Abstract
Innate immunity is a rapidly evolving field with novel cell types and molecular pathways being discovered and paradigms changing continuously. Innate and adaptive immune responses are traditionally viewed as separate from each other, but emerging evidence suggests that they overlap and mutually interact. Recently discovered cell types, particularly innate lymphoid cells and myeloid-derived suppressor cells, are gaining increasing attention. Here, we summarize and highlight current concepts in the field, focusing on innate immune cells as well as the inflammasome and DNA sensing which appear to be critical for the activation and orchestration of innate immunity, and may provide novel therapeutic opportunities for treating autoimmune, autoinflammatory, and infectious diseases.
Keywords: Host defense, Immune response, Neutrophils, Pattern recognition receptors, Phagocytosis, Toll-like receptor, Innate Immunity, Myeloid-derived suppressor cells, Innate lymphoid cells, Macrophages
Innate Immunity
The innate immune system is an evolutionarily conserved host defense system with key features being shared between plants, invertebrates, and mammals [1, 2]. Innate immune defenses in mammals encompass virtually all tissues, particularly barrier surfaces such as the skin or the mucosal surfaces of the respiratory and gastrointestinal tract. Specialized myeloid and lymphoid sensor and effector cells [3], but also nonhematopoietic cells, can initiate and exert innate defense mechanisms and become activated in response to tissue damage, infection, or genotoxic stress. The innate immune system can “sense” such situations through germline-encoded receptors (e.g. pattern recognition receptors [PRRs] such as toll-like receptors [TLRs]). Innate immune responses can be mediated through cell-dependent mechanisms (e.g. phagocytosis and cytotoxicity) or secreted factors, including antimicrobial peptides (AMPs) [4, 5, 6], complement factors [7, 8, 9], alarmins [10, 11], cytokines/chemokines [12], chitinases/chitinase-like proteins [13], acute-phase proteins, proteases, and other less-categorized molecules. Innate immune responses are typically rapid and can be triggered without the selective events that underlie adaptive immunity, which is characterized by antigen-specificity and immunological memory. In contrast, innate responses are traditionally described as lacking in memory, and instead become activated gradually, depending on the PRR activation of the respective microbial threat independent of previous exposure to pathogens. However, this paradigm has been recently challenged with the concept of “trained immunity” [14], and the demonstration of memory responses of natural killer (NK) cells and innate lymphoid cells (ILCs) [[15, 16, 17]; for a review, see [18]]. At host-environment contact interfaces, such as the skin, the gut or the airways, microbial stimuli initially elicit the secretion of generic antimicrobial peptides (AMPs, such as defensins or cathelicidin/LL-37) as well as organ-specific mediators (such as dermcidin for the skin [19, 20]). Innate immune PRRs, prototypically TLRs [21, 22], expressed by resident (epithelial) or recruited (hematopoietic) cells, sense pathogen-associated molecular patterns (PAMPs) or, in the case of nonpathogenic microbes, more broadly termed “microbe-associated molecular patterns” (MAMPs), and trigger downstream effector programs. Besides TLRs, other innate PRRs include NOD-like receptors, complement receptors, scavenger receptors (e.g. CD36, MARCO, SR-A, LOX-1, and dSR-C), intracellular nucleic acid-sensing receptors (e.g. AIM2, MDA-5, RIG-I, and cyclic GMP-AMP synthetase [cGAS]) and C-type lectin receptors (CLRs, such as mannose-binding proteins, dectin-1, dectin-2, and DC-SIGN) [23, 24, 25, 26, 27]. Upon microbial exposure, these receptors induce the secretion of cytokines and chemoattractants, such as IL-8 (CXCL8). Attracted by chemokine gradients, innate immune cells migrate into the infected target organ [28]. Chemokines recruit innate immune cells through cognate G-protein-coupled chemokine receptors to sites of inflammation, and are termed according to their first cysteine residues into C, C-C, C-X-C, and CX3C chemokines, with C-C and C-X-C as the largest families [12, 29]. Chemokines can be further classified as homeostatic and inflammatory chemokines, depending on whether they play a role in basal homeostatic immune-cell trafficking or in inflammation. Neutrophils represent the earliest innate immune cells recruited to the site of inflammation through chemokine gradients, followed by monocytes and dendritic cells (DCs), which can then interact with tissue-resident myeloid and lymphoid cells. In this review, we will provide an update on the key cell types of the innate immune system. In addition, we discuss the current concepts of the mechanisms of DNA sensing and the function of inflammasomes, which appear to be critical for the initiation and orchestration of multicellular innate immune responses.
Innate Immune Cells
Innate immune cells comprise a broad and expanding range of myeloid and lymphoid cell types. Common to the majority of these cell types is that they originate from the hematopoietic system (with exceptions, e.g. epithelial cells), lack somatically recombined antigen-receptors and conventional immunological memory (see above and [14]), and exert antimicrobial or tissue-protective functions. In the following section, we will review our current understanding of the development and functions of (i) neutrophils, (ii) macrophages, (iii) myeloid-derived suppressor cells (MDSCs), and (iv) ILCs.
Neutrophils
Traditionally, neutrophils are regarded as short-lived and terminally differentiated phagocytes without considerable gene expression and lacking regulatory roles in adaptive immunity. However, these classical views of neutrophil biology and functions have recently been challenged by several observations in the field [30]: (i) in vivo tracing in healthy volunteers revealed an average circulatory neutrophil lifespan of 5.4 days, at least 10 times longer than previously reported [31]; (ii) neutrophils were found to extrude their own nuclear or mitochondrial DNA as neutrophil extracellular traps (NETs), a phenomenon termed “beneficial suicide” [32]; (iii) neutrophils can act as MDSCs [33], thereby suppressing adaptive T cell functionalities, such as T cell proliferation or cytokine production; (iv) neutrophils move in the interstitial space in swarm-like formations [34, 35], and (v) neutrophil ageing is modulated by the gut microbiome [36]. Collectively, these novel findings raise a variety of questions about neutrophil functions, heterogeneity, plasticity [30], and interspecies differences.
The fastest response of neutrophils to microbial exposure is phagocytosis, which occurs within minutes and is frequently followed by phagocytosis-induced cell death. While opsonization and phagocytosis are primarily relevant for the engulfment of smaller bacteria, larger microbes, including bacteria and fungi, elicit neutrophilic granule release. Granule release occurs sequentially in neutrophils [37]: first, secretory vesicles which contain surface receptors are mobilized, followed by the release of tertiary granules containing matrix metalloproteases to facilitate migration through the extracellular matrix. Secondary and primary granules are mainly extruded in response to strong sterile or infectious stimuli leading to the release of AMPs and proteases that can degrade bacterial and fungal proteins efficiently. After prolonged pathogen contact, neutrophils undergo specific forms of cell death, including apoptosis, necroptosis, or NET formation [38, 39]. These distinct neutrophil fates shape the further process of innate immune cell activation.
NET formation was initially described as an extracellular form of antimicrobial host defense against bacteria [40]. Emerging evidence indicates that the relevance of NETosis reaches far beyond microbial killing to autoimmune/rheumatic and autoinflammatory disease conditions [41, 42, 43]. NETs contain DNA, histones, and distinct granule proteins, which can be used as markers for NET formation. Several aspects of NETosis, however, remain enigmatic and are under controversial discussion in the field [44]: (i) Are mainly nuclear or also mitochondrial NETs released? [45] (ii) Is NET formation another form of cell death, or is it, as recently suggested by mainly murine studies, also executed by living cells? [46, 47, 48] (iii) Do extruded NETs really kill microbes or just immobilize them? (iv) Besides microbial size [49], which criteria determine whether neutrophils form NETs?
When viewing murine and human data in combination, the results obtained on NET formation seem to critically depend on the species as well as the assays and the quantitative read-outs used. Simply measuring free DNA is, however, unable to distinguish NET-derived DNA from necrosis-derived DNA. In vivo imaging studies are essential to understand the kinetics and biodistribution of neutrophils in the living body, but frequently lack high-resolution proof of bona fide NET strands. Another layer of complexity is added by interspecies differences. While murine neutrophils were initially reported to generate NETs after a substantially longer period of stimulation compared to their human counterparts [50], recent studies on live-cell NET formation challenge this concept and suggest that neutrophils can simultaneously chemotax, perform NET formation and phagocytosis in a collaborative manner [47, 51]. When viewed in combination, the mechanisms underlying NET formation and the pathophysiological relevance for human disease conditions remain complex and multifaceted and need to be defined in future studies.
Macrophages
More than 100 years ago, the Nobel Prize laureate Elie Metchnikoff described macrophages as “the phagocytic component of the immune system.” In the last century, these cells were fundamental to the understanding of the basic principles of the innate immune response and host defense, with their canonical functions including the phagocytosis of microorganisms, the engulfment of apoptotic/dead cells, and the production of inflammatory cytokines. However, it is now well accepted that macrophages have several additional key functions. They continuously scan the tissue in which they reside, and actively participate in maintaining homeostasis and integrity [52]. Due to this key role, it is not surprising that abnormal macrophage behavior has been implicated in the pathophysiology of several human disease conditions, including cancer, atherosclerosis, inflammatory bowel disease, rheumatoid arthritis, fibrosis, neurodegenerative disorders, and chronic inflammatory lung diseases (e.g. asthma, COPD, cystic fibrosis, and fibrosis) [53, 54, 55]. The variety of plasma membrane and intracellular receptors expressed by distinct subsets of macrophages explains their capacity to sense the surrounding environment and respond promptly to environmental cues (e.g. MAMPs/PAMPs/microorganisms, DAMPs, pH, and oxygen concentrations) [52, 55]. Once activated, macrophages are armed with several mechanisms to negatively regulate the innate immune response, triggering anti-inflammatory pathways and facilitating the phagocytosis of apoptotic cells, events required to reestablish tissue homeostasis [56, 57]. Furthermore, macrophages coordinate the stress response through crosstalk with other neighboring cell types in the surrounding tissue. Alveolar macrophages, for example, abundantly secrete cytokines, chemokines, and growth factors, as well as microvesicles containing anti-inflammatory mediators (e.g. SOCS1) [58]; this ensures a rapid and effective paracrine communication with epithelial, stromal, dendritic, and T regulatory cells in the pulmonary environment [59, 60]. Macrophages also establish direct cell-to-cell contacts through receptors and gap junction formation with the surrounding respiratory epithelium. This enables the distribution of specific signals throughout the tissue and helps to coordinate the tissue response to insults and injury [61, 62].
It is estimated that humans have approximately 0.2 trillion macrophages throughout the body which can be identified in almost every tissue compartment [63]. The tissue-resident macrophages have a slower turnover rate under steady-state conditions, but this population rapidly and dynamically changes during tissue stress (e.g. infection). The conventional concept is that tissue-resident macrophages originate from blood-circulating monocytes, which arise from a myeloid-committed precursor in adult bone marrow (BM). However, this traditional concept was recently challenged. The current revised understanding is that distinct tissue-resident macrophages (e.g. in the lungs, liver, and brain) are established prior to birth during the embryonic and fetal waves of hematopoiesis. Even more striking, it is now clear that these macrophages are capable of self-renewing themselves inside the tissue. However, if the local population is completely depleted (e.g. by irradiation), recruited circulating monocytes can seed the tissues and adopt a resident macrophage phenotype [64]. The self-renewal capability is not conserved by all macrophages. In fact, macrophages derived from the intestine, pancreas, dermis, and heart seem to be continuously replaced by circulating monocytes (circulating Ly6Chi monocytes) in a CCR2-dependent manner (review [65, 66]). Macrophages from different tissues have a well-defined epigenetic landscape and tissue-specific transcriptional profiles [67]. The epigenetic “imprinting” of tissue-resident macrophages is mainly programmed by the surrounding environment. If macrophages derived from the yolk sac, fetal liver, or adult BM are transferred into the lung alveolar space, they are reprogrammed to express alveolar-macrophage-specific sets of genes. However, once macrophages have been fully committed by their environment (e.g. Kupffer cells in the liver), they lose this “plasticity” and can no longer acquire a lung signature when exposed to the lung niche [64]. Tissue macrophage populations evolve continuously during the life of an organism. Each time they are exposed to stress, tissues are rapidly populated by waves of circulating monocytes [68]. These monocytes are first involved in mounting a robust proinflammatory response to fight the microorganism and then, in a time-dependent manner, they give rise to macrophages with high scavenging and anti-inflammatory capabilities. These macrophages will then facilitate the resolution of the inflammatory response, together with tissue repair and regeneration. Interestingly, a recent study suggests that circulating monocytes may not be the only source of macrophages during tissue stress. In fact, peritoneal-cavity macrophages also serve as a reservoir for mature macrophages in a murine model of sterile inflammation of the liver [69].
During stress, the local tissue microenvironment undergoes dynamic changes, including the presence of PAMPs, cytokines, growth factors, and alarmins. These changes dictate and orchestrate the phenotypic and functional adaptation of both recruited and resident macrophages. If the activation and priming processes for these cells are not controlled effectively, then persistent inflammation and aberrant repair processes can lead to tissue damage and fibrosis [70]. In in vitro settings, macrophages have been classified into 2 distinct main subphenotypes. M1 (or classically activated macrophages) are primed by Th1 cytokines, e.g. interferon (IFN)-γ and bacterial products. M2 (or alternatively activated macrophages) are primed by Th2 cytokines (e.g. IL-4 and IL-13). M1 macrophages are primarily relevant for antibacterial defense, while M2 macrophages are involved in anti-inflammatory, allergic, and tissue repair processes. However, this simplification of macrophage plasticity does not translate to in vivo studies, where macrophages are likely exposed to a cocktail of Th1 and Th2 cytokines and microbial products, at various concentrations and in a dynamic fashion [71, 72]. The currently emerging concept is that macrophage activation and priming gives rise to a continuous spectrum of phenotypes rather than a few distinct subsets, implying that different phenotypes are not mutually exclusive [73]. Moreover, comprehensive genomic studies have revealed an epigenetic mechanism by which macrophages fine-tune gene expression during activation and priming. Macrophages have also been shown to retain a certain degree of epigenetic plasticity after activation [74, 75]. Importantly, researchers have revised the conventional belief that, as part of the innate immune system, macrophages have only an unspecific response and do not acquire “memory.” Instead, macrophages exposed to bacterial products undergo epigenetic programming that establishes an “innate immune memory,” which will ultimately shapes the organism's immune response to subsequent external insults [76]. Given the key role of macrophages in maintaining tissue health under steady-state and stress conditions, strategies to target these cells may have potential for treating several diseases. Thanks to this prospect, enormous efforts are being dedicated to a better understanding of the complex biology of these fascinating cells of the innate immune system.
Myeloid-Derived Suppressor Cells
MDSCs are defined as cells of myeloid origin that suppress T cell responses. Phenotypically, MDSCs can be subdivided into neutrophilic/granulocytic and monocytic MDSCs [33, 77, 78]. In mice, both subsets can be phenotypically classified based on CD11b expression and Ly6C (monocytic) versus Ly6G (neutrophilic) surface markers, whereas the distinction in the human system is less precise. Both monocytic and neutrophilic/granulocytic MDSCs in humans express the myeloid markers CD11b and CD33. Human monocytic MDSCs are CD14-positive and have been described as expressing low amounts of/no MHC-II, while neutrophilic/granulocytic MDSCs are CD14-negative/low in CD14, express CD15 and CD66b, and have been described as having a lower-density gradient centrifugations (i.e. “low-density neutrophils/granulocytes” compared to their conventional “high-density” neutrophilic counterparts) [79, 80]. Since these surface markers are not restricted to MDSCs, having also been found on other immune cells, their bona fide distinction from other cells requires functional assays that demonstrate that MDSCs suppress T cell responses. For a more in-depth discussion of the MDSC nomenclature, phenotypes, and functionalities, we refer to a recently published review [81]. Controversy still exists around the hematopoietic lineage origin of MDSCs, particularly neutrophilic/granulocytic MDSCs. Are these merely immature neutrophils? Or postactivated neutrophils, as supported by the high expression of the secondary granule marker CD66b on their cell surface? Or do neutrophilic/granulocytic MDSCs represent an early distinct subtype of neutrophilic myeloid cells? For human MDSCs in particular, these questions remain to be answered in the future, possibly utilizing myeloid lineage tracing technologies.
A further aspect of MDSCs that needs to be defined is their precise cellular suppressive effector mode of action. While murine studies have involved a plethora of mechanisms and pathways underlying the generation of and suppressive functionalities of MDSCs, including GM-CSF, IL-6, signal transducer and activator of transcription 3 (STAT3), indole amine 2,3 dioxygenase (IDO), calcium-binding S100 proteins, IL-1β, high-mobility group box 1 (HMGB1), IL-6, arginase-1, inducible nitric oxide synthase (iNOS), and the production of nitric oxide (NO), reactive oxygen species (ROS), TNF-α, hypoxia-inducible factor 1 α (HIF-1α) (summarized in recent reviews specifically dedicated to MDSCs [78, 79, 82, 83, 84]), evidence of these putative mechanisms in humans is scarce. Initially only described as dampening T cell proliferation, follow-up studies extended this view by showing that MDSCs can also regulate NK, NKT, DC, and neutrophil responses. Thus, MDSCs can be regarded as a key type of innate immune cell that dampens the activation and function of adaptive (T cells) and innate (NK cells) immune cells. The therapeutic modulation of the generation of or effector functions of MDSCs could pave the way for novel approaches for shaping specific immune responses in cancer, infections, and autoimmune disorders. In cancer and certain types of infection [85], MDSCs could be targeted to empower T cell host defense (similar to the anti-CTLA-4/anti-PD-1 checkpoint blockade in cancer immunotherapies). Conversely, the induction of immunosuppressive MDSC activities could be harnessed to dampen overshooting autoimmune responses.
Innate Lymphoid Cells
In addition to the innate myeloid cells described above, many mucosal tissues harbor ILCs. These lineage-negative CD127+CD90+ cells can execute functional and transcriptional programs that were originally described in the context of adaptive Th cells [18, 86], but are considered “innate” with regard to their lack of somatically recombined antigen receptors. Secretion of the “helper” cytokines IFN-γ, IL-13 or IL-17, in a manner dependent on T-bet, GATA-3 or RORγt, respectively, led to the classification of ILCs. Group 1 ILCs consist of Tbet+ ILC1 and Tbet+ Eomes+ NK cells. GATA-3hi ILC2 produces IL-4, IL-5, and IL-13. Group 3 ILCs consist of RORγt+ IL-17- and IL-22-producing subsets of ILC3, as well as lymphoid tissue inducer cells (LTi) [86]. NF-κB-activating IL-1 family cytokine members, such as IL-1β, IL-18 or IL-33, in combination with STAT signaling pathways engaged via IL-23, IL-12, IFN-α/β or TSLP, efficiently trigger the production of “helper” cytokines in ILCs. Because adaptive and innate-like T cells can also execute these functions in both a cognate and noncognate manner, it is important to examine the redundant versus specific functions of ILCs [18, 87]. The major unanswered questions in the field pertain to nonredundant, activating, and inhibitory receptors that may regulate specific ILC functions. For example, it has been proposed that NKp46, an activating receptor expressed on NK cells, ILC1, and subsets of ILC3, plays a critical role in the sensing of adipose tissue “stress” [88]. One emerging theme is that ILCs interact with both local hematopoietic cells and nonhematopoietic stromal and epithelial cells, thereby contributing to the physiologic mechanisms of tissue homeostasis, metabolism, epithelial repair, and barrier function, e.g. through the secretion of amphiregulin, IL-4/IL-13, and IL-22 [89, 90, 91, 92] and reciprocal interactions with local macrophages [88, 93, 94]. Consistent with the idea of ILCs being local sentinels and keepers of tissue homeostasis, these cells are found most prominently in nonlymphoid tissues and at mucosal sites, where they exist as tissue-resident cells that may expand locally during acute inflammation [95, 96]. While these observations suggest that ILCs may self-renew within tissues, ILC progenitors in adult BM can reconstitute the ILC compartment upon transplantation and likely contribute to the regeneration of ILCs, e.g. during chronic inflammation [95, 97, 98]. Distinct from T cells, which exist in lymphoid organs as naïve cells and need to be “primed” to differentiate into the various helper subsets that gain effector function and can then locate to specific nonlymphoid tissues, it appears that ILCs acquire effector function and tissue localization developmentally [18, 99].
Interestingly, ILCs exhibit functional and developmental plasticity [100], which may have evolved as a mechanism to rapidly adopt the functions of resident lymphocyte pools without requiring the recruitment or differentiation of additional cells. These differences in acquisition of effector function and tissue localization raise the possibility that ILCs have critical functions early during immune responses, but also early in life. ILCs (similar to subsets of innate-like T cells) seed nonlymphoid organs early during ontogeny, and could then be the dominant providers of “helper” cytokines, while adoptive T cells will complement these peripheral lymphocyte pools later in life, raising the possibility of extensive collaboration and interactions between innate and adaptive lymphocytes [101, 102]. T cells may activate or regulate ILCs, e.g. by modulating the availability of IL-2 [103, 104]. Vice versa, it has been suggested that ILCs play a role in the primary and recall responses of adaptive T cells [105, 106, 107]. Interestingly, ILCs may also participate in the regulation of T cells through mechanisms reminiscent of myeloid or thymic epithelial cells. ILC3 can phagocytose and process antigens for MHC-II-mediated presentation, and mediate the negative selection of commensal bacteria-specific CD4+ T cells, preventing severe intestinal pathology in mice [102, 108]. Whether the phagocytic activity of ILCs is linked to pattern recognition or innate sensing is a matter currently under investigation. It is also not clear whether ILCs may contribute more generally to antigen presentation and T cell regulation, and where antigen-dependent interactions with T cells occur. Interestingly, ILCs have been identified in the BM and the secondary lymphoid organs, and have been proposed to migrate from the intestine to the mesenteric lymph nodes [109]. Recent demonstrations of memory-like features of NK cells [16, 17] and ILC2 [15] raise the possibility that the pool of ILCs is fundamentally altered through exposure to environmental challenges. A major question is whether ILCs, if long-lived, acquire epigenetic modifications consistent with the concept of “trained immunity” [76] or exhibit adaptive-like features of specificity as described for NK cells [16, 17]. Future studies will address how the functions of ILCs and their ability to modulate tissue homeostasis and innate and adaptive immunity can be targeted to treat inflammatory diseases. For additional discussion of ILCs, we refer to recently published in-depth reviews [100, 110, 111, 112].
Inflammasomes and the Activation of Innate Immunity
Inflammasomes are macromolecular platforms for the recruitment and activation of inflammatory caspases in the context of stress or danger signals [113]. Several distinct inflammasomes have been identified accordingly to the unique sensor molecule responsible for detecting a specific trigger (PAMPs or DAMPs). These intracellular receptors belong to the nucleotide-binding domain leucine-rich repeat containing (NLR) protein family or to AIM2-like receptor (ALR) members [114, 115]. These proteins have an N-terminal pyrin domain (PYD) in common, required for homotypic interaction with the PYD domain of the bipartite adaptor ASC, which contains a CARD domain essential for the recruitment of inactive caspases [116]. In particular, inflammatory caspase-1 is responsible for cleaving the immature pro-IL-1β and pro-IL-18 into their bioactive forms before they can be released outside the cells [113]. In addition, active caspase-1 triggers pyroptosis, an inflammatory form of cell death characterized by the rupture of cell membranes and the release of cytoplasmic contents [117, 118]. Human inflammatory caspase-4 and caspase-5 (corresponding to caspase-11 in mice) can trigger pyroptosis when engaged by direct binding of intracellular LPS to form a so called “non-canonical inflammasome” [119, 120]. Recently, the N-terminal-cleaved fragment of the intracellular protein gasdermin D was identified as a crucial driver of pyroptosis generated upon the activation of inflammatory caspases, but the cellular mechanism of death requires further investigation [121, 122]. Other noninflammatory caspases have been described in the context of inflammasome activation. In addition to apoptosis, caspase-8 can directly control IL-1β processing in the absence of caspase-1 or also modulate the expression of inflammasome components through a not well understood mechanism [123].
Among different NLR sensors, NLRP3 is probably the most well characterized inflammasome [124]. NLRP3 inflammasome activation has been considered to follow a 2-signal model: the first signal, also called the priming step, upregulates immature pro-IL-1β and pro-IL-18, together with NLRP3 protein. This process is typically regulated by LPS-mediated activation of the TLR4-MyD88-NF-κB pathway. Signal 1-induced deubiquitination and dephosphorylation of NLRP3 by BRCC3 and PTPN22, respectively, have been proposed to be necessary for the licensing of NLRP3 [125, 126], and Ca2+-sensitive cAMP has been shown to act as an intracellular inhibitor of NLRP3 [127]. The second signal is less defined, but involves potassium efflux, calcium mobilization from intracellular store compartments, and the production of ROS [124]. A novel “alternative inflammasome” pathway which does not require 2 signals has been recently described in human and porcine primary monocytic cells [128]. In these cells, engagement of TLR4 by extracellular LPS triggered IL-1β secretion without pyroptosis or the need for “signal 2.” This pathway, in addition to NLRP3, ASC and caspase-1, requires TRIF-, RIPK1-, and FADD-dependent caspase-8 activation and, interestingly, is not conserved in mice [129]. This pathway is distinct to the described intracellular sensing pathway for LPS by caspase-11/-4/-5 mechanism which also leads to inflammasome formation [122, 130]. How the different pathways, especially those upstream of NLRP3, relating to the activation of the NLRP3 inflammasome are explained on the molecular level, i.e. by involving additional binding partners, has remained largely elusive.
Recent discoveries relating to NLRP3 biology include Bruton's tyrosine kinase [131] and NEK7 [132], findings which may help to further unlock the molecular functioning of the NLRP3 inflammasome. Recent studies further highlight that reprogramming or perturbation of distinct metabolic pathways can trigger inflammasome activation, representing an emerging research area in the field of immunometabolism [133, 134, 135, 136, 137, 138]. Interestingly, while located in the cytoplasm of myeloid cells, NLRP3 can localize in the nucleus of Th2 cells, where it has been shown to act as a transcription factor essential for Th2 polarization and, in a complex with IRF4, to promote IL-4 production, relevant for asthma and melanoma [139].
Although the different inflammasomes and their components have been primarily been studied in myeloid lineages, their roles in lymphoid cells and nonhematopoietic cells are now beginning to emerge. In the epidermis, keratinocytes are the first nonimmune cells where NLRP3, NLRP1, and AIM2 inflammasomes expression and activity have been studied. UVB irradiation activates the NLRP3 inflammasome, with subsequent IL-1β secretion that is dependent on the release of calcium from intracellular stores [140]. In psoriasis, patients display increased concentrations of cytosolic DNA that activates the AIM2 inflammasome [141]. In the central nervous system, NLRP3 has also been shown to play a role. Inflammasome-deficient mice were protected from disease progression in experimental autoimmune encephalitis, a mouse model of multiple sclerosis [142]. More recently, T cell-intrinsic NLRP3-ASC activities have been shown to be important for caspase-8-mediated IL-1β secretion in the context of Th17-mediated neuroinflammation, extending the role of the inflammasome to the lymphoid cells [143].
Dysregulation of inflammasome activation drives pathological inflammation in the context of an increasing number of disease conditions. Prototypically, gain-of-function mutations in NLRP3 cause a human syndrome called CAPS (cryopyrin-associated periodic syndrome), characterized by spontaneous episodes of fever and sterile systemic autoinflammation in its milder form, but also manifesting with hearing loss and bone deformities in more severe cases [144]. In addition, inflammasomes contribute to the chronic inflammation typical of neurodegenerative diseases such as multiple sclerosis, Alzheimer disease, and Parkinson disease, and metabolic disorders including atherosclerosis, type 2 diabetes, and obesity, and therefore represent an excellent target for therapeutic approaches [145]. The clinical relevance of pyroptotic cell death has been demonstrated recently in the context of HIV infection where it drives CD4 T cell depletion in cells expressing chemokine CCR5 [146]. Recently, inflammasome activity was visualized in subcapsular sinus macrophages in vivo, which impacted neutrophils and NK cell recruitment in the draining lymph node upon infection with modified vaccinia Ankara (MVA) virus, a double-stranded DNA poxvirus evaluated as a recombinant vaccine vector [147]. These findings are consistent with the idea that inflammasome-dependent IL-18 production is critical for the orchestration of multicellular innate immune responses during viral or bacterial infection [148].
In summary, the recent discoveries of “non-canonical” and “alternative” inflammasome pathways have revitalized the field and added new layers of complexity to our understanding of extracellular versus intracellular, two-step versus one-step, and interspecies differences in inflammasome activation. A recently described inflammasome inhibitor, MCC950 [149], may aid the study of this interesting innate immune process in greater detail in vitro and in vivo.
DNA Sensing
Sensing of nucleic acids in different cellular compartments is a key mechanism of the innate immune system to mount an immune response against microbial pathogens. This phenomenon has been known for decades, but profound and significant progress in understanding its molecular mechanisms and potential disease implications has only recently been made. Recognition of nucleic acids induces the expression of IFN type I (IFN-I) and other inflammatory cytokines required for the host defense. In the endolysosomal compartment, TLRs recognize nucleic acids [150]. In the cytoplasm, dsRNA is recognized by MDA5, while RIG-I senses 5′ triphosphate RNA. Binding of these nucleic acid species leads to recruitment of MDA5 and RIG-I to mitochondrial MAVS and the downstream activation of IRF3, MAPK, and NF-κB [151]. Several cytosolic DNA sensors including IFI16, DIA, and DDX41 [152, 153, 154] have been described, but the mechanism of how these receptors mediate a DNA-dependent immune response is not completely understood and requires further investigation. dsDNA binds to AIM2 (absent in melanoma 2) in the cytoplasm, driving the activation of the inflammasome, the expression of IL-1β and IL-18, and the activation of pyroptosis in a caspase-1-dependent manner [155, 156].
Recently, it was discovered that cGAS is a major driver of IFN-I expression in response to dsDNA in the cytoplasm [157, 158]. Upon recognition of dsDNA, monomeric cGAS forms a dimer and undergoes a conformational change that allows the binding and enzymatic conversion of ATP and GTP into the second messenger, cyclic di-GMP-AMP (cGAMP) [159, 160, 161]. cGAMP then binds to STING which resides in the endoplasmatic reticulum. Interestingly, cGAS synthesizes cGAMP with unique 2′5′-3′5′ phosphodiester linkages and binds with higher affinity to STING than the bacterial cyclic dinucleotides that contain conventional phosphodiester linkages [160, 162, 163, 164]. Another interesting feature is the capability of cGAMP to bind to STING in neighboring cells after transfer via gap junctions, allowing antiviral responses in the cells in the absence of pathogen-derived nucleic acids [165]. In addition, cGAMP can be transferred through viral particles that deliver it to newly infected cells [166, 167]. cGAMP binding to STING dimers induces a conformational change, and STING then binds to TBK1 and translocates to the perinuclear Golgi region [168]. TBK1 phosphorylates STING, allowing the recruitment of IRF3 which, after binding to STING, gets phosphorylated and activated by TBK1 [169]. In addition, TBK1 also activates the NF-κB pathway [170].
Binding of DNA and activation of cGAS is sequence-independent, and a wide spectrum of viral/bacterial pathogens, but also endogenous DNA, induce IFN-I responses. Several DNA viruses including herpes simplex virus 1 (HSV-1), hepatitis B virus (HBV), vaccinia virus (VACV), adenovirus, and Kaposi's sarcoma-associated herpesvirus (KSHV) are recognized by the cGAS-STING pathway [171, 172, 173, 174, 175]. Bacterial infection with Mycobacterium tuberculosis, Listeria monocytogenes, Chlamydia trachomatis, and Francisella tularensis activates IFN-I expression that is dependent on cGAS activity and bacteria-produced cyclic dinucleotides that act directly on STING [176, 177, 178, 179, 180, 181, 182]. There is emerging evidence from monogenic interferonopathies and related mouse models that DNA sensing by the cGAS-STING pathway may be involved in the pathogenesis of autoinflammatory disorders. Mutations in the Trex1 gene, a 3′→5′ DNA-specific exonuclease that can clear the cytoplasm from self-DNA, have been identified in patients suffering from Aicardi-Goutières syndrome, who develop an inflammatory disorder with onset in early childhood, familial chilblain lupus, and systemic lupus erythematous [183]. In a mouse model of Aicardi-Goutières syndrome, Trex1 knockout mice developed severe multiorgan inflammation [184, 185, 186]. An additional deletion of either cGAS or STING prevented the induction of IFN-I, and the respective mice lacked signs of inflammation in different organs [184, 187, 188].
DNase II is a lysosomal DNase involved in the fragmentation of DNA of phagocytosed apoptotic cells. DNase-II-deficient mice are embryonic-lethal and become severely anemic during embryo development. This defect in erythropoiesis is a result of high IFN-I expression of macrophages unable to digest DNA from phagocytosed erythrocyte precursors [189, 190]. Consequently, mice that are deficient in IFNAR are rescued from DNase II deficiency-mediated lethality, but these mice develop chronic polyarthritis [191]. Lethality in DNase II knockout mice and anemia are prevented by the deletion of either cGAS or STING with protection from arthritis [187, 192]. Interestingly, STING-deficient mice crossed with lupus-prone MRL/Faslpr/lpr mice developed more severe disease [193]. This was characterized by higher levels of autoantibodies, increased expression of IFN-induced genes, accelerated mortality, and hyperresponsiveness to TLR signaling. The above-mentioned studies show the severe consequences of an uncontrolled activation of the IFN-I response mediated by the cGAS-STING pathway. Cells have adopted mechanisms to regulate the potent inflammatory IFN-I response, and 2 recent studies describe the inhibitory regulation of cGAS. Reversible glutamylation by tubulin tyrosine ligase-like (TTLL) glutamylases inhibits cGAS synthase and DNA-binding activity [194], and the phosphorylation of cGAS by Akt dampens its activity [195]. More regulatory mechanisms are likely to be discovered in the future.
In summary, DNA sensing by the cGAS-STING pathway is a potent inducer of IFN-I and other inflammatory cytokines. Therapeutically, cGAS and STING are interesting targets, and antagonizing cGAS or STING may allow the dampening of chronic inflammation in autoimmune diseases. Activating the cGAS-STING pathway may be beneficial in the context of infections and cancer.
Summary and Outlook
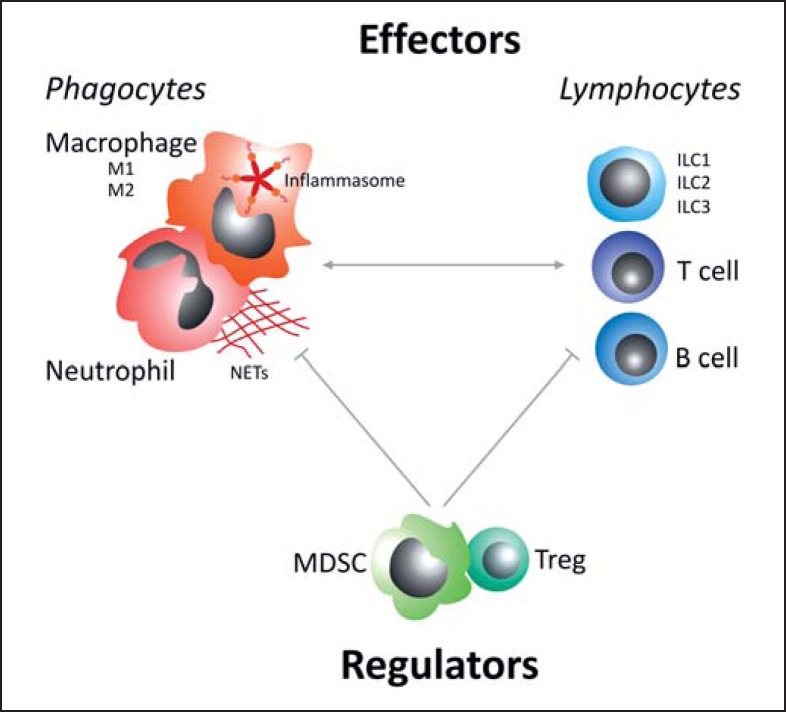
Our knowledge about innate immune cells and their functions is constantly evolving. Figure 1 summarizes key effector and counterregulatory immune-cell subsets. Of the phagocytes, macrophages are the principal cells generating inflammasome-derived proinflammatory cytokines, while neutrophils have a special potential to expel their own DNA, in the form of NETs, in order to capture and kill pathogens in the extracellular space. In addition to adaptive T and B cell subsets, lymphocytes encompass ILCs that belong to the innate immune system. In analogy to Th1/Th2, ILCs comprise at least 3 major groups of cells, termed ILC1, ILC2, and ILC3. The mutual interactions of these innate immune-cell types and other components of the immune system are still poorly understood [18]. While novel pathways and cell types have recently been discovered and studied in murine disease models, their role and therapeutic potential in human diseases remains largely to be defined. In this context, it will be important to understand how environmental factors, such as allergens and hazards, nutrition and lifestyle habits, and symbiotic microbiota shape the innate immune system. Several studies have demonstrated a close interaction between microbiota and innate immune-cell components [for reviews, see [196, 197, 198, 199, 200]]. The individual composition of the microbiota thus adds another layer of complexity in the regulation and function of the innate immune system [196, 201, 202, 203, 204] in both health and disease [196, 205, 206]. Seminal findings support an impact of the microbiome on (i) innate immune cells (neutrophils [36, 207], DCs [208, 209, 210, 211, 212, 213, 214, 215, 216, 217], macrophages [218, 219, 220], ILCs [198, 221, 222, 223, 224, 225], NK cells [226], and NKT cells [227]), (ii) the complement system [228], and (iii) defensins [196, 229, 230]. A major challenge in the field that remains is to define the critical microbiota-to-host interfaces that fine-tune the human immune system and to exploit their therapeutic potential.
Fig. 1.
Key immune effector cells (phagocytes and lymphocytes) and regulatory cells (T regulatory cells, Tregs; myeloid-derived suppressor cells, MDSCs). Macrophages show different phenotypes, with M1 and M2 being the main subtypes. Beyond this plasticity, macrophages are the main cells responsible for inflammasome activity (leading to proinflammatory IL-1β and IL-18 production). Specific to neutrophils is the release of their own DNA, called neutrophil extracellular traps (NETs), which can entrap and immobilize pathogens. In addition to traditional adaptive T and B cell subsets, lymphocytes also encompass the innate lymphoid cells (ILCs) that belong to the innate immune system. In analogy to Th1/Th2, ILCs comprise at least 3 different subtypes, termed ILC1, ILC2, and ILC3.
Disclosure Statement
There were no conflicts of interest.
Acknowledgements
This work was supported by the DFG Emmy Noether program and DFG priority program 1937 (G.G.). The authors thank Peter M. Weber (University of Tübingen, Tübingen, Germany) for the excellent illustration.
References
- 1.Buchmann K. Evolution of innate immunity: clues from invertebrates via fish to mammals. Front Immunol. 2014;5:459. doi: 10.3389/fimmu.2014.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryant CE, Monie TP. Mice, men and the relatives: cross-species studies underpin innate immunity. Open Biol. 2012;2:120015. doi: 10.1098/rsob.120015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galli SJ, Borregaard N, Wynn TA. Phenotypic and functional plasticity of cells of innate immunity: macrophages, mast cells and neutrophils. Nat Immunol. 2011;12:1035–1044. doi: 10.1038/ni.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilchie AL, Wuerth K, Hancock RE. Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nat Chem Biol. 2013;9:761–768. doi: 10.1038/nchembio.1393. [DOI] [PubMed] [Google Scholar]
- 5.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 6.Lai Y, Gallo RL. Amped up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Degn SE, Thiel S. Humoral pattern recognition and the complement system. Scand J Immunol. 2013;78:181–193. doi: 10.1111/sji.12070. [DOI] [PubMed] [Google Scholar]
- 8.de Cordoba SR, Tortajada A, Harris CL, Morgan BP. Complement dysregulation and disease: from genes and proteins to diagnostics and drugs. Immunobiology. 2012;217:1034–1046. doi: 10.1016/j.imbio.2012.07.021. [DOI] [PubMed] [Google Scholar]
- 9.Carroll MC, Isenman DE. Regulation of humoral immunity by complement. Immunity. 2012;37:199–207. doi: 10.1016/j.immuni.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan JK, Roth J, Oppenheim JJ, Tracey KJ, Vogl T, Feldmann M, Horwood N, Nanchahal J. Alarmins: awaiting a clinical response. J Clin Invest. 2012;122:2711–2719. doi: 10.1172/JCI62423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang D, de la Rosa G, Tewary P, Oppenheim JJ. Alarmins link neutrophils and dendritic cells. Trends Immunol. 2009;30:531–537. doi: 10.1016/j.it.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sokol CL, Luster AD. The chemokine system in innate immunity. Cold Spring Harb Perspect Biol. 2015;7:a016303. doi: 10.1101/cshperspect.a016303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee CG, Da Silva CA, Lee JY, Hartl D, Elias JA. Chitin regulation of immune responses: an old molecule with new roles. Curr Opin Immunol. 2008;20:684–689. doi: 10.1016/j.coi.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Meer JW, Joosten LA, Riksen N, Netea MG. Trained immunity: a smart way to enhance innate immune defence. Mol Immunol. 2015;68:40–44. doi: 10.1016/j.molimm.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Gonzalez I, Matha L, Steer CA, Ghaedi M, Poon GF, Takei F. Allergen-experienced group 2 innate lymphoid cells acquire memory-like properties and enhance allergic lung inflammation. Immunity. 2016;45:198–208. doi: 10.1016/j.immuni.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 16.O'Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat Immunol. 2006;7:507–516. doi: 10.1038/ni1332. [DOI] [PubMed] [Google Scholar]
- 17.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bedoui S, Gebhardt T, Gasteiger G, Kastenmuller W. Parallels and differences between innate and adaptive lymphocytes. Nat Immunol. 2016;17:490–494. doi: 10.1038/ni.3432. [DOI] [PubMed] [Google Scholar]
- 19.Burian M, Schittek B. The secrets of dermcidin action. Int J Med Microbiol. 2015;305:283–286. doi: 10.1016/j.ijmm.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 20.Schittek B. The multiple facets of dermcidin in cell survival and host defense. J Innate Immun. 2012;4:349–360. doi: 10.1159/000336844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 22.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 23.Pluddemann A, Mukhopadhyay S, Gordon S. Innate immunity to intracellular pathogens: macrophage receptors and responses to microbial entry. Immunol Rev. 2011;240:11–24. doi: 10.1111/j.1600-065X.2010.00989.x. [DOI] [PubMed] [Google Scholar]
- 24.Mukhopadhyay S, Gordon S. The role of scavenger receptors in pathogen recognition and innate immunity. Immunobiology. 2004;209:39–49. doi: 10.1016/j.imbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Taylor PR, Martinez-Pomares L, Stacey M, Lin HH, Brown GD, Gordon S. Macrophage receptors and immune recognition. Annu Rev Immunol. 2005;23:901–944. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- 26.Hornung V, Hartmann R, Ablasser A, Hopfner KP. OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat Rev Immunol. 2014;14:521–528. doi: 10.1038/nri3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornung V, Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- 28.Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol. 2014;32:659–702. doi: 10.1146/annurev-immunol-032713-120145. [DOI] [PubMed] [Google Scholar]
- 29.Gerard C, Rollins BJ. Chemokines and disease. Nat Immunol. 2001;2:108–115. doi: 10.1038/84209. [DOI] [PubMed] [Google Scholar]
- 30.Kruger P, Saffarzadeh M, Weber AN, Rieber N, Radsak M, von Bernuth H, Benarafa C, Roos D, Skokowa J, Hartl D. Neutrophils: between host defence, immune modulation, and tissue injury. PLoS Pathog. 2015;11:e1004651. doi: 10.1371/journal.ppat.1004651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pillay J, den Braber I, Vrisekoop N, Kwast LM, de Boer RJ, Borghans JA, Tesselaar K, Koenderman L. In vivo labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days. Blood. 2010;116:625–627. doi: 10.1182/blood-2010-01-259028. [DOI] [PubMed] [Google Scholar]
- 32.Brinkmann V, Zychlinsky A. Beneficial suicide: why neutrophils die to make nets. Nat Rev Microbiol. 2007;5:577–582. doi: 10.1038/nrmicro1710. [DOI] [PubMed] [Google Scholar]
- 33.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lammermann T. In the eye of the neutrophil swarm - navigation signals that bring neutrophils together in inflamed and infected tissues. J Leukoc Biol. 2016;100:55–63. doi: 10.1189/jlb.1MR0915-403. [DOI] [PubMed] [Google Scholar]
- 35.Lammermann T, Afonso PV, Angermann BR, Wang JM, Kastenmuller W, Parent CA, Germain RN. Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature. 2013;498:371–375. doi: 10.1038/nature12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang D, Chen G, Manwani D, Mortha A, Xu C, Faith JJ, Burk RD, Kunisaki Y, Jang JE, Scheiermann C, Merad M, Frenette PS. Neutrophil ageing is regulated by the microbiome. Nature. 2015;525:528–532. doi: 10.1038/nature15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hager M, Cowland JB, Borregaard N. Neutrophil granules in health and disease. J Intern Med. 2010;268:25–34. doi: 10.1111/j.1365-2796.2010.02237.x. [DOI] [PubMed] [Google Scholar]
- 38.Yipp BG, Kubes P. NETosis: how vital is it? Blood. 2013;122:2784–2794. doi: 10.1182/blood-2013-04-457671. [DOI] [PubMed] [Google Scholar]
- 39.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 40.Zychlinsky A. Neutrophil extracellular traps. Eur J Clin Invest. 2009;39:27–27. [Google Scholar]
- 41.Lightfoot YL, Kaplan MJ. Disentangling the role of neutrophil extracellular traps in rheumatic diseases. Curr Opin Rheumatol. 2016 doi: 10.1097/BOR.0000000000000357. E-pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grayson PC, Kaplan MJ. At the bench: neutrophil extracellular traps (NETs) highlight novel aspects of innate immune system involvement in autoimmune diseases. J Leukoc Biol. 2016;99:253–264. doi: 10.1189/jlb.5BT0615-247R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnado A, Crofford LJ, Oates JC. At the bedside: neutrophil extracellular traps (NETs) as targets for biomarkers and therapies in autoimmune diseases. J Leukoc Biol. 2016;99:265–278. doi: 10.1189/jlb.5BT0615-234R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sorensen OE, Borregaard N. Neutrophil extracellular traps - the dark side of neutrophils. J Clin Invest. 2016;126:1612–1620. doi: 10.1172/JCI84538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yousefi S, Mihalache C, Kozlowski E, Schmid I, Simon HU. Viable neutrophils release mitochondrial DNA to form neutrophil extracellular traps. Cell Death Differ. 2009;16:1438–1444. doi: 10.1038/cdd.2009.96. [DOI] [PubMed] [Google Scholar]
- 46.Pilsczek FH, Salina D, Poon KK, Fahey C, Yipp BG, Sibley CD, Robbins SM, Green FH, Surette MG, Sugai M, Bowden MG, Hussain M, Zhang K, Kubes P. A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J Immunol. 2010;185:7413–7425. doi: 10.4049/jimmunol.1000675. [DOI] [PubMed] [Google Scholar]
- 47.Yipp BG, Petri B, Salina D, Jenne CN, Scott BN, Zbytnuik LD, Pittman K, Asaduzzaman M, Wu K, Meijndert HC, Malawista SE, de Boisfleury Chevance A, Zhang K, Conly J, Kubes P. Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat Med. 2012;18:1386–1393. doi: 10.1038/nm.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. 2013;13:159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 49.Branzk N, Lubojemska A, Hardison SE, Wang Q, Gutierrez MG, Brown GD, Papayannopoulos V. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat Immunol. 2014;15:1017–1025. doi: 10.1038/ni.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ermert D, Urban CF, Laube B, Goosmann C, Zychlinsky A, Brinkmann V. Mouse neutrophil extracellular traps in microbial infections. J Innate Immun. 2009;1:181–193. doi: 10.1159/000205281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peschel A, Hartl D. Anuclear neutrophils keep hunting. Nat Med. 2012;18:1336–1338. doi: 10.1038/nm.2918. [DOI] [PubMed] [Google Scholar]
- 52.Okabe Y, Medzhitov R. Tissue biology perspective on macrophages. Nat Immunol. 2016;17:9–17. doi: 10.1038/ni.3320. [DOI] [PubMed] [Google Scholar]
- 53.Byrne AJ, Mathie SA, Gregory LG, Lloyd CM. Pulmonary macrophages: key players in the innate defence of the airways. Thorax. 2015;70:1189–1196. doi: 10.1136/thoraxjnl-2015-207020. [DOI] [PubMed] [Google Scholar]
- 54.Murray PJ, Wynn TA. Obstacles and opportunities for understanding macrophage polarization. J Leukoc Biol. 2011;89:557–563. doi: 10.1189/jlb.0710409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rothlin CV, Carrera-Silva EA, Bosurgi L, Ghosh S. Tam receptor signaling in immune homeostasis. Annu Rev Immunol. 2015;33:355–391. doi: 10.1146/annurev-immunol-032414-112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anwar MA, Basith S, Choi S. Negative regulatory approaches to the attenuation of toll-like receptor signaling. Exp Mol Med. 2013;45:e11. doi: 10.1038/emm.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bourdonnay E, Zaslona Z, Penke LR, Speth JM, Schneider DJ, Przybranowski S, Swanson JA, Mancuso P, Freeman CM, Curtis JL, Peters-Golden M. Transcellular delivery of vesicular SOCS proteins from macrophages to epithelial cells blunts inflammatory signaling. J Exp Med. 2015;212:729–742. doi: 10.1084/jem.20141675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jenkins SJ, Ruckerl D, Cook PC, Jones LH, Finkelman FD, van Rooijen N, MacDonald AS, Allen JE. Local macrophage proliferation, rather than recruitment from the blood, is a signature of Th2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soroosh P, Doherty TA, Duan W, Mehta AK, Choi H, Adams YF, Mikulski Z, Khorram N, Rosenthal P, Broide DH, Croft M. Lung-resident tissue macrophages generate Foxp3+ regulatory T cells and promote airway tolerance. J Exp Med. 2013;210:775–788. doi: 10.1084/jem.20121849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westphalen K, Gusarova GA, Islam MN, Subramanian M, Cohen TS, Prince AS, Bhattacharya J. Sessile alveolar macrophages communicate with alveolar epithelium to modulate immunity. Nature. 2014;506:503–506. doi: 10.1038/nature12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hussell T, Bell TJ. Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol. 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 63.Lee GR, Bithell TC, Foerster J, Athens JW, Lukens JN, editors. ed 9. Philadelphia: Lea & Febiger; 1993. Wintrobe's Clinical Hematology. [Google Scholar]
- 64.van de Laar L, Saelens W, De Prijck S, Martens L, Scott CL, Van Isterdael G, Hoffmann E, Beyaert R, Saeys Y, Lambrecht BN, Guilliams M. Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity. 2016;44:755–768. doi: 10.1016/j.immuni.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 65.Perdiguero EG, Geissmann F. The development and maintenance of resident macrophages. Nat Immunol. 2016;17:2–8. doi: 10.1038/ni.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ginhoux F, Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity. 2016;44:439–449. doi: 10.1016/j.immuni.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 67.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Auffray C, Fogg DK, Narni-Mancinelli E, Senechal B, Trouillet C, Saederup N, Leemput J, Bigot K, Campisi L, Abitbol M, Molina T, Charo I, Hume DA, Cumano A, Lauvau G, Geissmann F. CX3CR1+ CD115+ CD135+ common macrophage/DC precursors and the role of CX3CR1 in their response to inflammation. J Exp Med. 2009;206:595–606. doi: 10.1084/jem.20081385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J, Kubes P. A reservoir of mature cavity macrophages that can rapidly invade visceral organs to affect tissue repair. Cell. 2016;165:668–678. doi: 10.1016/j.cell.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 70.Wynn TA, Vannella KM. Macrophages in tissue repair, regeneration, and fibrosis. Immunity. 2016;44:450–462. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mantovani A, Biswas SK, Galdiero MR, Sica A, Locati M. Macrophage plasticity and polarization in tissue repair and remodelling. J Pathol. 2013;229:176–185. doi: 10.1002/path.4133. [DOI] [PubMed] [Google Scholar]
- 73.Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, De Nardo D, Gohel TD, Emde M, Schmidleithner L, Ganesan H, Nino-Castro A, Mallmann MR, Labzin L, Theis H, Kraut M, Beyer M, Latz E, Freeman TC, Ulas T, Schultze JL. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Turner M, Galloway A, Vigorito E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat Immunol. 2014;15:484–491. doi: 10.1038/ni.2887. [DOI] [PubMed] [Google Scholar]
- 75.Glass CK, Natoli G. Molecular control of activation and priming in macrophages. Nat Immunol. 2016;17:26–33. doi: 10.1038/ni.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Netea MG, Joosten LA, Latz E, Mills KH, Natoli G, Stunnenberg HG, O'Neill LA, Xavier RJ. Trained immunity: a program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Greten TF, Manns MP, Korangy F. Myeloid derived suppressor cells in human diseases. Int Immunopharmacol. 2011;11:802–807. doi: 10.1016/j.intimp.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mantovani A. The growing diversity and spectrum of action of myeloid-derived suppressor cells. Eur J Immunol. 2010;40:3317–3320. doi: 10.1002/eji.201041170. [DOI] [PubMed] [Google Scholar]
- 79.Brandau S, Moses K, Lang S. The kinship of neutrophils and granulocytic myeloid-derived suppressor cells in cancer: cousins, siblings or twins? Semin Cancer Biol. 2013;23:171–182. doi: 10.1016/j.semcancer.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 80.Dumitru CA, Moses K, Trellakis S, Lang S, Brandau S. Neutrophils and granulocytic myeloid-derived suppressor cells: immunophenotyping, cell biology and clinical relevance in human oncology. Cancer Immunol Immunother. 2012;61:1155–1167. doi: 10.1007/s00262-012-1294-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, Mandruzzato S, Murray PJ, Ochoa A, Ostrand-Rosenberg S, Rodriguez PC, Sica A, Umansky V, Vonderheide RH, Gabrilovich DI. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nagaraj S, Gabrilovich DI. Myeloid-derived suppressor cells. Adv Exp Med Biol. 2007;601:213–223. doi: 10.1007/978-0-387-72005-0_22. [DOI] [PubMed] [Google Scholar]
- 83.Youn JI, Gabrilovich DI. The biology of myeloid-derived suppressor cells: the blessing and the curse of morphological and functional heterogeneity. Eur J Immunol. 2010;40:2969–2975. doi: 10.1002/eji.201040895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gantt S, Gervassi A, Jaspan H, Horton H. The role of myeloid-derived suppressor cells in immune ontogeny. Front Immunol. 2014;5:387. doi: 10.3389/fimmu.2014.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ost M, Singh A, Peschel A, Mehling R, Rieber N, Hartl D. Myeloid-derived suppressor cells in bacterial infections. Front Cell Infect Microbiol. 2016;6:37. doi: 10.3389/fcimb.2016.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Diefenbach A, Colonna M, Koyasu S. Development, differentiation, and diversity of innate lymphoid cells. Immunity. 2014;41:354–365. doi: 10.1016/j.immuni.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rankin LC, Girard-Madoux MJ, Seillet C, Mielke LA, Kerdiles Y, Fenis A, Wieduwild E, Putoczki T, Mondot S, Lantz O, Demon D, Papenfuss AT, Smyth GK, Lamkanfi M, Carotta S, Renauld JC, Shi W, Carpentier S, Soos T, Arendt C, Ugolini S, Huntington ND, Belz GT, Vivier E. Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nat Immunol. 2016;17:179–186. doi: 10.1038/ni.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wensveen FM, Jelencic V, Valentic S, Sestan M, Wensveen TT, Theurich S, Glasner A, Mendrila D, Stimac D, Wunderlich FT, Bruning JC, Mandelboim O, Polic B. Nk cells link obesity-induced adipose stress to inflammation and insulin resistance. Nat Immunol. 2015;16:376–385. doi: 10.1038/ni.3120. [DOI] [PubMed] [Google Scholar]
- 89.Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CGK, Doering TA, Angelosanto JM, Laidlaw BJ, Yang CY, Sathaliyawala T, Kubota M, Turner D, Diamond JM, Goldrath AW, Farber DL, Collman RG, Wherry EJ, Artis D. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol. 2011;12:1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lindemans CA, Calafiore M, Mertelsmann AM, O'Connor MH, Dudakov JA, Jenq RR, Velardi E, Young LF, Smith OM, Lawrence G, Ivanov JA, Fu YY, Takashima S, Hua G, Martin ML, O'Rourke KP, Lo YH, Mokry M, Romera-Hernandez M, Cupedo T, Dow LE, Nieuwenhuis EE, Shroyer NF, Liu C, Kolesnick R, van den Brink MR, Hanash AM. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528:560–564. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.von Moltke J, Ji M, Liang HE, Locksley RM. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature. 2016;529:221–225. doi: 10.1038/nature16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee MW, Odegaard JI, Mukundan L, Qiu Y, Molofsky AB, Nussbaum JC, Yun K, Locksley RM, Chawla A. Activated type 2 innate lymphoid cells regulate beige fat biogenesis. Cell. 2015;160:74–87. doi: 10.1016/j.cell.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Molofsky AB, Nussbaum JC, Liang HE, Van Dyken SJ, Cheng LE, Mohapatra A, Chawla A, Locksley RM. Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J Exp Med. 2013;210:535–549. doi: 10.1084/jem.20121964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, Merad M. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 2014;343:1249288. doi: 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gasteiger G, Fan X, Dikiy S, Lee SY, Rudensky AY. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science. 2015;350:981–985. doi: 10.1126/science.aac9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moro K, Kabata H, Tanabe M, Koga S, Takeno N, Mochizuki M, Fukunaga K, Asano K, Betsuyaku T, Koyasu S. Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat Immunol. 2016;17:76–86. doi: 10.1038/ni.3309. [DOI] [PubMed] [Google Scholar]
- 97.Klose CS, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, Fabiunke C, Pfeifer D, Sexl V, Fonseca-Pereira D, Domingues RG, Veiga-Fernandes H, Arnold SJ, Busslinger M, Dunay IR, Tanriver Y, Diefenbach A. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 98.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shih HY, Sciume G, Mikami Y, Guo L, Sun HW, Brooks SR, Urban JF, Jr, Davis FP, Kanno Y, O'Shea JJ. Developmental acquisition of regulomes underlies innate lymphoid cell functionality. Cell. 2016;165:120–133. doi: 10.1016/j.cell.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gronke K, Kofoed-Nielsen M, Diefenbach A. Innate lymphoid cells, precursors and plasticity. Immunol Lett. 2016;179:9–18. doi: 10.1016/j.imlet.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 101.Gasteiger G, Rudensky AY. Interactions between innate and adaptive lymphocytes. Nat Rev Immunol. 2014;14:631–639. doi: 10.1038/nri3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hepworth MR, Sonnenberg GF. Regulation of the adaptive immune system by innate lymphoid cells. Curr Opin Immunol. 2014;27:75–82. doi: 10.1016/j.coi.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oliphant CJ, Hwang YY, Walker JA, Salimi M, Wong SH, Brewer JM, Englezakis A, Barlow JL, Hams E, Scanlon ST, Ogg GS, Fallon PG, McKenzie AN. MHCII-mediated dialogue between group 2 innate lymphoid cells and CD4(+) T cells potentiates type 2 immunity and promotes parasitic helminth expulsion. Immunity. 2014;41:283–295. doi: 10.1016/j.immuni.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gasteiger G, Hemmers S, Bos PD, Sun JC, Rudensky AY. IL-2-dependent adaptive control of NK cell homeostasis. J Exp Med. 2013;210:1179–1187. doi: 10.1084/jem.20122571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Halim TY, Hwang YY, Scanlon ST, Zaghouani H, Garbi N, Fallon PG, McKenzie AN. Group 2 innate lymphoid cells license dendritic cells to potentiate memory Th2 cell responses. Nat Immunol. 2016;17:57–64. doi: 10.1038/ni.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Halim TY, Steer CA, Matha L, Gold MJ, Martinez-Gonzalez I, McNagny KM, McKenzie AN, Takei F. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 2014;40:425–435. doi: 10.1016/j.immuni.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Withers DR, Gaspal FM, Mackley EC, Marriott CL, Ross EA, Desanti GE, Roberts NA, White AJ, Flores-Langarica A, McConnell FM, Anderson G, Lane PJ. Cutting edge: lymphoid tissue inducer cells maintain memory CD4 T cells within secondary lymphoid tissue. J Immunol. 2012;189:2094–2098. doi: 10.4049/jimmunol.1201639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hepworth MR, Fung TC, Masur SH, Kelsen JR, McConnell FM, Dubrot J, Withers DR, Hugues S, Farrar MA, Reith W, Eberl G, Baldassano RN, Laufer TM, Elson CO, Sonnenberg GF. Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4(+) T cells. Science. 2015;348:1031–1035. doi: 10.1126/science.aaa4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mackley EC, Houston S, Marriott CL, Halford EE, Lucas B, Cerovic V, Filbey KJ, Maizels RM, Hepworth MR, Sonnenberg GF, Milling S, Withers DR. CCR7-dependent trafficking of RORγ(+) ILCs creates a unique microenvironment within mucosal draining lymph nodes. Nat Commun. 2015;6:5862. doi: 10.1038/ncomms6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Serafini N, Vosshenrich CA, Di Santo JP. Transcriptional regulation of innate lymphoid cell fate. Nat Rev Immunol. 2015;15:415–428. doi: 10.1038/nri3855. [DOI] [PubMed] [Google Scholar]
- 111.Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348:aaa6566. doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McKenzie AN, Spits H, Eberl G. Innate lymphoid cells in inflammation and immunity. Immunity. 2014;41:366–374. doi: 10.1016/j.immuni.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 113.Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 114.Kanneganti TD. The inflammasome: firing up innate immunity. Immunol Rev. 2015;265:1–5. doi: 10.1111/imr.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rathinam VA, Fitzgerald KA. Inflammasome complexes: emerging mechanisms and effector functions. Cell. 2016;165:792–800. doi: 10.1016/j.cell.2016.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, Egelman EH. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Man SM, Kanneganti TD. Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat Rev Immunol. 2016;16:7–21. doi: 10.1038/nri.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 120.Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, Zhang J, Lee WP, Roose-Girma M, Dixit VM. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 121.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 122.Kayagaki N, Stowe IB, Lee BL, O'Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, Liu PS, Lill JR, Li H, Wu J, Kummerfeld S, Zhang J, Lee WP, Snipas SJ, Salvesen GS, Morris LX, Fitzgerald L, Zhang Y, Bertram EM, Goodnow CC, Dixit VM. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 123.Gurung P, Kanneganti TD. Novel roles for caspase-8 in IL-1β and inflammasome regulation. Am J Pathol. 2015;185:17–25. doi: 10.1016/j.ajpath.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jo EK, Kim JK, Shin DM, Sasakawa C. Molecular mechanisms regulating NLRP3 inflammasome activation. Cell Mol Immunol. 2016;13:148–159. doi: 10.1038/cmi.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Py BF, Kim MS, Vakifahmetoglu-Norberg H, Yuan J. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol Cell. 2013;49:331–338. doi: 10.1016/j.molcel.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 126.Spalinger MR, Kasper S, Gottier C, Lang S, Atrott K, Vavricka SR, Scharl S, Gutte PM, Grutter MG, Beer HD, Contassot E, Chan AC, Dai X, Rawlings DJ, Mair F, Becher B, Falk W, Fried M, Rogler G, Scharl M. NLRP3 tyrosine phosphorylation is controlled by protein tyrosine phosphatase PTPN22. J Clin Invest. 2016;126:1783–1800. doi: 10.1172/JCI83669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lee GS, Subramanian N, Kim AI, Aksentijevich I, Goldbach-Mansky R, Sacks DB, Germain RN, Kastner DL, Chae JJ. The calcium-sensing receptor regulates the NLRP3 inflammasome through CA2+ and cAMP. Nature. 2012;492:123–127. doi: 10.1038/nature11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gaidt MM, Ebert TS, Chauhan D, Schmidt T, Schmid-Burgk JL, Rapino F, Robertson AA, Cooper MA, Graf T, Hornung V. Human monocytes engage an alternative inflammasome pathway. Immunity. 2016;44:833–846. doi: 10.1016/j.immuni.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 129.Gaidt MM, Ebert TS, Chauhan D, Schmidt T, Schmid-Burgk JL, Rapino F, Robertson AAB, Cooper MA, Graf T, Hornung V. Human monocytes engage an alternative inflammasome pathway. Immunity. 2016;44:833–846. doi: 10.1016/j.immuni.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 130.Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszynski A, Forsberg LS, Carlson RW, Dixit VM. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341:1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- 131.Ito M, Shichita T, Okada M, Komine R, Noguchi Y, Yoshimura A, Morita R. Bruton's tyrosine kinase is essential for NLRP3 inflammasome activation and contributes to ischaemic brain injury. Nat Commun. 2015;6:7360. doi: 10.1038/ncomms8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.He Y, Zeng MY, Yang D, Motro B, Nunez G. Nek7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature. 2016;530:354–357. doi: 10.1038/nature16959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wolf AJ, Reyes CN, Liang W, Becker C, Shimada K, Wheeler ML, Cho HC, Popescu NI, Coggeshall KM, Arditi M, Underhill DM. Hexokinase is an innate immune receptor for the detection of bacterial peptidoglycan. Cell. 2016;166:624–636. doi: 10.1016/j.cell.2016.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.O'Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ravindran R, Loebbermann J, Nakaya HI, Khan N, Ma H, Gama L, Machiah DK, Lawson B, Hakimpour P, Wang YC, Li S, Sharma P, Kaufman RJ, Martinez J, Pulendran B. The amino acid sensor GCN2 controls gut inflammation by inhibiting inflammasome activation. Nature. 2016;531:523–527. doi: 10.1038/nature17186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, Gardet A, Tong Z, Jany SS, Corr SC, Haneklaus M, Caffrey BE, Pierce K, Walmsley S, Beasley FC, Cummins E, Nizet V, Whyte M, Taylor CT, Lin H, Masters SL, Gottlieb E, Kelly VP, Clish C, Auron PE, Xavier RJ, O'Neill LA. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Guo C, Xie S, Chi Z, Zhang J, Liu Y, Zhang L, Zheng M, Zhang X, Xia D, Ke Y, Lu L, Wang D. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity. 2016;45:944. doi: 10.1016/j.immuni.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 138.Mills EL, Kelly B, Logan A, Costa AS, Varma M, Bryant CE, Tourlomousis P, Dabritz JH, Gottlieb E, Latorre I, Corr SC, McManus G, Ryan D, Jacobs HT, Szibor M, Xavier RJ, Braun T, Frezza C, Murphy MP, O'Neill LA. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell. 2016;167:457–470. doi: 10.1016/j.cell.2016.08.064. e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bruchard M, Rebe C, Derangere V, Togbe D, Ryffel B, Boidot R, Humblin E, Hamman A, Chalmin F, Berger H, Chevriaux A, Limagne E, Apetoh L, Vegran F, Ghiringhelli F. The receptor NLRP3 is a transcriptional regulator of Th2 differentiation. Nat Immunol. 2015;16:859–870. doi: 10.1038/ni.3202. [DOI] [PubMed] [Google Scholar]
- 140.Feldmeyer L, Keller M, Niklaus G, Hohl D, Werner S, Beer HD. The inflammasome mediates UVB-induced activation and secretion of interleukin-1β by keratinocytes. Curr Biol. 2007;17:1140–1145. doi: 10.1016/j.cub.2007.05.074. [DOI] [PubMed] [Google Scholar]
- 141.Dombrowski Y, Peric M, Koglin S, Kammerbauer C, Goss C, Anz D, Simanski M, Glaser R, Harder J, Hornung V, Gallo RL, Ruzicka T, Besch R, Schauber J. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci Transl Med. 2011;3:82ra38. doi: 10.1126/scitranslmed.3002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shaw PJ, Lukens JR, Burns S, Chi H, McGargill MA, Kanneganti TD. Cutting edge: critical role for PYCARD/ASC in the development of experimental autoimmune encephalomyelitis. J Immunol. 2010;184:4610–4614. doi: 10.4049/jimmunol.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Martin BN, Wang C, Zhang CJ, Kang Z, Gulen MF, Zepp JA, Zhao J, Bian G, Do JS, Min B, Pavicic PG, Jr, El-Sanadi C, Fox PL, Akitsu A, Iwakura Y, Sarkar A, Wewers MD, Kaiser WJ, Mocarski ES, Rothenberg ME, Hise AG, Dubyak GR, Ransohoff RM, Li X. T cell-intrinsic ASC critically promotes Th17-mediated experimental autoimmune encephalomyelitis. Nat Immunol. 2016;17:583–592. doi: 10.1038/ni.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kuemmerle-Deschner JB. CAPS - pathogenesis, presentation and treatment of an autoinflammatory disease. Semin Immunopathol. 2015;37:377–385. doi: 10.1007/s00281-015-0491-7. [DOI] [PubMed] [Google Scholar]
- 145.Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21:677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Doitsh G, Galloway NL, Geng X, Yang Z, Monroe KM, Zepeda O, Hunt PW, Hatano H, Sowinski S, Munoz-Arias I, Greene WC. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505:509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sagoo P, Garcia Z, Breart B, Lemaitre F, Michonneau D, Albert ML, Levy Y, Bousso P. In vivo imaging of inflammasome activation reveals a subcapsular macrophage burst response that mobilizes innate and adaptive immunity. Nat Med. 2016;22:64–71. doi: 10.1038/nm.4016. [DOI] [PubMed] [Google Scholar]
- 148.Kastenmuller W, Torabi-Parizi P, Subramanian N, Lammermann T, Germain RN. A spatially-organized multicellular innate immune response in lymph nodes limits systemic pathogen spread. Cell. 2012;150:1235–1248. doi: 10.1016/j.cell.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Coll RC, Robertson AA, Chae JJ, Higgins SC, Munoz-Planillo R, Inserra MC, Vetter I, Dungan LS, Monks BG, Stutz A, Croker DE, Butler MS, Haneklaus M, Sutton CE, Nunez G, Latz E, Kastner DL, Mills KH, Masters SL, Schroder K, Cooper MA, O'Neill LA. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med. 2015;21:248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 151.Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 152.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 153.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic gmp-amp synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, Tuschl T, Patel DJ. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li X, Shu C, Yi G, Chaton CT, Shelton CL, Diao J, Zuo X, Kao CC, Herr AB, Li P. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity. 2013;39:1019–1031. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, Hornung V. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bridgeman A, Maelfait J, Davenne T, Partridge T, Peng Y, Mayer A, Dong T, Kaever V, Borrow P, Rehwinkel J. Viruses transfer the antiviral second messenger cGAMP between cells. Science. 2015;349:1228–1232. doi: 10.1126/science.aab3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gentili M, Kowal J, Tkach M, Satoh T, Lahaye X, Conrad C, Boyron M, Lombard B, Durand S, Kroemer G, Loew D, Dalod M, Thery C, Manel N. Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science. 2015;349:1232–1236. doi: 10.1126/science.aab3628. [DOI] [PubMed] [Google Scholar]
- 168.Ishikawa H, Ma Z, Barber GN. Sting regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, Chen ZJ. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 170.Abe T, Barber GN. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J Virol. 2014;88:5328–5341. doi: 10.1128/JVI.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Dai P, Wang W, Cao H, Avogadri F, Dai L, Drexler I, Joyce JA, Li XD, Chen Z, Merghoub T, Shuman S, Deng L. Modified vaccinia virus Ankara triggers type I IFN production in murine conventional dendritic cells via a cGAS/STING-mediated cytosolic DNA-sensing pathway. PLoS Pathog. 2014;10:e1003989. doi: 10.1371/journal.ppat.1003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Dansako H, Ueda Y, Okumura N, Satoh S, Sugiyama M, Mizokami M, Ikeda M, Kato N. The cyclic GMP-AMP synthetase-STING signaling pathway is required for both the innate immune response against HBV and the suppression of HBV assembly. FEBS J. 2016;283:144–156. doi: 10.1111/febs.13563. [DOI] [PubMed] [Google Scholar]
- 173.Lam E, Stein S, Falck-Pedersen E. Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade. J Virol. 2014;88:974–981. doi: 10.1128/JVI.02702-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wu JJ, Li W, Shao Y, Avey D, Fu B, Gillen J, Hand T, Ma S, Liu X, Miley W, Konrad A, Neipel F, Sturzl M, Whitby D, Li H, Zhu F. Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host Microbe. 2015;18:333–344. doi: 10.1016/j.chom.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Collins AC, Cai H, Li T, Franco LH, Li XD, Nair VR, Scharn CR, Stamm CE, Levine B, Chen ZJ, Shiloh MU. Cyclic GMP-AMP synthase is an innate immune DNA sensor for Mycobacterium tuberculosis. Cell Host Microbe. 2015;17:820–828. doi: 10.1016/j.chom.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Dey B, Dey RJ, Cheung LS, Pokkali S, Guo H, Lee JH, Bishai WR. A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat Med. 2015;21:401–406. doi: 10.1038/nm.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hansen K, Prabakaran T, Laustsen A, Jorgensen SE, Rahbaek SH, Jensen SB, Nielsen R, Leber JH, Decker T, Horan KA, Jakobsen MR, Paludan SR. Listeria monocytogenes induces IFNβ expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J. 2014;33:1654–1666. doi: 10.15252/embj.201488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Storek KM, Gertsvolf NA, Ohlson MB, Monack DM. cGAS and IFI204 cooperate to produce type I IFNs in response to Francisella infection. J Immunol. 2015;194:3236–3245. doi: 10.4049/jimmunol.1402764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Watson RO, Bell SL, MacDuff DA, Kimmey JM, Diner EJ, Olivas J, Vance RE, Stallings CL, Virgin HW, Cox JS. The cytosolic sensor cGAS detects Mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host Microbe. 2015;17:811–819. doi: 10.1016/j.chom.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhang Y, Yeruva L, Marinov A, Prantner D, Wyrick PB, Lupashin V, Nagarajan UM. The DNA sensor, cyclic GMP-AMP synthase, is essential for induction of IFN-β during Chlamydia trachomatis infection. J Immunol. 2014;193:2394–2404. doi: 10.4049/jimmunol.1302718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Crow YJ, Manel N. Aicardi-Goutières syndrome and the type I interferonopathies. Nat Rev Immunol. 2015;15:429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 184.Gall A, Treuting P, Elkon KB, Loo YM, Gale M, Jr, Barber GN, Stetson DB. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, Daly G, Lindahl T, Barnes DE. Gene-targeted mice lacking the Trex1 (DNase III) 3′->5′ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gao D, Li T, Li XD, Chen X, Li QZ, Wight-Carter M, Chen ZJ. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc Natl Acad Sci USA. 2015;112:E5699–E5705. doi: 10.1073/pnas.1516465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gray EE, Treuting PM, Woodward JJ, Stetson DB. Cutting edge: cGAS is required for lethal autoimmune disease in the Trex1-deficient mouse model of Aicardi-Goutières syndrome. J Immunol. 2015;195:1939–1943. doi: 10.4049/jimmunol.1500969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kawane K, Fukuyama H, Kondoh G, Takeda J, Ohsawa Y, Uchiyama Y, Nagata S. Requirement of DNase II for definitive erythropoiesis in the mouse fetal liver. Science. 2001;292:1546–1549. doi: 10.1126/science.292.5521.1546. [DOI] [PubMed] [Google Scholar]
- 190.Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat Immunol. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]
- 191.Kawane K, Ohtani M, Miwa K, Kizawa T, Kanbara Y, Yoshioka Y, Yoshikawa H, Nagata S. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature. 2006;443:998–1002. doi: 10.1038/nature05245. [DOI] [PubMed] [Google Scholar]
- 192.Ahn J, Gutman D, Saijo S, Barber GN. STING manifests self DNA-dependent inflammatory disease. Proc Natl Acad Sci USA. 2012;109:19386–19391. doi: 10.1073/pnas.1215006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sharma S, Campbell AM, Chan J, Schattgen SA, Orlowski GM, Nayar R, Huyler AH, Nundel K, Mohan C, Berg LJ, Shlomchik MJ, Marshak-Rothstein A, Fitzgerald KA. Suppression of systemic autoimmunity by the innate immune adaptor STING. Proc Natl Acad Sci USA. 2015;112:E710–E717. doi: 10.1073/pnas.1420217112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Xia P, Ye B, Wang S, Zhu X, Du Y, Xiong Z, Tian Y, Fan Z. Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat Immunol. 2016;17:369–378. doi: 10.1038/ni.3356. [DOI] [PubMed] [Google Scholar]
- 195.Seo GJ, Yang A, Tan B, Kim S, Liang Q, Choi Y, Yuan W, Feng P, Park HS, Jung JU. AKT kinase-mediated checkpoint of cGAS DNA sensing pathway. Cell Rep. 2015;13:440–449. doi: 10.1016/j.celrep.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Caballero S, Pamer EG. Microbiota-mediated inflammation and antimicrobial defense in the intestine. Annu Rev Immunol. 2015;33:227–256. doi: 10.1146/annurev-immunol-032713-120238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sonnenberg GF, Artis D. Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity. 2012;37:601–610. doi: 10.1016/j.immuni.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Mavrommatis B, Young GR, Kassiotis G. Counterpoise between the microbiome, host immune activation and pathology. Curr Opin Immunol. 2013;25:456–462. doi: 10.1016/j.coi.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 200.Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Donia MS, Fischbach MA. Human microbiota. Small molecules from the human microbiota. Science. 2015;349:1254766. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.McKenney PT, Pamer EG. From hype to hope: the gut microbiota in enteric infectious disease. Cell. 2015;163:1326–1332. doi: 10.1016/j.cell.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Salzman NH. The role of the microbiome in immune cell development. Ann Allergy Asthma Immunol. 2014;113:593–598. doi: 10.1016/j.anai.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 206.Zeevi D, Korem T, Segal E. Talking about cross-talk: the immune system and the microbiome. Genome Biol. 2016;17:50. doi: 10.1186/s13059-016-0921-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O'Leary CE, Oliver PM, Kolls JK, Weiser JN, Worthen GS. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli k1 sepsis in neonatal mice. Nat Med. 2014;20:524–530. doi: 10.1038/nm.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Niess JH, Reinecker HC. Dendritic cells in the recognition of intestinal microbiota. Cell Microbiol. 2006;8:558–564. doi: 10.1111/j.1462-5822.2006.00694.x. [DOI] [PubMed] [Google Scholar]
- 209.Niess JH, Kurachi K, Go XB, Vyas J, Ploegh HL, Fox JG, Reinecker HC. The intestinal microbiota drives the development of a lamina propria dendritic cell system with intestine specific differentiation. Gastroenterology. 2004;126:A34–A34. [Google Scholar]
- 210.Fujiwara D, Wei B, Presley LL, Brewer S, McPherson M, Lewinski MA, Borneman J, Braun J. Systemic control of plasmacytoid dendritic cells by CD8(+) T cells and commensal microbiota. J Immunol. 2008;180:5843–5852. doi: 10.4049/jimmunol.180.9.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Fujiwara D, Wei B, McPherson M, Braun J. Commensal microbiota and CD8(+)T cells link to control systemic levels of plasmacytoid dendritic cells. Gastroenterology. 2007;132:A397–A397. [Google Scholar]
- 212.Ng SC, Benjamin JL, McCarthy NE, Hedin CRH, Koutsoumpas A, Plamondon S, Price CL, Hart AL, Kamm MA, Forbes A, Knight SC, Lindsay JO, Whelan K, Stagg AJ. Relationship between human intestinal dendritic cells, gut microbiota, and disease activity in Crohn's disease. Inflamm Bowel Dis. 2011;17:2027–2037. doi: 10.1002/ibd.21590. [DOI] [PubMed] [Google Scholar]
- 213.Mueller C, Katepalli M, Steinmeyer S, Jayaraman A, Alaniz R. A role for microbiota metabolites in generation of mucosal dendritic cells. J Immunol. 2013;190:61–65. [Google Scholar]
- 214.Young JA, He TH, Reizis B, Winoto A. Commensal microbiota are required for systemic inflammation triggered by necrotic dendritic cells. Cell Rep. 2013;3:1932–1944. doi: 10.1016/j.celrep.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ruane D, Chorny A, Lee H, Faith J, Pandey G, Shan M, Simchoni N, Rahman A, Garg A, Weinstein EG, Oropallo M, Gaylord M, Ungaro R, Cunningham-Rundles C, Alexandropoulos K, Mucida D, Merad M, Cerutti A, Mehandru S. Microbiota regulate the ability of lung dendritic cells to induce IgA class-switch recombination and generate protective gastrointestinal immune responses. J Exp Med. 2016;213:53–73. doi: 10.1084/jem.20150567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Han D, Walsh MC, Cejas PJ, Dang NN, Kim YF, Kim J, Charrier-Hisamuddin L, Chau L, Zhang Q, Bittinger K, Bushman FD, Turka LA, Shen H, Reizis B, DeFranco AL, Wu GD, Choi Y. Dendritic cell expression of the signaling molecule TRAF6 is critical for gut microbiota-dependent immune tolerance. Immunity. 2013;38:1211–1222. doi: 10.1016/j.immuni.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Hagerbrand K, Westlund J, Yrlid U, Agace W, Johansson-Lindbom B. Myd88 signaling regulates steady-state migration of intestinal CD103(+) dendritic cells independently of TNF-α and the gut microbiota. J Immunol. 2015;195:2888–2899. doi: 10.4049/jimmunol.1500210. [DOI] [PubMed] [Google Scholar]
- 218.Matsuoka K, Sheikh SZ, Li FL, Uno JK, Plevy SE. Macrophage tolerance to the enteric microbiota is mediated by IL-10 at the chromatin level. Gastroenterology. 2009;136:A256–A256. [Google Scholar]
- 219.Lopes MM, Carneiro MBH, dos Santos LM, Vaz LG, Martins FD, Vieira LQ. Absence of microbiota impairs macrophage microbicide activity and production of nitric oxide and reative species of oxygen. Free Radical Bio Med. 2012;53:S79–S79. [Google Scholar]
- 220.Allen RG, Lafuse WP, Galley JD, Ali MM, Ahmer BMM, Bailey MT. The intestinal microbiota are necessary for stressor-induced enhancement of splenic macrophage microbicidal activity. Brain Behav Immun. 2012;26:371–382. doi: 10.1016/j.bbi.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, Mantegazza AR, Ma HL, Crawford A, Angelosanto JM, Wherry EJ, Koni PA, Bushman FD, Elson CO, Eberl G, Artis D, Sonnenberg GF. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Berard M, Kleinschek M, Cua D, Di Santo JP, Eberl G. RORγt+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. 2011;12:U320–U371. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 223.Moro K, Koyasu S. Innate lymphoid cells, possible interaction with microbiota. Semin Immunopathol. 2015;37:27–37. doi: 10.1007/s00281-014-0470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Guo XH, Liang Y, Zhang Y, Lasorella A, Kee BL, Fu YX. Innate lymphoid cells control early colonization resistance against intestinal pathogens through ID2-dependent regulation of the microbiota. Immunity. 2015;42:731–743. doi: 10.1016/j.immuni.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Buela KAG, Omenetti S, Pizarro TT. Cross-talk between type 3 innate lymphoid cells and the gut microbiota in inflammatory bowel disease. Curr Opin Gastroen. 2015;31:449–455. doi: 10.1097/MOG.0000000000000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, Lienenklaus S, Weiss S, Staeheli P, Aichele P, Diefenbach A. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 227.Chen JN, Wei YF, He JQ, Cui GY, Zhu YN, Lu C, Ding YL, Xue RF, Bai L, Uede T, Li LJ, Diao HY. Natural killer T cells play a necessary role in modulating of immune-mediated liver injury by gut microbiota. Sci Rep. 2014;4:7259. doi: 10.1038/srep07259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Chehoud C, Rafail S, Tyldsley AS, Seykora JT, Lambris JD, Grice EA. Complement modulates the cutaneous microbiome and inflammatory milieu. Proc Natl Acad Sci USA. 2013;110:15061–15066. doi: 10.1073/pnas.1307855110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Salzman NH, Bos NA, Harmsen HJ, Welling GW, Paterson Y, Bevins CL. Regulation of the intestinal microbiota by Paneth cell defensins. Gastroenterology. 2005;128:A508–A508. [Google Scholar]
- 230.Salzman NH, Underwood MA, Bevins CL. Paneth cells, defensins, and the commensal microbiota: a hypothesis on intimate interplay at the intestinal mucosa. Semin Immunol. 2007;19:70–83. doi: 10.1016/j.smim.2007.04.002. [DOI] [PubMed] [Google Scholar]