silencing. Instead, the quantity of loop regions may contribute to silencing, as the 382-bp fabI fragment exhibited 20 loops compared to 4 to 16 loops of the other fragments (see Supplementary Material). Therefore, antisense silencing seems to depend significantly on fragment size and target region. Thus, the final effectiveness of the fragments is difficult to predict necessitating further examinations.
Conclusion
Our results demonstrate the crucial role of TetR and its major importance in negative regulation of repressor expression for episomal antisense silencing. However, to perform efficient antisense silencing, size and region of the antisense fragment complementary to the region of the target gene remain critical factors that need further detailed investigations. Plasmid pSN1 allowed antisense silencing of the fabI gene in an easy-to-handle one-plasmid setup. Due to the MCS directly downstream of the inducible Pxyl/tet promoter, genes of interest can easily be cloned into the plasmid. Moreover, the two-tetO Pxyl/tet version exerted a tight repression of the gene of interest in the uninduced state, thus there is no necessity of chromosomal tetR integration prior to use. Also other SCV characteristic (e.g., hemolysis behavior, coagulase and protease production, as well as antibiotic resistance) can be analyzed with this system as already shown for antisense silencing of hla (encodes for α-toxin) (Ji et al., 1999). Owing to induction with ATc, pSN1 mediated antisense silencing may not only be applicable to different S. aureus backgrounds, but also in cell culture studies (Ji et al., 1999; Bateman et al., 2001). Therefore, pSN1 mediated antisense silencing may not only help to identify SCV phenotype-related genes but also to analyse the consequences of the switching process to the host cells and organs.
Data Availability
All relevant data for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
KB and AK designed the study concept. NS, AK, and BB designed the experiments. NS performed the laboratory work, evaluated the data, and drafted and wrote the manuscript. UK contributed to the data evaluation and writing of the manuscript. SF and JS contributed to the laboratory work. RP provided the scientific support regarding SCVs and interpreted the data. All authors have read and approved the final draft of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Ralph Bertram from the Department of Microbial Genetics, University of Tübingen, Tübingen, Germany for the generous gift of plasmid pRAB11. Furthermore, we are grateful to Melanie Bach and Daniela Kuhn for excellent technical assistance. This manuscript is part of the Ph.D. thesis entitled “Staphylococcus aureus Small-Colony Variants – Insights in Formation and Antibiotic Susceptibilities” by NS (2019).
Footnotes
Funding. This research was supported in part by a grant to KB (BE 2546/1-2) from the Deutsche Forschungsgemeinschaft (DFG).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02044/full#supplementary-material
References
- Abu-Qatouseh L., Chinni S., Seggewiß J., Proctor R. A., Brosius J., Rozhdestvensky T. S., et al. (2010). Identification of differentially expressed small non-protein-coding RNAs in Staphylococcus aureus displaying both the normal and the small-colony variant phenotype. J. Mol. Med. 88 565–575. 10.1007/s00109-010-0597-2 [DOI] [PubMed] [Google Scholar]
- Augustin J., Götz F. (1990). Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol. Lett. 66 203–207. 10.1111/j.1574-6968.1990.tb03997.x [DOI] [PubMed] [Google Scholar]
- Ballhausen B., Kriegeskorte A., Schleimer N., Peters G., Becker K. (2014). The mecA Homolog mecC Confers Resistance against β-Lactams in Staphylococcus aureus irrespective of the genetic strain background. Antimicrob. Agents Chemother. 58 3791–3798. 10.1128/AAC.02731-2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balwit J. M., van Langevelde P., Vann J. M., Proctor R. A. (1994). Gentamicin-resistant menadione and hemin auxotrophic Staphylococcus aureus persist within cultured endothelial cells. J. Infect. Dis. 170 1033–1037. 10.1093/infdis/170.4.1033 [DOI] [PubMed] [Google Scholar]
- Band L., Yansura D. G., Henner D. J. (1983). Construction of a vector for cloning promoters in Bacillus subtilis. Gene 26 313–315. 10.1016/0378-1119(83)90204-90204 [DOI] [PubMed] [Google Scholar]
- Bateman B. T., Donegan N. P., Jarry T. M., Palma M., Cheung A. L. (2001). Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 69 7851–7857. 10.1128/IAI.69.12.7851-7857.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D. M., von Eiff C., McNamara P. J., Peters G., Yeaman M. R., Bayer A. S., et al. (2003). Staphylococcus aureus menD and hemB mutants are as infective as the parent strains, but the menadione biosynthetic mutant persists within the kidney. J. Infect. Dis. 187 1654–1661. 10.1086/374642 [DOI] [PubMed] [Google Scholar]
- Bazaid A. S., Forbes S., Humphreys G. J., Ledder R. G., O’Cualain R., McBain A. J. (2018). Fatty acid supplementation reverses the small colony variant phenotype in triclosan-adapted Staphylococcus aureus: genetic. proteomic and phenotypic analyses. Sci. Rep. 8:3876. 10.1038/s41598-018-21925-21926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C. F., Mutzel R., Barbé J., Müller W. (1982). A multifunctional gene (tetR) controls tn10-encoded tetracycline resistance. J. Bacteriol. 150 633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K., Al-Laham N., Fegeler W., Proctor R. A., Peters G., von Eiff C. (2006). Fourier-transform infrared spectroscopic analysis is a powerful tool for studying the dynamic changes in Staphylococcus aureus small-colony variants. J. Clin. Microbiol. 44 3274–3278. 10.1128/JCM.00847-846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram R., Hillen W. (2008). The application of Tet repressor in prokaryotic gene regulation and expression. Microb. Biotechnol. 1 2–16. 10.1111/j.1751-7915.2007.00001.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui L. M. G., Hoffmann P., Turnidge J. D., Zilm P. S., Kidd S. P. (2015). Prolonged growth of a clinical Staphylococcus aureus strain selects for a stable small-colony-variant cell type. Infect. Immun. 83 470–481. 10.1128/IAI.02702-2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui L. M. G., Kidd S. P. (2015). A full genomic characterization of the development of a stable small colony variant cell-type by a clinical Staphylococcus aureus strain. Infect. Genet. Evol. 36 345–355. 10.1016/J.MEEGID.2015.10.011 [DOI] [PubMed] [Google Scholar]
- Cao S., Huseby D. L., Brandis G., Hughes D. (2017). Alternative evolutionary pathways for drug-resistant small colony variant mutants in Staphylococcus aureus. mBio 8:e00358-17. 10.1128/mBio.00358-317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee I., Kriegeskorte A., Fischer A., Deiwick S., Theimann N., Proctor R. A., et al. (2008). In vivo mutations of thymidylate synthase (encoded by thyA) are responsible for thymidine dependency in clinical small-colony variants of Staphylococcus aureus. J. Bacteriol. 190 834–842. 10.1128/JB.00912-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan R. M., Foster T. J. (2009). An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid 61 126–129. 10.1016/j.plasmid.2008.10.001 [DOI] [PubMed] [Google Scholar]
- Dean M. A., Olsen R. J., Long S. W., Rosato A. E., Musser J. M. (2014). Identification of point mutations in clinical Staphylococcus aureus strains that produce small-colony variants auxotrophic for menadione. Infect. Immun. 82 1600–1605. 10.1128/IAI.01487-1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenkolb J., Takahashi M., Ellestad G. A., Hillen W. (1991). Structural requirements of tetracycline-Tet repressor interaction: determination of equilibrium binding constants for tetracycline analogs with the Tet repressor. Antimicrob. Agents Chemother. 35 1591–1595. 10.1128/AAC.35.8.1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval B. D., Mathew A., Satola S. W., Shafer W. M. (2010). Altered growth, pigmentation, and antimicrobial susceptibility properties of Staphylococcus aureus due to loss of the major cold shock gene cspB. Antimicrob. Agents. Chemother. 54 2283–2290. 10.1128/AAC.01786-1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrt S., Guo X. V., Hickey C. M., Ryou M., Monteleone M., Riley L. W., et al. (2005). Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33:e21. 10.1093/nar/gni013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Chua K., Davies J. K., Newton H. J., Seemann T., Harrison P. F., et al. (2010). Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog. 6:e1000944. 10.1371/journal.ppat.1000944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaupp R., Schlag S., Liebeke M., Lalk M., Götz F. (2010). Advantage of upregulation of succinate dehydrogenase in Staphylococcus aureus Biofilm. J. Bacteriol. 192 2385–2394. 10.1128/JB.01472-1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissendörfer M., Hillen W. (1990). Regulated expression of heterologous genes in Bacillus subtilis using the Tn10 encoded tet regulatory elements. Appl. Microbiol. Biotechnol. 33 657–663. 10.1007/BF00604933 [DOI] [PubMed] [Google Scholar]
- Giulieri S. G., Baines S. L., Guerillot R., Seemann T., Gonçalves da Silva A., Schultz M., et al. (2018). Genomic exploration of sequential clinical isolates reveals a distinctive molecular signature of persistent Staphylococcus aureus bacteraemia. Genome. Med. 10:65. 10.1186/s13073-018-0574-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer N. D., Reniere M. L., Cassat J. E., Zhang Y., Hirsch A. O., Indriati Hood M., et al. (2013). Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. mBio 4:e00241-13. 10.1128/mBio.00241-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. (1983). Studies on Transformation of Escherichia coli with plasmids. J. Mol. Biol. 166 557–580. 10.1016/S0022-2836(83)80284-80288 [DOI] [PubMed] [Google Scholar]
- Helle L., Kull M., Mayer S., Marincola G., Zelder M.-E., Goerke C., et al. (2011). Vectors for improved Tet repressor-dependent gradual gene induction or silencing in Staphylococcus aureus. Microbiology 157 3314–3323. 10.1099/mic.0.052548-0 [DOI] [PubMed] [Google Scholar]
- Herbert S., Ziebandt A.-K., Ohlsen K., Schäfer T., Hecker M., Albrecht D., et al. (2010). Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect. Immun. 78 2877–2889. 10.1128/IAI.00088-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordanescu S., Surdeanu M. (1976). Two restriction and modification systems in Staphylococcus aureus NCTC8325. J. Gen. Microbiol. 96 277–281. 10.1099/00221287-96-2-277 [DOI] [PubMed] [Google Scholar]
- James K. L., Mogen A. B., Brandwein J. N., Orsini S. S., Ridder M. J., Markiewicz M. A., et al. (2019). Interplay of nitric oxide synthase (NOS) and SrrAB in modulation of Staphylococcus aureus metabolism and virulence. Infec. Immun. 87:e00570-18. 10.1128/IAI.00570-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana M., Luong T.-T., Komatsuzawa H., Shigeta M., Lee C. Y. (2000). A method for demonstrating gene essentiality in Staphylococcus aureus. Plasmid 44 100–104. 10.1006/PLAS.2000.1473 [DOI] [PubMed] [Google Scholar]
- Ji Y., Marra A., Rosenberg M., Woodnutt G. (1999). Regulated antisense RNA eliminates alpha-toxin virulence in Staphylococcus aureus infection. J. Bacteriol. 181 6585–6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Yin D., Fox B., Holmes D. J., Payne D., Rosenberg M. (2004). Validation of antibacterial mechanism of action using regulated antisense RNA expression in Staphylococcus aureus. FEMS Microbiol. Lett. 231 177–184. 10.1016/S0378-1097(03)00931-935 [DOI] [PubMed] [Google Scholar]
- Kahl B. C., Becker K., Löffler B. (2016). Clinical Significance and pathogenesis of Staphylococcal small colony variants in persistent infections. Clin. Microbiol. Rev. 29 401–427. 10.1128/CMR.00069-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köser C. U., Holden M. T. G., Ellington M. J., Cartwright E. J. P., Brown N. M., Ogilvy-Stuart A. L., et al. (2012). Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N. Engl. J. Med. 366 2267–2275. 10.1056/NEJMoa1109910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte A., Grubmüller S., Huber C., Kahl B. C., von Eiff C., Proctor R. A., et al. (2014). Staphylococcus aureus small colony variants show common metabolic features in central metabolism irrespective of the underlying auxotrophism. Front. Cell. Infect. Microbiol. 4:414. 10.3389/fcimb.2014.00141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte A., König S., Sander G., Pirkl A., Mahabir E., Proctor R. A., et al. (2011). Small colony variants of Staphylococcus aureus reveal distinct protein profiles. Proteomics 11 2476–2490. 10.1002/pmic.201000796 [DOI] [PubMed] [Google Scholar]
- Lannergård J., von Eiff C., Sander G., Cordes T., Seggewiß J., Peters G., et al. (2008). Identification of the genetic basis for clinical menadione-auxotrophic small-colony variant isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 52 4017–4022. 10.1128/AAC.00668-668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-T., Tsai J.-C., Yamamoto T., Chen H.-J., Hung W.-C., Hsueh P.-R., et al. (2016). Emergence of a small colony variant of vancomycin-intermediate Staphylococcus aureus in a patient with septic arthritis during long-term treatment with daptomycin. J. Antimicrob. Chemother. 71 1807–1814. 10.1093/jac/dkw060 [DOI] [PubMed] [Google Scholar]
- Lutz R., Bujard H. (1997). Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25 1203–1210. 10.1093/nar/25.6.1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean K., Holmes E. A., Penewit K., Lee D. K., Hardy S. R., Ren M., et al. (2019). Artificial selection for pathogenicity mutations in Staphylococcus aureus identifies novel factors relevant to chronic infection. Infec. Immun. 87 e00884-18. 10.1128/IAI.00884-818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I., Wray L. V., Hillen W. (1988). Differential regulation of the Tn10-encoded tetracycline resistance genes tetA and tetR by the tandem tet operators O1 and O2. EMBO J. 7 567–572. 10.1002/J.1460-2075.1988.TB02846.X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell G., Fugère A., Pépin Gaudreau K., Brouillette E., Frost E. H., Cantin A. M., et al. (2013). SigB Is a dominant regulator of virulence in Staphylococcus aureus small-colony variants. PLoS One. 8:e65018. 10.1371/journal.pone.0065018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisan H., Brouillette E., Jacob C. L., Langlois-Bégin P., Michaud S., Malouin F. (2006). Transcription of virulence factors in Staphylococcus aureus small-colony variants isolated from cystic fibrosis patients is influenced by SigB. J. Bacteriol. 188 64–76. 10.1128/JB.188.1.64-76.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. (1967). Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33 155–166. 10.1016/0042-6822(67)90105-90105 [DOI] [PubMed] [Google Scholar]
- Painter K. L., Strange E., Parkhill J., Bamford K. B., Armstrong-James D., Edwards A. M. (2015). Staphylococcus aureus adapts to oxidative stress by producing H2O2-resistant small-colony variants via the SOS response. Infect. Immun. 83 1830–1844. 10.1128/IAI.03016-3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J. B., Frank M. W., Jackson P., Subramanian C., Rock C. O. (2014). Incorporation of extracellular fatty acids by a fatty acid kinase-dependent pathway in Staphylococcus aureus. Mol. Microbiol. 92 234–245. 10.1111/mmi.12556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J. B., Frank M. W., Rosch J. W., Rock C. O. (2013). Staphylococcus aureus fatty acid auxotrophs do not proliferate in mice. Antimicrob. Agents Chemother. 57 5729–5732. 10.1128/AAC.01038-1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J. B., Frank M. W., Subramanian C., Saenkham P., Rock C. O. (2011). Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proc. Natl. Acad. Sci. U.S.A. 108 15378–15383. 10.1073/pnas.1109208108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor R. A., Kriegeskorte A., Kahl B. C., Becker K., Löffler B., Peters G. (2014). Staphylococcus aureus small colony variants (SCVs): a road map for the metabolic pathways involved in persistent infections. Front. Cell. Infect. Microbiol. 4:99. 10.3389/fcimb.2014.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor R. A., von Eiff C., Kahl B. C., Becker K., McNamara P., Herrmann M., et al. (2006). Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4 295–305. 10.1038/nrmicro1384 [DOI] [PubMed] [Google Scholar]
- Schaaff F., Bierbaum G., Baumert N., Bartmann P., Sahl H.-G. (2003). Mutations are involved in emergence of aminoglycoside-induced small colony variants of Staphylococcus aureus. Int. J. Med. Microbiol. 293 427–435. 10.1078/1438-4221-00282 [DOI] [PubMed] [Google Scholar]
- Schleimer N., Kaspar U., Drescher M., Seggewiß J., von Eiff C., Proctor R. A., et al. (2018). The energy-coupling factor transporter module EcfAA’T, a novel candidate for the genetic basis of fatty acid-auxotrophic small-colony variants of Staphylococcus aureus. Front. Microbiol. 9:1863 10.3389/fmicb.2018.01863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seggewiß J., Becker K., Kotte O., Eisenacher M., Yazdi M. R. K., Fischer A., et al. (2006). Reporter metabolite analysis of transcriptional profiles of a Staphylococcus aureus strain with normal phenotype and its isogenic hemB mutant displaying the small-colony-variant phenotype. J. Bacteriol. 188 7765–7777. 10.1128/JB.00774-776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stary E., Gaupp R., Lechner S., Leibig M., Tichy E., Kolb M., et al. (2010). New architectures for Tet-on and Tet-off regulation in Staphylococcus aureus. Appl. Environ. Microbiol. 76 680–687. 10.1128/AEM.02416-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan A., Tonelli A., Schwarz F., Hochkoeppler A. (2008). Artificial antisense RNAs silence lacZ in E. coli by decreasing target mRNA concentration. BMB Rep. 41 568–574. 10.5483/bmbrep.2008.41.8.568 [DOI] [PubMed] [Google Scholar]
- Tinsley E., Khan S. A. (2007). A Bacillus anthracis-based in vitro system supports replication of plasmid pXO2 as well as rolling-circle-replicating plasmids. Appl. Environ. Microbiol. 73 5005–5010. 10.1128/AEM.00240-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuchscherr L., Bischoff M., Lattar S. M., Noto Llana M., Pförtner H., Niemann S., et al. (2015). Sigma factor SigB is crucial to mediate Staphylococcus aureus adaptation during chronic infections. PLoS Pathog. 11:e1004870. 10.1371/journal.ppat.1004870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuchscherr L., Medina E., Hussain M., Völker W., Heitmann V., Niemann S., et al. (2011). Staphylococcus aureus phenotype switching: an effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol. Med. 3 129–141. 10.1002/emmm.201000115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaudaux P., Kelley W. L., Lew D. P. (2006). Staphylococcus aureus small colony variants: difficult to diagnose and difficult to treat. Clin. Infect. Dis. 43 968–970. 10.1086/507643 [DOI] [PubMed] [Google Scholar]
- Vestergaard M., Nøhr-Meldgaard K., Ingmer H. (2018). Multiple pathways towards reduced membrane potential and concomitant reduction in aminoglycoside susceptibility in Staphylococcus aureus. Int. J. Antimicrob. Agents. 51 132–135. 10.1016/J.IJANTIMICAG.2017.08.024 [DOI] [PubMed] [Google Scholar]
- von Eiff C., Heilmann C., Proctor R., Woltz C., Peters G., Götz F. (1997). A site-directed Staphylococcus aureus hemB Mutant is a small-colony variant which persists intracellularly. J. Bacteriol. 179 4706–4712. 10.1128/jb.179.15.4706-4712.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeman C. A., Hammer N. D., Stauff D. L., Attia A. S., Anzaldi L. L., Dikalov S. I., et al. (2012). Menaquinone biosynthesis potentiates haem toxicity in Staphylococcus aureus. Mol. Microbiol. 86 1376–1392. 10.1111/mmi.12063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland K.-P., Wieland B., Götz F. (1995). A promoter-screening plasmid and xylose-inducible, glucose-repressible expression vectors for Staphylococcus carnosus. Gene 158 91–96. 10.1016/0378-1119(95)00137-U [DOI] [PubMed] [Google Scholar]
- Wissmann A., Meier I., Hillen W. (1988). Saturation mutagenesis of the Tn10-encoded tet operator O1: identification of base-pairs involved in Tet repressor recognition. J. Mol. Biol. 202 397–406. 10.1016/0022-2836(88)90273-2 [DOI] [PubMed] [Google Scholar]
- Yang H.-L., Zubay G., Levy S. B. (1976). Synthesis of an R plasmid protein associated with tetracycline resistance is negatively regulated. Proc. Natl. Acad. Sci. U.S.A. 73 1509–1512. 10.1073/PNAS.73.5.1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Fan F., Palmer L. M., Lonetto M. A., Petit C., Voelker L. L., et al. (2000). Regulated gene expression in Staphylococcus aureus for identifying conditional lethal phenotypes and antibiotic mode of action. Gene 255 297–305. 10.1016/S0378-1119(00)00325-325 [DOI] [PubMed] [Google Scholar]
- Zhang P., Wright J. A., Osman A. A., Nair S. P. (2017). An aroD Ochre mutation results in a Staphylococcus aureus small colony variant that can undergo phenotypic switching via two alternative mechanisms. Front. Microbiol. 8:1001. 10.3389/fmicb.2017.01001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31 3406–3415. 10.1093/NAR/GKG595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data for this study are included in the manuscript and/or the Supplementary Files.