Abstract
Cholera epidemics are caused by Vibrio cholerae serogroups O1 and O139, whereas strains collectively known as non-O1/non-O139 V. cholerae are found in cases of extraintestinal infections and bacteremia. The mechanisms and factors influencing the occurrence of bacteremia and survival of V. cholerae in normal human serum have remained unclear. We found that naturally occurring IgG recognizing V. cholerae outer membrane protein U (OmpU) mediates a serum-killing effect in a complement C1q-dependent manner. Moreover, outer membrane vesicles (OMVs) containing OmpU caused enhanced survival of highly serum-sensitive classical V. cholerae in a dose-dependent manner. OMVs from wild-type and ompU mutant V. cholerae thereby provided a novel means to verify by extracellular transcomplementation the involvement of OmpU. Our data conclusively indicate that loss, or reduced expression, of OmpU imparts resistance to V. cholerae towards serum killing. We propose that the difference in OmpU protein levels is a plausible reason for differences in serum resistance and the ability to cause bacteremia observed among V. cholerae biotypes. Our findings provide a new perspective on how naturally occurring antibodies, perhaps induced by members of the microbiome, may play a role in the recognition of pathogens and the provocation of innate immune defense against bacteremia.
Key Words: Vibrio cholerae, Outer membrane vesicles, Outer membrane protein U, Serum resistance, Naturally occurring IgG antibodies, C1q
Introduction
Vibrio cholerae is the causal organism of the potentially life-threatening gastrointestinal disease cholera and is commonly transmitted by contaminated water and food. The cholera-causing serogroups O1 and O139 of V. cholerae very rarely cause extraintestinal infections. In contrast, the non-O1 non-O139 V. cholerae (NOVC) isolates are commonly associated with septicemia in patients with underlying liver disorders and have a fatality rate exceeding 50% [1].
Bacterial outer membrane proteins (OMPs) are essential components of the outer membrane of Gram-negative bacteria and are involved in multiple processes, such as nutrient transport, antimicrobial resistance and responses to environmental signals. In V. cholerae, outer membrane protein U (OmpU) and OmpT play important roles in bacterial physiology. The expression of OmpU and OmpT is under the control of the ToxR regulon, a master regulator of virulence genes in V. cholerae. ToxR negatively regulates the expression of OmpT, while OmpU is positively regulated by ToxR [2]. OmpU plays a role in resistance against bile acids and antimicrobial peptides [3]. However, the potential role of V. cholerae OMPs in host immunomodulation is poorly characterized.
In general, pathogenic invasive bacteria are capable of effectively evading immune responses. One of the strategies of invading bacteria is the ability to avoid the bactericidal activity of serum [4]. In earlier studies, it was demonstrated that NOVC can cause bacteremia [5]. The underlying mechanism(s) of V. cholerae bacteremia was less studied.
Bacterial pathogens have evolved a number of excreted and membrane-bound proteins that interfere with several steps of the complement cascade in order to survive in the human host. The complement system can be activated by the classical, alternative and mannose-binding lectin (MBL) pathways [6]. The binding of complement regulators to OMPs plays an important role in modulating complement activation among several Gram-negative pathogens [7]. While some OMPs can antagonize complement action, others may be targets for the initiation of complement. OmpC, a major immunogen on the surface of Escherichia coli, promotes the deposition of C1q and induction of the antibody-dependent classical complement pathway [8]. The direct activation of the classical pathway or antibody-independent binding of C1q was demonstrated recently for Streptococcus pneumoniae[9].
During physiological conditions, human serum contains naturally occurring antibodies of the IgG, IgM and IgA isotypes, which presumably are produced in response to endogenously occurring antigens or after the sporadic introduction of foreign antigens [10]. Several functions have been proposed for natural antibodies, including the neutralization of microbes as a consequence of their cross-reactivity to microbial antigens [11]. In earlier studies, it was shown that naturally occurring antibodies to the endotoxin core of E. coli can protect against Pseudomonas aeruginosa septicemia [12]. Recently, natural IgG was shown to bind to ficolin and MBL, soluble pattern recognition receptors that interact with pattern-associated molecular patterns at bacterial surfaces. Natural IgG-lectin immunocomplexes mediated bacterial clearance during infection/inflammatory conditions [13, 14].
In this study, we found that natural IgG directly recognizes OmpU of V. cholerae, thereby mediating C1q binding on to the bacterial surface via IgG resulting in complement-mediated serum sensitivity of V. cholerae. Furthermore, we demonstrated that OmpU on released bacterial outer membrane vesicles (OMVs) may divert IgG binding and the C1q recruitment away from the bacteria, contributing to serum resistance.
Materials and Methods
Bacterial Strains, Culture Conditions and Plasmids
The strains and plasmids used in this study are listed in table 1. Bacterial strains were grown overnight at 37°C in Luria-Bertani (LB) broth. One hundred microliters of overnight culture was inoculated into 10 ml of LB broth in a water bath shaker at 37°C and bacterial growth was monitored by optical density (OD600 nm) measurement. When needed, kanamycin (50 µg/ml) or carbenicillin (100 µg/ml) was added to the culture media.
Table 1.
The strains and plasmids used in this study
| Strain/plasmid | Description/relevant characteristics | Reference No./source |
|---|---|---|
| E. coli strains | ||
| SM10λpir | thi thr leu tonA lacY supE recA:: RP4-2 TC:: Mu Km λpir | 35 |
| MC4100 | araD139Δ(lac)U169 strA thi | 36 |
| ΔompC | ΔompC derivative of MC4100 | 37 |
| V. cholerae strains | ||
| A1552 | O1 El Tor, Inaba, RifR | 30 |
| C6706 | O1 El Tor, Inaba, StrR | 38 |
| 569B | O1 Classical, Inaba, RifR | 39 |
| O395 | O1 Classical, Inaba, StrR | 40 |
| V:1/05 | Non-O1 Non-O139, clinically isolated (2005) | Swedish Institute of Infectious Diseases, Sweden |
| V:5/04 | Non-O1 Non-O139, clinically isolated (2004) | Swedish Institute of Infectious Diseases, Sweden |
| V:10/04 | Non-O1 Non-O139, clinically isolated (2004) | Swedish Institute of Infectious Diseases, Sweden |
| V:7/04 | Non-O1 Non-O139, environmentally isolated (2004) | Swedish Institute of Infectious Diseases, Sweden |
| ΔompU | ΔompU derivative of A1552 | This study |
| ΔompT | ΔompT derivative of A1552 | 41 |
| ΔompA | ΔompA derivative of A1552 | 30 |
| ΔtoxR | ΔtoxR derivative of A1552 | 41 |
| Plasmids | ||
| pBAD18 | Arabinose-inducible vector, CbR | 42 |
| pMMB66HE | IPTG inducible vector, CbR | 43 |
| pJET1.2 | Cloning vector, CbR | Fermentas |
| pBR322 | Cloning vector, CbR, TcR | 44 |
| pJET-ompU | pJET1.2, ompU gene from A1552 | This study |
| pMM-ompU | pMMB66HE, ompU gene from A1552 | This study |
| pMM-ompUD114A | pMM-ompU, D114A substitution | This study |
Construction of the ΔompU Mutant
The ompU deletion mutant was constructed using procedures that have been described previously [15]. The oligonucleotide primers used are listed in table 2.
Table 2.
The primers used in this study (recognition sites for restriction enzymes are marked by bold italic letters)
| Primer | Sequence (5′ to 3′) | Restriction site | Used for construction of |
|---|---|---|---|
| Primers for deletion mutant construction | |||
| TIS-56 | CGCTCTAGAAATAAAAAATTTCCCAACATC | XbaI | ΔompU |
| TIS-57 | CCCATCCACTAAACTTAAACAGTCCATAAATTTGATTTTTG | ΔompU | |
| TIS-58 | TGTTTAAGTTTAGTGGATGGGTTCTAATTGTTGACTTCAGG | ΔompU | |
| TIS-59 | CGCTCTAGACAAATCGCCCTCAATCCTAC | XbaI | ΔompU |
| Primers for cloning | |||
| ompUup1 | CGCGTCGACGCTTGATGCATCACCTATTTCG | SalI | OmpU clone |
| ompUdo2 | CGAATTCGTGAGCAGGCGTTTGGCGTGTG | EcoRI | OmpU clone |
| Primers for site-directed mutagenesis | |||
| ompUmutup | GGTAAAAACGCGTCTAACAACAGCCTAGTCAACCGTTATACCTACG | D to A introduce HincII | OmpU amino acid substitution mutant clone |
| ompUmutdo | CGTAGGTATAACGGTTGACTAGGCTGTTGTTAGACGCGTTTTTACC | D to A introduce HincII | OmpU amino acid substitution mutant clone |
Preparation of Human Sera, Complement C1q (Active) Protein, and C1q and IgG Antisera
Normal human serum (NHS) was pooled from healthy volunteers and stored at −80°C. Heat-inactivated serum (HIS) was prepared by heating NHS at 56°C for 30 min. The NHS and HIS were diluted to 20% with sterile phosphate-buffered saline (PBS) for the serum-killing assay. Purified human C1q protein, C1q antiserum and IgG antiserum were purchased from antibodies-online (Aachen, Germany) and Dakopatts (Dako, Glostrup, Denmark).
This study was carried out in accordance with the recommendations of the local ethical committee (Regionala etikprövningsnämnden i Umeå). Ethics committee approval for the use of rabbit red blood cells for this study was issued with the permit number Dnr: A76-12. The present research has been performed in accordance with the Swedish Act (2003:460) on ‘Ethical Review of Research Involving Humans’ from the Ministry of Education (issued June 5, 2003; http://www.riksdagen.se/sv/Dokument-Lagar/Lagar/Svenskforfattningssamling/Lag-2003460-om-etikprovning_sfs-2003-460/).
EGTA/Mg2+ Treatment of NHS and IdeS Enzyme Treatment
To block classical pathway activation, NHS was treated with 5 mM MgCl2 and 10 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA/Mg2+) for 30 min as described previously [16]. To degrade the IgG in the NHS, IgG-degrading IdeS enzyme was used [17, 18].
Serum-Killing Assay
Bacterial cells were grown to OD600 nm 2.0 in LB broth and 50 µl of bacterial culture was mixed with 50 µl of 20% NHS or HIS and incubated with the sample at 37°C for 1 h. Colony-forming units (CFU) at OD600 nm 2.0 were determined with samples of each bacterial strain: A1552 (1.40 × 109 CFU/ml), C6706 (1.46 × 109 CFU/ml), 569B (1.13 × 109 CFU/ml), O395 (1.20 × 109 CFU/ml), V1:05 (1.33 × 109 CFU/ml), V5:04 (1.27 × 109 CFU/ml), V10:04 (1.26 × 109 CFU/ml) and V7:04 (1.47 × 109 CFU/ml). To block the classical pathway activation, 50 µl of 20% NHS was pretreated with 10 mM EGTA and 5 mM MgCl2 for 30 min at 37°C. For IgG cleavage, 50 µl of 20% NHS was treated with 2 μg of IgG-degrading enzyme of Streptococcus pyogenes (IdeS) at 37°C for 2 h.
For the serum-killing assay with OMVs, a 50-µl aliquot of wild-type V. cholerae 569B strain was mixed with 50 µl of OMVs isolated from the wild-type V. cholerae strain A1552 or its omp mutant derivatives and 50 µl of 20% NHS or HIS. For the serum-killing assay, after blocking the classical pathway, IdeS-treated or EGTA-MgCl2-treated NHS was used. The reaction mixtures were incubated at 37°C for 1 h. Subsequently, viable cell counts were determined by plating serial dilutions onto LB agar plates. Results of the serum-killing assay are reported as the percent survival, which was calculated by dividing the CFU/ml recovered after NHS or HIS incubation by the CFU/milliliter recovered from the sample without serum incubation.
For the MBL-depleted serum preparation, NHS was passed through a 5-ml column of manose-agarose beads (Sigma-Aldrich, St. Louis, Mo., USA) equilibrated in 10 mM HEPES, 140 mM NaCl, 5 mM CaCl2, pH 7.4 at 4°C, and then used in the serum-killing assay. The alternative pathway has been shown to be inhibited by the absorption of properdin with bentonite [8]. Bentonite (10 mg) was washed with sterile PBS three times and incubated with NHS or HIS at 37°C for 10 min to absorb properdin.
For the neutralization assay, 20% NHS was preincubated with 1:5,000 dilutions of polyclonal rabbit antisera against OmpU [19] or 1:500 dilutions of mouse monoclonal anti-human complement C1q antibody or 1:1,000 dilutions of anti-IgG rabbit anti-serum (Dako) or 1:1,000 dilutions of anti-Hcp polyclonal antiserum [20] at 37°C for 30 min, followed by the serum-killing assay.
For the bacterial washing experiment, samples from cultures at OD600 nm 2.0 of WT A1552 and WT 569B were washed three times with PBS and followed by serum-killing assay. The results were compared to serum-killing assay with nonwashed bacteria.
Electron Microscopy
The bacterial cells were negatively stained and examined by transmission electron microscopy as described previously [21]. For ultrathin sectioning and ferritin labeling, bacteria were grown on LB agar plates overnight at 37°C, harvested and washed once with cacodylate buffer (0.1 M, pH 7.0). Bacterial cells were fixed with 5% glutaraldehyde in cacodylate buffer for 2 h at room temperature. Fixed bacteria were washed and resuspended in cacodylate buffer containing 1 mg of polycationic ferritin (Sigma-Aldrich) per milliliter. After 30 min at room temperature, samples were diluted 1:10 with the same buffer. The ferritin-labeled bacterial cells were washed three times and fixed with 4% agar. After fixation, the sample was treated for 2 h with 1% osmium tetroxide and then washed three times. Samples were dehydrated in a graded series (30–100%) of ethanol solutions, washed twice with propylene oxide and embedded in Epon by a rapid-embedding method. Thin sections were cut and placed on 300-mesh formvar/carbon grids, stained with uranyl acetate and lead citrate, and examined under a JEOL EX transmission electron microscope operating at an accelerating voltage of 100 kV.
Western Blot Analysis
Bacteria cells were grown to OD600 nm 2.0 in LB broth in a water bath shaker at 37°C. Cells were harvested by centrifugation at 14,000 rpm for 3 min at 4°C. The pellets were resuspended in 20 mM Tris-HCl buffer, pH 8.0, and used as protein samples. Protein samples were denatured in sample buffer containing 10% glycerol, 0.05% bromophenol blue, 2% SDS, 5% 2-mercaptoethanol and 10 mM Tris-HCl, pH 6.8, and resolved by 13.5% SDS-PAGE with a discontinuous buffer system at a constant voltage of 60 V for the stacking gel and 120 V for the resolving gel [22]. The proteins in the gel were transferred to a polyvinylidene difluoride membrane (Millipore, Billerica, Mass., USA) in standard transfer buffer with a Bio-Rad semidry transfer system at 23 V for 35 min. Proteins with known molecular masses (Thermo Scientific, Waltham, Mass., USA) were used as molecular mass markers. After the transfer was completed, the membrane was blocked with 5% skimmed milk in PBS (pH 7.2) containing 0.05% Tween-20 (PBST) at 4°C overnight. The immunoblot membrane was incubated with 1:5,000 dilutions of polyclonal antisera against OmpU [19] as a primary antibody for 1 h. Goat anti-rabbit HRP-conjugated IgG antibody (AgriSera AB, Vännäs, Sweden) was used as a secondary antibody at a final dilution of 1:25,000. The ECL+ chemiluminescence system (GE Healthcare) was used to detect immunoreaction bands that were recorded using a Fluor-S MultiImager (Bio-Rad, Hercules, Calif., USA) and by autoradiography.
Far Western Analyses
For the detection of indirect binding of C1q on the OmpU by Far Western analysis, following blocking of the immunoblot membrane with 5% milk, the membrane was incubated with 20% NHS for 2 h at room temperature and subsequently the membrane was rinsed with PBST. The complement C1q protein (10 μg/ml) in PBST was overlaid on the membrane and incubated for 2 h at room temperature. After washing four times with PBS, the membrane was incubated with a mouse anti-human complement C1q mAb (Aachen) diluted 1:1,000 in PBST for 1 h. After four further washes in PBST, the membrane was incubated in an anti-rabbit horseradish peroxidase-conjugated pAb preparation (Dako) at a final dilution of 1:10,000 in PBST-M for 1 h. Detection was performed as described above.
Isolation of OMVs from E. coli and V. cholerae
OMVs from V. cholerae WT-A1552, E. coli WT-MC4100 and their omp mutants were isolated from the bacterial cultures as previously described [23]. Briefly, bacteria were inoculated into a 200-ml culture flask containing LB and incubated for 16 h at 37°C with shaking. Bacterial cells were removed from the culture fluid by centrifugation at 5,000 g for 30 min. The supernatant was sterile filtered through a 0.22-μm PVDF membrane filter (Millipore). The cell-free supernatant was centrifuged at 100,000 g for 2 h at 4°C in a 45 Ti rotor (Beckman) to pellet the vesicles. The OMVs were suspended in 20 mM of Tris-HCl (pH 8.0) and adjusted to equal the total protein concentrations measured by a NanoDrop 1000 Spectrophotometer (Thermo Scientific). The protein concentration of the OMV samples was estimated using a bicinchoninic acid assay kit (Thermo Scientific Pierce, Rockford, Ill., USA). To determine the number of OMVs, the samples (5 mg/ml) were subjected to analysis by Nanosight NS300 (Malvern) and nanoparticle tracking analysis, as described earlier [24]. The OMV sample obtained from the ΔompA mutant contains 1.45 × 1011 particles/µl, whereas the wild-type strain A1552, ΔompU, ΔompT and ΔtoxR mutant contain 1.51 × 1010, 1.62 × 1010, 1.66 × 1010 and 1.49 × 1010 OMVs/µl, respectively.
Construction of ompU and ompU D114A Clone
For the construction of the wild-type ompU clone, a fragment of the ompU gene (VC0633) was PCR amplified from chromosomal DNA isolated from strain A1552 using the primers ompUup1 and ompUdo2 with additional EcoRI or SalI restriction endonuclease recognition sites at the ends. The PCR amplification was performed using Kapa HiFi HotStart Polymerase (Kapa Biosystems) according to the manufacturer's manual. The PCR product was cloned into pJET1.2 for sequencing and amplification purposes. A correct plasmid construct, pJET-ompU, was digested with EcoRI and SalI, and the gel-purified ompU fragment was cloned into EcoRI and SalI digested pMMB66HE, resulting in plasmid pMM-ompU.
For the construction of the ompU D114A mutant clone, pJET-ompU was used as a template to replace aspartic acid (D114) for alanine (A) in the ompU gene sequence by site-directed mutagenesis. The primer pair introducing the alanine residue, OmpUmutup and OmpUmutdo, creates a HincII site instead of the bases coding for the aspartic acid. Site-directed mutagenesis was performed using 250 ng of pJET-ompU DNA and Kapa HiFi HotStart Polymerase (Kapa Biosystems) in a 25-µl reaction according to the manufacturer's manual (58°C anneal 30 s, 69°C extension 9 min, 30 cycles). The template plasmid was then digested by DpnI (fast digest; Fermenta) for 1 h at 37°C, and afterwards the enzyme was inactivated at 80°C for 5 min followed by transformation into CaCl2-competent DH5α. Successful mutagenesis was verified by sequencing. The mutated ompU fragment was excised by digestion with EcoRI and SalI, gel-purified, and cloned into EcoRI and SalI digested pMMB66HE, resulting in plasmid pMM-ompUD114A (ΔompU/D114A).
Determination of anti-OmpU IgG in NHS by ELISA
High-binding 96-well polystyrene plates (Thermo Scientific) were coated with 50 µl of OMVs isolated from V. cholerae or E. coli and 150 µg/ml BSA in 0.1 M carbonate-bicarbonate buffer, pH 9.5. The plates were incubated for 1 h at 37°C and then overnight at 4°C, and were washed three times in PBST. Nonspecific binding was blocked with 5% milk diluted in PBS for 2 h at 37°C. After washing three times with PBST, 100 µl of 20% NHS from 9 donors in PBST was added into the first well and diluted serially 2-fold. PBS was used instead of 20% NHS as a negative control. The plate was incubated for 2 h at 37°C. After washing three times with PBST, the 1:5,000-diluted peroxidase-conjugated anti-human IgG antibodies (AgriSera AB) were added, followed by 1 h of incubation at 37°C and six additional washes with PBST. Detection was performed using the TMB substrate Kit (Thermo Scientific) as recommended by the manufacturer.
Statistical Analyses
All statistical analyses were performed with Student's t test. p < 0.05 was considered significant.
Results
Analysis of Serum Resistance in Different V. cholerae Serogroups
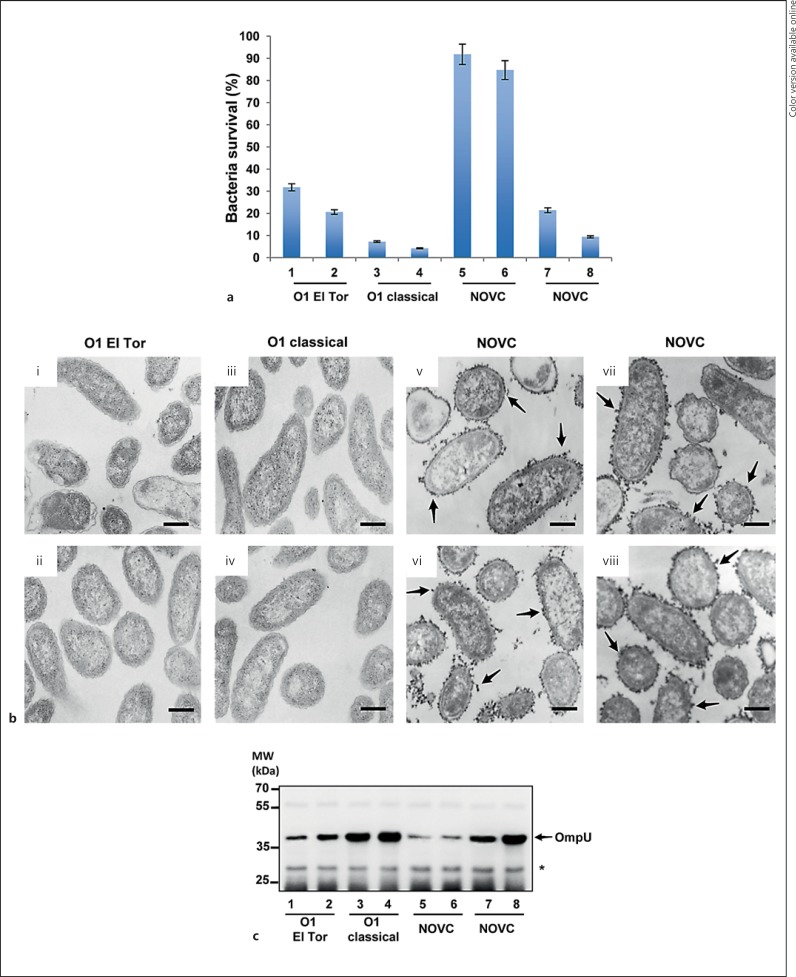
Limited information is available on the phenotypic differences of various V. cholerae serogroups in relation to serum resistance. To investigate the levels of serum resistance among isolates of different V. cholerae serogroups, serum-killing assays were performed with NHS. We performed serum-killing assays using different concentrations of serum, i.e. 10, 20 and 30%. We found no significant difference in bactericidal activity among these concentrations and, therefore, decided to use a 20% serum concentration for further experiments. Wild-type V. cholerae O1 El Tor strains A1552 and C6706, the wild-type O1 classical strains 569B and O395, and the wild-type NOVC strains V:1/05, V:5/04, V:10/04 and V:7/04 were tested for serum sensitivity (fig. 1a). HIS was used as a negative control. The O1 El Tor strains A1552 and C6706 showed 31.8 and 20.6% survival, respectively (fig. 1a, columns 1 and 2), whereas the O1 classical strains 569B and O395 were more susceptible to serum killing, with only 7.2 and 4.2% survival, respectively (fig. 1a, columns 3 and 4). The V. cholerae NOVC strains V:1/05 and V:5/04 displayed a quite high resistance level with 91.8 and 84.7% survival, respectively (fig. 1a, columns 5 and 6). In contrast, the wild-type NOVC strains V:10/04 and V7:04 had 21.4 and 9.4% survival, respectively (fig. 1a, columns 7 and 8). All strains showed a similar capacity to survive in the HIS (data not shown). This initial screening suggested that serum resistance among V. cholerae varies both within and among different serogroups.
Fig. 1.
Serum sensitivity, electron microscopic analysis and Western blot analysis of different V. cholerae serogroups. a Serum sensitivity of different V. cholerae serogroups. O1 El Tor V. cholerae strains A1552 and C6706 (columns 1 and 2, respectively), O1 classical V. cholerae strains 569B and O395 (columns 3 and 4, respectively), and NOVC strains V:1/05, V:5/04, V:10/04 and V7:04 (columns 5-8, respectively). The survival rate (percentage) is indicated. Values represent the mean ± SD of three independent experiments. b Electron micrograph of ultrathin sections of polycationic ferritin-labeled bacterial cells. O1 El Tor strains A1552 and C6706 (i and ii, respectively), O1 classical strains 569B and O139 (iii and iv, respectively), and NOVC strains V:1/05, V:5/04, V:10/04 and V7:04 (v-viii, respectively). The arrows show ferritin particles surrounding the bacterial cells. Scale bars = 250 nm. c Immunoblots of whole-cell lysates isolated from different V. cholerae serogroups. The immunoblot membrane was probed with polyclonal anti-OmpU. The arrow shows the OmpU reaction band. The asterisk shows nonspecific reaction bands to estimate the amount of sample loading. O1 El Tor strains A1552 and C6706 (lanes 1 and 2, respectively), O1 classical strains 569B and O139 (lanes 3 and 4, respectively) and NOVC strains V:1/05, V:5/04, V:10/04 and V7:04 (lanes 5-8, respectively).
Role of OmpU but Not Capsular Polysaccharide Production in V. cholerae Serum Resistance
Encapsulation is an important virulence factor for a number of bacterial species, contributing protection against host defenses including serum killing and phagocytosis [25]. To determine whether the presence of capsular polysaccharide at the surface of V. cholerae would correlate to the relative serum resistance, ultrathin sections of bacterial cells of the different strains were stained with polycationic ferritin and examined by transmission electron microscopy (fig. 1b). The moderately serum-resistant V. cholerae O1 El Tor strains A1552 and C6706 (fig. 1b, panels i and ii) and the serum-sensitive O1 classical strains 569B and O395 (fig. 1b, panels iii and iv) seemed to lack a capsular layer. In the case of the serum-resistant NOVC strains V:1/05 and V:5/04 (fig. 1b, panels v and vi) and the serum-sensitive NOVC strains V:10/04 and V:7/04 (fig. 1b, panels vii and viii), we observed an electron-dense layer completely surrounding the cells. Therefore, our data indicated that the presence of a capsule per se was not responsible for serum resistance.
Some OMPs have been shown to play a role in resistance to complement-mediated serum killing of Gram-negative bacteria [7]. In order to investigate if the major OMPs could possibly be involved in serum resistance of V. cholerae, the OmpU and OmpT protein expression levels were analyzed by Western blot. As shown in figure 1c, OmpU expression was inversely correlated with serum resistance of the different V. cholerae serogroups. The most serum-resistant V. cholerae strains, V:1/05 and V:5/04 (lanes 5 and 6), expressed the lowest level of OmpU, while much higher levels of OmpU were observed in the most serum-sensitive V. cholerae classical strains (lanes 3 and 4). These observations suggest that the level of OmpU expression in V. cholerae may determine the level of serum resistance in V. cholerae.
OmpU-Mediated Bactericidal Effect of Serum on V. cholerae via the Classical Complement Pathway
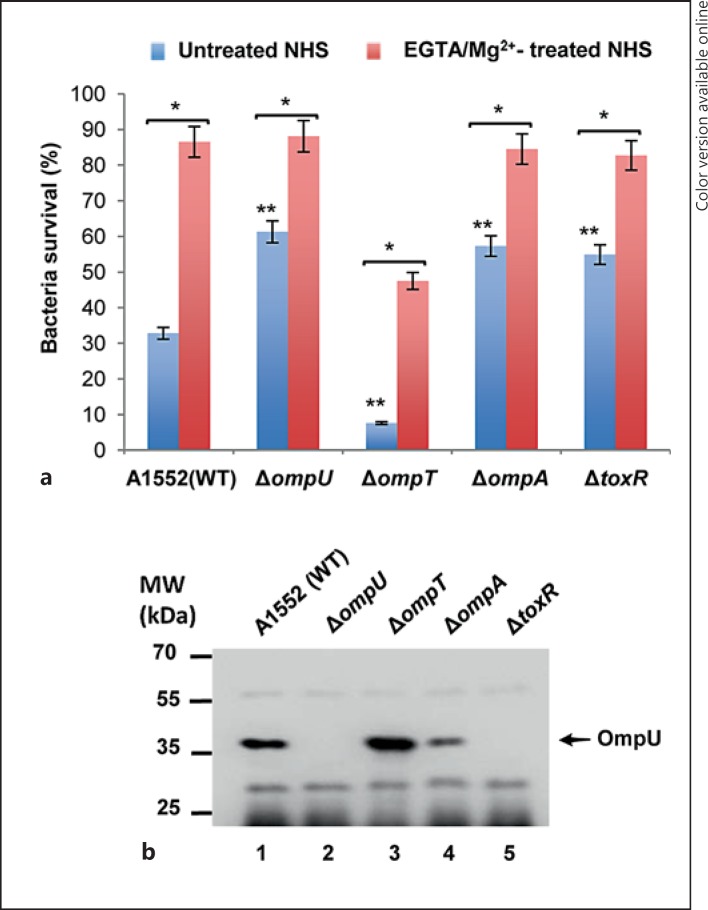
To further analyze the role of the OmpU protein in serum resistance of V. cholerae, we studied a selection of mutant derivatives of the moderately serum-resistant V. cholerae strain A1552. In-frame deletions in the ompU, ompT, ompA and toxR genes were constructed in the A1552 strain as described in Materials and Methods. The serum resistance of these mutants was compared with that of the wild-type strain A1552. The survival rates of ΔompU, ΔompA and ΔtoxR mutants were 61.3, 57.3 and 54.9%, respectively, whereas the wild-type strain A1552 had a 32.8% survival rate (fig. 2a, blue columns). Interestingly, the ΔompT mutant had a clearly lower survival rate (7.6%) when compared with the wild-type A1552 and the other tested mutants (fig. 2a, blue columns). The survival rates of the wild-type strain A1552 and its mutant derivatives were similar in HIS (data not shown), and there was no difference in the growth rate indicating that the mutations did not impair growth (see online suppl. fig. S1; see www.karger.com/doi/10.1159/000443646 for all online suppl. material). The OmpU expression level was determined in the different strains by immunoblot analysis (fig. 2b). As expected, the ΔtoxR strain produced no OmpU (fig. 2b, lane 5) since ToxR is a positive regulator of OmpU expression [2]. Interestingly, in the ΔompA strain, OmpU expression was lower than that of the wild type (fig. 2b, lanes 1 and 4), which correlated with reduced serum sensitivity of the ΔompA strain in comparison to the wild type. In addition, we observed that the survival of the ΔompA and ΔtoxR mutants were similar, although in contrast to the case of the ΔompA mutant there was no detectable level of OmpU in the ΔtoxR mutant. It might indicate that some additional factor(s) could be involved in serum resistance in the ΔompA mutant. Similarly, the higher serum sensitivity observed for the ΔompT mutant was correlated with an increased level of OmpU protein in that mutant (fig. 2b, lanes 3). By comparing the wild-type strain A1552 and these OMP mutant derivatives, we conclude that there was a strong correlation between higher levels of OmpU and higher serum sensitivity.
Fig. 2.
Serum sensitivity assay of the wild-type (WT) strain V. cholerae A1552 and its outer membrane protein-deficient derivatives. a The bacterial survival percentage after incubation with untreated NHS is shown with blue columns. The red columns represent the survival percentage of bacterial strains incubated with NHS pretreated with EGTA/Mg2+ to inhibit the classical pathway of complement activation (colors refer to the online version only). Values represent the mean ± SD of three independent experiments. The single asterisks indicate EGTA/Mg2+-treated NHS versus untreated NHS (p < 0.005). The double asterisks indicate untreated NHS wild-type A1552 versus its mutant derivatives (p < 0.005). b Immunoblot of whole-cell lysates from the wild-type V. cholerae strain A1552 and its mutant derivatives. The arrow indicates immunoblot reactivity to OmpU. The molecular weight markers are shown at the left of the membrane.
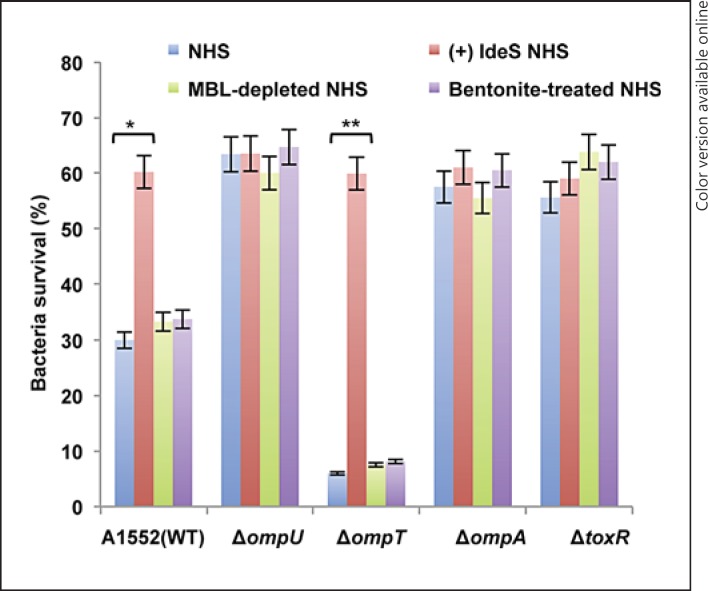
Complement activation may proceed through one or more of three pathways: the classical pathway, the MBL pathway or the alternative pathway [6]. To investigate which complement pathway was involved in the OmpU-mediated bactericidal activity of NHS, we performed serum bactericidal assays on the wild-type strain A1552 and its mutant derivatives using serum treated with EGTA/Mg2+, which inhibits the classical complement pathway. The bactericidal activity of EGTA/Mg2+-treated NHS was significantly lower when compared to that of untreated NHS (fig. 2a, compare the blue and red columns). The ΔompT strain showed 47.5% survival in EGTA/Mg2+-treated serum, whereas the survival rates of the wild-type strain A1552 and the ΔompU, ΔompA and ΔtoxR mutants in EGTA/Mg2+-treated serum were all above 80% (fig. 2a, red columns). In all cases the survival was clearly lower in untreated NHS (fig. 2a, blue columns). In order to assess whether activation of the lectin pathway or the alternative pathway would be involved in OmpU-mediated serum bactericidal activity, serum-killing assays were performed using MBL-depleted NHS or bentonite-absorbed NHS. The alternative pathway was shown to be inhibited by the absorption of properdin with bentonite [8, 26, 27]. The effect on the viability of the wild-type strain A1552 and its mutant derivatives remained similar to that of NHS without such treatments (fig. 3, compare the green and violet columns with the blue column). All strains showed a similar ability to survive in HIS (data not shown). These results suggested that the OmpU-mediated bactericidal effect does not involve the lectin pathway or the alternative pathway. Taken together, these findings suggested that the classical complement pathway is the primary reason behind the OmpU-mediated serum killing of V. cholerae by NHS.
Fig. 3.
Serum sensitivity assay with V. cholerae using NHS treated with IgG-degrading enzyme (IdeS), MBL-depleted NHS or bentonite-treated NHS. The blue, red, green and violet columns represent the percentage of bacterial surviving after incubation with NHS, IdeS-treated NHS, MBL-depleted NHS and bentonite-treated NHS, respectively (colors refer to the online version only). Values represent the mean ± SD of three independent experiments. The single asterisk indicates wild-type V. cholerae A1552 in NHS without IdeS versus NHS after treatment with IdeS (p < 0.05). The double asterisk indicates the ΔompT mutant in NHS without IdeS versus with IdeS (p < 0.05).
The classical pathway of the complement system can be triggered by the binding of either IgG or IgM antibody to antigens. To investigate whether IgG was involved in the OmpU-mediated bactericidal effect, the serum-killing assay was performed using NHS with or without treatment with IdeS, an IgG-specific cleavage protease of S. pyogenes [17, 18]. When serum was pretreated with IdeS, survival of the wild-type A1552 and the ΔompT mutant were about 60%, i.e. similar to that of the ΔompU, ΔompA and ΔtoxR mutants in NHS (fig. 3). The ΔompU, ΔompA and ΔtoxR mutants had similar survival rates in serum with or without IdeS treatment. These data suggested that we need to consider that OmpU and OmpT are involved in antibody-dependent bactericidal activity of complement primarily via IgG and not via IgM.
V. cholerae OmpU Involved in C1q Binding via IgG
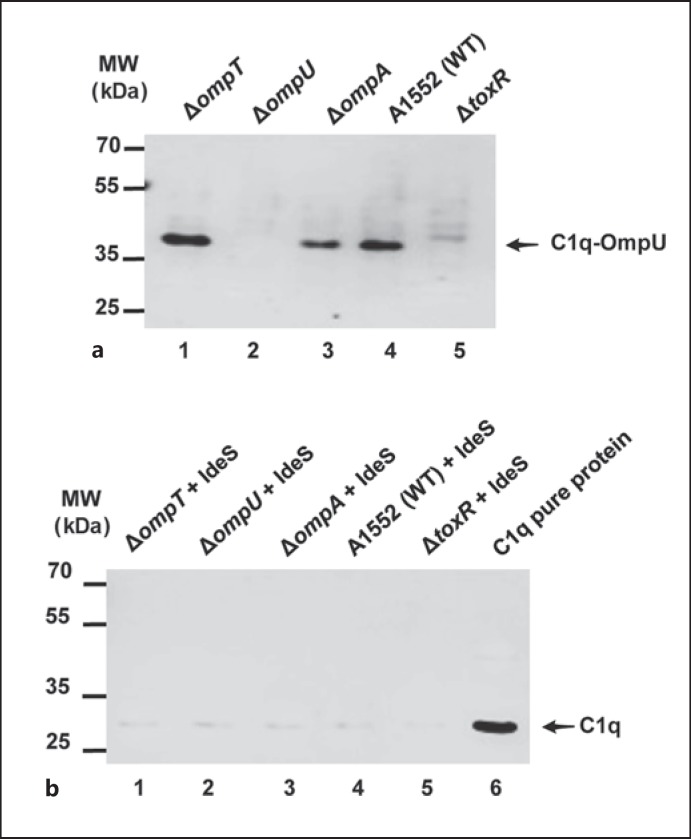
Antibody-antigen binding exposes a binding site on the antibody for the C1 complex of the complement system. The C1 complex is composed of one C1q molecule, two C1r and two C1s molecules. The C1q molecule was shown to be responsible for binding to the antibody [28]. To define the role of OmpU in serum sensitivity of V. cholerae, we performed the Far Western analysis using NHS instead of purified C1q as described in Materials and Methods. The detection using anti-C1q antibody indicated that there was C1q binding as represented by a band on the membrane at the expected position corresponding to the size of OmpU, which was clearly observed in the case of the wild-type strain A1552 (fig. 4a, lane 4). Furthermore, there was no similar C1q binding detected with the samples from the ΔompU or ΔtoxR mutant strains (fig. 4a, lanes 2 and 5), in line with the hypothesis that there would be an OmpU-dependent C1q binding to V. cholerae. Consistently, this analysis indicated a slightly higher binding efficiency of C1q in the case of the sample from the ΔompT mutant that had a higher expression level of OmpU than the wild-type strain (fig. 4a, lane 1). In respect to this lower level of OmpU expressed, the ΔompA mutant appeared to bind somewhat less C1q (fig. 4a, lane 3). From these results we hypothesized that antibodies in NHS that were directed against OmpU would promote the interaction of C1q with the OmpU protein of V. cholerae. To further confirm whether the presumed interaction of C1q and OmpU was mediated by some anti-OmpU antibody present in NHS, the ligand overlay assay was performed after depleting the IgG of NHS using the enzyme IdeS. There was no evidence of C1q binding to OmpU in any of the samples from strains tested in the ligand overlay assay after IdeS pretreatment of NHS (fig. 4b).
Fig. 4.
Far Western analysis to detect C1q binding to the major Omps of V. cholerae. a SDS-PAGE and immunoblot analysis to detect indirect binding of C1q to OmpU. Lane 1, ΔompT; lane 2, ΔompU; lane 3, ΔompA; lane 4, wild-type A1552; lane 5, ΔtoxR. The arrow shows the C1q signal on the OmpU protein. Molecular weight markers are shown to the left of the membrane. b SDS-PAGE and immunoblot analysis of antibody-dependent binding of C1q on OmpU after IdeS treatment. Lane 1, ΔompT; lane 2, ΔompU; lane 3, ΔompA; lane 4, wild-type A1552; lane 5, ΔtoxR. Lane 6 was loaded with 100 ng of purified C1q protein. The arrow shows the immunoreaction band of C1q protein. Molecular weight markers are shown to the left of the membrane.
OMVs Protect V. cholerae against Serum Killing by Binding to the IgG-C1q Complex
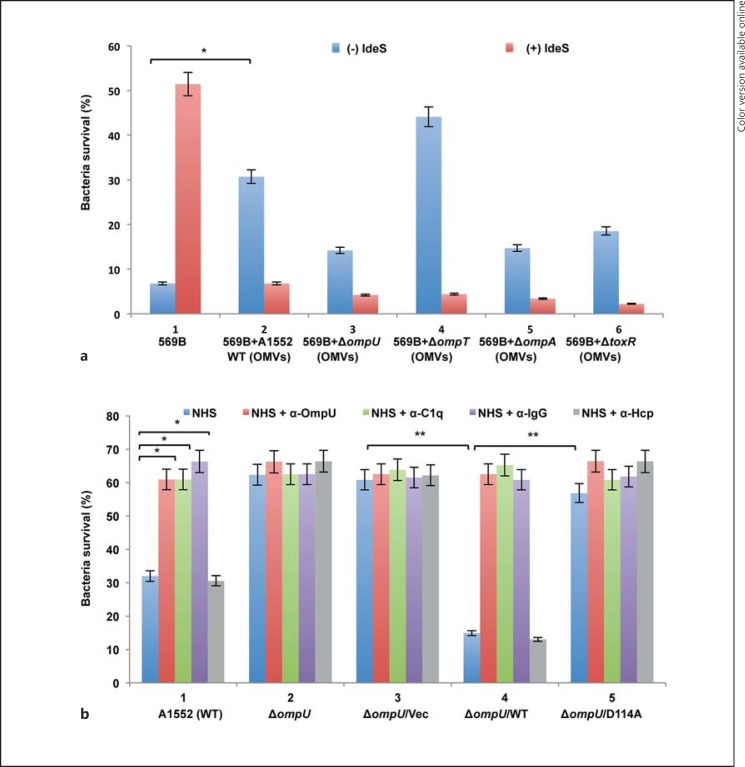
Recently, we discovered that OMVs isolated from V. cholerae may protect the bacteria against antimicrobial peptides by sequestering and quenching the effect of antimicrobial peptides through binding on the OMV surfaces [29]. In order to test if OMVs could also be involved in the serum resistance of V. cholerae, OMVs were isolated from the wild-type strain A1552 and from each of the ΔompU, ΔompA, ΔompT and ΔtoxR mutants. Using the bicinchoninic acid assay kit, the total protein concentration of each OMV sample was measured. All samples had similar protein concentrations except the OMV sample isolated from the ΔompA mutant [wild-type strain A1552 (5.12 mg/ml), ΔompU (4.61 mg/ml), ΔompA (48.98 mg/ml), ΔompT (4.54 mg/ml) and ΔtoxR (5.39 mg/ml)]. The protein concentration of the OMV sample obtained from the ΔompA mutant was 10 times higher in comparison with other samples. In our earlier studies, we have shown that the ΔompA mutant released a larger amount of OMVs when compared to the wild-type strain A1552 [30]. Furthermore, the number of OMV particles was measured by nanoparticle tracking analysis using the Nanosight equipment as described in Materials and Methods. Our data show that the ΔompA mutant released 10 times more OMVs in comparison with the other strains. For the serum-killing protection assay, we used OMV samples in a volume of 50 µl containing 5 × 1010 OMV particles. The classical V. cholerae wild-type strain 569B was used in the serum-killing protection assay since this strain showed very low serum resistance among strains of the different V. cholerae serogroups tested (fig. 1). As shown in figure 5a, the survival rates of V. cholerae strain 569B in NHS and in the presence of OMVs isolated from the wild-type strain A1552 were significantly increased from 6.8 to 30.7% (fig. 5a, panels 1 and 2, blue columns). In addition, the survival rate of the wild-type V. cholerae strain 569B was increased to 51% when IdeS-treated NHS was used in the killing assay (fig. 5a, panel 1, red column). When we tested the survival of strain 569B incubated with OMV+IdeS-treated NHS, the survival of 569B was nevertheless not recovered (fig. 5a, panel 2, red column). This effect might be due to both inhibition of IgG-C1q binding on the bacterial cells and IdeS degradation of OMV-associated IgG. It might indicate that some OMV-associated additional factor(s) could be involved. We further tested the role of OmpU in OMV-mediated protection against serum killing of V. cholerae using OMVs isolated from the ΔompU, ΔompT, ΔompA and ΔtoxR mutants. The highest survival rate of strain 569B (44.1%) was observed when the NHS treatment was performed in the presence of OMVs isolated from the ΔompT mutant (fig. 5a, panel 4, blue column). Much lower survival rates (14.2, 14.7 and 18.5%) were observed when the serum treatment was performed in the presence of OMVs isolated from the ΔompU, ΔompA and ΔtoxR mutants, respectively (fig. 5a, panels 3, 5 and 6, blue columns). These results indicated that OmpU-associated OMVs could efficiently quench the complement activity and we hypothesized that C1q in the NHS would be sequestered through an IgG-C1q complex binding on the surface of the OMVs via the OmpU protein. As described above, the ΔompA mutant showed increased serum resistance at the same level as observed in the case of the ΔtoxR mutant in comparison with the wild-type A1552. Increased release of OMVs from the ΔompA mutant might contribute to a higher serum resistance of bacteria. In the serum-killing assay, we used a 50-µl bacterial suspension which contains OMVs spontaneously released from the bacterial cells during growth. In order to test if these released OMVs can affect the serum-killing activity, we performed the serum-killing assay using washed V. cholerae wild-type strains A1552 and 569B. However, as shown in online supplementary figure S2, there was no difference in survival between washed and unwashed bacteria.
Fig. 5.
Serum sensitivity assay with V. cholerae monitoring the effects of IdeS, OMVs and antisera raised against OmpU, C1q, IgG or Hcp. a Panel 1: blue column, percent survival of V. cholerae strain 569B in NHS; red column, percent survival of V. cholerae strain 569B in NHS treated with IdeS. Panel 2: blue column, percent survival of 569B in NHS and OMVs from wild-type strain A1552. Panel 3: blue column, percent survival of 569B in NHS with OMVs from the ΔompU mutant. Panel 4: blue column, percent survival of 569B in NHS with OMVs from the ΔompT mutant. Panel 5: blue column, percent survival of 569B in NHS with OMVs from ΔompA mutant. Panel 6: blue column, percent survival of 569B in NHS with OMVs from ΔtoxR mutant. Panels 2-6: red columns represent percent survival of the same strains in the presence of NHS+OMVs+IdeS (colors refer to the online version only). Values represent the mean ± SD of three independent experiments. The asterisk indicates the survival percentage of V. cholerae 569B in NHS treated with OMVs versus NHS without treatment with OMVs (p < 0.05). b Blue columns represent the survival of V. cholerae wild-type strain A1552 and its derivatives in NHS alone. Red columns show the percent survival of A1552 and its derivatives treated in NHS and anti-OmpU antiserum. Green columns represent the survival percentage of A1552 and its derivatives in NHS and anti-C1q antiserum. Violet columns show the percent survival of A1552 and its derivatives in NHS and anti-IgG antibody. Gray columns show the percent survival of A1552 and its derivatives in NHS and anti-Hcp antibody (colors refer to the online version only). The strain genotypes are indicated below the columns. Values represent the mean ± SD of three independent experiments. Single asterisks indicate wild-type V. cholerae A1552 in NHS versus NHS pretreated with anti-C1q antibody, anti-IgG antibody or anti-OmpU antibody (p < 0.05). Double asterisks indicate ΔompU mutant in NHS versus the ΔompU mutant harboring either vector control or the wild-type ompU clone or ompU/D114A clone in NHS (p < 0.05).
Role of OmpU D114 Amino Acid Residue in the Serum Sensitivity of V. cholerae
In earlier studies, it was shown that a negatively charged residue D116 of V. cholerae OmpU influenced the permeation and binding of hydrophilic compounds, including β-lactam antibiotics and Na-deoxycholate bile salt [31]. In the wild-type V. cholerae strain A1552 the aspartic acid residue is at position 114 instead of 116. To test if D114 has a role in the OmpU-mediated serum sensitivity of V. cholerae, we performed site-directed mutagenesis to generate an ompU plasmid clone with a D114A substitution. Serum-killing assays were performed using the ΔompU mutant and the ΔompU mutant harboring either the wild-type ompU+ clone (ΔompU/WT), the D114A substitution clone (ΔompU/D114A) or the vector control (ΔompU/Vec). The ΔompU mutant strain, which had a 62.3% serum survival rate, became significantly serum sensitive (14.9% survival) when the wild-type ompU+ plasmid clone was introduced (fig. 5b, panels 2 and 4, blue columns), whereas the introduction of the D114A substitution, as in the derivative ΔompU/D114A, did not alter serum sensitivity and it remained at the same level as that of the ΔompU mutant (fig. 5b, panels 2 and 5, blue columns). All strains showed a similar ability to survive in HIS (data not shown). The expression levels of OmpU protein were similar in the ΔompU/WT and the ΔompU/D114A derivatives (online suppl. fig. S3, lanes 4 and 5).
Role of the OmpU-IgG-C1q-Mediated Classical Pathway in Bactericidal Activity
To test the effect of the OmpU-IgG-C1q-mediated activation of the classical pathway on killing OmpU-expressing V. cholerae, we performed neutralization assays using anti-C1q, anti-IgG and anti-OmpU antisera. After 1 h of incubation of V. cholerae in NHS preincubated with anti-C1q, anti-IgG or anti-OmpU antibodies, survival of the V. cholerae wild-type strain A1552 increased significantly (fig. 5b, panel 1; compare the blue column with the red, green and violet columns). The neutralization effect of antibodies recognizing OmpU, IgG or C1q was also observed in the case of the ΔompU/WT derivative (fig. 5b, panel 4; compare the blue column with the red, green and violet columns). On the basis of the results in figure 5b and those reported above, we propose that the serum killing of OmpU-expressing V. cholerae was mediated mainly by the anti-OmpU antibody (IgG) and the C1q-dependent classical pathway present in NHS, and that the ompU mutant bacteria survived by escaping this complement killing mechanism. The antiserum generated against an unrelated protein, the type VI secretion system protein Hcp, was included as a negative control. As shown in figure 5b (gray columns), anti-Hcp antiserum could not neutralize the serum-killing activity of NHS.
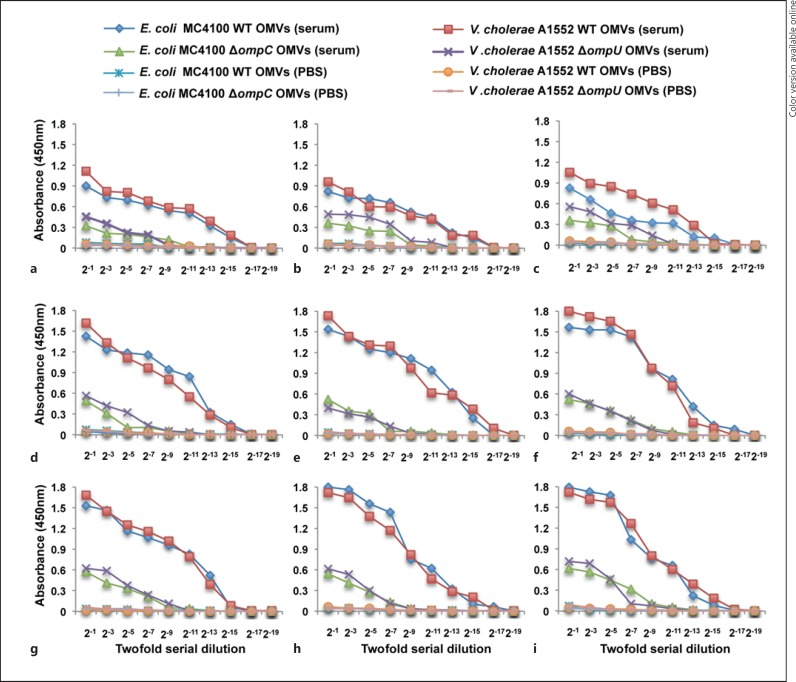
Evidence for Antibodies Recognizing V. cholerae OmpU Protein in Serum of Healthy Humans
Among Gram-negative bacterial species, immunogenic cross-reactions are very common, especially for the antigenic OMPs from various bacteria [32]. The presence of anti-OmpU antibodies in NHS might be due to an immunogenic cross-reaction between the OMPs of intestinal commensal bacteria, such as E. coli and OmpU of V. cholerae. To determine to what extent anti-OmpU antibody is present in NHS, the IgG titers in 9 different serum samples were determined by ELISA. PBS was used as a negative control to test for nonspecific binding of peroxidase-coated IgG to the OMVs. Anti-OmpU IgG was detected in all the sera tested (fig. 6, red squares). When OMVs isolated from the V. cholerae ΔompU mutant were used as bate, consistently lower IgG levels were detected (fig. 6, purple crosses). This was presumably due to the presence of other IgG-binding proteins on the surface of the OMVs from the ΔompU mutant.
Fig. 6.
a-i Serological analysis of IgG. ELISA to measure the levels of IgG against OmpU of V. cholerae and OmpC of E. coli in NHS. Sera were isolated from the healthy donors. OMVs isolated from the wild-type V. cholerae strain A1552 and its ΔompU mutant, and the wild-type E. coli strain MC4100 and its ompC mutants were used as antigens for ELISA. Red squares and purple crosses show the IgG titers in sera from the donor against OMVs of the wild-type strain A1552 and that of the ΔompU mutant, respectively. Blue squares and green triangles show the IgG titer in sera from the donor against OMVs of the wild-type E. coli strain MC4100 and that of the ΔompC mutant, respectively. Orange circles, pink lines, blue crosses and blue lines represent negative controls (colors refer to the online version only).
We considered that a plausible reason for the presence of ‘anti-OmpU’ antibodies in serum from healthy donors, without any known previous exposure to cholera, would be that these IgG antibodies had been developed against similar proteins in the outer membrane of commensals such as the OmpC protein of E. coli, known as a functional homolog of OmpU. We isolated OMVs from the wild-type E. coli strain MC4100 and the ΔompC mutant E. coli derivative, and used these vesicles as the antigens in ELISA. With all serum samples we observed a higher titer of IgG against the OMVs containing E. coli OmpC (fig. 6, compare blue squares and green triangles).
Discussion
In this study, we elucidated a mechanism of how serum resistance may occur in V. cholerae. Serum sensitivity and complement-mediated killing of different V. cholerae isolates belonging to O1 El Tor, O1 classical and NOVC serogroups were found to differ considerably. Despite not producing cholera toxin, non-O1, non-O139 V. cholerae can be clearly pathogenic, causing clinical syndromes ranging from mild diarrheal illness to septicemia, primarily in vulnerable hosts [5].
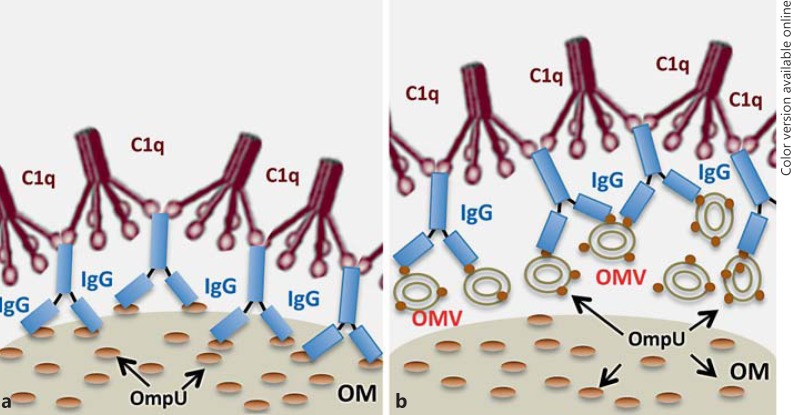
We suggest that the presence of natural IgG antibodies in NHS, most likely raised against some OmpU homologs of the commensal flora, can contribute to the defense against invading V. cholerae pathogens. Such IgG can bind on the surface of bacteria and attract C1q molecules, leading to activation of the complement system and clearance of bacterial invaders in the blood. Furthermore, OMVs released from bacterial cells and containing OmpU can also trap the IgG-C1q complex, thereby protecting the bacterial cells against C1q-IgG-mediated serum killing. An illustration of the OmpU-IgG-C1q-mediated serum-killing effect on bacterial cells and the mechanism by which bacteria can evade this system, by releasing and trapping the complexes, is presented in figure 7. Recently, the recognition of the pattern recognition receptors ficolin and MBL by natural IgG was shown to mediate protection from experimental infection of mice with P. aeruginosa[13]. Here we demonstrate a lectin-independent direct immunorecognition of bacterial surface proteins by natural IgGs, mediating activation of the classical complement cascade and serum sensitivity of V. cholerae.
Fig. 7.
An illustration showing the deposition of the OmpU-IgG-C1q complex on a bacterial cell surface and OMV-mediated protection against OmpU-IgG-C1q-dependent serum killing. a IgG-C1q complex binding to OmpU protein of bacterial outer membrane leads to a complement-mediated serum killing effect. b OmpU-associated OMVs released from the bacterial cell can bind C1q through IgG leading to the inactivation of serum killing. OM = Outer membrane.
Interestingly, the level of OmpU was significantly different among V. cholerae isolates. Our findings led to the conclusion that this major outer membrane porin protein was a primary factor in the serum-sensitive phenotype of V. cholerae. Decreased expression of OmpU in V. cholerae may be advantageous, especially for NOVC V. cholerae strains associated with invasive infection such as bacteremia. The serum sensitivity of V. cholerae was reduced by adding a classical complement pathway blocker (EGTA and MgCl2) to the serum. The results reported here confirm the importance of the classical pathway in serum killing of V. cholerae strains and extend the previous knowledge of the interaction between this bacterium and the classical complement pathway. Using defined OMP mutant strains and a mutant deficient in regulating the expression of OMPs, we obtained results clearly consistent with the notion that Clq can bind to OmpU via IgG. The molecular basis of C1q binding to bacterial porins has not been clarified. Earlier studies suggested that residue D116 (D114 in our strain) of OmpU protein might bind to membrane-targeting antibiotics and, thus, confer antibiotic resistance to V. cholerae[31]. We anticipated that the homologous pore-exposed residue D114 may control channel properties or ligand binding of the OmpU porins of V. cholerae. To test this hypothesis, we engineered a substitution mutation in OmpU at the selected residue D114. To further analyze details of the mechanism of interactions between OmpU and its ligand C1q, the wild-type ompU gene was introduced into the highly serum-resistant ΔompU mutant. The resulting strain became very serum sensitive (ΔompU/WT), but introduction of the mutant OmpU (D114A) could not alter serum sensitivity of the ΔompU mutant (ΔompU/D114A).
It has been clearly demonstrated that almost all Gram-negative bacteria release detectable amounts of OMVs during normal growth. The main components of OMVs are major OMPs. We hypothesized that if OmpU-containing OMVs are released, these OMVs might sequester and quench C1q when the bacterial cells are exposed to NHS.
We tested whether OMVs isolated from the wild-type V. cholerae strain A1552 can quench C1q present in NHS. Interestingly, OMVs from the V. cholerae wild-type strain A1552 could protect the classical serotype strain 569B against the bactericidal effect of NHS while OMVs isolated from the ΔompU mutant could not prevent serum killing. We suggest that OmpU-containing OMVs can efficiently quench the activity of C1q in serum by binding of the complex with IgG and C1q to OmpU on the surface of the OMVs, thus providing serum resistance to the 569B strain.
In our study, a surprisingly high titer of IgG against the OmpU protein of V. cholerae was observed in the tested samples of human serum. Detection of the IgG-recognizing OmpC protein of E. coli, homolog of V. cholerae OmpU, suggested that these antibodies might have developed against the outer membrane of commensal E. coli bacteria.
The immune system-microbiota organization can allow the induction of a protective immune response to pathogenic microorganisms and the maintenance of tolerance to inoffensive or harmless antigens [33]. Interestingly, it was recently demonstrated that a novel symbiont molecule, polysaccharide A, PSA, from the human commensal bacteria Bacteroides fragilis could protect mice from experimental colitis induced by Helicobacter hepaticus, a commensal bacterium with pathogenic potential, by promoting regulatory immune responses [34]. Based on the complexity and abundance of the commensal flora, it can be speculated that opportunities for cross-reaction between commensals and pathogens may be high. The results obtained in this work highlight the potential of V. cholerae to utilize a differential expression of OMPs to evade host immunity.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgements
This work was performed within the Umeå Centre for Microbial Research (UCMR) Linnaeus Programme and was supported by grants from the Swedish Research Council [S.N.W.: 2013-2392 (VR-M), 2014-4401 (VR-NT); B.E.U.: 2012-4638 (VR-NT); K.R.: K2015-57X-03163-43-4] and the Faculty of Medicine at Umeå University. We thank Dr. M. Iwanaga and Dr. C. Toma for providing OmpU antiserum.
References
- 1.Blake PA, Weaver RE, Hollis DG. Diseases of humans (other than cholera) caused by vibrios. Annu Rev Microbiol. 1980;34:341–367. doi: 10.1146/annurev.mi.34.100180.002013. [DOI] [PubMed] [Google Scholar]
- 2.Provenzano D, Lauriano CM, Klose KE. Characterization of the role of the ToxR-modulated outer membrane porins OmpU and OmpT in Vibrio cholerae virulence. J Bacteriol. 2001;183:3652–3662. doi: 10.1128/JB.183.12.3652-3662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duperthuy M, Schmitt P, Garzon E, Caro A, Rosa RD, Le Roux F, Lautredou-Audouy N, Got P, Romestand B, de Lorgeril J, Kieffer-Jaquinod S, Bachere E, Destoumieux-Garzon D. Use of ompU porins for attachment and invasion of Crassostrea gigas immune cells by the oyster pathogen Vibrio splendidus. Proc Natl Acad Sci USA. 2011;108:2993–2998. doi: 10.1073/pnas.1015326108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miajlovic H, Cooke NM, Moran GP, Rogers TR, Smith SG. Response of extraintestinal pathogenic Escherichia coli to human serum reveals a protective role for Rcs-regulated exopolysaccharide colanic acid. Infect Immun. 2014;82:298–305. doi: 10.1128/IAI.00800-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson AM, Varkey JB, Petti CA, Liddle RA, Frothingham R, Woods CW. Non-O1 Vibrio cholerae septicemia: case report, discussion of literature, and relevance to bioterrorism. Diagn Microbiol Infect Dis. 2004;49:295–297. doi: 10.1016/j.diagmicrobio.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 6.Walport MJ. Complement. First of two parts. N Engl J Med. 2001;344:1058–1066. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 7.Hallstrom T, Riesbeck K. Haemophilus influenzae and the complement system. Trends Microbiol. 2010;18:258–265. doi: 10.1016/j.tim.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Liu YF, Yan JJ, Lei HY, Teng CH, Wang MC, Tseng CC, Wu JJ. Loss of outer membrane protein C in Escherichia coli contributes to both antibiotic resistance and escaping antibody-dependent bactericidal activity. Infect Immun. 2012;80:1815–1822. doi: 10.1128/IAI.06395-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agarwal V, Ahl J, Riesbeck K, Blom AM. An alternative role of C1q in bacterial infections: facilitating Streptococcus pneumoniae adherence and invasion of host cells. J Immunol. 2012;191:4235–4245. doi: 10.4049/jimmunol.1300279. [DOI] [PubMed] [Google Scholar]
- 10.Avrameas S. Natural autoantibodies: from ‘horror autotoxicus’ to ‘gnothi seauton’. Immunol Today. 2012;12:154–159. doi: 10.1016/S0167-5699(05)80045-3. [DOI] [PubMed] [Google Scholar]
- 11.Kaveri SV. Intravenous immunoglobulin: exploiting the potential of natural antibodies. Autoimmun Rev. 2012;11:792–794. doi: 10.1016/j.autrev.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Pollack M, Huang AI, Prescott RK, Young LS, Hunter KW, Cruess DF, Tsai CM. Enhanced survival in Pseudomonas aeruginosa septicemia associated with high levels of circulating antibody to Escherichia coli endotoxin core. J Clin Invest. 1983;72:1874–1881. doi: 10.1172/JCI111150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panda S, Zhang J, Tan NS, Ho B, Ding JL. Natural IgG antibodies provide innate protection against ficolin-opsonized bacteria. EMBO J. 2013;32:2905–2919. doi: 10.1038/emboj.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panda S, Ding JL. Natural antibodies bridge innate and adaptive immunity. J Immunol. 2015;194:13–20. doi: 10.4049/jimmunol.1400844. [DOI] [PubMed] [Google Scholar]
- 15.Vaitkevicius K, Lindmark B, Ou G, Song T, Toma C, Iwanaga M, Zhu J, Andersson A, Hammarstrom ML, Tuck S, Wai SN. A Vibrio cholerae protease needed for killing of Caenorhabditis elegans has a role in protection from natural predator grazing. Proc Natl Acad Sci USA. 2006;103:9280–9285. doi: 10.1073/pnas.0601754103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fine DP, Marney SR, Jr, Colley DG, Sergent JS, Des Prez., RM C3 shunt activation in human serum chelated with EGTA. J Immunol. 1972;109:807–809. [PubMed] [Google Scholar]
- 17.von Pawel-Rammingen U, Johansson BP, Bjorck L. IdeS, a novel streptococcal cysteine proteinase with unique specificity for immunoglobulin G. EMBO J. 2002;21:1607–1615. doi: 10.1093/emboj/21.7.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vindebro R, Spoerry C, von Pawel-Rammingen., U Rapid IgG heavy chain cleavage by the streptococcal IgG endopeptidase IdeS is mediated by IdeS monomers and is not due to enzyme dimerization. FEBS Lett. 2013;587:1818–1822. doi: 10.1016/j.febslet.2013.04.039. [DOI] [PubMed] [Google Scholar]
- 19.Nakasone N, Iwanaga M. Characterization of outer membrane protein OmpU of Vibrio cholerae O1. Infect Immun. 1998;66:4726–4728. doi: 10.1128/iai.66.10.4726-4728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishikawa T, Rompikuntal PK, Lindmark B, Milton DL, Wai SN. Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS One. 2009;4:e6734. doi: 10.1371/journal.pone.0006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wai SN, Mizunoe Y, Takade A, Kawabata S, Yoshida S. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl Environ Microbiol. 1998;64:3648–3655. doi: 10.1128/aem.64.10.3648-3655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Wai SN, Lindmark B, Soderblom T, Takade A, Westermark M, Oscarsson J, Jass J, Richter-Dahlfors A, Mizunoe Y, Uhlin BE. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell. 2003;115:25–35. doi: 10.1016/s0092-8674(03)00754-2. [DOI] [PubMed] [Google Scholar]
- 24.Rompikuntal PK, Vdovikova S, Duperthuy M, Johnson TL, Åhlund M, Lundmark R, Oscarsson J, Sandkvist M, Uhlin BE, Wai SN. Outer membrane vesicle-mediated export of processed PrtV protease from Vibrio cholerae. PLoS One. 2015;10:e0134098. doi: 10.1371/journal.pone.0134098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rautemaa R, Meri S. Complement-resistance mechanisms of bacteria. Microbes Infect. 1999;1:785–794. doi: 10.1016/s1286-4579(99)80081-1. [DOI] [PubMed] [Google Scholar]
- 26.Joens LA, Nuessen ME. Bactericidal effect of normal swine sera on Treponema hyodysenteriae. Infect Immun. 1986;51:282–285. doi: 10.1128/iai.51.1.282-285.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steele NP, Munson RS, Jr, Granoff DM, Cummins JE, Levine RP. Antibody-dependent alternative pathway killing of Haemophilus influenzae type b. Infect Immun. 1984;44:452–458. doi: 10.1128/iai.44.2.452-458.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaboriaud C, Juanhuix J, Gruez A, Lacroix M, Darnault C, Pignol D, Verger D, Fontecilla-Camps JC, Arlaud GJ. The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J Biol Chem. 2003;278:46974–46982. doi: 10.1074/jbc.M307764200. [DOI] [PubMed] [Google Scholar]
- 29.Duperthuy M, Sjostrom AE, Sabharwal D, Damghani F, Uhlin BE, Wai SN. Role of the Vibrio cholerae matrix protein Bap1 in cross-resistance to antimicrobial peptides. PLoS Pathog. 2013;9:e1003620. doi: 10.1371/journal.ppat.1003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song T, Mika F, Lindmark B, Liu Z, Schild S, Bishop A, Zhu J, Camilli A, Johansson J, Vogel J, Wai SN. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol Microbiol. 2008;70:100–111. doi: 10.1111/j.1365-2958.2008.06392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pagel M, Simonet V, Li J, Lallemand M, Lauman B, Delcour AH. Phenotypic characterization of pore mutants of the Vibrio cholerae porin OmpU. J Bacteriol. 2007;189:8593–8600. doi: 10.1128/JB.01163-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu C, Wang S, Zhaoxia Z, Peng X. Immunogenic cross-reaction among outer membrane proteins of Gram-negative bacteria. Int Immunopharmacol. 2005;5:1151–1163. doi: 10.1016/j.intimp.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 33.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 35.Miller VL, Taylor RK, Mekalanos JJ. Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 36.Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 37.Benson SA, Decloux A. Isolation and characterization of outer membrane permeability mutants in Escherichia coli K-12. J Bacteriol. 1985;161:361–367. doi: 10.1128/jb.161.1.361-367.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thelin KH, Taylor RK. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chatterjee S, Mondal AK, Begum NA, Roychoudhury S, Das J. Ordered cloned DNA map of the genome of Vibrio cholerae 569B and localization of genetic markers. J Bacteriol. 1998;180:901–908. doi: 10.1128/jb.180.4.901-908.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song T, Sabharwal D, Wai SN. VrrA mediates Hfq-dependent regulation of OmpT synthesis in Vibrio cholerae. J Mol Biol. 2010;400:682–688. doi: 10.1016/j.jmb.2010.05.061. [DOI] [PubMed] [Google Scholar]
- 42.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furste JP, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 44.Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW, Crosa JH, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data