Abstract
Defence against invading DNA occurs in both mammals and bacteria. Recognition of stray DNA can initiate responses to infection, but may also protect against potentially mutagenic integration of transposons or retrotransposons into the genome. Double-stranded DNA detected in the cytosol of mammalian macrophages can elicit inflammatory cytokines and cell death following assembly of the AIM2 inflammasome. Amongst eukaryotes, responses to cytosolic DNA have so far only been detected in mammals, and AIM2 is mammalian restricted. In protecting genome integrity, we reasoned that pathways recognising invading DNA should be fundamental to cellular life, and that cell death would be an appropriate response to an overwhelming foreign DNA burden. We found that Drosophila S2 cells were killed by transfection of DNA from a range of natural sources. Unlike with mammalian cells, responses were not prevented by DNA denaturation. There was an element of sequence specificity, as synthetic single-stranded homopolymers were not toxic, whilst mixed-base synthetic DNA caused significant cell death. Death occurred with rapid loss of membrane integrity, and without the characteristic features of apoptosis. We have defined a novel defence against invading DNA in Drosophila. An active necrotic pathway has not previously been described in insects.
Key Words: Cytosolic DNA, Drosophila, Cell death, Necrosis, Host defence
Introduction
Defence against invading DNA occurs in both mammals and bacteria. Bacteria utilise both restriction-modification systems and CRISPRs (clustered regularly interspaced short palindromic repeats) to identify and defend against foreign DNA [1]. In eukaryotes, recognition of stray DNA can initiate responses to infection, but may also protect against potentially mutagenic integration of transposons or retrotransposons into the genome. Eukaryotic responses to cytosolic DNA have so far only been characterised in human and mouse cells. In mammalian cells, double-stranded DNA (dsDNA) abnormally located in the cytosol is detected by several receptor systems, leading to type I interferon (IFN), inflammatory cytokines and cell death [2, 3, 4]. The receptor for cytosolic DNA eliciting type I IFN production was recently identified as cyclic GMP-AMP synthase (cGAS), which, in response to DNA binding, catalyses formation of a cyclic dinucleotide from GTP and ATP. This acts as a second messenger and is sensed by STING, which then initiates signalling leading to type I IFN transcription.
A second mammalian response to cytosolic DNA is the assembly of the AIM2 inflammasome, a multiprotein complex that promotes activation of caspase-1 and caspase-8. Caspase-1 cleaves the precursors of IL-1β and IL-18 prior to their release from the cell, and also induces a rapid lytic programmed cell death, termed pyroptosis in mammalian macrophages [3, 4, 5, 6, 7]. AIM2-activated caspase-8 induces a parallel program of apoptosis [8]. AIM2 is restricted to mammals and, interestingly, not even all mammals seem to have AIM2; no gene can be found in bats, and the presence of AIM2 pseudogenes in elephant, cow, llama and dog, amongst other species, suggest it has been lost from genomes several times in evolution [9].
Detection of foreign nucleic acid on the basis of unusual structure, sequence or location provides a versatile means of identifying diverse infections, and we would expect this to have been exploited as a strategy by the immune systems of many different species. Cytosolic DNA could be a result not only of infection, but also of high activity of transposons or retrotransposons, defects in DNA repair, genome instability or an impaired DNA degradation pathway. In protecting genome integrity, we reasoned that pathways recognising invading DNA should be fundamental to cellular life. Cell death is an appropriate response to an overwhelming foreign DNA burden, whether it is perceived as an indication of infection or as mutagenic danger. Given that AIM2 is a mammalian protein, we hypothesised that responses to foreign DNA have evolved independently in different lineages. We therefore examined insect cells for cytosolic DNA-induced cell death. Drosophila S2 cells were killed by transfection of DNA from a range of natural sources, but the structures and sequences of DNA that induced death were quite distinct from mammalian AIM2 responses. The mode of death was rapid and lytic, and clearly distinct from apoptosis. We have defined a novel defence against invading DNA in Drosophila. A mode of death resembling programmed necrosis has not previously been described in insects.
Materials and Methods
Cell Culture
S2 cells [10] were obtained from ATCC and propagated at 25°C in Schneider's medium supplemented with 10% fetal calf serum, 50 U/ml penicillin and 50 µg/ml streptomycin (medium components from Life Technologies). Bone marrow-derived macrophages (BMMs) from C57BL/6 mice were obtained by flushing cells from femurs and tibias, and culturing with macrophage growth factor CSF-1 for 7 days, as previously described [11]. Mice were used under approval from the University of Queensland Animal Ethics Committee.
Nucleic Acids
Calf thymus (CT) DNA, salmon sperm DNA and Escherichia coli DNA were purchased from Sigma Aldrich and purified as previously described [12]. Polyinosinic:polycytidylic acid [poly(I:C)] was obtained from InvivoGen. S2 DNA was extracted using Proteinase K (ThermoFisher Scientific) according to the manufacturer's protocol, and purified using phenol-chloroform extraction followed by diethyl ether extraction. Poly(dA), poly(dT) and poly(dC) single-stranded DNA (ssDNA) were synthesized using terminal deoxynucleotidyl transferase (TdT; New England Biolabs). Oligo(dA), oligo(dT) and oligo(dC) primers (20 mers at 0.3 μM) were incubated with TdT at 1 unit/μl and corresponding triphosphodeoxynucleotides (1 mM for dATP and 4 mM for dTTP and dCTP) at 37°C overnight. Purification was done using phenol-chloroform extraction followed by ethanol precipitation. The length of the synthesized ssDNA was assessed by annealing the synthetic homopolymers to complementary 20-mer primers and visualising by standard agarose gel electrophoresis. Poly(dA:dT) heteroduplex was obtained by annealing purified poly(dA) and poly(dT) in a 1:1 ratio at 37°C overnight. Double-stranded status was confirmed with S1 nuclease. Synthetic random ssDNA was obtained using TdT elongation of oligo(dA)20 in the presence of dATP, dCTP, dGTP and with or without dTTP (2:3:3:6 molar ratio, due to the preference of TdT for incorporation of individual bases). All synthesised DNA were purified with phenol-chloroform extraction. Randomly synthesized DNAs were further purified using Amicon Ultra-4 Centrifugal Filter Unit with Ultracel-30 membrane (Merck Millipore). Determination of concentration of random polymers assumed equal representation of all bases. pBlueScript (pBS) DNA was purified using an EndoFree Plasmid Maxi Kit (QIAGEN) from overnight E. coli culture. Methylation of pBS was done using CpG-methyltransferase M. SssI (New England Biolabs), followed by enzyme inactivation for 20 min at 65°C. DNA concentrations were either estimated by A260 or PicoGreen assay (Life Technologies), which specifically measures dsDNA. Cyclic dinucleotides were prepared by Zhao-Xun Liang, Nanyang Technological University, Singapore [13].
Electroporation
Drosophila melanogaster S2 cells, haemocytes and mouse BMM were electroporated with various nucleic acids or cyclic dinucleotides in 400 µl of growth medium at 400 V, 500 µF (S2 cells and haemocytes) or 240 V, 1000 µF (BMM) using a BioRad GenePulser MX. Cells were incubated at room temperature with nucleic acid for 10 min prior to electroporation. 200,000 S2 cells or 100,000 BMMs were then plated in 96-well tissue culture plates.
Chemical Transfection
For chemical transfection, 250,000 S2 cells were plated per well in 100 µl of Schneider's media without FCS or antibiotics, and transfected with nucleic acids or cyclic dinucleotides complexed with Lipofectamine 2000 (Life Technologies). Ratios of Lipofectamine to nucleic acid of 2:1 (v/w) for nucleic acids and 1:1 (v/w) for cyclic dinucleotides were used. Plates containing cells and Lipofectamine complexes were centrifuged at 500 g for 10 min to enhance transfection, and incubated at 25°C for 18 h.
MTT Assay for Viability
Cleavage of tetrazolium dye MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] was used as an indicator of cell viability [14]. MTT is cleaved by a mitochondrial enzyme succinate dehydrogenase and produces an insoluble blue product. MTT was added to a final concentration 1 mg/ml, and cells were incubated for 1 h, or 40 min in the case of BMMs. After incubation, cells were solubilised overnight by using an equal volume of MTT solubilisation solution (10% Triton X-100 and 0.1 N HCl in isopropanol), and absorbance was measured at 570 nm.
Flow Cytometric Analysis of Cell Death
For measurement of annexin V/propidium iodide (PI) staining by flow cytometry, 200,000-300,000 S2 cells were centrifuged at 500 g for 5 min, washed with 1 ml of PBS, and resuspended in 100 μl of 1× binding buffer (10 mM HEPES, 140 mM NaCl and 2.5 mM CaCl2, pH 7.4) with 1 µl of annexin V-Alexa Fluor 488 (Life Technologies) per sample. Samples were incubated on ice for 20 min in the dark. Next, 400 µl of 1× binding buffer with PI (final concentration 1 µg/ml) was added and the samples were analysed by flow cytometry (Accuri C6; BD Bioscience). The threshold levels for annexin V positivity were set based on the annexin V staining of the membrane-permeable PI-positive cells, which have full exposure of phosphatidylserine to annexin V. PI single staining was used to assess the time course of cell death. After electroporation (as described above), 200 μl of cell suspension was diluted with 200 μl of full Schneider's media with PI (final concentration 1 µg/ml). Cell suspension was analysed by flow cytometry every 5 min after electroporation, and during analysis the samples were at room temperature. Pan-caspase inhibitor Z-VAD-FMK (Calbiochem) was used to test the involvement of caspases in DNA-induced cell death after electroporation. Actinomycin D (Boehringer-Mannheim) was used for inducing apoptosis in S2 cells.
SubG0/G1 DNA Analysis
106 cells were centrifuged at 500 g for 5 min, washed with PBS, resuspended in 50 μl of PBS, and fixed with 1 ml of 100% ice-cold ethanol. The samples were stored at 4°C overnight. Cells were centrifuged at 800 g for 5 min, washed with PBS with 10% FCS, centrifuged again, and resuspended in citrate buffer (38 mM trisodium citrate) with 70 μM of PI and RNase A (10 μg/ml). Samples were analysed by flow cytometry (FACS Canto II, BD Biosciences) [15].
Haemocyte Extraction
For haemocyte collection, the fly line CG-GAL4 that expresses GFP in cells of the immune system (haemocyte, lymph gland and fat body) was used. The method was based on the work of Lanot et al. [16]. Each larva was washed to remove food particles in water followed by 1× PBS, and then sterilized in 70% ethanol for 2–3 min. Traces of ethanol were removed by washing in water and 1× PBS. Then the larvae were placed onto a clean surface in a drop of sterile 1× PBS. Thin forceps were used to pull apart the cuticle and the larvae were left in the drop for 3–5 min to collect the haemolymph, which was stored on ice. The average number of larvae used in each experiment was 30–50.
Real Time PCR
Expression of IFN-β and HPRT mRNAs were determined by real time PCR as previously described [3], using mouse IFN-β primers: CCACAGCCCTCTCCATCAAC, TGAAGTCCGCCCTGTAGGTG.
Statistics
To compensate for any cell number differences between experiments, MTT assay data is presented normalised to the control sample. Statistical significance was determined prior to normalisation by paired t test, one-tailed in figure 1, and two-tailed in figure 2. Analysis was done using R statistical software.
Fig. 1.
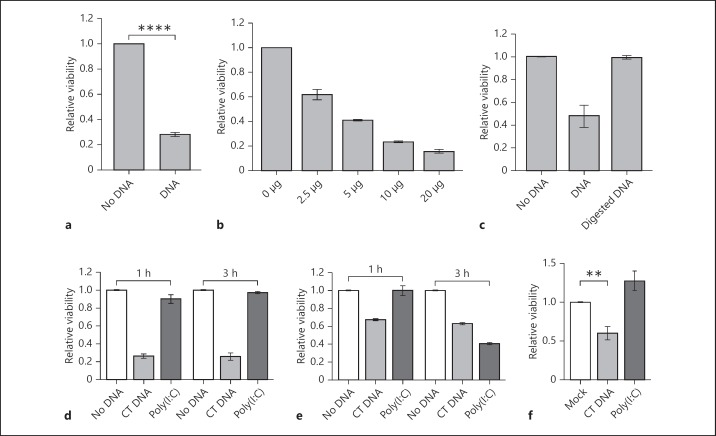
Transfected DNA rapidly kills Drosophila S2 cells. The viability of S2 cells was measured by MTT cleavage at 1 h after electroporation, unless otherwise stated. a DNA-dependent cell death of S2 cells after electroporation with 10 µg of CT DNA. The bars represent data from 40 experiments (mean ± SEM) normalised to the no-DNA sample for each experiment. **** p < 10−15. b Dose-dependent response of S2 cells after transfection with the indicated amounts of CT DNA. A representative of 2 experiments is shown, with error bars showing the range of results for duplicate electroporations. c DNA-specific response of S2 cells. Cells were electroporated with 10 µg of CT DNA that was untreated or treated with DNase I. The bars represent data from 3 experiments (mean ± SEM), normalised to the no-DNA sample for each experiment. d S2 cells are not killed by synthetic dsRNA poly(I:C). S2 cells were electroporated with 10 µg of CT DNA or poly(I:C), and cleavage of MTT was measured after 1 and 3 h. The bars represent data from 3 experiments (mean ± SEM), normalised to the no-DNA samples. e Mouse BMM responses to DNA and dsRNA transfection. BMMs were electroporated as per panel d. Data are from a representative experiment and error bars show the range of duplicate electroporations. f DNA-dependent cytotoxic effect in S2 cells after transfection with Lipofectamine 2000. Cells in 100 µl were transfected with 2 µg of CT DNA or poly(I:C), with 4 µl of transfection reagent or 4 µl of transfection reagent alone (mock). Cleavage of MTT was measured at 18 h after transfection. Data are from 5 experiments (mean ± SEM), normalised to the mock sample (** p = 0.013).
Fig. 2.
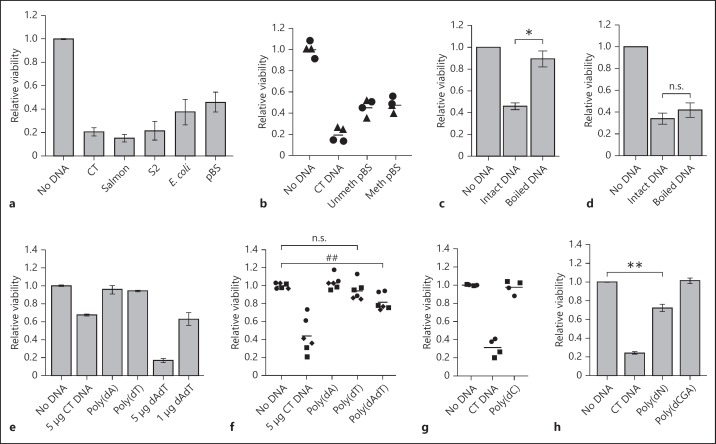
Response of S2 cells and BMMs to different types of DNA. a Cytotoxic effect of DNA from different sources on S2 cells; 10 µg of CT DNA or DNA from salmon sperm, S2 cells, E. coli and pBS plasmid were used in electroporations. Data are from 2 experiments (mean ± range). b Cytotoxic effect of DNA on S2 cells is not dependent on methylation. Cells were electroporated with 10 µg of CT DNA or pBS DNA, either methylated or unmethylated on CpG sequences. Each dot represents a separate electroporated sample, with different symbols denoting 2 different experiments. Response of BMM (c) and S2 cells (d) to boiled DNA; 10 µg of CT DNA was left intact or boiled for 10 min, transferred to ice and immediately used for electroporation. The bars represent data from 3 (c) and 5 (d) experiments (mean ± SEM), normalised to the no-DNA sample (* p = 0.029). e Cytotoxic effect of synthetic ssDNA and dsDNA in BMM; 5 µg of CT DNA or synthetic DNA [poly(dA), poly(dT) or poly(dA:dT)] or 1 µg of poly(dA:dT) was electroporated. f Cytotoxic effect of synthetic DNAs in S2 cells, as per panel e. Each dot represents a separate electroporated sample, with the different symbols denoting the 3 different experiments (n = 3, ## p = 0.054). g Absence of cytotoxic effect of poly(dC) on S2 cells; 5 µg of CT DNA or synthetic DNA was used for electroporation. Each dot represents a separate electroporated sample, with the different symbols denoting the different experiments. h Toxic effect of synthetic mixed-base DNA with or without thymidine [poly(dN) and poly(dGCA)] on S2 cells; 10 μg of DNA was used for electroporation. Data are from 5 experiments [CT DNA and poly(dN), mean ± SEM; ** p = 0.0085], or 2 experiments [poly(dGCA), mean ± range].
Results
S2 Cells Die Rapidly following DNA Transfection
We electroporated S2 cells [10], a macrophage-like cell line derived from the late embryonic stage of D. melanogaster, with CT DNA. S2 cells can be transfected with plasmids for expression of recombinant proteins, thus we expected only limited DNA-dependent death in this immortal cell line. Assessment of cell viability at 1 h after electroporation revealed that cytosolic DNA induced rapid dose-dependent cell death (fig. 1a, b). DNase I-treated CT DNA did not induce cell death, confirming that the effect is due to DNA, and not a contaminant in the CT DNA preparation (fig. 1c). Poly(I:C), a synthetic dsRNA, served as a control for the presence of a negatively charged polymer as a death-inducing factor. S2 cells were not sensitive to poly(I:C), even at 3 h after electroporation (fig. 1d). For comparison, similar experiments were performed with mouse BMMs. Both dsDNA and dsRNA were toxic to mouse cells, but with differing time courses for cell death (fig. 1e). Electroporated dsDNA kills BMM via AIM2-dependent pyroptosis within 1 h [8], but with poly(I:C) cell viability was reduced only at 3 h after electroporation. Analysis of nuclear morphology confirmed that the dsRNA induced apoptosis of BMM (data not shown), explaining the slower response. Thus, mouse BMM and Drosophila S2 cells both showed rapid death in response to DNA, but had contrasting responses to dsRNA.
As another method of nucleic acid delivery, S2 cells were transfected with a cationic liposome reagent. At 18 h after transfection with CT DNA, cell viability was reduced compared to the mock-transfected controls (fig. 1f). Poly(I:C) was used in this experiment as a control polyanion capable of forming a similar complex with transfection reagent. As for electroporation, the observed toxicity was DNA specific, and not observed with dsRNA. Differences between the time of cell death in electroporated and chemically transfected S2 cells were presumably due to the mechanisms of DNA introduction into the cells. Electroporation permeabilises the plasma membrane, which allows direct and synchronous DNA delivery into the cytosol. Using a transfection reagent requires uptake of the DNA-reagent complex and escape from the endosomal pathway, which would take longer to reach a toxic threshold of DNA.
Effects of DNA Source, Sequence and Strandedness on S2 Cell Viability
dsDNA from other sources was used to show that the observed cell death is not specific to bovine DNA. CT DNA, salmon sperm DNA, DNA isolated from S2 cells, E. coli genomic DNA or plasmid DNA (pBS) all induced cell death (fig. 2a). E. coli DNA, plasmid DNA and Drosophila DNA all lack the methylation of cytosine in CpG motifs seen in vertebrates, although some other insects and invertebrates exhibit CpG methylation [17]. Since E. coli and plasmid DNAs were the least toxic, we examined the role of CpG methylation. Plasmid DNA was treated with CpG-methyltransferase M.SssI, and the reaction was shown to be complete by lack of digestion by methylation-sensitive restriction enzyme HpaII (data not shown). Electroporation of S2 cells with equal amounts of plasmid DNA or CpG-methylated plasmid DNA revealed that CpG methylation status had no effect on the toxicity of plasmid DNA (fig. 2b).
In mammalian macrophages, cell death follows recognition of cytosolic DNA by AIM2, which mediates responses only to dsDNA, and not ssDNA [3, 18]. To investigate the dependence of S2 cell death on DNA strandedness, we denatured CT by boiling and transferring the DNA to ice for less than 2 min prior to electroporation into cells to minimise reannealing. Boiled DNA had little toxic effect on BMMs, which is consistent with the known specificity of AIM2 responses for dsDNA [3, 18] (fig. 2c). However, S2 cells were sensitive to both boiled and intact CT DNA (fig. 2d), suggesting that ssDNA was still toxic. We used synthetic DNA to investigate the effect of strandedness further. In our previous work on mouse macrophage cytosolic DNA responses, synthetic ssDNA, poly(dA), was inactive, whilst a duplex of annealed homopolymers, poly(dA):(dT), was the most toxic DNA identified [3]. Because long poly(dA) and poly(dT) homopolymers are no longer commercially available, they were generated using TdT. The lengths of synthesized homopolymers were approximately 2.5 kb (data not shown). Transfection of BMM with these single- and double-stranded homopolymers confirmed previous results of high poly(dA):(dT) toxicity [3], indicating a successful synthesis (fig. 2e). In contrast, poly(dA:dT) had low but measurable toxicity in S2 cells (fig. 2f). There was no death of S2 cells with poly(dA) or poly(dC), and a minor effect of poly(dT), which did not reach significance (fig. 2f, g).
To investigate the effect of base composition, we generated two different mixed-base DNAs with TdT, one contained all nucleotides (pN) while the other lacked thymidine nucleotides (pGCA). The role of thymine was tested because amongst the homopolymers only poly(dT) gave any suggestion of toxicity (fig. 2f). Whilst we thought that the procedure would generate random-base ssDNA, digestion with S1 nuclease suggested the presence of significant double-stranded regions (data not shown). Nevertheless, this showed that DNA of random sequence and mixed single-/double-stranded characteristics could achieve some toxicity, but the presence of thymine bases was required for DNA toxicity (fig. 2h). Overall, the results suggest that Drosophila cells respond to both ssDNA and dsDNA, but in a sequence-dependent manner such that homopolymers are not toxic.
DNA-Induced S2 Cell Death Is Necrotic-Like and Not Apoptotic
In order to characterise the mechanism of cell death, we stained cells with annexin V and the membrane impermeable DNA stain, PI. Annexin V binds to phosphatidylserine, and early apoptotic cells are identified as annexin V positive/PI negative [8]. In contrast, cells dying via necrosis or pyroptosis rapidly lose membrane integrity and stain with PI and annexin V simultaneously. S2 cells were stained with annexin V and PI 30 min after electroporation. DNA-treated cells rapidly lost their membrane integrity, and no significant population of cells displayed an apoptotic annexin V-positive/PI-negative profile (fig. 3a). In apoptosis, loss of membrane integrity is not seen within 30 min of the stimulus [8]. As a control, staining was conducted on S2 cells that were treated with actinomycin D for 6 h, revealing the expected population of annexin V-positive/PI-negative apoptotic cells (fig. 3a). In summary, DNA-induced cell death proceeds via a mechanism involving rapid loss of membrane integrity, lacking an annexin V-positive/PI-negative population and, hence, is distinct from apoptosis.
Fig. 3.
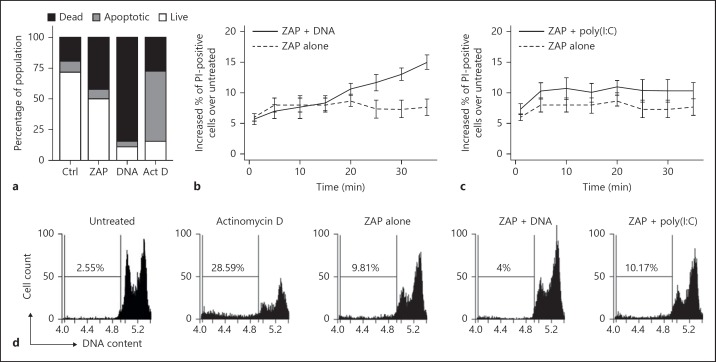
DNA induces cell death in S2 cells through a non-apoptotic mechanism. a Rapid loss of membrane integrity in response to DNA transfection. S2 cells were either untreated (Ctrl), electroporated with no DNA (ZAP) or electroporated with 10 µg of CT DNA (DNA) and stained after 30 min. As a positive control for apoptosis, S2 cells were incubated in 1 µM of actinomycin D for 6 h (Act D). Annexin V-/PI-stained cells were analysed by flow cytometry. Apoptotic cells are defined as annexin V positive/PI negative, necrotic (dead) cells as PI positive, and live cells as double negative. Data are representative of at least 3 experiments. b, c Time course of S2 cell permeability to PI, in response to electroporation of 10 μg of CT DNA (b) or poly(I:C) (c). Cells were incubated at room temperature, and compared to those electroporated with no addition (ZAP alone). After electroporation, cells were stained with PI and analysed by flow cytometry. Data from 3 experiments are shown (mean ± SEM), with results for PI-positive cells in an untreated sample subtracted. Data represent the increase in PI-positive cells relative to untreated cells. d Analysis of DNA content is consistent with non-apoptotic death. S2 cells were electroporated with 10 µg of CT DNA or poly(I:C), and then incubated for 18 h at 25°C. As a control for apoptosis, S2 cells were treated with 1 µM of actinomycin D for 18 h. Plots show the DNA content of cells based on PI staining intensity measured by flow cytometry, with the percentage of particles with a sub-G₀/G1 level of DNA indicated. Data represent 1 of 3 independent experiments.
We examined the kinetics of loss of membrane integrity. S2 cells were electroporated with and without CT DNA, then incubated at room temperature with PI and analysed by flow cytometry every 5 min to differentiate live and dead cells (fig. 3b). After 5 min there was a modest increase in the percentage of dead cells in electroporated samples compared to untreated (unelectroporated) cells, and this was relatively constant over the following 30 min of incubation. Electroporation with DNA resulted in an increase in PI-positive cells that first became apparent from 20 min after electroporation. The incubation temperature of cells was 4°C lower than in figure 3a, and this is likely to affect the rate of death. Electroporation with the dsRNA analogue poly(I:C) (fig. 3c) did not result in a progressive increase in dead cells, confirming that the response was specific to dsDNA (fig. 3b).
To confirm the non-apoptotic nature of this cell death, we analysed the DNA content of cells by flow cytometry. Cells undergoing later stages of apoptosis can be detected as nuclei with sub-G₀/G1 DNA due to apoptotic internucleosomal DNA cleavage, and loss of small fragments of DNA during fixation [15]. S2 cells were electroporated with CT DNA or poly(I:C), or treated with actinomycin D as a positive control, and incubated for 18 h prior to processing. Whilst nuclei with sub-G₀/G1 DNA are clearly present in actinomycin D-treated samples (fig. 3d), DNA-induced cell death did not produce the sub-G₀/G1 cell fraction that is characteristic of apoptosis. Electroporation alone induced a degree of apoptosis, as evident by the appearance of sub-G₀/G1 DNA; however, the inclusion of DNA in the electroporation apparently reduced such apoptosis, presumably as the cells had already rapidly died by another route. These results confirmed that cytosolic DNA induces a type of cell death that is phenotypically distinct from apoptosis.
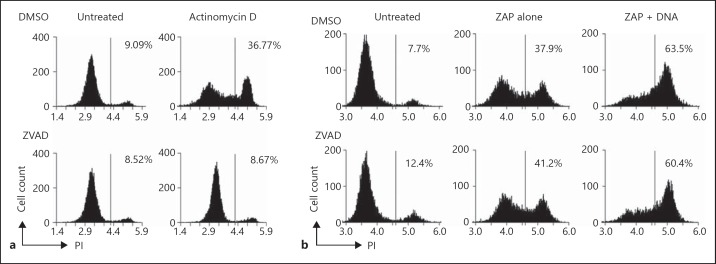
Activation of caspases initiates apoptosis in both flies and mammals. Z-VAD-FMK, a pan-caspase inhibitor, was employed to investigate the caspase dependence of DNA-dependent cell death. S2 cells were pretreated with Z-VAD-FMK or DMSO as a control for 1 h before subsequent treatments and flow cytometric analysis with PI to measure cell death. Z-VAD-FMK prevented death induced by actinomycin D (fig. 4a), which proceeds by an apoptotic route (fig. 3b). In contrast, Z-VAD-FMK did not prevent DNA-induced cell death measured at 30 min after electroporation (fig. 4b), suggesting a lack of requirement for typical apoptotic caspases. Overall, DNA-induced death exhibits no characteristics of apoptosis, and the rapid loss of membrane integrity demonstrates a necrotic style of death previously undescribed in insects.
Fig. 4.
DNA-induced rapid cell death is not prevented by caspase inhibition. a Actinomycin D-induced apoptosis is prevented by Z-VAD-FMK. S2 cells were treated with or without actinomycin D with either 50 µM of Z-VAD-FMK or DMSO vehicle control for 6 h, and analysed for loss of membrane integrity by PI staining and flow cytometry. b DNA-induced cell death in S2 cells is not affected by Z-VAD-FMK. S2 cells were pretreated with 50 µM of Z-VAD-FMK or DMSO vehicle control for 1 h prior to electroporation with 10 µg of CT DNA. After 30 min they were stained with PI and analysed by flow cytometry. Data are representative of 2 experiments.
Known Pathways Do Not Explain S2 Cell Death
Since pathways can be reused for new purposes in evolution, we considered whether a STING-dependent pathway could contribute to Drosophila DNA-induced death. Mammalian STING recognises bacterial second messengers cyclic di-GMP and cyclic di-AMP in addition to the ligand generated by cGAS following DNA recognition. An uncharacterised STING homologue is present in the fly genome. Cyclic di-GMP and cyclic di-AMP [13] were introduced into S2 cells using electroporation and chemical transfection, but had no toxic effect on S2 cells (fig. 5a, b). Under similar conditions, cyclic di-GMP transfected into BMM strongly induced IFN-β mRNA (fig. 5c). We conclude that DNA-induced cyclic dinucleotides and a STING-dependent pathway are unlikely to play a role in Drosophila DNA-induced cell death.
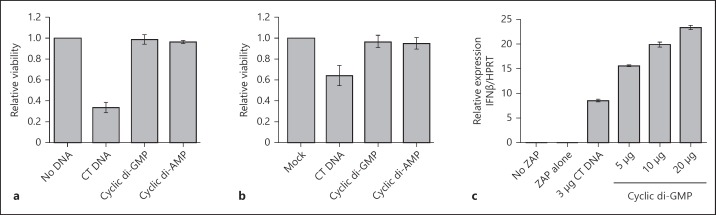
Fig. 5.
Cyclic dinucleotides have no toxic effect on S2 cells, and are not likely to be second messengers in the cell death response to DNA. a Electroporation of S2 cells with CT DNA and cyclic dinucleotides. Cells were electroporated with 10 µg of CT DNA or 20 µg of cyclic di-GMP or cyclic di-AMP, and MTT cleavage was measured 1 h after treatment. Results show the mean and range of duplicate electroporations. b Transfection of S2 cells with CT DNA and cyclic dinucleotides using Lipofectamine 2000; 2 µg of CT DNA and 4 µg of cyclic dinucleotides were transfected into S2 cells complexed with 4 µl of Lipofectamine 2000, and MTT cleavage was measured 18 h after transfection. The bars show data from 4 experiments (mean ± SEM). c Induction of IFN-β mRNA in BMM either untreated (no ZAP) or 2 h after electroporation with either no addition, CT DNA or various amounts of cyclic di-GMP. The real time PCR results shown are mean and range of duplicate assays of IFN-β mRNA relative to HPRT.
Drosophila Primary Haemocytes Display DNA-Dependent Cell Death
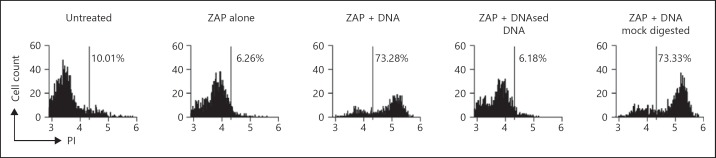
Responses to foreign DNA have been predominantly characterised in mammalian macrophages. The phagocytic innate immune cells in D. melanogaster are haemocytes. We isolated haemocytes and tested their responses to transfected DNA. Haemocytes were electroporated with either CT DNA, DNase I-treated or mock DNase I-treated CT DNA. Flow cytometric analysis revealed that the haemocytes died in response to transfected DNA (fig. 6), and that DNase I treatment abolished this sensitivity. Electroporation with DNA that had been mock DNase I-treated retained toxicity, confirming that components of the digest did not prevent DNA toxicity. This establishes that the DNA-induced death observed in S2 cells is relevant to primary Drosophila haemocytes.
Fig. 6.
Primary haemocytes are sensitive to cytosolic DNA-induced death. Primary haemocytes were untreated or electroporated with no additions (ZAP alone), with 10 μg of CT DNA, CT DNA digested with DNase I, or CT DNA mixed with digestion buffer and no enzyme (mock digest). After 30 min they were stained with PI and analysed by flow cytometry. Data are representative of 2 (DNase-treated sample) or 5 (CT DNA) experiments.
Discussion
Recognition of cytosolic foreign DNA is of demonstrated importance in a number of bacterial and viral infections of mouse cells [19, 20], and even prokaryotes have restriction-modification and CRISPR systems for defence against foreign DNA. Thus, responses to invading DNA might be anticipated to be widespread in nature. The insect cell death in response to electroporated DNA observed here was of similar sensitivity and rapidity to AIM2-mediated death of mouse macrophages [3]. AIM2 responses mediate protection against both viruses and cytosolic bacteria [20], and our work suggests insect cells have a similar DNA-mediated defence. A DNA virus naturally infecting Drosophila species has been recently identified [21], but recent work showing that positive-strand RNA viruses generate viral cDNA using endogenous reverse transcriptases [22] extends the relevance of DNA recognition pathways.
The insect cell recognition of DNA was demonstrably different from a mammalian AIM2 response. AIM2 recognises only dsDNA, and responses are lost with freshly boiled DNA [18]. In contrast, denatured DNA retained activity on Drosophila cells. In addition, the strongest ligand we have found for murine AIM2 is dsDNA obtained by the annealing of two synthetic homopolymers to give poly(dA):(dT) [3]. This double-stranded homopolymer is known to have an altered helical pitch compared to normal B-form DNA [23]. This may be favourable for AIM2 recognition, but was only a weak ligand for Drosophila cell death. No synthetic DNA approached the toxicity of natural DNAs, suggesting that particular structures or sequences not frequent in a random sequence may be involved. The omission of thymine from randomly synthesised DNA suggested that this base was essential for the Drosophila responses.
For many years apoptosis was the only described route of programmed cell death. Apoptosis is caspase-dependent cell death characterised by cell shrinkage and blebbing, nuclear condensation and fragmentation, with nuclear DNA being cleaved in internucleosomal regions [24]. An early marker of apoptosis is the exposure of phosphatidylserine on the outer surface of the cell membrane, which acts as an ‘eat-me’ signal for phagocytes. If not phagocytosed, apoptotic cells eventually undergo secondary necrosis and lose membrane integrity. It is now apparent that apoptosis is not the only ‘purposeful’ way for a cell to die, with the description of programmed necrosis (necroptosis) and caspase 1-dependent pyroptosis [25, 26]. Unlike apoptosis, necrosis and pyroptosis both involve rapid loss of membrane integrity, leading to an inflammatory response. Here we found that apoptotic features were absent in DNA-dependent Drosophila cell death, and the cells rapidly lost membrane integrity. Apoptosis is well characterized in both mammals and flies, whereas necroptosis and pyroptosis are not described in insects. Direct orthologues of genes encoding proteins involved in necroptosis (RIPK1, RIPK3, MLKL) or pyroptosis (caspase-1) are not found in the Drosophila genome (Drosophila initiator caspases are more closely related to mammalian caspases-8 and 9 [27]). Thus, the mechanism through which DNA induces rapid necrotic-type death remains to be established. The use of a pan-caspase inhibitor failed to prevent death, suggesting a lack of caspase involvement. Experimental infection of Drosophila with a lepidopteran baculovirus suggests apoptosis is a host defence [28, 29], but viral studies have not yet identified rapid lytic insect cell death. Cell death is an important defence against intracellular pathogens, and simultaneous induction of multiple cell death pathways is likely as a host response to pathogen evasion strategies.
The DNA-induced death pathway described here for Drosophila may be more widely relevant; we have recently found that DNA-induced death in chicken macrophages is similarly sensitive to denatured DNA and independent of AIM2 [Vitak et al., in preparation]. Elucidation of the molecular details of this pathway cannot readily exploit the use of Drosophila mutant libraries, since a high-throughput screen for response to transfected DNA is not feasible. However, direct affinity purification of cytosolic DNA-binding proteins can be pursued [3], and candidate genes assessed with knockouts or knockdown.
Despite a lack of characterised DNA receptor, there is some evidence suggesting other responses to non-chromosomal DNA in Drosophila. DNase II is a lysosomal DNase involved in the degradation of phagocytosed DNA [30]. DNA within apoptotic cells is cut into nucleosomal units by caspase-activated DNase, and further DNase II-dependent degradation occurs following uptake into a macrophage phagolysosome [30, 31]. A mutation in the Drosophila DNase II orthologue that leads to diminished enzyme activity results in accumulation of DNA in apoptotic cells in the ovary, increased susceptibility to bacterial infections, a reduced number of haemocytes, and elevated levels of the antimicrobial peptides diptericin and attacin A [32, 33]. The induction of antimicrobial peptides was further increased in DNase II mutant flies without functional Drosophila caspase-activated DNase [32]. This presents an interesting parallel to the DNase II knockout mouse, which has an elevated expression of type I IFNs, causing embryonic lethality [34]. The mouse phenotype is independent of Toll-like receptors that might respond to DNA, but is dependent on STING, implicating the involvement of the cGAS pathway of recognition of cytosolic DNA leading to IFN-β production [2, 35, 36]. Thus, it is likely that accumulated DNA in the phagolysosome of DNase II-deficient mouse or Drosophila cells escapes to the cytosol for recognition. A role for STING has not been investigated in the DNase II mutant fly, but we showed that it is unlikely to play a role in DNA-induced cell death. However, given the likely escape of undigested DNA into the cytosol of DNase II mutant cells, the cell death pathway we have described is consistent with the documented low level of haemocytes in the DNase II mutant flies [33].
Apart from response to infection, DNA-induced death could equally well have evolved to protect the genome from invading (retro)transposons. Although newly acquired DNA can drive evolution, the too-rapid accumulation of DNA insertions could lead to loss of viability. DNA insertions into the germ line have obvious consequences for organism viability and evolution, but insertions in somatic cells can drive tumourigenesis or lead to a loss of function during the life of the animal. Consequently, organisms should limit the integration of exogenous DNA, or the reintegration of endogenous retroelements, into their genome. The nuclease Trex1 (DNase III) that is involved in the degradation of cDNA of invading retroviruses and endogenous retroelements [37, 38] may provide one level of defence. However, if the burden of unintegrated DNA remains high, cell death may protect the organism from replicating retrotransposons, or may protect unicellular organisms at the population level. Elucidation of the DNA recognition pathway, as well as the specific DNA sequences that drive insect cell death, will aid the understanding of the evolution of these responses.
Disclosure Statement
The authors have no financial conflicts of interest relating to the results of this work.
Acknowledgements
This project was funded by the Australian Research Council (ARC) Grant DP120103332. K.J.S. was supported by ARC and NHMRC fellowships FT0991576 and 1059729. Cyclic dinucleotides were kindly provided by Zhao-Xun Liang, Nanyang Technological University, Singapore.
References
- 1.Gasiunas G, Sinkunas T, Siksnys V. Molecular mechanisms of CRISPR-mediated microbial immunity. Cell Mol Life Sci. 2014;71:449–465. doi: 10.1007/s00018-013-1438-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, Hume DA, Stacey KJ. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 4.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 7.Miao EA, Rajan JV, Aderem A. Caspase-1-induced pyroptotic cell death. Immunol Rev. 2011;243:206–214. doi: 10.1111/j.1600-065X.2011.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sagulenko V, Thygesen SJ, Sester DP, Idris A, Cridland JA, Vajjhala PR, Roberts TL, Schroder K, Vince JE, Hill JM, Silke J, Stacey KJ. AIM2 and NLRP3 inflammasomes activate both apoptotic and pyroptotic death pathways via ASC. Cell Death Differ. 2013;20:1149–1160. doi: 10.1038/cdd.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cridland JA, Curley EZ, Wykes MN, Schroder K, Sweet MJ, Roberts TL, Ragan MA, Kassahn KS, Stacey KJ. The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC Evol Biol. 2012;12:140. doi: 10.1186/1471-2148-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- 11.Costelloe EO, Stacey KJ, Antalis TM, Hume DA. Regulation of the plasminogen activator inhibitor-2 (PAI-2) gene in murine macrophages: demonstration of a novel pattern of responsiveness to bacterial endotoxin. J Leukoc Biol. 1999;66:172–182. doi: 10.1002/jlb.66.1.172. [DOI] [PubMed] [Google Scholar]
- 12.Stacey KJ, Young GR, Clark F, Sester DP, Roberts TL, Naik S, Sweet MJ, Hume DA. The molecular basis for the lack of immunostimulatory activity of vertebrate DNA. J Immunol. 2003;170:3614–3620. doi: 10.4049/jimmunol.170.7.3614. [DOI] [PubMed] [Google Scholar]
- 13.Rao F, Pasunooti S, Ng Y, Zhuo W, Lim L, Liu AW, Liang ZX. Enzymatic synthesis of c-di-GMP using a thermophilic diguanylate cyclase. Anal Biochem. 2009;389:138–142. doi: 10.1016/j.ab.2009.03.031. [DOI] [PubMed] [Google Scholar]
- 14.Denizot F, Lang R. Rapid colorimetric assay for cell growth and survival: modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J Immunol Methods. 1986;89:271–277. doi: 10.1016/0022-1759(86)90368-6. [DOI] [PubMed] [Google Scholar]
- 15.Riccardi C, Nicoletti I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat Protoc. 2006;1:1458–1461. doi: 10.1038/nprot.2006.238. [DOI] [PubMed] [Google Scholar]
- 16.Lanot R, Zachary D, Holder F, Meister M. Postembryonic hematopoiesis in Drosophila. Dev Biol. 2001;230:243–257. doi: 10.1006/dbio.2000.0123. [DOI] [PubMed] [Google Scholar]
- 17.Raddatz G, Guzzardo PM, Olova N, Fantappie MR, Rampp M, Schaefer M, Reik W, Hannon GJ, Lyko F. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc Natl Acad Sci U S A. 2013;110:8627–8631. doi: 10.1073/pnas.1306723110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stacey KJ, Ross IL, Hume DA. Electroporation and DNA-dependent cell death in murine macrophages. Immunol Cell Biol. 1993;71:75–85. doi: 10.1038/icb.1993.8. [DOI] [PubMed] [Google Scholar]
- 19.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, Fitzgerald KA. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unckless RL. A DNA virus of Drosophila. PLoS One. 2011;6:e26564. doi: 10.1371/journal.pone.0026564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goic B, Vodovar N, Mondotte JA, Monot C, Frangeul L, Blanc H, Gausson V, Vera-Otarola J, Cristofari G, Saleh MC. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat Immunol. 2013;14:396–403. doi: 10.1038/ni.2542. [DOI] [PubMed] [Google Scholar]
- 23.Nelson HC, Finch JT, Luisi BF, Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987;330:221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- 24.Strasser A, Cory S, Adams JM. Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. Embo J. 2011;30:3667–3683. doi: 10.1038/emboj.2011.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aachoui Y, Sagulenko V, Miao EA, Stacey KJ. Inflammasome-mediated pyroptotic and apoptotic cell death, and defense against infection. Curr Opin Microbiol. 2013;16:319–326. doi: 10.1016/j.mib.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanlangenakker N, Vanden Berghe T, Vandenabeele P. Many stimuli pull the necrotic trigger, an overview. Cell Death Differ. 2012;19:75–86. doi: 10.1038/cdd.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denton D, Aung-Htut MT, Kumar S. Developmentally programmed cell death in Drosophila. Biochim Biophys Acta. 2013;1833:3499–3506. doi: 10.1016/j.bbamcr.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 28.Lamiable O, Imler JL. Induced antiviral innate immunity in Drosophila. Curr Opin Microbiol. 2014;20C:62–68. doi: 10.1016/j.mib.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu B, Behura SK, Clem RJ, Schneemann A, Becnel J, Severson DW, Zhou L. P53-mediated rapid induction of apoptosis conveys resistance to viral infection in Drosophila melanogaster. PLoS Pathog. 2013;9:e1003137. doi: 10.1371/journal.ppat.1003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans CJ, Aguilera RJ. DNase II: genes, enzymes and function. Gene. 2003;322:1–15. doi: 10.1016/j.gene.2003.08.022. [DOI] [PubMed] [Google Scholar]
- 31.Nagata S, Nagase H, Kawane K, Mukae N, Fukuyama H. Degradation of chromosomal DNA during apoptosis. Cell Death Differ. 2003;10:108–116. doi: 10.1038/sj.cdd.4401161. [DOI] [PubMed] [Google Scholar]
- 32.Mukae N, Yokoyama H, Yokokura T, Sakoyama Y, Nagata S. Activation of the innate immunity in Drosophila by endogenous chromosomal DNA that escaped apoptotic degradation. Genes Dev. 2002;16:2662–2671. doi: 10.1101/gad.1022802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seong CS, Varela-Ramirez A, Aguilera RJ. DNase II deficiency impairs innate immune function in Drosophila. Cell Immunol. 2006;240:5–13. doi: 10.1016/j.cellimm.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Lethal anemia caused by interferon-β produced in mouse embryos carrying undigested DNA. Nat Immunol. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]
- 35.Ahn J, Gutman D, Saijo S, Barber GN. STING manifests self DNA-dependent inflammatory disease. Proc Natl Acad Sci U S A. 2012;109:19386–19391. doi: 10.1073/pnas.1215006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okabe Y, Kawane K, Akira S, Taniguchi T, Nagata S. Toll-like receptor-independent gene induction program activated by mammalian DNA escaped from apoptotic DNA degradation. J Exp Med. 2005;202:1333–1339. doi: 10.1084/jem.20051654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]