Abstract
Viruses are known to induce pathological cellular states that render infected cells susceptible or resistant to immune recognition. Here, we characterize an MHC-I-independent natural killer (NK) cell recognition mechanism that involves modulation of inhibitory NKR-P1B:Clr-b receptor-ligand interactions in response to mouse cytomegalovirus (MCMV) infection. We demonstrate that mouse Clr-b expression on healthy cells is rapidly lost at the cell surface and transcript levels in a time- and dose-dependent manner upon MCMV infection. In addition, cross-species infections using rat cytomegalovirus (RCMV) infection of mouse fibroblasts and MCMV infection of rat fibroblasts suggest that this response is conserved during host-pathogen interactions. Active viral infection appears to be necessary for Clr-b loss, as cellular stimulation using UV-inactivated whole virus or agonists of many innate pattern recognition receptors failed to elicit efficient Clr-b downregulation. Notably, Clr-b loss could be partially blocked by titrated cycloheximide treatment, suggesting that early viral or nascent host proteins are required for Clr-b downregulation. Interestingly, reporter cell assays suggest that MCMV may encode a novel Clr-b-independent immunoevasin that functionally engages the NKR-P1B receptor. Together, these data suggest that Clr-b modulation is a conserved innate host cell response to virus infection that is subverted by multiple CMV immune evasion strategies.
Key Words: Natural killer cell, Mouse cytomegalovirus, MCMV, Nkrp1:Clr, Missing-self recognition, Host response
Introduction
Natural killer (NK) cells are innate cytotoxic lymphocytes capable of recognizing pathological target cells via multiple germline-encoded receptor-ligand interactions. They can detect and eliminate cancerous, virus-infected, transplanted, antibody-opsonized and ‘stressed’ cells [1]. Their effector function is tightly regulated by receptor-ligand interactions, inhibitory or stimulatory in nature, which are collectively integrated to enable target cell recognition [1, 2]. These interactions follow the tenets of ‘missing-self’ and ‘induced-self’ recognition [3], respectively, which allow NK cells to be self-tolerant yet responsive to ‘altered-self’ pathologies and ‘non-self’ foreign entities [2]. Initially, missing-self recognition was shown to be dependent on the expression of polymorphic MHC class I (MHC-I) molecules [3], recognized by inhibitory self-MHC-I-specific receptors (rodent Ly49 or human KIR). More recently, however, numerous receptors have been identified that recognize non-MHC-I ligands [4].
In rodents, most NK cell receptors consist of type-II transmembrane C-type lectin-like proteins encoded within the NK gene complex (NKC). These include the Ly49 receptors that recognize classical MHC-I molecules, the CD94/NKG2 heterodimeric receptors that recognize nonclassical MHC-I molecules, the NKG2D homodimer that recognizes MHC-I-related stress-induced ligands and the NKR-P1 receptors that recognize other genetically linked C-type lectin-related (Clr) proteins [5, 6].
At least five members are known to exist in the mouse NKR-P1 family, three predicted stimulatory isoforms (NKR-P1A, C and F) and two inhibitory isoforms (NKR-P1B and G) [6, 7]. NKR-P1C, which encodes the well-known NK1.1 antigen in the B6 mouse strain, directly stimulates NK cell function. Yet, similar to NKR-P1A, NKR-P1C remains an orphan receptor. On the other hand, the NKR-P1F and NKR-P1G receptors recognize an overlapping yet distinct set of ligands, whereby NKR-P1F recognizes Clr-c, d and g, and NKR-P1G recognizes Clr-d, f and g in reporter cell assays [8, 9]. Recently, it was shown that NKR-P1G and Clr-f play an important role in mucosal immunity [10], and that the NKR-P1F/G receptors are expressed and functional on rat NK cells [11]. The remaining receptor, NKR-P1B, and its likely allelic variant, NKR-P1D, recognize Clr-b (i.e. Ocil/Clec2d) [12, 13]. Clr-b expression is quite broad and mirrors MHC-I expression on hematopoietic cells, yet it is commonly downregulated during cellular pathologies, including on cancerous cell lines [12], in response to genotoxic stress [14] and during viral infection [15, 16, 17]. During viral infection, mouse Clr-b and rat Clr-11 expression are rapidly lost at both the transcript and protein levels during poxvirus (Vaccinia virus, VV; Ectromelia virus, ECTV) and rat cytomegalovirus (RCMV-E; MHV8) infection, respectively [15, 16, 17].
Cytomegaloviruses (CMV) are members of the Betaherpesvirinae subfamily and are highly species-specific, possessing restricted host tropisms and requirements for productive infection. They are double-stranded DNA viruses with large genomes (≥200 kb) that frequently accommodate numerous immunoevasin genes. While CMV infections are generally asymptomatic in immune-competent hosts, uncontrolled infections can occur in immunocompromised individuals and in newborns, where infection can lead to severe complications and congenital defects. NK cells play a pivotal role in responding to and controlling acute CMV infections, and, as such, considerable coevolution at this host-pathogen interface has led to reciprocal strategies to maintain balance [18]. In fact, numerous CMV-encoded immunoevasin genes are known to modulate NK cell receptor-ligand interactions and effector function. Mouse CMV (MCMV) encodes stealth proteins (m138, m145, m152 and m155) that prevent induced-self maturation of NKG2D ligands (Mult1, Rae-1 and H60), cloaking ligands (m04 and m06) that regulate MHC-I recognition by T and NK cells, at least one MHC-like decoy ligand (m157) that is directly recognized by host NK cell receptors (inhibitory Ly49I and stimulatory Ly49H), and presumably others (e.g. m144) [19, 20, 21, 22, 23, 24]. In addition, the English isolate of rat CMV (RCMV-E) encodes an immunoevasin, RCTL, that targets the rat NKR-P1B:Clr-11 axis during host-pathogen interactions in vivo [16, 25].
Here, we demonstrate that there is striking evolutionary conservation in the host cell response to pathogen-induced regulation of Clr/Clec2d expression. Both mouse Clr-b (Clec2d) and rat Clr-11 (Clec2d11) are acutely modulated at the transcript and protein levels in response to reciprocal cross-species MCMV or RCMV infection of rat or mouse host cells, respectively. Downregulation of Clr-b requires live virus infection, as it does not occur using whole inactivated virus or simple agonists of innate pattern recognition receptor (PRR) pathways. Moreover, loss of cell surface Clr-b can be partially blocked by inhibition of nascent protein synthesis and the ubiquitin (Ub)-proteasome degradation pathway, but not using inhibitors of late viral replication. We also provide evidence of a novel Clr-b-independent immunoevasin capable of engaging the mouse NKR-P1B receptor upon MCMV infection of fibroblasts in vitro.
Materials and Methods
Cells
NIH3T3 fibroblast and J774A.1 monocyte-like cells were obtained from the American Type Culture Collection (ATCC). Wild-type C57BL/6 (WT B6) mouse embryonic fibroblasts (MEFs) were obtained from T.W. Mak (University of Toronto, Canada). Cells were cultured in complete DMEM-HG, supplemented with 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 50 μg/ml gentamicin, 110 μg/ml sodium pyruvate, 50 μM 2-mercaptoethanol, 10 mM HEPES and 10–20% FCS.
Virus Infections
MCMV (Smith strain) was provided by Dr. A. Makrigiannis (University of Ottawa, Canada). MCMV-GFP was provided by Dr. S. Vidal (McGill University, Canada) and has been previously described [26]. Wild-type RCMV-E and a ΔRCTL-mutant have been previously described [16]. Viruses were passaged on MEF or rat embryonic fibroblast (REF) monolayers. Plaque assays were used to determine viral titers as described [27], without centrifugation during infection. For in vitro infections, fibroblasts were seeded to subconfluent monolayers, virus was added [multiplicity of infection (MOI) ∼0.5 PFU/cell], then cells were centrifuged at 800 g for 30 min at 37°C (effective MOI ∼10 PFU/cell with centrifugation) and incubated at 37°C, 5% CO2 for the indicated times.
RNA Isolation, cDNA Synthesis and qRT-PCR
Total RNA was isolated using a mirVana RNA isolation kit (LifeTechnologies), and reverse transcription reactions were carried out using Superscript III cDNA synthesis kits (LifeTechnologies). Quantitative real-time RT-PCR was performed on a CFX-96 real-time PCR detection System (BioRad) using 20–50 ng of cDNA, Sso-Fast EvaGreen Supermix (BioRad) and gene-specific primers designed using Primer-Blast (www.ncbi.nlm.nih.gov/tools/primer-blast), selected to span at least one intron and possess minimal reactivity with other genes (Clec2d/Clr-b: 5′-AGC TCC TCA GCT CTG AGA TGT GTG, 5′- AGG GGA GAT GGT TCC GTG CCT TT; Tbp: 5′-AGA GCC ACG GAC AAC TGC GTT G, 5′-CTG GGA AGC CCA ACT TCT GCA C). Data were analyzed using CFX Manager software (BioRad).
Flow Cytometry
Cells were stained in flow buffer (HBSS, 0.5% BSA and 0.03% NaN3) on ice with primary monoclonal antibodies (mAb) for 25–30 min, or secondary streptavidin conjugates for 15–20 min, washed between incubations and then analyzed using a FACSCalibur flow cytometer (BD Biosciences). Cells were gated by FSC/SSC, and propidium iodide exclusion for viability. Data were analyzed using FlowJo software (Treestar). Biotinylated 4A6 mAb (mClr-b; rat IgMκ) and R3A8 mAb (rClr-11, RCMV-E RCTL; mouse IgMκ) were described previously [12, 28]. Biotin-conjugated mAb specific for H-2KbDb (28-8-6, mouse IgG2aκ), H-2DqLq (KH117, mouse IgG2aκ) were purchased from BD Pharmingen, purified mouse pan-Rae-1 mAb (186107, rat IgG2a) was purchased from R&D Systems, CD71 mAb (RI7 217.1.4) was purchased from eBioscience, SA-APC was purchased from LifeTechnologies and all other mAb and secondary reagents were purchased from BD Biosciences, e-Bioscience or LifeTechnologies. HT29 mAb was developed in the laboratory of Dr. K. Wonigeit (Hannover Medical School). Briefly, intraperitoneal alloimmunization was performed using NKC-congenic LEW donor (Clr-11LEW) splenocytes into LEW.TO-NKC recipient rats (Clr-11TO); hybridoma supernatants were screened by indirect immunofluorescence and flow cytometry for expression on LEW-strain IL-2 lymphokine-activated killer (NK-LAK) cells, but not LEW.TO-NKC NK-LAK cells and 293 cells transfected with Clr-11LEW but not Clr-11TO.
BWZ Reporter Cell Assays
BWZ.CD3ζ/NKR-P1BB6/129 reporter cells were generated as described [28, 29]. Stimulator cells (mock- or MCMV-infected fibroblasts) were cultured in flat, 96-well plates in 3-fold dilutions, reporter cells (5 × 104/well) were then added and cocultures were incubated overnight at 37°C. Purified blocking antibody was added at 10 μg/ml. Control reporter cells were stimulated with 10 ng/ml PMA plus 0.5 μM ionomycin. Cells were washed, resuspended in 100 μl of 1X CPRG buffer (90 mg/l chlorophenol-red-β-D-galactopyranoside (Roche), 9 mM MgCl2, 0.1% NP-40, in PBS), incubated at room temperature, and then analyzed using a Varioskan microplate reader (Thermo Scientific), using OD595-655.
Agonists, Inhibitors and Cytokines
Various Toll-like receptor (TLR), Nod-like receptor (NLR) and RIG-I-like receptor (RLR) agonists were obtained from Invivogen (San Diego, Calif., USA). Additional NLR agonists were provided by Dr. D.J. Philpott (University of Toronto, Canada). 3′,5′-c-di-AMP was purchased from Invivogen and 2′,5-3,5′-cGAMP was purchased from Biolog (Bremen, Germany). Actinomycin D (ActD), cycloheximide (CHX), phosphonoacetic acid (PAA), cytosine arabinoside (AraC), adenosine monophosphate (AMP), adenosine diphosphate (ADP), adenosine triphosphate (ATP) and nigericin were purchased from Sigma-Aldrich (St. Louis, Mo., USA). All reagents were dissolved in DMSO, water, serum-free DMEM or PBS according to the manufacturer's directions. Cells were treated for 24 h with reagents or solvent controls in parallel.
Statistical Analysis
Data were analyzed using GraphPad Prism 5, employing either a paired Student's two-tailed t test, or one-way or two-way ANOVA, with Bonferroni's correction, where applicable (see figure legends). All graphs show mean values ± SEM; * p < 0.05, ** p < 0.01, *** p < 0.001. All data are representative of at least 3 independent experiments.
Results
MCMV Infection Promotes Cell-Intrinsic mClr-b Downregulation in vitro
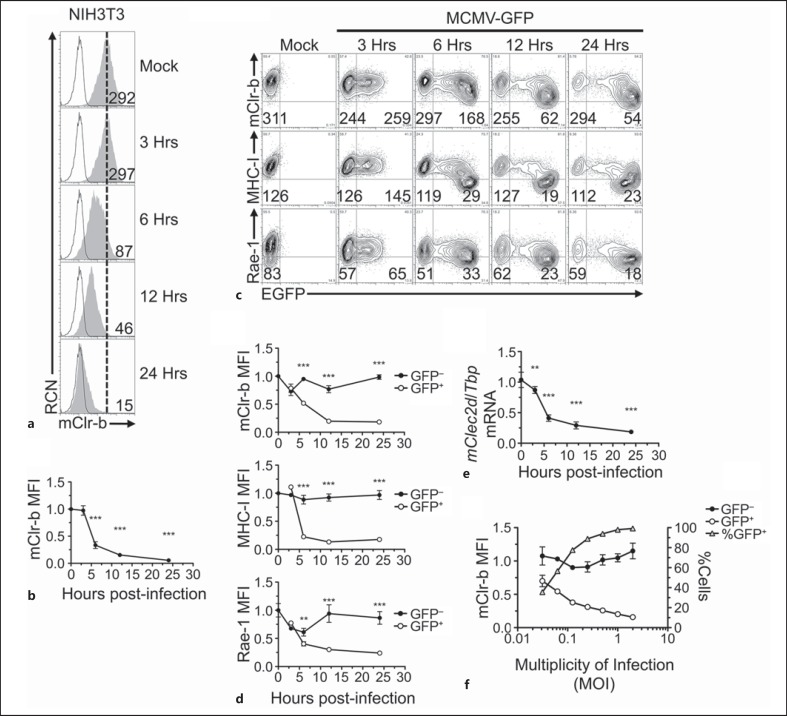
We have previously shown that RCMV-E infection of REFs results in a rapid loss of the rat Clr-b homolog, rClec2d11 (rClr-11), by 24 h postinfection (h.p.i.) [16]. To extend these observations to MCMV infection of mouse fibroblasts, we infected mouse NIH3T3 fibroblasts with purified MCMV-Smith virus (MOI of 0.5 PFU/cell by plaque assay; effective MOI ∼10 with centrifugal enhancement) and assessed Clr-b cell surface expression. mClr-b was temporally regulated during MCMV infection, with a pronounced loss by 6–24 h.p.i. (fig. 1a, b). However, the use of WT MCMV could not discern whether mClr-b downregulation was a direct or an indirect consequence of virus infection at the single cell level.
Fig. 1.
MCMV infection promotes rapid Clr-b downregulation. a NIH3T3 cells were infected with MCMV-Smith virus at a MOI of 0.5 PFU/cell over a 24-hour time course, and were analyzed by flow cytometric analysis for the cell surface expression of mClr-b. 4A6 Clr-b mAb (shaded histogram); secondary reagent alone (black line); reference for mock-treated control median fluorescence intensity (MFI) level (dotted vertical line). Numbers indicate MFI values. b Quantitation of mClr-b MFI levels in a normalized to mock-treated control levels (mean ± SEM). Statistical significance is shown between mock treatment and a given time point. c Time course of Clr-b, MHC-I and Rae-1 expression upon NIH3T3 cell infection using recombinant MCMV-GFP (MOI = 0.5 PFU/cell). Numbers represent MFI levels of the respective markers gated on GFP− (uninfected) or GFP+ (MCMV-infected) cells. d Quantitation of mClr-b, MHC-I and Rae-1 MFI levels on MCMV-GFP-infected cells in c gated by GFP expression and normalized to mock-treated controls. Statistical significance is shown between GFP− and GFP+ population. e Time course quantitation of Clr-b (Clec2d) transcript levels following MCMV infection at MOI of 0.5 PFU/cell (indexed relative to Tbp levels and normalized relative to mock-treated controls). Statistical significance is shown between mock treatment and a given time point. f Quantitation of mClr-b MFI levels and % of GFP+ cells upon MCMV-GFP infections at different MOI ratios analyzed at 22 h.p.i. Black and white circles represent GFP− and GFP+ cell subsets (corresponding with the left y-axis) and gray triangles represent % of GFP+ cells (right y-axis). Graphs show mean ± SEM. Experiments were analyzed using ANOVA with Bonferroni's post hoc analysis. * p < 0.05, ** p < 0.01, *** p < 0.001. All data are representative of at least 3 independent experiments.
Consequently, we repeated NIH3T3 infections using a recombinant MCMV-GFP virus that ectopically expresses enhanced GFP driven by an immediate-early (IE) gene promoter [26]. The MCMV-GFP was generated previously by homologous integration of EGFP under the control of a native MCMV ie1/3 gene promoter into MCMV-Smith (VR-194), between the ie1/3 and ie2 genes, and proximal to the native ie1/3 enhancer elements. It does not appear to exhibit any differences in growth kinetics in cell culture in vitro or in infected salivary glands in vivo [26]. MCMV-GFP infections distinguished infected (GFP-positive, GFP+) from uninfected (GFP-negative, GFP−) cells as early as 3 h.p.i., with prominent mClr-b loss ensuing by 6–24 h.p.i. (fig. 1c, d). In parallel, we examined cell surface expression of MHC-I ligands for the Ly49 receptors (H-2q) and Rae-1 ligands for the NKG2D receptor . Similar to mClr-b, MHC-I and Rae-1 levels were also reduced by 6–24 h.p.i., likely due to the action of several known MCMV-encoded immunoevasins (e.g. m04, m06, m138, m145, m152 and m155) [18, 21, 22, 23, 24, 30]. As a control, MCMV-GFP infection had no effect on cell surface levels of the transferrin receptor (CD71, online suppl. fig. 1; for all online suppl. material, see www.karger.com/doi/10.1159/000382032). Importantly, the restricted loss of mClr-b on infected (GFP+) cells suggests that this response occurs by an MCMV infection-dependent cell-intrinsic mechanism.
Notably, experiments using MCMV culture supernatants revealed a pronounced upregulation of Clr-b surface expression on uninfected (GFP−) ‘bystander’ cells, likely mediated by soluble factors such as interferons (C.L.K., in preparation); however, this bystander mClr-b upregulation was not observed using purified virus, which only promoted infection-dependent mClr-b downregulation (fig. 1c, d). In addition, similar losses of mClr-b expression on the infected GFP+ but not the uninfected GFP− subsets were also observed for several cell types, including primary WT B6 MEFs and adult ear fibroblasts (AEFs), S17 bone marrow stromal cells, J774 and RAW264.7 monocyte/macrophage cells and fetal ILC-like MNK-3 cells (data not shown; see below).
It has previously been shown that RCMV-E infection of rat fibroblasts results in a loss of rClr-11 at both the transcript and cell surface protein levels [16]. Similarly, quantitative real-time RT-PCR analysis of MCMV-infected mouse NIH3T3 fibroblasts revealed that mClr-b surface protein downregulation was mirrored by a time-dependent loss of mClr-b (Clec2d) transcripts by 6–24 h.p.i. (normalized to Tbp; fig. 1e). Interestingly, similar losses were observed for other mouse mClr family members, including mClr-a/c/f/g (Clec2e/f/h/i; data not shown). Titration of MCMV infections using 2-fold MOI dilutions demonstrated that mClr-b loss correlated with viral dosage and was restricted to infected GFP+ cells (fig. 1f). Collectively, these results indicate that MCMV promotes an infection-dependent cell-intrinsic loss of mClr-b (Clec2d) at the transcript and cell surface protein levels.
MCMV-Mediated mClr-b Downregulation Is Partially Blocked by Translational Inhibition
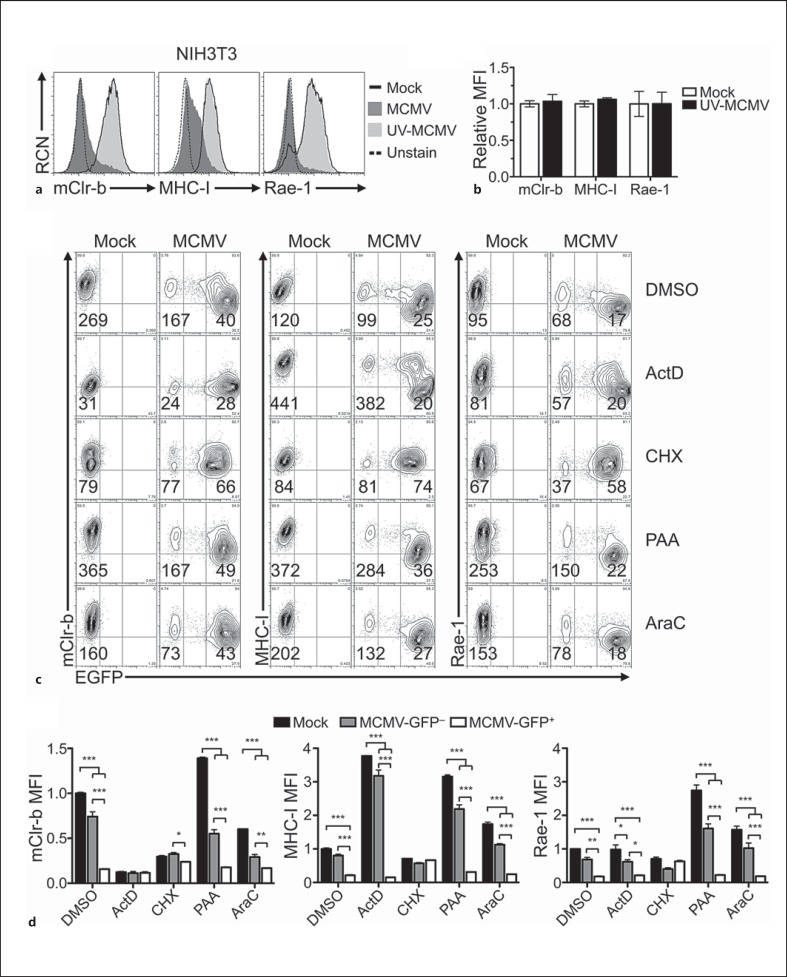
Since mClr-b downregulation was triggered early in the MCMV replication cycle and only in infected cells, we next assessed the contribution of de novo gene expression to mClr-b downregulation. To this end, we repeated MCMV infections in the presence of transcriptional, translational and viral replication inhibitors. It is important to note that some of these inhibitors can also prevent host gene expression and/or induce cellular stress responses, resulting in mClr-b downregulation in the absence of infection. In addition, cell surface Clr-b exhibits fast turnover kinetics, with a half-life of about 3 h (online suppl. fig. 2). Thus, we titrated these chemicals where possible to minimize the extent of mClr-b downregulation by chemical treatment alone, and then tested their ability to block MCMV-mediated mClr-b downregulation.
UV-inactivated MCMV failed to promote mClr-b downregulation on NIH3T3 cells (fig. 2a, b). As expected, MHC-I and Rae-1 levels did not change using UV-inactivated MCMV [31], suggesting that active or productive viral infection are required for MCMV-mediated loss of mClr-b, MHC-I and Rae-1 ligands. In contrast, inhibition of transcription and expression of IE (α) genes using ActD resulted in a substantial loss of basal mClr-b surface expression (even on mock-infected and uninfected GFP− cells; fig. 2c, d). This implies either that mClr-b undergoes rapid turnover in the absence of nascent transcripts (blocked by ActD; online suppl. fig. 2) or that ActD may also promote stress-mediated mClr-b downregulation [14, 15]. Thus, we attempted to titrate ActD levels to achieve only partial Clr-b loss, while maintaining partial inhibition of IE genes; however, such an optimal dose was not possible, making inferences on IE gene blockade inconclusive (online suppl. fig. 3).
Fig. 2.
MCMV-mediated mouse Clr-b downregulation is prevented by UV-inactivation and inhibition of early viral and host protein synthesis. a Characterization of mClr-b, MHC-I and Rae-1 expression upon treatment with UV-inactivated MCMV-GFP. Infected NIH3T3 cells were analyzed for surface expression of mClr-b (4A6 mAb), MHC-I (H-2LqDq) and Rae-1 (α-ε). b Quantitation of mClr-b, MHC-I and Rae-1 MFI levels in a relative to mock-treated controls. c Characterization of mClr-b, MHC-I and Rae-1 expression upon MCMV-GFP infections in the presence of various chemical inhibitors. NIH3T3 cells were pretreated with DMSO (controls), ActD (10 nM), CHX (10 μg/ml), PAA (400 μg/ml) or AraC (50 μg/ml), and then infected with MCMV-GFP and analyzed by flow cytometry at 24 h.p.i. d Quantitation of mClr-b, MHC-I and Rae-1 MFI levels in c normalized to DMSO-treated mock MFI levels. Graphs show mean ± SEM. Experiments were analyzed using ANOVA with Bonferroni's post hoc analysis. * p < 0.05, ** p < 0.01, *** p < 0.001. All data are representative of at least 3 independent experiments.
MCMV early (β) gene expression can be inhibited using the translational inhibitor, CHX. Notably, pretreatment of NIH3T3 cells with a low dose of CHX alone caused some basal mClr-b loss on uninfected cells (compare mock DMSO vs. CHX; fig. 2c, d; online suppl. fig. 2). Interestingly, however, CHX treatment rendered the expression profiles of infected (GFP+) versus uninfected (GFP−) cells nearly indistinguishable, suggesting that CHX was sufficient to prevent MCMV infection-mediated loss of mClr-b. Notably, higher CHX doses blocked mClr-b expression on uninfected cells, similar to ActD treatment (online suppl. fig. 3). Similarly, MHC-I and Rae-1 downregulation, mediated by known early viral immunoevasin genes, was also blocked by CHX treatment. Nonetheless, CHX is capable of inhibiting the translation of both viral IE and host gene products; thus, IE to early viral proteins and/or de novo cellular protein synthesis appear to be involved in regulating mClr-b expression during infection.
Late (γ) viral gene expression is enhanced by viral genome replication, and can be blocked by viral DNA polymerase inhibitors like PAA or AraC. However, viral replication appeared to be dispensable for the MCMV-mediated downregulation of mClr-b, MHC-I and Rae-1 ligands (fig. 2c, d). Collectively, these results suggest that MCMV-mediated Clr-b downregulation requires IE to early events in active MCMV infection, but not late events nor viral genome replication per se.
We next tested the effects of the proteasomal inhibitor, MG132, on MCMV infection-mediated Clr-b loss, as we previously showed that inhibition of the Ub-proteasome degradation pathway could prevent genotoxic stress- mediated Clr-b downregulation [14, 15]. MG132 was effective at blocking MCMV-mediated Clr-b loss, albeit incompletely even at high doses (online suppl. fig. 4). However, MG132 also affects numerous Ub-dependent cellular processes apart from proteasomal degradation, including autophagy and Ub-dependent signaling pathways. These results suggest that the Ub-proteasomal degradation pathway may be partially responsible for Clr-b loss induced by MCMV infection, perhaps by accelerating Clr-b turnover.
RCMV-E-Mediated Clr-11 Loss and RCTL Induction Are Blocked by CHX Treatment
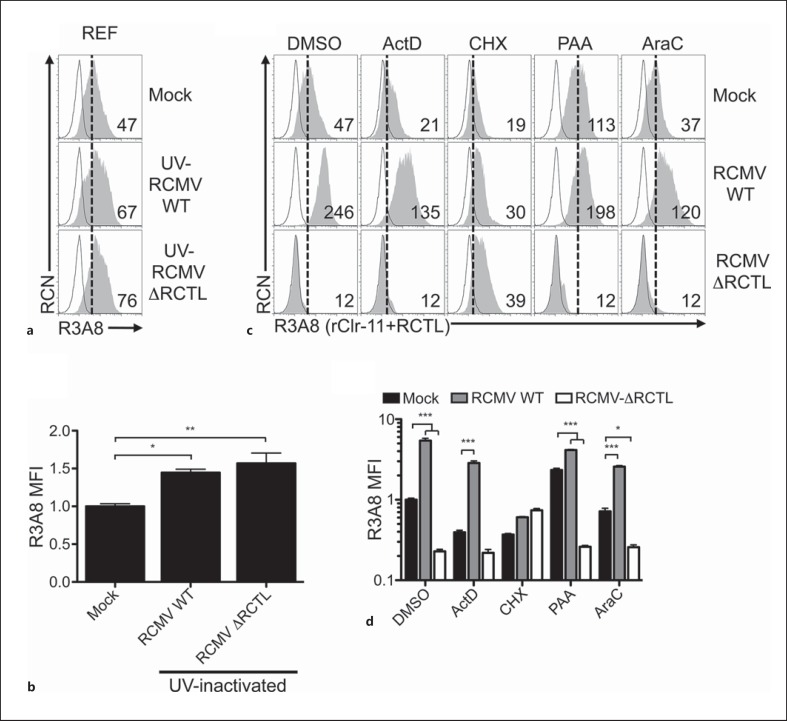
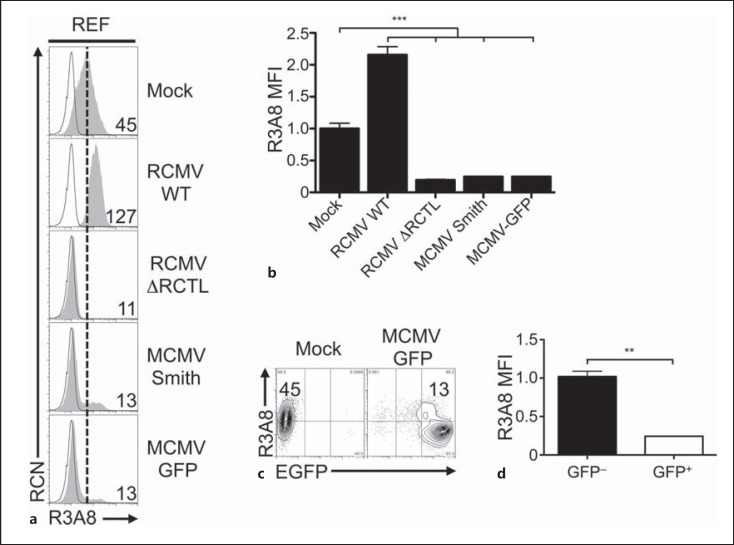
Using new reagents and cell lines, we conducted a closer examination of the requirements for RCMV-E-mediated rClr-11 downregulation and RCTL immunoevasin expression in the rat system. First, we repeated RCMV-E infections using both REFs (fig. 3) [16] and primary rat AEFs (online suppl. fig. 5). Since R3A8 antibody is dual-specific (cross-reactive for both the RCTL decoy and host rClr-11) [16, 28], we infected REFs using either WT RCMV-E (RCMV WT) or an RCTL-deficient virus (RCMV ΔRCTL) in order to distinguish rClr-11 from RCTL. Interestingly, infections using UV-inactivated RCMV viruses appeared to slightly increase R3A8 staining on REFs (fig. 3a, b), which may have been due to weak host rClr-11 induction in the absence of active infection, or perhaps weak cross reactivity with another unidentified host rClr family member. Nonetheless, RCMV WT infection of REFs results in an increase in R3A8 staining (due to RCTL), versus mock-treated control cells (rClr-11 alone; fig. 3c, d). In contrast, ΔRCTL-mutant infection of REFs promotes a loss of R3A8 staining (rClr-11 alone) versus mock-treated control cells (rClr-11 alone). Taken together, RCTL appears to replace host Clr-11 during RCMV infection. Similar data were observed for rClr-11hi rat AEFs (online suppl. fig. 5B); however, R3A8 staining decreases here due to a much larger loss of rClr-11 not fully compensated by RCTL upregulation. Second, we used a novel rClr-11 mAb, HT29, in parallel with R3A8 mAb. Importantly, HT29 mAb does not cross-react with RCTL, yet possesses robust reactivity with rClr-11; this allowed us to observe loss of rClr-11 in isolation upon infection of REFs and AEFs using RCMV WT (online suppl. fig. 5A/B).
Fig. 3.
RCMV-mediated rat Clr-11 downregulation and RCTL immunoevasin induction are prevented by UV-inactivation and inhibition of early viral and host protein synthesis. a Characterization of rClr-11 expression on RCMV-E-infected REFs upon treatment with UV-inactivated RCMV WT or ΔRCTL-mutant virus. Treated cells were analyzed by flow cytometry for rClr-11/RCTL (R3A8 mAb) expression (numbers indicate MFI values). b Quantitation of R3A8 MFI levels shown in a relative to mock-treated controls. c Characterization of rClr-11/RCTL (R3A8 mAb) expression upon RCMV WT or ΔRCTL-mutant virus infections in the presence of various chemical inhibitors. REFs were pretreated with DMSO (controls), ActD (10 nM), CHX (10 μg/ml), PAA (400 μg/ml) or AraC (50 μg/ml), and then infected with RCMV WT or ΔRCTL-mutant virus and analyzed by flow cytometry at 24 h.p.i. for R3A8 expression. R3A8 rClr-11/RCTL mAb (shaded histogram); secondary reagent alone (black line); reference for mock-treated control MFI level (dotted vertical line). Numbers indicate MFI values. d Quantitation of rClr-11/RCTL (R3A8 mAb) MFI levels in c normalized to DMSO-treated mock control MFI levels. Graphs show mean ± SEM. Experiments were analyzed using ANOVA with Bonferroni's post hoc analysis. * p < 0.05, ** p < 0.01, *** p < 0.001. All data are representative of at least 3 independent experiments.
Next, the effects of inhibitors were examined. Similar to the results in mouse NIH3T3 cells, ActD treatments alone resulted in a substantial infection-independent loss of basal rClr-11 from the cell surface of rat AEFs and REFs, albeit incomplete (compare mock DMSO and ActD; fig. 3c, d; online suppl. fig. 5A/B). However, this titrated ActD concentration failed to prevent expression of the rctl gene product (RCMV WT vs. ΔRCTL-mutant). Nor did ActD prevent infection-mediated downregulation of host rClr-11 (mock vs. ΔRCTL-mutant). This RCMV-mediated induction of viral RCTL and loss of host rClr-11 were also confirmed using HT29 versus R3A8 mAb (see ActD; online suppl. fig. 5B). Notably, as seen in the mouse system, higher ActD doses were required to block RCTL induction upon RCMV-E infection, and these doses completely blocked basal rClr-11 expression on uninfected cells, precluding usage of an optimal ActD dose (online suppl. fig. 6).
Again, similar to the mouse system, treatment with a titrated CHX dose resulted in decreased basal rClr-11 cell surface expression (mock DMSO vs. CHX; fig. 3c, d; online suppl. fig. 5A/B). More importantly, however, CHX rendered the R3A8 expression profiles of the two infected populations indistinguishable (RCMV WT vs. ΔRCTL-mutant) and blocked infection-mediated loss of host rClr-11 visualized using both R3A8 and HT29 mAb (mock vs. RCMV). These data suggest that CHX is sufficient to prevent both infection-mediated loss of rClr-11 as well as RCTL induction, and are in agreement with previous data suggesting that rctl is an early gene [16].
Notably, similar to UV-inactivated RCMV (fig. 3a, b), the subtle increase in R3A8 staining observed upon RCMV infection in the presence of CHX treatment (using both RCMV WT and ΔRCTL-mutant; fig. 3c, d) may result from weak rClr-11 induction in the absence of productive infection (in the presence of CHX) or perhaps weak cross reactivity with another unidentified host rClr family member; however, this was not observed using another rClr-11 mAb, HT29 (online suppl. fig. 5A/B). Of note, treatments with PAA or AraC were insufficient to prevent rClr-11 downregulation or RCTL induction. Collectively, these results demonstrate that only translational inhibition using CHX abrogates RCMV-mediated host rClr-11 downregulation, at the same time preventing RCTL immunoevasin induction. Thus, host rClr-11 downregulation is a phenomenon induced by active RCMV infection and is countered by the rctl gene product.
Cross-Species CMV Infection Promotes Host mClr-b and rClr-11 Downregulation
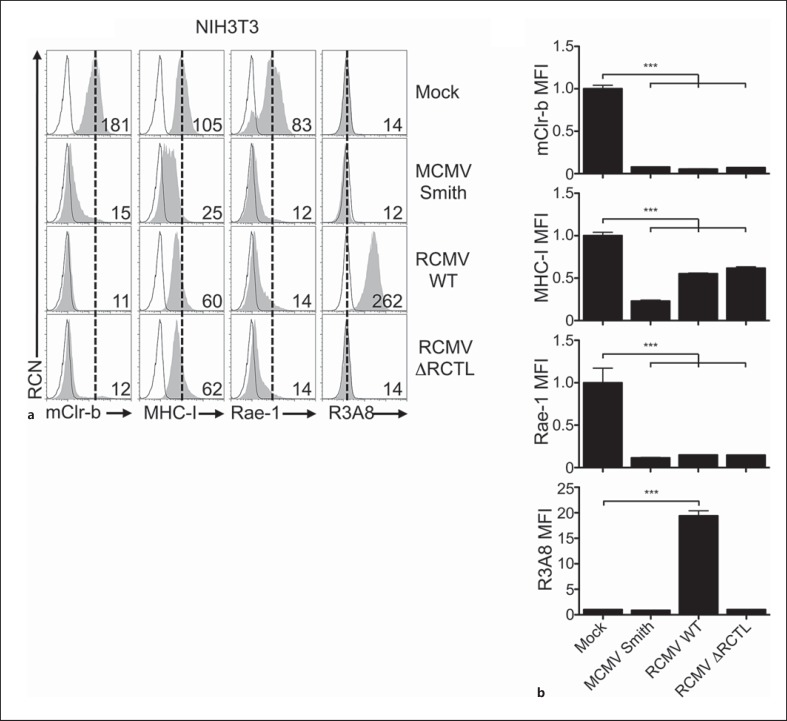
The similar results obtained using MCMV infection of mouse fibroblasts and RCMV infection of rat fibroblasts prompted us to investigate whether host mClr-b/rClr-11 downregulation was a conserved cellular response to infection or whether virus-specific (and potentially host-adapted) immunoevasins were involved. Using cross-species infections, host mClr-b expression can also be monitored without RCTL cross reactivity (using 4A6 mAb), while RCTL expression can be monitored without rClr-11 cross reactivity (using R3A8 mAb). We thus infected mouse NIH3T3 cells with MCMV or RCMV-E and measured cell surface expression of mClr-b, MHC-I and Rae-1 (fig. 4a, b). Strikingly, mClr-b downregulation was observed using MCMV or RCMV viruses (∼15-fold reduction); interestingly, however, mClr-b loss was consistently more robust using cross-species RCMV infection. In contrast, MHC-I molecules showed a smaller <2-fold reduction upon cross-species RCMV infection versus a >4-fold reduction using MCMV infection. Rae-1 downregulation was similar using MCMV or RCMV infection (∼6-fold reduction). To monitor the RCTL immunoevasin in isolation, we also stained infected mouse NIH3T3 cells using the rat-specific R3A8 mAb; as expected, RCTL was only detected using RCMV WT virus. Notably, no cross contamination of the viruses could be detected (data not shown), and RCMV itself is noncytopathic in NIH3T3 cells, although it can induce cytomegaly.
Fig. 4.
Xenogeneic RCMV-E infection promotes mouse Clr-b loss on mouse fibroblasts. a NIH3T3 were infected with MCMV-Smith or RCMV-E viruses and then analyzed by flow cytometry at 24 h.p.i. for mClr-b, MHC-I, Rae-1 and R3A8 (RCTL) expression. Primary mAb stain (shaded histograms); secondary reagent alone (black line); reference for mock-treated control MFI level (dotted vertical line). Numbers indicate MFI values. b Quantitation of MFI levels in (a) normalized to mock-treated control MFI levels. Graphs show mean ± SEM. Experiments were analyzed using one-way ANOVA with Bonferroni's post hoc analysis. * p < 0.05, ** p < 0.01, *** p < 0.001. All data are representative of at least 3 independent experiments.
To reciprocate these observations, we infected REFs with RCMV-E or MCMV viruses and measured cell surface expression of rClr-11 (±RCTL) using R3A8 mAb (fig. 5a, b). Interestingly, cross-species MCMV infection of REFs also promoted rClr-11 downregulation (∼4-fold); however, as observed for mClr-b using mouse fibroblasts, rClr-11 loss was slightly more robust using RCMV-E virus (ΔRCTL-mutant) versus cross-species MCMV viruses. Upon further investigation using MCMV-GFP, the infected (GFP+) REFs were observed to downregulate rClr-11 (R3A8) expression, whereas uninfected (GFP−) REFs did not (fig. 5c, d). Similar findings were also observed using Clr-11hi primary rat AEFs (data not shown). These data suggest that host Clr (mClr-b/rClr-11) downregulation is a conserved cellular response to CMV infection, and that RCTL is an RCMV-E-specific immunoevasin.
Fig. 5.
Xenogeneic MCMV infection promotes rat Clr-11 loss on rat fibroblasts. a REFs were infected with MCMV or RCMV-E viruses and analyzed by flow cytometry at 24 h.p.i. for R3A8 (rClr-11/RCTL) expression. R3A8 stain (shaded histograms); secondary reagent alone (black line); reference for mock-treated control MFI level (dotted vertical line). Numbers indicate MFI values. b Quantitation of MFI levels in a normalized to mock-treated control MFI levels. c Representative R3A8 (rClr-11) flow cytometric analysis of MCMV-GFP-infected REFs. d Quantitation of R3A8 MFI levels in c gated by MCMV-GFP expression. Graphs show mean ± SEM. Experiments were analyzed using one-way ANOVA with Bonferroni's post hoc analysis (except d: Student's t test). * p < 0.05, ** p < 0.01, *** p < 0.001. All data are representative of at least 3 independent experiments.
Modulation of mClr-b by PRR Agonists
Both viral and bacterial infections lead to the detection of numerous pathogen-/danger-associated molecular patterns (PAMP/DAMP) via host PRRs; such PAMP include both extracellular moieties and intracellular (vesicular or cytosolic) metabolites. Thus, we sought to determine whether CMV-mediated mClr-b downregulation could be recapitulated in isolation upon cellular exposure to defined PAMP or, alternatively, if Clr-b loss may require active viral infection. Since whole UV-inactivated CMV is insufficient to promote mClr-b loss, and viral replication is dispensable, while the expression of IE to early viral genes and/or host protein translation are required, it is likely that intermediates of virus infection, perhaps signaling metabolites, nucleic acids or unidentified viral gene products, may be required to mimic CMV infection. To this end, we treated cell lines with a panel of known PRR agonists targeting various innate sensing pathways.
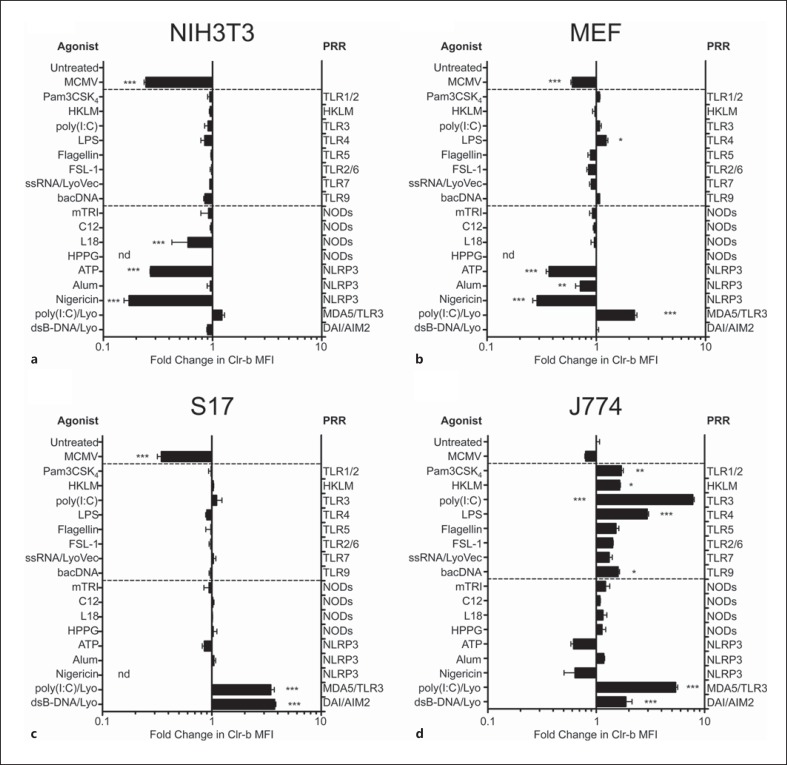
Initially, mouse fibroblast cell lines (NIH3T3) were treated with various PRR agonists targeting TLRs 1–9, NLRs or cytosolic nucleic acid receptors including RLRs or AIM2-like receptors (fig. 6a, b). However, no significant mClr-b downregulation comparable to MCMV infection was observed upon treatment of fibroblasts using these PRR agonists, including intracellular double-stranded RNA (polyI:C) and double-stranded immunostimulatory DNA (dsB-DNA). However, upon investigation of the known inducers of the Nlrp3 inflammasome, extracellular ATP was the sole metabolite that autonomously downregulated mClr-b on all cell lines, while nigericin (a K+ ionophore) was effective for some cell lines, and inflammasome activation by alum salts was inconclusive. We also tested cyclic dinucleotides (c-di-AMP and c-GAMP), recently shown to serve as bacterial PAMP or second messengers of host cytosolic dsDNA sensing (via cGAS [32, 33, 34] and the mitochondrial STING sensor), but failed to observe any changes in mClr-b expression (data not shown). In addition, we repeated the above treatments using J774 macrophages (as a hematopoietic immune cell lineage); however, ATP and nigericin treatments were less robust at inducing mClr-b downregulation on these cells (fig. 6c, d). Moreover, cytosolic nucleic acids actually promoted mClr-b upregulation on many cells, and numerous PRR agonists induced mClr-b on J774 cells. Collectively, these results suggest that single PRR engagement in isolation is not sufficient to mimic MCMV-mediated mClr-b downregulation; however, the role of ATP in promoting mClr-b loss is intriguing (see below).
Fig. 6.
Modulation of mouse Clr-b levels on various mouse cell lines treated with simple PRR agonists. a NIH3T3 cells. b MEFs. c S17 stromal cells. d J774 macrophage cells were treated with various PRR agonists and then analyzed for mouse Clr-b levels by flow cytometry. Clr-b MFI levels were quantitated relative to untreated cells and represented as fold-change in MFI values. Synthetic triacylated Pam3CSK4 lipoprotein (Pam3CSK4) 1 μg/ml; heat-killed L. monocytogenes (HKLM) 104; poly(I:C) 100 ng/ml; lipopolysaccharide (LPS) 1 μg/ml; flagellin 1 μg/ml; synthetic FSL-1 diacylated lipoprotein (FSL-1) 1 μg/ml; single-stranded RNA complexed with LyoVec (ssRNA/LyoVec) 100 ng/ml; E. coli bacterial DNA (bacDNA) 1 μg/ml; mTRI-DAP muramyl tripeptide (mTRI) 1 μg/ml; acylated iE-DAP dipeptide (C12) 1 μg/ml; muramyl dipeptide with a C18 fatty acid chain (L18) 1 μg/ml; H. pylori proteoglycan (HPPG) 10 μg/ml; ATP 25 mM; aluminum potassium sulphate (Alum) 10 μg/ml, nigericin (50 μM), poly(I:C) complexed with LyoVec liposomes [poly(I:C)/Lyo] 100 ng/ml; dsB-DNA complexed with LyoVec liposomes (dsB-DNA/Lyo) 100 ng/ml and MCMV-GFP virus. The labels represent agonist treatments (left side) and the PRR targets of the agonists (right side). Graphs show mean ± SEM. Experiments were analyzed using one-way ANOVA with Bonferroni's post hoc analysis. * p < 0.05, ** p < 0.01, *** p < 0.001. All data are representative of at least 3 independent experiments.
To further investigate the role of PRR in MCMV- mediated mClr-b downregulation, we obtained mutant fibroblasts deficient in various sensing pathways and infected them with MCMV-GFP, to monitor mClr-b expression on infected versus uninfected cells. Infection of Ifnar1−/-, Irf3−/-, Irf3−/-/Irf7−/- and Tbk1−/-/Ikke−/- primary fibroblasts demonstrated that these cells retained mechanisms to downregulate mClr-b upon MCMV-GFP infection (online suppl. fig. 7), albeit to different extents. Similarly, fibroblasts deficient in several innate immune sensors (Pkr−/-, RnaseL−/-, Pkr−/-/RnaseL−/-, Asc−/-, Caspase 1−/-, Nlrp3−/-, Aim2−/-, Mavs−/- and Dai/Zbp-1−/-) all retained the ability to downregulate mClr-b upon MCMV-GFP infection. It should be noted that initial experiments using primary Dai/Zbp-1−/- AEFs revealed a significant block in mClr-b downregulation by MCMV-GFP; however, this significance was lost upon extended cell culture, suggesting that complex mechanistic redundancy may also be possible. Interestingly, the MCMV-dependent loss of mClr-b on inflammasome mutants (Asc−/-, Caspase-1−/-, Nlrp3−/- and Aim2−/-) and nucleic acid sensing mutants (Pkr−/-/RNaseL−/-, Mavs−/- and Dai/Zbp-1−/-) suggests that the autonomous action of ATP in promoting mClr-b loss may occur via a distinct mechanism other than inflammasome activation or strict PRR signaling. In other experiments, transduction of NIH3T3 cells using lentiviral shRNA vectors to knockdown Dai/Zbp-1 or Sting expression failed to prevent mClr-b downregulation upon MCMV-GFP infection (data not shown). Together, these results remain inconclusive, but suggest that there may be redundancy in innate mechanisms that trigger CMV infection-mediated mClr-b downregulation.
The observation that extracellular ATP induced a potential inflammasome-independent Clr-b loss on a number of cell lines prompted further investigation. To extend this finding, we treated NIH3T3 cells with ATP, ADP or AMP at various concentrations. Interestingly, Clr-b downregulation was observed above ∼5 mM ATP, while ADP and AMP were even more effective at lower doses; notably, the ADP effects were also bimodal, such that Clr-b loss could be titrated out at either low or high doses (online suppl. fig. 8). It should be noted that ATP treatment failed to promote loss of Clr-b nascent transcripts (data not shown), suggesting a mechanism distinct from MCMV infection.
MCMV Infection Downregulates mClr-b but Induces a Clr-b-Independent NKR-P1B Ligand
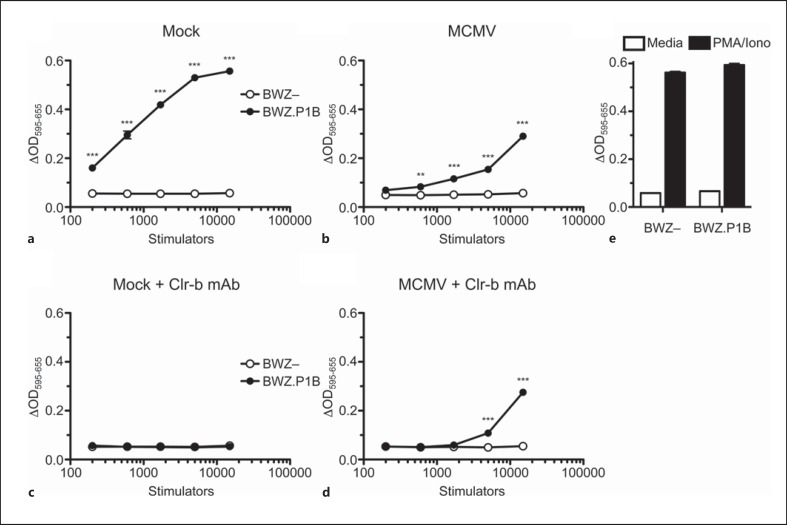
To assess the functional consequences of MCMV-mediated mClr-b loss on NKR-P1B receptor recognition, we utilized BWZ reporter cell assays [28, 29]. Briefly, BWZ.36 cells possess a LacZ gene under the control of tandem NFAT enhancer elements, such that TCR-like signals delivered via NKR-P1/CD3ζ-fusion receptors induce β- galactosidase expression, which in turn can be detected spectrophotometrically using a colorimetric substrate, CPRG. Both mAb-dependent and cellular ligand-dependent signals can be measured semiquantitatively using this assay. Here, BWZ.36 reporter cells bearing a CD3ζ/NKR-P1B chimeric fusion receptor (BWZ.P1B cells) were used to interrogate NKR-P1B-dependent ligand function when incubated with MCMV-infected cells. As previously shown, coculture of BWZ.P1B reporter cells with titrated doses of NIH3T3 stimulator cells yielded a strong NKR-P1B-dependent response, in comparison to parental BWZ.36 (BWZ-) cells lacking the NKR-P1B receptor (fig. 7a). Upon MCMV infection of NIH3T3 stimulator cells, the signal observed for BWZ.P1B reporter cells was greatly diminished, as expected, due to MCMV-mediated mClr-b downregulation (fig. 7b). To confirm that the response of uninfected NIH3T3 cells was mClr-b specific, we utilized blocking mClr-b (4A6) mAb to abrogate the NKR-P1B:Clr-b interaction (fig. 7c). Interestingly, however, 4A6 mAb failed to block residual NKR-P1B-dependent stimulation of MCMV-infected NIH3T3 cells (fig. 7d).
Fig. 7.
BWZ.CD3ζ/NKR-P1B reporter cell analysis of ligand expression on MCMV-infected NIH3T3 fibroblasts. NIH3T3 cells were infected with MCMV at a MOI ∼0.5 PFU/cell. Infection was allowed to proceed overnight (∼18 h), and then infected cells were used as stimulator cells upon coculture with CD3ζ/NKR-P1B receptor-bearing BWZ reporter cells (vs. parental BWZ.36 cells), subsequently analyzed for β-galactosidase activity 20 h later. Stimulator cell titrations included: mock-treated control NIH3T3 cells (a), MCMV-infected NIH3T3 cells (b), mock-treated control NIH3T3 cells with 4A6 blocking Clr-b mAb (10 μg/ml; c) or MCMV-infected NIH3T3 cells with blocking 4A6 Clr-b mAb (d). e Values of experimental controls. Graphs show mean ± SEM. Experiments were analyzed using ANOVA with Bonferroni's post hoc analysis. * p < 0.05, ** p < 0.01, *** p < 0.001. All data are representative of at least 3 independent experiments. BWZ- = parental BWZ.36 cells; BWZ.P1B = BWZ cells bearing the CD3ζ/NKR-P1B ectodomain.
In parallel, we tested whether MCMV infection could induce stimulation of BWZ reporter cells bearing other CD3ζ/NKR-P1-family receptors; however, no signal was observed using reporter cells bearing NKR-P1A, NKR-P1C, NKR-P1F or NKR-P1G chimeric receptors (data not shown). These results suggest that MCMV may encode an immunoevasin that interacts with the mouse NKR-P1B receptor, similar in function to RCTL encoded by RCMV-E.
Discussion
Previous work has demonstrated that rat Clr-11 transcripts and cell surface expression are rapidly lost in response to RCMV-E infection, yet RCTL functionally replaces rClr-11 as an NKR-P1B ligand [16]. In agreement with this, similar findings were reported for mClr-b in a mouse model of orthopoxvirus (VV, ECTV) infection [15]. Here, we provide evidence for a conserved mechanistic response for both mClr-b and rClr-11 in response to cross-species CMV infection. Studies initiated using MCMV-Smith virus to infect mouse and rat fibroblasts revealed a striking loss of mClr-b, similar to the rClr-11 downregulation observed in REFs using RCMV-E infection. Further investigation using MCMV-GFP virus revealed that only infected cells (GFP+), and not uninfected ‘bystander’ cells (GFP−), downregulate mClr-b/rClr-11 cell surface expression at the single cell level. This effect was observed as early as 6 h.p.i., a time point that represents an early event in the course of CMV infection. Since new infectious MCMV virions are usually produced within 16–24 h.p.i., and given that AraC and PAA treatments used to block late CMV gene expression had no effect on mClr-b downregulation, viral replication appears to be dispensable for mClr-b loss. In addition, attempts to investigate a role for IE expression were inconclusive, as ActD treatments alone abrogated host mClr-b/rClr-11 expression on uninfected cells.
However, observations using CHX as well as UV-inactivated whole CMV virions suggest that interference with expression of IE to early viral genes and/or host genes during MCMV infection is sufficient to prevent mClr-b loss, yet active viral infection is necessary to promote mClr-b downregulation. These findings demonstrate a pivotal role of IE to early gene CMV infection events in the initiation of the cellular ‘missing-self’ Clr-b response. Notably, we have observed a similar response using other viruses in vitro, including RNA viruses (data not shown); thus, loss of inhibitory Clr is a conserved response to infection by many types of viruses (and cellular pathologies, including genotoxic stress and oncogenic transformation) [14, 15, 16]. Since both RCMV and MCMV appear to have evolved functional surrogate ligands for NKR-P1B, immunoevasins that counteract the infection-mediated loss of mClr-b/rClr-11, the ‘missing-self’ phenomenon observed may be a host response; however, we cannot rule out a complex role for necessary viral genes in eliciting host Clr downregulation.
On this point, we have attempted here to expand current understanding of the host intracellular innate immune response during CMV infection. We hypothesized that PRR engagement, e.g. via TLR, NLR or cytosolic nucleic acid sensors, might mimic the innate immune response to MCMV infection, evident by the observed phenotype of mClr-b downregulation. However, studies using single PRR agonists in isolation and MCMV-GFP infection studies using mutant fibroblasts suggest that simple PRR engagement may not be sufficient. These findings are perhaps not entirely surprising, since engagement of some PRR in isolation may be detrimental to the host immune response, in that uninfected bystander cells may become targets recognized by NK cells. A more refined hypothesis hints that viral PAMP or ‘infection-stress’ intermediates may be sensed intracellularly to enable a cell-autonomous ‘missing-self’ mClr-b response in infected cells alone. This response may be opposed in uninfected ‘bystander’ cells by maintaining or even upregulating ‘healthy-self’ NK cell inhibition via mClr-b (e.g. via paracrine cytokines such as interferons). In fact, plasmid DNA transfection occasionally results in subtle mClr-b loss (on both transfected and untransfected cells), while soluble factors in CMV culture supernatants reproducibly result in mClr-b upregulation on uninfected (GFP−) cells (data not shown).
However, our cytosolic PRR agonists also failed to replicate MCMV-mediated mClr-b downregulation. Indeed, a combination of PRR agonists may be required to invoke mClr-b downregulation; for example, signal-1 associated with TLR ligation (e.g. via TLR3, 7, 8 and 9), followed by signal-2 associated with inflammasome activation (e.g. via Nlrp3 or AIM2) [35]. However, attempts to provide TLR stimulation in combination with cytosolic DNA agonists resulted in no alterations in mClr-b expression (data not shown). Alternatively, cytosolic DNA may be detected by one sensor, yet further processing or signaling intermediates may be generated by another cofactor, such as the synergy observed between cGAS and Ddx41 [32, 33, 34]. Nonetheless, transfection of cyclic-di-AMP or c-GAMP into NIH3T3 cells alone resulted in no changes in mClr-b expression (data not shown). Hence, our efforts to find a single PRR trigger have proven ineffective.
Another possibility is that signal initiation may occur in the nucleus. Intricate details of interplay during host-pathogen interactions continue to be discovered, such as recent findings using HCMV, whereby host IFI16 directly recognizes HCMV genomic DNA, yet HCMV circumvents viral detection via pUL83 [36]. It remains to be determined whether a nuclear sensor or repressor (host or viral) may be involved in the cessation of mClr-b expression, but estimates of surface mClr-b turnover put the half-life at ∼3 h, in line with the observed rapid CMV-mediated mClr-b loss.
Perhaps not surprisingly, cellular responses to stimulation using PRR agonists also appeared to be cell-type specific. For example, responses of J774 macrophages and S17 stromal cells deviated from observations using NIH3T3 and primary fibroblast cell lines, in that mClr-b was found to be upregulated in response to many PRR agonists; however, some of these effects may be due to differential sensitivity to or production of type-I IFN (data not shown). Nonetheless, even when induced upon MCMV-GFP infection (e.g. J774 macrophages), mClr-b levels still remained lower on infected (GFP+) versus uninfected (GFP−) cells. Of relevance, however, is that rodent Clr induction on hematopoietic cells during infection is reminiscent of observations regarding the closest human homolog, LLT-1 (CLEC2D; ligand for human inhibitory NKR-P1A/KLRB1); LLT1 is induced at the cell surface upon activation of immune cells [15, 16, 37, 38]. This opposing regulation could be due to differential PRR expression, cell-type-specific ligand regulation in immune versus somatic cells or inherent differences between the mouse and human homologs. Further work is required to elucidate the in vivo significance of mClr-b and other Clec2d family members with respect to viral infections.
In contrast to other PRR agonists, ATP (and perhaps nigericin, in certain cells) appears to be uniquely capable of invoking mClr-b loss in the absence of other treatments. The significance of this remains unknown, although these compounds may induce signaling events that intersect with pathways that promote mClr-b downregulation, such as cellular transformation or stress responses [14]. Extracellular ATP and nigericin can induce inflammasome activation [35]; however, all of the inflammasome-mutant MEF investigated (Asc−/-, Caspase-1−/-, Nlrp3−/- and Aim2−/-) retained the ability to downregulate mClr-b in response to MCMV-GFP infection; therefore, further work is required to decipher the role of inflammasome activation and the specific role of the adenosine phosphates (ATP, ADP and AMP). An intriguing possibility is AMP kinase, which is regulated by all three forms.
The conserved mClr-b and rClr-11 downregulation observed in response to MCMV and RCMV infection of mouse and rat fibroblasts, along with the observation that cross-species infection and other virus infections promote the same phenotype, suggests that this phenomenon may be an innate host response to viral infection, in order to signal recognition by NK cells. Notably, the mClr-b (Clec2d) and rClr-11 (Clec2d11) gene products both contain discontinuous hammerhead ribozyme (HHR) motifs in their 3′UTR [39, 40] capable of mediating posttranscriptional regulation of transcript size and abundance in cis, akin to micro-RNA (miR)-mediated regulation in trans. We and others [L. Horan, pers. commun.] have confirmed that the presence of the HHR motif does affect mRNA stability and abundance (data not shown); however, whether it provides a constitutive turnover mechanism (to enable rapid shutoff) or if it is differentially regulated under conditions of health versus stress has yet to be resolved. Of note, the human LLT1 (CLEC2D) gene product lacks the HHR motif; however, it is still subject to posttranscriptional regulation via alternative splicing [39, 41] (O.A.A., unpubl. results).
Importantly, these studies also provide evidence that there is conservation of CMV strategies to subvert NK receptor-ligand interactions, since RCMV-E infection of mouse fibroblasts invokes downregulation of mClr-b, MHC-I molecules and Rae-1 isoforms. This implies that RCMV-E may encode immunoevasin genes that counterregulate, retain intracellularly or facilitate the degradation of Clr, MHC-I and NKG2D ligands, as shown for several MCMV and HCMV gene products. In addition, MCMV is capable of cross-species downregulation of rClr-11, and may encode a viral immunoevasin that functionally engages the mouse NKR-P1B inhibitory receptor akin to the RCMV-E RCTL gene product that engages rat NKR-P1A/B alleles [16, 25]. As MCMV infection stimulated the engagement of NKR-P1B-bearing BWZ reporter cells that was not blocked using mClr-b (4A6) mAb, this phenomenon appears to be mClr-b independent. It has been previously been shown that MCMV encodes gene products that target inhibitory receptors on NK cells by mimicking or stabilizing MHC-I expression (m157, m144 and m04) [18, 20, 42]. In light of our findings, MCMV may also target MHC-I-independent ‘missing-self’ recognition. Mining the MCMV genome will undoubtedly reveal the nature of this mechanism, and this work is currently underway using mutant mice and MCMV strains. Further investigations into the expression and modulation of human NKR-P1A:LLT1 (KLRB1:CLEC2D) interactions by HCMV and poxviruses [15, 16, 37, 38] are also warranted. Together, these results further reveal the importance of the NKR- P1:Clr receptor-ligand system in innate self-nonself discrimination, particularly in the context of viral infection.
Disclosure Statement
We declare that none of the authors have any competing financial interests.
Supplementary Material
Supplementary data
Acknowledgements
We would like to thank Dr. A.P. Makrigiannis for providing MCMV-Smith virus and Dr. S.M. Vidal for providing MCMV-GFP virus. This work was supported by an operating grant from the Canadian Institutes of Health Research (CIHR 106491), an Early Researcher Award from the Ontario Ministry of Research and Innovation and an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. O.A.A. was supported by a post-graduate scholarship-doctoral award from the Natural Sciences and Engineering Research Council of Canada (NSERC). A.M. was supported by a Vanier Canada graduate scholarship doctoral award from the CIHR. J.R.C. was supported by a CIHR New Investigator Award and an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
References
- 1.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 2.Raulet DH, Vance RE. Self-tolerance of natural killer cells. Nat Rev Immunol. 2006;6:520–531. doi: 10.1038/nri1863. [DOI] [PubMed] [Google Scholar]
- 3.Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 4.Kumar V, McNerney ME. A new self: MHC-class-I-independent natural-killer-cell self-tolerance. Nat Rev Immunol. 2005;5:363–374. doi: 10.1038/nri1603. [DOI] [PubMed] [Google Scholar]
- 5.Kirkham CL, Carlyle JR. Complexity and diversity of the NKR-P1:Clr (Klrb1:Clec2) recognition systems. Front Immunol. 2014;5:214. doi: 10.3389/fimmu.2014.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlyle JR, Mesci A, Fine JH, Chen P, Belanger S, Tai LH, Makrigiannis AP. Evolution of the Ly49 and NKRP1 recognition systems. Semin Immunol. 2008;20:321–330. doi: 10.1016/j.smim.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Mesci A, Ljutic B, Makrigiannis AP, Carlyle JR. NKR-P1 biology: from prototype to missing self. Immunol Res. 2006;35:13–26. doi: 10.1385/IR:35:1:13. [DOI] [PubMed] [Google Scholar]
- 8.Chen P, Belanger S, Aguilar OA, Zhang Q, St-Laurent A, Rahim MM, Makrigiannis AP, Carlyle JR. Analysis of the mouse 129-strain Nkrp1-Clr gene cluster reveals conservation of genomic organization and functional receptor-ligand interactions despite significant allelic polymorphism. Immunogenetics. 2011;63:627–640. doi: 10.1007/s00251-011-0542-8. [DOI] [PubMed] [Google Scholar]
- 9.Kveberg L, Dai KZ, Inngjerdingen M, Brooks CG, Fossum S, Vaage JT. Phylogenetic and functional conservation of the NKR-P1F and NKR-P1G receptors in rat and mouse. Immunogenetics. 2011;63:429–436. doi: 10.1007/s00251-011-0520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leibelt S, Friede ME, Rohe C, Gutle D, Rutkowski E, Weigert A, Kveberg L, Vaage JT, Hornef MW, Steinle A. Dedicated immunosensing of the mouse intestinal epithelium facilitated by a pair of genetically coupled lectin-like receptors. Mucosal Immunol. 2015;8:232–242. doi: 10.1038/mi.2014.60. [DOI] [PubMed] [Google Scholar]
- 11.Kveberg L, Sudworth A, Todros-Dawda I, Inngjerdingen M, Vaage JT. Functional characterization of a conserved pair of NKR-P1 receptors expressed by NK cells and T lymphocytes in liver and gut. Eur J Immunol. 2015;45:501–512. doi: 10.1002/eji.201444710. [DOI] [PubMed] [Google Scholar]
- 12.Carlyle JR, Jamieson AM, Gasser S, Clingan CS, Arase H, Raulet DH. Missing self-recognition of Ocil/Clr-b by inhibitory NKR-P1 natural killer cell receptors. Proc Natl Acad Sci USA. 2004;101:3527–3532. doi: 10.1073/pnas.0308304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iizuka K, Naidenko OV, Plougastel BF, Fremont DH, Yokoyama WM. Genetically linked C-type lectin-related ligands for the NKRP1 family of natural killer cell receptors. Nat Immunol. 2003;4:801–807. doi: 10.1038/ni954. [DOI] [PubMed] [Google Scholar]
- 14.Fine JH, Chen P, Mesci A, Allan DS, Gasser S, Raulet DH, Carlyle JR. Chemotherapy-induced genotoxic stress promotes sensitivity to natural killer cell cytotoxicity by enabling missing-self recognition. Cancer Res. 2010;70:7102–7113. doi: 10.1158/0008-5472.CAN-10-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams KJ, Wilson E, Davidson CL, Aguilar OA, Fu L, Carlyle JR, Burshtyn DN. Poxvirus infection-associated downregulation of C-type lectin-related-b prevents NK cell inhibition by NK receptor protein-1b. J Immunol. 2012;188:4980–4991. doi: 10.4049/jimmunol.1103425. [DOI] [PubMed] [Google Scholar]
- 16.Voigt S, Mesci A, Ettinger J, Fine JH, Chen P, Chou W, Carlyle JR. Cytomegalovirus evasion of innate immunity by subversion of the NKR-P1b:Clr-b missing-self axis. Immunity. 2007;26:617–627. doi: 10.1016/j.immuni.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Ettinger J, Geyer H, Nitsche A, Zimmermann A, Brune W, Sandford GR, Hayward GS, Voigt S. Complete genome sequence of the English isolate of rat cytomegalovirus (Murid herpesvirus 8) J Virol. 2012;86:13838. doi: 10.1128/JVI.02614-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanier LL. Evolutionary struggles between NK cells and viruses. Nat Rev Immunol. 2008;8:259–268. doi: 10.1038/nri2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SH, Girard S, Macina D, Busa M, Zafer A, Belouchi A, Gros P, Vidal SM. Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat Genet. 2001;28:42–45. doi: 10.1038/ng0501-42. [DOI] [PubMed] [Google Scholar]
- 20.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 21.Lodoen M, Ogasawara K, Hamerman JA, Arase H, Houchins JP, Mocarski ES, Lanier LL. NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J Exp Med. 2003;197:1245–1253. doi: 10.1084/jem.20021973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lodoen MB, Abenes G, Umamoto S, Houchins JP, Liu F, Lanier LL. The cytomegalovirus m155 gene product subverts natural killer cell antiviral protection by disruption of H60-NKG2D interactions. J Exp Med. 2004;200:1075–1081. doi: 10.1084/jem.20040583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krmpotic A, Hasan M, Loewendorf A, Saulig T, Halenius A, Lenac T, Polic B, Bubic I, Kriegeskorte A, Pernjak-Pugel E, Messerle M, Hengel H, Busch DH, Koszinowski UH, Jonjic S. Nk cell activation through the NKG2D ligand Mult-1 is selectively prevented by the glycoprotein encoded by mouse cytomegalovirus gene m145. J Exp Med. 2005;201:211–220. doi: 10.1084/jem.20041617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenac T, Budt M, Arapovic J, Hasan M, Zimmermann A, Simic H, Krmpotic A, Messerle M, Ruzsics Z, Koszinowski UH, Hengel H, Jonjic S. The herpesviral Fc receptor Fcr-1 down-regulates the NKG2D ligands Mult-1 and H60. J Exp Med. 2006;203:1843–1850. doi: 10.1084/jem.20060514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voigt S, Sandford GR, Ding L, Burns WH. Identification and characterization of a spliced C-type lectin-like gene encoded by rat cytomegalovirus. J Virol. 2001;75:603–611. doi: 10.1128/JVI.75.2.603-611.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henry SC, Schmader K, Brown TT, Miller SE, Howell DN, Daley GG, Hamilton JD. Enhanced green fluorescent protein as a marker for localizing murine cytomegalovirus in acute and latent infection. J Virol Methods. 2000;89:61–73. doi: 10.1016/s0166-0934(00)00202-0. [DOI] [PubMed] [Google Scholar]
- 27.Brune W, Hengel H, Koszinowski UH. A mouse model for cytomegalovirus infection. Curr Protoc Immunol. 2001 doi: 10.1002/0471142735.im1907s43. chapt 19:unit 19.7. [DOI] [PubMed] [Google Scholar]
- 28.Mesci A, Carlyle JR. A rapid and efficient method for the generation and screening of monoclonal antibodies specific for cell surface antigens. J Immunol Methods. 2007;323:78–87. doi: 10.1016/j.jim.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Chen P, Mesci A, Carlyle JR. Cellisa: reporter cell-based immunization and screening of hybridomas specific for cell surface antigens. Methods Mol Biol. 2011;748:209–225. doi: 10.1007/978-1-61779-139-0_15. [DOI] [PubMed] [Google Scholar]
- 30.Wagner M, Gutermann A, Podlech J, Reddehase MJ, Koszinowski UH. Major histocompatibility complex class I allele-specific cooperative and competitive interactions between immune evasion proteins of cytomegalovirus. J Exp Med. 2002;196:805–816. doi: 10.1084/jem.20020811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tokuyama M, Lorin C, Delebecque F, Jung H, Raulet DH, Coscoy L. Expression of the RAE-1 family of stimulatory NK-cell ligands requires activation of the PI3K pathway during viral infection and transformation. PLoS Pathog. 2011;7:e1002265. doi: 10.1371/journal.ppat.1002265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, Liu ZJ, Modlin RL, Liu YJ, Cheng G. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol. 2012;13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM. Differential activation of the inflammasome by caspase-1 adaptors ASC and IPAF. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 36.Li T, Chen J, Cristea IM. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe. 2013;14:591–599. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Germain C, Meier A, Jensen T, Knapnougel P, Poupon G, Lazzari A, Neisig A, Hakansson K, Dong T, Wagtmann N, Galsgaard ED, Spee P, Braud VM. Induction of lectin-like transcript 1 (LLT1) protein cell surface expression by pathogens and interferon-gamma contributes to modulate immune responses. J Biol Chem. 2011;286:37964–37975. doi: 10.1074/jbc.M111.285312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosen DB, Cao W, Avery DT, Tangye SG, Liu YJ, Houchins JP, Lanier LL. Functional consequences of interactions between human NKR-P1A and its ligand LLT1 expressed on activated dendritic cells and B cells. J Immunol. 2008;180:6508–6517. doi: 10.4049/jimmunol.180.10.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–902. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott WG, Martick M, Chi YI. Structure and function of regulatory RNA elements: ribozymes that regulate gene expression. Biochim Biophys Acta. 2009;1789:634–641. doi: 10.1016/j.bbagrm.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 41.Germain C, Bihl F, Zahn S, Poupon G, Dumaurier MJ, Rampanarivo HH, Padkjaer SB, Spee P, Braud VM. Characterization of alternatively spliced transcript variants of Clec2d gene. J Biol Chem. 2010;285:36207–36215. doi: 10.1074/jbc.M110.179622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Babic M, Pyzik M, Zafirova B, Mitrovic M, Butorac V, Lanier LL, Krmpotic A, Vidal SM, Jonjic S. Cytomegalovirus immunoevasin reveals the physiological role of ‘missing self’ recognition in natural killer cell-dependent virus control in vivo. J Exp Med. 2010;207:2663–2673. doi: 10.1084/jem.20100921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data