Abstract
Upon binding to RNA structures from invading viruses, RIG-I and MDA5 are recruited to mitochondria to interact with VISA and initiate antiviral type I interferon (IFN) responses. How this process is mediated is less understood. In this report, we demonstrate that ECSIT is an essential scaffolding protein that mediates the association of VISA and RIG-I or MDA5. Overexpression of ECSIT potentiated virus-triggered activation of IFN-regulatory factor 3 (IRF3) and expression of IFNB1, whereas knockdown of ECSIT impaired viral infection-induced activation of IRF3 and expression of IFNB1 as well as cellular antiviral responses. Mechanistically, ECSIT was associated with VISA on mitochondria and important for bridging RIG-I and MDA5 to VISA. Our findings suggest that ECSIT mediates virus-triggered type I IFN induction by bridging RIG-I and MDA5 to the VISA complex, and provide new insights into the molecular events of innate antiviral immune responses.
Key Words: ECSIT, Innate immunity, Interferon, RIG-I, Signaling, Virus, VISA
Introduction
The germ line-encoded pattern recognition receptors recognize invading viral RNAs or their replication intermediates, and initiate a series of signaling events leading to the induction of hundreds of effector proteins that are involved in the inhibition of viral replication and clearance or apoptosis of infected cells [1, 2, 3]. Various pattern recognition receptors have so far been identified, such as membrane-bound Toll-like receptors (TLRs), cytoplasmic nucleic acid sensors RIG (retinoic acid-inducible gene)-I and MDA5 (melanoma differentiation-associated gene 5), Lsm14A, cyclic GMP-AMP synthase (cGAS) and RNA polymerase III [4, 5, 6, 7, 8]. RIG-I and MDA5 are collectively called RIG-I-like receptors (RLRs), which contain two N-terminal tandem caspase activation and recruitment domains (CARDs) and a C-terminal DexD/H box RNA helicase domain [9]. The RNA helicase domains of RLRs bind to viral RNAs, which leads to conformational changes and translocation of RLRs to the mitochondria, where RLRs interact with the mitochondrial adaptor VISA (virus-induced signaling adaptor, also known as MAVS, IPS-1 and Cardif) by their CARDs. VISA further recruits multiple proteins to form a signalosome, leading to the activation of IKK kinase family members, such as TBK1 (TANK-binding kinase 1) and IKKβ (IκB kinase β), the downstream transcription factors interferon (IFN)-regulatory factor 3 (IRF3), nuclear factor (NF)-κB and the induction of type I IFNs [10, 11, 12, 13].
It has been demonstrated that the translocation of RLRs to mitochondria for the association with VISA represents an essential step in type I IFN induction upon viral infection, and several proteins have been reported to mediate this process. The mitochondrial targeting chaperon protein 14-3-3ε interacts with RIG-I and TRIM25 to form a ‘translocon’ complex that directs RIG-I and TRIM25 to MAM (mitochondria-associated membrane), where 14-3-3ε stabilizes RIG-I-TRIM25 interactions and facilitates K63-linked ubiquitination of RIG-I by TRIM25 [14, 15]. In addition, the assembly and up-regulation of actin and tubulin on mitochondria is required for the translocation of RIG-I to mitochondria and the interaction between RIG-I and VISA [16]. On the other hand, gC1qR (receptor for globular head domain of complement component C1q) disrupts the RLR-VISA interaction on mitochondria, thereby inhibiting virus-triggered type I IFN signaling [17]. Thus, whether and how other proteins regulate the RLR-VISA interaction is of great interest.
ECSIT (evolutionarily conserved signaling intermediate in Toll pathways) is a component critically involved in Toll/IL-1-induced activation of MEKK1 (MEK kinase 1) and NF-κB and required for BMP signaling and mesoderm formation during mouse embryogenesis [18, 19]. It has been demonstrated that ECSIT localized to mitochondria through its N-terminal domain and it is implicated in the assembly and stability of the mitochondrial respiratory complex [20]. Recently, ECSIT has been found to induce mitochondrial reactive oxygen species (mROS) generation downstream of TLR1, TLR2 and TLR4, which is critical for mouse macrophage bactericidal activity [21]. Although it has been reported that TRIM59 inhibits VISA-induced activation of IRF3 and NF-κB by interacting with ECSIT [22], it is unclear how ECSIT is involved in virus-triggered type I IFN signaling. In this report, we found that ECSIT acted as an essential scaffolding protein that bridged RIG-I and MDA5 to VISA, and thereby plays an essential role in RLR-mediated type I IFN induction and cellular antiviral responses. Our findings provide new information on the molecular mechanisms of RLR-mediated cellular antiviral responses.
Materials and Methods
Constructs
ISRE, NF-κB and IFN-β promoter luciferase reporter plasmids, mammalian expression plasmids for BID-GFP, Sec61β-GFP, HA- or Flag-tagged RIG-I, MDA5, VISA, TLR3, TRAF3, TRAF6, MITA and their truncations were previously described [8, 23, 24]. Cherry-, Flag- or HA-tagged cGAS, ECSIT and its mutants were constructed by standard molecular biology techniques.
Reagents and Antibodies
Recombinant human IFN-β (PeproTech), mouse monoclonal antibodies against Flag, HA and β-actin (Sigma), rabbit monoclonal antibody against p-IRF3 (S396; CST), mouse monoclonal antibody against p-IκBα (CST), mouse antibody against β-tubulin (Invitrogen), mouse polyclonal antibody against LMNB1 and rabbit polyclonal antibody against IRF3 (Santa Cruz Biotechnology) were purchased from the indicated manufacturers. Rabbit anti-ECSIT, rabbit anti-VISA, mouse anti-IκBα, mouse anti-RIG-I and mouse anti-MDA5 antisera were raised against recombinant human ECSIT, VISA, IκBα, RIG-I and MDA5, respectively. Sendai virus (SeV), vesicular stomatitis virus (VSV), GFP-Newcastle disease virus (NDV) and GFP-VSV [8, 25, 26] and the herpes simplex virus-1 (HSV-1) D305A, a DNA virus, were described previously [27]; HSV-1 D305A was provided by Dr. Chunfu Zheng (Soochow University).
Transfection and Reporter Gene Assays
Human embryonic kidney 293 (HEK293) cells (∼1 × 105) were seeded on 24-well plates and transfected on the following day using the standard calcium phosphate precipitation method. In the same experiment, empty control plasmid was added to ensure that each transfection receives the same amount of total DNA. To normalize for transfection efficiency, 0.01 μg of pRL-TK Renilla luciferase reporter plasmid was added to each transfection. Luciferase assays were performed using a dual-specific luciferase assay kit (Promega), and firefly luciferase activities were normalized based on Renilla luciferase activities [26].
Co-Immunoprecipitation and Western Blot Analysis/Cell Fractionation Experiments
These experiments were performed as described [23, 24, 26, 28].
Real-Time PCR
Total RNA was isolated from cells using TRIzol reagent (Takara, Japan) and subjected to real-time PCR analysis to determine mRNA expression. mRNA levels of specific genes were normalized to GAPDH mRNA. Gene-specific primer sequences were as listed in the following:
IFNB1: TTGTTGAGAACCTCCTGGCT (forward),
TGACTATGGTCCAGGCACAG (reverse);
Rantes: GGCAGCCCTCGCTGTCATCC (forward),
GCAGCAGGGTGTGGTGTCCG (reverse);
ISG15: AGGACAGGGTCCCCCTTGCC (forward),
CCTCCAGCCCGCTCACTTGC (reverse);
IL-8: AGGACAGGGTCCCCCTTGCC (forward),
CCTCCAGCCCGCTCACTTGC (reverse);
MCP1: AGGACAGGGTCCCCCTTGCC (forward),
CCTCCAGCCCGCTCACTTGC (reverse);
GAPDH: GAGTCAACGGATTTGGTCGT (forward),
GACAAGCTTCCCGTTCTCAG (reverse).
VSV Plaque Assays
HEK293 cells were grown in 24-well plates and transfected with the indicated plasmids. Twenty-four hours after transfection, cells were transfected with Lipofectamine with polyinosinic-polycytidylic acid [poly(I:C)] and B-DNA (1 μg) or control buffer for another 24 h prior to VSV infection. One hour after infection, cells were washed with warm PBS and then fresh medium was added. The supernatant was harvested 24 h later and used to infect confluent cultured Vero cells. Plaque assays were then performed as described [23, 24, 26].
Virus Manipulation
Cells were grown in 24-well plates and transfected with the indicated plasmids prior to virus infection. The culture medium was replaced by serum-free DMEM containing NDV-GFP or VSV-GFP virus (multiplicity of infection, MOI = 0.1). One hour later, cells were washed with PBS and then supplemented with DMEM containing 10% FBS. NDV-GFP or VSV-GFP replication was visualized by monitoring GFP expression using fluorescence microscopy [26].
RNAi
Double-strand oligonucleotides corresponding to the target sequences were cloned into the pSuper.Retro RNAi plasmid (Oligoengine Inc.). The following sequences were targeted for human ECSIT:
No. 1: GCAACAAGTGTGTGTATTA;
No. 2: GTGTATTACCACATCCTCA;
No. 3: GCCACATTGACTTCATCTA.
Results
Overexpression of ECSIT Potentiates Virus-Triggered IFN-β Induction
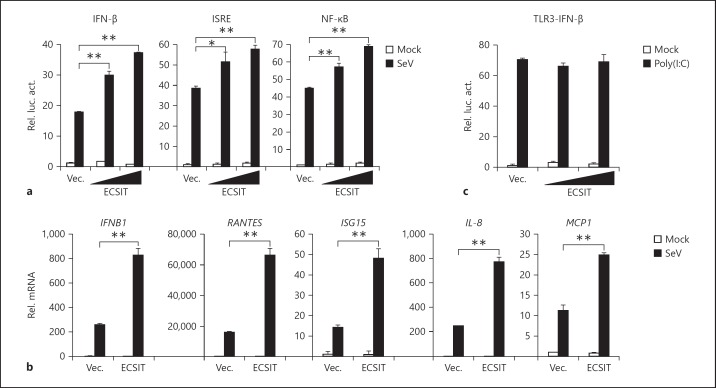
To examine whether ECSIT is involved in the regulation of RLR-mediated IFN induction, we performed reporter assays. We found that overexpression of human ECSIT potentiated SeV-triggered activation of the IFN-β promoter in HEK293 cells (fig. 1a). Since activation of the IFN-β promoter requires coordinated and cooperative activation of IRF3 and NF-κB, we further determined whether ECSIT potentiated virus-induced activation of ISRE and NF-κB. The results indicated that overexpression of ECSIT also potentiated SeV-triggered activation of ISRE and NF-κB (fig. 1a). Results from real-time PCR experiments indicated that overexpression of ECSIT potentiated SeV-triggered transcription of downstream genes, including IFNB1, CCL5, ISG15, IL-8 and MCP1 (fig. 1b). In contrast, ECSIT did not potentiate TLR3-mediated activation of the IFN-β promoter (fig. 1c). These results suggest that overexpression of ECSIT potentiates virus-triggered IFN induction.
Fig. 1.
Overexpression of ECSIT potentiates virus-triggered IFN-β induction. a ECSIT potentiates SeV-induced activation of ISRE and the IFN-β promoter in a dose-dependent manner. HEK293 cells (1 × 105) were transfected with the IFN-β promoter, ISRE (0.1 μg) and NF-κB (0.01 μg) luciferase reporter and the indicated amounts of ECSIT expression plasmids. Twenty hours after transfection, cells were infected with SeV or left uninfected for 10 h before reporter assays were performed. b ECSIT potentiates SeV-induced expression of IFNB1 and downstream genes. HEK293 cells (4 × 105) were transfected with control or ECSIT expression plasmids (1 μg) for 20 h. Cells were then infected with SeV or left uninfected for 10 h before real-time PCR analysis was performed. c ECSIT does not affect TLR3-mediated activation of IFN-β promoter. 293-TLR3 cells (1 × 105) were transfected with IFN-β promoter luciferase reporter plasmids (0.1 μg) and the indicated amounts of ECSIT expression plasmids. Twenty hours after transfection, cells were left untreated or treated with poly(I:C) (25 μg/ml) for 12 h before luciferase assays were performed. Means ± SD, n = 3. * p < 0.05, ** p < 0.01.
ECSIT Is Required for Virus-Triggered Expression of IFNB1
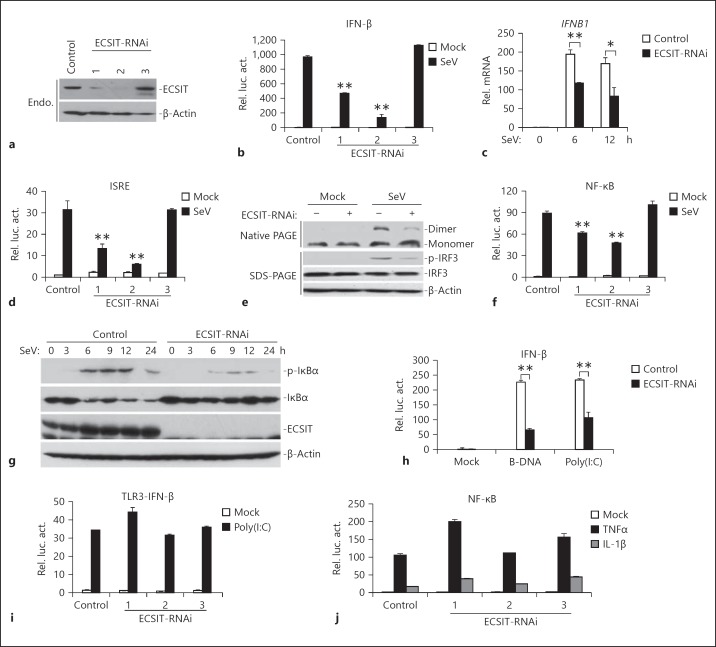
Because overexpression of ECSIT potentiated virus-triggered induction of IFNB1, we next determined whether endogenous ECSIT is required for virus-triggered IFN-β induction. We constructed three RNAi plasmids (pSuper-ECSIT-RNAi Nos. 1–3) targeting ECSIT mRNA and detected their effects on knockdown of ECSIT expression. As shown in figure 2a, the ECSIT-RNAi-1 and -2 constructs could inhibit endogenous expression of ECSIT in HEK293 cells, whereas the ECSIT-RNAi-3 had no marked effect on ECSIT expression. In reporter assays, ECSIT-RNAi-1 and -2 but not ECSIT-RNAi-3 constructs could significantly inhibit SeV-triggered activation of the IFN-β promoter, which was correlated with their effects on ECSIT expression (fig. 2b; we selected the ECSIT-RNAi-2 construct for additional experiments described below; similar results were observed with ECSIT-RNAi-1). Knockdown of ECSIT also inhibited GFP-VSV- and GFP-NDV-triggered activation of the IFN-β promoter (online suppl. fig. 1; for all online suppl. material, see www.karger.com/doi/10.1159/000365971). Consistently, results from real-time PCR experiments confirmed that knockdown of ECSIT inhibited virus-triggered transcription of IFNB1 and downstream genes (fig. 2c and data not shown). In addition, we also found that ECSIT-RNAi significantly inhibited SeV-induced ISRE activation, IRF3 phosphorylation and dimerization, which are hallmarks of IRF3 activation (fig. 2d, e). Furthermore, knockdown of ECSIT markedly inhibited SeV-induced NF-κB activation in reporter assays in HEK293 cells (fig. 2f). Consistently, knockdown of ECSIT also impaired virus-triggered IκBα phosphorylation and degradation (fig. 2g). In reporter assays, ECSIT-RNAi also inhibited activation of the IFN-β promoter triggered by cytosolic poly(I:C) and B-DNA, which were recognized by RIG-I in the cytosol (fig. 2h). In contrast, ECSIT-RNAi had minimal effects on TLR3-mediated or the DNA virus HSV-1 (D305A)-triggered activation of the IFN-β promoter as well as TNFα- and IL-1β-triggered activation of NF-κB (fig. 2i, j; online suppl. fig. S2A). Collectively, these results suggest that endogenous ECSIT plays a key role in the induction of virus-triggered activation of IRF3 and type I IFNs.
Fig. 2.
ECSIT is required for virus-triggered expression of IFNB1. a Effects of ECSIT-RNAi plasmids on the expression of transfected ECSIT. HEK293 cells (4 × 105) were transfected with control or the indicated ECSIT-RNAi plasmids (2 μg each). Thirty-six hours later, cells were lysed and the cell lysates were analyzed by immunoblot with the indicated antibodies. b Effects of ECSIT-RNAi plasmids on SeV-induced activation of the IFN-β promoter. HEK293 cells (1 × 105) were transfected with the indicated ECSIT-RNAi (0.5 μg) and IFN-β reporter (0.1 μg) plasmids. Thirty-six hours after transfection, cells were left uninfected or infected with SeV for 10 h before luciferase assays were performed. c Effects of ECSIT-RNAi plasmids on SeV-induced expression of IFNB1 gene. HEK293 cells (4 × 105) were transfected with control or ECSIT-RNAi plasmids (22 μg). Thirty-six hours after transfection, cells were left uninfected or infected with SeV for the indicated time points before real-time PCR analysis was performed. d Effects of ECSIT-RNAi plasmids on SeV-induced activation of the ISRE. HEK293 cells (1 × 105) were transfected with the indicated ECSIT-RNAi (0.5 μg) and ISRE (0.1 μg) plasmids. Thirty-six hours after transfection, cells were left uninfected or infected with SeV for 10 h before luciferase assays were performed. e Effects of ECSIT-RNAi plasmids on SeV-induced activation IRF3. HEK293 cells (4 × 105) were transfected with control or ECSIT-RNAi plasmids (2 μg). Thirty-six hours after transfection, cells were left uninfected or infected with SeV for 6 h. Cell lysates were separated by native (top) or SDS (bottom three panels) PAGE and analyzed with the indicated antibodies. f Effects of ECSIT-RNAi plasmids on SeV-induced activation of NF-κB. HEK293 cells (1 × 105) were transfected with the indicated ECSIT-RNAi (0.5 μg) and NF-κB reporter (0.02 μg) plasmids. Thirty-six hours after transfection, cells were left uninfected or infected with SeV for 10 h before luciferase assays were performed. g Effects of ECSIT-RNAi plasmids on SeV-induced phosphorylation and degradation of IκBα. HEK293 cells (4 × 105) were transfected with control or ECSIT-RNAi (2 μg). Thirty-six hours after transfection, cells were left uninfected or infected with SeV for the indicated time points. Cell lysates were separated by SDS-PAGE and analyzed with the indicated antibodies. h Effects of ECSIT-RNAi plasmids on cytoplasmic B-DNA- and poly(I:C)-induced activation of IFN-β promoter. HEK293 cells (1 × 105) were transfected with ECSIT-RNAi (0.5 μg) and IFN-β reporter (0.1 μg) plasmids. Thirty-six hours after transfection, cells were mock-transfected or transfected with B-DNA and poly(I:C) (1 μg) with Lipofectamine 2000 for 12 h before luciferase assays were performed. i Effects of ECSIT-RNAi plasmids on TLR3-mediated activation of IFN-β promoter. The 293-TLR3 cells (1 × 105) were transfected with control or ECSIT-RNAi plasmids (0.5 μg) and the IFN-β reporter plasmids (0.1 μg). Thirty-six hours after transfection, cells were left untreated or treated with poly(I:C) (25 μg/ml) for 12 h before luciferase assays were performed. j Effects of ECSIT-RNAi plasmids on TNFα- and IL-1β-induced NF-κB activation. HEK293 cells (1 × 105) were transfected with ECSIT-RNAi (0.5 μg each) and NF-κB luciferase reporter (0.02 μg) plasmids. Thirty-six hours after transfection, cells were left untreated or treated with TNFα (20 ng/ml) and IL-1β (10 ng/ml) for 10 h before luciferase assays were performed. Means ± SD, n = 3. * p < 0.05, ** p < 0.01.
ECSIT Is Essential for Cellular Antiviral Responses
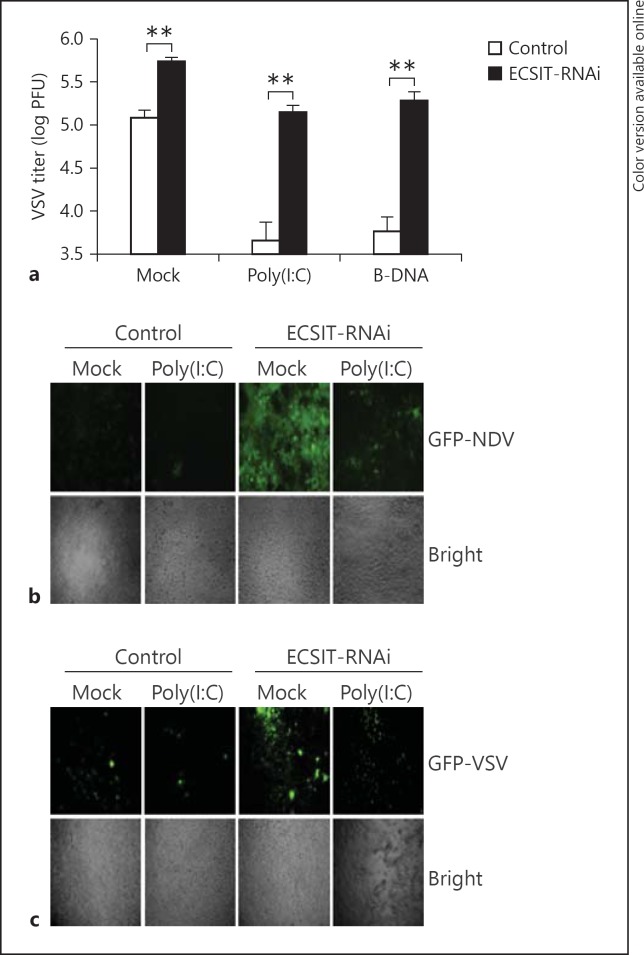
Because ECSIT plays a key role in virus-triggered type I IFN expression, we next determined whether ECSIT was involved in the regulation of cellular antiviral responses. In plaque assays with VSV, we observed that higher VSV titers were produced from ECSIT knockdown compared with control cells. In addition, transfection of cytosolic poly(I:C) and B-DNA enhanced cellular antiviral responses and led to a reduction in VSV titers in the control but not ECSIT knockdown cells (fig. 3a). To directly detect viral replication, we infected ECSIT-RNAi or control cells with GFP-tagged NDV or VSV, and found that knockdown of ECSIT greatly increased GFP signals of infected cells (fig. 3b, c). Collectively, these data suggest that ECSIT is essential for cellular antiviral responses.
Fig. 3.
ECSIT is essential for cellular antiviral response. a Effects of ECSIT-RNAi on VSV replication. HEK293 cells (1 × 105) were transfected with ECIST-RNAi plasmid (0.5 μg). Thirty-six hours later, cells were mock transfected or transfected with B-DNA and poly(I:C) (1 μg) for 16 h and then infected with VSV (MOI = 0.05). The supernatants were harvested 24 h after infection for standard plaque assays. b Effects of ECSIT-RNAi on NDV-GFP replication. HEK293 (1 × 105) cells were transfected with ECSIT-RNAi plasmid (0.5 μg). Thirty-six hours later, cells were infected with GFP-NDV (MOI = 0.2) for 24 h and imaged by microscopy. c Effects of ECSIT-RNAi on VSV-GFP replication. HEK293 (1 × 105) cells were transfected with ECSIT-RNAi plasmid (0.5 μg). Thirty-six hours later, cells were infected with GFP-VSV (MOI = 0.1) for 24 h and imaged by microscopy. Means ± SD, n = 3. ** p < 0.01.
ECSIT Localizes to Mitochondria
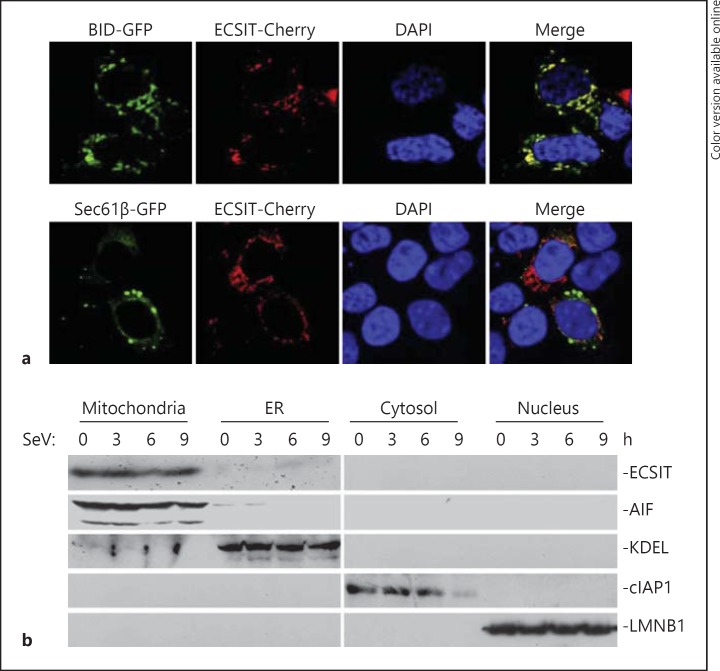
Having established that ECSIT is crucial for virus-triggered type I IFN induction and cellular antiviral responses, we further examined how ECSIT was spatially related in virus-triggered signaling. We analyzed subcellular localizations of ECSIT by confocal microscopy and cell fractionation assays. As shown in figure 4a, ECSIT-Cherry overlapped with mitochondria but not endoplasmic reticulum (ER). In cell fractionation experiments, we observed that ECSIT was mostly detected in the mitochondrial fraction but not in other fractions, such as ER, cytosol and nucleus, and this distribution was not significantly changed after virus infection (fig. 4b). These data suggested that ECSIT localized to mitochondria to regulate virus-triggered signaling.
Fig. 4.
ECIST localized to the mitochondria. a Colocalization of ECSIT with mitochondria. HEK293 cells (1 × 105) were transfected with Cherry-ECSIT (0.1 μg) and the indicated GFP-tagged organelle marker plasmids (0.1 μg). Twenty-four hours after transfection, cells were fixed with 4% (wt/vol) paraformaldehyde and then subjected to confocal microscopy. BID-GFP = Mitochondrial marker; Sec61β-GFP = ER marker. b Immunoblot analysis of the subcellular fractions. HEK293 cells (4 × 106) were infected with SeV for the indicated time points. Cell fractionations were performed and the fractions were analyzed by immunoblotting with the indicated antibodies.
ECSIT Regulates Virus-Triggered Signaling at RLR-VISA Level
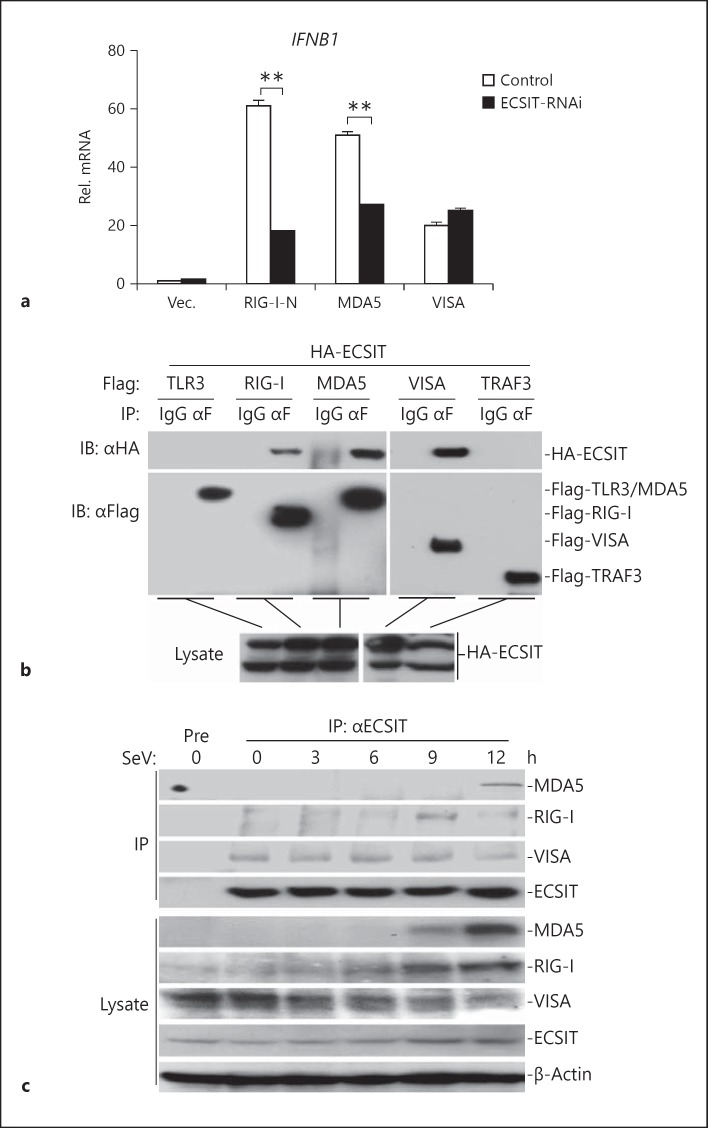
Because ECSIT localized to mitochondria and potentiated virus-triggered IFN induction, we further examined the relationships between ECSIT and VISA-associated components in virus-triggered signaling pathways. In real-time PCR experiments, we found that ECSIT knockdown markedly inhibited RIG-I- and MDA5-mediated IFNB1 expression, but had a minor effect on VISA-mediated IFNB1 expression (fig. 5a), indicating that ECSIT acted upstream of VISA but downstream of RIG-I and MDA5. In transient transfection and co-immunoprecipitation experiments, we found that ECSIT interacted with RIG-I, MDA5 and VISA, but not TLR3 and TRAF3 (fig. 5b). In similar experiments, ECSIT did not interact with cGAS or MITA, which is a critical sensor or adaptor for DNA virus-triggered signaling (online suppl. fig. S2B). Furthermore, overexpression or knockdown of ECSIT did not markedly affect cGAS- and MITA-mediated activation of the IFN-β promoter (online suppl. fig. S2C, D). These data suggest that ECSIT was not involved in DNA sensor-mediated IFN induction.
Fig. 5.
ECSIT regulates virus-triggered signaling at the RLR-VISA level. a Knockdown of ECSIT inhibits RIG-I- and MDA5- but not VISA-mediated IFNB1 gene expression. HEK293 cells (4 × 105) were transfected with control or ECSIT-RNAi plasmids (2 μg). Thirty-six hours later, cells were further transfected with the indicated plasmids. Twenty hours later, real-time PCR experiments were performed. b ECSIT interacts with RIG-I, MDA5 and VISA. HEK293 cells (2 × 106) were transfected with the indicated plasmids. Twenty hours after transfection, co-immunoprecipitation was performed with anti-Flag or control IgG. The immunoprecipitates were analyzed by immunoblot with anti-HA or anti-Flag (upper panel). The lysates were analyzed by immunoblotting with anti-HA (lower panels). c Endogenous ECSIT interacts with RIG-I, MDA5 and VISA. HEK293 cells (1 × 107) were left uninfected or infected with SeV for the indicated time points. Cells were lysed and the lysates were immunoprecipitated with anti-ECSIT. The immunoprecipitates were analyzed by immunoblotting with the indicated antibodies (upper panel). Expression levels of endogenous ECIST, RIG-I, MDA5, VISA and β-actin were analyzed by immunoblotting (lower panels). Means ± SD, n = 3. ** p < 0.01. IP = Immunoprecipitation; IB = immunoblotting.
Since ECSIT is specifically involved in RLR-mediated signaling pathways, we examined endogenous associations between ECSIT and VISA, RIG-I or MDA5. The results indicated that ECSIT was constitutively associated with VISA but not RIG-I and MDA5 in the absence of virus infection. However, the interaction between ECSIT and RIG-I or MDA5 could be detected after virus infection for different times, whereas the interaction between ECSIT and VISA was not changed after virus infection (fig. 5c). These results suggest that ECSIT is constitutively associated with VISA, whereas its association with RIG-I and MDA5 is dependent on viral infection.
It has been reported that high molecular weight (HMW) poly(I:C) is a ligand for MDA5, whereas low molecular weight (LMW) poly(I:C) is a ligand for RIG-I [29]. We examined the effects of ECSIT knockdown on the poly(I:C)-HMW- and poly(I:C)-LMW-induced activation of the IFN-β promoter. As shown in online suppl. fig. S3, knockdown of ECSIT markedly inhibited IFN-β promoter activation triggered by both HMW and LMW poly(I:C). These data suggest that ECSIT plays an important role in both RIG-I- and MDA5-mediated IFN induction.
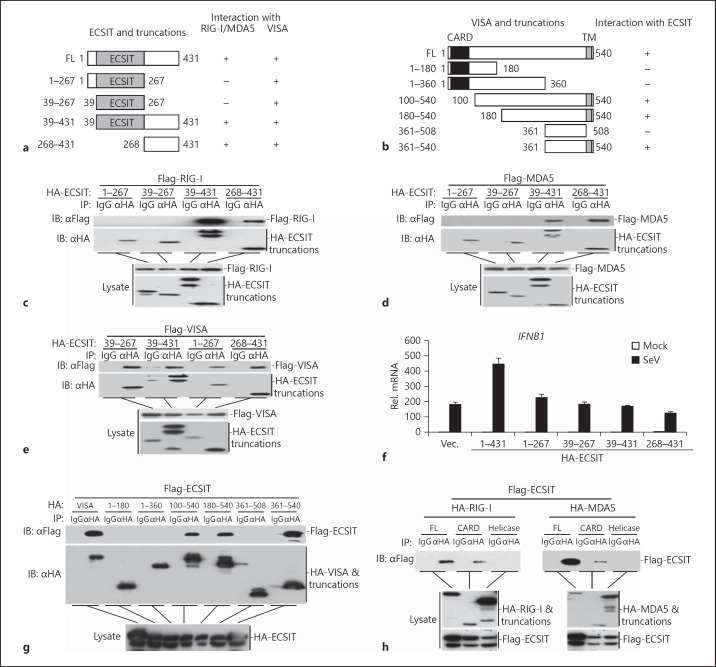
ECSIT Interacts at Different Domains
To further evaluate the domains of ECSIT responsible for ECSIT-associated complex formation, we generated a series of truncation mutants of ECSIT and VISA (fig. 6a, b). The results indicated that the C-terminal region of ECSIT (amino acids 268–431) was required and sufficient for its ability to interact with RIG-I and MDA5 (fig. 6c, d). In contrast, we observed that all the truncation mutants of ECSIT interacted with VISA (fig. 6e). Results from real-time PCR experiments suggested that only full-length but not ECSIT truncation mutants potentiated SeV-triggered expression of IFNB1 and downstream genes (fig. 6f and data not shown). These data suggest that the C-terminal region of ECSIT is sufficient for its association with RIG-I and MDA5, and the structural integrity of ECSIT is required for its function in virus-triggered signaling.
Fig. 6.
Domain mapping of the ECSIT interaction. a A schematic presentation of full-length ECSIT, its mutants and their abilities to interact with RIG-I, MDA5 and VISA. b A schematic presentation of full-length VISA, its mutants and their abilities to interact with ECSIT. c Interaction between full-length RIG-I and ECSIT truncations. HEK293 cells (2 × 106) were transfected with the indicated plasmids. Twenty hours after transfection, co-immunoprecipitation was performed with anti-HA or control IgG. The immunoprecipitates were analyzed by immunoblotting with anti-Flag or anti-HA (upper panel). The lysates were analyzed by immunoblotting with anti-Flag or anti-HA (lower panels). d Interaction between full-length MDA5 and ECSIT truncations. The experiments were performed as in c. e Interaction between full-length VISA and ECSIT truncations. The experiments were performed as in c. f Effects of wild-type or truncated ECSIT on SeV-induced transcription of the IFNB1 gene. HEK293 cells (4 × 105) were transfected with the indicated expression plasmids (2 μg each) for 20 h. Cells were then infected with SeV or left uninfected for 10 h before real-time PCR analysis was performed. g Interaction between full-length ECSIT and VISA truncations. HEK293 cells (2 × 106) were transfected with the indicated plasmids. Twenty hours after transfection, co-immunoprecipitation was performed with anti-HA or control IgG. The immunoprecipitates were analyzed by immunoblotting with anti-Flag or anti-HA (upper panel). The lysates were analyzed by immunoblotting with anti-HA (lower panels). h Interaction between full-length ECSIT and RIG-I or MDA5 truncations. The experiments were performed as in g.
We further mapped the domains of VISA that interact with ECSIT. As shown in figure 6g, only full-length VISA and the mutants containing the C-terminal transmembrane domain interacted with ECSIT, indicating that VISA interacts with ECSIT through its transmembrane domain. RIG-I and MDA5 contained an N-terminal CARD followed by a C-terminal helicase domain. As shown in figure 6h, ECSIT interacted with CARD but not the helicase domain of RIG-I or MDA5. These data together suggest that ECSIT interacts with the CARDs of RLRs and the transmembrane domain of VISA.
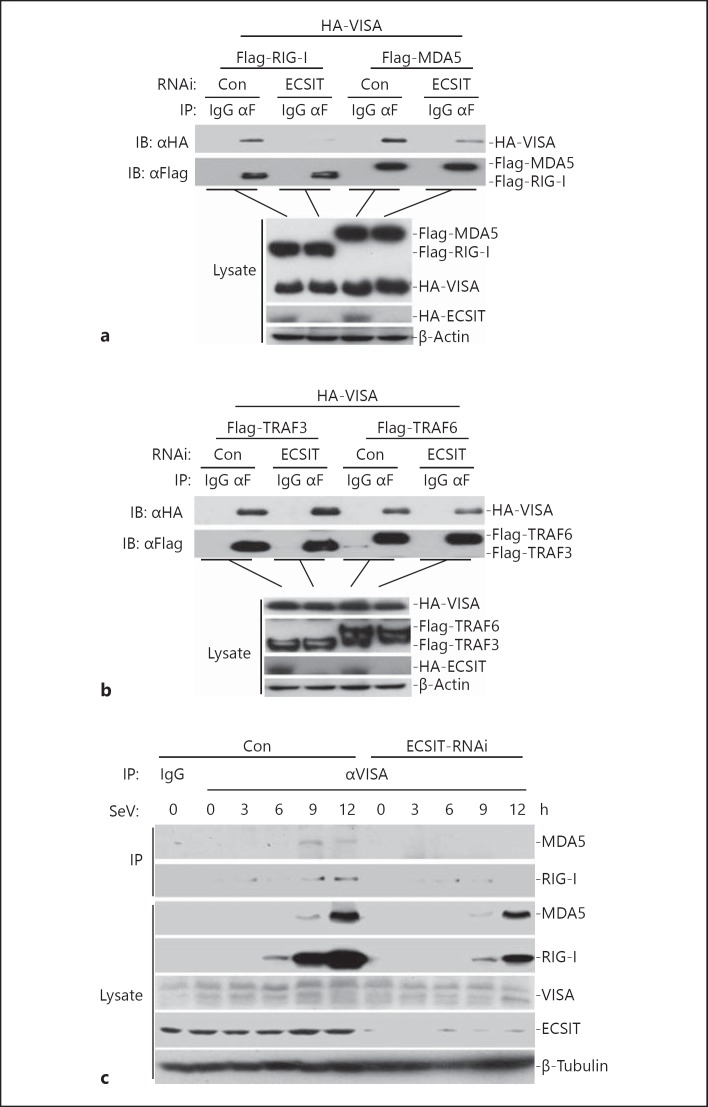
ECSIT Is Essential for the RLR-VISA Association
Because ECSIT interacts with RLRs and VISA, and functions downstream of RLRs and upstream of VISA, we hypothesized that ECSIT might mediate RLR-VISA interaction. Results from transient transfection and co-immunoprecipitation assays indicated that knockdown of ECSIT impaired RIG-I- or MDA5-VISA interaction (fig. 7a). In similar experiments, knockdown of ECSIT had minor effects on TRAF3- or TRAF6-VISA interaction, an interaction involved in RLR-mediated signaling downstream of VISA (fig. 7b). Furthermore, virus-induced VISA-RIG-I or VISA-MDA5 interaction was severely impaired in ECSIT knockdown cells (fig. 7c). These data indicate that ECSIT mediates the RLR-VISA interaction.
Fig. 7.
ECSIT is essential for the RLR-VISA interaction. a Knockdown of ECSIT impaired RIG-I- or MDA5-VISA interaction. HEK293 cells (2 × 106) were transfected with control or ECSIT-RNAi plasmids (12 μg). Thirty-six hours after transfection, cells were further transfected with the indicated plasmids by Lipofactamine 2000. Co-immunoprecipitation was performed with anti-Flag or control IgG. The immunoprecipitates were analyzed by immunoblotting with anti-HA or anti-Flag (upper panel). The lysates were analyzed by immunoblotting with anti-Flag or anti-HA (lower panels). b Knockdown of ECSIT did not affect TRAF3- or TRAF6-VISA interaction. The experiments were performed as in a. c Knockdown of ECSIT impaired SeV-triggered RIG-I- or MDA5-VISA interaction. ECSIT-RNAi stable cells were left uninfected or infected with SeV for the indicated time points. Cells were lysed and the lysates were immunoprecipitated with anti-VISA. The immunoprecipitates were analyzed by immunoblotting with the indicated antibodies (upper panel). Expression levels of the endogenous ECIST, RIG-I, MDA5, VISA and β-tublin were analyzed by immunoblotting (lower panels).
Discussion
Upon recognition of viral dsRNAs, RIG-I and MDA5 undergo conformational changes and are recruited to mitochondria to interact with the adaptor protein VISA, leading to the formation of the VISA signalsome. This process is critical for the initiation of virus-triggered induction of type I IFNs. In the present study, we identified ECSIT as an essential scaffolding protein mediating recruitment of RIG-I and MDA5 to the VISA complex on the mitochondria.
Overexpression of ECSIT potentiated virus-triggered activation of the IFN-β promoter and induction of the endogenous IFNB1 gene, whereas knockdown of ECSIT had opposite effects, suggesting that ECSIT is an important component of virus-triggered type I IFN induction pathways. Several lines of evidence suggest that ECSIT functions by bridging the recruitment of RIG-I and MDA5 to the mitochondrial-associated VISA complex. Firstly, ECSIT mostly localized to the mitochondria, and this localization was not significantly changed after viral infection. Second, ECSIT is constitutively associated with VISA, whereas its association with RIG-I and MDA5 occurs after viral infection. Moreover, knockdown of ECSIT impaired the RIG-I- or MDA5-VISA interaction. Third, knockdown of ECSIT inhibited RIG-I and MDA5- but not VISA-mediated induction of the IFNB1 gene.
Although it has been reported that SeV RNA is sensed by RIG-I rather than MDA5 [30], in several later studies MDA5 also played an important role in SeV-triggered signaling [31, 32, 33]. It is likely that SeV infection activates RIG-I-mediated signaling, which induces the expression of MDA5, and the accumulation of MDA5 further amplifies antiviral signaling by recruiting VISA signalosomes. In our study, we found that ECSIT was required for the RIG-I-VISA or MDA5-VISA interaction. Thus, it is possible that ECSIT mediates SeV-triggered signaling by promoting the recruitment of RIG-I or MDA5 to VISA signalosomes at the early and late phases of infection, respectively.
The mitochondrial targeting chaperon 14-3-3ε is reported to interact with TRIM25 and RIG-I to form a translocon that directs RIG-I to mitochondria upon viral infection. In light of our new observations, it is likely that 14-3-3ε directs RIG-I to mitochondria, while ECSIT directs mitochondrial RIG-I to VISA, which represents a stepwise activation of the RLR-VISA complex.
ECSIT has been reported to regulate mROS induction after bacterial infection and facilitates macrophage bactericidal activity. In this study, we found that ECSIT was critical for virus-triggered type I IFN induction and cellular antiviral responses by bridging RLRs to VISA. Whether and how ECSIT-induced mROS is involved in this process requires further investigation. Nonetheless, our study established a crucial role of ECSIT in the regulation of RLR-mediated IFN induction and provided new insight into the complicated molecular mechanisms of innate antiviral immune responses.
Disclosure Statement
The authors declare that they have no conflict of interest related to this work.
Supplementary Material
Supplementary data
Acknowledgments
This work was supported by grants from the Chinese Ministry of Science and Technology (2012CB910201 and 2014CB542601), the National Science Foundation of China (91029302, 31130020, 30921001 and 31371427), the Fundamental Research Funds for the Central Universities (204-274098) and the Academic Award for Excellent PhD Candidates from the Ministry of Education of China.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Hiscott J. Convergence of the NF-kappaB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev. 2007;18:483–490. doi: 10.1016/j.cytogfr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 4.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 6.Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Chen R, Zhou Q, Xu Z, Li C, Wang S, Mao A, Zhang X, He W, Shu HB. LSm14A is a processing body-associated sensor of viral nucleic acids that initiates cellular antiviral response in the early phase of viral infection. Proc Natl Acad Sci USA. 2012;109:11770–11775. doi: 10.1073/pnas.1203405109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoneyama M, Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 2008;29:178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 11.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 12.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 13.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 15.Liu HM, Loo YM, Horner SM, Zornetzer GA, Katze MG, Gale M., Jr The mitochondrial targeting chaperone 14-3-3ε regulates a RIG-I translocon that mediates membrane association and innate antiviral immunity. Cell Host Microbe. 2012;11:528–537. doi: 10.1016/j.chom.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohman T, Rintahaka J, Kalkkinen N, Matikainen S, Nyman TA. Actin and RIG-I/MAVS signaling components translocate to mitochondria upon influenza A virus infection of human primary macrophages. J Immunol. 2009;182:5682–5692. doi: 10.4049/jimmunol.0803093. [DOI] [PubMed] [Google Scholar]
- 17.Xu L, Xiao N, Liu F, Ren H, Gu J. Inhibition of RIG-I and MDA5-dependent antiviral response by gC1qR at mitochondria. Proc Natl Acad Sci USA. 2009;106:1530–1535. doi: 10.1073/pnas.0811029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kopp E, Medzhitov R, Carothers J, Xiao C, Douglas I, Janeway CA, Ghosh S. ECSIT is an evolutionarily conserved intermediate in the Toll/IL-1 signal transduction pathway. Genes Dev. 1999;13:2059–2071. doi: 10.1101/gad.13.16.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao C, Shim JH, Kluppel M, Zhang SS, Dong C, Flavell RA, Fu XY, Wrana JL, Hogan BL, Ghosh S. Ecsit is required for Bmp signaling and mesoderm formation during mouse embryogenesis. Genes Dev. 2003;17:2933–2949. doi: 10.1101/gad.1145603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogel RO, Janssen RJ, van den Brand MA, Dieteren CE, Verkaart S, Koopman WJ, Willems PH, Pluk W, van den Heuvel LP, Smeitink JA, Nijtmans LG. Cytosolic signaling protein Ecsit also localizes to mitochondria where it interacts with chaperone NDUFAF1 and functions in complex I assembly. Genes Dev. 2007;21:615–624. doi: 10.1101/gad.408407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West AP, Brodsky IE, Rahner C, Woo DK, Erdjument-Bromage H, Tempst P, Walsh MC, Choi Y, Shadel GS, Ghosh S. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo T, Watanabe M, Hatakeyama S. TRIM59 interacts with ECSIT and negatively regulates NF-kappaB and IRF-3/7-mediated signal pathways. Biochem Biophys Res Commun. 2012;422:501–507. doi: 10.1016/j.bbrc.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 23.Lei CQ, Zhong B, Zhang Y, Zhang J, Wang S, Shu HB. Glycogen synthase kinase 3β regulates IRF3 transcription factor-mediated antiviral response via activation of the kinase TBK1. Immunity. 2010;33:878–889. doi: 10.1016/j.immuni.2010.11.021. [DOI] [PubMed] [Google Scholar]
- 24.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Hu MM, Wang YY, Shu HB. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem. 2012;287:28646–28655. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei CQ, Zhang Y, Xia T, Jiang LQ, Zhong B, Shu HB. FoxO1 negatively regulates cellular antiviral response by promoting degradation of IRF3. J Biol Chem. 2013;288:12596–12604. doi: 10.1074/jbc.M112.444794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Wang K, Lin R, Zheng C. Herpes simplex virus 1 serine/threonine kinase US3 hyperphosphorylates IRF3 and inhibits beta interferon production. J Virol. 2013;87:12814–12827. doi: 10.1128/JVI.02355-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Lei CQ, Hu YH, Xia T, Li M, Zhong B, Shu HB. Kruppel-like factor 6 is a co-activator of NF-kappaB that mediates p65-dependent transcription of selected downstream genes. J Biol Chem. 2014;289:12876–12885. doi: 10.1074/jbc.M113.535831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 31.Diao F, Li S, Tian Y, Zhang M, Xu LG, Zhang Y, Wang RP, Chen D, Zhai Z, Zhong B, Tien P, Shu HB. Negative regulation of MDA5- but not RIG-I-mediated innate antiviral signaling by the dihydroxyacetone kinase. Proc Natl Acad Sci USA. 2007;104:11706–11711. doi: 10.1073/pnas.0700544104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gitlin L, Benoit L, Song C, Cella M, Gilfillan S, Holtzman MJ, Colonna M. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 2010;6:e1000734. doi: 10.1371/journal.ppat.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H, Li Y, Zhang J, Ran Y, Wei J, Yang Y, Shu HB. RAVER1 is a coactivator of MDA5-mediated cellular antiviral response. J Mol Cell Biol. 2013;5:111–119. doi: 10.1093/jmcb/mjt006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data