ABSTRACT
This study aimed to investigate the effect of circ_0000950/miR-103 network on regulating neuron apoptosis, neurite outgrowth and inflammation in Alzheimer’s disease (AD). Cellular AD model of rat pheochromocytoma cell line PC12 cells and cellular AD model of rat cerebral cortex neurons were constructed, and the effect of circ_0000950 on apoptosis, neurite outgrowth and inflammation in both cellular AD models was determined through upregulation and knockdown of circ_0000950 expression by transfection. Compensation experiments and luciferase assay were further performed to validate the sponging effect of circ_0000950 on miR-103 as well as the mechanisms of circ_0000950/miR-103 on regulating apoptosis, neurite outgrowth and inflammation in both cellular AD models. Circ_0000950 reduced miR-103 expression and increased prostaglandin-endoperoxide synthase 2 (PTGS2) expression in both two cellular AD models. And circ_0000950 overexpression promoted neuron apoptosis, suppressed neurite outgrowth and elevated IL-1β, IL-6 and TNF-α levels compared with overexpression control, whereas circ_0000950 knockdown inhibited neuron apoptosis, enhanced neurite outgrowth and lowered IL-1β, IL-6 and TNF-α levels compared with shRNA control in both two cellular AD models. Compensation experiments along with luciferase reporter assay validated that circ_0000950 promoted cell apoptosis, suppressed neurite outgrowth and elevated inflammatory cytokines levels via directly sponging miR-103. In conclusion, circ_0000950 promotes neuron apoptosis, suppresses neurite outgrowth and elevates inflammatory cytokines levels through directly sponging miR-103 in AD.
KEYWORDS: Circ_0000950, apoptosis, Alzheimer’s disease
Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder in the brain that is characterized by impaired cognition, memory and language, as well as dementia [1]. The incidence of AD is around 6.25 cases per 1000 person-years in China, and the prevalence of disease greatly increases with age [2]. The cause of AD is not completely clear until now, but it has been proposed for over 20 years that abnormal accumulation of Aβ peptides and microtubule-associated protein tau is the main cause of neuron degeneration in AD [3]. Although there are approved therapeutic drugs for AD, such as acetylcholine esterase inhibitor, the efficacy of these drugs is moderate, and there are currently no agents that could halt or reverse the disease [4]. Moreover, it takes a long duration for the disease to take course and patients eventually require complete nursing home care, which places a huge emotional and financial burden on patients, families and society [4]. Therefore, discovery and validation of novel treatment targets to develop new therapeutic methods are necessary for improving AD patients’ well-being and alleviating burdens of society.
Circular RNAs (circRNAs) are a class of non-coding RNAs consisting of circular configuration through 3ʹ to 5ʹ-phosphodiester bond, which makes them more stable in cells compared with other types of non-coding RNAs [5]. Researches investigating molecular functions of circRNAs exhibit that circRNAs act as sponges for miRNAs thereby prevent targeted messenger RNAs from being degraded by miRNAs, and growing evidence also presents that circRNAs exert both positive and negative regulation on the transcription of parental genes [6–8]. In addition, the biological functions, as well as implications of circRNAs in neuronal diseases, cardiovascular diseases and cancers, have been increasingly reported [9–12].
In our previous study, we observed that miR-103 promoted neurite outgrowth and suppress cell apoptosis by targeting prostaglandin-endoperoxide synthase 2 (PTGS2) in two cellular models of AD (PC12 cellular AD model and cellular AD model of rat cerebral cortex neurons), however, the upstream regulator of miR-103 in AD is still unclear [13]. According to the data retrieved from miRanda Database (http://www.microrna.org/microrna/home.do) and Tissue-Specific CircRNA Database (http://gb.whu.edu.cn/TSCD/), we observed several circRNAs including circ_0000950 that were potential regulators of miR-103 in AD that might share binding sites with miR-103, while our preliminary experiments showed that only circ_0000950 reversely regulated miR-103 in cellular AD models. Thus, we hypothesized that circ_0000950 might promote AD progression via targeting miR-103, and we conducted this study to investigate the effect of circ_0000950/miR-103 network on regulating neuron apoptosis, neurite outgrowth and inflammation in AD.
Materials and methods
Cells sources
Rat pheochromocytoma cell line PC12 and human embryonic kidney cells 293T cell line were purchased from Cell Resource Center of Shanghai Institute of Life Sciences, Chinese Academy of Sciences (Shanghai, China). Primary cerebral cortex neurons from rat embryo were acquired according to the methods as described in our previous study [13].
Cells culture
PC12 cells were cultured in 85% Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, USA) supplemented with 10% horse serum (Gibco, USA) and 5% fetal bovine serum (FBS) (Gibco, USA) under 95% air and 5% CO2 at 37 °C. Primary cerebral cortex neurons were cultured in 98% Neurobasal medium (Gibco, USA) supplemented with 2% B27 (Gibco, USA) under 95% air and 5% CO2 at 37 °C. 293T cells were cultured in dulbecco’s modified eagle medium (DMEM) (Gibco, USA) supplemented with 10% FBS (Gibco, USA) under 95% air and 5% CO2 at 37 °C.
Animal ethics statement
The study was approved by the Animal Ethics Committee of the Fourth Affiliated Hospital of Harbin Medical University, and all related experiments were conducted according to the “Code for the Care and Use of Animals for Scientific Purposes” statement and under the principles of 3R (replacing, refining and reducing).
Construction of cellular AD model
Two AD models including PC12 cellular AD model and cellular AD model of cerebral cortex neurons were constructed according to the methods described in our previous study [13]. In brief, (1) Aβ1-42 (Sigma, USA) was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 1 mM and pre-incubated at 37°C for 7 days to promote aggregation and then diluted in medium, then oligomerized Aβ1-42 peptides (equivalent to 1 mM peptides) were prepared for Aβ1-42 insult experiments. (2) For PC12 cellular AD model construction, PC12 cells were firstly cultured in 20 ng/ml nerve growth factor (NGF) (Sigma, USA) and 10% FBS (Gibco, USA) for 72 h at 37°C with 95% air and 5% CO2 to promote PC12 cells differentiation, and then 1 uM of oligomerized Aβ1-42 peptides were added for 24 h to build PC12 cellular AD models. (3) For cellular AD model of cerebral cortex neurons, 1 mM of oligomerized Aβ1-42 peptides was added in primary cerebral cortex neurons for 24 h to build cellular AD model of cerebral cortex neurons. (4) For validation of cellular AD model construction, 3 – (4, 5-dimethylthiazol-2-yl) – 2, 5 – diphenyl – 2H – tetrazolium bromide (MTT) assay (Invitrogen, USA) was performed to detect the activity of cells viability after Aβ1-42 insult, and cells treated without oligomerized Aβ1-42 peptides were regarded as controls. (5) In addition, expression of circ_0000950 was measured by quantitative polymerase chain reaction (qPCR) after Aβ1-42 insult.
Measurement of the effect of circ_0000950 on apoptosis, neurite outgrowth and inflammation of cellular AD models
Empty overexpression plasmids, circ_0000950 overexpression plasmids, empty shRNA plasmids and circ_0000950 shRNA plasmids were constructed by Shanghai GenePharma Bio-Tech Company (Shanghai, China) using pEX-2 and pGPU6 plasmids and then transfected into cellular AD models (both PC12 cellular AD model and cellular AD model of cerebral cortex neurons) as NC(+), Circ(+), NC(-) and Circ(-) groups. Subsequently, circ_0000950 expression and miR-103 expression were measured by qPCR, and PTGS2 expression was measured by qPCR and Western Blot at 24 h post-transfection. Cells apoptosis rate was measured by Hoechst/propidium iodide (PI) (Sigma, USA), and apoptotic markers Cleaved-Caspase3 (C-Caspase3) as well as Bcl-2 expressions were measured by Western Blot at 24 h post-transfection. Then, neurite outgrowth was measured using a microscope (Nikon, Japan) at 48 h and 72 h post-transfection. In addition, expressions of key inflammatory cytokines including interleukin (IL)-1β, IL-6 and tumor necrosis factor (TNF)-α were measured by qPCR and Western Blot at 24 h post-transfection.
Compensation effect of miR-103 on circ_0000950 functions in cellular AD models
To determine whether circ_0000950 regulated AD cells function by targeting miR-103, compensation experiments were performed. In brief, Empty shRNA plasmids, miR-103 shRNA plasmids, circ_0000950 shRNA plasmids, and circ_0000950&miR-103 shRNA plasmids were constructed by Shanghai GenePharma Bio-Tech Company (Shanghai, China) using pEX-2 and pGPU6 plasmids and then transfected into both PC12 cellular AD model and cellular AD model of cerebral cortex neurons as NC(-), MiR(-), Circ(-) and Circ(-)/miR(-) groups. Subsequently, circ_0000950 expression and miR-103 expression were measured by qPCR at 24 h post-transfection. Cells apoptosis rate was measured by Hoechst/PI (Sigma, USA), and apoptotic markers C-Caspase3 as well as Bcl-2 expressions were measured by Western Blot at 24 h post-transfection. Then, neurite outgrowth was measured using a microscope (Nikon, Japan) at 48 h post-transfection. In addition, expressions of key inflammatory cytokines including IL-1β, IL-6 and TNF-α were measured by qPCR and Western Blot at 24 h post-transfection.
Luciferase reporter assay
In order to validate the direct binding site between circ_0000950 and miR-103, luciferase reporter assay was performed as follows: (1) luciferase reporter plasmids of circ_0000950 with wild type (WT) and mutant type (Mut), as well as miR-103 overexpression and empty overexpression plasmids, were constructed by Shanghai GenePharma Bio-Tech Company (Shanghai, China). (2) These plasmids were transfected into 293T cells accordingly as WT/miR-NC, WT/miR-103, Mut/miR-NC and Mut/miR-103 groups, and then luciferase activity was examined by a dual-luciferase reporter assay system (Promega, USA) according to the instructions of the manufacturer.
MTT
Cells viability was measured by MTT reagent (Sigma, USA) according to the instructions of the manufacturer. In brief, 100 mg of MTT was dissolved in 20 ml (phosphate buffer solution) PBS to make a 5 mg/ml solution, then 10 μl of MTT solution was added to each well, which contained 100 μl culture medium, and incubated at 37°C for 4 h. Subsequently, the culture medium in the wells was sucked, and 100 μl of DMSO (Dimethyl sulfoxide) was added to each well, the solution was dissolved for 10 min. Finally, the plates were analyzed using a microplate reader (Molecular Devices, USA) at 490 nm.
Western blot
Total proteins were extracted from cells of each group with RIPA buffer (Thermo Fisher Scientific, USA). The protein concentration in each sample was then measured using the bicinchoninic acid (BCA) kit (Pierce Biotechnology, USA) and compared with the standard curve, the mean of two measurements was calculated for each sample. 20 μg protein samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride membranes (Millipore, USA). After blocking with 5% skim milk for 2 h, membranes were incubated with the corresponding primary antibody overnight at 4°C. Then, membranes were incubated with the appropriate secondary antibody for 1 h at room temperature. The bands were visualized using Novex™ ECL Chemiluminescent Substrate Reagent Kit (Invitrogen, USA) followed by exposure to X-ray film (Kodak, USA). The antibodies used in this study are summarized in Table 1.
Table 1.
Antibodies applied in Western Blot.
| Antibody | Company | Dilution |
|---|---|---|
| Primary antibody | ||
| Rabbit polyclonal to Caspase 3 | Abcam (UK) | 1:1000 |
| Rabbit polyclonal to Cleaved Caspase 3 | Abcam (UK) | 1:1000 |
| Rabbit polyclonal to Bcl 2 | Abcam (UK) | 1:1000 |
| Rabbit polyclonal to IL1B | Abcam (UK) | 1:1000 |
| Rabbit polyclonal to IL6 | Abcam (UK) | 1:1000 |
| Rabbit polyclonal to TNF alpha | Abcam (UK) | 1:1000 |
| Rabbit polyclonal to PTGS2 | Abcam (UK) | 1:1000 |
| Rabbit monoclonal to GAPDH | Abcam (UK) | 1:1000 |
| Secondary antibody | ||
| Goat Anti-Rabbit IgG H&L (HRP) | Abcam(HK) | 1:2000 |
qPCR
For CircRNA detection, cells were digested using 0.25% Trypsin (Gibco, USA) and collected, followed by extraction of total RNA using TRIzol Reagent (Invitrogen, USA). Then, 1 ug total RNA was used for linear RNAs digestion using RNase R (Epicentre, USA), followed by reverse transcription using PrimeScript™ RT reagent Kit (Takara, Japan). qPCR was performed using TB Green™ Fast qPCR Mix (Takara, Japan) for measurement of circRNA expression. For miRNA and mRNA detection, 1 ug total RNA was reversely transcribed to cDNA using PrimeScript™ RT reagent Kit (Takara, Japan), and qPCR was carried out using TB Green™ Fast qPCR Mix (Takara, Japan). The primers used in qPCR are listed in Table 2, and the result was calculated using 2−△△Ct with GAPDH or U6 as internal references.
Table 2.
Primers applied in qPCR.
| Gene | Forward primer (5ʹ–3ʹ) | Reverse primer (5ʹ–3ʹ) |
|---|---|---|
| Circ_0000950 | ATACTCAACTCAGCAGGCAAGAT | TTACTACATCAGCAGTCGCAAGA |
| IL1B | CGAATCTCCGACCACCACTAC | CACCACTTGTTGCTCCATATCC |
| IL-6 | AGTAGTGAGGAACAAGCCAGAG | CGCAGAATGAGATGAGTTGTCA |
| TNFA | TGTTCCTCAGCCTCTTCTCCTT | CTCTCAGCTCCACGCCATTG |
| PTGS2 | TGACCAGAGCAGGCAGATGA | CCAGTAGGCAGGAGAACATATAACA |
| GAPDH | GAGTCCACTGGCGTCTTCAC | ATCTTGAGGCTGTTGTCATACTTCT |
| MiR-103 | ACACTCCAGCTGGGGGCTTCTTTACAGTGCTG | TGTCGTGGAGTCGGCAATTC |
| U6 | CGCTTCGGCAGCACATATACTA | ATGGAACGCTTCACGAATTTGC |
Hoechst/PI
Cells death was measured by double nuclear staining with fluorescent dyes Hoechst 33342 (Sigma, USA) and PI (Sigma, USA). Briefly, Hoechst 33342 (λex350 nm, λem461 nm) and PI (λex535 nm, λem617 nm) were added to the cultured medium at final concentrations of 8 M and 1.5 M, respectively, and cultured at 37°C for 30 min. Images were collected by inversion fluorescence microscope (Nikon, Japan). The total cells and apoptotic cells were counted, and the percentage of apoptotic cells was calculated.
Morphology detection and total neurite outgrowth calculation
Cellular morphology was detected by microscope (Nikon, Japan). Imaging software Presage (Advanced Imaging Concepts, Inc., USA) was used to quantify the total neurite outgrowth per cell which was defined as total length of neurite outgrowth of cells divided by number of cells included [13].
Statistics
Statistics were carried out using SPSS 22.0 Software (IBM, USA) and graphs were made using GraphPad 6.01 Software (GraphPad Software, USA). Data were presented as mean ± standard deviation, and comparison was determined by t test. P value < 0.05 was considered as significant in this study.
Results
Cellular AD models construction
Cell viability was reduced in Aβ-42 insult group compared with a control group in NGF stimulated PC 12 cells (Figure 1(a)) and primary cerebral cortex neurons from rat embryo cells (Figure 1(b)), indicating successes in the construction of cellular AD models. Then, we detected whether circ_0000950 was affected by Aβ-42 insult, and observed that circ_0000950 expression was similar in Aβ-42 insult group compared with control in the two cellular AD models (Figure 1(c,d)), which suggested that Aβ-42 insult did not affect the expression of circ_0000950 in PC 12 cells or rat cerebral cortex neurons.
Figure 1.
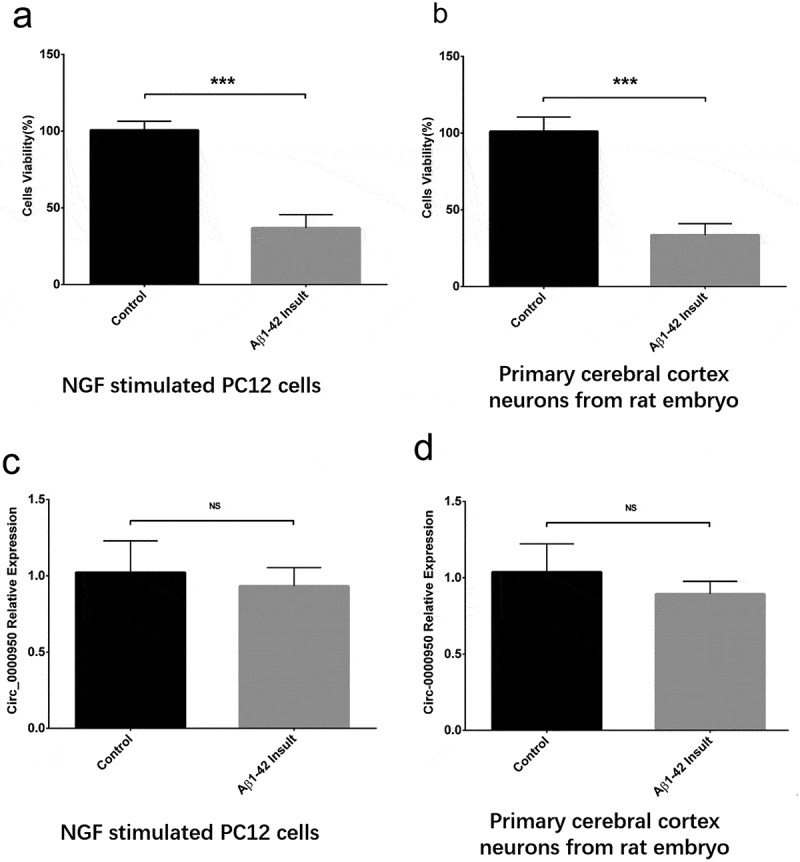
Change in cell viability and circ_0000950 expression after Aβ-42 insult. Aβ-42 insult reduced cell viability in NGF stimulated PC 12 cells (a) and primary cerebral cortex neurons from rat embryo cells (b), whereas circ_0000950 expression was not influenced by Aβ-42 insult in both NGF stimulated PC 12 cells (c) and primary cerebral cortex neurons from rat embryo cells (d). Comparison of cell viability and circ_0000950 expression was determined by t test. ***P< 0.001, NS, non-significant. P value < 0.05 was considered significant.
Effect of circ_0000950 on miR-103 and PTGS2 expression in cellular AD models
After transfection of plasmids, circ_0000950 relative expression was increased in Circ(+) group compared with NC(+) group but decreased in Circ(-) group compared with NC(-) group in both PC12 cellular AD model (Figure 2(a)) and cellular AD model of cerebral cortex neurons, indicating successful transfection (Figure 2(b)). MiR-103 expression was reduced in Circ(+) group compared with NC(+) group but promoted in Circ(-) group compared with NC(-) group in the two cellular AD models (Figure 2(c,d)). Also, PTGS2 mRNA and protein expression were both increased in Circ(+) group compared with NC(+) group but decreased in Circ(-) group compared with NC(-) group in the two cellular AD models (Figure 2(e-h)).
Figure 2.
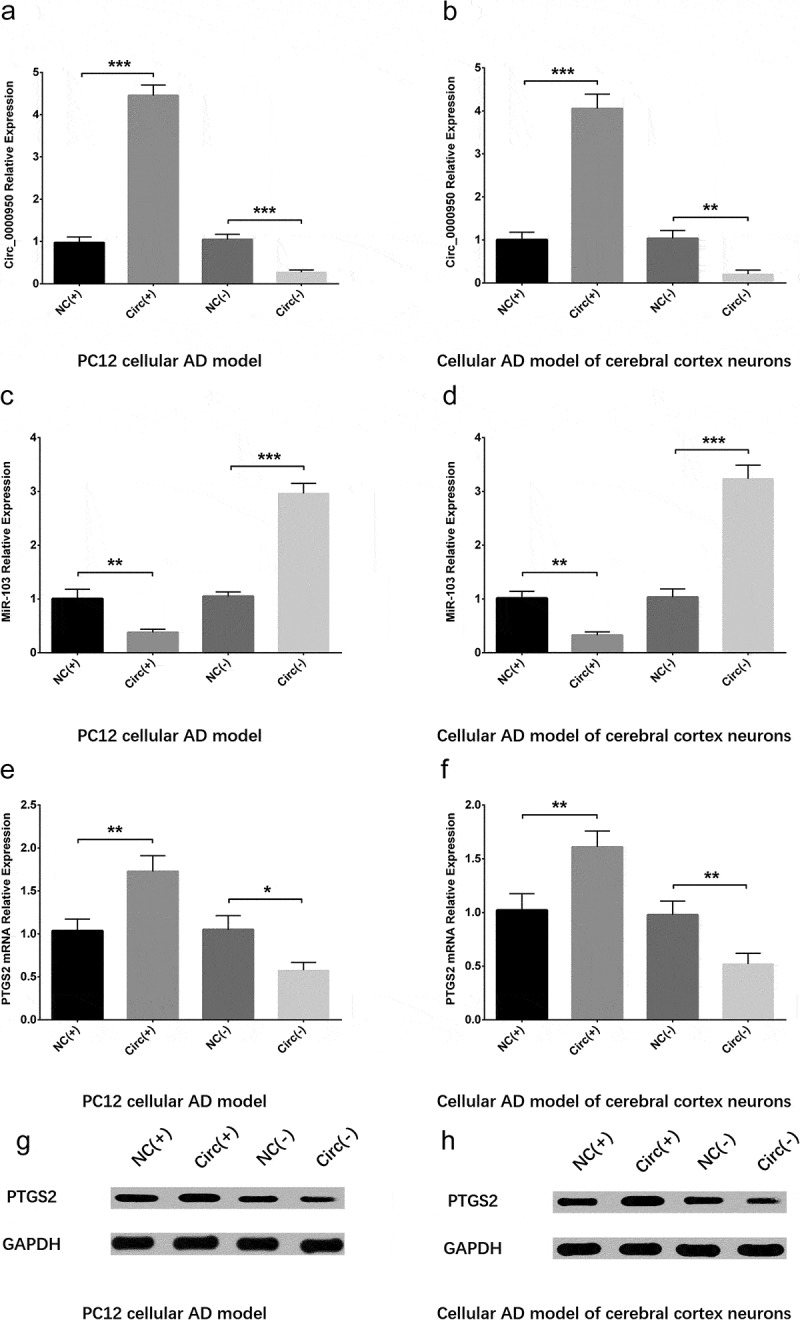
Circ_0000950 downregulated miR-103 and upregulated PTGS2. Circ_0000950 relative expression was promoted by circ_0000950 overexpression plasmids but suppressed by circ_0000950 shRNA plasmids in PC12 cellular AD model (a) and cellular AD model of cerebral cortex neurons (b). MiR-103 expression was decreased by circ_0000950 overexpression plasmids but increased by circ_0000950 shRNA plasmids in PC12 cellular AD model (c) and cellular AD model of cerebral cortex neurons (d). PTGS2 mRNA expression was inhibited by circ_0000950 overexpression plasmids but enhanced by circ_0000950 shRNA plasmids in PC12 cellular AD model (e, g) and cellular AD model of cerebral cortex neurons (f, h). Comparison of circ_0000950, miR-103 and PTGS2 mRNA expressions was determined by t test. ***P< 0.001, **P< 0.01, *P< 0.05. P value < 0.05 was considered significant.
Effect of circ_0000950 on cell apoptosis in cellular AD models
Hoechst/PI assay was used to measure cell apoptosis at 24 h after plasmid transfection, which exhibited that cell apoptosis rate was increased in Circ(+) group compared with NC(+) group but decreased in Circ(-) group compared with NC(-) group in both PC12 cellular AD model (Figure 3(a,c)) and cellular AD model of cerebral cortex neurons (Figure 3(b,d)). Western blot assay displayed that apoptotic marker C-Caspase 3 was enhanced in Circ(+) group compared with NC(+) group but decreased in Circ(-) group compared with NC(-) group in both two cellular AD models, whereas Bcl-2 expression was suppressed in Circ(+) group compared with NC(+) group but promoted in Circ(-) group compared with NC(-) group in the two cellular AD models (Figure 3(e,f)). These suggested that circ_0000950 promoted neuron apoptosis in AD.
Figure 3.
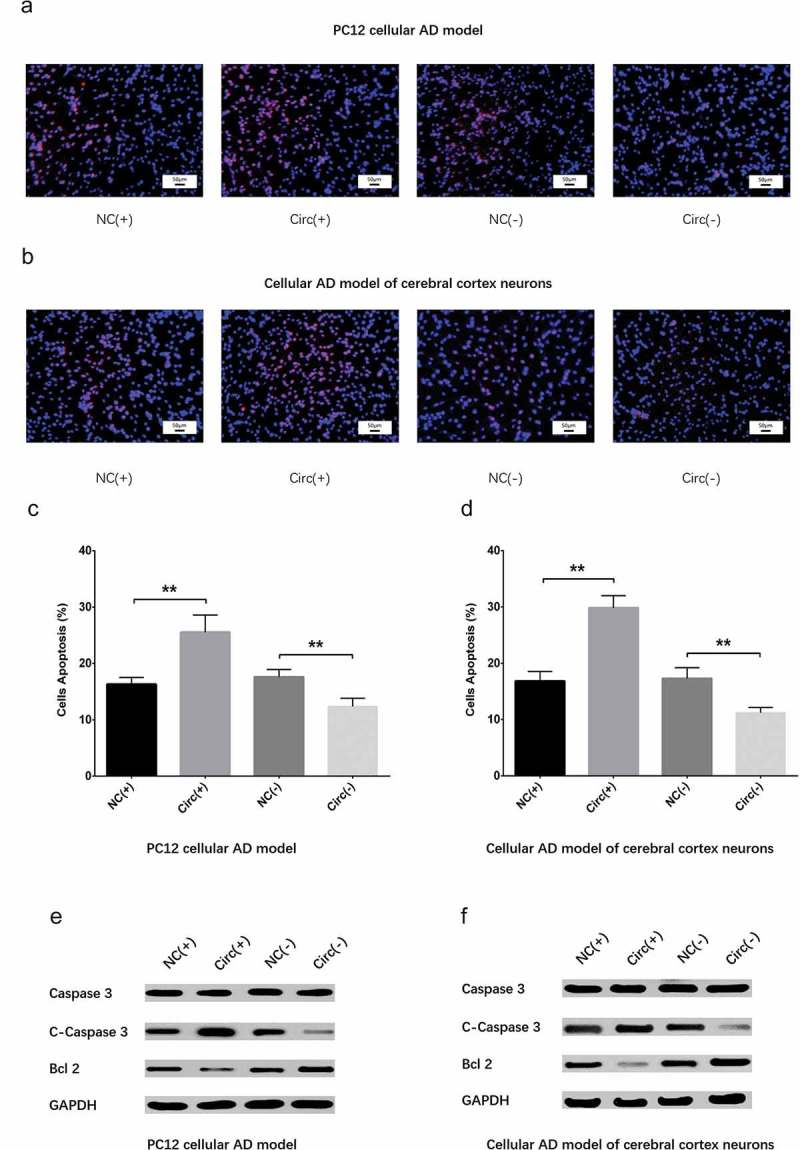
Circ_0000950 promoted cell apoptosis in cellular AD models. Cell apoptosis rate was promoted by circ_0000950 overexpression plasmids but suppressed by circ_0000950 shRNA plasmids in PC12 cellular AD model (a, c) and cellular AD model of cerebral cortex neurons (b, d). Apoptotic marker C-Caspase 3 expression was increased by circ_0000950 overexpression plasmids but decreased by circ_0000950 shRNA plasmids in both PC12 cellular AD model and cellular AD model of cerebral cortex neurons (e). Bcl-2 expression was suppressed by circ_0000950 overexpression plasmids but promoted by circ_0000950 shRNA plasmids in PC12 cellular AD model and cellular AD model of cerebral cortex neurons (F). Comparison of cell apoptosis rate was determined by t test. **P< 0.01. P value < 0.05 was considered significant.
Effect of circ_0000950 on neurite outgrowth in cellular AD models
Neurite outgrowth was measured using microscope (Nikon, Japan) at 48 h post-transfection, which disclosed that total neurite outgrowth was inhibited in Circ(+) group compared with NC(+) group but promoted in Circ(-) group compared with NC(-) group in both PC12 cellular AD model (Figure 4(a,c)) and cellular AD model of cerebral cortex neurons (Figure 4(b,d)), which indicated that circ_0000950 suppressed neurite outgrowth in AD. In addition, the observation time was extended from 48 h to 72 h, and a similar pattern was observed to that observed at 48 h in both PC cellular AD model and cellular AD model of cerebral cortex neurons (Figure 14(a,b)).
Figure 4.
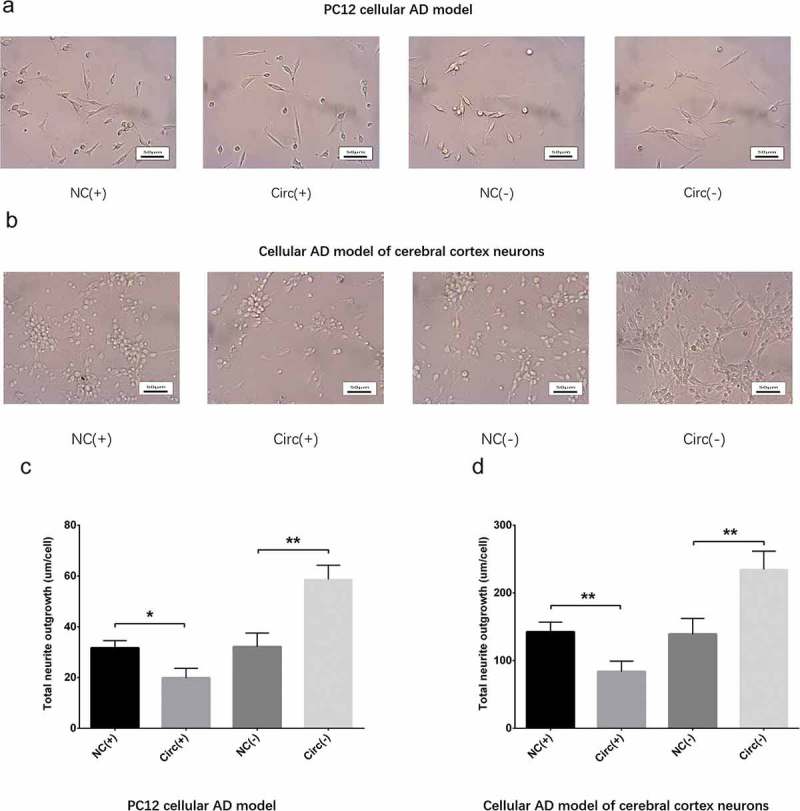
Circ_0000950 inhibited neurite growth in cellular AD models (48 h). Total neurite outgrowth was suppressed by circ_0000950 overexpression plasmids but enhanced by circ_0000950 shRNA plasmids in PC12 cellular AD model (a, c) and cellular AD model of cerebral cortex neurons (b, d). Comparison of total neurite growth was determined by t test. **P< 0.01, *P< 0.05. P value < 0.05 was considered significant.
Figure 14.
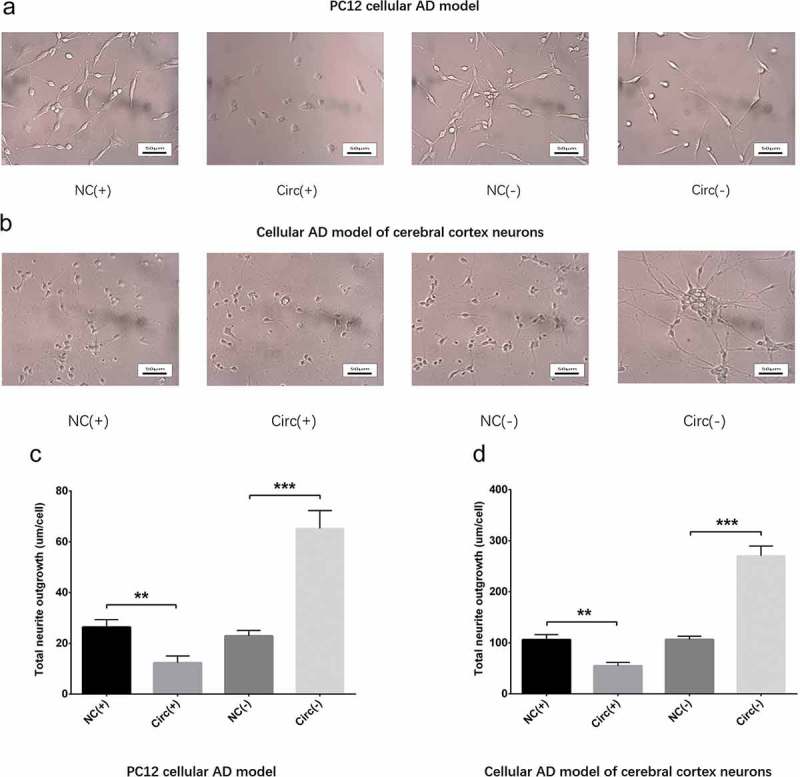
Circ_0000950 inhibited neurite growth in cellular AD models (72 h). Total neurite outgrowth was suppressed by circ_0000950 overexpression plasmids but enhanced by circ_0000950 shRNA plasmids in PC12 cellular AD model (a, c) and cellular AD model of cerebral cortex neurons (b, d). Comparison of total neurite growth was determined by t test. **P< 0.01, ***P< 0.001. P value < 0.05 was considered significant.
Effect of circ_0000950 on inflammatory cytokines levels in cellular AD models
Expressions of key inflammatory cytokines including IL-1β, IL-6 and TNF-α were measured by qPCR and Western Blot at 24 h post-transfection. In PC12 cellular AD model, IL-1β (Figure 5(a,d)), IL-6 (Figure 5(b,d)) and TNF-α (Figure 5(c,d)) expressions were increased in Circ(+) group compared with NC(+) group but decreased in Circ(-) group compared with NC(-) group. In cellular AD model of cerebral cortex neurons, IL-1β (Figure 5(e,h)), IL-6 (Figure 5(f,h)) and TNF-α (Figure 5(g,h)) expressions were promoted in Circ(+) group compared with NC(+) group but suppressed in Circ(-) group compared with NC(-) group as well. These implied that circ_0000950 elevated inflammation in AD.
Figure 5.
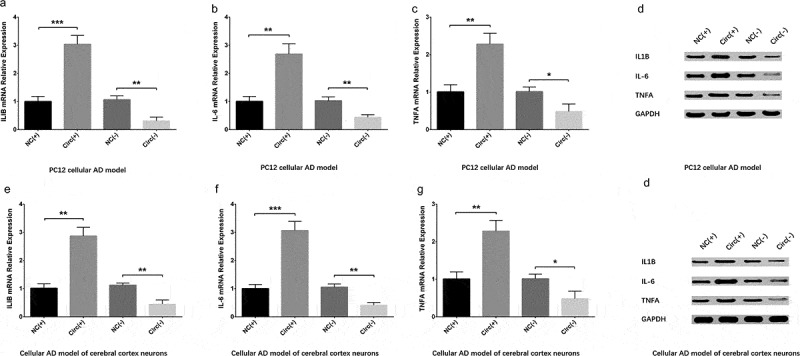
Circ_0000950 increased inflammatory cytokine levels in cellular AD models. IL-1β (a, b), IL-6 (b, d) and TNF-α (c, d) levels were elevated by circ_0000950 overexpression plasmids but decreased by circ_0000950 shRNA plasmids in PC12 cellular AD model. Also, IL-1β (e, h), IL-6 (f, h) and TNF-α (g, h) expressions were promoted by circ_0000950 overexpression plasmids but suppressed by circ_0000950 shRNA plasmids in cellular AD model of cerebral cortex neurons. Comparison of inflammatory cytokines levels was determined by t test. ***P< 0.001, **P< 0.01, *P< 0.05. P value < 0.05 was considered significant.
Interaction between circ_0000950 and miR-103 in cellular AD models
In order to validate the targeting effect of circ_0000950 on miR-103, compensation experiments were performed in PC12 cellular AD model and cellular AD model of cerebral cortex neurons. Circ_0000950 expression was reduced in Circ(-) group compared with NC(-) group but similar between NC(-) group and MiR(-) groups as well as between Circ(-) group and Circ(-)/MiR(-) group (Figures 6(a), 11(a)). MiR-103 expression was promoted in Circ(-) group compared with NC(-) group, but it was decreased in MiR(-) group compared with NC(-) group as well as in Circ(-)/MiR(-) group compared with Circ(-) group (Figures 6(b), 11(b)). These indicated that circ_0000950 adversely targeted miR-103, whereas miR-103 did not affect circ_0000950 expression in AD.
Figure 6.
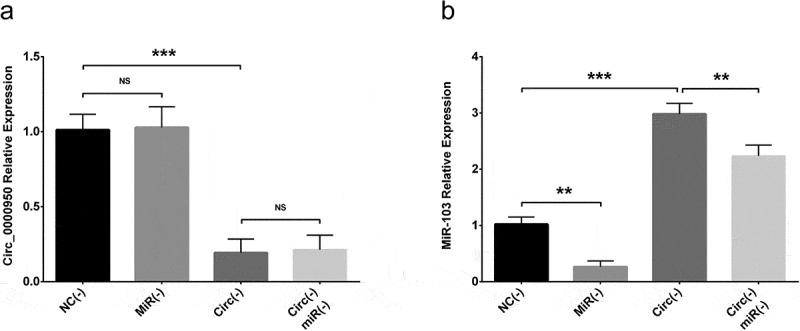
The targeting effect of circ_0000950 on miR-103 in PC12 cellular AD model. Circ_0000950 expression was not influenced by miR-103 shRNA plasmids, and its inhibition by circ_0000950 shRNA plasmids was not affected by circ_0000950&miR-103 shRNA plasmids (a). MiR-103 expression was reduced by miR-103 shRNA plasmids, and its elevation by circ_0000950 overexpression plasmids was decreased by circ_0000950&miR-103 shRNA plasmids (b). Comparison of circ_0000950 and miR-103 expressions was determined by t test. ***P< 0.001, **P< 0.01, NS, non-significant. P value < 0.05 was considered significant.
Figure 11.
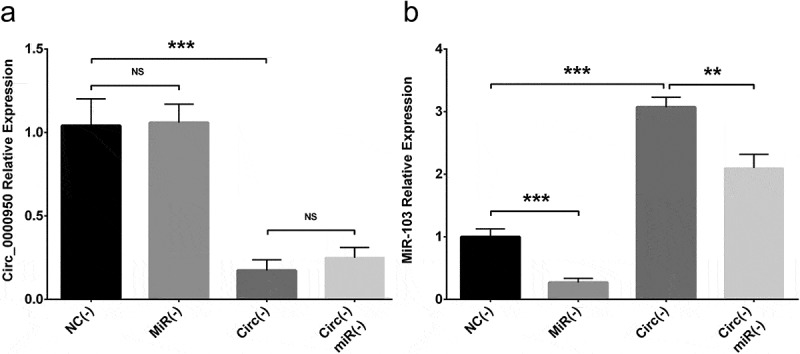
The targeting effect of circ_0000950 on miR-103 in cellular AD model of cerebral cortex neurons. Circ_0000950 expression was not influenced by miR-103 shRNA plasmids, and its inhibition by circ_0000950 shRNA plasmids was not affected by circ_0000950&miR-103 shRNA plasmids (a). MiR-103 expression was reduced by miR-103 shRNA plasmids, and its elevation by circ_0000950 overexpression plasmids was decreased by circ_0000950&miR-103 shRNA plasmids (b). Comparison of circ_0000950 and miR-103 expressions was determined by t test. ***P< 0.001, **P< 0.01, NS, non-significant. P value < 0.05 was considered significant.
Circ_0000950 promoted cell apoptosis by targeting miR-103 in cellular AD models
Compensation experiments were also carried out to determine whether circ_0000950 regulated AD cells function by targeting miR-103. Cell apoptosis rate was increased in MiR(-) group compared with NC(-) group as well as in Circ(-)/MiR(-) group compared with Circ(-) group (Figures 7(a,b), 12(a,b)). In addition, C-caspase 3 expression was elevated in MiR(-) group compared with NC(-) group as well as in Circ(-)/MiR(-) group compared with Circ(-) group, whereas Bcl-2 expression was reduced in MiR(-) group compared with NC(-) group as well as in Circ(-)/MiR(-) group compared with Circ(-) group (Figures 7(c), 12(c)). These evidence suggested that circ_0000950 promoted cell apoptosis by targeting miR-103 in AD.
Figure 7.
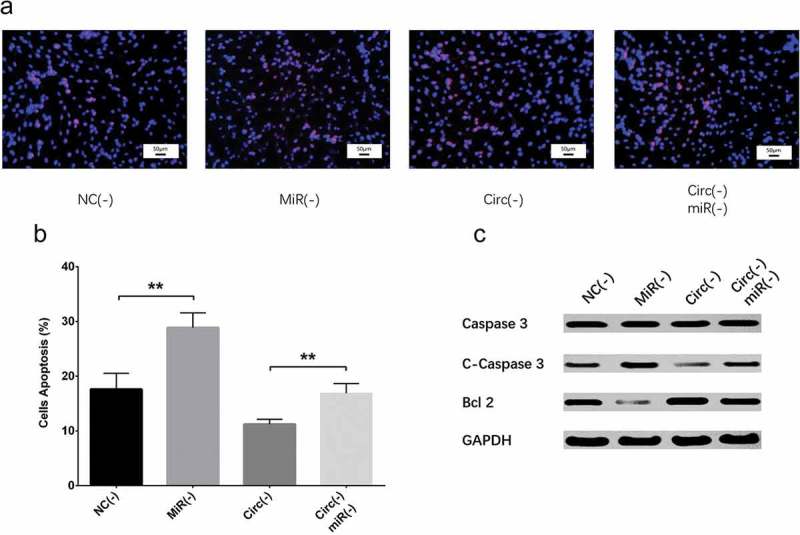
Effect of circ_0000950 and miR-103 on cell apoptosis in compensation experiment (PC12 cellular AD model). Cell apoptosis rate was facilitated by miR-103 shRNA plasmids, and reduction in cell apoptosis rate by circ_0000950 shRNA plasmids was attenuated by circ_0000950&miR-103 shRNA plasmids (a, b). C-caspase 3 expression was increased by miR-103 shRNA plasmids, and the reduction in C-caspase 3 expression by circ_0000950 shRNA plasmids was attenuated by circ_0000950&miR-103 shRNA plasmids. Bcl-2 expression was suppressed by miR-103 shRNA plasmids, and the increased in Bcl-2 expression by circ_0000950 shRNA plasmids was attenuated by circ_0000950&miR-103 shRNA plasmids (C). Comparison of cell apoptosis rate was determined by t test. **P< 0.01. P value < 0.05 was considered significant.
Figure 12.
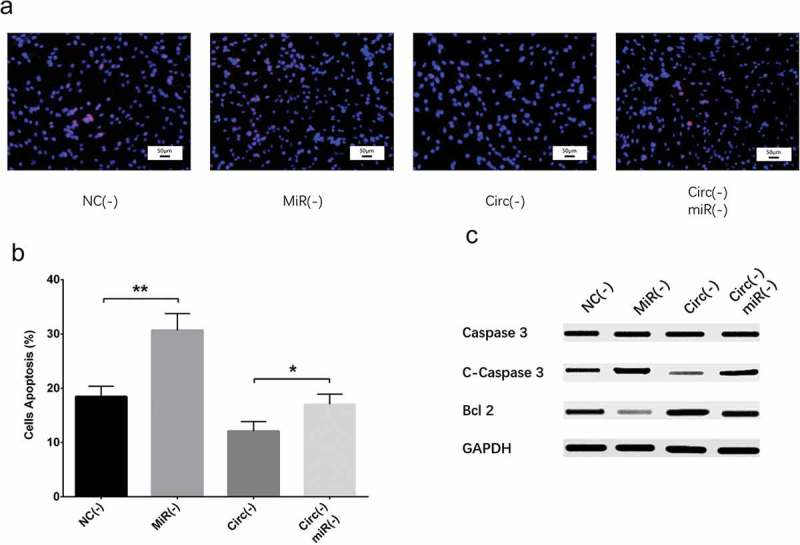
Effect of circ_0000950 and miR-103 on cell apoptosis in compensation experiment (cellular AD model of cerebral cortex neurons). Cell apoptosis rate was facilitated by miR-103 shRNA plasmids, and reduction in cell apoptosis rate by circ_0000950 shRNA plasmids was attenuated by circ_0000950&miR-103 shRNA plasmids (a, b). C-caspase 3 expression was increased by miR-103 shRNA plasmids, and the reduction in C-caspase 3 expression by circ_0000950 shRNA plasmids was attenuated by circ_0000950&miR-103 shRNA plasmids. Bcl-2 expression was suppressed by miR-103 shRNA plasmids, and the increased in Bcl-2 expression by circ_0000950 shRNA plasmids was attenuated by circ_0000950&miR-103 shRNA plasmids (C). Comparison of cell apoptosis rate was determined by t test. *P< 0.05, **P< 0.01. P value < 0.05 was considered significant.
Circ_0000950 suppressed neurite outgrowth by targeting miR-103 in cellular AD models
Total neurite outgrowth was reduced in MiR(-) group compared with NC(-) group as well as in Circ(-)/MiR(-) group compared with Circ(-) group (Figures 8(a,b), 13(a,b)), implying that circ_0000950 suppressed neurite outgrowth by targeting miR-103 in AD.
Figure 8.
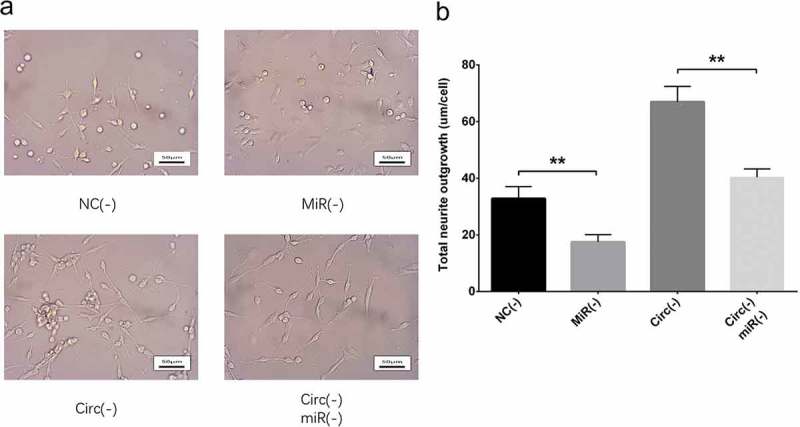
Effect of circ_0000950 and miR-103 on neurite growth in compensation experiment (PC12 cellular AD model). Total neurite outgrowth was inhibited by miR-103 shRNA plasmids, and the increase in neurite growth by circ_0000950 shRNA plasmids was attenuated by circ_0000950&miR-103 shRNA plasmids (a, b). Comparison of total neurite growth was determined by t test. **P< 0.01. P value < 0.05 was considered significant.
Figure 13.
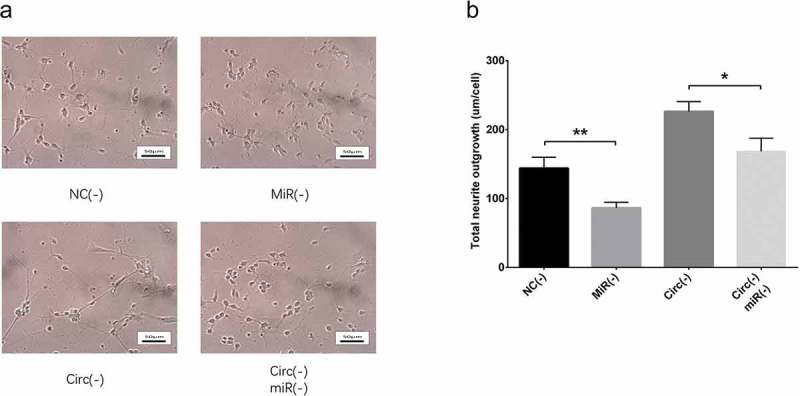
Effect of circ_0000950 and miR-103 on neurite growth in compensation experiment (cellular AD model of cerebral cortex neurons). Total neurite outgrowth was inhibited by miR-103 shRNA plasmids, and the increase in neurite growth by circ_0000950 shRNA plasmids was attenuated by circ_0000950&miR-103 shRNA plasmids (a, b). Comparison of total neurite growth was determined by t test. *P< 0.05, **P< 0.01. P value < 0.05 was considered significant.
Circ_0000950 enhanced inflammatory cytokines levels by targeting miR-103 in cellular AD models
IL-1β (Figures 9(a,d), 15(a,d)), IL-6 (Figures 9(b,d), 15(b,d)) and TNF-α (Figures 9(c,d), 15(c,d)) expressions were increased in MiR(-) group compared with NC(-) group as well as in Circ(-)/MiR(-) group compared with Circ(-) group, which indicated that circ_0000950 enhanced inflammation by targeting miR-103 in AD.
Figure 9.
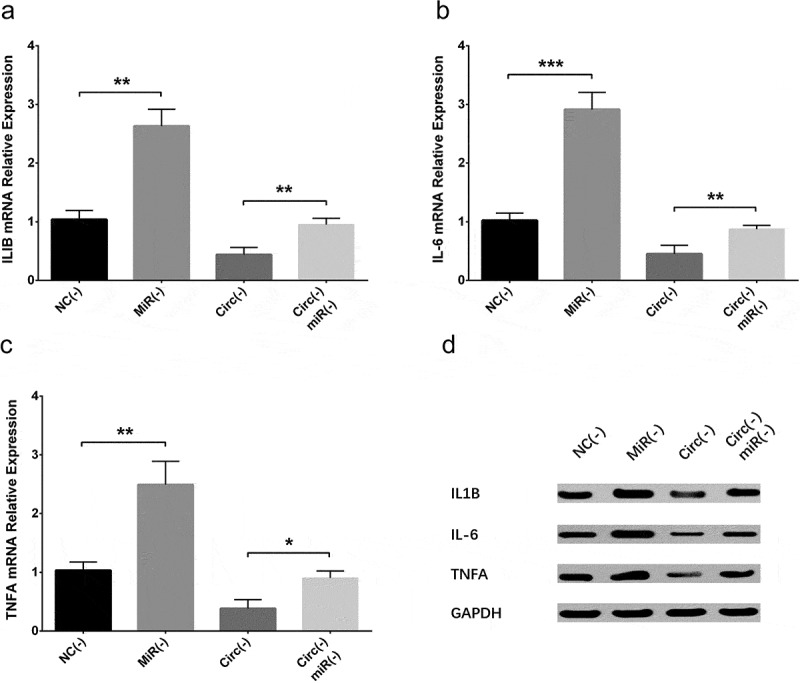
Effect of circ_0000950 and miR-103 on inflammatory cytokine levels in compensation experiment (PC12 cellular AD model). IL-1β (a, d), IL-6 (b, d) and TNF-α (c, d) expressions were raised by miR-103 shRNA plasmids, and the reduction in IL-1β, IL-6 and TNF-α expressions by circ_0000950 shRNA plasmids was attenuated by circ_0000950&miR-103 shRNA plasmids. Comparison of inflammatory cytokines levels was determined by t test. ***P< 0.001, **P< 0.01, *P< 0.05. P value < 0.05 was considered significant.
Figure 15.
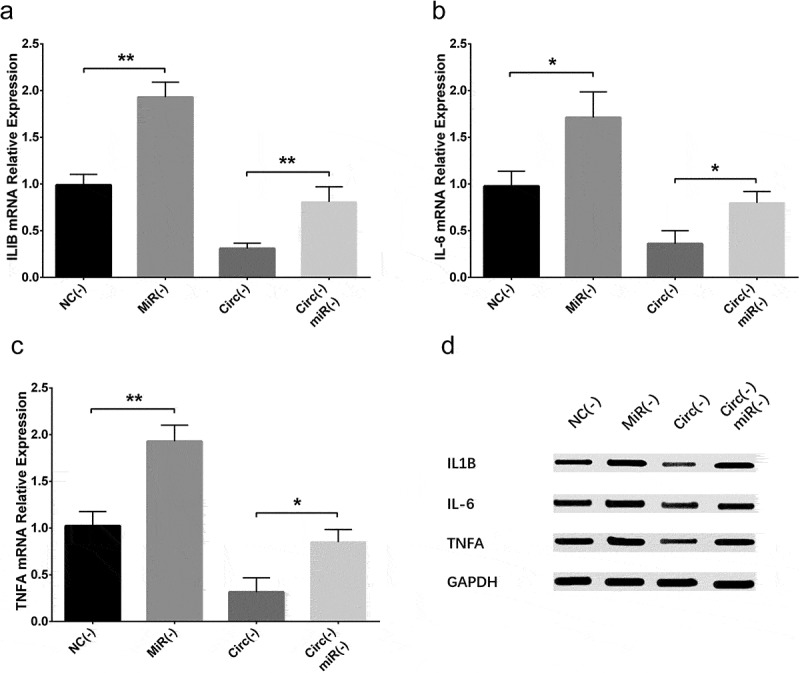
Effect of circ_0000950 and miR-103 on inflammatory cytokine levels in compensation experiment (cellular AD model of cerebral cortex neurons). IL-1β (a, d), IL-6 (b, d) and TNF-α (c, d) expressions were raised by miR-103 shRNA plasmids, and the reduction in IL-1β, IL-6 and TNF-α expressions by circ_0000950 shRNA plasmids was attenuated by circ_0000950&miR-103 shRNA plasmids. Comparison of inflammatory cytokines levels was determined by t test. **P< 0.01, *P< 0.05. P value < 0.05 was considered significant.
Circ_0000950 targeted miR-103 by direct binding
In order to validate the direct binding site between circ_0000950 and miR-103, luciferase reporter assay was performed. Sequence of miR-103 as well as its possible binding site with circ_0000950 were displayed in Figure 10(a). For wild type circ_0000950, the relative luciferase activity was lower in the miR-103 group compared with the miR-NC group, whereas for mutant circ_0000950, the relative luciferase activity was similar between miR-103 group and miR-NC group (Figure 10(b)). These elucidated that circ_0000950 targeted miR-103 by direct binding.
Figure 10.
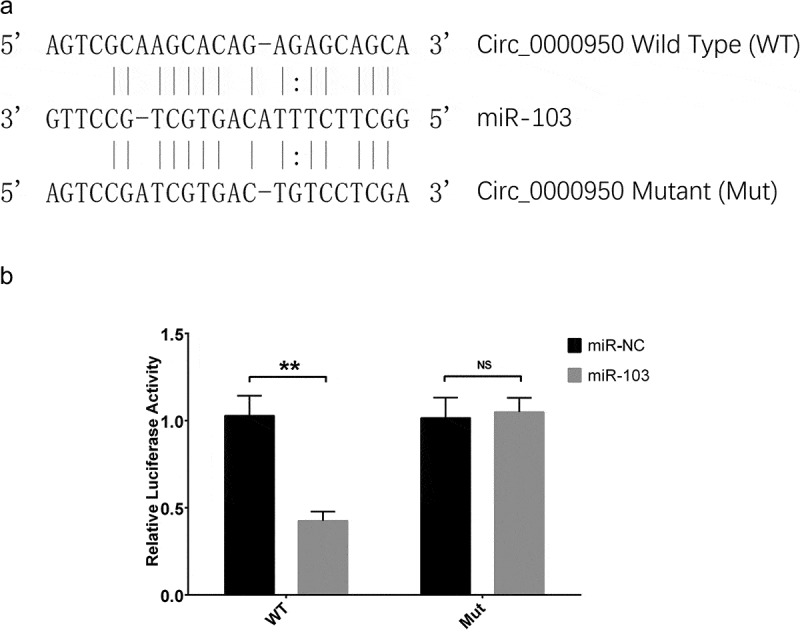
Luciferase reporter assay of circ_0000950. MiR-103 sequence as well as binding sites between circ_0000950 and miR-103 were shown (a). The relative luciferase activity for wild type circ_0000950 was reduced in the miR-103 group compared to a miR-NC group, while the relative luciferase activity for mutant circ_0000950 was unchanged in the miR-103 group compared to miR-NC group (b). Comparison of luciferase activity was determined by t test. **P< 0.01, NS, non-significant. P value < 0.05 was considered significant.
Discussion
In this study, we discovered that (1) circ_0000950 reduced miR-103 but increased PTGS2 expression, and it promoted neuron apoptosis, suppressed neurite outgrowth and enhanced inflammatory cytokines levels in AD. (2) Further compensation experiments and luciferase reporter assay disclosed that circ_0000950 facilitated neuron apoptosis, inhibited neurite outgrowth and increased inflammatory cytokines levels by directly sponging miR-103 in AD.
AD is featured as one of the most prevalent neurodegenerative disorders that leads to change in brain structure and impairment in brain function [14]. Neuroanatomical studies illuminate that the destruction of axons initially arises from certain brain areas at the onset of AD then spread widely at the end stage, and neuronal cell death as well as loss of synapses are prominent components of AD etiology [15,16]. Also, neuroinflammation in response to injury or amyloid plaque also aggravates dysregulation of glia cells, contributing to the deterioration of brain function, and several inflammatory cytokines including IL-1β, IL-6, IL-4, TNF-α and interferon γ (IFNγ) are associated with increased risk of AD [17,18]. These evidence highlights the pathogenesis of AD, which encourages research and development of potential treatment approaches for AD. Although the mechanisms of AD are under extensive investigation, its underlying genetic mechanisms are still not well understood.
CircRNAs, one of the breakthrough findings in RNA researches, have provided newer and deeper insight into various human diseases including neurodegenerative diseases [19]. CircRNAs are illustrated to share binding sites with specific miRNAs, which enables them to act as sponges for miRNAs and inhibited the regulatory function of these miRNAs [8]. For instance, ciRS-7 serves as an endogenous sponge for miR-107 and quenches the normal function of miR-107, thereby alters levels of proteins such as ubiquitin protein ligase A (UBE2A), which is essential for clearance of amyloid peptides in AD [18]. Also, ciRS-7 participates in the pathogenesis of Parkinson’s disease by sponging miR-7 and regulating ɑ-synuclein [5]. In addition, knockdown of circ-zip-2 reduces aggregation of ɑ-synuclein protein by sponging miR-60 in C. elegans model of Parkinson’s disease [20]. These studies indicate that circRNAs play critical roles in neurodegenerative diseases and may have therapeutic potential for these diseases.
A number of cellular models have been established to investigate the mechanism of AD, and proteomic/transcriptomic analysis of cells from these models greatly facilitate with identifying novel biomarkers or therapeutic targets [21]. In our previous study, two cellular AD models were constructed by Aβ1-42 insult and validated by cell viability assessment, and we discovered that miR-103 inhibited neuron apoptosis, promoted neurite outgrowth by adversely regulating PTGS2 gene in AD, which was involved in inflammation [13]. Considering that circRNAs served as sponges to adversely regulate miRNAs function, we were interested in whether miR-103, which was investigated in our previous study, would be regulated by certain circRNAs. Then, we used miRanda Database (http://www.microrna.org/microrna/home.do) and Tissue-Specific CircRNA Database (http://gb.whu.edu.cn/TSCD/) to search for potential circRNAs (including circ_0000950) that possibly regulated miR-103 by sharing binding sites, and our preliminary experiments exhibited that circ_0000950 repressed miR-103 and promoted PTGS2 expression (a proinflammatory gene reversely regulated by miR-103) in PC12 cellular AD model and cellular AD model of rat cerebral cortex neurons. Therefore, we hypothesized that circ_0000950 might affect neuron apoptosis, neurite outgrowth and inflammation via targeting miR-103 in AD and conducted this present study. Interestingly, we indeed observed that in PC12 cellular AD model and cellular AD model of rat cerebral cortex neurons, circ_0000950 promoted neuron apoptosis, reduced total neurite growth as well as increased IL-1β, IL-6 and TNF-α levels. This implied that circ_0000950 involved in the pathogenesis of AD by regulating neuron apoptosis, differentiation and inflammation, which partially validated out hypothesis. Although there was currently no study about the function of circ_0000950 in AD that supported our result, our data could be explained by that overexpression of circ_0000950 might inhibit expression of neurogenerative or anti-inflammatory miRNAs (possibly miR-103) and promote neurotoxic or pro-inflammatory genes that were targeted by those miRNAs, thereby altered cell activities as well as promoted inflammation in AD.
MiRNAs, as described in our previous study, participate in the epigenetic regulation of protein synthesis in various diseases via binging to 3ʹ UTR of mRNAs and suppressing the translation of mRNAs [22]. As for AD, a group of miRNAs (such as miR-107, miR-181c, miR-29b, miRNA-9, miRNA-125b, miRNA-146a, and miRNA-155) are pathogenically dysregulated and suppress mRNAs that are key regulatory genes for neuroinflammation, synaptogenesis and so on [23,24]. They also play important roles in biological pathways including DNA methylation and histone modification in neuronal as well as glial cells to alter cell functions in AD [23]. Among these miRNAs, miR-103 is reduced in an animal model of AD as well as in cerebrospinal fluid of AD patients, and its downregulation facilitates with pathogenesis of AD via increasing cyclin-dependent kinase 5 activity [25,26]. Considering that the activity of miRNAs is commonly known to be regulated by circRNAs who have high affinity to miRNAs via base pairing, it would be meaningful to explore the circRNA/miRNA regulatory pathway in AD and better understand the genetic mechanism of AD pathogenesis [27].
In order to further testify that circ_0000950 promoted AD progression via targeting miR-103 in AD, compensation experiments were carried out in both PC 12 cellular AD model and cellular AD model of cerebral cortex neurons. We observed that circ_0000950 shRNA plasmids transfection promoted miR-103 expression, but this effect was attenuated by circ_0000950&miR-103 shRNA plasmids transfection, indicating that circ_0000950 targeted miR-103 by adverse regulation in both cellular AD models. Subsequently, the effect of circ_0000950 shRNA plasmids transfection on inhibiting neuron apoptosis, promoting neurite outgrowth as well as reducing IL-1β, IL-6 and TNF-α levels were attenuated by circ_0000950&miR-103 shRNA plasmids transfection as well. Additionally, luciferase reporter assay displayed direct interaction between circ_0000950 and miR-103. Gathering the data above, circ_0000950 promoted neuron apoptosis, suppressed neurite outgrowth and elevated IL-1β, IL-6 and TNF-α levels through directly sponging miR-103 in cellular AD models, which validated our hypothesis. This might due to that circ_0000950 directly bound miR-103 (validated by luciferase reporter assay), hence upregulated the subsequent genes that were neurotoxic or pro-inflammatory, such as PTGS2 (a proinflammatory gene that was shown to be adversely regulated by miR-103 in our previous study), thereby increased neuron apoptosis, reduced neurite outgrowth and promoted inflammation in AD. Our present data might provide additional information for understanding about genetic mechanisms of AD pathogenesis and serve as references for identification and development of novel therapeutic targets for AD.
In conclusion, circ_0000950 promotes neuron cell apoptosis, suppresses neurite outgrowth and elevates inflammatory cytokines levels through directly sponging miR-103 in AD.
Funding Statement
This study was supported by the Fundamental Research Funds for the Provincial Universities.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].De Strooper B, Karran E.. The cellular phase of Alzheimer’s disease. Cell. 2016. February 11;164(4):603–615. PubMed PMID: 26871627. [DOI] [PubMed] [Google Scholar]
- [2].Chan KY, Wang W, Wu JJ, et al. Epidemiology of Alzheimer’s disease and other forms of dementia in China, 1990-2010: a systematic review and analysis. Lancet. 2013. June 8;381(9882):2016–2023. PubMed PMID: 23746902. [DOI] [PubMed] [Google Scholar]
- [3].Scheltens P, Blennow K, Breteler MM, et al. Alzheimer’s disease. Lancet. 2016. July 30;388(10043):505–517. PubMed PMID: 26921134. [DOI] [PubMed] [Google Scholar]
- [4].Huang Y, Mucke L.. Alzheimer mechanisms and therapeutic strategies. Cell. 2012. March 16;148(6):1204–1222. PubMed PMID: 22424230; PubMed Central PMCID: PMCPMC3319071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hsiao KY, Sun HS, Tsai SJ. Circular RNA - New member of noncoding RNA with novel functions. Exp Biol Med (Maywood). 2017. June;242(11):1136–1141. PubMed PMID: 28485684; PubMed Central PMCID: PMCPMC5478007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014. October 2;56(1):55–66. PubMed PMID: 25242144. [DOI] [PubMed] [Google Scholar]
- [7].Belousova EA, Filipenko ML, Kushlinskii NE. Circular RNA: new regulatory molecules. Bull Exp Biol Med. 2018. April;164(6):803–815. PubMed PMID: 29658072. [DOI] [PubMed] [Google Scholar]
- [8].Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Natur. 2013. March 21;495(7441):384–388. PubMed PMID: 23446346. [DOI] [PubMed] [Google Scholar]
- [9].Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017. February;24(2):357–370. PubMed PMID: 27886165; PubMed Central PMCID: PMCPMC5299715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017. May 4;14(5):514–521. PubMed PMID: 26649774; PubMed Central PMCID: PMCPMC5449088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Boeckel JN, Jae N, Heumuller AW, et al. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ Res. 2015. October 23;117(10):884–890. PubMed PMID: 26377962. [DOI] [PubMed] [Google Scholar]
- [12].Holdt LM, Stahringer A, Sass K, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016. August 19;7:12429 PubMed PMID: 27539542; PubMed Central PMCID: PMCPMC4992165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yang H, Wang H, Shu Y, et al. miR-103 promotes neurite outgrowth and suppresses cells apoptosis by targeting prostaglandin-endoperoxide synthase 2 in cellular models of Alzheimer’s disease. Front Cell Neurosci. 2018;12:91 PubMed PMID: 29674956; PubMed Central PMCID: PMCPMC5895658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ballard C, Gauthier S, Corbett A, et al. Alzheimer’s disease. Lancet. 2011. March 19;377(9770):1019–1031. PubMed PMID: 21371747. [DOI] [PubMed] [Google Scholar]
- [15].Overk C, Masliah E. Perspective on the calcium dyshomeostasis hypothesis in the pathogenesis of selective neuronal degeneration in animal models of Alzheimer’s disease. Alzheimers Dement. 2017. February;13(2):183–185. PubMed PMID: 28130011; PubMed Central PMCID: PMCPMC5935121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Absalon S, Kochanek DM, Raghavan V, et al. MiR-26b, upregulated in Alzheimer’s disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. J Neurosci. 2013. September 11;33(37):14645–14659. PubMed PMID: 24027266; PubMed Central PMCID: PMCPMC3810537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Denver P, McClean PL. Distinguishing normal brain aging from the development of Alzheimer’s disease: inflammation, insulin signaling and cognition. Neural Regen Res. 2018. October;13(10):1719–1730. PubMed PMID: 30136683; PubMed Central PMCID: PMCPMC6128051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lukiw WJ. Circular RNA (circRNA) in Alzheimer’s disease (AD). Front Genet. 2013;4:307 PubMed PMID: 24427167; PubMed Central PMCID: PMCPMC3875874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kumar L, Shamsuzzama, Haque R, et al. Circular RNAs: the emerging class of non-coding RNAs and their potential role in human neurodegenerative diseases. Mol Neurobiol. 2017. November;54(9):7224–7234. PubMed PMID: 27796758. [DOI] [PubMed] [Google Scholar]
- [20].Kumar L, Shamsuzzama, Jadiya P, et al. Functional characterization of novel circular RNA molecule, circzip-2 and its synthesizing gene zip-2 in C. elegans model of Parkinson’s disease. Mol Neurobiol. 2018. August;55(8):6914–6926. PubMed PMID: 29363043. [DOI] [PubMed] [Google Scholar]
- [21].Gotz J, Ittner LM. Animal models of Alzheimer’s disease and frontotemporal dementia. Nat Rev Neurosci. 2008. July;9(7):532–544. PubMed PMID: 18568014. [DOI] [PubMed] [Google Scholar]
- [22].Zhao J, Yue D, Zhou Y, et al. The role of MicroRNAs in abeta deposition and tau phosphorylation in Alzheimer’s disease. Front Neurol. 2017;8:342 PubMed PMID: 28769871; PubMed Central PMCID: PMCPMC5513952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fransquet PD, Ryan J. Micro RNA as a potential blood-based epigenetic biomarker for Alzheimer’s disease. Clin Biochem. 2018. August;58:5–14. PubMed PMID: 29885309. [DOI] [PubMed] [Google Scholar]
- [24].Lukiw WJ, Andreeva TV, Grigorenko AP, et al. Studying micro RNA function and dysfunction in Alzheimer’s disease. Front Genet. 2012;3:327 PubMed PMID: 23390425; PubMed Central PMCID: PMCPMC3565163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Huynh RA, Mohan C. Alzheimer’s disease: biomarkers in the genome, blood, and cerebrospinal fluid. Front Neurol. 2017;8:102 PubMed PMID: 28373857; PubMed Central PMCID: PMCPMC5357660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Moncini S, Lunghi M, Valmadre A, et al. The miR-15/107 Family of microRNA Genes Regulates CDK5R1/p35 with implications for Alzheimer’s disease pathogenesis. Mol Neurobiol. 2017. August;54(6):4329–4342. PubMed PMID: 27343180. [DOI] [PubMed] [Google Scholar]
- [27].Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014. July 29;15(7):409 PubMed PMID: 25070500; PubMed Central PMCID: PMCPMC4165365. [DOI] [PMC free article] [PubMed] [Google Scholar]


