Abstract
Coxsackievirus strain B serotype 3 (CVB3)-induced myocarditis is an important human disease that causes permanent tissue damage and can lead to death from acute infection or long-term morbidity caused by chronic inflammation. The timing and magnitude of immune activation following CVB3 infection can mediate a positive host outcome or increase tissue pathology. To better elucidate the role of endosomal Toll-like receptor (TLR) signaling in acute CVB3 infection, we studied mice with a loss-of-function mutation, known as Letr for ‘loss of endosomal TLR response’, in Unc93b1, which is a chaperone protein for TLR3, TLR7 and TLR9. Using Unc93b1Letr/Letr mice, we determined that Unc93b1-dependent TLR activation was essential for the survival of acute CVB3-induced myocarditis. We also determined that a lack of endosomal TLR signaling was associated with a higher viral load in target organs and that it increased inflammation, necrosis and fibrosis in cardiac tissue. Loss of Unc93b1 function was also associated with increased cardiac expression of Ifn-b and markers of tissue injury and fibrosis including Lcn2 and Serpina3n early after CVB3 infection. These observations establish a significant role for Unc93b1 in the regulation of the host inflammatory response to CVB3 infection and also reveal potential mediators of host tissue damage that merit further investigation in acute viral myocarditis.
Key Words: Coxsackievirus strain B serotype 3, Myocarditis, Unc93b1, Endosomal Toll-like receptor, Fibrosis, Gene expression array
Introduction
Myocarditis is characterized by cardiomyocyte necrosis adjacent to tissue inflammation [1, 2]. In the Western world, coxsackievirus strain B serotype 3 (CVB3) is implicated in 20–40% of human viral myocarditis cases [1, 2]. CVB3, a positive-sense single-stranded RNA virus of the Picornaviridae family, causes an infection via the fecal-oral route that is usually asymptomatic or can result in a mild febrile or gastrointestinal illness [1, 2]. Occasionally, this infection can trigger more severe sequelae including meningitis, pancreatitis or myocarditis. The true incidence of CVB3-induced myocarditis is unknown due to its variable presentation and a lack of sensitive and specific diagnostic techniques [2]. Patients with viral myocarditis may rapidly succumb to acute heart failure or develop dilated cardiomyopathy (DCM), a chronic inflammatory form of the disease that occurs in 20–30% of cases and frequently requires a heart transplant [1, 2]. Poor outcome following CVB3 infection has been linked to several factors including a patient's sex, nutritional status, age, and the host immune response [1, 2].
During the initial phase of the infection, CVB3 virus replication causes direct cardiomyocyte damage and cell lysis that can contribute to worsening disease [3, 4]. Innate immune cell infiltration, including macrophages and NK cells, occurs 4–5 days after infection and is followed several days later by an adaptive T and B cell response [1, 2]. Immune activation in the heart following CVB3 infection has been associated with both protective and pathological features of disease [2]. Therefore, a delicate balance in the timing and magnitude of the host response is required to successfully clear the virus from the heart without the induction of severe and permanent immunopathology. Myocardial cell damage caused by viral or immune mechanisms is particularly deleterious as cardiomyocytes do not regenerate easily, and permanent structural changes that severely compromise cardiac muscle contractility and function can occur following host sensing of damage and stimulation of fibrotic repair mechanisms [2, 5].
Infection with medically relevant viruses from the Picornaviridae family leads to the expression of type I interferon (IFN) [6]. In acute CVB3-induced myocarditis, studies of IFN-β-or IFN-α/β receptor (IFNAR)-deficient mice demonstrated a protective role for type I IFN [7, 8], and a subsequent clinical study in DCM patients also demonstrated an improvement in symptoms and cardiovascular damage in response to exogenous IFN-β administration [9]. Detection of CVB3 infection and rapid IFN induction is mediated by a variety of host pattern recognition receptors, including membrane-associated Toll-like receptors (TLRs) and cytoplasmic RIG-like receptors (RLRs) [10]. Mice lacking TLR3 are more susceptible to acute CVB3-induced myocarditis and have increased tissue damage and viral load [11, 12]. Conversely, the transgenic expression of TLR3 in mice lacking IFNAR leads to a higher expression of type II IFN during CVB3 infection that is associated with a reduction in mortality, tissue damage and viral titer [11]. Genetic analysis of a cohort of patients suffering from viral myocarditis revealed a rare variant and significantly increased occurrence of a common TLR3 polymorphism compared to controls [13]. In vitro expression of mutated TLR3 abrogated activation of the type I IFN pathway, leading to increased viral replication [13]. TIR-domain-containing adaptor molecule 1 (TICAM1), a protein associated with TLR3 and TLR4 signaling, has also been shown to be important in the host survival of acute CVB3-induced myocarditis [12]. Nevertheless, the precise role of TICAM1 in type I IFN activation during acute viral myocarditis is not well understood as TICAM1-deficient mice exhibited a dynamic pattern of cardiac IFN-β expression following CVB3 infection [12].
In addition to the TLR3/TICAM1 pathway, CVB3 is also recognized by TLR4, 7, 8 and 9 [14, 15]. TLR4-deficient mice challenged with CVB3 have significantly reduced levels of myocarditis, viral replication and cardiac IL-1β/IL-18 expression [16]. CVB3 infection of human cardiomyocytes or TLR-transfected HEK293T cells can trigger immune activation through TLR7 and TLR8 and, to a lesser extent, TLR4 [15]. Furthermore, TLR9 expression is upregulated in the wild-type heart whereas TLR9 deficiency reduces cardiac inflammation and immune cell infiltration in a mouse model of acute CVB3-induced myocarditis [14]. Similarly, mice that lack myeloid differentiation primary response gene 88 (MyD88), an adaptor for all TLRs except TLR3, are protected from mortality following CVB3 infection with a decrease in viral load and inflammation in multiple organs [17]. Interestingly, in both the TLR9- and MyD88-deficient models of acute-CVB3 induced myocarditis, cardiac IFN-β is elevated at day 7 after infection; this highlights the complexity of type I IFN regulation [14, 17]. The potentially pathogenic role of immune activation via TLR9/MyD88 in acute CVB3-induced myocarditis contrasts with the protective effect of TLR3/TICAM1-mediated immune activation [11, 12, 14, 17] and seems to demonstrate opposing roles for endosomal TLR signaling during CVB3 infection.
Cytoplasmic recognition of Picornaviridae was first demonstrated in encephalomyocarditis virus infection [18, 19]. Mice lacking the RIG-I-like receptor melanoma differentiation-associated protein 5 (MDA5) were found to be susceptible to lethal encephalomyocarditis virus infection that was associated with higher cardiac viral titers, focal necrosis, depressed contractility and lower serum levels of type I IFN and inflammatory cytokines [18]. Subsequently, mice lacking MDA5 or its adaptor MAVS were shown to be highly susceptible to lethal CVB3 infection [20, 21]. The expression of type I IFN was significantly lower in MAVS-deficient mice at day 2 after infection [21]; this supports a direct role for this pathway in the antiviral response. Recently, the double-stranded replicative form of CVB3 and several other picornavirus strains was shown to directly bind MDA5, leading to its activation and the synthesis of type I IFN [22]. The relative contribution of the TLR3/TICAM1 and MDA5/MAVS pathways during CVB3-induced myocarditis has not been definitively resolved, although it has been shown that the TLR3/TICAM1 pathway plays an indispensable role in IFN production and a dominant role in host protection compared to the MDA5/MAVS pathway in poliovirus infection [11, 21, 23]. Finally, a very recent study demonstrated that the NLRP3-dependent inflammasome is also activated during CVB3 infection and contributes to enhanced viral myocarditis [24].
In an effort to further parse the complex immune signaling mechanisms triggered by CVB3 infection, we have taken advantage of a recently developed loss-of-function mutation of Unc93b1, and aimed to study its role in immune activation following CVB3 infection. UNC93B1 is a transmembrane chaperone protein that trafficks endosomal TLRs (TLR3, TLR7 and TLR9) from the endoplasmic reticulum to the endosome to permit downstream immune signaling [25, 26, 27]. Using N-ethyl-N-nitrosourea mutagenesis, we created a novel mutation in Unc93b1 that results in an in-frame deletion of exon 4 following transcript splicing [28]. Mice with this mutation have a complete lack of immune activation via TLR3, TLR7 and TLR9 in vitro and in vivo following specific agonist stimulation with polyinosinic:polycytidylic acid (poly I:C), imiquimod and CpG DNA, respectively [28]. We termed this mutation Letr for ‘loss of endosomal TLR response’, and recently used this model to identify a specific role for Unc93b1 in early immune activation following influenza A/PR/8/34 (H1N1) infection [28]. A previously derived ENU-induced loss-of-function mutation known as 3d has also been used to implicate Unc93b1 in immune protection during other viral infections including a mouse model of neuroadapted Sindbis virus (NSV), mouse Cytomegalovirus (MCMV) and herpes simplex virus (HSV) [27, 29, 30]. Finally, a role for Unc93b1 has been demonstrated in protection against the development of encephalitis following human HSV infection [31].
In this study, we used Unc93b1Letr/Letr mice to study the consequences of a complete loss of endosomal TLR signaling in acute CVB3-induced myocarditis. This approach allowed us to specifically analyze the role of endosomal TLRs in the presence of functional MyD88 and TICAM1 adaptors that can also mediate immune signaling initiated by inflammatory cytokines and alternative pattern recognition receptors. Our data demonstrate that the loss of Unc93b1 function dramatically increased susceptibility to acute CVB3-induced myocarditis, was associated with enhanced viral replication in multiple organs and increased cardiac inflammation and necrosis that led to greater myocardial fibrosis. Global cardiac transcriptional profiling determined that the Unc93b1Letr/Letr mice had an overall increase in the expression of inflammatory response genes but significantly less expression of the genes involved in antigen processing and the presentation of exogenous peptide antigen via major histocompatibility complex (MHC) class II than Unc93b1+/+ mice. More detailed analysis of the gene expression array revealed a specific upregulation of interferon-beta (Ifn-b), lipocalin2(Lcn2) and serine (or cysteine) peptidase inhibitor, clade A, member 3N(Serpina3n) in the cardiac tissue of Unc93b1Letr/Letr mice during the early response to infection. This study contributes to an improved understanding of the essential role of Unc93b1 in the host response to CVB3 infection and identifies several interesting mediators that are activated downstream of endosomal TLRs and may be associated with severe cardiac pathology.
Materials and Methods
Ethics Statement
All animal experiments were conducted by following the Canadian Council on Animal Care guidelines and were approved by the McGill University Animal Care Committee, Montréal, Canada.
Mice, Virus and Infection
Unc93b1 +/+ mice and Unc93b1Letr/Letr mice on a C57BL/6 background were housed in a specific pathogen-free facility and used at 8 weeks of age. The CVB3-CG virus was administered intraperitoneally at 400 PFU/g in PBS (Wisent, St.-Bruno, Canada). They were monitored daily for signs of illness and were sacrificed humanely if they appeared moribund.
Viral Quantification by Plaque Assay
HeLa cells were grown at 37°C and 5% CO2 in DMEM (Wisent) supplemented with 10% FBS (Wisent) and 1% penicillin/streptomycin (Life Technologies, Burlington, Canada). The mice were humanely sacrificed at days 2, 4 and 8 after infection and the hearts were perfused with 10 ml of PBS. The apex of the heart, a single liver lobe and half of the spleen were placed in Lysing Matrix D tubes (MP Biomedicals, Solon, Ohio, USA) containing 1-ml aliquots of DMEM. Samples were homogenized at 7,000 rpm for 40 s using a MagNA Lyser (Roche Applied Science, Laval, Que., Canada) and subjected to 3 freeze/thaw cycles. Following 10-fold serial dilutions in DMEM, 200 μl of supernatant was added in triplicate to a confluent monolayer of HeLa cells on a 12-well plate (Greiner Bio-One, Frickenhausen, Germany) and incubated for 1 h at 37°C and 5% CO2. A 50:50 mixture of 0.5% agarose (Life Technologies) and DMEM supplemented with 20% FBS and 2% penicillin/streptomycin (1.5 ml) was added and the plates were then incubated at 37°C and 5% CO2. After 48 h, 1 ml of 10% phosphate-buffered formalin (Fisher Scientific, Toronto, Canada) was added and then incubated at room temperature overnight. Agarose plugs and formalin were then removed and plaques were stained with 100 μl of a 0.5% crystal violet solution (Sigma-Aldrich, Oakville, Canada) in 70% ethanol (Commercial Alcohols, Brampton, Canada).
Histology
The mice were humanely sacrificed at day 8 after infection and the hearts were cut in the sagittal plane, embedded in optimal cutting temperature compound (Tissue-Tek, VWR, Ville Mont-Royal, Canada) and then frozen in liquid nitrogen-chilled isopentane. Tissue sections (5 μm) were stained with hematoxylin and eosin (H&E) at the Histology Facility of the University of Ottawa, Ottawa, Canada, or with Masson's trichrome at the Histology Facility of the Goodman Cancer Research Centre, McGill University, Montréal, Canada. Representative photographs of the heart sections were visualized at ×4 and ×10 magnification on a BX51 microscope (Olympus, Richmond Hill, Canada) with a QICAM Fast 1394 digital CCD camera (QImaging, Surrey, Canada) and Image-Pro Plus, version 7.0.1.658 (Media Cybernetics, Rockville, Md., USA). The slides were blindly scored by two independent observers using a previously published scoring system that classifies samples based on a composite of observed cardiac inflammation and necrosis [32]. Inflammation is represented by clumps of cells with nuclei stained blue by hematoxylin while areas of clearance illustrate necrosis. Normal cardiac muscle is visualized as eosin-stained red cells. A score of ‘0’ represents a healthy heart that is free from myocarditis while a score of ‘4’ indicates extensive necrosis and inflammation observed in the majority of the tissue.
RNA Extraction and Reverse Transcription
Hearts from female Unc93b1+/+ and Unc93b1Letr/Letr mice at day 0 and day 2 after infection were aseptically removed, and the apex was snap-frozen in RNAlater (Life Technologies). RNA was extracted using the RNeasy fibrous tissue mini kit (Qiagen, Toronto, Canada) following the manufacturer's instructions, quantified with a NanoDrop (Fisher Scientific) and stored at −80°C. Reverse transcription was performed using the ABI high-capacity cDNA reverse transcription kit with RNase inhibitor according to the manufacturer's instructions (Life Technologies).
Microarray
Gene expression analyses of total RNA from cardiac tissue were performed using the Mouse WG-6 Expression BeadChips (Illumina, San Diego, Calif., USA) at the McGill University and Genome Quebec Innovation Centre, Montréal, Canada. For the microarray, 3 biological replicates were analyzed for each genotype at day 0 and day 2 after infection. The expression data from the microarray were analyzed using FlexArray, version 1.6.2 [33]. Data were normalized using the robust spline normalization of the lumi algorithm. To obtain lists of differentially expressed genes between Unc93b1+/+ and Unc93b1Letr/Letr hearts at day 2 after infection, the Cyber-T test was used with a Benjamini-Hochberg false discovery rate correction to account for multiple testing. Genes with >2-fold differential expression and an adjusted p value (padj) <0.05 were used for further pathway and ontology analysis, and a cut-off of >5-fold differential expression and a padj <0.05 was used to select genes for confirmation of array expression data by quantitative reverse transcription PCR (qRT-PCR).
Analysis of Differentially Expressed Genes
A principal component analysis (PCA) plot was used to visualize the changes in total normalized gene expression. The Mouse Genome Informatics (MGI) database, version 5.15 [34] (The Jackson Laboratory, Bar Harbor, Me., USA) was used to annotate the unassigned targets that represented gene transcripts in the expression microarray. Pathway, gene ontology (GO) biological process and GO cellular component analyses of gene expression data were conducted using the Database for Annotation, Visualization, and Integrated Discovery (DAVID), version 6.7 [35, 36] (http://david.abcc.ncifcrf.gov; National Institute of Allergy and Infectious Diseases, National Institutes of Health, Frederick, Md., USA). Differentially expressed genes that were not recognized by either the MGI or DAVID annotation databases were excluded from further analysis, as a role for these unassigned targets in CVB3 infection could not be inferred. Hierarchical clustering with complete linkage was conducted using the MultiExperiment Viewer, version 4.9 [37] (http://www.tm4.org/mev.html; Dana-Farber Cancer Institute, Boston, Mass., USA). The AmiGO database, version 1.8 [38] and the GO biological process term for immune response (GO:0006955; http://amigo.geneontology.org/cgi-bin/amigo/go.cgi; the Gene Ontology Consortium) were used to define immune genes on the expression array.
Quantitative Reverse Transcription PCR
To confirm differential gene expression, qRT-PCR was conducted using the ABI 7500 real-time PCR system and accompanying software, version 2.0.3 (Life Technologies). TaqMan probes were used for Gapdh (Mm99999915_g1), Ifn-b (Mm00439552_s1), Lcn2 (Mm01324470_m1) and Serpina3n (Mm00776439_m1). Each reaction consisted of 10 μl of TaqMan genotyping master mix (Life Technologies), 5 μl of water (Wisent), 1 μl of TaqMan probe (Life Technologies) and 80 ng of cDNA in 4 μl of water. The PCR cycling conditions were 95°C for 10 min followed by 40 cycles of 95°C for 15 s/60°C for 1 min. Analysis was performed using the comparative Ct method (2−ΔΔCt) [39]. Gapdh was used as an endogenous control and expression at day 2 is relative to expression in uninfected hearts.
Statistical Analysis
Statistical analyses, excluding the analysis of microarray data, were conducted using GraphPad Prism, version 4 (GraphPad Software, La Jolla, Calif., USA). Survival analysis was performed using the log-rank test. An unpaired Student t test was used for comparisons of viral load, myocarditis score and relative gene expression measured by qRT-PCR.
Results
Loss of Unc93b1 Function Increases Mortality and Delays Viral Clearance in CVB3 Infection
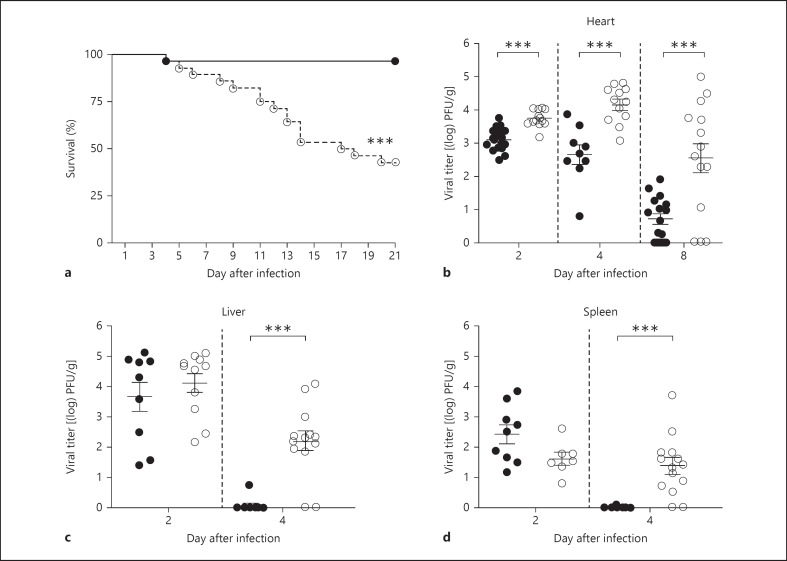
Endosomal TLR signaling via TICAM1 or MyD88 in acute CVB3-induced myocarditis has been associated with host protection or disease progression, respectively. To investigate the effect of a complete loss of endosomal TLR signaling on infection outcome, we infected Unc93b1+/+ and Unc93b1Letr/Letr mice with CVB3 and followed the clinical and virological evolution of disease. All experiments were conducted using mice on a C57BL/6 genetic background as this strain can effectively clear the virus from the heart following acute infection without developing DCM and present with disease pathology that is similar to humans [2]. After intraperitoneal infection with 400 PFU/g of the CVB3-CG strain, the Unc93b1Letr/Letr mice succumbed more rapidly with an overall mortality of 57% compared to 4% in Unc93b1+/+ mice, during an acute 21-day infection period (fig. 1a). The increased susceptibility observed in the Unc93b1Letr/Letr mice was associated with a significantly elevated cardiac viral load at days 2, 4 and 8 after infection compared to the Unc93b1+/+ mice (fig. 1b). Both genotypes had a substantial viral titer in the liver and spleen at day 2, and this was sustained in the Unc93b1Letr/Letr mice at day 4 when compared to the controls (fig. 1c, d). Detectable virus was cleared from the liver and spleen by day 8 after infection in both genotypes (data not shown).
Fig. 1.
Unc93b1-mediated immune activation is protective during acute CVB3 infection. a Survival of Unc93b1+/+ mice (black circles, solid line) and Unc93b1Letr/Letr mice (white circles, dashed line; n ≥27 in each group). Viral load in the heart (b), liver (c) and spleen (d) at days 2, 4 and 8 after infection in Unc93b1+/+ mice (black circles) and Unc93b1Letr/Letr mice (white circles; n ≥7 in each group). Data are representative of 2 independent experiments. *** p < 0.001, log-rank test (a) or 2-tailed unpaired Student's t test (b-d).
Increased Cardiac Inflammation and Cell Necrosis during Acute CVB3-Induced Myocarditis in the Absence of Functional Unc93b1
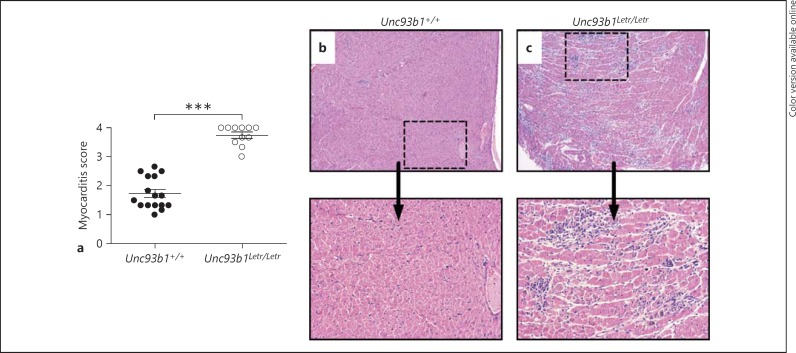
Along with virus-induced mechanisms of pathology, inflammatory cells that are recruited to the heart to eliminate CVB3 can trigger cell necrosis and lead to permanent structural changes in the heart that result in a prolonged host recovery time and increased infection-related morbidity and mortality [2]. To determine if a functional deficiency of Unc93b1 causes increased cardiac damage and inflammation, we analyzed tissue sections stained with HE at day 8 after infection, a time point at which both necrosis and inflammation can be visualized [32]. The Unc93b1Letr/Letr mice had significantly increased myocarditis compared to the Unc93b1+/+ mice when both inflammation and necrosis were quantified using a previously established scoring system [32] (fig. 2a). Representative histological sections of infected heart tissue clearly demonstrate more foci of inflammation (clusters of cells that stain blue) and substantially more cell necrosis (areas of clearance) in the hearts of the Unc93b1Letr/Letr mice than in the Unc93b1+/+ controls (fig. 2b, c).
Fig. 2.
Loss of Unc93b1 function increases cardiac damage and inflammation during acute CVB3 infection. a Myocarditis score, a composite of cardiac inflammation and necrosis, for Unc93b1+/+ hearts (black circles) and Unc93b1Letr/Letr hearts (white circles) at day 8 after infection. A score of ‘0’ indicates the absence of myocarditis while ‘4’ denotes pervasive necrosis and inflammation throughout the heart section (n ≥11 in each group). *** p < 0.001, 2-tailed unpaired Student's t test. b, c Representative histological heart sections. HE. ×4 (top panel), ×10 (bottom panel). Inflammation is visualized as condensed areas of nucleated cells (stained blue) while necrosis causes the appearance of areas of clearance within normal heart tissue (stained red) [32].
Loss of Unc93b1 Function Causes Dysregulated Early Cardiac Gene Expression in Acute CVB3-Induced Myocarditis
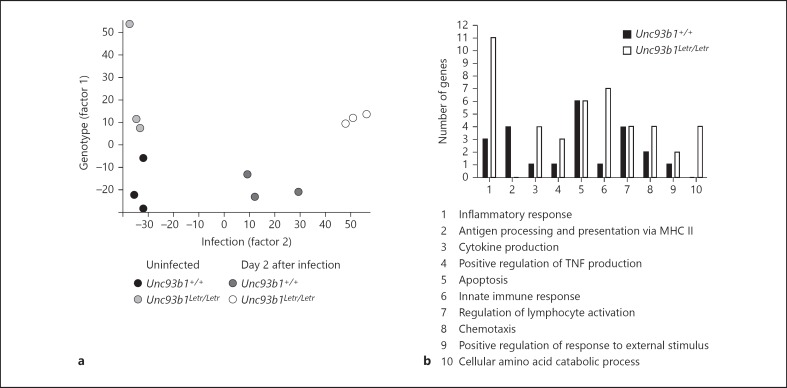
As Unc93b1Letr/Letr mice were significantly more susceptible to infection and had more cardiac damage, we analyzed how the loss of Unc93b1 function affected cardiac gene expression during CVB3 infection, and sought to determine if the pattern of early transcriptional activation could be correlated with host outcome. Accordingly, we performed a global gene transcriptional analysis on biological replicates of heart tissue from Unc93b1+/+ mice and Unc93b1Letr/Letr mice at day 0 and day 2 after infection. Day 2 was examined as TLR activation can occur rapidly in primary human aortic muscle cells following infection [15] and early changes in cardiac signaling can predispose the host to a deleterious outcome as the infection progresses [40]. In addition, significant immune cell infiltration to the heart has not occurred by 48 h after infection, so that gene expression at this time point predominantly reflects the response of resident immune cells and cardiomyocytes. PCA of normalized global expression data from this array clustered heart samples based on 2 experimental factors, i.e. factor 1: genotype and factor 2: infection, with only minor variations between the assayed biological replicates being observed (fig. 3a). PCA further demonstrated that the infection (factor 2) caused a more dramatic change in gene expression in the Unc93b1Letr/Letr hearts than in the Unc93b1+/+ hearts.
Fig. 3.
Loss of Unc93b1 function causes dysregulated cardiac gene expression in CVB3 infection. a A PCA plot of normalized gene expression from Unc93b1+/+ and Unc93b1Letr/Letr hearts at days 0 and 2. b Enriched GO biological process clusters of differentially expressed genes at day 2, stratified based on increased gene expression in the Unc93b1+/+ or Unc93b1Letr/Letr hearts.
FlexArray [33] analysis of gene expression data identified 182 genes with >2-fold differential expression and a padj <0.05 between Unc93b1+/+ and Unc93b1Letr/Letr hearts at day 2 after infection (online suppl. table 1; for all online suppl. material, see www.karger.com/doi/10.1159/000369342). Using this gene list, GO biological process cluster analysis was performed in order to uncover terms specifically related to the biological function of the genes that were significantly enriched in this gene list and to provide a better understanding of the differential gene activation occurring between genotypes that may be linked to differential function and therefore contribute to the observed differential infection outcome [35]. Gene transcripts within these significantly enriched GO clusters were subsequently segregated based on increased expression in the Unc93b1+/+ or Unc93b1Letr/Letr hearts at day 2 after infection (fig. 3b). Visual inspection revealed that differentially expressed genes associated with the term antigen processing and presentation via MHC class II appear to be more highly expressed in Unc93b1+/+ hearts than in Unc93b1Letr/Letr hearts. Conversely, differentially expressed genes associated with the GO terms inflammatory response, cytokine production, positive regulation of TNF production, innate immune response, chemotaxis and cellular amino acid catabolic process appear to be more highly expressed in Unc93b1Letr/Letr than in Unc93b1+/+ hearts at day 2 after infection.
Of the 182 differentially expressed genes at day 2, 49 were more highly expressed in the Unc93b1+/+ hearts whereas the other 133 were more highly expressed in the Unc93b1Letr/Letr hearts (online suppl. table 1). A subsequent analysis was conducted on each gene list to determine the GO biological process, GO cellular component and pathway enrichment terms that were significant to each genotype. These results were in agreement with several findings from the preliminary GO analysis (fig. 3b), and demonstrated that genes associated with the GO biological process term antigen processing and presentation of exogenous peptide antigen via MHC class II and the GO cellular component term MHC class II protein complex were significantly enriched in Unc93b1+/+ mice at day 2 whereas the GO biological process term inflammatory response was significantly enriched in Unc93b1Letr/Letr hearts (table 1). Pathway analysis determined that immune genes involved in the TLR signaling pathway were significantly enriched in the Unc93b1Letr/Letr hearts whereas the pathway terms for intestinal immune network for IgA production, asthma and systemic lupus erythematosus were significantly enriched in the hearts of the Unc93b1+/+ mice (table 1).
Table 1.
Significant term enrichment following CVB3 infection in the heart
| Term | Genes | Fold enrichment | padj value |
|---|---|---|---|
| Significantly enriched terms in Unc93b1+/+ hearts at day 2 | |||
| Antigen processing and presentation of exogenous peptide antigen via MHC class II | |||
| GO:0019886 | H2-EB1, H2-AA, H2-AB1, CD74 | 92.3 | 3.43E-03 |
| MHC class II protein complex | |||
| GO:0042613 | H2-EB1, H2-AA, H2-AB1 | 125.23 | 1.39E-02 |
| Intestinal immune network for IgA production | |||
| mmu04672 | H2-EB1, H2-AA, H2-AB1, CXCL12 | 21.18 | 2.56E-02 |
| Asthma | |||
| mmu05310 | H2-EB1, H2-AA, H2-AB1 | 27.38 | 4.27E-02 |
| Systemic lupus erythematosus | |||
| mmu05322 | C8B, H2-EB1, H2-AA, H2-AB1 | 11.64 | 4.77E-02 |
| Significantly enriched terms in Unc93b1Letr/Letr hearts at day 2 | |||
| Inflammatory response | |||
| GO:0006954 | CCL11, SERPINA3N, MYD88, SAA1, SPHK1,CXCL9, TLR3, C2, IDO1, LBP, CXCL10 | 6.70 | 1.06E-03 |
| TLR signaling pathway | |||
| mmu04620 | FOS, MYD88, IFNB1, CXCL9, TLR3, LBP, CXCL10 | 8.93 | 7.23E-03 |
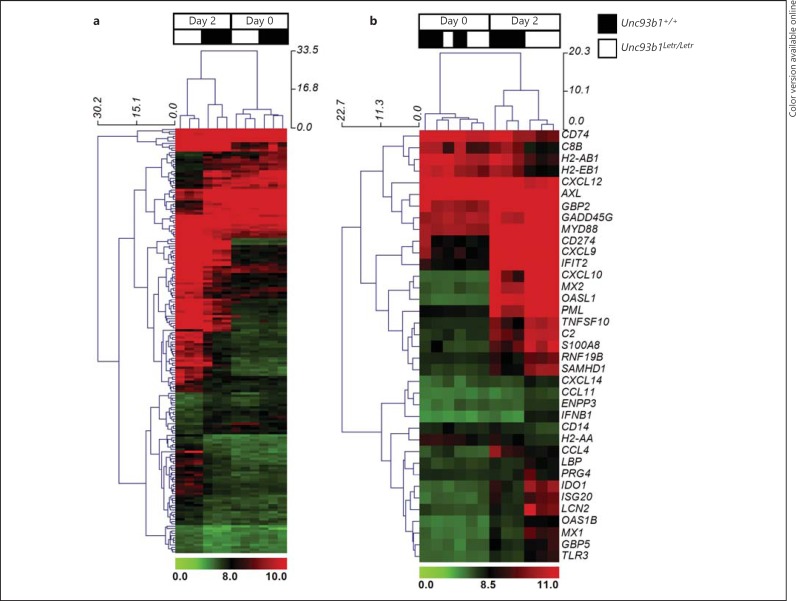
Hierarchical clustering of normalized data for the 182 differentially expressed genes was performed to order biological replicates based on gene expression similarities (fig. 4a; biological replicates for Unc93b1+/+ and Unc93b1Letr/Letr samples are indicated by black and white, respectively, at the top of each heat map). Hierarchical clustering successfully ranked the biological replicates first by infection (day 0 vs. day 2 after infection) and then by genotype. This analysis demonstrated substantial differences between Unc93b1+/+ and Unc93b1Letr/Letr cardiac gene expression at day 2 (fig. 4a, left columns) that were not seen between the 2 genotypes at day 0 (fig. 4a, right columns). Further analysis of the 37 genes classified under the GO biological process term immune response (GO:0006955) successfully clustered the biological replicates by infection, but was only able to cluster samples by genotype at day 2 (fig. 4b, right columns). Clustering by genotype in uninfected hearts at day 0 (fig. 4b, left columns) was less precise, an observation that was not surprising given that the uninfected Unc93b1+/+ and Unc93b1Letr/Letr hearts were not expected to have baseline differences in immune gene expression. This analysis indicated a greater induction of immune genes in the Unc93b1Letr/Letr than in the Unc93b1+/+ hearts at day 2 (fig. 4b), which may be have been related to the higher cardiac viral load observed at this time point. Gene clustering on the y-axis indicates similarity in gene expression patterns and may also indicate similarity in gene regulation at the biological level [41].
Fig. 4.
Differential gene expression clustering in Unc93b1+/+ and Unc93b1Letr/Letr hearts following CVB3 infection. Hierarchical clustering with complete linkage of normalized gene expression data from all 182 differentially expressed genes (a) or the differentially expressed immune response genes (b) at day 0 and day 2 following infection in Unc93b1+/+ and Unc93b1Letr/Letr hearts. The day and genotype are indicated at the top of each heat map. Red/green signatures denote the normalized gene expression profile, with red representing gene upregulation and green denoting gene downregulation. Dendrograms on the x-axis denote sample clustering while the y-axis indicates gene clustering. Clusters were calculated by hierarchical clustering using MeV [37]. Genes clustered together suggest a degree of similarity in expression, which may relate to similarities in gene regulation at the biological level.
At day 0, there were 21 genes that showed >2-fold differential expression and a padj <0.05 between the Unc93b1+/+ and Unc93b1Letr/Letr hearts (online suppl. table 2). Eight of these genes also showed differential expression at day 2; only one (c8b) was significantly clustered in the secondary microarray analysis (table 1). These findings suggest that the basal gene expression did not play a significant role in the expression differences observed between Unc93b1+/+ and Unc93b1Letr/Letr hearts at day 2 following CVB3 infection.
Loss of Unc93b1 Function Increased Cardiac Expression of Ifn-b, Lcn2 and Serpina3n in Acute CVB3-Induced Myocarditis
To identify the genes with dramatically differential expression between Unc93b1+/+ and Unc93b1Letr/Letr hearts during acute CVB3-induced myocarditis that would be amenable to confirmatory analysis by qRT-PCR, the microarray analysis was restricted to transcripts that had >5-fold differential expression and a padj <0.05. This more rigorous threshold revealed five genes, only one of which (Dbp) was more significantly upregulated in the Unc93b1+/+ hearts while the other four (Cdo1, Ifn-b, Lcn2, Serpina3n) were more significantly upregulated in the Unc93b1Letr/Letr hearts (table 2). Consistent with our findings using a cut-off of >2-fold differential expression; (online suppl. table 1), there were also a greater number of upregulated transcripts in the Unc93b1Letr/Letr hearts than in the Unc93b1+/+ hearts, using a cut-off of >5-fold differential expression.
Table 2.
Genes differentially expressed >5-fold at day 2 following infection
| Gene | Gene name | Fold enrichment | padj value |
|---|---|---|---|
| Upregulatedgenes in Unc93b1+/+ hearts | |||
| DBP | D site albumin-promoter-binding protein | 5.22 | 5.02E-8 |
| Upregulated genes in Unc93b1Letr/Letr hearts | |||
| CDO1 | cysteine dioxygenase 1, cytosolic | 0.109 | 7.22E-8 |
| IFN-B1 | interferon beta 1, fibroblast | 0.143 | 3.77E-7 |
| LCN2 | lipocalin 2 | 0.198 | 1.53E-5 |
| SERPINA3N | serine (or cysteine) peptidase inhibitor, clade A, member 3N | 0.157 | 3.09E-5 |
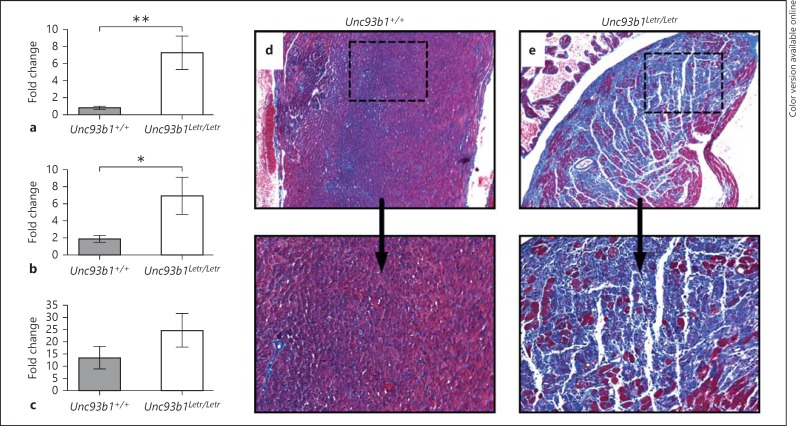
To verify expression data from the microarray, Ifn-b, Lcn2 and Serpina3n were analyzed by qRT-PCR. These genes were selected based on the literature, which describes their importance in CVB3 infection or a previous association with cardiac pathology. We confirmed a significantly higher induction of Ifn-b and Lcn2 in the Unc93b1Letr/Letr hearts than in the Unc93b1+/+ hearts (fig. 5a, b) with a trend towards increased expression of Serpina3n in the Unc93b1Letr/Letr hearts at day 2 after infection (fig. 5c). A previous study demonstrated that the exogenous addition of recombinant Serpin A 3N (rSerpin) increased cardiac remodeling and fibrosis in a mouse model of CVB3-induced autoimmune myocarditis caused by an injection of CVB3 in combination with cardiac myosin protein [40]. Therefore, to determine if the extent of cardiac remodeling was affected in the Unc93b1Letr/Letr mice, we analyzed the degree of fibrosis in heart sections from infected Unc93b1+/+ and Unc93b1Letr/Letr mice at day 8 after infection using a Masson's trichrome stain. Representative histological sections clearly demonstrated a substantially greater amount of cardiac fibrosis (visualized as blue-staining collagen) in the Unc93b1Letr/Letr hearts (fig. 5e) compared to the Unc93b1+/+ hearts (fig. 5d), a finding that correlates with the increased inflammation and necrosis observed at the same time point.
Fig. 5.
Functional deficiency of Unc93b1 increases Ifn-b, Lcn2 and Serpina3n expression during CVB3 infection. qRT-PCR analysis of Ifn-b (a), Lcn2 (b) and Serpina3n (c) expression at day 2 after infection in Unc93b1+/+ (grey) and Unc93b1Letr/Letr (white) hearts. Target gene expression was normalized to endogenous Gapdh expression and is presented relative to target gene expression in uninfected mice (n ≥5 in each group). * p < 0.05, ** p < 0.01, 1-tailed unpaired Student's t test. d, e Representative heart sections were stained with Masson's trichrome to measure fibrosis (blue) at day 8 after infection. ×4 (top panel), ×10 (bottom panel).
Discussion
CVB3-induced myocarditis is a serious and often underdiagnosed condition. The outcome of infection is determined by a complex interaction of host and viral factors that can result in an asymptomatic and self-limited infection, sudden death during acute myocarditis or DCM due to persistent inflammation [1, 2]. In this report, we used mice with a chemically induced loss-of-function mutation in Unc93b1[28] on a resistant C57BL/6 genetic background, in order to study the role of endosomal TLR signaling in acute CVB3-induced myocarditis. While previous studies have examined the role of immune signaling in mice lacking a single TLR or adaptor, this is the first report examining how a complete loss of endosomal TLR activation affects host outcome following CVB3 infection. We demonstrated that a functional deficiency of Unc93b1 led to increased mortality during acute CVB3-induced myocarditis, and this correlated with a significantly elevated viral load in the heart, liver and spleen as well as increased cardiac inflammation and necrosis. This differential host susceptibility was accompanied by a significantly dysregulated expression of genes associated with several biological processes including antigen processing and presentation of exogenous peptide antigen via MHC class II and the inflammatory response, with a specific increase in Ifn-b, Lcn2 and Serpina3n expression in Unc93b1Letr/Letr hearts at day 2 after infection. These results demonstrate that endosomal TLR signaling is crucial for survival and recovery from an acute CVB3 infection, and further reveal potentially interesting mediators that may be associated with pathology in acute CVB3-induced myocarditis.
Increased mortality due to defective Unc93b1 function is consistent with previous studies of Tlr3- or Ticam1-deficient mice, both of which have increased mortality, inflammation and cardiac viral load following CVB3 infection [11, 12]. In addition, the transgenic expression of TLR3 was shown to protect mice from lethal CVB3 infection, even in the absence of type I IFN signaling, via TICAM1-dependent induction of type II IFN [11]. In contrast to the TLR3/TICAM1 pathway, signaling via TLR9/MyD88 contributes to disease pathogenesis in acute CVB3-induced myocarditis [14, 17]. Specifically, MyD88 deficiency improved host survival, decreased cardiac and pancreatic tissue damage and reduced viral load in multiple organs during acute CVB3 infection [17]. Interestingly, our report demonstrates that the simultaneous loss of protective immune responses through TLR3/TICAM1 and pathogenic responses through TLR9/MyD88 increased the overall susceptibility to acute CVB3-induced myocarditis. We observed comparable IFN-γ production and expression of type II IFN-inducible genes between Unc93b1+/+ and Unc93b1Letr/Letr hearts (online suppl. fig. 1) suggesting that the susceptibility of Unc93b1Letr/Letr mice to early viral replication and myocarditis is not attributable to the loss of this pathway. Our findings further demonstrate that immune activation through MDA5/MAVS, a cytoplasmic pathway that is essential for host survival following CVB3 infection [20, 21], and other viral recognition mechanisms in Unc93b1Letr/Letr mice are not sufficient to compensate for the absence of endosomal TLR signaling in acute CVB3-induced myocarditis.
Microarray analysis at day 2 following CVB3 infection revealed an increase in total gene expression as well as the expression of genes associated with the GO biological process term inflammatory response in Unc93b1Letr/Letr hearts when compared to Unc93b1+/+ hearts. This finding suggests that the loss of Unc93b1-dependent endosomal TLR signaling did not dampen the host response, but instead led to greater immune activation. TICAM1-deficient mice exhibited a similar response characterized by increased TNF-α and IL-1β with a concomitant increase in activated T-cell, B-cell and macrophage infiltration at day 7 after infection, despite an increased susceptibility to CVB3 infection [12]. GO analysis also showed that genes associated with antigen processing and presentation of exogenous peptide antigen via MHC class II were more highly expressed in Unc93b1+/+ hearts than in Unc93b1Letr/Letr hearts at day 2. Previous studies have noted that MHC class II can be constitutively expressed by resident antigen-presenting cells in the heart and activated on cardiomyocytes following IFN-γ expression [42, 43]. Effective antigen presentation via MHC class II is a crucial mechanism for priming CD4+ T lymphocytes that, in turn, facilitate the continued activation of cytotoxic CD8+ T cells and B-cell activation to eliminate infected cells and produce strain-specific antibodies, respectively. MHC class II may play an important role in mediating host resistance against CVB3 as it was found that infection of MHC class II-deficient mice led to significantly worse cardiac disease that manifested as persistent immune cell infiltration, fibrosis and sustained viral load at day 21 after infection, although no survival difference was reported [44]. In the original paper by Tabeta et al. [27] that established the function of Unc93b1 in endosomal TLR activation, a second independent role for Unc93b1 in antigen presentation via MHC class II as well as antigen cross-presentation was proposed. Conversely, recent studies using experimental conditions similar to in Tabeta et al. [27] were unable to reproduce this finding, and therefore proposed that a TLR-independent role for Unc93b1 in antigen presentation did not exist [45]. Further data from Toxoplasma gondii and Trypanosoma cruzi infections have demonstrated that the combined loss of multiple endosomal TLRs recapitulated the susceptibility phenotype seen in infected Unc93b13d/3d mice [46, 47]. Taken together, these observations suggest that the early decrease in gene expression associated with antigen processing and presentation of exogenous peptide antigen via MHC class II in Unc93b1Letr/Letr mice may be attributable to the loss of endosomal TLR activation in the heart rather than due to an independent functional role for Unc93b1.
Pathway analysis at day 2 revealed that genes involved in the intestinal immune network for IgA production, asthma and systemic lupus erythematosus were significantly overrepresented in the Unc93b1+/+ compared to the Unc93b1Letr/Letr hearts. All of these pathways have previously been associated with the upregulation of Th2-mediated immune signaling, which suggests that Unc93b1Letr/Letr mice activate an immune response with a Th1/Th2 imbalance following CVB3 infection. In particular, increased expression of Th2-associated genes in Unc93b1+/+ hearts may indicate a bias towards Th1-mediated immune activation in Unc93b1Letr/Letr hearts that could contribute to immune-mediated cardiac pathology.
Identification of elevated Ifn-b expression in the hearts of susceptible Unc93b1Letr/Letr mice compared to resistant Unc93b1+/+ mice was unexpected, as a protective role for IFN-β has been demonstrated in both experimental and natural studies of acute viral myocarditis and DCM. Both IFN-β- and IFNAR-deficient mice are highly susceptible to acute CVB3-induced myocarditis and exhibit increased splenic and hepatic necrosis [7, 8]. Research has also demonstrated that the exogenous addition of IFN-β to human myocardial fibroblasts in vitro or mice in vivo reduced viral load and cell damage [48, 49] and a clinical trial of IFN-β treatment over a 6-month period improved disease outcome and reduced viral titer in chronically infected DCM patients [9]. However, not all studies have demonstrated that elevated IFN-β correlates with protection in acute CVB3-induced myocarditis. For example, analysis of resistant B10.A and susceptible A/J mice revealed a relationship between higher cardiac viral titer in the first 4 days after infection and increased serum IFN-β expression [50], a finding that is similar to our observations in the Unc93b1Letr/Letr model. TLR3 deficiency significantly increased mortality and serum viral load, but did not affect cardiac IFN-β expression at day 3 after infection [11]. The expression of TLR7 and TLR8 has been associated with IFN-β induction following CVB3 infection of a HEK-293T cell line as well as human primary aortic cells [15]; however, a specific role for TLR7 in IFN-β expression during CVB3 infection has not been determined in vivo. Accordingly, during CVB3 infection of Unc93b1Letr/Letr mice, the specific influence of defective TLR7-mediated immune activation on Ifn-b induction is difficult to predict.
As endosomal TLRs are nonfunctional in Unc93b1Letr/Letr mice, one or more alternative pathways must mediate the cardiac Ifn-b upregulation in this model. In vitro studies have shown that increased MAVS expression is correlated with increased Ifn-b expression and decreased viral RNA [51], and both in vitro and in vivo analyses of Unc93b1Letr/Letr mice revealed residual cytokine responsiveness following poly I:C stimulation, an agonist for both TLR3 and MDA5 [28]. Taken together, these data suggest that the early upregulation of Ifn-b in Unc93b1Letr/Letr hearts may occur, at least in part, through the MDA5/MAVS pathway. Furthermore, despite a lack of functional TLR3 in the Unc93b1Letr/Letr mice, its intracellular adaptor TICAM1 remains functional and can be recruited by both cell membrane-localized TLR4 and a cytoplasmic DDX sensor composed of the DDX1, DDX21 and DHX36 helicases [52]. TLR4 deficiency is associated with decreased myocarditis in CVB3-infected mice, suggesting that its activation may contribute to disease in Unc93b1Letr/Letr mice [16]. In myeloid cells, the cytoplasmic DDX sensor activates an early type I IFN response in response to poly I:C, a synthetic analog of dsRNA [52]. Triggering of this pathway leads to the subsequent induction of other pattern recognition receptors, such as MDA5, that further amplify type I IFN production [52]. Therefore, it is reasonable to speculate that the early elevation in Ifn-b expression in the hearts of Unc93b1Letr/Letr mice may be attributable to the MDA5/MAVS pathway and/or one or more TICAM1-dependent but endosomal TLR-independent pathways. As there was no difference on the cardiac gene array at day 2 (data not shown) between Unc93b1+/+ and Unc931b1Letr/Letr hearts in the expression of Mavs or Ticam1, further study is required to determine if one or more of these pathways is indeed responsible for the elevated Ifn-b in the Unc931b1Letr/Letr mice. Despite the known protective effect of IFN-β in CVB3 infection, the Unc93b1Letr/Letr mice had significantly higher mortality than the Unc93b1+/+ mice, suggesting that the observed Ifn-b increase may have been transient or that the elevated Ifn-b expression in Unc93b1Letr/Letr hearts was not sufficient to compensate for the loss of protective immune mechanisms activated by endosomal TLR signaling.
Lcn2, a 25-kDa small molecule transporter and a member of the lipocalin family [53], was also significantly upregulated in Unc93b1Letr/Letr hearts at day 2 after infection. LCN2 protects the host during bacterial infection by binding and sequestering iron-scavenging bacterial siderophores to limit the iron acquisition required for bacterial growth [54]. Indeed, Lcn2-deficient mice are more susceptible to Escherichia coli-induced sepsis and Klebsiella pneumoniae infection than wild-type mice [55, 56]. In a model of West Nile virus-induced encephalitis however, Lcn2 deficiency did not affect the host's ability to mount an appropriate cellular immune response, to clear brain viral load or to survive the disease [57], suggesting that it may be dispensable in certain viral infections. Lcn2 has also been proposed as a clinical marker of acute kidney injury as it has low basal expression that is significantly elevated in both plasma and urine samples following renal inflammation or injury [58]. Recent work suggests that Lcn2 may also be a marker of cardiac injury as its expression is elevated in patients with coronary artery disease and heart failure [59]. Based on these studies, the upregulation of Lcn2 in Unc93b1Letr/Letr hearts at day 2 may reflect a response to damage and act as an early indicator of the increased cardiac tissue injury observed at day 8. Interestingly, differential cardiac expression of Lcn2 was also seen during the acute phase of CVB3-induced autoimmune myocarditis that was caused by injection of a combination of CVB3 and cardiac myosin protein in susceptible male Balb/c mice, suggesting that its expression in the heart may predict progression to DCM [40]. Our finding of an early increase in cardiac Lcn2 expression in a model of CVB3 infection alone complements this study and further suggests that there is a potential utility for Lcn2 as an informative biomarker of tissue damage in acute as well as chronic myocarditis.
At day 2, we also observed a trend towards increased expression of Serpina3n in Unc93b1Letr/Letr hearts by qRT-PCR.Serpina3n can bind and inactivate serine proteases that normally degrade the extracellular matrix [60, 61]. Patients with atherosclerosis have been found to have significantly increased Serpina3n expression in cardiac cells compared to healthy controls, suggesting that increased expression of Serpina3n may be linked to more severe symptoms in cardiovascular disease [62]. Elevated Serpina3n expression has also been seen in patients with heart failure, further supporting an association of Serpina3n expression with increased cardiac damage and repair [63]. Similar to findings with Lcn2, the induction of experimental CVB3-induced autoimmune myocarditis causes elevated expression of Serpina3n in the acute phase of disease, correlating with an increased propensity to progress to chronic DCM [40]. In this study, elevated cardiac Serpina3n began at day 2 in susceptible mice and was sustained until day 10 [40]. Serpina3n expression was elicited from non-CD11b+ cells, suggesting that resident cardiac cells, rather than immune cells, are the predominant cellular source following infection [40]. Finally, the exogenous addition of rSerpin to mice with CVB3-induced autoimmune myocarditis significantly increased fibrosis, implicating Serpina3n expression in cardiac remodeling [40]. Collectively, these findings suggest that the substantial fibrosis observed in the Unc93b1Letr/Letr hearts at day 8 may have been due, at least in part, to the early increase in Serpina3n expression. Though elevated Serpina3n has been linked to increased pathology in CVB3-induced autoimmune myocarditis, a notable difference between our study and this previous report is that there was no observed difference in viral load between resistant and susceptible Balb/c mice in the latter [40]. This may be because the 2 studies used different models of virus-induced myocarditis, but it is also possible that additional virus-mediated mechanisms of damage contributed to significantly worse fibrosis in the Unc93b1Letr/Letr mice during acute CVB3-induced myocarditis.
In summary, our studies using mice with a complete loss-of-function mutation of Unc93b1 have shown that endosomal TLR signaling is essential for survival of acute CVB3-induced myocarditis, and that compensatory immune signaling via MDA5/MAVS or alternative host defense mechanisms in Unc93b1Letr/Letr mice are insufficient for recovery. This data extends the results of previous studies in mouse MCMV, HSV and NSV infection that also demonstrated an essential role for Unc93b1-mediated immune activation in infection survival [27, 29, 30]. Our studies confirm the indispensable role of TLR3-mediated immune activation in host protection during CVB3 infection, and further demonstrate that the concomitant loss of pathological immune activation mediated by MyD88 does not protect Unc93b1Letr/Letr mice against worse disease following infection. It is currently unknown whether the protection attributed to MyD88 deficiency in acute CVB3-induced myocarditis [17] is due to the loss of IL-1 receptor (IL-1R)-mediated immune activation, which also uses the MyD88 adaptor [64], or is related to the loss of endosomal TLR7- and TLR9-mediated immune activation. In experimental models of CVB3-induced autoimmune myocarditis, the exogenous addition of IL-1β induced or worsened inflammation [65] whereas in a model of acute CVB3-induced myocarditis, the inhibition of IL-1-mediated immune activation with an IL-1R antagonist significantly decreased mortality, cardiac viral load and tissue damage [66]. Therefore IL-1R-mediated immune activation can increase host pathology, and its absence may be contributing to the enhanced host protection observed in MyD88-deficient mice [17]. As no in vivo studies on the role of TLR7 following CVB3 infection have been conducted to date, it is challenging to predict the relative contribution of this pathway to host pathology or protection. The use of Unc93b1Letr/Letr mice, which maintain intact MyD88 and TICAM1 adaptors, allowed us to clearly define the role of endosomal TLRs in acute CVB3-induced myocarditis without the confounding effects of the loss of immune activation through other signaling pathways. Identification of significantly elevated Lcn2 expression and a trend towards increased Serpina3n expression in the Unc93b1Letr/Letr hearts complements previous studies that have demonstrated a correlation between the expression of these genes and worsened cardiac damage and infection outcome in CVB3-induced autoimmune myocarditis, and extends these findings to a model of acute viral myocarditis. This is the first study to demonstrate a potential link between the activation of these genes and increased susceptibility using an immunodeficient mouse model, and it suggests that further investigation of Lcn2 and Serpina3n may confirm their potential as markers of cardiac damage during acute as well as chronic viral myocarditis.
Disclosure Statement
The authors declare no current or potential conflicts of interest.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Acknowledgements
The authors would like to acknowledge the technical support of Jennifer Marton, Patricia D'Arcy, and Geneviève Perrault and Dr. Patricia Tonin and François Lefebvre for guidance conducting microarray analysis. This work was supported by a grant from the Fonds de recherche du Québec-Santé to the Research Institute of the McGill University Health Centre, a CIHR team grant (CTP-87520) and a Canada Research Chair in Host Resistance to Respiratory Infections (to S.T.Q.).
References
- 1.Corsten MF, Schroen B, Heymans S. Inflammation in viral myocarditis: friend or foe? Trends Mol Med. 2012;18:426–437. doi: 10.1016/j.molmed.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Esfandiarei M, McManus BM. Molecular biology and pathogenesis of viral myocarditis. Annu Rev Pathol. 2008;3:127–155. doi: 10.1146/annurev.pathmechdis.3.121806.151534. [DOI] [PubMed] [Google Scholar]
- 3.Chow LH, Beisel KW, McManus BM. Enteroviral infection of mice with severe combined immunodeficiency. Evidence for direct viral pathogenesis of myocardial injury. Lab Invest. 1992;66:24–31. [PubMed] [Google Scholar]
- 4.McManus BM, Chow LH, Wilson JE, Anderson DR, Gulizia JM, Gauntt CJ, Klingel KE, Beisel KW, Kandolf R. Direct myocardial injury by enterovirus: a central role in the evolution of murine myocarditis. Clin Immunol Immunopathol. 1993;68:159–169. doi: 10.1006/clin.1993.1113. [DOI] [PubMed] [Google Scholar]
- 5.Tam PE. Coxsackievirus myocarditis: interplay between virus and host in the pathogenesis of heart disease. Viral Immunol. 2006;19:133–146. doi: 10.1089/vim.2006.19.133. [DOI] [PubMed] [Google Scholar]
- 6.Feng Q, Langereis MA, van Kuppeveld FJ. Induction and suppression of innate antiviral responses by picornaviruses. Cytokine Growth Factor Rev. 2014;25:577–585. doi: 10.1016/j.cytogfr.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deonarain R, Cerullo D, Fuse K, Liu PP, Fish EN. Protective role for interferon-beta in coxsackievirus B3 infection. Circulation. 2004;110:3540–3543. doi: 10.1161/01.CIR.0000136824.73458.20. [DOI] [PubMed] [Google Scholar]
- 8.Wessely R, Klingel K, Knowlton KU, Kandolf R. Cardioselective infection with coxsackievirus B3 requires intact type I interferon signaling: implications for mortality and early viral replication. Circulation. 2001;103:756–761. doi: 10.1161/01.cir.103.5.756. [DOI] [PubMed] [Google Scholar]
- 9.Kuhl U, Pauschinger M, Schwimmbeck PL, Seeberg B, Lober C, Noutsias M, Poller W, Schultheiss HP. Interferon-beta treatment eliminates cardiotropic viruses and improves left ventricular function in patients with myocardial persistence of viral genomes and left ventricular dysfunction. Circulation. 2003;107:2793–2798. doi: 10.1161/01.CIR.0000072766.67150.51. [DOI] [PubMed] [Google Scholar]
- 10.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Negishi H, Osawa T, Ogami K, Ouyang X, Sakaguchi S, Koshiba R, Yanai H, Seko Y, Shitara H, Bishop K, Yonekawa H, Tamura T, Kaisho T, Taya C, Taniguchi T, Honda K. A critical link between Toll-like receptor 3 and type II interferon signaling pathways in antiviral innate immunity. Proc Natl Acad Sci U S A. 2008;105:20446–20451. doi: 10.1073/pnas.0810372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riad A, Westermann D, Zietsch C, Savvatis K, Becher PM, Bereswill S, Heimesaat MM, Lettau O, Lassner D, Dorner A, Poller W, Busch M, Felix SB, Schultheiss HP, Tschope C. TRIF is a critical survival factor in viral cardiomyopathy. J Immunol. 2011;186:2561–2570. doi: 10.4049/jimmunol.1002029. [DOI] [PubMed] [Google Scholar]
- 13.Gorbea C, Makar KA, Pauschinger M, Pratt G, Bersola JL, Varela J, David RM, Banks L, Huang CH, Li H, Schultheiss HP, Towbin JA, Vallejo JG, Bowles NE. A role for Toll-like receptor 3 variants in host susceptibility to enteroviral myocarditis and dilated cardiomyopathy. J Biol Chem. 2010;285:23208–23223. doi: 10.1074/jbc.M109.047464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riad A, Westermann D, Escher F, Becher PM, Savvatis K, Lettau O, Heimesaat MM, Bereswill S, Volk HD, Schultheiss HP, Tschope C. Myeloid differentiation factor-88 contributes to TLR9-mediated modulation of acute coxsackievirus B3-induced myocarditis in vivo. Am J Physiol Heart Circ Physiol. 2010;298:H2024–H2031. doi: 10.1152/ajpheart.01188.2009. [DOI] [PubMed] [Google Scholar]
- 15.Triantafilou K, Orthopoulos G, Vakakis E, Ahmed MA, Golenbock DT, Lepper PM, Triantafilou M. Human cardiac inflammatory responses triggered by coxsackie B viruses are mainly Toll-like receptor (TLR) 8-dependent. Cell Microbiol. 2005;7:1117–1126. doi: 10.1111/j.1462-5822.2005.00537.x. [DOI] [PubMed] [Google Scholar]
- 16.Fairweather D, Yusung S, Frisancho S, Barrett M, Gatewood S, Steele R, Rose NR. Il-12 receptor beta 1 and Toll-like receptor 4 increase IL-1 beta- and IL-18-associated myocarditis and coxsackievirus replication. J Immunol. 2003;170:4731–4737. doi: 10.4049/jimmunol.170.9.4731. [DOI] [PubMed] [Google Scholar]
- 17.Fuse K, Chan G, Liu Y, Gudgeon P, Husain M, Chen M, Yeh WC, Akira S, Liu PP. Myeloid differentiation factor-88 plays a crucial role in the pathogenesis of coxsackievirus B3-induced myocarditis and influences type I interferon production. Circulation. 2005;112:2276–2285. doi: 10.1161/CIRCULATIONAHA.105.536433. [DOI] [PubMed] [Google Scholar]
- 18.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of MDA-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 20.Huhn MH, McCartney SA, Lind K, Svedin E, Colonna M, Flodstrom-Tullberg M. Melanoma differentiation-associated protein-5 (MDA-5) limits early viral replication but is not essential for the induction of type 1 interferons after coxsackievirus infection. Virology. 2010;401:42–48. doi: 10.1016/j.virol.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Wang JP, Cerny A, Asher DR, Kurt-Jones EA, Bronson RT, Finberg RW. MDA5 and MAVS mediate type I interferon responses to coxsackie B virus. J Virol. 2010;84:254–260. doi: 10.1128/JVI.00631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Q, Hato SV, Langereis MA, Zoll J, Virgen-Slane R, Peisley A, Hur S, Semler BL, van Rij RP, van Kuppeveld FJ. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Reports. 2012;2:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oshiumi H, Okamoto M, Fujii K, Kawanishi T, Matsumoto M, Koike S, Seya T. The TLR3/TICAM-1 pathway is mandatory for innate immune responses to poliovirus infection. J Immunol. 2011;187:5320–5327. doi: 10.4049/jimmunol.1101503. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Gao B, Xiong S. Involvement of NLRP3 inflammasome in CVB3-induced viral myocarditis. Am J Physiol Heart Circ Physiol. 2014;307:H1438–H1447. doi: 10.1152/ajpheart.00441.2014. [DOI] [PubMed] [Google Scholar]
- 25.Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim YM. The interaction between the ER membrane protein Unc93b and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol. 2007;177:265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. Unc93b1 delivers nucleotide-sensing Toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 27.Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, Mudd S, Mann N, Sovath S, Goode J, Shamel L, Herskovits AA, Portnoy DA, Cooke M, Tarantino LM, Wiltshire T, Steinberg BE, Grinstein S, Beutler B. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 28.Lafferty EI, Flaczyk A, Angers I, Homer R, d'Hennezel E, Malo D, Piccirillo CA, Vidal SM, Qureshi ST. An ENU-induced splicing mutation reveals a role for Unc93b1 in early immune cell activation following influenza A H1N1 infection. Genes Immun. 2014;15:320–332. doi: 10.1038/gene.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esen N, Blakely PK, Rainey-Barger EK, Irani DN. Complexity of the microglial activation pathways that drive innate host responses during lethal alphavirus encephalitis in mice. ASN Neuro. 2012;4:207–221. doi: 10.1042/AN20120016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang JP, Bowen GN, Zhou S, Cerny A, Zacharia A, Knipe DM, Finberg RW, Kurt-Jones EA. Role of specific innate immune responses in herpes simplex virus infection of the central nervous system. J Virol. 2012;86:2273–2281. doi: 10.1128/JVI.06010-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casrouge A, Zhang SY, Eidenschenk C, Jouanguy E, Puel A, Yang K, Alcais A, Picard C, Mahfoufi N, Nicolas N, Lorenzo L, Plancoulaine S, Senechal B, Geissmann F, Tabeta K, Hoebe K, Du X, Miller RL, Heron B, Mignot C, de Villemeur TB, Lebon P, Dulac O, Rozenberg F, Beutler B, Tardieu M, Abel L, Casanova JL. Herpes simplex virus encephalitis in human Unc-93b deficiency. Science. 2006;314:308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 32.Aly M, Wiltshire S, Chahrour G, Osti JC, Vidal SM. Complex genetic control of host susceptibility to coxsackievirus B3-induced myocarditis. Genes Immun. 2007;8:193–204. doi: 10.1038/sj.gene.6364374. [DOI] [PubMed] [Google Scholar]
- 33.Blazejczyk M, Miron M, Nadon R. Flexarray: a statistical data analysis software for gene expression microarrays. Montreal, Genome Quebec. 2007 [Google Scholar]
- 34.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. The mouse genome database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 2012;40:D881–D886. doi: 10.1093/nar/gkr974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 38.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. Amigo: Online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(t) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 40.Coronado MJ, Brandt JE, Kim E, Bucek A, Bedja D, Abston ED, Shin J, Gabrielson KL, Mitzner W, Fairweather D. Testosterone and interleukin-1 beta increase cardiac remodeling during coxsackievirus B3 myocarditis via serpin A 3n. Am J Physiol Heart Circ Physiol. 2012;302:H1726–H1736. doi: 10.1152/ajpheart.00783.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonner F, Borg N, Burghoff S, Schrader J. Resident cardiac immune cells and expression of the ectonucleotidase enzymes CD39 and CD73 after ischemic injury. PLoS One. 2012;7:e34730. doi: 10.1371/journal.pone.0034730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li WM, Liu W, Gao C, Zhou BG. Immunoregulatory effects of atorvastatin on experimental autoimmune myocarditis in Lewis rats. Immunol Cell Biol. 2006;84:274–280. doi: 10.1111/j.1440-1711.2006.01430.x. [DOI] [PubMed] [Google Scholar]
- 44.Leipner C, Borchers M, Merkle I, Stelzner A. Coxsackievirus B3-induced myocarditis in MHC class II-deficient mice. J Human Virol. 1999;2:102–114. [PubMed] [Google Scholar]
- 45.Deguine J, Lee BL, Newman ZR, Barton GM. No antigen-presentation defect in Unc93b1(3d/3d) (3d) mice. Nat Immunol. 2013;14:1101–1102. doi: 10.1038/ni.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andrade WA, Souza Mdo C, Ramos-Martinez E, Nagpal K, Dutra MS, Melo MB, Bartholomeu DC, Ghosh S, Golenbock DT, Gazzinelli RT. Combined action of nucleic acid-sensing Toll-like receptors and TLR11/TLR12 heterodimers imparts resistance to Toxoplasma gondii in mice. Cell Host Microbe. 2013;13:42–53. doi: 10.1016/j.chom.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caetano BC, Carmo BB, Melo MB, Cerny A, dos Santos SL, Bartholomeu DC, Golenbock DT, Gazzinelli RT. Requirement of Unc93b1 reveals a critical role for TLR7 in host resistance to primary infection with Trypanosoma cruzi. J Immunol. 2011;187:1903–1911. doi: 10.4049/jimmunol.1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heim A, Stille-Seigener M, Pring-Akerblom P, Grumbach I, Brehm C, Kreuzer H, Figulla HR. Recombinant interferons beta and gamma have a higher antiviral activity than interferon-alpha in coxsackievirus B3-infected carrier state cultures of human myocardial fibroblasts. J Interferon Cytokine Res. 1996;16:283–287. doi: 10.1089/jir.1996.16.283. [DOI] [PubMed] [Google Scholar]
- 49.Wang YX, da Cunha V, Vincelette J, White K, Velichko S, Xu Y, Gross C, Fitch RM, Halks-Miller M, Larsen BR, Yajima T, Knowlton KU, Vergona R, Sullivan ME, Croze E. Antiviral and myocyte protective effects of murine interferon-beta and -{alpha}2 in coxsackievirus B3-induced myocarditis and epicarditis in Balb/c mice. Am J Physiol Heart Circ Physiol. 2007;293:H69–H76. doi: 10.1152/ajpheart.00154.2007. [DOI] [PubMed] [Google Scholar]
- 50.Wiltshire SA, Leiva-Torres GA, Vidal SM. Quantitative trait locus analysis, pathway analysis, and consomic mapping show genetic variants of Tnni3k, Fpgt, or H28 control susceptibility to viral myocarditis. J Immunol. 2011;186:6398–6405. doi: 10.4049/jimmunol.1100159. [DOI] [PubMed] [Google Scholar]
- 51.Zhang QM, Song WQ, Li YJ, Qian J, Zhai AX, Wu J, Li AM, He JM, Zhao JY, Yu X, Wei LL, Zhang FM. Over-expression of mitochondrial antiviral signaling protein inhibits coxsackievirus B3 infection by enhancing type-I interferon production. Virol J. 2012;9:312. doi: 10.1186/1743-422X-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Z, Kim T, Bao M, Facchinetti V, Jung SY, Ghaffari AA, Qin J, Cheng G, Liu YJ. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34:866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding L, Hanawa H, Ota Y, Hasegawa G, Hao K, Asami F, Watanabe R, Yoshida T, Toba K, Yoshida K, Ogura M, Kodama M, Aizawa Y. Lipocalin-2/neutrophil gelatinase B-associated lipocalin is strongly induced in hearts of rats with autoimmune myocarditis and in human myocarditis. Circ J. 2010;74:523–530. doi: 10.1253/circj.cj-09-0485. [DOI] [PubMed] [Google Scholar]
- 54.Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 55.Chan YR, Liu JS, Pociask DA, Zheng M, Mietzner TA, Berger T, Mak TW, Clifton MC, Strong RK, Ray P, Kolls JK. Lipocalin 2 is required for pulmonary host defense against Klebsiella infection. J Immunol. 2009;182:4947–4956. doi: 10.4049/jimmunol.0803282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berger T, Togawa A, Duncan GS, Elia AJ, You-Ten A, Wakeham A, Fong HE, Cheung CC, Mak TW. Lipocalin 2-deficient mice exhibit increased sensitivity to Escherichia coli infection but not to ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2006;103:1834–1839. doi: 10.1073/pnas.0510847103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nocon AL, Ip JP, Terry R, Lim SL, Getts DR, Muller M, Hofer MJ, King NJ, Campbell IL. The bacteriostatic protein lipocalin 2 is induced in the central nervous system of mice with West Nile virus encephalitis. J Virol. 2014;88:679–689. doi: 10.1128/JVI.02094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singer E, Marko L, Paragas N, Barasch J, Dragun D, Muller DN, Budde K, Schmidt-Ott KM. Neutrophil gelatinase-associated lipocalin: pathophysiology and clinical applications. Acta Physiol (Oxf) 2013;207:663–672. doi: 10.1111/apha.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cruz DN, Gaiao S, Maisel A, Ronco C, Devarajan P. Neutrophil gelatinase-associated lipocalin as a biomarker of cardiovascular disease: a systematic review. Clin Chem Lab Med. 2012;50:1533–1545. doi: 10.1515/cclm-2012-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horvath AJ, Irving JA, Rossjohn J, Law RH, Bottomley SP, Quinsey NS, Pike RN, Coughlin PB, Whisstock JC. The murine orthologue of human antichymotrypsin: a structural paradigm for clade A3 serpins. J Biol Chem. 2005;280:43168–43178. doi: 10.1074/jbc.M505598200. [DOI] [PubMed] [Google Scholar]
- 61.Sipione S, Simmen KC, Lord SJ, Motyka B, Ewen C, Shostak I, Rayat GR, Dufour JM, Korbutt GS, Rajotte RV, Bleackley RC. Identification of a novel human granzyme B inhibitor secreted by cultured Sertoli cells. J Immunol. 2006;177:5051–5058. doi: 10.4049/jimmunol.177.8.5051. [DOI] [PubMed] [Google Scholar]
- 62.Wagsater D, Johansson D, Fontaine V, Vorkapic E, Backlund A, Razuvaev A, Mayranpaa MI, Hjerpe C, Caidahl K, Hamsten A, Franco-Cereceda A, Wilbertz J, Swedenborg J, Zhou X, Eriksson P. Serine protease inhibitor a3 in atherosclerosis and aneurysm disease. Int J Mol Med. 2012;30:288–294. doi: 10.3892/ijmm.2012.994. [DOI] [PubMed] [Google Scholar]
- 63.Asakura M, Kitakaze M. Global gene expression profiling in the failing myocardium. Circ J. 2009;73:1568–1576. doi: 10.1253/circj.cj-09-0465. [DOI] [PubMed] [Google Scholar]
- 64.Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 65.Lane JR, Neumann DA, Lafond-Walker A, Herskowitz A, Rose NR. Interleukin 1 or tumor necrosis factor can promote coxsackie B3-induced myocarditis in resistant b10.A mice. J Exp Med. 1992;175:1123–1129. doi: 10.1084/jem.175.4.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lim BK, Choe SC, Shin JO, Ho SH, Kim JM, Yu SS, Kim S, Jeon ES. Local expression of interleukin-1 receptor antagonist by plasmid DNA improves mortality and decreases myocardial inflammation in experimental coxsackieviral myocarditis. Circulation. 2002;105:1278–1281. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data