Abstract
Infection with hepatitis C virus (HCV) results in chronic and progressive liver disease. Persistency rates add up to 85%. Despite recognition of the virus by the human host in peripheral blood and in the liver, immune response appears to be ineffective in clearing infection. The ability to spontaneously eradicate the virus as well as the outcome of infection upon therapy with human recombinant interferon-α (IFN-α) was found to correlate most closely with genetic variations within the region encoding the IFN-λ genes, as revealed by genome-wide association studies on main ethnic populations in 2009. This review summarizes the induction of type I and type III IFN genes and their effectors, the IFN-stimulated genes. It focusses on the in vivo situation in chronic HCV infection in man both in the peripheral blood compartment and in the liver. It also addresses the impact of genetic polymorphisms in the region of type III IFN genes on their activation. Finally, it discusses how antiviral drugs (i.e. IFN-α, ribavirin and the direct-acting antivirals) may complementarily control the activation of endogenous IFNs and succeed in combatting infections.
Key Words: Host defense, Pattern recognition receptors, Virology
Introduction
Interferon (IFN) was discovered in 1957 as an entity which interferes with viral replication [1]. It can be induced either by noninfectious inactivated virus particles or by the infectious live virus. The activating microbial compounds are highly conserved unique molecular signatures - so-called pathogen-associated molecular patterns (PAMPs). In the case of viral infections they may comprise single-stranded (ss) RNA, double-stranded (ds) RNA or an uncapped exposed 5′-ppp (5′-triphosphate) moiety on an RNA molecule, tagging it as non-self [reviewed in [2, 3]].
Sensors of PAMPs generally comprise a set of germline-encoded pathogen recognition receptors which divide into the membranous toll-like receptors (TLRs) and into the cytoplasmic members of the RIG-1-like receptor (RLR) family [reviewed in [4]]. While the TLRs sense pathogens extracellularly (e.g. microbial membrane components via TLR1, TLR2, TLR4, TLR5, TLR6) or after having entered the endosomal compartment in the course of phagocytosis (e.g. nucleic acids via TLR3, TLR7, TLR8, TLR9), the RLR family members detect replicating viruses intracellularly within the cytosol. After recognizing their respective PAMPs, pathogen recognition receptors activate signaling pathways, resulting in the induction of proinflammatory cytokines. The nucleic acid-sensing TLRs as well as the RIG-like receptors (RLR) additionally promote the activation of IFNs.
Hepatitis C virus (HCV) is a hepatotropic virus which was discovered in 1989 as the etiological agent of nonA-nonB or posttransfusion hepatitis in man [5]. It is an enveloped ssRNA virus with a positive sense genome of about 9.6 kb in length. It replicates in hepatocytes via minus-strand RNA intermediates after having entered them in a multistep process that involves the low-density lipoprotein receptor as well as four essential cell surface molecules [reviewed in [6]]. HCV is noncytopathic and it is excreted from the liver by way of blood. Thus, the infected human host senses circulating intact viral particles within the peripheral blood compartment as well as replicating viruses within hepatocytes.
However, most of the estimated 160 million people worldwide infected by HCV do not clear the virus but develop a chronic, life-long infection in the liver [7]. Persistent infection leads to varying degrees of liver disease and might end in liver failure or hepatocellular carcinoma. The mechanisms underlying the high persistency rates are still not fully understood. The importance of innate immunity in the control of HCV infection is underscored by the fact that HCV has evolved ways to inactivate innate immune signaling adaptor proteins and by genome-wide association studies, showing that the host's genetic background in the region of the type III IFN genes determines the outcome of infection.
Interferons
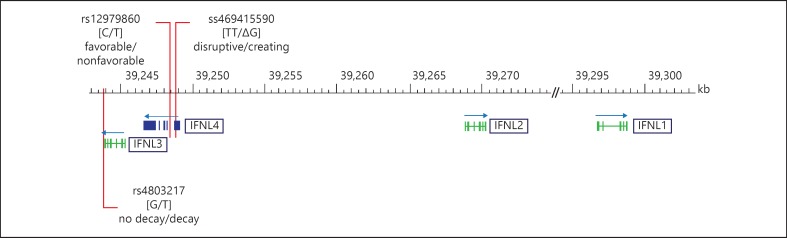
Since its discovery a bundle of IFN subtypes have been identified. Today they are divided into three families - type I, type II and type III IFNs. In humans, type I IFNs comprise 13 closely related IFN-α subtypes, a single IFN-β and the more distantly related IFN-κ, IFN-ω and IFN-ε which can be secreted by many cell types in response to infection. The expression of the only type II IFN, IFN-γ, is far less ubiquitous and restricted to T cells and NK and NKT cells. By mediating the clearance of viral infections, IFN-γ is a functional homolog in terms of exhibiting broader and nonredundant biological activities than the other types of IFNs, i.e. by activating macrophages and by promoting adaptive immune responses. Aiming at identifying further IFN-like cytokines, more recently type III IFNs were discovered by two independent genomic screening approaches [8, 9]. They are phylogenetically close to the type I IFNs. They are clustered on chromosome 19 and have been named IFN-λ1, IFN-λ2 and IFN-λ3 or IL-29, IL-28A and IL-28B, respectively. Like type I IFNs, they are induced by viral infections and confer antiviral activity by activating various antiviral effectors, the so-called IFN-stimulated genes (ISGs) [10, 11]. Type I and type III IFNs do, however, utilize distinct receptor complexes for signaling. While IFN-α receptor expression is ubiquitous, the expression of the IFN-λ receptor is much more limited and restricted to epithelial cells and hepatocytes [10, 11]. Only recently, a novel type III IFN was discovered - termed IFN-λ4. IFN-λ4 is encoded by the IFNL4 gene, which is located upstream of IFNL3 [12]. For their genomic organization see figure 1.
Fig. 1.
Genomic organization of the IFNL genes. In man, multiexon type III IFNs are organized in a single gene cluster spanning approximately 55 kb on chromosome 19, with IFNL1 and IFNL2 in opposite orientation to IFNL3 and IFNL4. Genome-wide association studies have identified rs12979860 to be associated with viral clearance. It is located upstream of IL28B/IFNL3 and was thus termed an IL28B SNP. However, with the discovery of IFNL4, rs12979860 was found to be located within IFNL4 intron 1 [12]. Protein expression of IFNL4 is controlled by a dinucleotide polymorphism within exon 1, ss469415590. While the TT allele introduces a frameshift and disrupts the open reading frame, only the variant allele ΔG creates IFNL4. The minor allele of a further SNP within exon 2, rs117648444, causes an amino acid substitution (P70S), leading to a functionally impaired IFNL4 protein. Transcription of IFNL3 has been shown to be related to rs12979860 [58] and ss469415590 [57], as well as to an SNP within the 3′-UTR which was shown to mediate miRNA-driven mRNA decay [60].
Induction of IFNs and ISGs in Chronic HCV Infection: The Peripheral Blood Mononuclear Cell Compartment
In peripheral blood mononuclear cells (PBMCs) from patients with HCV infection the induction of an antiviral state was shown at first indirectly by quantifying the human myxovirus resistance protein 1 (MxA) in cell lysates which, owing to high induction by IFN, is frequently used as a marker for the IFN-induced antiviral response [13]. Its expression is highly regulated by type I and type III IFNs, and it is not inducible directly by viruses or other stimuli. It was found to be significantly augmented in PBMCs from chronic hepatitis C patients compared to basal levels from healthy volunteers [14]. Moreover, the patients' basal levels of MxA rose in the course of a 6-month IFN-α therapy, returning to baseline during posttreatment follow-up.
Also on the mRNA level, PBMCs from chronic hepatitis C patients were found to have higher MxA gene activation than PBMCs from healthy controls [15, 16], further rising during an IFN-α therapy [15]. A gene expression profiling of PBMCs revealed an elevated expression of ISGs in chronic hepatitis patients [17], and a recent transcriptome analysis of peripheral blood monocytes resumed a significant ISG mRNA induction during IFN-α treatment [18].
Direct measurements of IFN gene transcripts were carried out in RNA preparations from PBMCs derived from chronic hepatitis C patients and from noninfected individuals [16]. IFN-α gene activation was measured both by assays specific for single species as well as by an assay covering all subtypes (IFN-αn). IFN-λ gene activation was quantified by applying an assay specific for IFN-λ1 and one covering the highly conserved sequences of the paralogs IFN-λ2 and IFN-λ3 (IFN-λ2/3). Analysis revealed significant gene activation of IFN-α, IFN-β, IFN-λ1, and IFN-λ2/3 in chronic HCV infection [16].
Taken together, it is well accepted that HCV is sensed within the peripheral blood compartment, provoking the activation of both IFN genes and ISGs. Recognition most probably takes place by plasmacytoid dendritic cell precursors (pDCs), which are the professional IFN producers in peripheral blood [19]. This specialized cell type is characterized by the expression of TLR7 sensing ssRNA. Recognition might also take place by subsets of myeloid DCs, e.g. BDCA3+ (blood dendritic cell antigen 3) DCs, which have been shown to be responsive to the TLR3 agonist polyIC, a synthetic analog of dsRNA, or to HCV in vitro in terms of IFN-λ1, IFN-λ2 and IFN-λ3 production [20, 21].
Induction of IFNs and ISGs in Chronic HCV Infection: The Liver Compartment
The situation in the human liver, however, presents differently. While MxA and many other ISGs were found to be increased in chronic hepatitis C compared to, for example, nonviral liver diseases and while the human liver responds to IFN-α therapy by an upregulation of ISGs, particularly in responders [22, 23], the IFN genes could not be demonstrated to be activated beyond the constitutive levels of control tissue [16]. Evidence for this also came from microarray analyses comparing HCV-associated liver cirrhosis tissue to nondiseased liver tissue [24] or from a comparison of HCV-associated hepatocellular carcinoma specimens to a noninfected nontumor liver [25], where the activation of ISGs but not of IFNs was easily detectable. Moreover, an analysis of a subtracted library (transcriptome of an HCV-infected liver tissue minus that of noninfected tissues with similar histopathological changes) also revealed an enhanced expression of ISGs but not that of type I or type III IFN genes themselves [26]. This is distinct from the situation in chimpanzees, who do show induction of IL-29 and IL-28 early in HCV infection [27, 28].
The finding of a lack of hepatic IFN inducibility by HCV in humans is in line with the discovery that the HCV protease NS3/4A targets and cleaves two crucial adaptor proteins in viral RNA sensing [29, 30]. Inactivation of the adaptors leads to a block in the activation of IRF-3, a transcription factor that is essential for IFN induction. One adaptor, MAVS (mitochondrial antiviral signaling protein), is an essential component of the RIG-I- and Mda5-mediated cytosolic sensory pathway, while the other one, TRIF (toll-IL-1 receptor domain-containing adaptor inducing IFN-β), mediates recognition of HCV dsRNA intermediates via TLR3 [31, 32]. As a measure of MAVS integrity, its intracellular location could be demonstrated with mitochondrial membrane proteins in specimens from patients with nonviral liver diseases. In patients with chronic hepatitis C, in contrast, its distribution failed to associate with mitochondria, indicating proteolysis and cellular redistribution [33]. Moreover, cleavage of MAVS in liver biopsy samples from patients with chronic hepatitis C was also demonstrated by means of Western blotting, discriminating between full-length and cleaved forms [34]. This latter study also showed a correlation between MAVS cleavage and a reduced ISG activation.
Regarding the noninfected, nonparenchymal liver cell compartment, HCV might be sensed by TLR7 expressing pDCs. This sensing might take place following the uptake of HCV RNA from the extracellular environment either by phagocytosis or by autophagy from dying infected cells, or by a pathway that was uncovered only recently, namely, the exosomal transfer of viral nucleic acids from infected cells to pDCs [35, 36, 37]. Common to all three pathways is that they do not require active viral replication, and pDCs are not productively infected with HCV. Notably, these pathways have been investigated in vitro. In chronic hepatitis C, the number and function of pDCs have been found to be impaired [reviewed in [38, 39]]. As type I IFN has been shown to negatively control pDC numbers in various in vitro infection models, it was thus suggested that type I IFN might negatively regulate pDC numbers during HCV infection. A recent immunohistochemistry analysis in fact failed to detect any pDCs both in pretreatment and end-of-treatment human liver biopsies [40].
Kupffer cells, the resident macrophages in the liver, represent the largest population of innate immune cells in the liver. Physiologically, they capture gut-derived pathogens or their products that enter the liver via the portal vein by phagocytosis. They might sense HCV viral particles from the blood on the one hand, and they might be exposed to viral nucleic acids by phagocytosis of infected hepatocytes on the other. In chronic hepatitis C, they do respond to infection in many various respects [reviewed in [41]]. Little is known, however, about their ability to contribute to an IFN response. Low levels of IFN-β protein were demonstrated recently in Kupffer cells in liver sections, specifically from nonresponding chronic hepatitis C patients (n = 2) [42], though whether Kupffer cells contribute to IFN production needs further investigation.
Despite the overall lack of hepatic IFN gene induction in chronic HCV infection beyond its constitutive expression, a clear so-called ‘activation of the endogenous IFN system’ is consistently observed, meaning a broad mRNA induction of ISGs [43]. The pattern of ISGs detected in patients with chronic hepatitis C corresponds to a type I or type III IFN receptor signaling signature [28, 43]. Seemingly paradoxical, the transcript baseline activity of the IFN effectors is inversely linked to the outcome of infection - a robust preactivation of ISGs associates with a poor outcome of infection, both with regard to spontaneous resolution and to an IFN-α-based antiviral treatment regimen [22, 23, 44, 45].
The following two puzzling findings: (1) a pronounced ISG activation in spite of only constitutive IFN gene expression in chronic HCV infection, and (2) the activation of ISGs being unfavorable with regard to the outcome of infection, might be linked to a recently identified new member of the IFN-λ family, namely, IFN-λ4/IFNL4.
The Novel IFNL4
IFNL4 was uncovered through an RNA sequencing approach on a 150-kb region spanning the area of the IFN-λ genes only recently. Prokunina-Olsson et al. [12] found a transient activation of a novel transcribed region located in-between IFNL3 and IFNL2 in primary human hepatocytes that had been stimulated with polyIC for different time periods to mimic an RNA virus infection. The transcript from this region harbors a dinucleotide polymorphism ss469415590 (refSNP No. rs368234815), the major allele TT of which causes a frameshift and disrupts the IFNL4 ORF, while only the minor allele ΔG enables IFN-λ4 protein expression. The major allele TT thus results in a loss of function, meaning that IFN-λ4 is distinguished from other IFN subtypes in that only a subpopulation of human beings is predisposed to express a functional protein [46] (fig. 1). In fact, the IFNL4 gene, which is evolutionarily conserved in mammals, is recognized as turning into a polymorphic pseudogene in humans [47]. Pseudogenization is inferred to be targeted by a positive selection of various strengths rising in dimension from Africa to Europe and the New World and reaching near fixation in East Asia.
This dinucleotide polymorphism is in linkage disequilibrium (LD) with rs12979860, which is located about 3 kb upstream of IL28B/IFNL3 and which was most convincingly identified in genome-wide association studies to be associated with the outcome of an HCV infection in patients of European and African ancestry [48, 49]. LD between ss469415590 and rs12979860 was found to be perfect in Asians and Japanese, good in Europeans and moderate in Africans [12]. The dinucleotide polymorphism is less closely correlated to rs8099917 [12], the second SNP that was found to be associated with the outcome of HCV infection in cohorts of European [50, 51] and Asian [52] ancestry, located about 8 kb upstream of IL28B/IFNL3. As both SNPs differ in terms of frequency geographically, they also predict clearance differently in different populations. Intriguingly, the disrupting allele TT associates with the favorable rs12979860 allele C, meaning that obviously the absence of IFNL4 correlates with low ISG baseline activity and a favorable outcome of infection [12, 53].
The identification of this novel IFN prompted us to investigate intrahepatic IFNL4 transcripts in human liver biopsy specimens [54]. By applying a 5′-nuclease assay designed to quantify IFNL4 transcripts (i.e. JN806234 and JN806227) on a broad panel of liver tissue samples (from patients with nonviral liver diseases of various etiology, from patients with chronic hepatitis B or hepatitis C and from patients with healthy livers), IFNL4 mRNA was exclusively detected in specimens from patients with chronic HCV infection. The activation of the IFNL4 gene, therefore, was found to be related to chronic HCV infection, yielding in vivo relevance for the finding by Prokunina-Olsson et al. [12] that IFNL4 is inducible in primary human hepatocytes. With this, it constitutes the most prominently (if not the sole) activated IFN subtype in the liver of chronic hepatitis C patients. Hence, its expression sufficiently could explain the activation of hepatic ISGs driven by HCV infection. Accordingly, mRNA expression of ISGs (MxA and p44) was found to be positively related to IFNL4 transcript expression [54]. In PBMCs, in contrast, we have not been able to detect any IFNL4 transcripts, either in samples from hepatitis C patients or in those from healthy individuals (own unpublished observation). The reason for this pronounced hepatic inducibility of the IFNL4 subtype in chronic HCV infection while the other subtypes may be subject to viral protease-driven suppression is unknown. Recently, Odendall et al. [55] showed a differential RLR/MAVS-mediated activation of type I and type III IFNs by distinct intracellular organelles, i.e. mitochondria and peroxisomes. The authors identified MAVS that is associated with peroxisomes as a major mediator of an IRF-1-driven type III IFN expression in various cell types and by various viruses. Whether this pathway might also be of relevance in chronic HCV infection warrants further investigation.
Nonetheless, it remains enigmatic that the activation of the only IFN subtype in the liver of chronic hepatitis C patients relates to virus persistence. This might point to a specific antagonistic mode of action of IFNL4, e.g. by continuous stimulation of negative regulators of the immune response. The concept that the ss469415590 ΔG allele governing IFNL4 expression constitutes the primary variant for impaired viral clearance is supported by the recent finding that a missense variation encoding for an altered IFNL4 protein (P70S) with an attenuated antiviral activity (fig. 1) associates with an improved prognosis for HCV-infected patients [56]. Alternatively, the genetic predisposition to encode IFNL4 might tag further functional variations.
Further Proposed Roles of IFNL SNPs
IFNL4 ss469415590
The same genetic polymorphism, ss469415590, which has been shown to determine the ability to encode for IFNL4 in primary human hepatocytes, intriguingly, has been shown to relate to IFNL3 transcript expression in PBMCs, the promoter region in which it is likewise located (fig. 1). Bibert et al. [57] demonstrated this SNP to create a methylation motif in a CpG island and to correlate with IFNL3 mRNA expression in polyIC-stimulated PBMCs from healthy blood donors and from chronic hepatitis C patients. Of note, the authors applied an assay discriminating between the highly homologous IFNL2 and IFNL3 subtypes, thus specifically assessing IFNL3 mRNA expression. Intriguingly, the IFNL4 disrupting allele TT associates with high IFNL3 mRNA expression in PBMCs (table 1).
Table 1.
Major and minor allele phenotypes of IFNL gene polymorphisms
| SNP | Alleles | Major allele phenotype | Minor allele phenotype | Ref. No. |
|---|---|---|---|---|
| IFNL4 ss469415590 | TT/AG | IFNL4 disruptive | IFNL4 creating | 12 |
| IFNL4 rs117648444 | C/T | Wild-type IFNL4 protein | Impaired IFNL4 protein (P70S) | 56 |
| IFNL4 ss469415590 | TT/AG | ↑ IFNL3 mRNA in PBMCs | ↓ IFNL3 mRNA in PBMCs | 57 |
| IL28B/IFNL4 rs12979860 | C/T | ↑ IFNL3 mRNA in PBMCs | ↓ IFNL3 mRNA in PBMCs | 58 |
| IFNL3 rs4803217 | G/T | No hepatic IFNL3 mRNA decay | Hepatic IFNL3 mRNA decay | 60 |
| IL28B/IFNL4 rs12979860 | C/T | ↓ Hepatic IFNLR1 | ↑ Hepatic IFNLR1 | 59 |
IL28B/IFNL3 rs12979860
Similarly, PBMCs from healthy donors homozygous for the IL28B/IFNL3 rs12979860 unfavorable minor T allele were found to express significantly less IFNL3 upon a viral trigger (cytomegalovirus) compared to major C allele homozygotes. Here too, a specific design ensured differentiation between IFNL3 and IFNL2 transcripts. The cohort analyzed by Egli et al. [58] was not genotyped for ss469415590. However, as in the Canadian population IL28B/IFNL3 rs12979860 and IFNL4 ss469415590 are in close LD, data might still reflect an association between the IFNL4 ss469415590 TT allele and an increased IFNL3 expression. These results are thus consistent with the aforementioned findings by Bibert et al. [57].
Duong et al. [59] recently proposed hepatic expression of the IFN-λ receptor subunit 1 (IFN-λR1) as a missing link between IFNL genotypes and the associated phenotype of ISG activation and treatment outcome. According to their findings on human liver biopsy specimens, samples from carriers of the unfavorable T allele presented higher hepatic IFN-λR1 expression and higher ISG activation. Their data point to a higher sensitivity towards type III IFN species in patients carrying the risk alleles. Furthermore, they provide complementary insights into how IFNL genotypes might translate into ISG activation.
IFNL3 rs4803217
McFarland et al. [60], moreover, uncovered a mechanism by which two microRNAs (MIR208B and MIR499A) which are induced by HCV mediate the decay of IFNL3 transcripts. This decay was shown to depend on a polymorphism in the 3′-UTR (untranslated region) of the IFNL3 mRNA, rs4803217, which is also in close LD with IL28B/IFNL3 rs12979860. The polymorphism was shown to influence the binding of HCV-induced microRNAs during experimental in vitro infection, e.g. in hepatoma cells. The favorable allele was shown to escape mRNA decay, meaning again that an IFNL4 disruptive genotype should associate with a higher relative abundance of IFNL3 transcripts, this time in the liver compartment.
Taken together, the phenotype of a favorable outcome of HCV infection associates with the inability to encode IFNL4 and a low hepatic expression of its receptor IFN-λR1 on the one hand, and with a capability of PBMCs to respond to stimuli with an IFNL3 induction (in vitro) on the other (table 1).
Lessons from Antivirals
Given that the HCV protease NS3/4A inactivates adaptor proteins and thereby blocks IFN induction, antiviral drugs might not only inhibit viral replication but also restore innate antiviral defense by diminishing the load of viral proteins intracellularly, thereby retrieving adaptor efficiency and allowing reactivation of the endogenous IFN system. In addition, they might act by enhancing sensitivity towards endogenous or exogenous IFNs.
This concept is supposed for all compounds that affect HCV replication, including ribavirin (RBV), direct-acting antivirals (DAAs) inhibiting the NS5B RNA-dependent RNA polymerase or the nonstructural protein 5A (NS5A) or those which directly target NS3/4A protease. It has been proven true for RBV, which exerts only a modest antiviral activity in chronic hepatitis C [61]. In a cohort of patients who received a 4-week treatment with RBV, or not, before combining it with IFN-α, pretreatment with RBV was found to lower the hepatic baseline of ISGs and to augment ISG induction by a following single dose of PEG-IFN-α. RBV was thus suggested to reset IFN responsiveness in the HCV-infected liver.
A first microarray gene expression profiling has been conducted on paired liver biopsies from patients before and at the end of an IFN-free antiviral treatment regimen with sofosbuvir/RBV [40]. In the mostly responding patients, hepatic ISG expression was found to be downregulated when liver specimens taken at the end of treatment were compared to those taken before. The downregulation of hepatic ISGs included USP18 (ubiquitin carboxy-terminal hydrolase 18), which is a negative regulator of IFN-α signaling [62]. USP18 has been reported to be elevated in the liver of chronic hepatitis C patients, particularly in those who do not respond to IFN therapy [63]. At the same time, the favorable outcome in the sofosbuvir/RBV-treated patients was found to be accompanied by a relative decrease of basal levels of some of the type III IFNs and a relative increase of type I IFN-α2 subtype. Interestingly, the IFNLR1 was downregulated too. These data thus provide evidence for a restoration of hepatic type I activation by the end of the treatment. Moreover, they fit into the concept that the activation of IFNL4 and its receptor, which correlates with the stimulation of ISGs [54, 59], is disadvantageous for viral clearance.
The assumption that antivirals restore defense mechanisms is in line with empirical clinical knowledge. RBV-IFN-α combination therapy is known to be superior to IFN-α monotherapy [64], primarily by suppressing a relapse of HCV upon cessation of therapy. Moreover, triple therapy with novel DAAs outperforms former IFN-based regimens [65] and renders IFN nonresponders responsive to IFN treatment.
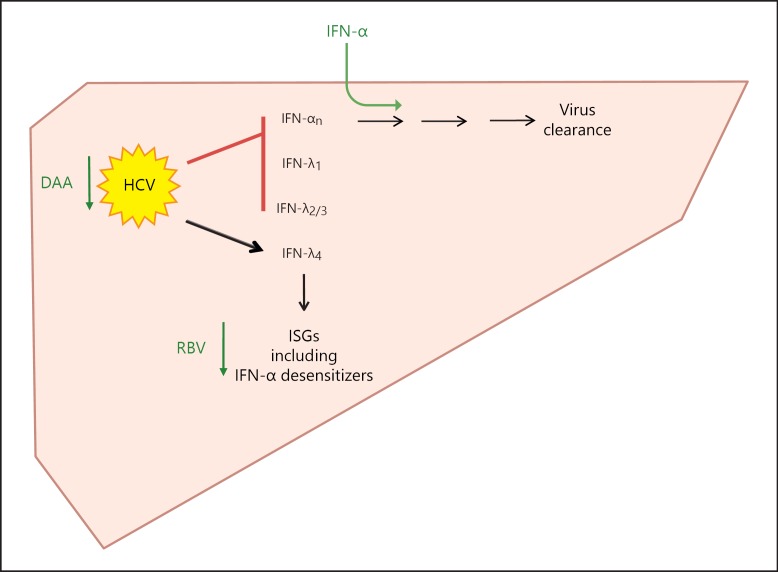
Based on current knowledge, the relation between HCV infection and the activation of the IFN system in the human liver might be represented as depicted in figure 2. By its protease NS3/4A, HCV actively suppresses TLR- and RLR-driven type I and type III IFN induction, apparently with the exception of IFNL4 [54]. Hepatic viral load relates to the number of hepatic IFNL4 transcripts [54]. Not affected by the inhibiting action of USP18 on IFN-α signaling, hepatic ISG activation in chronic HCV infection might be run by type III IFNs (IFNL4), thereby decreasing sensitivity towards endogenous and exogenous IFN-α by activating negative regulators of IFN-α signaling. This is based on close positive correlations between the hepatic stimulation of ISGs and IFNL4 [54] and its receptor [59]. ISGs include inhibitors that interfere with type I IFN signaling [40].
Fig. 2.
Proposed relation between HCV infection and the activation of the IFN system in the human liver. In the human liver, HCV actively suppresses TLR- and RLR-driven type I and type III IFN induction by its protease NS3/4A, apparently with the exception of IFNL4. Hepatic viral load relates to the number of hepatic IFNL4 transcripts. Based on a close positive correlation, hepatic stimulation of ISGs is supposed to be driven by IFNL4 and its receptor. ISGs include inhibitors that interfere with type I IFN signaling. According to this model, the efficacy of antiviral drugs relies on their interference with the hepatic endogenous IFN system (see text for details).
According to this model, the efficacy of IFN-α depends, on the one hand, on simply providing it to the liver as the endogenous source is blocked and, on the other, on its balance with IFN-α desensitizing ISGs. The efficacy of RBV in combination with IFN-α might rely on its ability to lower hepatic ISGs [61]. The efficacy of DAAs relies on their strong inhibitory activity on viral replication, which might decrease the activation of type III IFNs (IFNL4) and ISGs [40]. Moreover, a decreasing load of HCV protease restores virus sensing by retrieving adaptor integrity and reactivates endogenous type I IFNs [40].
Conclusion
HCV is sensed quite well in peripheral blood in terms of type I and type III IFN gene induction and ISG activation. The constitutive IFN gene activation in the livers of chronic hepatitis C patients might be due to the ability of the virus to abate its recognition by cleaving pivotal signaling adaptor molecules in infected hepatocytes. The induction of IFNL4 apparently is an exception as it appears not to be as vigorously affected by this suppression as the other IFN subtypes. The underlying mechanism, however, needs to be elucidated.
The ability to encode IFNL4 is genetically determined and limited to IFNL4 ss469415590 ΔG allele homozygous and IFNL4 ss469415590 ΔG/TT heterozygous individuals. IFNL4 might drive hepatic ISG expression, which might antagonize IFN-α efficacy. Its role in the peripheral blood compartment needs to be investigated. Further polymorphisms which are in close LD with IFNL4 ss469415590 might affect IFNL3 activation in PBMCs and might also contribute to virus eradication.
Complementary effects of the following: (1) strong direct antiviral activity, (2) a reconstitution of the full sensing of infection, and (3) enhancing sensitivity towards endogenous or exogenous IFN, could explain the superior efficacy of (multiple) drug combinations comprising DAAs, RBV and IFNs.
References
- 1.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- 2.Baum A, Garcia-Sastre A. Induction of type I interferon by RNA viruses: cellular receptors and their substrates. Amino Acids. 2010;38:1283–1299. doi: 10.1007/s00726-009-0374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horner SM. Activation and evasion of antiviral innate immunity by hepatitis C virus. J Mol Biol. 2014;426:1198–1209. doi: 10.1016/j.jmb.2013.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011;3:920–940. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 6.Lindenbach BD, Rice CM. The ins and outs of hepatitis C virus entry and assembly. Nat Rev Microbiol. 2013;11:688–700. doi: 10.1038/nrmicro3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect. 2011;17:107–115. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 8.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, Ostrander C, Dong D, Shin J, Presnell S, Fox B, Haldeman B, Cooper E, Taft D, Gilbert T, Grant FJ, Tackett M, Krivan W, McKnight G, Clegg C, Foster D, Klucher KM. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 9.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-λs mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 10.Kotenko SV. IFN-λs. Curr Opin Immunol. 2011;23:583–590. doi: 10.1016/j.coi.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermant P, Michiels T. Interferon-λ in the context of viral infections: production, response and therapeutic implications. J Innate Immun. 2014;6:563–574. doi: 10.1159/000360084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I, Chen S, Brand N, Tarway M, Liu L, Sheikh F, Astemborski J, Bonkovsky HL, Edlin BR, Howell CD, Morgan TR, Thomas DL, Rehermann B, Donnelly RP, O'Brien TR. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haller O, Kochs G. Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. J Interferon Cytokine Res. 2011;31:79–87. doi: 10.1089/jir.2010.0076. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez M, Quiroga JA, Martin J, Herrero M, Pardo M, Horisberger MA, Carreno V. In vivo and in vitro induction of MxA protein in peripheral blood mononuclear cells from patients chronically infected with hepatitis C virus. J Infect Dis. 1999;180:262–267. doi: 10.1086/314859. [DOI] [PubMed] [Google Scholar]
- 15.Meier V, Mihm S, Ramadori G. MxA gene expression in peripheral blood mononuclear cells from patients infected chronically with hepatitis C virus treated with interferon-α. J Med Virol. 2000;62:318–326. [PubMed] [Google Scholar]
- 16.Mihm S, Frese M, Meier V, Wietzke-Braun P, Scharf JG, Bartenschlager R, Ramadori G. Interferon type I gene expression in chronic hepatitis C. Lab Invest. 2004;84:1148–1159. doi: 10.1038/labinvest.3700135. [DOI] [PubMed] [Google Scholar]
- 17.Bolen CR, Robek MD, Brodsky L, Schulz V, Lim JK, Taylor MW, Kleinstein SH. The blood transcriptional signature of chronic hepatitis C virus is consistent with an ongoing interferon-mediated antiviral response. J Interferon Cytokine Res. 2013;33:15–23. doi: 10.1089/jir.2012.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou J, Groothuismink ZM, Koning L, Roomer R, van IWF, Kreefft K, Liu BS, Janssen HL, de Knegt RJ, Boonstra A. Analysis of the transcriptome and immune function of monocytes during IFNα-based therapy in chronic HCV revealed induction of TLR7 responsiveness. Antiviral Res. 2014;109:116–124. doi: 10.1016/j.antiviral.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 19.Yin Z, Dai J, Deng J, Sheikh F, Natalia M, Shih T, Lewis-Antes A, Amrute SB, Garrigues U, Doyle S, Donnelly RP, Kotenko SV, Fitzgerald-Bocarsly P. Type III IFNs are produced by and stimulate human plasmacytoid dendritic cells. J Immunol. 2012;189:2735–2745. doi: 10.4049/jimmunol.1102038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauterbach H, Bathke B, Gilles S, Traidl-Hoffmann C, Luber CA, Fejer G, Freudenberg MA, Davey GM, Vremec D, Kallies A, Wu L, Shortman K, Chaplin P, Suter M, O'Keeffe M, Hochrein H. Mouse CD8α+ DCs and human BDCA3+ DCs are major producers of IFN-λ in response to polyIC. J Exp Med. 2010;207:2703–2717. doi: 10.1084/jem.20092720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshio S, Kanto T, Kuroda S, Matsubara T, Higashitani K, Kakita N, Ishida H, Hiramatsu N, Nagano H, Sugiyama M, Murata K, Fukuhara T, Matsuura Y, Hayashi N, Mizokami M, Takehara T. Human blood dendritic cell antigen 3 (BDCA3)+ dendritic cells are a potent producer of interferon-λ in response to hepatitis C virus. Hepatology. 2013;57:1705–1715. doi: 10.1002/hep.26182. [DOI] [PubMed] [Google Scholar]
- 22.Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, Filipowicz W, Heim MH. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci U S A. 2008;105:7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feld JJ, Nanda S, Huang Y, Chen W, Cam M, Pusek SN, Schweigler LM, Theodore D, Zacks SL, Liang TJ, Fried MW. Hepatic gene expression during treatment with peginterferon and ribavirin: identifying molecular pathways for treatment response. Hepatology. 2007;46:1548–1563. doi: 10.1002/hep.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shackel NA, McGuinness PH, Abbott CA, Gorrell MD, McCaughan GW. Insights into the pathobiology of hepatitis C virus-associated cirrhosis: analysis of intrahepatic differential gene expression. Am J Pathol. 2002;160:641–654. doi: 10.1016/S0002-9440(10)64884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iizuka N, Oka M, Yamada-Okabe H, Mori N, Tamesa T, Okada T, Takemoto N, Tangoku A, Hamada K, Nakayama H, Miyamoto T, Uchimura S, Hamamoto Y. Comparison of gene expression profiles between hepatitis B virus- and hepatitis C virus-infected hepatocellular carcinoma by oligonucleotide microarray data on the basis of a supervised learning method. Cancer Res. 2002;62:3939–3944. [PubMed] [Google Scholar]
- 26.Patzwahl R, Meier V, Ramadori G, Mihm S. Enhanced expression of interferon-regulated genes in the liver of patients with chronic hepatitis C virus infection: detection by suppression-subtractive hybridization. J Virol. 2001;75:1332–1338. doi: 10.1128/JVI.75.3.1332-1338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park H, Serti E, Eke O, Muchmore B, Prokunina-Olsson L, Capone S, Folgori A, Rehermann B. IL-29 is the dominant type III interferon produced by hepatocytes during acute hepatitis C virus infection. Hepatology. 2012;56:2060–2070. doi: 10.1002/hep.25897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas E, Gonzalez VD, Li Q, Modi AA, Chen W, Noureddin M, Rotman Y, Liang TJ. HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology. 2012;142:978–988. doi: 10.1053/j.gastro.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foy E, Li K, Wang C, Sumpter R, Jr, Ikeda M, Lemon SM, Gale M., Jr Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- 30.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Jr, Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang N, Liang Y, Devaraj S, Wang J, Lemon SM, Li K. Toll-like receptor 3 mediates establishment of an antiviral state against hepatitis C virus in hepatoma cells. J Virol. 2009;83:9824–9834. doi: 10.1128/JVI.01125-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li K, Li NL, Wei D, Pfeffer SR, Fan M, Pfeffer LM. Activation of chemokine and inflammatory cytokine response in hepatitis C virus-infected hepatocytes depends on toll-like receptor 3 sensing of hepatitis C virus double-stranded RNA intermediates. Hepatology. 2012;55:666–675. doi: 10.1002/hep.24763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, Fujita T, Saito T, Lee WM, Hagedorn CH, Lau DT, Weinman SA, Lemon SM, Gale M., Jr Viral and therapeutic control of IFN-β promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellecave P, Sarasin-Filipowicz M, Donze O, Kennel A, Gouttenoire J, Meylan E, Terracciano L, Tschopp J, Sarrazin C, Berg T, Moradpour D, Heim MH. Cleavage of mitochondrial antiviral signaling protein in the liver of patients with chronic hepatitis C correlates with a reduced activation of the endogenous interferon system. Hepatology. 2010;51:1127–1136. doi: 10.1002/hep.23426. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi K, Asabe S, Wieland S, Garaigorta U, Gastaminza P, Isogawa M, Chisari FV. Plasmacytoid dendritic cells sense hepatitis C virus-infected cells, produce interferon, and inhibit infection. Proc Natl Acad Sci U S A. 2010;107:7431–7436. doi: 10.1073/pnas.1002301107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenhill C. Hepatitis: new route of HCV transmission. Nat Rev Gastroenterol Hepatol. 2013;10:504. doi: 10.1038/nrgastro.2013.148. [DOI] [PubMed] [Google Scholar]
- 37.Ramakrishnaiah V, Thumann C, Fofana I, Habersetzer F, Pan Q, de Ruiter PE, Willemsen R, Demmers JA, Stalin Raj V, Jenster G, Kwekkeboom J, Tilanus HW, Haagmans BL, Baumert TF, van der Laan LJ. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci U S A. 2013;110:13109–13113. doi: 10.1073/pnas.1221899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albert ML, Decalf J, Pol S. Plasmacytoid dendritic cells move down on the list of suspects: in search of the immune pathogenesis of chronic hepatitis C. J Hepatol. 2008;49:1069–1078. doi: 10.1016/j.jhep.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 39.Pelletier S, Bedard N, Said E, Ancuta P, Bruneau J, Shoukry NH. Sustained hyperresponsiveness of dendritic cells is associated with spontaneous resolution of acute hepatitis C. J Virol. 2013;87:6769–6781. doi: 10.1128/JVI.02445-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meissner EG, Wu D, Osinusi A, Bon D, Virtaneva K, Sturdevant D, Porcella S, Wang H, Herrmann E, McHutchison J, Suffredini AF, Polis M, Hewitt S, Prokunina-Olsson L, Masur H, Fauci AS, Kottilil S. Endogenous intrahepatic IFNs and association with IFN-free HCV treatment outcome. J Clin Invest. 2014;124:3352–3363. doi: 10.1172/JCI75938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boltjes A, Movita D, Boonstra A, Woltman AM. The role of Kupffer cells in hepatitis B and hepatitis C virus infections. J Hepatol. 2014;61:660–671. doi: 10.1016/j.jhep.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 42.Lau DT, Negash A, Chen J, Crochet N, Sinha M, Zhang Y, Guedj J, Holder S, Saito T, Lemon SM, Luxon BA, Perelson AS, Gale M., Jr Innate immune tolerance and the role of Kupffer cells in differential responses to interferon therapy among patients with HCV genotype 1 infection. Gastroenterology. 2013;144:402–413. doi: 10.1053/j.gastro.2012.10.044. e412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Metz P, Reuter A, Bender S, Bartenschlager R. Interferon-stimulated genes and their role in controlling hepatitis C virus. J Hepatol. 2013;59:1331–1341. doi: 10.1016/j.jhep.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 44.Asselah T, Bieche I, Narguet S, Sabbagh A, Laurendeau I, Ripault MP, Boyer N, Martinot-Peignoux M, Valla D, Vidaud M, Marcellin P. Liver gene expression signature to predict response to pegylated interferon plus ribavirin combination therapy in patients with chronic hepatitis C. Gut. 2008;57:516–524. doi: 10.1136/gut.2007.128611. [DOI] [PubMed] [Google Scholar]
- 45.Chen L, Borozan I, Feld J, Sun J, Tannis LL, Coltescu C, Heathcote J, Edwards AM, McGilvray ID. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128:1437–1444. doi: 10.1053/j.gastro.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 46.Booth D, George J. Loss of function of the new interferon IFN-λ4 may confer protection from hepatitis C. Nat Genet. 2013;45:119–120. doi: 10.1038/ng.2537. [DOI] [PubMed] [Google Scholar]
- 47.Key FM, Peter B, Dennis MY, Huerta-Sanchez E, Tang W, Prokunina-Olsson L, Nielsen R, Andres AM. Selection on a variant associated with improved viral clearance drives local, adaptive pseudogenization of interferon-λ4 (IFNL4) PLoS Genet. 2014;10:e1004681. doi: 10.1371/journal.pgen.1004681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 49.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E, Riordan S, Sheridan D, Smedile A, Fragomeli V, Muller T, Bahlo M, Stewart GJ, Booth DR, George J. IL28B is associated with response to chronic hepatitis C interferon-α and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 51.Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J, Colombo S, Cerny A, Dufour JF, Furrer H, Gunthard HF, Heim M, Hirschel B, Malinverni R, Moradpour D, Mullhaupt B, Witteck A, Beckmann JS, Berg T, Bergmann S, Negro F, Telenti A, Bochud PY, Swiss Hepatitis C Cohort Study; Swiss HIV Cohort Study Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–1345. doi: 10.1053/j.gastro.2009.12.056. 1345.e1-e7. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, Ito Y, Mita E, Tanaka E, Mochida S, Murawaki Y, Honda M, Sakai A, Hiasa Y, Nishiguchi S, Koike A, Sakaida I, Imamura M, Ito K, Yano K, Masaki N, Sugauchi F, Izumi N, Tokunaga K, Mizokami M. Genome-wide association of IL28B with response to pegylated interferon-α and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 53.O'Brien T, Pfeiffer R, Tang W, Kuhs K, Chen S, Howell C, Prokunina-Olsson L. P1306 comparison of IFNL4 ΔG and IFNL3 3′UTR rs4803217 genotypes for association with HCV treatment response. J Hepatol. 2014;60:S530. [Google Scholar]
- 54.Amanzada A, Kopp W, Spengler U, Ramadori G, Mihm S. Interferon-λ4 (IFNL4) transcript expression in human liver tissue samples. PLoS One. 2013;8:e84026. doi: 10.1371/journal.pone.0084026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Odendall C, Dixit E, Stavru F, Bierne H, Franz KM, Durbin AF, Boulant S, Gehrke L, Cossart P, Kagan JC. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat Immunol. 2014;15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terczyńska-Dyla E, Bibert S, Duong FHT, Krol I, Jørgensen S, Collinet E, Kutalik Z, Aubert V, Cerny A, Kaiser L, Malinverni R, Mangia A, Moradpour D, Müllhaupt B, Negro F, Santoro R, Semela D, Semmo N, Heim MH, Bochud P-Y, Hartmann R. 75: a variant of IFNλ4 displaying decreased activity is associated with better hepatitis C virus clearance and reduced ISG expression. Cytokine. 2014;70:45–46. [Google Scholar]
- 57.Bibert S, Roger T, Calandra T, Bochud M, Cerny A, Semmo N, Duong FH, Gerlach T, Malinverni R, Moradpour D, Negro F, Mullhaupt B, Bochud PY, Swiss Hepatitis C Cohort Study IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med. 2013;210:1109–1116. doi: 10.1084/jem.20130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Egli A, Levin A, Santer DM, Joyce M, O'Shea D, Thomas BS, Lisboa LF, Barakat K, Bhat R, Fischer KP, Houghton M, Tyrrell DL, Kumar D, Humar A. Immunomodulatory function of interleukin 28B during primary infection with cytomegalovirus. J Infect Dis. 2014;210:717–727. doi: 10.1093/infdis/jiu144. [DOI] [PubMed] [Google Scholar]
- 59.Duong FH, Trincucci G, Boldanova T, Calabrese D, Campana B, Krol I, Durand SC, Heydmann L, Zeisel MB, Baumert TF, Heim MH. IFN-λ receptor 1 expression is induced in chronic hepatitis C and correlates with the IFN-λ3 genotype and with nonresponsiveness to IFN-α therapies. J Exp Med. 2014;211:857–868. doi: 10.1084/jem.20131557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McFarland AP, Horner SM, Jarret A, Joslyn RC, Bindewald E, Shapiro BA, Delker DA, Hagedorn CH, Carrington M, Gale M, Jr, Savan R. The favorable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus-induced microRNAs. Nat Immunol. 2014;15:72–79. doi: 10.1038/ni.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rotman Y, Noureddin M, Feld JJ, Guedj J, Witthaus M, Han H, Park YJ, Park SH, Heller T, Ghany MG, Doo E, Koh C, Abdalla A, Gara N, Sarkar S, Thomas E, Ahlenstiel G, Edlich B, Titerence R, Hogdal L, Rehermann B, Dahari H, Perelson AS, Hoofnagle JH, Liang TJ. Effect of ribavirin on viral kinetics and liver gene expression in chronic hepatitis C. Gut. 2014;63:161–169. doi: 10.1136/gutjnl-2012-303852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dill MT, Makowska Z, Duong FH, Merkofer F, Filipowicz M, Baumert TF, Tornillo L, Terracciano L, Heim MH. Interferon-γ-stimulated genes, but not USP18, are expressed in livers of patients with acute hepatitis C. Gastroenterology. 2012;143:777–786. doi: 10.1053/j.gastro.2012.05.044. e771-e776. [DOI] [PubMed] [Google Scholar]
- 64.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Jr, Haussinger D, Diago M, Carosi G, Dhumeaux D, Craxi A, Lin A, Hoffman J, Yu J. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 65.Feeney ER, Chung RT. Antiviral treatment of hepatitis C. BMJ. 2014;348:G3308. doi: 10.1136/bmj.g3308. [DOI] [PMC free article] [PubMed] [Google Scholar]