ABSTRACT
Various drug treatments including doxorubicin (DOX) have been proved efficient in the suppression of breast cancer. Nonetheless, drug resistance became an obstacle in the therapeutic process. According to recent literatures, breast cancer stem cells (BCSCs) were considered contributing to drug resistance, besides, microRNAs (miRNAs) could regulate proteins associated with drug resistance in human breast cancer. To further understand the inner mechanism of drug resistance in breast cancer and look for remedy methods, we referred to bioinformatic analysis and predicted that signal transducer and activator of transcription 3 (STAT3) and miR-124 was overexpressed in MCF7-R cells (MCF7 cells resistant to DOX) compared with MCF cells. Expression levels of RNA and protein were separately determined by qRT-PCR and western blot. Dual luciferase assay was performed to verify the targeting relationship between STAT3 and miR-124. Optical density (OD) values and apoptotic rates of cells were respectively determined via MTT assays and flow cytometric analysis. Cell invasion was detected to verify drug resistance. Results of above assays indicated that STAT3 was highly expressed in MCF7-R cells than in MCF7 cell lines and affected doxorubicin resistance of BCSCs, and miR-124 reversed the doxorubicin resistance of breast cancer stem cells through targeting STAT3 to control the HIF-1 signaling pathway. To conclude, this research may be valuable for the treatment of breast cancer as the restoration of miR-124 and inhibition of STAT3 could be applied to therapeutic strategy and help overcome drug resistance.
KEYWORDS: Mir-124, STAT3, breast cancer stem-like cells, doxorubicin resistance, HIF-1 signaling pathway
Introduction
Breast cancer (BC) is one of the most frequently occurring carcinomas in women worldwide [1]. Doxorubicin (DOX is a natural anthracycline antibiotic that is an essential component of many treatment regimens for solid and blood tumors. At the same time, it is well accepted as the most active single agent available for breast cancer treatment [2]. Over time, resistance to doxorubicin has frequently appeared in reports on breast cancer [3,4]. Thus, refined investigation of the key molecular mechanisms of chemoresistance in breast cancer will provide novel targets for advancing anticancer therapy.
Considering that cancer stem cells serve vital roles in carcinogenesis and has become a significant focus in cancer study [5], breast cancer stem cells (BCSCs) were taken into our research. BCSCs are a subpopulation of BCs that are known to act crucially in tumor formation, growth, drug resistance and recurrence [6]. According to a recent review, BCSCs are a pivotal target for clinical therapy to overcome resistance and recurrence [7]. Furthermore, effective agents targeting BCSCs may offer a therapeutic promise [8].
MicroRNAs are a family of small noncoding RNA with a length of approximately 22 nucleotides. In addition, they post-transcriptionally regulate gene expression [8]. Currently, a large amount of miRNAs, including miR-34c, have been validated to regulate expressions of target genes and modulate signaling pathways, such as the Wnt/β-catenin signal pathway, in BCSCs [9]. MicroRNA-124 (miR-124) has been shown to participate in BC progression, including cell proliferation, invasion and migration [10,11]. Only a few studies have explored and drawn incomplete conclusions regarding how miR-124 affects cancer stem cells. In a recent study by Wang X et al., improvement in chemotherapies can be obtained from properties of cancer stem cell-like via the miR-124/STAT3 axis [12]. In fact, no study has announced the function of miR-124 in BCSCs.
Signal Transducer and Activator of Transcription (STAT) is a group of transcription factors that function in various cellular functions. The STAT family has been elucidated to participate in every stage of mammary gland development as well as breast tumorigenesis. STAT3, a member of the STAT family, has been found to have low levels of expression and transcription in patients with BC [13]. In the study by Sirkisoon, S. R et al., STAT3 undergoes protein-protein interactions with glioma oncogene homolog 1 (GLI1) and (truncated GLI1) tGLI1, which promote gene activation and a stem-like phenotype of breast cancer [14]. Furthermore, the target relationship between miR-124 and STAT3 has been revealed in human epidermal growth factor receptor 2 (HER2)-positive breast cancer cells [15]. How STAT3 interacts with miR-124 in BCSCs is still unclear.
As the main transcription factor, hypoxia-inducible factor-1 (HIF-1) is involved in the cell response to hypoxia. At the same time, it participates in the modulation of genes related to cancer aggressiveness via different mechanisms. HIF-1α and HIF-1β are the two subunits of HIF-1 [16]. The HIF-1 signaling pathway has been previously observed in breast cancer. Yang, J et al. found that HIF-1α overexpression promotes breast tumor growth and resistance to Tamoxifen treatment [17]. More importantly, it has been confirmed that HIF-1α is required for BCSC enrichment [18]. For a better understanding of HIF-1 in breast cancer, it is indispensable to carry out discussions and investigations.
The goal of our research is to uncover the underlying relationship of the miR-124/STAT3/HIF-1 signaling pathway. In addition, we aim to elucidate the coeffects of the miR-124/STAT3/HIF-1 signaling pathway on doxorubicin-resistant BCSCs. Finally, we validate STAT3 expression and the function on the HIF-1 signaling pathway in doxorubicin-resistant BCSCs. Our research indicated that miR-124 was involved in BCSC resistance to doxorubicin through the STAT3/HIF-1 signaling pathway. This result is the most important finding in the present study and may open a new avenue and benefit further investigations on overcoming drug resistance in other human cancers.
Materials and methods
Bioinformatics analysis
To identify the differentially expressed mRNAs of multidrug resistance genes in putative cancer stem cells, we used GSE24460 based on GPL571 for microarray analysis. There were 2 groups in GSE24460, including 2 parental controls and 2 drug-resistant MCF7 cells. Statistical software R (version 3.5.0, https://www.r-project.org/) and packages of limma were utilized to analyze differentially expressed genes (DEGs) between the two groups. The fold change (FC) threshold was set to 2, and the P value (adjusted by the BH method) was set to less than 0.05 for screening out the DEGs. Then, the DEGs were uploaded to the DAVID website (https://david.ncifcrf.gov/) to perform KEGG enrichment analysis.
Cell culture
The MCF7 cell line was purchased from BeNa Culture Collection (http://www.bnbio.com/). Cells were incubated in DMEM with high glucose (BeNa Culture Collection, Beijing, China) and supplemented with 10% FBS (Gibco, Grand Island, NY, USA). In a 5% CO2 humidified incubator, cells were maintained at 37°C. Paclitaxel was purchased from Molecular Probes Invitrogen.
BCSC division
MCF7 cells were collected and enzymatically dissociated into a single-cell suspension. The cell suspension was centrifuged at 300 × g for 10 minutes, and the cell pellet was resuspended in 40 μL suspension buffer (~10 [7] total cells). The cells were then incubated with CD24 Microbead Kit and CD44 Microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) for 15 minutes in a refrigerator (4°C), washed and resuspended in 500 μL buffer, followed by magnetic separation. The CD44+CD24− cells were then collected as the BCSCs.
Cell transfection
MicroRNA-124 mimics and the nonspecific miRNA control were synthesized by GenePharma, Shanghai, China. STAT3 siRNA and control siRNA were purchased from Thermo Fisher Scientific, Waltham, MA, USA. The pcDNA3.1-STAT3 plasmid was derived from GenePharma. MCF7 cells were grown in 6-well plates to confluence and were transfected using Lipofectamine TM 2000 (Invitrogen Co., Carlsbad, CA), based on the product instructions.
Cell viability assay
MCF7 cells (4 × 103) were plated in each well of 96-well plates and transfected with RNAs and plasmids. Twenty-four hours after transfection, TRAIL, doxorubicin, or cisplatin was added to each well. After 48 hours, cell viability was evaluated via MTT assay. Relative absorbance was read at 450 nm using a Bio-Rad microplate reader (Bio-Rad, Hercules, CA, USA).
Dual luciferase reporter assay
The STAT3 3′ UTR containing the putative miR-124 binding site was analyzed by TargetScan (www.targetscan.org), and this miRNA site was inserted downstream of the firefly luciferase reporter gene (Promega, Madison, WI, USA). The cultures were transiently transfected together with 50 nM miR-124 mimic and 600 ng dual-luciferase vectors (containing either wild type or mutant 3′ UTR). Twenty-four hours after transfection, firefly luciferase activity was measured with the Dual Luciferase Assay Kit (Promega) and normalized to the Renilla luciferase reference plasmid.
Western blot
After cell lysis, the protein concentrations were quantified using a BCA Pierce Assay Kit (Pierce Chemical Co.). Protein samples (20 mg/lane) were resolved by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were blocked with 5% nonfat dry milk for 1 hour. GAPDH served as a control. The membrane was co-incubated with the primary antibodies overnight at 4°C. After being washed at least three times, the membrane was incubated with the secondary antibody.
The primary antibodies were as followed: rabbit anti-STAT3 (1:2000, ab68153, Abcam), rabbit anti-STAT3 (phosphor-STAT3, 1:1000, ab30647, Abcam), rabbit anti-ALDH1 (1:1000, ab52492, Abcam), rabbit anti-SOX2 (1 µg/mL, ab97959, Abcam), rabbit anti-OCT4 (1 µg/mL. ab18976, Abcam), rabbit anti-HIF-1α (1:500. ab51608, Abcam), rabbit anti-GAPDH (1:2500, ab9485, Abcam). The secondary antibody was goat anti-rabbit IgG H&L (HRP) (ab6721, 1:2000, Abcam).
Quantitative real-time reverse transcription PCR (qRT-PCR) analysis
RNA from cells was extracted with TRIzol reagent following the manufacturer’s instructions (Invitrogen, Gaithersburg, MD, USA). qRT-PCR was conducted by the SYBR Select Master Mix in an ABI Prism 7000 Sequence Detection. To determine the RNA levels of STAT3 miR-124, and total RNA, RNAs were reverse transcribed using RT Reagent Kit (Vazyme, Nanjing, China). The relative quantification (2−ΔΔCt) was used to assess STAT3, miR-124 and total RNAs levels. The internal controls were U6 and GAPDH. Primers are shown in Table 1.
Table 1.
Primer Sequences for qRT-PCR and transfection.
| Genes | Sequences |
|---|---|
| STAT3 | F: 5ʹ-CAGCAGCTTGACACACGGTA-3’ |
| miR-124 | R: 5ʹ-AAACACCAAAGTGGCATGTGA-3ʹ F: 5ʹ-GCAGCGTGTTCACAGC-3ʹ R: 5ʹ-TCCAGTTTTTTTTTTTTTTTATCAAGGT-3’ |
| GAPDH | F: 5ʹ-CAGGTGGTCTCCTCTGACTT-3’ |
| R: 5ʹ-CCAAATTCGTTGTCATACCA-3’ | |
| U6 | F: 5ʹ-CGCTTCGGCAGCACATATAC-3’ |
| R: 5ʹ-AAA ATATGGAACGCTTCACGA-3’ | |
| STAT3-1 | 5ʹ-CAGCTCTACAGTGACAGCTTCCCAA-3’ |
| STAT3-2 | 5ʹ-CACATGCCACTTTGGTGTTTCATAA-3’ |
| STAT3-3 | 5ʹ-GAGGCGGCAACAGATTGCCTGCATT-3’ |
F: forward primer; R: reverse primer
Flow cytometry apoptosis assay
Cell apoptosis was detected by the Annexin V-PE Apoptosis Detection Kit (Beyotime, Shanghai, China). Cells were added to 6-well plate and grown to 60–80% confluence before transfection. After 48 hours, cells were washed twice in PBS (Sangon Biotech, Shanghai, China) before resuspension in 500 μL 1× Binding Buffer. Next, cells were mixed with 5 μL annexin V-FITC and 5 μL propidium iodide (PI) in the dark at 37°C for 30 min, followed by adding 1× Binding Buffer to each tube. Stained cells were measured by flow cytometry (Bio-Rad, CA, USA) using the Cell Quest Pro software (BD, USA). The data were analyzed via FlowJo 9.1 software.
Cell cycle analysis
The cells were first harvested after 72 hours of transfection, and the cell suspension was then digested. Afterwards, the cells were fixed with ethanol (75%) for 4 h at 4°C, the supernatant was then discarded, and the cells were incubated with an RNA enzyme containing iodide (PI, 40%, Sigma-Aldrich). Cells were washed three times with PBS. The cell cycle was subsequently detected by using FACS Calibur (BD Biosciences, USA), and data analysis was conducted using FACS Diva (BD Biosciences).
Transwell assay
Cells were cultured with 50 μL madrigal diluted with serum-free medium. The cell suspensions were obtained after transfection. Then, 200 μL cell suspension (5 × 103 cells each well) was placed in the top compartment, and 500 μL RPMI 164 medium with 10% FBS was mixed in the bottom compartment. After washing twice with PBS, the cells were fixed using 4% paraformaldehyde for 20 min and stained with 0.1% crystal violet for 30 min, followed by being washed twice with PBS. The cells were photographed under a microscope.
Statistical analysis
All histogram data are presented as the mean ± SD. Student’s t-test was used for comparisons between groups. A two-way Student’s t-test was used to analyze experimental data. P < 0.05 was regarded as statistically significant. All the experiments were conducted in triplicate.
Results
MCF7 presented resistance to doxorubicin, paclitaxel, 5-fluorouracil, and cisplatin
At the beginning of our study, we cultured four drug-resistant MCF7 cell lines. MTT assays were utilized to detect the inhibition rate of cell lines under the treatment with different drug concentrations. According to the inhibition rate, we calculated the half maximal inhibitory concentration (IC50) and the resistance factor. The results listed in Table 2 show that the resistance factor was the greatest in the MCF7-DOX-R cell lines; thus, we chose DOX, MCF7 and MCF7-R (MCF7 cells resistant to DOX) in our subsequent studies.
Table 2.
Resistance of gastric cancer cell line MCF-7 to different chemotherapeutic drugs ().
| Chemotherapeutics | IC50 (mg/L) |
RF | |
|---|---|---|---|
| MCF7 | MCF7-R | ||
| DOX | 0.28 ± 0.17 | 12.91 ± 1.14 | 46.11 |
| PTX | 0.22 ± 0.04 | 9.58 ± 0.11 | 43.55 |
| 5-FU | 0.68 ± 0.27 | 14.72 ± 0.12 | 21.65 |
| DDP | 1.40 ± 0.07 | 48.51 ± 2.18 | 34.65 |
SD: standard deviation; RF: resistance factor; DOX: doxorubicin; PTX: paclitaxel; 5-FU: 5-fluorouracil; DDP: cisplatin.
STAT3 was highly expressed in MCF7/ADR cells in GSE24460
We used the Gene Expression Omnibus to perform the bioinformatic analysis. There were 2990 differentially expressed mRNAs under the following screening condition: log2FC>1&adjusted P value<0.05 (Figure 1A). Figure 1B shows the top 10 upregulated and downregulated mRNAs. Then, we used the DAVID website to perform KEGG enrichment analysis. As shown in Table 3, 12 pathways were enriched, and the HIF-1 signaling pathway was among them. Putting the differentially expressed genes together with the genes in the HIF-1 signaling pathway, we noticed that the mRNA of STAT3 was upregulated (Figure 1C), and the protein-protein interaction networks in the HIF-1 signaling pathway were visualized using the STRING website (Figure 1D).
Figure 1.
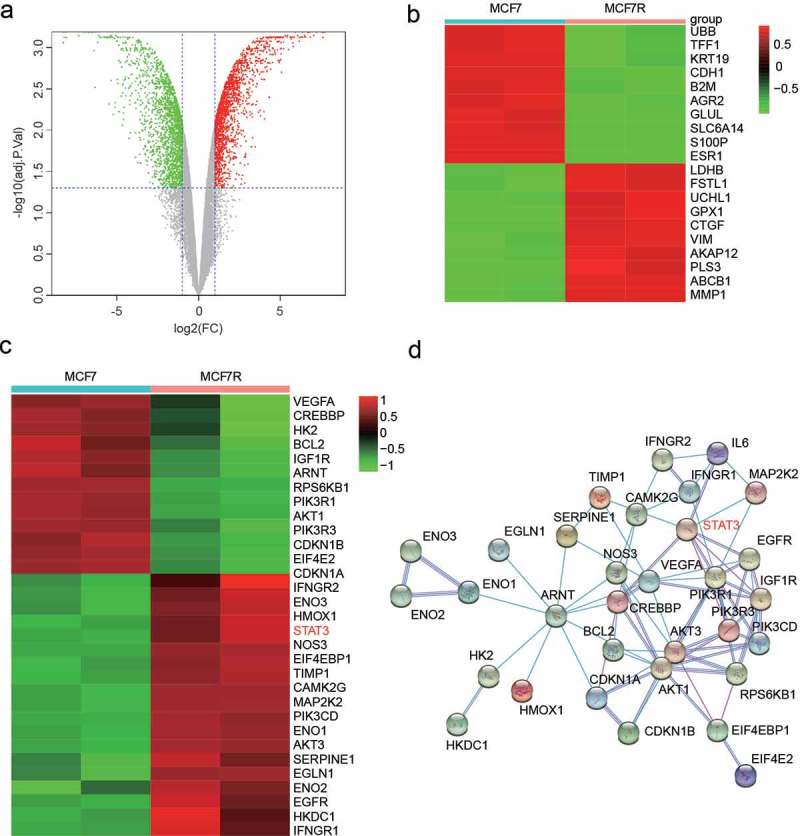
STAT3 was highly expressed in MCF7-R cells. (a) A total of 2990 differentially expressed mRNAs under the screening condition log2FC>1&adjusted P < 0.05 are shown as a Volcano Plot. (b) Heatmap showing the 20 most differentially regulated genes in MCF7 and MCF7-R cells identified by total RNA sequencing analysis. (c) Differentially expressed genes enriched in the HIFα signaling pathway. (d) The protein-protein interaction networks in the HIF-1 signaling pathway were visualized using the STRING website.
Table 3.
The KEGG results of DAVID online bioinformatics analysis.
| Category | Term | P-Value |
|---|---|---|
| KEGG_PATHWAY | PI3K-Akt signaling pathway | 4.20E-04 |
| KEGG_PATHWAY | ErbB signaling pathway | 1.60E-03 |
| KEGG_PATHWAY | HIF-1 signaling pathway | 2.60E-03 |
| KEGG_PATHWAY | Sphingolipid signaling pathway | 5.60E-03 |
| KEGG_PATHWAY | AMPK signaling pathway | 7.40E-03 |
| KEGG_PATHWAY | FoxO signaling pathway | 1.10E-02 |
| KEGG_PATHWAY | VEGF signaling pathway | 1.20E-02 |
| KEGG_PATHWAY | mTOR signaling pathway | 1.50E-02 |
| KEGG_PATHWAY | Hippo signaling pathway | 1.60E-02 |
| KEGG_PATHWAY | MAPK signaling pathway | 2.60E-02 |
| KEGG_PATHWAY | p53 signaling pathway | 3.30E-02 |
| KEGG_PATHWAY | cGMP-PKG signaling pathway | 4.30E-02 |
KEGG: Kyoto Encyclopedia of Genes and Genomes, a bioinformatics resource for linking genomes to life and the environment
Role of STAT3 in regulating stemness of BCSCs
A previous study [19] reported that CD24−CD44+ cells derived from primary breast cancer samples and cell lines displayed stem cell-like features. Thus, we identified the percentage of CD24−CD44+ cells from both MCF7 and MCF7-R cell lines. As shown in Figure 2A, there were nearly 25% CD24−CD44+ cells in MCF7-R cell lines and 10% CD24−CD44+ cells in MCF7 cell lines (P < 0.01). Therefore, drug-resistant cell lines showed greater dryness. After purification, STAT3 expression in CD24−CD44+ BCSCs was evaluated by qRT-PCR and western blot. Greater levels of mRNA and protein expression were shown in MCF7-R cells than in MCF7 cell lines (Figure 2B-C). Furthermore, the protein level of p-STAT3 was significantly greater in MCF7-R cell lines, which indicated that the phosphorylation of STAT3 may play a key regulatory role in doxorubicin-induced chemoresistance (Supplementary Figure 1). Moreover, the transfection efficiency of siRNAs and pcDNA-STAT3 was verified through qRT-PCR (Figure 2D-E).
Figure 2.
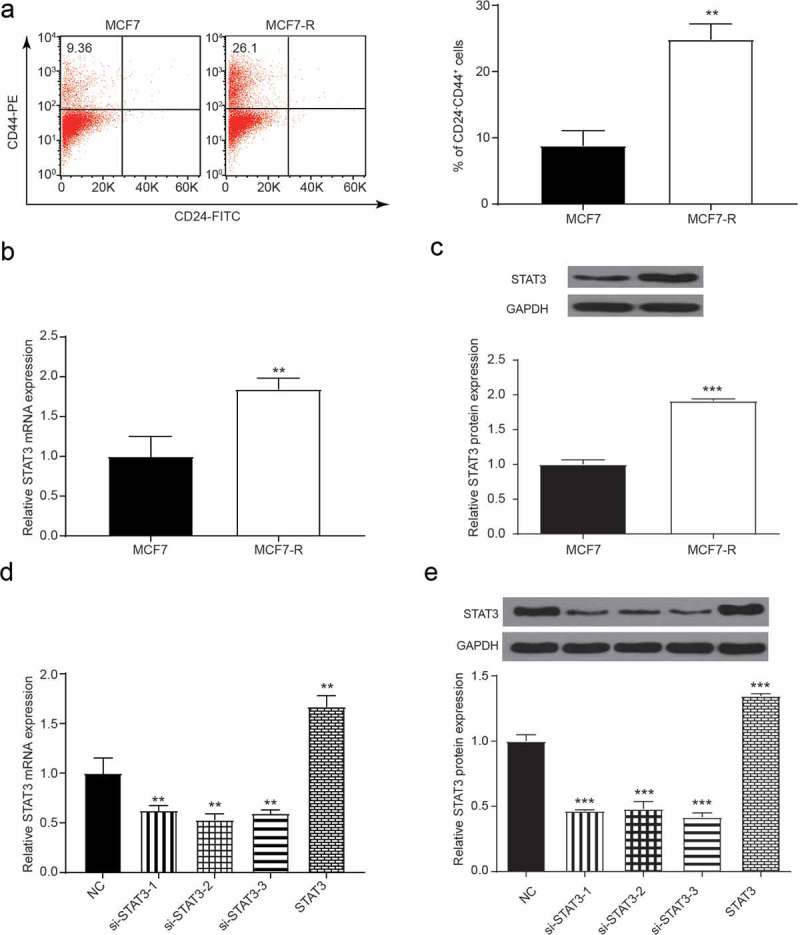
The expression of STAT3 in MCF7 and MCF7-R. (a) Flow cytometry analysis showed the percentage of CD44+CD24− cells. (b) The expression of STAT3 was measured by qRT-PCR, and the expression in the MCF7-R group was greater than that in the MCF7 group (**P < 0.01). (c) Western blot assay showed that the expression of the STAT3 protein in BCSCs increased in the MCF7-R group (**P < 0.01). (d) qRT-PCR results showed that the expression of STAT3 was increased by pc-STAT3 and inhibited by si-STAT3 (**P < 0.01). (e) Western blot results showed that the expression of STAT3 was increased by pc-STAT3 and inhibited by si-STAT3 (**P < 0.01).
STAT3 affects doxorubicin resistance of breast cancer stem cells through the HIF-1 signaling pathway
To demonstrate the inhibitory effects of STAT3 on CD24−CD44+ BCSCs, the pcDNA3.1-STAT3 vector plasmid was transfected into these cells. The pcDNA3.1-STAT3 vector plasmid encodes the full-length coding sequence of the STAT3 domain without the 3ʹ UTR, and the si-STAT3 was also transfected for the loss-of-function assay. As shown in Figure 3A, doxorubicin-resistant CD24−CD44+ BCSCs showed a much greater IC50. The transfection of STAT3 further increased the resistance while si-STAT3 reduced the drug resistance, making the expression closer to that of normal CD24−CD44+ BCSCs. In the same manner, the migration of CD24−CD44+ cells in MCF7-R cell lines was increased by STAT3 and decreased by si-STAT3 (Figure 3B). The knockdown of STAT3 impeded the cell cycle in the G0 phase, whereas overexpressed STAT3 accelerated the cell cycle period into the S or M phase (Figure 3C, P < 0.05). To test the doxorubicin resistance of STAT3 in BCSCs, we used flow apoptosis assays and transwell invasion assays to examine the effect of STAT3 on the doxorubicin resistance of BCSCs. Comparing with the MCF7-R group, the si-STAT3 group had a greater apoptotic rate and the pc-group had a reverse consequence at both concentrations of doxorubicin. Moreover, the NC group had a greater apoptotic rate than did the MCF7 group (Figure 3D, P < 0.05). Compared with the NC group, the number of invaded BCSCs decreased in the si-STAT3 group but increased in the STAT3 group (Figure 3E, P < 0.05).
Figure 3.
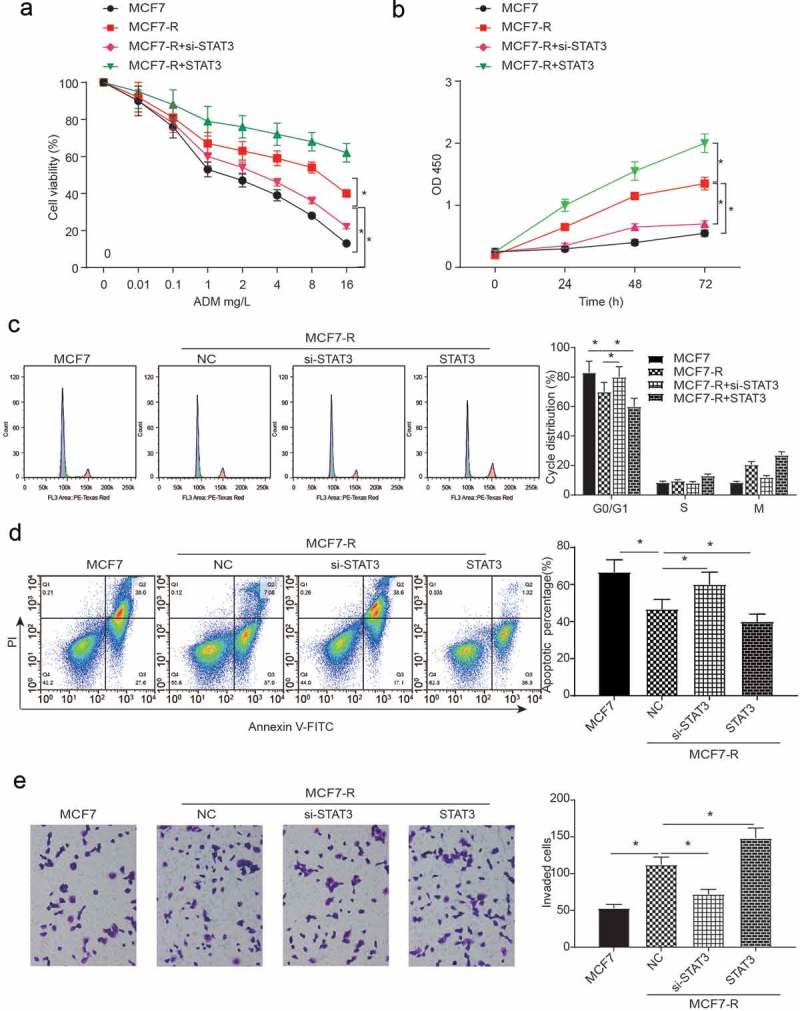
The flow apoptosis assays and cell invasion of breast cancer stem cells. (a) The cell viability of all groups decreased when the concentration of DOX increased. Compared with the MCF7-R group, the knockdown of STAT3 inhibited cell viability while the overexpression of STAT3 could promote cell viability (*P < 0.05). (b) An MTT assay was used to test the OD value at different concentrations of doxorubicin (*P < 0.05). (c) The results of the cell cycle analysis at different concentrations of doxorubicin showed that compared with the MCF-R group, the knockdown of STAT3 impeded the cell cycle in the si-STAT3 group, whereas the overexpression of STAT3 arrested the cell cycle in the STAT3 group (*P < 0.05). (d) The percentage of apoptotic cells was determined by flow cytometric analysis. The apoptosis rate was calculated by counting the percentage of early apoptotic and late apoptotic cells. The NC group had a greater apoptotic rate than the MCF7 group. Compared with the NC group, the si-STAT3 group had a greater rate of apoptosis, while the STAT3 group had a reverse consequence at both concentrations of doxorubicin. (*P < 0.05). (e) Compared with the MCF7 group, the invasion of the MCF7-R group decreased in the si-STAT3 group, whereas the invasion increased with the transfection of STAT3 (*P < 0.05).
As for the western blot, we tested some genes, including ALDH1, SOX2 and OCT4, that are related to the expression of cancer stem cells. All genes had a greater expression in the pc-STAT3 group than in the MCF7-R group. The relative expression level of these genes was lower in the si-STAT3 group than in the NC group (Figure 4A, P < 0.05). Then, we used western blots to verify whether STAT3 would affect the BCSCs through the HIF-1 signaling pathway. Figure 4B shows that HIF-1α had a greater expression in the pc-STAT3 group, and the HIF-1 signaling pathway was activated (P < 0.01).
Figure 4.
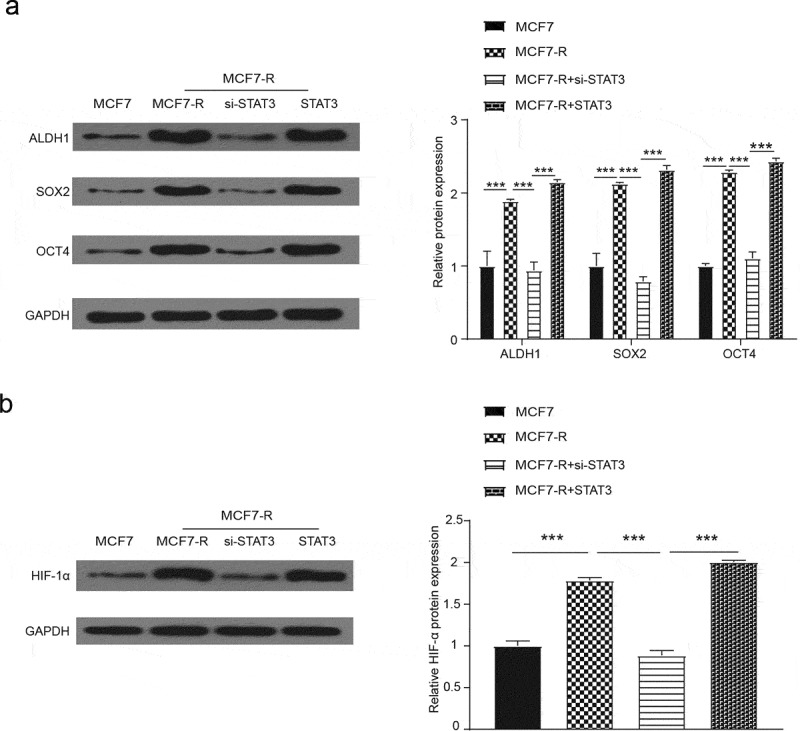
Western blot was used to measure the relative expression when STAT3 was overexpressed or knocked down. (a) With the treatment of doxorubicin, the expression of ALDH1, SOX2 and OCT4 in the MCF7-R-STAT3 group was greater than the MCF7-R-NC group, while the relative expression was inverse with the transfection of si-STAT3 (*P < 0.05). (b) Western blot was used to assess the activation of the HIF-1 pathway, and the relative expression of HIF-1α was downregulated by si-STAT3 but upregulated by STAT3 (*P < 0.05).
miR-124 reversed the doxorubicin resistance of breast cancer stem cells through STAT3
We evaluated whether miR-124 could reverse the doxorubicin resistance of breast cancer stem cells through STAT3. First, the target relationship between miR-124 and STAT3 was predicted by TargetScan. The luciferase reporter assay for the miR-124 in the presence of STAT3 is shown in Figure 5A, and the result showed that the STAT3 wild-type reporter was strongly suppressed by miR-124 (P < 0.05). The transfection efficiency of miR-124 mimics and inhibitor was quantified by qRT-PCR (Figure 5B, P < 0.05). The protein level of STAT3 in the MCF7-R group was lower than that in the inhibitor group, and the mimics group had an opposite effect (Figure 5C, P < 0.05). To verify the doxorubicin resistance of the combined action of miR-124 and STAT3 in breast cancer cell lines, we used flow cytometry, MTT assay and cell cycle analysis to test the co-effect of miR-124 and STAT3 on BCSCs. Compared with the MCF7-R group, the transfection of miR-124 mimics inhibited cell viability, while the co-transcription of miR-124 and STAT3 could reserve the effect. The cell viability of all groups decreased when the concentration of DOX increased (Figure 5D, P < 0.05). As shown in Figure 5E, the MCF7-R group had the greatest OD value. Compared with the MCF7 group, the mimics groups had a greater OD value (P < 0.05). The overexpression of miR-124 facilitated the cell cycle, whereas the co-transcription of miR-124 and STAT3 had a similar cell cycle as the MCF7-R group (Figure 5F, P < 0.05). It can be seen from Figure 5G that the miR-124 overexpression group had a higher apoptotic percentage than that of the MCF7-R group, while the co-transcription of miR-124 mimics and pc-STAT3 had a similar apoptotic percentage as that of the MCF7-R group.
Figure 5.
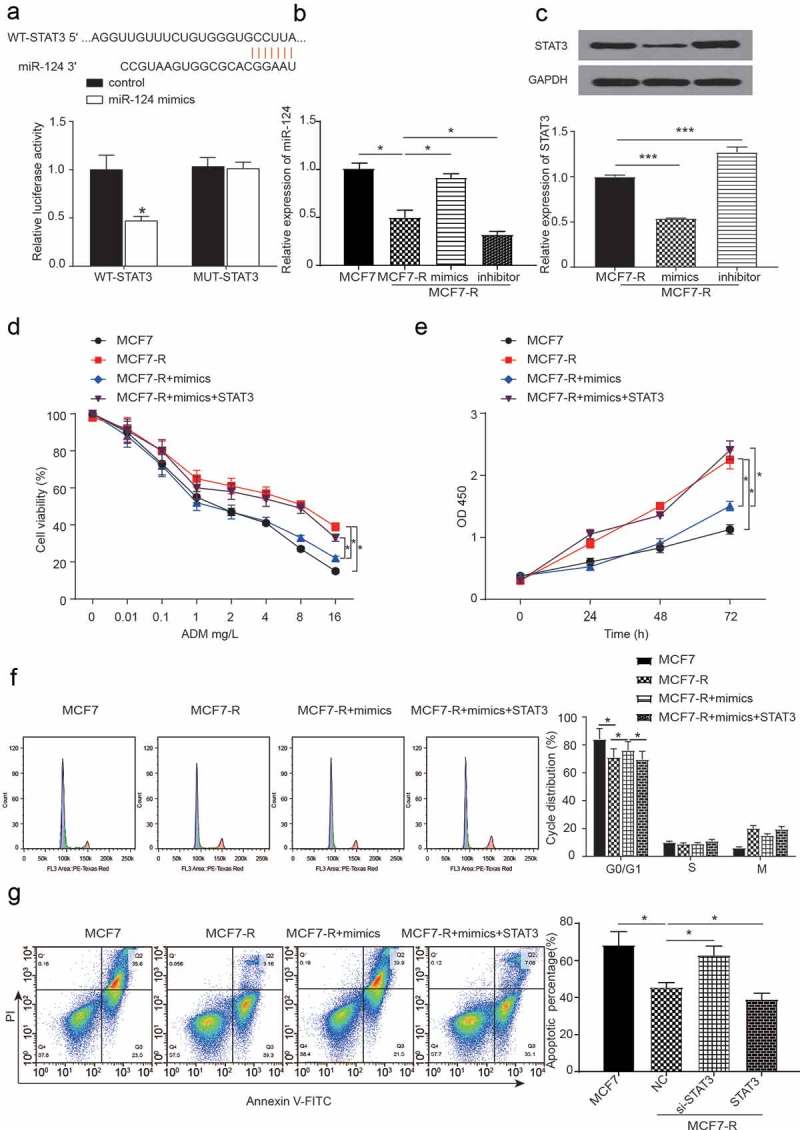
The expression of miR-124 and the coefficients of STAT3 and miR-124 regulate the doxorubicin resistance of breast cancer stem cells. (a) The 3ʹ UTR regions of STAT3 were found to harbor a binding site for miR-124. The dual luciferase assay was used to analyze the target between the STAT3 and miR-124 groups (*P < 0.05). (b) The expression of miR-124 with or without the transfection of miR-124 mimics or inhibitor in MCF7 and MCF7-R cell lines was analyzed by qRT-PCR. (c) Western blots were used to analyze the expression of STAT3 with the overexpression or knockdown of miR-124 in the MCF7-R cell line (*P < 0.05). (d) The cell viability of all groups decreased when the concentration of DOX increased. Compared with the MCF7-R group, the knockdown of miR-124 decreased the cell viability while the co-transfection of mimics and STAT3 has a similar tendency compared to the NC group in the MCF7-R cell line (*P < 0.05). (e) An MTT assay was used to test the OD value at different concentrations of doxorubicin (*P < 0.05). (f) Overexpression of miR-124 facilitated the cell cycle whereas the co-transfection of miR-124 and STAT3 had similar effects on the cell cycle compared with the MCF7-R group (*P < 0.05). (g) The overexpression of miR-124 group had a greater apoptotic percentage than the MCF-R group, while the co-transfection of the miR-124 mimics and STAT3 has a similar apoptotic percentage compared with the MCF7-R group (*P < 0.05).
In addition, the western blot and transwell invasion assays were also used to measure doxorubicin resistance of the combined action of miR-124 and STAT3 in breast cancer cell lines. As shown in Figure 6A, the MCF7-R group had the greatest number of invaded cells, and the MCF7 group had the lowest number. In addition, when compared with the MCF7-R group, the mimics group showed poor invasiveness. As shown in Figure 6B,C, compared with the MCF7-R group, the protein levels of ALDH1, SOX2 and OCT4 were lower in the miR-124 mimics group, and co-transfection with STAT3 reversed the effect of the miR-124 mimics. Furthermore, the miR-124 mimics were proven to inhibit the HIF-1 signaling pathway.
Figure 6.
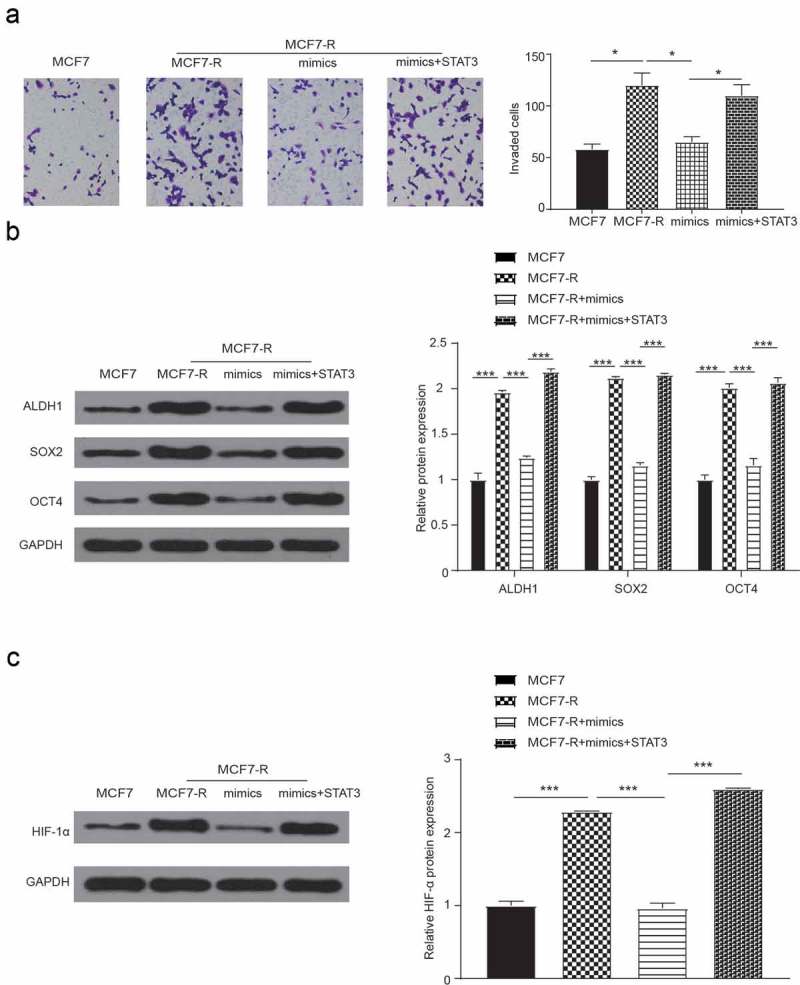
Cell invasion and western blot verify the effect of miR-124 on the drug resistance of breast cancer stem cells. (a) Compared with the MCF7-R-NC group, cell invasion decreased in the mimics group and increased in the co-transfection with mimics and STAT3 group (*P < 0.05). (b) The expression levels of ALDH1, SOX2 and OCT4 were analyzed by western blot (*P < 0.05). (c) The expression of HIF-1α was analyzed by western blot (*P < 0.05).
Discussion
Our results showed that STAT3 was upregulated in DOX-resistant BCSCs. According to a previous report, STAT3 activation could contribute to DOX resistance in breast cancer [20]. We observed the function of STAT3 in breast cancer cells and DOX-resistant BCSCs. Cell viability was modestly impaired with the overexpression of STAT3 when the DOX gradient increased. Furthermore, we tested the role of STAT3 in the cell cycle. Similarly, the overexpression of STAT3 benefited the progression of the cell cycle. Despite its contribution to cell viability and proliferation, STAT3 had a positive effect on promoting invasion and decreasing apoptosis in DOX-resistant BCSCs. Taken together, STAT3 partly resulted in DOX resistance and promoted the progression of BCSCs. In comparison with previous reports, we obtained similar results. Seo, H. S et al. found that inhibition of STAT3 suppressed drug resistance of breast cancer cells [21]. In a study by Soleymani Abyaneh, H et al., STAT3 was important for mediating the hypoxia-induced chemoresistance in triple negative breast cancer [22].
Recently, Lee, J. H et al. demonstrated that STAT3 functions as a transcription activator of ALDH1. In addition, inhibition of STAT3 led to the decline of ALDH1 levels [23]. Cancer cells with high levels of ALDH1 will present with CSCs-like properties, such as self-renewal and tumorigenicity [24]. In a study by Chhabra, R, suppression of SOX2 resulted in the decrease of the CSC population [25]. OCT4 directly modulates stemness in human germ cell tumors and enhances radio-resistance and migration activities of rectal cancer cells [26,27]. To confirm the positive effect of STAT3 on relevant markers in cancer stem cells, western blot was used to measure the expression levels of ALDH1, SOX2 and OCT4. The expression levels of ALDH1, SOX2 and OCT4 proteins were distinctly promoted when STAT3 was overexpressed. Therefore, STAT3 was associated with the occurrence and progression of BCSCs.
The cooperation of STAT3 and HIF-1 has been explored and confirmed in prior studies. He M. et al. found that HIF-1α downregulates miR-17/20a to release STAT3 in myeloid leukemia cell differentiation [28]. In addition, STAT3 cooperates with HIF-1α to activate HIF-1 target genes in a breast cancer cell line [29]. To validate their correlation, we determined the levels of HIF-1α. On the basis of protein expression analysis, HIF-1 was upregulated when STAT3 was aberrantly expressed. Hence, we confirmed that STAT3 affected BCSCs through the HIF-1 signaling pathway. It has been proved that the HIF-1 signal is involved in NOTCH signaling pathway which subsequently influences the process of epithelial-to-mesenchymal transition [30]. As BCSCs are invasive [31] and cell invasion, migration in breast cancer is closely related to tumor metastasis and prognosis [32], we performed the transwell assay and found that cell invasion in MCF7-R cells was remarkably increased compared to that in the MCF7 cell line.
It has been proved that STAT3 negatively interacted with miR-124 [33]. To confirm this result, we conducted a luciferase reporter assay to observe miR-124 expression. Our results of the luciferase reporter assay were identical to the results of the previous report. The inhibitory role of miR-124 on breast cancer has been previously identified in different mechanisms. For instance, miR-124 regulates SNAI2 to restrict the proliferation and migration of breast cancer cells [34]. Interestingly, miR-124 downregulation contributes to the proliferation of breast cancer via lncRNA-MALAT1 regulation and CDK4/E2F1 signal activation [35]. In addition, transwell results indicated that cell invasion in MCF7-R cells was inhibited by the treatment of miR-124 mimics and enhanced with additional STAT3, which proved that miR-124 reversed the cell invasion induced by doxorubicin resistance in BCSCs through STAT3.
In a recent study by Wang, M et al., miR-124 directly inhibited STAT3 to suppress growth and to enhance radiation-induced apoptosis in non-small cell lung cancer [36]. In a study by Sun, Y and colleagues, miR-124 was proved to inhibit the occurrence of pro-inflammatory cytokines to guarantee the anti-inflammatory effect of cholinergic [37]. To investigate the effect of miR-124 in BCSCs, we conducted MTT assays, cell cycle analyses and western blot assays. In the detection of cell viability, the role of miR-124 was completely inverse that of STAT3. The overexpression of STAT3 impaired the strong power of the upregulated miR-124. In addition, miR-124 generally dampened the progression of the cell cycle as the cell cycle was arrested in the G0/G1 phase in most DOX-resistant BCSCs. Thus, it is reasonable that miR-124 accelerated cell apoptosis. Cell invasive capacity distinctly weakened when miR-124 was upregulated. The expression of ALDH1, SOX2 and OCT4 was correspondingly inhibited by miR-124, indicating that miR-124 inhibits the progression of BCSCs.
Although the roles of miR-124 and the STAT3 and HIF-1 signaling pathways have been discussed in diverse human cancers, their cooperation in breast cancer and association with drug resistance have not been described. This research announced the role of miR-124 in BCSCs via the STAT3/HIF-1 signaling pathway and its effect on overcoming DOX resistance, which could be valuable for the improvement of therapeutic strategy in the future. Apart from our novel findings, some limitations must be considered. On the one hand, we selected one of the phenotypes of cancer stem cells (CSCs). Whether miR-124 still sustains its prosperity in other types of CSCs needs to be explored in further studies. On the other hand, the influence of miR-124 on cell migration is still elusive as we did not conduct migrant detection.
In summary, we identified a new mechanism of miR-124 on BCSC resistance to DOX as follows: miR-124 sensitizes DOX-resistant BCSCs to DOX by modulating the STAT3 and HIF-1 signaling pathways. Furthermore, our results provide new insight into the mechanisms underlying chemotherapeutic resistance in BCSCs. The restoration of miR-124 and inhibition of STAT3 may be a potential and useful strategy for overcoming drug resistance.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- [1].Woolston C. Breast cancer. Nature. 2015;527:S101. [DOI] [PubMed] [Google Scholar]
- [2].AbuHammad S, Zihlif M. Gene expression alterations in doxorubicin resistant MCF7 breast cancer cell line. Genomics. 2013;101:213–220. [DOI] [PubMed] [Google Scholar]
- [3].Greife A, Tukova J, Steinhoff C, et al. Establishment and characterization of a bladder cancer cell line with enhanced doxorubicin resistance by mevalonate pathway activation. Tumour Biol. 2015;36:3293–3300. [DOI] [PubMed] [Google Scholar]
- [4].Arora HC, Jensen MP, Yuan Y, et al. Nanocarriers enhance Doxorubicin uptake in drug-resistant ovarian cancer cells. Cancer Res. 2012;72:769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Peiris-Pages M, Martinez-Outschoorn UE, Pestell RG, et al. Cancer stem cell metabolism. BCR. 2016;18:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hong D, Fritz AJ, Finstad KH, et al. Suppression of breast cancer stem cells and tumor growth by the RUNX1 transcription factor. Mol Cancer Res. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bai X, Ni J, Beretov J, et al. Cancer stem cell in breast cancer therapeutic resistance. Cancer Treat Rev. 2018;69:152–163. [DOI] [PubMed] [Google Scholar]
- [8].Troschel FM, Bohly N, Borrmann K, et al. miR-142-3p attenuates breast cancer stem cell characteristics and decreases radioresistance in vitro. Tumour Biol. 2018;40:1010428318791887. [DOI] [PubMed] [Google Scholar]
- [9].Zhang Y, Xu B, Zhang XP. Effects of miRNAs on functions of breast cancer stem cells and treatment of breast cancer. Onco Targets Ther. 2018;11:4263–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Li W, Zang W, Liu P, et al. MicroRNA-124 inhibits cellular proliferation and invasion by targeting Ets-1 in breast cancer. Tumour Biol. 2014;35:10897–10904. [DOI] [PubMed] [Google Scholar]
- [11].Li L, Luo J, Wang B, et al. Microrna-124 targets flotillin-1 to regulate proliferation and migration in breast cancer. Mol Cancer. 2013;12:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang X, Li Y, Dai Y, et al. Sulforaphane improves chemotherapy efficacy by targeting cancer stem cell-like properties via the miR-124/IL-6R/STAT3 axis. Sci Rep. 2016;6:36796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wu HT, Liu J, Li GW, et al. The transcriptional STAT3 is a potential target, whereas transcriptional STAT5A/5B/6 are new biomarkers for prognosis in human breast carcinoma. Oncotarget. 2017;8:36279–36288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sirkisoon SR, Carpenter RL, Rimkus T, et al. Interaction between STAT3 and GLI1/tGLI1 oncogenic transcription factors promotes the aggressiveness of triple-negative breast cancers and HER2-enriched breast cancer. Oncogene. 2018;37:2502–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fu Y, Xiong J. MicroRNA-124 enhances response to radiotherapy in human epidermal growth factor receptor 2-positive breast cancer cells by targeting signal transducer and activator of transcription 3. Croat Med J. 2016;57:457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].De Francesco EM, Lappano R, Santolla MF, et al. HIF-1alpha/GPER signaling mediates the expression of VEGF induced by hypoxia in breast cancer associated fibroblasts (CAFs). Breast Cancer Res. 2013;15:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yang J, AlTahan A, Jones DT, et al. Estrogen receptor-alpha directly regulates the hypoxia-inducible factor 1 pathway associated with antiestrogen response in breast cancer. Proc Natl Acad Sci U S A. 2015;112:15172–15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Samanta D, Gilkes DM, Chaturvedi P, et al. Hypoxia-inducible factors are required for chemotherapy resistance of breast cancer stem cells. Proc Natl Acad Sci U S A. 2014;111:E5429–38. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [19].Ge G, Zhou C, Ren Y, et al. Enhanced SLC34A2 in breast cancer stem cell-like cells induces chemotherapeutic resistance to doxorubicin via SLC34A2-Bmi1-ABCC5 signaling. Tumour Biol. 2016;37:5049–5062. [DOI] [PubMed] [Google Scholar]
- [20].VanKlompenberg MK, Leyden E, Arnason AH, et al. APC loss in breast cancer leads to doxorubicin resistance via STAT3 activation. Oncotarget. 2017;8:102868–102879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Seo HS, Ku JM, Lee HJ, et al. SH003 reverses drug resistance by blocking signal transducer and activator of transcription 3 (STAT3) signaling in breast cancer cells. Biosci Rep. 2017;37(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Soleymani Abyaneh H, Gupta N, Radziwon-Balicka A, et al. STAT3 but Not HIF-1alpha is important in mediating hypoxia-induced chemoresistance in MDA-MB-231, a triple negative breast cancer cell line. Cancers (Basel). 2017;9(10): 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lee JH, Choi SI, Kim RK, et al. Tescalcin/c-Src/IGF1Rbeta-mediated STAT3 activation enhances cancer stemness and radioresistant properties through ALDH1. Sci Rep. 2018;8:10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wu S, Xue W, Huang X, et al. Distinct prognostic values of ALDH1 isoenzymes in breast cancer. Tumour Biol. 2015;36:2421–2426. [DOI] [PubMed] [Google Scholar]
- [25].Chhabra R. let-7i-5p, miR-181a-2-3p and EGF/PI3K/SOX2 axis coordinate to maintain cancer stem cell population in cervical cancer. Sci Rep. 2018;8:7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shao M, Bi T, Ding W, et al. OCT4 potentiates radio-resistance and migration activity of rectal cancer cells by improving epithelial-mesenchymal transition in a ZEB1 dependent manner. Biomed Res Int. 2018;2018:3424956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Song B, Kim DK, Shin J, et al. OCT4 directly regulates stemness and extracellular matrix-related genes in human germ cell tumours. Biochem Biophys Res Commun. 2018;503:1980–1986. [DOI] [PubMed] [Google Scholar]
- [28].He M, Wang QY, Yin QQ, et al. HIF-1alpha downregulates miR-17/20a directly targeting p21 and STAT3: a role in myeloid leukemic cell differentiation. Cell Death Differ. 2013;20:408–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pawlus MR, Wang L, Hu CJ. STAT3 and HIF1alpha cooperatively activate HIF1 target genes in MDA-MB-231 and RCC4 cells. Oncogene. 2014;33:1670–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].De Francesco EM, Maggiolini M, Musti AM. Crosstalk between Notch, HIF-1alpha and GPER in Breast Cancer EMT. Int J Mol Sci. 2018;19: 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Calcagno AM, Salcido CD, Gillet JP, et al. Prolonged drug selection of breast cancer cells and enrichment of cancer stem cell characteristics. J Natl Cancer Inst. 2010;102:1637–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol. 2017;11:28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li W, Huang H, Su J, et al. miR-124 acts as a tumor suppressor in glioblastoma via the inhibition of signal transducer and activator of transcription 3. Mol Neurobiol. 2017;54:2555–2561. [DOI] [PubMed] [Google Scholar]
- [34].Du S, Li H, Sun X, et al. MicroRNA-124 inhibits cell proliferation and migration by regulating SNAI2 in breast cancer. Oncol Rep. 2016;36:3259–3266. [DOI] [PubMed] [Google Scholar]
- [35].Feng T, Shao F, Wu Q, et al. miR-124 downregulation leads to breast cancer progression via LncRNA-MALAT1 regulation and CDK4/E2F1 signal activation. Oncotarget. 2016;7:16205–16216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang M, Meng B, Liu Y, et al. MiR-124 inhibits growth and enhances radiation-induced apoptosis in non-small cell lung cancer by inhibiting STAT3. Cell Physiol Biochem. 2017;44:2017–2028. [DOI] [PubMed] [Google Scholar]
- [37].Sun Y, Li Q, Gui H, et al. MicroRNA-124 mediates the cholinergic anti-inflammatory action through inhibiting the production of pro-inflammatory cytokines. Cell Res. 2013;23:1270–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


