Abstract
Reports have shown that the antimicrobial peptide LL-37 is abundantly expressed but has limited bactericidal effect in Streptococcus pyogenes infections. At sub-inhibitory concentrations, LL-37 has been reported to alter virulence gene expression. Here, we explored the interaction of S. pyogenes strains with LL-37, focusing on bacterial growth, cell surface alterations and pro-inflammatory responses. Bioscreen turbidity measurements of strain 5448 cultured in the presence or absence of LL-37 confirmed the poor antimicrobial effect, and revealed a significant increase in turbidity of bacterial cultures exposed to sub-inhibitory concentrations of LL-37. However, this was not linked to increased bacterial counts. Electron microscopy of LL-37-exposed bacteria revealed the presence of vesicle-like structures on the bacterial surface. The vesicles stained positive for LL-37 and were released from the bacterial surface. Concentrated supernatants enriched in these structures had a broader protein content, including several virulence factors, compared to supernatants from untreated bacteria. The supernatants from LL-37-exposed bacteria were pro-inflammatory and elicited resistin and myeloperoxidase release from neutrophils. This is the first report on S. pyogenes extracellular vesicle-like structures formed at the bacterial surface in response to LL-37. The associated increased pro-inflammatory activity further implicates LL-37 as a potential factor involved in S. pyogenes pathogenesis.
Key Words: Streptococcus pyogenes, LL-37, Extracellular vesicles, Inflammation
Introduction
The Gram-positive bacterium Streptococcus pyogenes (group A streptococcus) is an important human pathogen with an estimated 500,000 deaths yearly, placing it number 9 on the list of global killer pathogens [1]. It frequently colonises human skin and mucosal surfaces, in particular the oropharynx, and can cause a variety of diseases ranging from superficial skin and throat infections to life-threatening streptococcal toxic shock syndrome and necrotizing soft tissue infections [1]. Its success as a human coloniser and pathogen is largely attributed to its ability to adapt to the human host and to counteract the host defence system, for which it has evolved numerous immune evasion strategies (review [2]). These strategies include its anti-phagocytic properties, the degradation of neutrophil extracellular traps and the proteolytic degradation and inactivation of effector molecules such as complement and chemotactic factors, antibodies and antimicrobial peptides (AMPs). In the case of resistance to AMPs, many virulence factors have been implicated, including the streptococcal cysteine protease SpeB with activity particularly against LL-37 [3, 4], the streptococcal inhibitor of complement (SIC) which inhibits both LL-37 and defensins [5], streptokinase that confers resistance to LL-37 via the activation of plasminogen to plasmin [6] and M1-protein, proposed to confer resistance to LL-37 [7].
The cathelicidin LL-37 is a cationic AMP expressed by many different cell types, including epithelial cells, neutrophils, monocytes and natural killer cells [8]. It exhibits a broad range of antimicrobial activities against bacteria, viruses and fungi. Aside from its antimicrobial properties, immunomodulatory roles in pro-inflammatory and chemotactic responses as well as in wound healing and angiogenesis have been attributed to it [8, 9]. LL-37 has been shown to be upregulated in response to cutaneous injury [10] and to play an important role in the defence against S. pyogenes infections in murine skin and soft-tissue models [11]. Analyses of skin and soft-tissue biopsies from patients with S. pyogenes infections revealed high expression of LL-37 in infected tissue, and the levels were positively correlated to bacterial load [12]. The data suggested that despite a high concentration of LL-37 present at the infected tissue site, it did not contribute substantially to bacterial killing.
Additionally, it has been proposed that LL-37 influence bacterial virulence properties; sub-inhibitory concentrations of LL-37 resulted in up-regulated virulence gene expression in S. pyogenes [13]. LL-37 was found to alter virulence gene expression through the activation of the CovRS (also referred to as CsrRS) two-component regulatory system, which influences the expression of 15% of the S. pyogenes genome [13]. LL-37 induced what the authors refer to as an ‘ invasive phenotype’ of the bacteria, with up-regulation of genes encoding virulence factors associated with invasive properties including streptodornase D, streptolysin O, streptokinase and the IL-8 protease spyCEP [14]. In addition, Velarde et al. [15] recently demonstrated a direct interaction with LL-37 and CsrS that contributed to the signal transduction.
In this study, we explored how the interaction with LL-37 influences S. pyogenes growth, bacterial surface architecture and virulence properties. Our study demonstrates that LL-37, even at sub-inhibitory concentrations, elicits profound alterations of the bacterial surface architecture, including the formation of vesicle-like structures, which results in an enhanced pro-inflammatory activity.
Materials and Methods
Bacterial Strains and Culture Conditions
The S. pyogenes strains 5448 (M1T1) [16] and its natural isogenic covS mutant 5448AP (animal-passaged) [17], 8157 (M1T1), 8003 (M3), 5626 (M3), all from patients with streptococcal toxic shock syndrome and/or necrotizing fasciitis, were collected during an active surveillance study of invasive group A streptococcal infections in Ontario, Canada. Strain 90-226 (M1) was originally isolated from the blood of a septic patient [18]. Strains 581 (M49) and 591 (M49) were isolated from patients with severe skin infections and were obtained from the Reference Laboratory for Streptococci in Prague, Czech Republic. The covS-deficient mutant on a 581 chromosomal background was generated previously by insertional inactivation of the covS gene [19]. The bacteria were cultured at 37°C in Todd-Hewitt broth supplemented with 1.5% (w/v) yeast extract (THY) culture media. An Escherichia coli strain isolated from a patient with sepsis (strain ID: h1 [20]) was used as a control, and was cultured in LB medium.
LL-37, RI-10 and Vasoactive Intestinal Peptide
Synthesized LL-37, biotinylated LL-37, RI-10 and vasoactive intestinal peptide were all purchased from Innovagen (Lund, Sweden). All peptides were dissolved in 0.1% trifluoroacetic acid (TFA; Fluka BioChemika), aliquoted and stored at −20°C until use.
Antimicrobial Testing
The minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of LL-37 against the different clinical isolates [0.6-2 × 106 colony-forming units (CFU)/ml] were determined using bacteria from the stationary phase diluted 1:50 (Streptococcus) or 1:180 (E. coli). LL-37 was added in different concentrations achieved by 2-fold dilutions with 0.1% TFA in concentrations ranges of 0-16 and 8-128 μM for the E. coli and streptococcal strains, respectively. The bacterial cultures were incubated with the peptide for 3 h and then plated in serial dilutions on blood agar plates. The number of CFU were analysed after 18 h of incubation at 37°C. The MIC was defined as the lowest concentration of LL-37 that significantly could reduce the bacterial CFU related to the peptide-free control cultures [21]. MBC was defined as the peptide concentration where no growth (99.9% killing) was detected in comparison to the peptide-free control [21].
Bioscreen Assay
The bacteria were cultured as described above and the streptococcal strains were used at a dilution of 1:50. Initial experiments with 1:50 and 1:100 dilutions of stationary-phase bacteria revealed similar results. The peptides were used in the concentration range of 0-10 μM. As a negative control, THY broth supplemented with TFA at the same concentrations as present in the peptide samples was used. Bacterial growth was measured in the Bioscreen C MBR instrument (Oy Growth Curves AB Ltd.), which allows turbidometrical measurements at an optical density (OD) of 420-580 nm in a precision incubator (37°C) equipped with a linear shaker. Measurements were made every 15 min over a time period of 10 h. OD curves were generated by plotting turbidity versus time. Each sample was analysed in duplicate. At 10 h, the CFU were determined by plating serial dilutions on blood agar plates.
Confocal Microscopy
Stationary-phase cultures of strain 5448 were diluted 1:50 in THY broth and incubated with biotinylated LL-37 (1 and 10 μM) for 30 min at 37°C. The cultures were stained with live/dead BacLight™ (Invitrogen) according to the manufacturer's instructions. The stained cells were pelleted, resuspended in PBS and transferred onto glass slides (Diagnostic Microscope Slides, Erie Scientific). After 10 min of adherence at room temperature, the cells were fixed with freshly prepared 2% formaldehyde for 15 min. After washing with PBS, the cells were blocked with 2% FCS for 5 min at 37°C, followed by washes with 0.2% saponin, 2% HEPES in PBS and a subsequent blocking step with 0.1% BSA-c (Sigma-Aldrich, St. Louis, Mo., USA) blocking solution for 30 min at room temperature. Streptavidin-conjugated Alexa-647 (Molecular Probes, Eugene, Oreg., USA) in BSA blocking solution was added for 30 min at room temperature, after which the slides were washed and mounted with SlowFade® gold antifade reagent with DAPI (Prolong Gold, Life Technologies, Carlsbad, Calif., USA). For evaluation, a Nikon A1R confocal microscope was used (Nikon Instruments, Amstelveen, The Netherlands). Z-stacks were acquired for 3-5 fields of view at 0.175 μm per slice by sequential scanning, using a 100× oil Plan Apo objective (pixel size; 0.12 μm).
Preparation of Concentrated Bacterial Supernatants
Bacteria were exposed to LL-37 as described above. Through an initial centrifugation (for 10 min at 1,200 rpm), the supernatants were separated from the bacterial pellet and sterile-filtered (0.2 μM). The supernatants were concentrated through an additional centrifugation at 30,000 rpm for 2.5 h at 4°C, and the pellets were resuspended in PBS to achieve a 10-fold concentration.
Electron Microscopy
Field Emission Scanning Electron Microscopy. For field emission scanning electron microscopy (FESEM) analysis, the bacteria or concentrated supernatants were fixed with 1% formaldehyde in 0.1 M HEPES buffer (0.1 M HEPES, 0.01 M CaCl2, 0.01 M MgCl2 and 0.09 M sucrose; pH 6.9). Samples were washed with TE buffer (10 mM TRIS and 2 mM EDTA; pH 6.9), absorbed onto poly-L-lysine-coated coverslips, fixed with 1% glutaraldehyde at room temperature for 5 min, and then dehydrated in a graded series of acetone (10, 30, 50, 70, 90 and 100%) with 15 min of incubation on ice for each step. Following critical-point drying with liquid CO2 (CPD 30, Bal-Tec, Liechtenstein, or CPD 300, Leica), samples were coated with a thin gold-palladium film (SCD 500, Bal-Tec) and examined in a field emission scanning electron microscope Zeiss Merlin (Zeiss, Oberkochen, Germany) using the Everhart-Thornley SE-detector and Inlens SE-detector in a 25:75 ratio at an acceleration voltage of 5 kV and SmartSEM software v5.05. Contrast and brightness of the images were adjusted with Adobe Photoshop CS5.
Negative Staining of LL-37-Treated Streptococci. Concentrated supernatants were absorbed onto carbon film and washed with TE buffer and distilled water. Subsequently, samples were negatively stained with 1% aqueous uranyl acetate. Carbon film was picked up with 300-mesh nickel grids, blotted onto filter paper, air-dried and then examined in a Zeiss TEM 910 at an acceleration voltage of 80 kV and at calibrated magnifications. Images were recorded digitally with a Slow-Scan CCD camera (ProScan, 1,024 × 1,024, Scheuring, Germany) using ITEM-software (Olympus Soft Imaging Solutions, Münster, Germany). Contrast and brightness were adjusted with Adobe Photoshop CS5.
Immuno-Gold Labelling of LL-37 on Streptococci. Stationary-phase bacteria exposed to LL-37 (0, 1 and 10 μM) for 10 h were fixed with 1% formaldehyde in HEPES and washed repeatedly with TRIS buffer containing 10 μM glycine to quench free aldehyde groups. A rabbit polyclonal anti-LL-37 antibody (produced in the laboratory of Birgitta Agerberth, Karolinska Institutet) was added for 1 h at 30°C with occasional shaking followed by repeated washes with PBS. The pellet was resuspended in 100 µl PBS, and for visualization of bound antibodies, 5 µl of 15 nm protein A- or protein A/G-coated nanogold particles from the stock solution (BB International, Cardiff, UK) were added and incubated for 30 min at 30°C with occasional shaking. After washes with PBS, samples were fixed with 2% glutaraldehyde in TRIS buffer for 10 min at room temperature and absorbed onto carbon-coated butvar 300-mesh copper grids. After air-drying, samples were examined in a Zeiss Merlin with the HE-SE2 detector at an acceleration voltage of 10 kV.
Protein Extraction and Digestion
Concentrated bacterial supernatants were resuspended in 0.1% ProteaseMax (Promega, Fitchburg, Mass., USA), 50 mM ammonium bicarbonate and 10% acetonitrile. Protein concentrations were determined using the BCA kit (Pierce, ThermoFisher, Waltham, Mass., USA) and showed protein yields of between 1.6 and 1.9 µg/µl. Five µl of each sample were incubated for 30 min at 50°C followed by 10 min bath sonication at room temperature. Samples were centrifuged and the supernatant was directly subjected to a tryptic digestion protocol carried out by a liquid-handling robot (MultiProbe II, Perkin Elmer).
Liquid Chromatography Tandem Mass Spectrometry
Tryptic peptides were cleaned with C18 StageTips and the resulting peptide mixture was injected into an Ultimate system in-line coupled to a Q Exactive mass spectrometer (all from Thermo Scientific). The chromatographic separation of the peptides was achieved using an in-house packed column (C18-AQ ReproSil-Pur®, Dr. Maisch GmbH, Germany) with the following gradient: 5–35% acetonitrile for 89 min, 48–80% ACN for 5 min and 80% ACN for 8 min, all at a flow rate of 300 nl/min. The mass spectrometry acquisition method was comprised of one survey full scan ranging from 300 to 1,650 m/z acquired with a resolution of R = 70,000 at 400 m/z, followed by data-dependent HCD scans from the 10 most intense precursor ions.
Protein Identification and Quantitation
Tandem mass spectra were extracted using Raw2MGF (in-house software), and the resulting mascot generic files were searched against the concatenated SwissProt protein database using the Mascot Deamon 2.4.1 search engine (Matrix Science Ltd.) and a taxonomy filter for Firmicutes Gram-positive bacteria. Parameters were chosen as follows: up to two missed cleavage sites for trypsin, peptide mass tolerance 10 ppm and 0.05 Da for the HCD fragment ions. Carbamidomethylation of cysteine was specified as a fixed modification, whereas oxidation of methionine and deamidation of asparagine and glutamine were defined as variable modifications. A search against the human protein database confirmed that there was no residual LL-37 present in the concentrated supernatants.
SDS Gel and Silver Staining
Concentrated bacterial supernatants were incubated with NuPage SDS loading buffer (Life Technologies) at 70°C for 10 min. Proteins were separated using a 4–12% NuPage Novex Bis-Tris gel (Life Technologies). The gels were stained using the LiverQuest™ silver staining kit (Invitrogen) according to the basic staining protocol. Precision Plus Protein Dual Color (Bio-Rad) was used as molecular weight standard.
Isolation and Stimulation of Primary Neutrophils
Human neutrophils were isolated from healthy blood donors using PolymorphPrep (Axis Shield PoC AS, Oslo, Norway) density gradient centrifugation. The cells were stimulated essentially as previously detailed [20]. In brief, 5 × 105 cells/ml were stimulated with different concentrations (1:5 and 1:10) of the concentrated bacterial supernatants. The cells were incubated for 2 h at 37°C, 5% CO2, after which the neutrophil supernatants were collected and stored at −20°C until further analysis.
Resistin and Myeloperoxidase Determination
Resistin and myeloperoxidase (MPO) levels in stimulated neutrophil supernatants were determined by ELISA (BioVendor, CR, and Karasek, R&D Systems) according to the manufacturers' instructions. The detection range for resistin was 0.012 to 50 ng/ml and 0.156 to 10 ng/ml for MPO. All samples were measured in duplicate. Triton-X (0.02%) lysates of neutrophils served as controls for the total content of resistin and MPO in unstimulated cells.
Statistical Analyses
Data were analysed by GraphPad Prism v6.0 (GraphPad software) and statistical significant differences were determined by Student's t test or ANOVA (Kruskal-Wallis test with Dunn's multiple comparison). A p value of 0.05 was used for statistical significance.
Results
Increased Turbidity in Bacterial Cultures after Exposure to LL-37
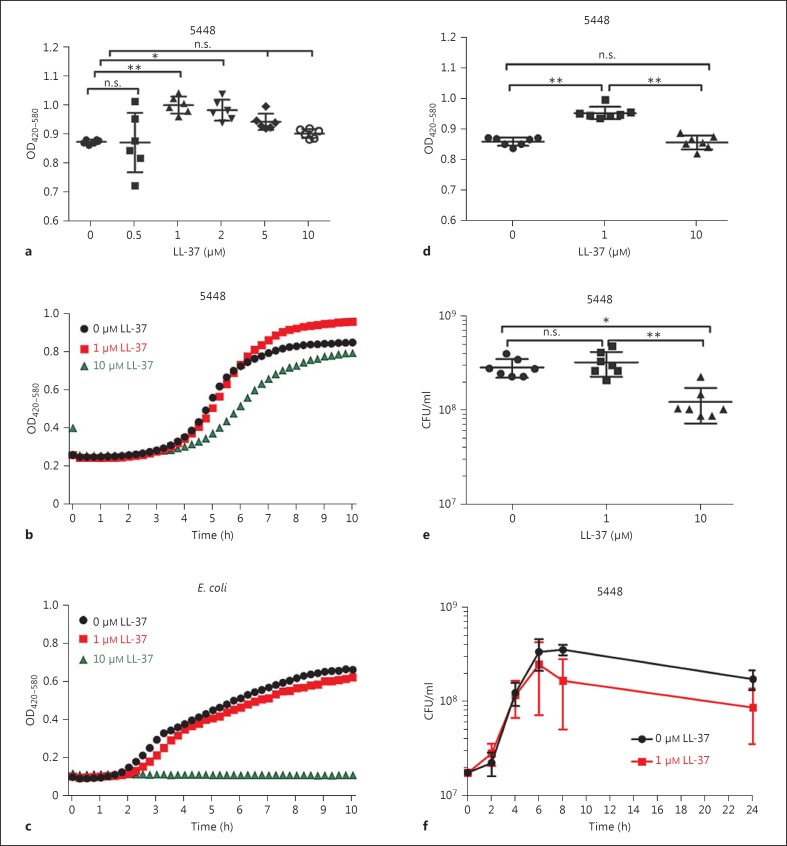
The S. pyogenes strain 5448 is used as a reference strain for the highly disseminated M1T1 clone associated with invasive infections worldwide [16]. To determine the susceptibility of the 5448 strain to LL-37, we conducted a dose-response experiment using LL-37 at concentrations ranging from 0 to 10 μM. Turbidity measurements of bacterial cultures over time were obtained by use of the Bioscreen C instrument. Comparison of the end OD after 10 h of incubation revealed a bell-shaped curve, with the highest turbidity evident in bacterial cultures exposed to 1 μM LL-37; at higher concentrations of LL-37, the turbidity starts to decline. The 1-μM LL-37 cultures had a significantly higher turbidity (OD) than the untreated control (p < 0.0018; fig. 1a). The OD curves over time showed typical bacterial growth curves with distinct lag, log and stationary phases (fig. 1b). Cultures of 5448 that were either untreated or exposed to 1 μM LL-37 showed almost overlapping curves during the first 5-6 h, but after this time, the untreated bacteria plateaued while the OD in the 1-μM LL-37-exposed 5448 continued to increase (fig. 1b). In contrast, 5448 cultures exposed to 10 μM LL-37 showed a flatter curve, resulting in a lower end OD. Exposure of a clinical E. coli strain isolated from a septic-shock patient showed a slight reduction in turbidity in cultures with 1 μM LL-37 but complete growth inhibition by 10 μM LL-37, evident from the flat line (fig. 1c). The results were congruent with the MIC and MBC values which were lower for the E. coli strain compared to the relatively resistant 5448 (table 1). Almost identical growth curves were noted when the control peptide, vasoactive intestinal peptide, which has similar structural properties to LL-37 (i.e. amphipathic, cationic and <50 aa), was added at 1 or 10 μM to either 5448 or E. coli cultures (online suppl. fig. S1a, b, s)ee www.karger.com/doi/10.1159/000441896).
Fig. 1.
Turbidity measurement and assessment of bacterial growth in the presence or absence of LL-37. a-d Turbidity measurements in bacterial cultures in the presence of different concentrations of LL-37 were done with the Bioscreen C instrument. OD values of duplicate cultures were determined every 15 min for a total of 10 h. a Dose-response experiment using LL-37 at concentrations ranging from 0 to 10 μM in cultures of S. pyogenes 5448. The scatter plot shows data obtained after 10 h of culture in 6 independent experiments. OD curves show measurements over time from 1 representative experiment of S. pyogenes 5448 (b) and of E. coli (c) cultures in the absence or presence of either 1 or 10 μM LL-37. d Comparison of OD values obtained after 10 h of 5448 culture in the absence or presence of indicated LL-37 concentrations of 1 and 10 μM after 10 h incubation (7 experiments). e Bacterial CFU/ml were determined immediately after the bioscreen assay had been completed. f After 10 h of 5448 culture in the absence or presence of LL-37, CFU/ml were determined over time (0-24 h). Mean ± SEM of 4 experiments at each time point is shown. Statistical significant differences were determined by ANOVA using the Kruskal-Wallis test and Dunn's multiple comparisons. n.s. = Not significant.
Table 1.
MIC and MBC of LL-37
| Strain | M-type | MIC, μM | MBC, μM |
|---|---|---|---|
| 5448 | M1 | <32 | >128 |
| 5448AP | M1 | <8 | 64 |
| 8157 | M1 | <16 | 128 |
| 90.226 | M1 | <16 | 128 |
| 5626 | M3 | <16 | 128 |
| 8003 | M3 | <16 | 128 |
| 581 | M49 | <8 | 32 |
| 581ΔcovS | M49 | <8 | <32 |
| E. coli h1 | - | <1 | 4 |
To determine whether these differences in turbidity of the cultures reflected variations in bacterial growth, bioscreen analyses were done on 5448 incubated with 0, 1 or 10 μM LL-37 (fig. 1d) and after 10 h of incubation, samples were collected for CFU determination (fig. 1e). The results revealed that, although 1-μM LL-37-exposed bacteria had significantly higher OD values (fig. 1d), there was no increase in bacterial CFU (fig. 1e). In fact, monitoring CFU over 24 h revealed slightly less CFU in the cultures exposed to 1 μM LL-37 (fig. 1f). The results did show a significant decrease in CFU in cultures exposed to 10 μM LL-37, indicating an antimicrobial effect at this concentration of LL-37 (fig. 1e).
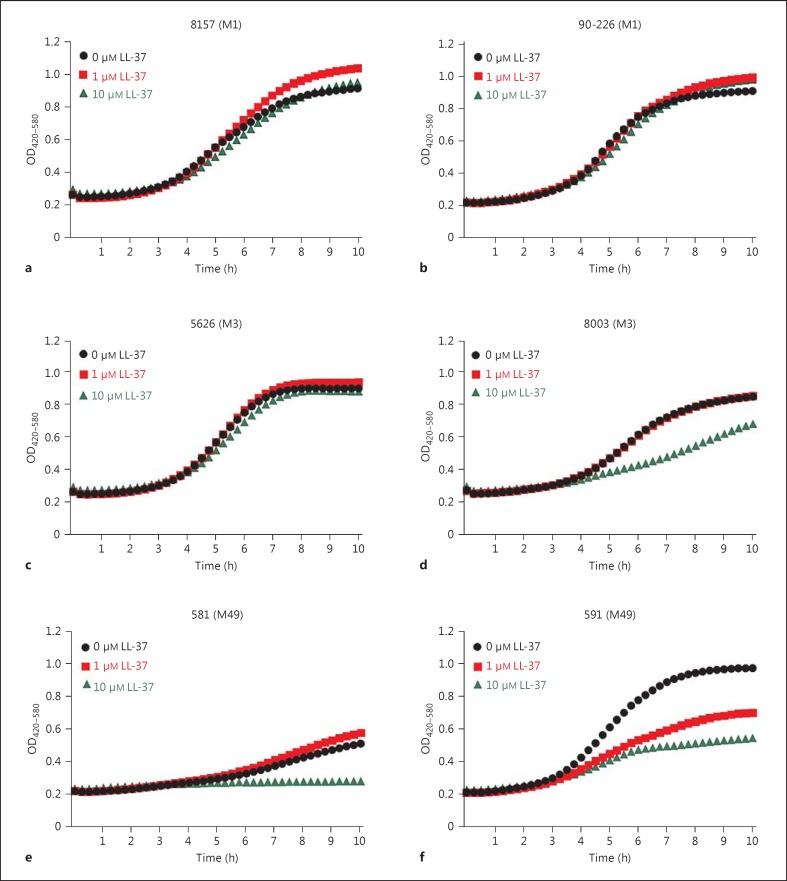
To test whether the noted increase in turbidity in the 1-μM LL-37 cultures is seen in strains other than 5448, we analysed additional S. pyogenes strains from patients with severe invasive infections. The data showed that the response to LL-37 varies between strains: both the M1T1 strains 8157 and 90-226 showed an increased turbidity when exposed to 1 μM LL-37 whereas with the 10-μM LL-37-exposed bacteria, it was either equal or higher than the control (fig. 2a, b). The M3T3 strains 5626 and 8003 demonstrated essentially similar curves in 1-μM LL-37-treated and untreated control cultures (fig. 2c, d). Both M49 strains demonstrated a marked antimicrobial susceptibility to LL-37 at 10 μM, but differed in response to 1 μM LL-37, with strain 581 having an increased turbidity whereas 591 exhibited reduced growth (fig. 2e, f). Thus, the data demonstrated inter-strain variability in response to LL-37, and the increased turbidity was noted in some, but not all, strains. There was no correlation between increased turbidity and the MIC and MBC for LL-37 with the different strains (table 1).
Fig. 2.
Varying response to LL-37 in different S. pyogenes strains. Turbidity measurements in bacterial cultures in the presence of different concentrations of LL-37 were done with the Bioscreen C instrument. OD values of duplicate cultures were determined every 15 min for a total of 10 h. OD curves show measurements over time in cultures of S. pyogenes strains of different types, i.e. M1 strains 8157 (a) and 90-226 (b), M3 strains 5626 (c) and 8003 (d) and M49 strains 581 (e) and 591 (f). The mean values of three separate experiments are shown.
LL-37 Interaction with Bacteria Results in Cell Surface Architectural Changes Including the Formation of Vesicle-Like Structures
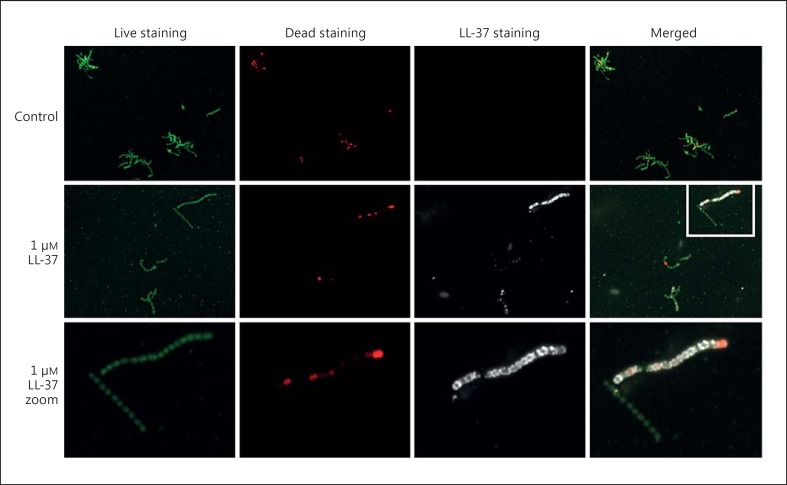
As the increase in turbidity was not due to increased bacterial growth, we employed confocal and electron microscopy to visualize LL-37 effects on bacterial cell morphology and cell surface architecture as well as the interaction between LL-37 and streptococci. We first used confocal microscopy to visualize the interaction between LL-37 and bacteria (fig. 3). After 30 min, LL-37 (1 μM) was found to bind extensively to the cell surface of some streptococcal chains, while other chains were negative for LL-37 (fig. 3). In agreement with the CFU results, the viability stain demonstrated that while some cocci stained positive for the dead cell marker (red), the majority of cocci with bound LL-37 stained green, and thus had an intact cell membrane indicating viability (fig. 3). In addition, the staining pattern suggested that the peptide accumulated at the plane of division (fig. 3, lowest panel).
Fig. 3.
LL-37 interaction with S. pyogenes and bacterial viability. Bacteria cultured for 30 min in the absence or presence of 1 μM biotinylated LL-37. Bacteria were detected using a bacterial viability stain that discriminates between viable (green) and dead bacteria (red). LL-37 was detected by the addition of streptavidin-Alexa 647 (white). The staining was analysed by confocal microscopy and representative images are shown. The lowest panel shows a zoom-in of the boxed area in the 1-μM LL-37-exposed bacteria image.
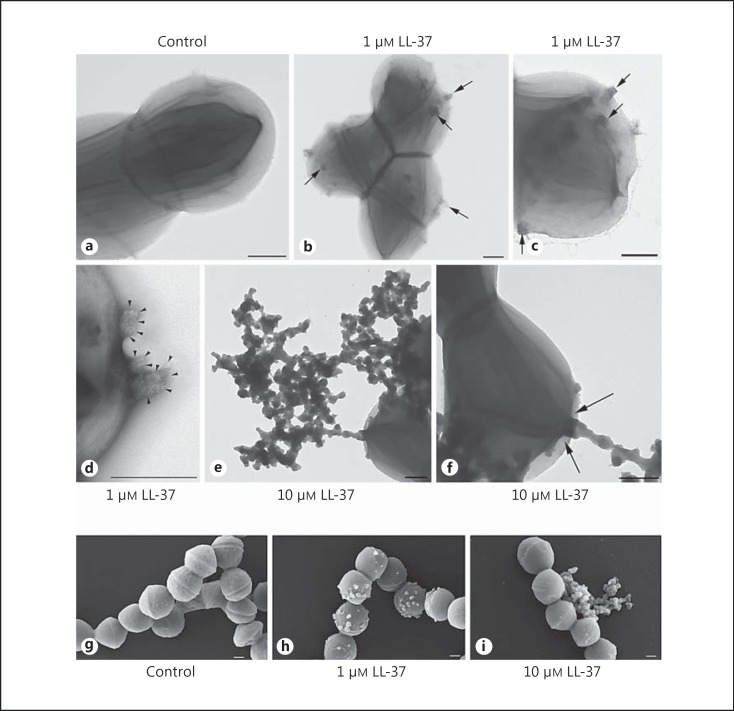
To gain a detailed insight into the interaction of the peptide with the bacterial cell surface, electron microscopy analysis was employed. First, the samples were negatively stained for examination in a transmission electron microscope and examined by FESEM. One of the most striking findings was the formation of vesicle-like structures on the cell surface, evident in LL-37-exposed bacteria but not in untreated bacteria (fig. 4, 5, 6). Negatively stained streptococci showed the formation of vesicle-like structures dispersed over the streptococcal cell wall when treated with 1 µM LL-37 (fig. 4b, c, d), whereas untreated bacteria exhibited a smooth cell wall (fig. 4a). Bacteria treated with 1 µM LL-37 exhibited several of these small structures (fig. 4b, c: arrows), which were protruding through the bacterial cell wall (fig. 4d: arrowheads). In contrast, streptococci treated with 10 µM LL-37 showed accumulation of large extracellular aggregates of vesicle-like structures (fig. 4e), demonstrating a more detrimental effect of higher LL-37 concentrations on the bacterial cell wall. Repeatedly, a connection of the outside aggregated vesicle-like structures with the cytoplasm of the bacteria could be observed (fig. 4f: arrows). Similarly, FESEM analyses confirmed the presence of vesicle-like structures on the bacterial surface in cultures exposed to 1 or 10 μM LL-37, with the latter showing large aggregates of extracellular components (fig. 4g, h, i). A time course experiment with 1-μM LL-37-exposed bacteria showed the presence of a few vesicles already at 30 min and an increase over time (data not shown).
Fig. 4.
Negative staining and FESEM analyses of LL-37-treated streptococci. After 10 h of culture, negatively stained, untreated streptococci show a smooth cell wall (a), whereas bacteria treated with 1 µM LL-37 exhibit the formation of several vesicle-like structures dispersed over the streptococcal cell wall (arrows, b, c), which protrude through the bacterial cell wall (arrowheads, d). e Bacteria treated with 10 μM LL-37 show accumulation of large aggregates of vesicle-like structures. Noteworthy: often a connection with the bacterial cytoplasm was observed (arrows, f). FESEM analyses were performed of 5448 incubated in the absence (g) or presence of LL-37 for 10 h, revealing vesicle-like structures attached to the bacterial surface after treatment with 1 µM LL-37 (h) and extrabacterial aggregates at 10 µM LL-37 (i). Scale bars: 200 nm.
Fig. 5.
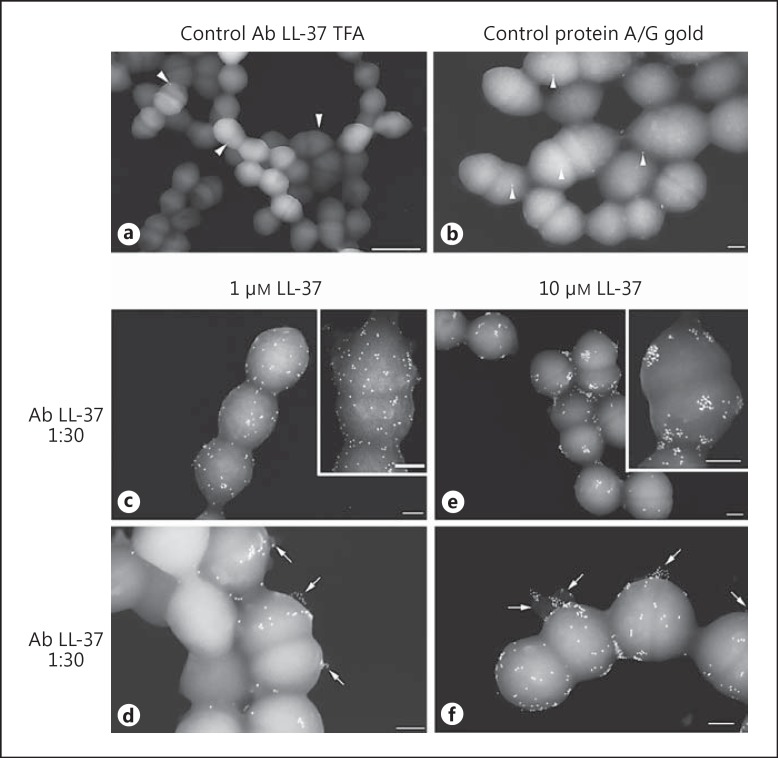
LL-37 interaction with the bacterial surface and presence on vesicle-like structures on the cell surface. Bacteria were cultured in the absence (a, b) or presence of 1 μM LL-37 (c, d) or 10 μM LL-37 (e, f) for 10 h. LL-37 was detected by immuno- gold labelling and the samples analysed by FESEM. a, b The arrowheads indicate single gold nanoparticles demonstrating the specificity of the labelling in c-f. Arrows in samples treated with LL-37 (d, f) indicate vesicle-like structures positively stained for LL-37. Scale bars: 1 µm (a), 200 nm (b-f) and insets (c, e).
Fig. 6.
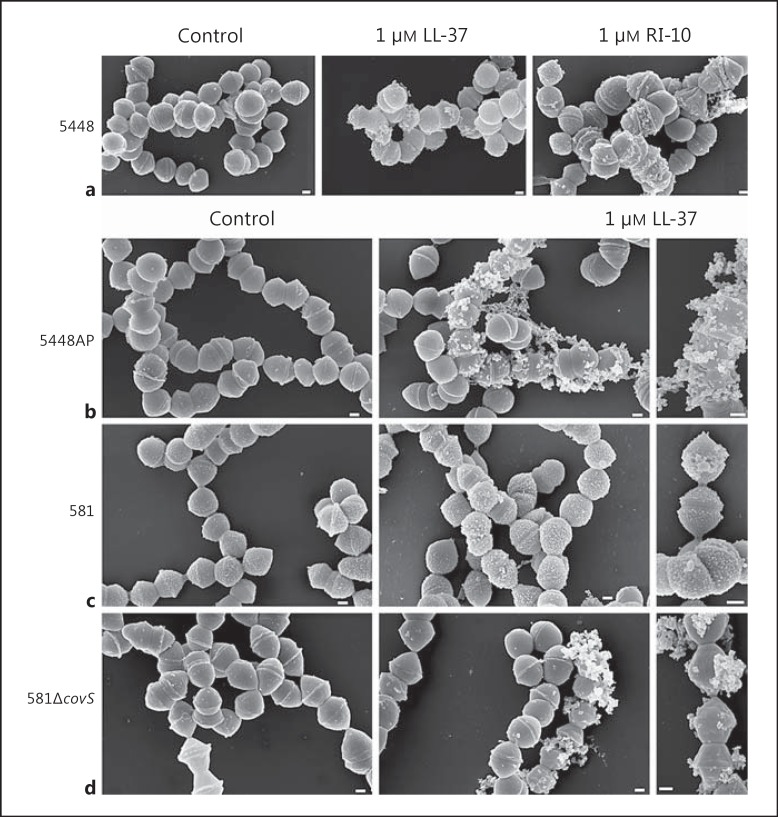
CovS involvement in LL-37 elicited vesicle formation in S. pyogenes. a FESEM analyses of 5448 incubated in the absence or presence of 1 μM LL-37 or 1 μM RI-10 for 10 h. 5448AP (b), 581 (c) and 581ΔcovS (d) strains incubated in the absence or presence of 1 μM LL-37 for 10 h were analysed by FESEM. Scale bars: 200 nm.
To explore the relation between vesicle formation and peptide binding, immuno-gold labelling of LL-37 with specific IgG antibodies and protein A- or protein A/G-coated gold nanoparticles was done. LL-37 was detected in several areas on the bacterial cell surface (fig. 5c, d, e, f), including the septum of streptococci (fig. 5d). Notably, LL-37-positive vesicle-like structures protruding from the bacterial membrane were clearly identifiable in bacteria exposed to either 1 or 10 μM of LL-37 (fig. 5d, f: arrows). In addition, LL-37 often appeared in clusters on the bacterial surface, particularly evident in the bacteria exposed to 10 μM as well as on the vesicles (inset in fig. 5c vs. 5e). No immuno-gold labelling was detectable on untreated streptococci (fig. 5a) or by adding protein A/G gold alone (fig. 5b); only rare, single gold nanoparticles were visible (arrowheads).
CovS Involvement in LL-37-Elicited Vesicle Formation
To seek mechanistic insight into the vesicle formation, we first addressed whether this was dependent on antimicrobial membrane permeation. A recent report [15] demonstrated that LL-37 signalling in S. pyogenes is due to a direct interaction with CovS. This study also identified that a 10-residue peptide RI-10, representing the minimal peptide structure of LL-37, was critical for signalling through CovS; this peptide lacked antimicrobial activity. We tested the RI-10 peptide and analysed the treated bacteria by FESEM, which revealed that RI-10 induced vesicle-like structures in strain 5448 to an extent similar to LL-37 (fig. 6a). In agreement with [15], our turbidity measurements showed no antimicrobial effect by RI-10 (data not shown); thus demonstrating that vesicle formation does not require membrane permeation.
Next, the covS mutants 5448AP, a natural covS variant with deficient signalling [17], and the 581ΔcovS deletion mutant [19] were analysed by FESEM. Although vesicle formation was evident in both strains in response to 1 μM LL-37, they displayed more extensive extracellular aggregations (fig. 6b, d) than their respective wild-type strain (fig. 6a, c). In fact, the large extracellular aggregates were similar to those seen in bacteria exposed to 10 μM LL-37, suggestive of a greater antimicrobial permeation effect when CovRS signalling is impaired. In line with this, both 5448AP and 581ΔcovS have lower MIC/MBCs than their wild-type strains (table 1).
Concentrated Supernatants Enriched in Vesicle-Like Structures Contain Bacterial Virulence Factors and Are Immune-Stimulatory
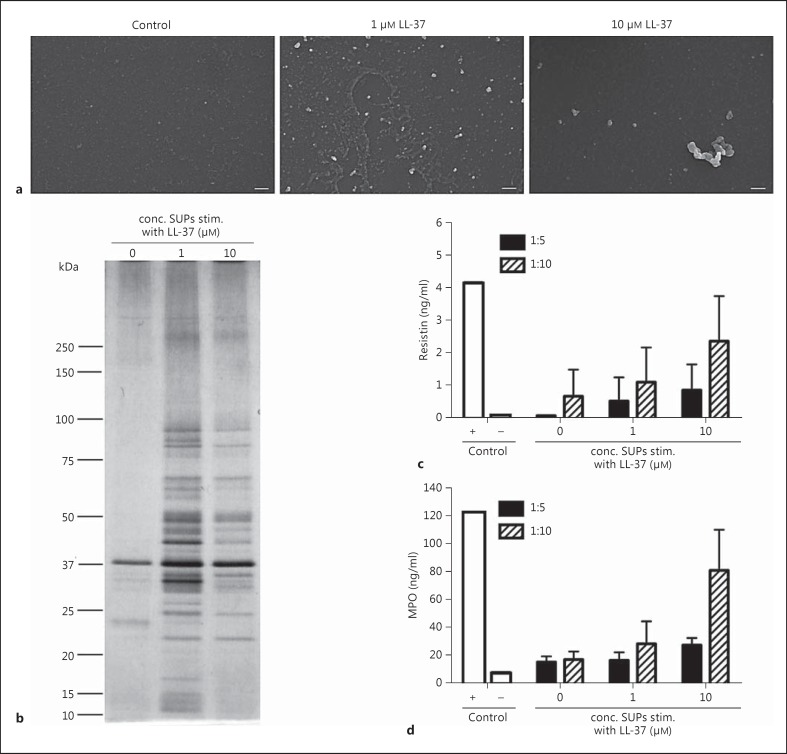
To further characterize the extracellular-like vesicles, cell-free concentrated supernatants were prepared by high-speed centrifugation. These were analysed by FESEM, which identified vesicle-like structures in the LL-37-exposed streptococcal samples (fig. 7a). Further analysis of the concentrated bacterial supernatants was done by SDS gel separation and silver staining, which revealed that the supernatants recovered from LL-37-exposed bacteria and thus, enriched in vesicle-like structures, contained more proteins than the untreated controls (fig. 7b). Similarly proteomic analysis using mass spectrometry identified more proteins present in the supernatants from LL-37-exposed bacteria, several of which are known virulence factors including M protein, streptolysin O, and streptokinase as well as moonlighting proteins such as glyceraldehyde 3-phosphate dehydrogenase (GAPDH), enolase and chaperone molecules (table 2).
Fig. 7.
Protein content and immunostimulatory properties of concentrated bacterial supernatants enriched in vesicle-like structures. 5448 was incubated in the absence or presence of 1 and 10 μM LL-37 for 10 h and cell-free, concentrated supernatants were prepared. a SEM analyses revealed the presence of vesicle-like structures in LL-37-exposed bacterial supernatants. Scale bar: 200 nm. b Proteins in the concentrated bacterial supernatants were separated on SDS gel and detected by silver staining. The same concentrated supernatants were added in different concentrations to cultures with primary neutrophils. d After 2 h of incubation, the neutrophil supernatants were analysed for resistin and MPO. Lysates of neutrophils served as a positive (+) and the supernatant of neutrophils incubated with cell culture media as negative (-) control (Ctr). The bars show the mean ± SD of 3-4 experiments. conc. SUPs stim. = Concentrated supernatants stimulated.
Table 2.
Bacterial proteins in concentrated supernatants enriched in vesicle-like structures after exposure to LL-37
| 5448 only | 5448 + 1 μM LL-37 | 5448 + 10 μM LL-37 |
|---|---|---|
| SpeB | SpeB | SpeB |
| Elongation factor Tu | elongation factor Tu | elongation factor Tu |
| ATP phosphoribosyltransferase | ATP phosphoribosyltransferase | |
| enolase | enolase | |
| M protein | M protein | |
| 60 kDa chaperonin | 60 kDa chaperonin | |
| GAPDH | GAPDH | |
| formate acetyltransferase | formate-tetrahydrofolate ligase | |
| 10 kDa chaperonin | chaperone protein DnaK | |
| streptolysin O | ||
| streptokinase A | ||
| putative septation protein SpoVG | ||
| phosphocarrier protein HPr | ||
| 5-methyltetrahydropteroyltriglutamatehomocysteine methyltransferase | ||
| DNA mismatch repair protein MutS | ||
| 50S ribosomal protein L7/L12 | ||
The protein content in concentrated supernatants was analysed by liquid chromatography tandem mass spectrometry.
As several of these virulence factors are known to be immuno-stimulatory, in particularly to neutrophils [20, 22], we explored neutrophil responses to the concentrated supernatants. The results showed that the concentrated supernatants of LL-37-exposed bacteria elicited the release of both resistin and MPO, with the highest levels detected in the cultures stimulated with the supernatant from bacteria exposed to 10 μM LL-37 (fig. 7c, d).
Discussion
There are reports that point towards a potentially harmful effect of LL-37 during severe S. pyogenes infection based on (1) findings in biopsies revealing a positive correlation between LL-37 levels and the severity of infection [12], (2) studies demonstrating the immuno-stimulatory properties of the peptide (review [23]) and (3) the ability of LL-37 to induce an altered bacterial virulence gene expression [13]. Here, we demonstrated that LL-37 induces the formation of extracellular vesicle-like structures on the streptococcal surface. These structures were released from the bacterial cell surface and analyses of the concentrated supernatants identified the presence of several streptococcal virulence factors with pro-inflammatory activity towards neutrophils. This novel finding that S. pyogenes forms extracellular vesicle-like structures in response to LL-37 is of interest, considering that severe invasive S. pyogenes tissue infections are hyper-inflammatory conditions characterized by a high bacterial load, infiltration of immune cells and the presence of LL-37 [24].
Formation and release of extracellular vesicles is a conserved feature of bacteria, eukaryotes and archaea. In bacteria, it has been extensively studied in Gram-negative bacteria where release of the outer membrane vesicles occurs constitutively during normal growth (review [25]). The vesicles derive from the cell surface and contain parts of the outer membrane, LPS, periplasmic and membrane-bound proteins, enzymes, toxins, DNA, RNA and peptidoglycan. The release of vesicles offers a controlled and protected secretion of these factors into the extracellular milieu, which has been shown to influence bacterial pathogenesis in several ways including the bacterial response to stress signals, bacterial communication, virulence and modulation of the host immune response (review [25, 26, 27]). Recently, extracellular vesicles with important functions for pathogenesis were also reported in Gram-positive bacteria, including in Staphylococcus aureus [28], Bacillus anthracis[29], Mycobacterium tuberculosis [30], Mycobacterium ulcerans[31] and Streptococcus pneumoniae[32]. Here, we report, for the first time, extracellular vesicle formation on the cell surface of S. pyogenes triggered in response to LL-37. Vesicle formation could be noted in several streptococcal invasive strains, including different M-types and strains with varying susceptibility to LL-37. Lauth et al. [7] reported that strains of the M1 type, such as strain 5448, are generally quite resistant to LL-37, which is of interest due to their association with large outbreaks of severe invasive infections. They also found higher MIC among invasive strains than non-invasive cases. LL-37 was identified as a critical factor in protection against necrotic tissue S. pyogenes infections using an in vivo murine model [11]. However, the report also highlighted the impact of the bacterial strain susceptibility, as the cathelicidin-resistant Streptococcus mutans demonstrated increased virulence.
In Gram-negative bacteria, studies have demonstrated that extracellular vesicles are formed as a protective mechanism against various stress signals, including AMPs and sub-inhibitory concentrations of antibiotics [33, 34]. In S. pyogenes, both polymyxin B and human defensins, all cationic AMPs, were reported to localize to single discrete sites of the S. pyogenes membrane, and further studies suggested that they targeted the ExPortal, a microdomain of the streptococcal membrane specialized for protein secretion [35, 36, 37, 38, 39, 40]. In contrast, here we demonstrate, by means of immuno-gold labelling, distinct clusters of LL-37 distributed all over the bacterial surface and not to a discrete site, and therefore likely not targeting the ExPortal. This is also in contrast to a recent report demonstrating that sub-lethal concentrations of penicillin resulted in the release of bacterial lipoproteins within vesicles [41]. The study demonstrated that vesicles were almost exclusively constituted by lipoproteins and were not representative of the bacterial membrane, but rather seemed to derive from the ExPortal. Due to the starkly different proteome composition of the penicillin-induced lipoprotein-rich membrane vesicles and the LL-37-triggered vesicle-like structures identified in this report, it seems that these are two discrete events.
To further understand the mechanisms underlying vesicle formation, we tested the RI-10 peptide, which was recently identified as the minimal peptide of LL-37 that retains the CovRS signalling ability [15]. The results revealed that this peptide elicited vesicle formation on the streptococcal surface to the same extent as seen with LL-37. This suggested that CovRS signalling is involved in the vesicle formation. Also, the protein content of the vesicle-enriched supernatants from 1-μM LL-37-exposed bacteria revealed, among others, streptokinase and streptolysin, which are among the genes reported to be upregulated in CovRS-mediated signalling in response to sub-inhibitory concentrations of LL-37 [13]. As RI-10 lacks antimicrobial activity, this showed that vesicle formation is independent of membrane permeation. Unexpectedly, also the covS mutants exposed to 1 μM LL-37 displayed vesicles but predominantly as large extracellular aggregates, which indicated greater antimicrobial membrane permeation in these bacteria. This suggests that other mechanisms, aside from CovS interaction, may attribute to the vesicle formation in response to LL-37.
It seems plausible that the extracellular vesicle formation triggered in response to LL-37 might serve as a protective mechanism against the AMP, both by the release of inhibitory bacterial proteins and/or by sequestering the AMP. The protein analyses revealed that the vesicle-enriched supernatants contained SpeB and the M1-protein, both of which have been reported to inhibit the action of LL-37 [3, 7]. However, the impact of these potential resistance mechanisms remains to be tested in future studies.
Supplementary Material
Supplementary data
Acknowledgement
The study was supported by grants from the Karolinska Institutet (A.N.T.), Stockholm county council (A.N.T.), the Swedish Research Council (A.N.T., L.J.), the Knut and Alice Wallenberg Foundation (A.N.T.) and the European Union Seventh Framework Program (FP7/2007-2013) under the grant agreement 305340. The Canadian strains were kindly provided by Dr. Donald E. Low, Mount Sinai Hospital and University of Toronto, and Dr. Malak Kotb (5448AP), University of North Dakota. We thank I. Schleicher for excellent help with the electron microscopic studies. The Proteomic Core Facility, Karolinska Institutet and Dorothea Rutishauser, Marie Ståhlberg and Carina Palmberg are gratefully acknowledged for excellent technical assistance with the proteomic part of the project.
References
- 1.Carapetis J, Steer AC, Mulholland EK, Weber M. The global burden of group a streptococcal diseases. Lancet Infect Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 2.Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, Sriprakash KS, Sanderson-Smith ML, Nizet V. Disease manifestations and pathogenic mechanisms of group A streptococcus. Clin Microbiol Rev. 2014;27:264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nyberg P, Rasmussen M, Björck L. Alpha2-macroglobulin-proteinase complexes protect Streptococcus pyogenes from killing by the antimicrobial peptide LL-37. J Biol Chem. 2004;279:52820–52823. doi: 10.1074/jbc.C400485200. [DOI] [PubMed] [Google Scholar]
- 4.Schmidtchen A, Frick IM, Andersson E, Tapper H, Björck L. Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol Microbiol. 2002;46:157–168. doi: 10.1046/j.1365-2958.2002.03146.x. [DOI] [PubMed] [Google Scholar]
- 5.Frick IM, Åkesson P, Rasmussen M, Schmidtchen A, Björck L. SIC, a secreted protein of Streptococcus pyogenes that inactivates antibacterial peptides. J Biol Chem. 2003;278:16561–16566. doi: 10.1074/jbc.M301995200. [DOI] [PubMed] [Google Scholar]
- 6.Hollands A, Gonzalez D, Leire E, Donald C, Gallo RL, Sanderson-Smith M, Dorrestein PC, Nizet V. A bacterial pathogen co-opts host plasmin to resist killing by cathelicidin antimicrobial peptides. J Biol Chem. 2012;287:40891–40897. doi: 10.1074/jbc.M112.404582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lauth X, von Kockritz-Blickwede M, McNamara CW, Myskowski S, Zinkernagel AS, Beall B, Ghosh P, Gallo RL, Nizet V. M1 protein allows group A streptococcal survival in phagocyte extracellular traps through cathelicidin inhibition. J Innate Immun. 2009;1:202–214. doi: 10.1159/000203645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durr UH, Sudheendra US, Ramamoorthy A. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim Biophys Acta. 2006;1758:1408–1425. doi: 10.1016/j.bbamem.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 9.Cederlund A, Gudmundsson GH, Agerberth B. Antimicrobial peptides important in innate immunity. FEBS J. 2011;278:3942–3951. doi: 10.1111/j.1742-4658.2011.08302.x. [DOI] [PubMed] [Google Scholar]
- 10.Dorschner RA, Pestonjamasp VK, Tamakuwala S, Ohtake T, Rudisill J, Nizet V, Agerberth B, Gudmundsson GH, Gallo RL. Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group a streptococcus. J Invest Dermatol. 2001;117:91–97. doi: 10.1046/j.1523-1747.2001.01340.x. [DOI] [PubMed] [Google Scholar]
- 11.Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, Pestonjamasp V, Piraino J, Huttner K, Gallo RL. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- 12.Johansson L, Thulin P, Sendi P, Hertzen E, Linder A, Åkesson P, Low DE, Agerberth B, Norrby-Teglund A. Cathelicidin LL-37 in severe Streptococcus pyogenes soft tissue infections in humans. Infect Immun. 2008;76:3399–3404. doi: 10.1128/IAI.01392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gryllos I, Tran-Winkler HJ, Cheng MF, Chung H, Bolcome R, 3rd, Lu W, Lehrer RI, Wessels MR. Induction of group A streptococcus virulence by a human antimicrobial peptide. Proc Natl Acad Sci USA. 2008;105:16755–16760. doi: 10.1073/pnas.0803815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran-Winkler HJ, Love JF, Gryllos I, Wessels MR. Signal transduction through csrRS confers an invasive phenotype in group a streptococcus. PLoS Pathog. 2011;7:e1002361. doi: 10.1371/journal.ppat.1002361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Velarde JJ, Ashbaugh M, Wessels MR. The human antimicrobial peptide LL-37 binds directly to csrS, a sensor histidine kinase of group A streptococcus, to activate expression of virulence factors. J Biol Chem. 2014;289:36315–36324. doi: 10.1074/jbc.M114.605394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatellier S, Ihendyane N, Kansal RG, Khambaty F, Basma H, Norrby-Teglund A, Low DE, McGeer A, Kotb M. Genetic relatedness and superantigen expression in group A streptococcus serotype M1 isolates from patients with severe and nonsevere invasive diseases. Infect Immun. 2000;68:3523–3534. doi: 10.1128/iai.68.6.3523-3534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kansal RG, Datta V, Aziz RK, Abdeltawab NF, Rowe S, Kotb M. Dissection of the molecular basis for hypervirulence of an in vivo-selected phenotype of the widely disseminated M1T1 strain of group A streptococcus bacteria. J Infect Dis. 2010;201:855–865. doi: 10.1086/651019. [DOI] [PubMed] [Google Scholar]
- 18.Zimmerlein B, Park HS, Li S, Podbielski A, Cleary PP. The M protein is dispensable for maturation of streptococcal cysteine protease speB. Infect Immun. 2005;73:859–864. doi: 10.1128/IAI.73.2.859-864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugareva V, Arlt R, Fiedler T, Riani C, Podbielski A, Kreikemeyer B. Serotype- and strain- dependent contribution of the sensor kinase covS of the covRS two-component system to Streptococcus pyogenes pathogenesis. BMC Microbiol. 2010;10:34. doi: 10.1186/1471-2180-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansson L, Linner A, Sunden-Cullberg J, Haggar A, Herwald H, Lore K, Treutiger CJ, Norrby-Teglund A. Neutrophil-derived hyperresistinemia in severe acute streptococcal infections. J Immunol. 2009;183:4047–4054. doi: 10.4049/jimmunol.0901541. [DOI] [PubMed] [Google Scholar]
- 21.Bergman P, Johansson L, Asp V, Plant L, Gudmundsson GH, Jonsson AB, Agerberth B. Neisseria gonorrhoeae downregulates expression of the human antimicrobial peptide LL-37. Cell Microbiol. 2005;7:1009–1017. doi: 10.1111/j.1462-5822.2005.00530.x. [DOI] [PubMed] [Google Scholar]
- 22.Nilsson M, Sorensen OE, Mörgelin M, Weineisen M, Sjöbring U, Herwald H. Activation of human polymorphonuclear neutrophils by streptolysin O from Streptococcus pyogenes leads to the release of proinflammatory mediators. Thromb Haemost. 2006;95:982–990. doi: 10.1160/TH05-08-0572. [DOI] [PubMed] [Google Scholar]
- 23.Nijnik A, Hancock RE. The roles of cathelicidin ll-37 in immune defences and novel clinical applications. Curr Opin Hematol. 2009;16:41–47. doi: 10.1097/moh.0b013e32831ac517. [DOI] [PubMed] [Google Scholar]
- 24.Johansson L, Thulin P, Low DE, Norrby- Teglund., A Getting under the skin: the immunopathogenesis of Streptococcus pyogenes deep tissue infections. Clin Infect Dis. 2010;51:58–65. doi: 10.1086/653116. [DOI] [PubMed] [Google Scholar]
- 25.Kaparakis-Liaskos M, Ferrero RL. Immune modulation by bacterial outer membrane vesicles. Nature Rev Immunol. 2015;15:375–387. doi: 10.1038/nri3837. [DOI] [PubMed] [Google Scholar]
- 26.Kuehn MJ, Kesty NC. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 2005;19:2645–2655. doi: 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- 27.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Ann Rev Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee EY, Choi DY, Kim DK, Kim JW, Park JO, Kim S, Kim S, Desiderio DM, Kim YK, Kim KP, Gho YS. Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics. 2009;9:5425–5436. doi: 10.1002/pmic.200900338. [DOI] [PubMed] [Google Scholar]
- 29.Rivera J, Cordero RJ, Nakouzi AS, Frases S, Nicola A, Casadevall A. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc Natl Acad Sci USA. 2010;107:19002–19007. doi: 10.1073/pnas.1008843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Athman JJ, Wang Y, McDonald DJ, Boom WH, Harding CV, Wearsch PA. Bacterial membrane vesicles mediate the release of Mycobacterium tuberculosis lipoglycans and lipoproteins from infected macrophages. J Immunol. 2015;195:1044–1053. doi: 10.4049/jimmunol.1402894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marsollier L, Brodin P, Jackson M, Korduláková J, Tafelmeyer P, Carbonnelle E, Aubry J, Milon G, Legras P, André JP, Leroy C, Cottin J, Guillou ML, Reysset G, Cole ST. Impact of Mycobacterium ulcerans biofilm on transmissibility to ecological niches and Buruli ulcer pathogenesis. PLoS Pathog. 2007;3:e62. doi: 10.1371/journal.ppat.0030062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olaya-Abril A, Prados-Rosales R, McConnell MJ, Martin-Pena R, Gonzalez-Reyes JA, Jimenez-Munguia I, Gomez-Gascon L, Fernandez J, Luque-Garcia JL, Garcia-Lidon C, Estevez H, Pachon J, Obando I, Casadevall A, Pirofski LA, Rodriguez-Ortega MJ. Characterization of protective extracellular membrane-derived vesicles produced by Streptococcus pneumoniae. J Proteom. 2014;106:46–60. doi: 10.1016/j.jprot.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 33.Dutta S, Iida K, Takade A, Meno Y, Nair GB, Yoshida S. Release of Shiga toxin by membrane vesicles in Shigella dysenteriae serotype 1 strains and in vitro effects of antimicrobials on toxin production and release. Microbiol Immunol. 2004;48:965–969. doi: 10.1111/j.1348-0421.2004.tb03626.x. [DOI] [PubMed] [Google Scholar]
- 34.Kadurugamuwa JL, Beveridge TJ. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol. 1995;177:3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campo N, Tjalsma H, Buist G, Stepniak D, Meijer M, Veenhuis M, Westermann M, Muller JP, Bron S, Kok J, Kuipers OP, Jongbloed JD. Subcellular sites for bacterial protein export. Mol Microbiol. 2004;53:1583–1599. doi: 10.1111/j.1365-2958.2004.04278.x. [DOI] [PubMed] [Google Scholar]
- 36.Hu P, Bian Z, Fan M, Huang M, Zhang P. Sec translocase and sortase A are colocalised in a locus in the cytoplasmic membrane of Streptococcus mutans. Arch Oral Biol. 2008;53:150–154. doi: 10.1016/j.archoralbio.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Rosch J, Caparon M. A microdomain for protein secretion in Gram-positive bacteria. Science. 2004;304:1513–1515. doi: 10.1126/science.1097404. [DOI] [PubMed] [Google Scholar]
- 38.Rosch JW, Caparon MG. The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol Microbiol. 2005;58:959–968. doi: 10.1111/j.1365-2958.2005.04887.x. [DOI] [PubMed] [Google Scholar]
- 39.Rosch JW, Hsu FF, Caparon MG. Anionic lipids enriched at the ExPortal of Streptococcus pyogenes. J Bacteriol. 2007;189:801–806. doi: 10.1128/JB.01549-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vega LA, Caparon MG. Cationic antimicrobial peptides disrupt the Streptococcus pyogenes ExPortal. Mol Microbiol. 2012;85:1119–1132. doi: 10.1111/j.1365-2958.2012.08163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biagini M, Garibaldi M, Aprea S, Pezzicoli A, Doro F, Becherelli M, Taddei AR, Tani C, Tavarini S, Mora M, Teti G, D'Oro U, Nuti S, Soriani M, Margarit I, Rappuoli R, Grandi G, Norais N. The human pathogen Streptococcus pyogenes releases lipoproteins as lipoprotein-rich membrane vesicles. Mol Cell Proteom. 2015;14:2138–2149. doi: 10.1074/mcp.M114.045880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data