Abstract
Macrophage-expressed gene 1 (MPEG1) encodes an evolutionarily conserved protein with a predicted membrane attack complex/perforin domain associated with host defence against invading pathogens. In vertebrates, MPEG1/perforin-2 is an integral membrane protein of macrophages, suspected to be involved in the killing of intracellular bacteria by pore-forming activity. Zebrafish have 3 copies of MPEG1; 2 are expressed in macrophages, whereas the third could be a pseudogene. The mpeg1 and mpeg1.2 genes show differential regulation during infection of zebrafish embryos with the bacterial pathogens Mycobacterium marinum and Salmonella typhimurium. While mpeg1 is downregulated during infection with both pathogens, mpeg1.2 is infection inducible. Upregulation of mpeg1.2 is partially dependent on the presence of functional Mpeg1 and requires the Toll-like receptor adaptor molecule MyD88 and the transcription factor NFκB. Knockdown of mpeg1 alters the immune response to M. marinum infection and results in an increased bacterial burden. In Salmonella typhimurium infection, both mpeg1 and mpeg1.2 knockdown increase the bacterial burdens, but mpeg1 morphants show increased survival times. The combined results of these two in vivo infection models support the anti-bacterial function of the MPEG1/perforin-2 family and indicate that the intricate cross-regulation of the two mpeg1 copies aids the zebrafish host in combatting infection of various pathogens.
Key Words: Membrane attack complex, Perforin, Perforin-2, mpeg1, Zebrafish embryo, Innate immunity, Macrophages, Infection, Salmonella, Mycobacterium
Introduction
Membrane attack complex/perforin (MACPF) proteins belong to a large superfamily of pore-forming molecules present in almost all living organisms [1, 2]. In vertebrates, MACPF proteins have crucial roles in immune defence against both extracellular and intracellular infections [3, 4]. The MACPF domain is present in the components of the terminal complement pathway (C6, C7, C8 and C9), which form the membrane attack complex (MAC) directly targeting gram-negative bacteria and certain pathogenic parasites by forming pores on their cell membranes [1]. The MACPF domain is also present in the perforins released by cytotoxic T lymphocytes and natural killer cells, which create pores in the plasma membranes of infected and transformed host cells, allowing the entry of cytolytic proteins [4].
In addition to the complement proteins and the perforins, there is another MACPF domain-containing membrane protein involved in the immune system, named macrophage-expressed gene 1 (MPEG1) or perforin-2 [3]. MPEG1 was first identified as a macrophage-specific gene in humans and mice, but how it contributes mechanistically to macrophage defence remains to be elucidated [3, 5]. Homologs of MPEG1 are found in marine metazoans, such as the sea sponge [6], the pacific oyster [7], and the abalone shellfish [8], and evolutionary reconstruction suggests that an ancestral MPEG1 gene gave rise to the vertebrate perforin genes [9]. The expression of MPEG1 homologs in the invertebrates is upregulated by viral and bacterial infections or by exposure to bacterial lipopolysaccharides (LPS) [6, 7, 8]. Furthermore, recombinant sea sponge Mpeg has been shown to inhibit bacterial growth in vitro [6]. In mice, Mpeg1 (also named perforin-2) is upregulated during prion infection and its expression could be induced in primary mouse embryonic fibroblasts by several types of bacterial infections [10, 11]. Mouse embryonic fibroblasts rapidly induce expression of Mpeg1 in response to infection with Mycobacterium smegmatis, and these fibroblasts lose their ability to kill this non-pathogenic mycobacterium species when Mpeg1 is knocked down with siRNA [11]. Furthermore, this study showed that M. smegmatis bacteria are sensitive to the bacteriolytic activity of lysozymes when they are recovered from Mpeg1-expressing fibroblasts but not when they are recovered from Mpeg1-deficient cells, suggesting physical attack of the bacterial membrane by the MACPF domain of Mpeg1 [11]. It has also been shown that Mpeg1 restricts the growth of Chlamydia trachomatis in macrophages, while studies in HeLa cells suggest that chlamydiae are protected from Mpeg1-mediated killing in epithelial cells by prevention of Mpeg1 transcription [12]. Following ectopic expression of Mpeg1 in HeLa cells, the protein concentrated around the chlamydia-containing vacuoles and inhibited chlamydial growth. It has been proposed that the MACPF domain of Mpeg1 is oriented inside membrane vesicles and that upon bacterial infection these vesicles traffic and fuse with bacteria-containing endosomes to mediate bacterial killing by pore formation [3].
The zebrafish genome contains an mpeg1 gene that, like its human and murine counterparts, is expressed by macrophages and encodes a protein with the conserved MACPF domain as well as the characteristic transmembrane region [13, 14]. There are several advantages of using zebrafish as a model to study host-pathogen interactions. Firstly, the free-living zebrafish embryos and early larval stages are optically transparent and in vivo visualization of infectious disease processes is facilitated by various transgenic lines, including reporter lines using the mpeg1 promoter to drive fluorescent protein expression in macrophages [14]. Secondly, genetic approaches are easily applicable, such as the efficient inactivation of gene functions achieved by injection of anti-sense morpholino oligonucleotides. Thirdly, it is possible to study the innate immune response in the absence of an adaptive immune contribution. Already at 1 day post-fertilisation (dpf), the innate immune system of the zebrafish embryo is capable of defence against microbial infections [15], while the adaptive immune system does not mature before 3 weeks of age [16]. Differentiated myeloid cells of the innate immune system are able to phagocytose apoptotic cell corpses [15] and adhere to and phagocytose bacteria injected into the blood [15, 17, 18]. These properties led to the development of zebrafish infection models for a wide variety of pathogens [19].
In this study, we used well-established zebrafish embryo models for M. marinum and Salmonella enterica serovar Typhimurium (S. typhimurium) infections to investigate the function of mpeg1 and one of its paralogues named mpeg1.2. We show that these two MPEG1 homologues are differentially regulated during infection and provide in vivo evidence for the function of both genes in anti-bacterial defence.
Materials and Methods
Zebrafish Lines and Handling of Embryos
Zebrafish were handled in compliance with local animal welfare regulations and maintained according to standard protocols (zfin.org). The culture was approved by the local animal welfare committee (DEC) of the University of Leiden and all protocols adhered to the international guidelines specified by EU Animal Protection Directive 2010/63/EU. Adult zebrafish were not sacrificed for this study. All experiments in this study were performed on embryos/larvae before the free-feeding stage and did not fall under the animal experimentation law according to EU Animal Protection Directive 2010/63/EU. The zebrafish lines used in this study included AB/TL, myd88hu3568[20], Tg(mpx:gfp)i114[21], Tg(mpeg1:EGFP)gl22[14], and Tg(mpeg1::mCherry-F)UMSF001[22]. Embryos were grown at 28.5-30°C in egg water (60 µg/ml Instant Ocean sea salts). For the duration of bacterial injections and imaging, the embryos were kept under anaesthesia in egg water containing 200 µg/ml tricaine (Sigma-Aldrich). Embryos used for stereo fluorescence imaging were kept in egg water containing 0.003% 1-phenyl-2-thiourea (Sigma-Aldrich) to prevent melanisation.
Morpholino Knockdown
Morpholino oligonucleotides (Gene Tools) were diluted to the desired concentration in 1× Danieau buffer [58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca(NO3)2, and 5.0 mM HEPES; pH 7.6] containing 1% phenol red (Sigma-Aldrich), and 1 ηl was injected into the yolk at the 1- to 2-cell stage using a Femtojet injector (Eppendorf). For knockdown of mpeg1 or mpeg1.2, two morpholinos each were used, i.e. one targeting the exon 1-intron 1 splice junction (mpeg1 Mo1: 5′ATTTTGTACTTACTTGAACCCGTGC3′, 0.3 mM; mpeg1.2 Mo1: 5′ACTTTTCTGTCTTACCTGAACTCGT3′, 0.1 mM) and the other targeting the intron 1-exon 2 splice junction (mpeg1 Mo2: 5′GGTTACGGACCTGAGAAACAAATTT3′, 0.1 mM; mpeg1.2 Mo2: 5′TGCGTACCTGAGAAGATAACACAAA3′, 0.1 mM). For knockdown of ptpn6, the splice junction morpholino 1 (ptpn6 Mo1: 5′ACTCATTCCTTACCCGATGCGGAGC3′) was used as described previously [23]. As a control, the standard control morpholino from Gene Tools was used at the same concentrations as the other morpholinos.
Chemical Treatments
NFκBactivation inhibitor (50 nM, 4 h total treatment, including 2 h pretreatment, No. 481406; Calbiochem) and copper sulphate (CuSO4, 10 µM, 2 h treatment, No. 1027910250; Merck) [24] were administered via the egg water. After copper sulphate treatment, the embryos were briefly washed 3 times with egg water and fixed in 4% paraformaldehyde in PBS.
Infection Experiments
M. marinum infections were performed using the Mma20 or ΔRD1 strains expressing mCherry in a pSMT3 vector [25, 40] or the M strain expressing GFP in a pMSP12 vector [26]. M. marinum was heat-killed by incubation in 80°C for 20 min and plated during injections to control for bacterial growth. S. typhimurium infections were performed using the S. typhimurium strain SL1027 and its isogenic LPS Ra mutant derivative SF1592, carrying the DsRed expression vector pGMDs3 [18], and Staphylococcus epidermidis infections were performed using strain O-47 [27]. Embryos were staged at 24 hpf by morphological criteria and manually dechorionated. Bacteria were prepared and injected into the blood circulation at 28 hpf, and PBS or 2% PVP was injected as a control [28]. The same conditions were used for injection of 1 nl of LPS from S. typhimurium dissolved in PBS (10 μg/ml, No. L6511; Sigma). For S. typhimurium plating assays, groups of 5 infected embryos (150 cfu) were homogenized using a Bullet Blender (Next Advance) for 3 min on speed 4 with five 1.0-mm zirconium oxide beads and a density of 5.5 g/cm3, and a dilutions series was plated at 1 and 16 hours post-infection (hpi).
Fluorescence-Activated Cell Sorting
Macrophages and neutrophils were isolated by FACS from 6-dpf Tg(mpeg1::mCherry-F)UMSF001[22] and Tg(mpx::egfp)i114[21] zebrafish larvae, respectively, and the RNA was extracted as described [13].
RNA Isolation, cDNA Synthesis, and Expression Analyses
Whole embryo RNA isolation, removal of residual genomic DNA, cDNA synthesis and quantitative RT-PCR (qPCR) analysis were performed as described by Stockhammer et al. [29]. qPCR results were analysed using the ΔΔCt method. All reactions were performed as technical duplicates and data were normalized to the expression of peptidylprolyl isomerase A-like (ppial) for whole embryo samples and to eif4 for cells isolated by FACS. The primer sequences for ppial and mpeg1 are described in the study of Zakrzewska et al. [13], and the primer sequences for mmp9 and il1b are described in the study of Stockhammer et al. [29]. The primer sequences for mpeg1.2 were: forward: 5′TCAGGCCAATGTGAACGACA3′; reverse: 5′GGTGACTCAGGAGTGCATGT3′. The primer sequences for eif4a1b were: forward: 5′TTCAGAAACTCAGTACTAGCATACA3′; reverse: 5′GTGACATCCAACACCTCTGC3′. Knockdown of mpeg1 with both morpholinos was verified by qPCR and, to validate the knockdown of mpeg1.2 with both morpholinos, the SuperScript® One-Step RT-PCR System (No. 10928-034; Invitrogen) was used with 50 ng DNAse-treated RNA template. The RT-PCR primers for mpeg1.2 Mo1 knockdown verification were: forward: 5′CTGCGCAATTTAGACGTGGG3′; reverse: 5′GACGTGTCGTTCACATTGGC3′. For Mo2 they were: forward: as Mo1 primer; reverse: 5′TTAATGCCTGCGGATGCAGA3′. The following adjustments were made to the PCR settings: 59.4°C for 30 s for the annealing step of the PCR amplification with 40 cycles.
RNA for deep sequencing analysis was isolated using QIAzol lysis reagent and purified using the RNeasy MinElute Cleanup Kit (QIAGEN Benelux B.V., Venlo, The Netherlands). RNA sequencing was performed as previously described [27]. The Gene Expression Omnibus (GEO) database accession numbers for RNASeq were GSE49188 and GSE54885.
Immunohistochemistry and Enzyme Histochemistry
For simultaneous identification of S. typhimurium, neutrophils and macrophages, a combination of immunolabelling and enzymatic staining was used. First, neutrophil-specific staining for myeloperoxidase (mpx) activity was performed. This was achieved by fixing embryos in 4% PFA at 4°C overnight, washing 3 times briefly in PBSTx (PBS with 0.05% Triton ×100; Sigma-Aldrich), washing briefly in amp diluent from the TSA Plus Fluorescein System Kit (Perkin-Elmer) and incubating in 1:50 of fluorescein in amp diluent at 28°C for 10 min. The embryos were then washed again 3 times in PBSTx and fixed in 4% PFA for 20 min at room temperature. Next, the embryos were immunolabelled with an anti-Salmonella polyclonal antibody with an Alexa 568-conjugated secondary antibody as described [30]. Finally, total leukocytes were immunolabelled with an L-plastin antibody and Alexa 633-conjugated secondary antibody [31]. Macrophages were identified as L-plastin-positive, mpx-negative cells.
Image Analysis
Fluorescence images were taken with a Leica MZ16FA stereo fluorescence microscope equipped with a DFC420C digital colour camera. In M. marinum infection experiments, bacterial pixel counts were obtained with stereo fluorescence images and analysed using dedicated pixel quantification software [32]. Each image contains 2 channels: bright field and fluorescence. Image recognition software identifies the location of each embryo (each image can contain up to 3 embryos) and links the outline and orientation per embryo in the bright field channel to the fluorescence channel. Each fluorescent pixel is quantified and allocated to a location in the embryo (the background is determined by a non-infected control) and the software provides the value of the total amount of fluorescent pixels per embryo. This is done for all groups of embryos and the results are written to a comma-separated file for statistical analysis. Leukocyte counts were performed on a Zeiss Axio Observer confocal microscope with an EC PlnN 10 × 0.3 NA objective, and all other confocal images were taken on a Leica TCS SPE with an HCX APO 40 × 0.8 NA objective. Maximum intensity projections of merged channel confocal images were made in Fiji.
Statistical Analysis
Data (mean ± SEM) were analysed (Prism 5.0; GraphPad Software, San Diego, Calif., USA) using unpaired, two-tailed t-tests for comparisons between 2 groups, and one-way ANOVA with Tukey's multiple comparisons method was used as a post hoc test for other data (* p < 0.05; ** p < 0.01; *** p < 0.001). Statistical significance in survival curves was determined by a log-rank test.
Results
The Zebrafish Genome Contains a Conserved Family of mpeg1 Genes
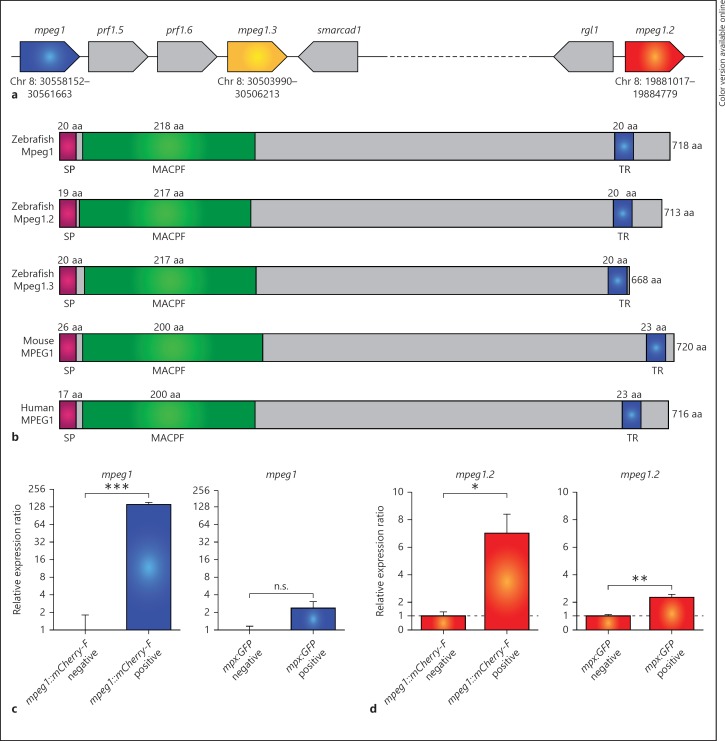
The zebrafish mpeg1 gene (chromosome 8: ENSDARG00000055290) has been previously described as a macrophage-specific marker [13], [14]. Inspection of the zebrafish reference genome sequence showed that 2 paralogues of the mpeg1 gene are present on the same chromosome (fig. 1a). In consultation with the zebrafish nomenclature committee (ZFIN), the 3 mpeg1 genes have been named mpeg1.1 (hereafter referred to as mpeg1 for consistency with previous publications and the nomenclature of the derived transgenic lines), mpeg1.2 (chromosome 8: ENSDARG00000043093) and mpeg1.3 (chromosome 8: ENSDARG00000078569). All 3 of the predicted Mpeg1 proteins contain conserved regions also found in mouse and human MPEG1, including a signal peptide, an MACPF domain, and a transmembrane region (fig. 1b). Amino acid alignment of Mpeg1 and Mpeg1.2 revealed that these domains are located in the same positions (online suppl. fig. 1; for all online suppl. material, see www.karger.com/doi/10.1159/000366103). The three zebrafish Mpeg1 isoforms show around 80–90% amino acid similarity (with ca. 60–70% identity) among each other, and their similarity with murine and human MPEG1 (overall and within the MACPF domain) is around 70–80% (with between 43 and 50% identity) (online suppl. table 1A, B).
Fig. 1.
Zebrafish mpeg1 genes encode proteins with conserved transmembrane and MACPF domains and are expressed in macrophages. a Schematic representation of the region containing the genes mpeg1, mpeg1.2 and mpeg1.3 on zebrafish chromosome 8 (Chr 8). The coding direction of the genes is indicated by the pointed end and the exact location is indicated in digits below the gene. b Comparison of the predicted Mpeg1, Mpeg1.2 and Mpeg1.3 protein structures with murine and human MPEG1. The signal peptide (SP), MACPF and transmembrane region (TR) are conserved domains detected by SMART analysis. c, d Expression of zebrafish mpeg1 and mpeg1.2 in macrophages (c) and neutrophils (d). qPCR analysis was performed on RNA from fluorescence-positive cell fractions obtained by cell sorting of 6-dpf larvae of transgenic reporter lines for macrophages [mpeg1: Tg(mpeg1::mCherry-F)] and neutrophils [mpx: Tg(mpx:EGFP)]. Expression was compared against the fluorescence-negative cell fraction of each transgenic line. Graphs show combined data of 3 biological replicates [log2 scale (c)]. n.s. = Not significant.
The mpeg1 promoter region has been successfully used to drive macrophage-specific expression in zebrafish transgenic reporter lines [14, 22]. We determined the RNAseq profiles of macrophages isolated from 5-day-old Tg(mpeg1::mCherry-F)UMSF001 zebrafish larvae by FACS and found the expression of both mpeg1 and mpeg1.2 to be enriched in the fluorescent macrophage cell fraction compared to the fluorescent-negative background (data not shown). The macrophage-specific expression of these two genes was confirmed by qPCR analysis on the same cell fractions and their expression was not enriched in neutrophils sorted from Tg(mpx::egfp)i114[21] transgenic larvae (fig. 1c, d). Expression of mpeg1.3 was not observed in any of our RNAseq deep sequencing data sets of larval leukocyte populations or of different bacterial infection experiments in various embryonic, larval, and adult stages, and it is well possible that this is a pseudogene. Therefore, we focused this study on mpeg1 and mpeg1.2.
Mpeg1-Expressing Macrophages Participate in the Formation of Granuloma-Like Structures
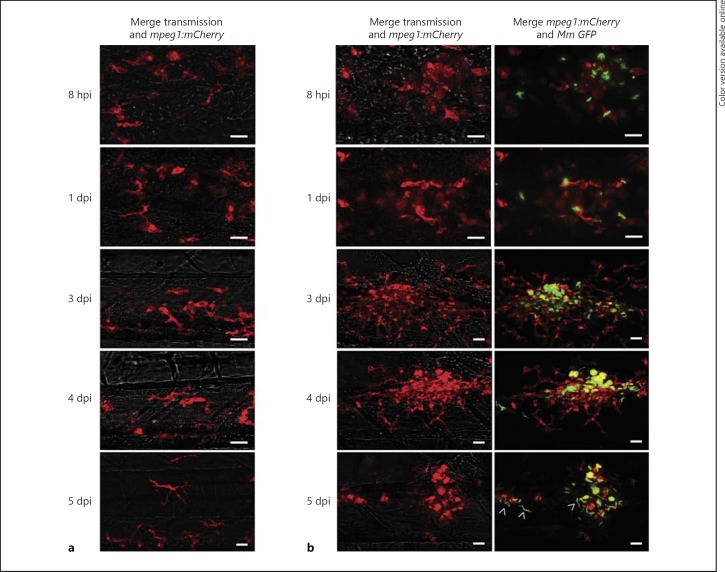
The development of mpeg1-driven transgenic lines has opened a new window of opportunity for detailed visualisation of host-pathogen interactions. We have previously shown that mpeg1-expressing macrophages are capable of phagocytosing both S. typhimurium and M. marinum bacteria [28]. The hallmark of infection with pathogenic mycobacteria is the clustering of infected macrophages into granuloma-like aggregates [17]. We used the Tg(mpeg1::mCherry-F)UMSF001 line to study the behaviour of mpeg1-expressing macrophages during infection with GFP-expressing M. marinum. Imaging of the same site of infection over a time course of 0–5 days post-infection (dpi) clearly showed that mpeg1-positive macrophages contribute to the formation of the tight granuloma-like structure (fig. 2). At 8 hpi, the majority of phagocytosed M. marinum bacteria were contained within mpeg1-positive cells. By 3 dpi, mpeg1-positive infected cells had formed a compact centre of a growing granuloma-like aggregate to which many uninfected mpeg1-macrophages were attracted. M. marinum-containing mpeg1-positive cells showed a rounded morphology at this stage, while mpeg1-positive cells attracted to the border of granulomas showed a branched morphology similar to that of mpeg1-positive cells in uninfected embryos. At 4 dpi, the granuloma structures had become more compact. At 5 dpi, cording of M. marinum was observed in granulomas, indicative of extracellular growth, while fewer mCherry-fluorescent macrophages were present compared to at 4 dpi, consistent with the occurrence of infected macrophage cell death at this stage [33]. The remaining mCherry-fluorescent cells all showed a rounded morphology markedly different from the extensively branched morphology of mCherry-fluorescent macrophages in uninfected embryos (fig. 2a). While these data do not exclude the possible presence of other macrophage subpopulations, they show that the majority of macrophages contributing to granuloma formation can be traced using the mpeg1 marker.
Fig. 2.
Tg(mpeg1)-positive macrophages are involved in the formation of M. marinum granuloma-like aggregates. GFP-expressing M. marinum M-strain (150 cfu) bacterial infection in Tg(mpeg1::mCherry-F) zebrafish embryos. Confocal z-stack projections showing identical locations in the posterior region of the caudal haematopoietic tissue in an uninfected embryo (a) and an infected embryo (b) developing a granuloma-like aggregate (same infection site followed over time). Images were taken at 8 hpi and at 1 dpi, 2 dpi (not shown), 3 dpi, 4 dpi and 5 dpi. Arrowheads indicate cording of M. marinum. Scale bar = 20 μm.
Opposite Regulation of mpeg1 and mpeg1.2 during Infection
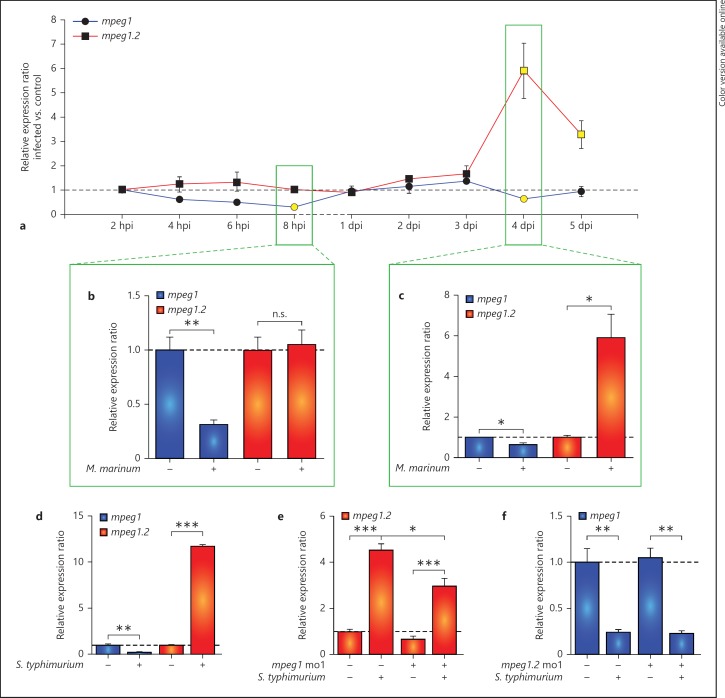
Analysis of the Tg(mpeg1::mCherry-F)UMSF001 reporter line indicated that the mpeg1 promoter version driving the expression of mCherry in this transgenic line is active over the entire time course of M. marinum infection, from the 1-day-old embryo up to the larval stage. However, in S. typhimurium infection of embryos we had previously observed mpeg1 expression to be downregulated [23, 29]. These observations prompted us to further investigate the regulation of mpeg1 expression during different infections and compare this with the expression of the closely related mpeg1.2 gene. First, we examined mpeg1 and mpeg1.2 expression by qPCR during a time course of M. marinum infection ranging from 2 hpi to 5 dpi (fig. 3a). Within several hours after the intravenous injection of M. marinum, the expression level of mpeg1 declined compared to the mock-injected controls, and this resulted in a significant downregulation of approximately 3-fold at 8 hpi (fig. 3b). A similar, though not significant, trend was observed at 8 hpi in embryos infected with the attenuated M. marinum ∆RD1 strain (online suppl. fig. 2A) as well as in embryos infected with heat-killed M. marinum (online suppl. fig. 2B). This indicates that the mpeg1 downregulation response results from the presence of M. marinum and is independent of the virulence or viability of the bacteria. Between 1 and 3 dpi, the level of expression returned to control levels, but at 4 dpi a minor but significant downregulation was observed again (fig. 3c). In contrast, the expression of mpeg1.2 was unaffected up to 3 dpi and was significantly upregulated at 4 and 5 dpi (fig. 3c).
Fig. 3.
mpeg1 is downregulated and mpeg1.2 is upregulated upon bacterial infection. a-cmpeg1 and mpeg1.2 expression during M. marinum infection. AB/TL embryos were injected with M. marinum Mma20 bacteria (200 cfu) or 2% PVP as a mock control. The expression of mpeg1 and mpeg1.2 was analysed by qPCR at 2, 4, 6 and 8 hpi and at 1, 2, 3, 4 and 5 dpi. The light colouring of the data points in a indicates that the expression in the infected embryos was significantly different from that in the uninfected controls, and in b and c the full data sets are shown for the time point indicated in boxes in a. dmpeg1 and mpeg1.2 expression in response to S. typhimurium SL1027 infection. AB/TL embryos were injected with 200 cfu of bacteria or mock injected with PBS, and qPCR was performed at 8 hpi. e, f Effect of mpeg1 and mpeg1.2 morpholino knockdown on each other's gene expression. AB/TL embryos were injected with mpeg1 (e) or mpeg1.2 (f) morpholino and subsequently infected with S. typhimurium SL1027 as in d. Note that mpeg1 knockdown had a reducing effect on the upregulation of mpeg1.2 expression by S. typhimurium infection, while mpeg1.2 knockdown did not affect the infection-dependent downregulation of mpeg1. Verification of the knockdown effects is shown in online supplementary figure 4A, B, C, D. qPCR results are presented as relative ratios of 3 biological replicates of the infected groups compared to the relevant mock injected control groups (n = 18 per group). n.s. = Not significant.
Next, we investigated if mpeg1 and mpeg1.2 were also differentially regulated during S. typhimurium infection. Intravenous S. typhimurium infection in zebrafish embryos is acute and lethal within 1 day [18]. We had previously reported a robust induction of proinflammatory gene expression at 8 hpi [23, 29] and therefore investigated mpeg1 and mpeg1.2 expression at this time point. While mpeg1 showed an approximately 4-fold downregulation, mpeg1.2 was upregulated by more than 10-fold (fig. 3d). The uninfected and S. typhimurium-infected embryos at 8 hpi contained similar numbers of macrophages (uninfected: 54 ± 4.5 s.e.m., n = 16 embryos; infected: 51 ± 3.6 s.e.m, n = 15 embryos). Furthermore, as previously shown, the expression of other markers for zebrafish embryonic macrophages, including csf1r, cxcr3.2 and mfap4, is unchanged at this time point of infection, indicating the specificity of alterations in mpeg1 and mpeg1.2 expression [23]. Infection with an attenuated LPS mutant of S. typhimurium (Ra) [18] also led to a significant downregulation of mpeg1, but mpeg1.2 showed only a slight and non-significant upregulation (online suppl. fig. 2C). We also observed downregulation of mpeg1 and upregulation of mpeg1.2 in infection models for S. epidermidis (online suppl. fig. 2D, E), indicating that the differential regulation of these genes is a more general phenomenon in response to bacterial infections.
The opposite regulation of mpeg1 and mpeg1.2 raised the question of whether these two genes might influence each other's expression levels. To address this question, we used the S. typhimurium infection model and knocked down mpeg1 or mpeg1.2 by injecting morpholinos specific for each gene (online suppl. fig. 3A, B, C, D). When mpeg1 was knocked down, the expression of mpeg1.2 was upregulated to a lower extent than in the control infected group (fig. 3e). In contrast, when mpeg1.2 was knocked down the downregulation of mpeg1 was unchanged during S. typhimurium infection (fig. 3f). Since the mpeg1 morpholino sequence did not overlap with the sequence of mpeg1.2, the mpeg1 knockdown effect on mpeg1.2 gene expression is unlikely to be due to a cross-reaction between the mpeg1 morpholino and the mpeg1.2 mRNA. Furthermore, we excluded that morpholino treatments might have affected the macrophage or neutrophil numbers (online suppl. fig. 3E). Therefore, our results suggest that infection-dependent upregulation of mpeg1.2 is partially dependent on the presence of a functional Mpeg1.
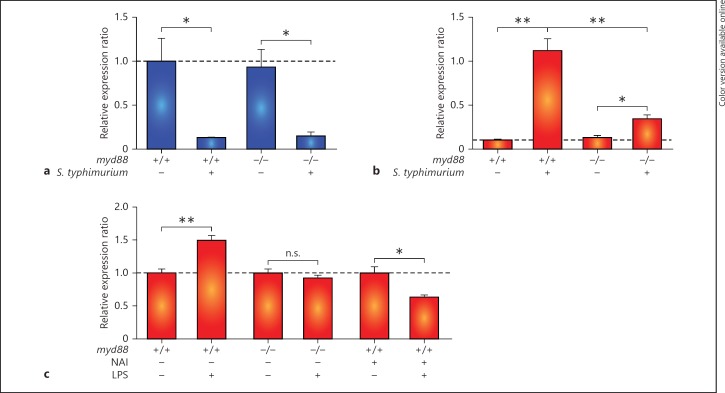
The innate immune response is initiated by the detection of microbes through pattern recognition receptors, including Toll-like receptors (TLR), which signal via both Myd88-dependent and Myd88-independent pathways [34]. Our previous analysis of an Myd88-deficient zebrafish mutant showed that a major part of the innate immune response of zebrafish embryos to bacterial infections is dependent on this central TLR signalling adaptor [20]. To determine whether the regulation of mpeg1 and mpeg1.2 in response to infection is Myd88 dependent, we injected S. typhimurium into myd88 mutants and their wild-type siblings. Downregulation of mpeg1 at 8 hpi was observed in myd88 mutants similar to the wild type (fig. 4a). In contrast, mpeg1.2 upregulation was reduced to approximately 30% of the level observed for wild-type infected embryos (fig. 4b). The TLR ligand LPS was sufficient to induce mpeg1.2 upregulation (fig. 4c) but did not lead to mpeg1 downregulation (data not shown). LPS-mediated upregulation of mpeg1.2 was abolished by myd88 mutation or by inhibition of the transcription factor NFκB, which functions downstream of TLR-Myd88 signalling (fig. 4c). In conclusion, upregulation of mpeg1.2 expression is partly dependent on MyD88-NFκB signalling, while downregulation of mpeg1 appears to be mediated by an Myd88-independent mechanism.
Fig. 4.
mpeg1.2 upregulation but not mpeg1 downregulation is Myd88 and NFκB dependent. a, b Effect of myd88 mutation on the response of mpeg1 and mpeg1.2 to S. typhimurium infection. Expression levels of mpeg1 (a) and mpeg1.2 (b) were determined by qPCR for myd88+/+ and myd88−/− embryos at 8 hpi after infection with S. typhimurium SL1027 bacteria (150 cfu) and mock PBS injected controls. c Effect of myd88 mutation and NFκB inhibition on the response of mpeg1.2 to LPS treatment. mpeg1.2 expression in myd88+/+, myd88−/− and NFκB activation inhibitor-treated embryos in response to injection with purified LPS (100 μg/ml) or PBS as a control was determined at 2 hpi. All graphs show data combined from 3 biological replicates (n = 20 per group, pooled per replicate). n.s. = Not significant.
Knockdown of mpeg1 Leads to Impaired Control of M. marinum Infection
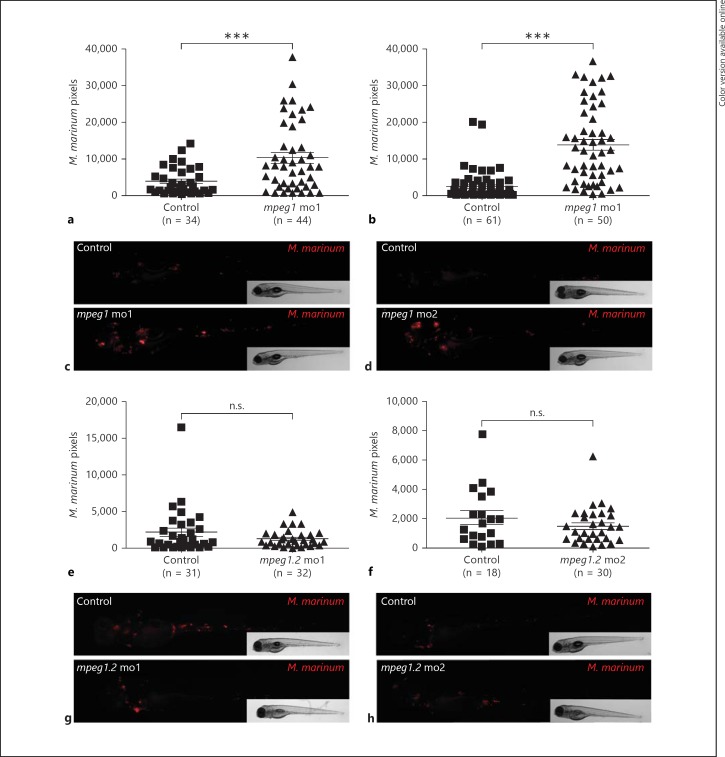
To study the function of Mpeg1 and Mpeg1.2 during bacterial infection, we used splice morpholinos targeting each gene. First, we investigated the effect of knocking down mpeg1 on M. marinum infection. The morphants and their controls were injected with mCherry-expressing M. marinum at 28 hpf and the bacterial fluorescent pixels per embryo were analysed at 4 dpi, when granuloma-like aggregates are formed (fig. 2). mpeg1 morphants showed higher levels of infection with M. marinum compared to their controls (fig. 5a, c). The higher bacterial load in the mpeg1 morphants was phenocopied with a second morpholino (fig. 5b, d). M. marinum infection experiments were terminated at 5 dpi before the larvae reached the free-feeding stage and mpeg1 morphants did not die from the increased bacterial load during this period. We did not expect to see an effect on the bacterial load in the mpeg1.2 morphants since the expression of this gene is barely detectable during the first days of development and only becomes induced at 4 dpi. As expected, mpeg1.2 knockdown with two different morpholinos did not have an effect on M. marinum infection (fig. 5 e-h). While mpeg1.2 may be important at later larval stages that cannot be analysed by morpholino knockdown, we conclude that only Mpeg1 plays an important role in controlling the early pathogenesis of M. marinum infection.
Fig. 5.
mpeg1 knockdown impairs control of M. marinum infection. AB/TL embryos were injected with 2 different splice-blocking morpholinos against mpeg1 (a-d) or mpeg1.2 (e-h) or with a control morpholino, and subsequently injected with mCherry-expressing M. marinum Mma20 strain; infected embryos were imaged at 4 dpi. a, b, e, f The bacterial burden was quantified by determining the number of fluorescent bacterial pixels using dedicated software, and representative stereo fluorescent images are shown below the graph of each experiment (c, d, g, h). Graphs show 1 representative result of 5 (a) or 3 (b, e, f) repeated independent experiments. Each data point represents an individual embryo. n.s. = Not significant; mo1 = morpholino 1; mo2 = morpholino 2.
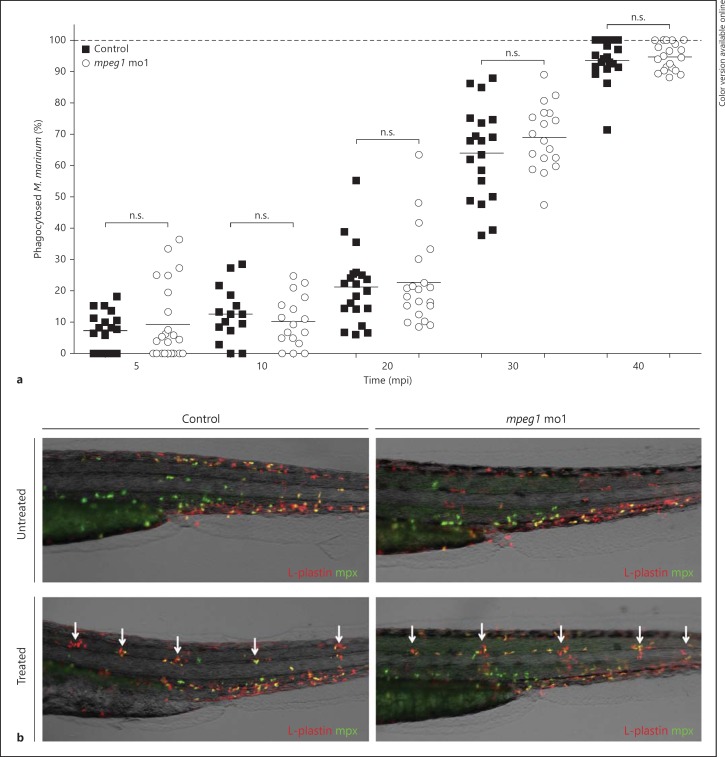
The observation that embryos deficient in Mpeg1 develop increased infection is consistent with the expected anti-bacterial function of Mpeg1 proteins as members of the MACPF superfamily. We next investigated if, other than this direct anti-bacterial role, Mpeg1 might be important for macrophage functions such as phagocytosis or migration. A phagocytosis assay was performed using both morpholinos targeting mpeg1. The morphants and their controls were injected with 180 cfu of mCherry-expressing Mma20 at 30 hpf and these embryos were fixed at the time points 5, 10, 20, 30 and 40 min post-infection. Leukocytes were immunolabelled with L-plastin antibody and Alexa488-conjugated secondary antibody [31] and all intracellular and extracellular Mma20 were counted over the yolk sac of each embryo to determine the percentage of phagocytosis. The yolk sac is an ideal location for analysing phagocytosis due to the superficial position of the blood circulation, which enables easy imaging, and this location is distant from the injection site, thereby minimizing possible wounding effects on the behaviour of macrophages. We observed normal levels of phagocytosis in both mpeg1 morphants at all stages post-infection (fig. 6a), demonstrating that Mpeg1 does not play a role in the phagocytosis of M. marinum. Since inflammation plays an important role in the M. marinum-granuloma environment, we also investigated whether mpeg1 deficiency might have a general effect on leukocyte recruitment. We therefore performed a chemically induced inflammation assay [24] on mpeg1 morphants and their controls. The copper-induced damage of neuromast hair cells in this assay was capable of attracting both macrophages and neutrophils in mpeg1 morphants similarly to the control group (fig. 6b). In summary, Mpeg1 does not affect the basal response of macrophages to infection, such as phagocytosis of M. marinum, or the migration of leukocytes towards local inflammation sites.
Fig. 6.
mpeg1 does not play a role in phagocytosis of M. marinum or in leukocyte migration towards local inflammation. a Quantification of M. marinum phagocytosis. mpeg1 morphants and their controls were injected with an mCherry-expressing M. marinum Mma20 strain (180 cfu), fixed at 5, 10, 20, 30 and 40 min post-infection, and stained with L-plastin Ab to label leukocytes. Intra- and extra-cellular bacteria were counted over the yolk sac and results are presented as percentages of phagocytosed Mma20. b Representative images of untreated and copper sulphate-treated control and mpeg1 morphant 3 dpf embryos. Embryos were immunolabelled with Ab against the general leukocyte marker L-plastin (red signal) in combination with a neutrophil-specific Mpx TSA staining (green signal). White arrows indicate accumulation of leukocytes at the local inflammation sites at the neuromasts. n.s. = Not significant; mo1 = morpholino 1.
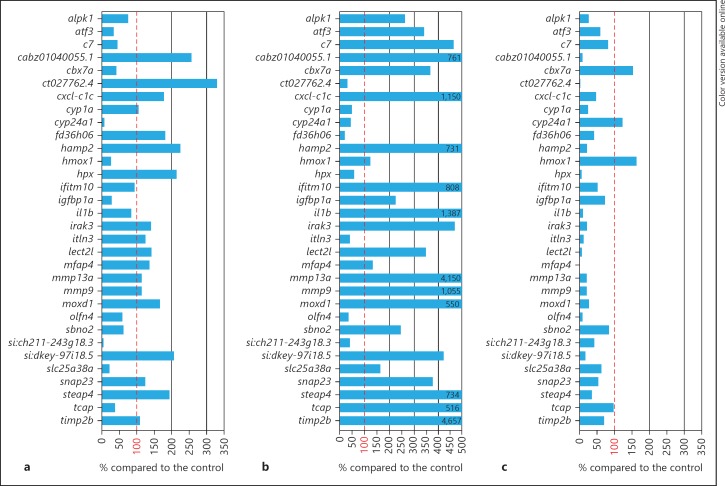
We next sought to identify whether mpeg1 morphants have a differently regulated immune response to M. marinum. To this end, we subjected pools of approximately 30 infected and uninfected mpeg1 morphants to RNA sequencing analysis at 4 dpi and compared the infection response with that of embryos treated with control morpholinos. We focused this comparison on a preselected set of 33 genes that showed robust and reproducible upregulation by M. marinum infection at 4 dpi in 4 independent experiments (online suppl. table 2). Approximately one third of the genes in this set, including pro-inflammatory markers such as il1b, mmp9 and mmp13a, showed a level of M. marinum-induced upregulation in mpeg1 morphants similar to that observed in control embryos (fig. 7a). Other genes showed an approximately 2- to 3-fold higher upregulation in the mpeg1 morphant group, including hamp2, hpx, steap4 and genes for a non-coding RNA (si:dkey-97i18.5) or uncharacterized proteins (cabz01040055.1 ct027762.4). Additionally, there was a group of genes with a notably lower expression in infected mpeg1 morphants (less than 28% of the expression level in infected control embryos), which included cyp24a1, hmox1, igfbp1a, slc25a38a and another non-coding RNA gene (si:ch211-243g18.3). The effect of mpeg1 knockdown on the M. marinum-induced gene expression profile was markedly different from the effects of 2 other genes that we analysed for comparison: ptpn6 and myd88. Knockdown of ptpn6, previously shown to function as a negative regulator of the innate immune response [23], led to a hyper-induction of approximately two thirds of the genes in the same gene set, including the inflammatory markers il1b, mmp9, and mmp13a (fig. 7b). In contrast, these inflammatory markers and the majority of the other genes in the gene set showed a strongly reduced expression in M. marinum-infected myd88 mutants (fig. 7c), consistent with the function of the MyD88 protein as an adaptor in Toll-like and interleukin receptor signalling [20]. We have previously shown that both the hyper-induced immune response in ptpn6 morphants and the immunodeficiency of myd88 mutants are associated with increased susceptibility to M. marinum [20, 23]. Since the mpeg1 knockdown effect on the M. marinum-dependent gene set is clearly different from both phenotypes, there is no indication that a general hyper-induction of the immune response or a general immunodeficiency could be the underlying cause of the increased bacterial burden under mpeg1 knockdown conditions.
Fig. 7.
Effect of mpeg1 knockdown on the innate immune response during M. marinum infection in comparison with known effects of ptpn6 and myd88 deficiencies. Graphs show the effects of mpeg1 mo1 knockdown (a), ptpn6 morpholino knockdown (b) andmyd88 mutation (c) on a set of genes that showed reproducible induction by M. marinum infection in control embryos. The expression level of these genes under conditions of mpeg1, ptpn6 or myd88 deficiency is expressed as the percentage of the expression level in the corresponding control. Results are based on RNAseq analysis of pools of 30 infected and 30 uninfected embryos for each group. Embryos were injected with M. marinum Mma20 (300 cfu) or mock injected with 2% PVP, and RNAseq analysis was performed at 4 dpi.
Mpeg1 and Mpeg1.2 both Function in Controlling S. typhimurium Infection
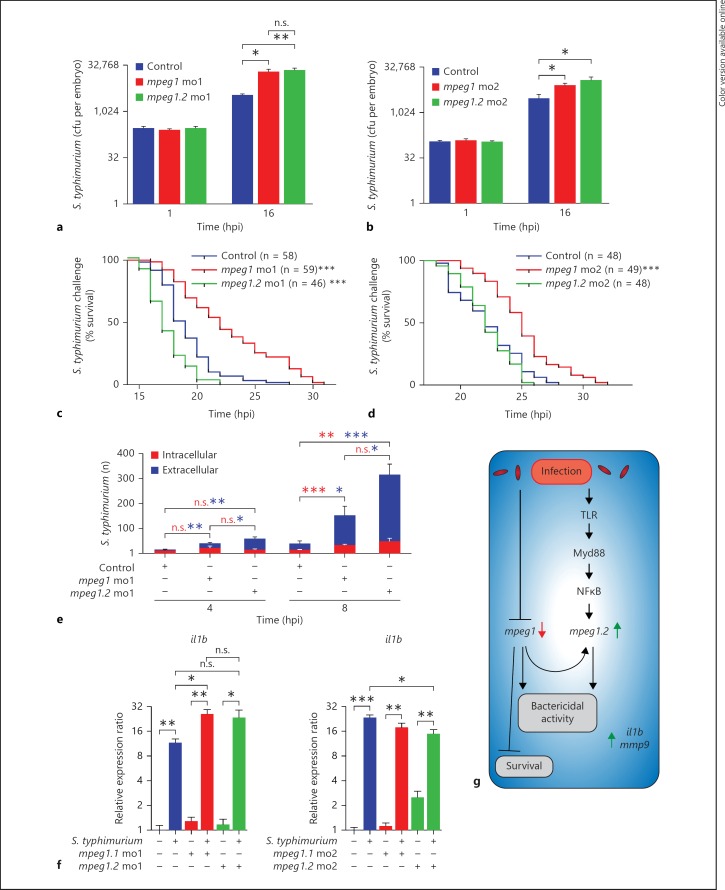
As described above, the more acute phenotype of S. typhimurium infection led to the rapid upregulation of mpeg1.2 gene expression, while mpeg1 was simultaneously downregulated (fig. 4a, b). To determine whether Mpeg1 and Mpeg1.2 play an anti-bacterial role during this acute infection, we injected S. typhimurium in mpeg1 and mpeg1.2 morphants at 28 hpf and assessed the bacterial burden based on cfu counts. Plating embryos for cfu counts showed that at 1 hpi all groups started with equal levels of S. typhimurium and at 16 hpi morphants of mpeg1 and mpeg1.2 both had approximately 6-fold higher cfu counts than the control group (fig. 8a). Increased cfu counts were also observed with a second set of morpholinos for mpeg1 and mpeg1.2 (fig. 8b). This striking difference in cfu counts indicates that both genes play a role in controlling S. typhimurium infection in zebrafish embryos.
Fig. 8.
mpeg1 and mpeg1.2 knockdown increase the bacterial burden and pro-inflammatory gene expression but have opposite effects on host survival during S. typhimurium infection. a, b Quantification of the bacterial burden. mpeg1 and mpeg1.2 morphants [morpholino 1 (a) and morpholino 2 (b)] and control embryos were injected with Ds-Red-expressing S. typhimurium SL1027 bacteria (200 cfu) and PBS as a mock control. Embryos were homogenized and plated at 1 and 16 hpi to determine S. typhimurium cfu counts (n = 5 per group, 2 biological replicates, log2 scale). c, d Survival rates. The percentage of survival of infected mpeg1 morphants, mpeg1.2 morphants [morpholino 1 (c) and morpholinos 2 (d)] and control embryos was determined over a time course of 32 hpi. DsRed-expressing S. typhimurium SL1027 bacteria were injected into the blood circulation (150 cfu). Survival curves of mpeg1 morphants, mpeg1.2 morphants and control embryos are shown (1 representative experiment of 3 individual experiments). e Quantification of intracellular and extracellular S. typhimurium over the duct of Cuvier in control embryos, mpeg1 morphants and mpeg1.2 morphants at 4 and 8 hpi (representative confocal images are shown in online suppl. fig. 4). Statistical significance is indicated for intracellular S. typhimurium (red asterisks) and extracellular S. typhimurium (blue asterisks) (calculated from confocal images of n = 6–8 embryos per group). f Pro-inflammatory gene expression of il1b under the same experimental conditions as in a and b was analysed by qPCR at 16 hpi (n = 15 per group, pooled per replicate, 3 biological replicates, log2 scale). g Schematic representation of mpeg1 and mpeg1.2 regulation and their function. n.s. = Not significant; mo1 = morpholino 1; mo2 = morpholino 2.
Next, we aimed to determine whether the higher cfu numbers in the morphants was associated with a decreased survival rate. Morphants of mpeg1 and mpeg1.2 and control embryos were injected with S. typhimurium, screened for equal levels of infection under a stereo fluorescence microscope directly after injection, and monitored for heart beat from 14 hpi onwards. mpeg1.2 deficiency caused a similar or decreased survival time compared to the control group (fig. 8c, d) and, unexpectedly, the mpeg1 morphants survived significantly longer than the control embryos (fig. 8c, d) despite the fact that these morphants showed increased cfu counts similarly to mpeg1.2 morphants. This opposing effect on survival could not be attributed to a difference in the intracellular or extracellular location of S. typhimurium between the two morphants because both mpeg1 and mpeg1.2 morphants showed a significantly higher number of extracellular S. typhimurium compared to the control embryos at 8 hpi (fig. 8e; online suppl. fig. 4). Furthermore, the opposing survival effect could also not be attributed to a difference in expression of the major S. typhimurium responsive pro-inflammatory genes il1b (fig. 8f) and mmp9 (online suppl. fig. 5), which showed clear induction in mpeg1 and mpeg1.2 morphants. While the increased survival rate of mpeg1 morphants remains unexplained, our results suggest that upregulation of mpeg1.2 with a concomitant downregulation of mpeg1 aids the host in combatting S. typhimurium infection (fig. 8g).
Discussion
The perforins of cytotoxic T cells and natural killer cells and the membrane attack complex proteins of the complement system share the ability to form pores in membranes [1, 2]. While the roles of these proteins in host defence are well understood, the function of the structurally related MPEG1/perforin-2 family in macrophages remains to be fully elucidated [3, 9]. In zebrafish, an mpeg1-expressing population of macrophages develops during the first day of embryogenesis [13]. The availability of well-established infection models for zebrafish embryos enabled us to study the function of mpeg1 in innate host defence. We showed that mpeg1 is downregulated during infection in a Myd88-independent manner, while a close homolog, i.e. mpeg1.2, is upregulated partially by an Myd88-NFκB-dependent mechanism. This opposite regulation pattern notwithstanding, we found that both mpeg1 and mpeg1.2 are important for controlling infections with intracellular bacterial pathogens.
The close chromosomal location of the different mpeg1 genes in zebrafish suggests that these are the result of recent gene duplication events, after which mpeg1 and mpeg1.2 appear to have developed specialized functions and mpeg1.3 may have become a pseudogene. A similar situation is found in mice and other rodents, where a gene named pore-forming protein-like (Pfpl) is most likely paralogous to Mpeg1 since it is located in close vicinity and encodes a protein with 66% amino acid identity. Whereas Mpeg1 is inducible by infection both in macrophages and other cell types [11, 12], the expression pattern of Pfpl in trophoblasts indicates a developmental function of this gene [35], and EST profiles in the NCBI database provide no indication of a significant expression of Pfpl in cells or organs related to the immune system. The EST profiles of the single MPEG1 gene in humans support its possible function in the immune system but, to the best of our knowledge, functional studies of the human gene have not yet been reported. Studies of murine Mpeg1 in cultured fibroblasts, macrophages and HeLa cells led to the hypothesis that, upon bacterial infection, Mpeg1/Perforin-2 vesicles traffic to and fuse with bacteria-containing compartments, where attack and pore formation on the bacterial surface are subsequently initiated [3, 11, 12]. To further investigate the correlation of MPEG1 with infectious diseases, we inspected curated microarray data sets in the NextBio database (http://www.nextbio.com/b/nextbioCorp.nb). This analysis showed that MPEG1 is frequently regulated during infection, both in studies using cultured macrophages and in studies using mouse infection models. For example, MPEG1 shows significant downregulation in human macrophages exposed to S. aureus[36] and in human monocytes infected with Francisella tularensis[37]. In contrast, upregulation has been observed in, for example, the lungs of mice infected with these pathogens [38, 39]. Thus, in humans and mice different infection conditions are associated either with up- or downregulation of MPEG1, while in zebrafish mpeg1 and mpeg1.2 are regulated in opposite directions under the same infection conditions.
Using the zebrafish-M. marinum infection model, we found that knockdown of mpeg1 results in an increased bacterial burden, consistent with the proposed bactericidal function of Mpeg1 as a pore-forming molecule [11]. However, there might be a broader effect as we observed an altered immune response expression signature in infected mpeg1 morphants that might also have an impact on the ability to control infection. The function of mpeg1.2, which has an extremely low basal expression level, could only be assessed using an S. typhimurium infection model, where this gene is rapidly upregulated. In this model, knockdown of mpeg1 attenuated the upregulation of mpeg1.2, indicating a broader effect of mpeg1 on the innate immune response, consistent with the results of M. marinum infection. Knockdown of either mpeg1 or the inducible mpeg1.2 gene led to an increased bacterial burden of S. typhimurium infection, providing in vivo support for the anti-bacterial function of both genes. Due to the toxicity effects of combining the morpholinos, we were unable to investigate a possible synergism between the anti-bacterial functions of mpeg1 and mpeg1.2. However, a remarkable difference between the separate knockdown of mpeg1 and mpeg1.2 was observed in that the survival time of mpeg1 morphants was prolonged despite the increase in cfu numbers. This suggests that the survival advantage of mpeg1 morphants could be due to an altered immune response and that, during the normal course of S. typhimurium infection, embryos might die from the host-damaging effects of the immune response rather than as a direct effect of the bacterial load. That S. typhimurium-infected zebrafish embryos die primarily from a host-damaging immune response is supported by our previous study of the regulatory phosphatase Ptpn6. A deficiency of Ptpn6 leads to a decreased survival rate during S. typhimurium infection, which correlates with a hyper-induced expression of many pro-inflammatory genes [23]. Two of the main pro-inflammatory marker genes, i.e. il1b and mmp9, were highly upregulated in both mpeg1 and mpeg1.2 morphants, similarly to infected control embryos. A difference in the expression of these marker genes therefore does not explain the survival advantage of mpeg1 morphants, and the underlying cause of this effect currently remains unknown. Together, this first study of the MPEG1/perforin-2 family in a whole organism model enables us to present a scheme that summarizes the regulatory mechanisms of both mpeg1 and mpeg1.2 and links this to the biological effects of the two genes (fig. 8g). The specialized function of the two mpeg1 paralogues in zebrafish indicated by our morpholino knockdown study can be seen as motivation to develop zebrafish or mouse knockout models for further investigation of the mechanisms via which Mpeg1/perforin-2 proteins exert their immunological function.
Disclosure Statement
The authors declare that there are no competing financial interests.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgements
The authors would like to thank Georges Lutfalla (University Montpellier 2) for the kind gift of Tg(mpeg1::mCherry-F) fish, Michiel van der Vaart for providing RNA of LPS-injected myd88 mutant embryos, Ulrike Nehrdich, Laura van Hulst and Davy de Witt for fish care, and other lab members for helpful discussions. E.B., A.E.N., F.J.V., H.P.S. and A.H.M. were supported by the Smart Mix Program of the Netherlands Ministry of Economic Affairs and the Ministry of Education, Culture and Science, P.I.R. was supported by a Marie Curie Intra-European Fellowship (PIEF-GA-2010-274389), and J.R. was supported by a Marie Curie Fellowship of the European 7th Framework Initial Training Network FishForPharma (PITG-GA-2011-289209).
References
- 1.Kondos SC, Hatfaludi T, Voskoboinik I, Trapani JA, Law RHP, Whisstock JC, Dunstone MA. The structure and function of mammalian membrane-attack complex/perforin-like proteins. Tissue Antigens. 2010;76:341–351. doi: 10.1111/j.1399-0039.2010.01566.x. [DOI] [PubMed] [Google Scholar]
- 2.Rosado CJ, Kondos S, Bull TE, Kuiper MJ, Law RH, Buckle AM, Voskoboinik I, Bird PI, Trapani JA, Whisstock JC, Dunstone MA. The MACPF/CDC family of pore-forming toxins. Cell Microbiol. 2008;10:1765–1774. doi: 10.1111/j.1462-5822.2008.01191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCormack R, de Armas L, Shiratsuchi M, Podack ER. Killing machines: three pore-forming proteins of the immune system. Immunol Res. 2013;57:268–278. doi: 10.1007/s12026-013-8469-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voskoboinik I, Dunstone MA, Baran K, Whisstock JC, Trapani JA. Perforin: structure, function, and role in human immunopathology. Immunol Rev. 2010;235:35–54. doi: 10.1111/j.0105-2896.2010.00896.x. [DOI] [PubMed] [Google Scholar]
- 5.Spilsbury K, O'Mara MA, Wu WM, Rowe PB, Symonds G, Takayama Y. Isolation of a novel macrophage-specific gene by differential cDNA analysis. Blood. 1995;85:1620–1629. [PubMed] [Google Scholar]
- 6.Wiens M. Innate immune defense of the sponge Suberites domuncula against bacteria involves a MyD88-dependent signaling pathway: induction of a perforin-like molecule. J Biol Chem. 2005;280:27949–27959. doi: 10.1074/jbc.M504049200. [DOI] [PubMed] [Google Scholar]
- 7.Renault T, Faury N, Barbosa-Solomieu V, Moreau K. Suppression substractive hybridisation (SSH) and real time PCR reveal differential gene expression in the pacific cupped oyster, Crassostrea gigas, challenged with ostreid herpesvirus 1. Dev Comp Immunol. 2011;35:725–735. doi: 10.1016/j.dci.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Kemp IK, Coyne VE. Identification and characterisation of the mpeg1 homologue in the South African abalone, Haliotis midae. Fish Shellfish Immunol. 2011;31:754–764. doi: 10.1016/j.fsi.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 9.D'Angelo ME, Dunstone MA, Whisstock JC, Trapani JA, Bird PI. Perforin evolved from a gene duplication of MPEG1, followed by a complex pattern of gene gain and loss within Euteleostomi. BMC Evol Biol. 2012;12:59. doi: 10.1186/1471-2148-12-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kopacek J, Sakaguchi S, Shigematsu K, Nishida N, Atarashi R, Nakaoke R, Moriuchi R, Niwa M, Katamine S. Upregulation of the genes encoding lysosomal hydrolases, a perforin-like protein, and peroxidases in the brains of mice affected with an experimental prion disease. J Virol. 2000;74:411–417. doi: 10.1128/jvi.74.1.411-417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCormack R, de Armas LR, Shiratsuchi M, Ramos JE, Podack ER. Inhibition of intracellular bacterial replication in fibroblasts is dependent on the perforin-like protein (perforin-2) encoded by macrophage-expressed gene 1. J Innate Immun. 2013;5:185–194. doi: 10.1159/000345249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fields KA, McCormack R, de Armas LR, Podack ER. Perforin-2 restricts growth of Chlamydia trachomatis in macrophages. Infect Immun. 2013;81:3045–3054. doi: 10.1128/IAI.00497-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zakrzewska A, Cui C, Stockhammer OW, Benard EL, Spaink HP, Meijer AH. Macrophage-specific gene functions in Spi1-directed innate immunity. Blood. 2010;116:e1–e11. doi: 10.1182/blood-2010-01-262873. [DOI] [PubMed] [Google Scholar]
- 14.Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. Mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood. 2010;117:e49–e56. doi: 10.1182/blood-2010-10-314120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herbomel P, Thisse B, Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999;126:3735–3745. doi: 10.1242/dev.126.17.3735. [DOI] [PubMed] [Google Scholar]
- 16.Trede NS, Langenau DM, Traver D, Look AT, Zon LI. The use of zebrafish to understand immunity. Immunity. 2004;20:367–379. doi: 10.1016/s1074-7613(04)00084-6. [DOI] [PubMed] [Google Scholar]
- 17.Davis JM, Clay H, Lewis JL, Ghori N, Herbomel P, Ramakrishnan L. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity. 2002;17:693–702. doi: 10.1016/s1074-7613(02)00475-2. [DOI] [PubMed] [Google Scholar]
- 18.van der Sar AM, Musters RJP, van Eeden FJM, Appelmelk BJ, Vandenbroucke-Grauls CMJE, Bitter W. Zebrafish embryos as a model host for the real time analysis of Salmonella typhimurium infections. Cell Microbiol. 2003;5:601–611. doi: 10.1046/j.1462-5822.2003.00303.x. [DOI] [PubMed] [Google Scholar]
- 19.Meijer AH, Spaink HP. Host-pathogen interactions made transparent with the zebrafish model. Curr Drug Targets. 2011;12:1000–1017. doi: 10.2174/138945011795677809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Vaart M, van Soest JJ, Spaink HP, Meijer AH. Functional analysis of a zebrafish myd88 mutant identifies key transcriptional components of the innate immune system. Dis Model Mech. 2013;6:841–854. doi: 10.1242/dmm.010843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, Whyte MK. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108:3976–3978. doi: 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- 22.Bernut A, Herrmann J, Kissaa K, Dubremetz J, Gaillard J, Lutfalla G, Kremera L. Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proc Natl Acad Sci USA. 2014;111:E943–E952. doi: 10.1073/pnas.1321390111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanwal Z, Zakrzewska A, den Hertog J, Spaink HP, Schaaf MJ, Meijer AH. Deficiency in hematopoietic phosphatase ptpn6/Shp1 hyperactivates the innate immune system and impairs control of bacterial infections in zebrafish embryos. J Immunol. 2013;190:1631–1645. doi: 10.4049/jimmunol.1200551. [DOI] [PubMed] [Google Scholar]
- 24.d'Alençon CA, Peña OA, Wittmann C, Gallardo VE, Jones RA, Loosli F, Liebel U, Grabher C, Allende ML. A high-throughput chemically induced inflammation assay in zebrafish. BMC Biol. 2010;8:151. doi: 10.1186/1741-7007-8-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Sar AM, Abdallah AM, Sparrius M, Reinders E, Vandenbroucke-Grauls CMJE, Bitter W. Mycobacterium marinum strains can be divided into two distinct types based on genetic diversity and virulence. Infect Immun. 2004;72:6306–6312. doi: 10.1128/IAI.72.11.6306-6312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cosma CL, Humbert O, Sherman DR, Ramakrishnan L. Trafficking of superinfecting mycobacterium organisms into established granulomas occurs in mammals and is independent of the Erp and ESX-1 mycobacterial virulence loci. J Infect Dis. 2008;198:1851–1855. doi: 10.1086/593175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veneman WJ, Stockhammer OW, de Boer L, Zaat SA, Meijer AH, Spaink HP. A zebrafish high throughput screening system used for Staphylococcus epidermidis infection marker discovery. BMC Genomics. 2013;14:255. doi: 10.1186/1471-2164-14-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benard EL, van der Sar AM, Ellett F, Lieschke GJ, Spaink HP, Meijer AH. Infection of zebrafish embryos with intracellular bacterial pathogens. J Vis Exp. 2012:p 3781. doi: 10.3791/3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stockhammer OW, Zakrzewska A, Hegedus Z, Spaink HP, Meijer AH. Transcriptome profiling and functional analyses of the zebrafish embryonic innate immune response to salmonella infection. J Immunol. 2009;182:5641–5653. doi: 10.4049/jimmunol.0900082. [DOI] [PubMed] [Google Scholar]
- 30.Meijer AH, van der Sar AM, Cunha C, Lamers GE, Laplante MA, Kikuta H, Bitter W, Becker TS, Spaink HP. Identification and real-time imaging of a myc-expressing neutrophil population involved in inflammation and mycobacterial granuloma formation in zebrafish. Dev Comp Immunol. 2008;32:36–49. doi: 10.1016/j.dci.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Mathias JR, Dodd ME, Walters KB, Rhodes J, Kanki JP, Look AT, Huttenlocher A. Live imaging of chronic inflammation caused by mutation of zebrafish Hai1. J Cell Sci. 2007;120:3372–3383. doi: 10.1242/jcs.009159. [DOI] [PubMed] [Google Scholar]
- 32.Nezhinsky AE, Verbeek FJ, Efficient and robust shape retrieval from deformable templates . Leveraging Applications of Formal Methods, Verification and Validation: Applications and Case Studies. In: Margaria T, Steffen B, Merten M, editors. vol 7610. Berlin: Springer; 2012. pp. pp 42–55. [Google Scholar]
- 33.Clay H, Volkman HE, Ramakrishnan L. Tumor necrosis factor signaling mediates resistance to mycobacteria by inhibiting bacterial growth and macrophage death. Immunity. 2008;29:283–294. doi: 10.1016/j.immuni.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 35.Hemberger M, Himmelbauer H, Ruschmann J, Zeitz C, Fundele R. cDNA subtraction cloning reveals novel genes whose temporal and spatial expression indicates association with trophoblast invasion. Dev Biol. 2000;222:158–169. doi: 10.1006/dbio.2000.9705. [DOI] [PubMed] [Google Scholar]
- 36.Koziel J, Maciag-Gudowska A, Mikolajczyk T, Bzowska M, Sturdevant DE, Whitney AR, Shaw LN, DeLeo FR, Potempa J. Phagocytosis of Staphylococcus aureus by macrophages exerts cytoprotective effects manifested by the upregulation of antiapoptotic factors. PLoS One. 2009;4:e5210. doi: 10.1371/journal.pone.0005210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butchar JP, Cremer TJ, Clay CD, Gavrilin MA, Wewers MD, Marsh CB, Schlesinger LS, Tridandapani S. Microarray analysis of human monocytes infected with Francisella tularensis identifies new targets of host response subversion. PLoS One. 2008;3:e2924. doi: 10.1371/journal.pone.0002924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frank KM, Zhou T, Moreno-Vinasco L, Hollett B, Garcia JG, Bubeck Wardenburg J. Host response signature to Staphylococcus aureus alpha-hemolysin implicates pulmonary Th17 response. Infect Immun. 2012;80:3161–3169. doi: 10.1128/IAI.00191-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walters KA, Olsufka R, Kuestner RE, Cho JH, Li H, Zornetzer GA, Wang K, Skerrett SJ, Ozinsky A. Francisella tularensis subsp. Tularensis induces a unique pulmonary inflammatory response: Role of bacterial gene expression in temporal regulation of host defense responses. PLoS One. 2013;8:e62412. doi: 10.1371/journal.pone.0062412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volkman HE, Clay H, Beery D, Chang JC, Sherman DR, Ramakrishnan L. Tuberculous granuloma formation is enhanced by a mycobacterium virulence determinant. PLoS Biol. 2004;2:e367. doi: 10.1371/journal.pbio.0020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data