Abstract
Guanylate binding protein (GBP) 5 belongs to the GBP family, which is involved in important cellular processes, including signal transduction, translation, vesicle trafficking, and exocytosis. Structurally, GBPs display a high degree of homology and share highly conserved GTP-binding or hydrolysis domains. GBP5 was reported to be a critical cellular factor in inflammasome assembly. However, little is known about its role in the host antiviral innate immune response. In this study, we found that GBP5 expression was significantly elevated in influenza patients and influenza A virus-infected A549 human lung epithelial cells. The overexpression of GBP5 inhibited virus replication by enhancing the expression of virus-induced interferon (IFN) and IFN-related effectors. Knockdown of GBP5 had the opposite effect. Moreover, GBP5 enhanced endogenous IFN expression by interacting with the NF-κB-essential modulator complex and stimulating NF-κB signaling. Additionally, the expression of proinflammatory factors, such as IL-6, IL-8, tumor necrosis factor-α, cyclooxygenase-2, and inducible nitric oxide synthase, was also activated by GBP5. Taken together, our results reveal that GBP5 inhibited virus replication through the activation of IFN signaling and proinflammatory factors.
Keywords: Cyclooxygenase-2, Guanylate binding protein 5, Influenza A virus, Interferon signaling, NF-κB-essential modulator
Introduction
Influenza A virus (IAV) is a negative-sense RNA virus of the Orthomyxoviridae family, which is the etiological agent of a contagious acute respiratory disease. Some seasonal and rare pandemic virus strains, such as the deadly 1918 H1N1 virus, belong to this family. These viruses are responsible for severe illness and considerable mortality worldwide. Annually, influenza viruses cause 3–5 million severe clinical infections and 250,000–500,000 fatal cases [1, 2, 3]. The outcome of IAV infections is largely determined by the complex interactions between the virus and the innate immune system; therefore, a detailed understanding of the molecular mechanisms involved in innate immune recognition and response can provide a valuable framework for the development of therapeutics [4].
During IAV infection, the first line of defense is formed by the innate immune system, which is quick but lacks specificity and memory. Innate immunity is formed by physical barriers and innate cellular immune responses [5]. IAV infection results in the recognition of PAMPs (pathogen-associated molecular patterns) by PRRs (pattern recognition receptors) and initiation of antiviral signaling cascades, eventually leading to the induction of interferons (IFNs), cytokines, and chemokines [6, 7]. So far, 3 types of IFNs have been identified [8]. Type I IFNs include IFN-α and IFN-β, which play an important role in limiting viral replication [9, 10]. Type I IFNs secreted by infected cells act on IFN-α/β receptors (IFN-α/βR) of the same cell or neighboring cells, activating an antiviral signaling cascade that is responsible for the transcription of over 300 genes that encode antiviral proteins, thus establishing an antiviral state in the cell to limit viral replication [11]. IFN-γ contributes to the establishment of an effective adaptive cytotoxic T cell (CTL) response against the influenza virus infection. IFN-γ regulates virus-specific CTL homeostasis in secondary lymph nodes and subsequent trafficking of CTLs to the site of infection [12]. Type III IFNs also control IAV infections in the lung [13].
Cyclooxygenase-2 (COX-2) is a critical inflammatory cytokine that plays a role in the response to IAV infection [14, 15]. COX-2 is an inducible COX isoform that is required for the biosynthesis of prostaglandins, such as PGE2, and is primarily involved in pathological processes, such as inflammation, fibrogenesis, and carcinogenesis. A variety of stimuli, including cytokines, mitogens, and growth factors, promote COX-2 induction at the transcriptional level [16, 17].
Guanine nucleotide binding proteins (GBPs) are involved in important cellular processes, including signal transduction, translation, vesicle trafficking, and exocytosis [18]. Structurally, GBPs have a molecular weight of 67–73 kDa, display a high degree of homology, and share highly conserved GTP-binding or hydrolysis domains [19, 20]. These GTPases bind phosphoinositides, cardiolipin, soluble NSF attachment protein receptor adaptor proteins, and other p47 IRGs (immunity-related guanosine triphosphatases) to direct their membrane regulatory activities against compartmentalized bacteria and protozoa [21, 22, 23]. Among them, guanylate binding protein 5 (GBP5) belongs to the family of IFN-γ-induced p65 GTPases, which has 7 members in humans [24]. This family of GBPs was originally identified by the ability of its members to bind to immobilized guanine nucleotides with similar affinities for GTP, GDP, and GMP [25, 26]. GBP5 was reported to be a critical cellular factor in inflammasome assembly. Furthermore, GBP5 has been reported to induce inflammatory cytokines, including IL-1β and IL-18, through the activation of NLRP3 and AIM2 [27, 28]. Recently, the research of Kirchhoff and colleagues [29] has shown that GBP5 is expressed in the primary target cells of HIV-1, where it impairs viral infectivity by interfering with the processing and virion incorporation of the viral envelope glycoprotein.
Murine GBPs have been confirmed to be coordinately regulated by IFN-γ and play an important role in host defense against bacterial pathogen infection [30], but few reports have mentioned the relationship between GBP5 and virus infection. Here, we investigated the role of GBP5 during viral infection to determine the mechanism of GBP5-mediated antiviral activity and the effects of cytokines regulated by GBP5.
Materials and Methods
Clinical Samples
Throat swab samples were collected from 19 IAV-infected patients and 18 healthy individuals in phosphate-buffered saline (PBS) and stored at 4°C for less than a week before use. These clinical samples were submitted to the Hubei Provincial Center for Disease Control and Prevention. The collection of clinical samples for this research was conducted according to the principles of the Declaration of Helsinki and approved by the Institutional Review Board of the College of Life Sciences, Wuhan University, in accordance with its guidelines for the protection of human subjects. Written informed consent was obtained from each participant in this study.
Cell Culture and Virus
Human lung epithelial cells (A549) were cultured in F12K medium. MRC5 and rhabdomyosarcoma (RD) cells were cultured in MEM medium. Human embryonic kidney (HEK-293T) and Huh7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM). All media were supplemented with 10% fetal bovine serum (FBS). All cell cultures were maintained at 37°C in a 5% CO2 incubator. The influenza virus strain A/Hong Kong/498/97 (H3N2) and A/Yamagata/120/86 (H1N1) used in this study was provided by the China Center for Type Culture Collection. The human enterovirus 71 (EV71) strain (GenBank accession No. JN230523.1) and hepatitis B virus (HBV) expression plasmid pHBV-1.3 (genotype D, serotype ayw, U95551) were reported previously [31, 32].
Isolation of PBMCs and ATII Cells
PBMCs (peripheral blood mononuclear cells) were isolated by density centrifugation diluted 1:1 in an isolation solution of human lymphocytes (TBD-science) as described previously [33]. PBMCs were washed twice with PBS and cultured in RPMI 1640 medium at 37°C in a 5% CO2 incubator. Human ATII (alveolar type II) cells were purchased from Wuhan PriCells Biotechnology & Medicine Co. Ltd., and cultured in RPMI 1640 medium. The ATII cells were infected with IAV at a multiplicity of infection (MOI) of 1 and harvested at 24 h postinfection (hpi) for real-time PCR.
Transfection and Luciferase Assay
Cells were plated at a density of 4 × 105 per plate and grown to 80% confluence prior to the transfection. They were transfected with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) for 24 h, serum starved for an additional 24 h, and then harvested for analysis. A Renilla luciferase reporter assay system (Promega, Madison, WI, USA) was used to measure the luciferase activity of each sample 48 h after transfection. Renilla luciferase activities were determined as internal controls for transfection efficiency.
Plasmids, Antibodies, Chemical Reagents, and Inhibitors
The coding regions of GBP5 were amplified from template cDNA from the Han Jiahuai laboratory of Xiamen University. The GBP5 fragment containing the KOZAK sequence was then inserted into the KpnI/BamHI sites of p3×FLAG-CMV10 to generate an expression plasmid. The I kappa B kinase (IKK) β, TBK1, NEMO (NF-κB-essential modulator), MDA5, RIG-I (retinoic acid-inducible gene I), and IRAK1 expression plasmids with an HA tag in pKH3 were generated in our lab. The NF-κB promoter, IFN-β promoter, and ISRE promoter were provided by Prof. Hongbing Shu of Wuhan University, China. A fragment of the GBP5 promoter (-800 to +200) was amplified from human genomic DNA and subcloned into the pGL3-promoter vector (Promega). COX-2 wild-type and mutant promoters in pGL3-Basic were previously described [34].
Antibodies against Influenza A NS1 antibody, OAS, PKR, IFN regulatory factor 3 (IRF3), NF-κB/p65, NF-κB/p50, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibodies against GBP5 (OriGene Technologies Inc., Rockville, MD, USA), EV71-VP1 (Abnova, Taipei City, Taiwan), lamin A (Epitomics), FLAG (MBL International Co. Ltd, Nagoya, Japan), HA (Sigma-Aldrich, St. Louis, MO, USA), and COX-2 (Cayman Chemical, Ann Arbor, MI, USA) were also used.
MTT Method
Methylthiazolyldiphenyl-tetrazolium bromide (5 mg/mL) was dissolved in PBS (0.01 M, pH 7.4) and stirred with a constant-temperature magnetic stirrer for 30 min. The solution was then passed through a 0.22-μm microfiltration membrane to remove bacteria and was kept for no longer than 2 weeks. A549 cells were seeded in a 96-well culture plate at a density of 104/cm2 and then 200 μL of DMEM containing 10% FBS was added to each well. After the cells had been cultured for 24 h, they were transfected with the GBP5 expression plasmid or vector. After 3 days, the cell culture plate was taken out for the MTT assay. We added 20 μL of the MTT reagent to each well and then cultured the cells for 4 h. Subsequently, the medium was removed and 150 μL of DMSO was added to each well to dissolve the formazan. The 96-well culture plate was agitated for 10 min on a shaker. Finally, a well with DMSO but without cells was used to adjust the zero value, and the optical density (OD) value of each well was determined at 490 nm.
Western Blot, Nuclear Extraction, and ELISA
Whole-cell lysates were prepared by lysing cells in PBS containing 0.01% Triton X-100, 0.01% EDTA, and 10% protease inhibitor mixture (Roche, Indianapolis, IN, USA). To separate and collect the cytosolic and nuclear protein fractions, cells were washed with ice-cold PBS and collected by centrifugation. The resulting pellets were resuspended in hypotonic buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 0.5 mM dithiothreitol [DTT], 10% protease mixture inhibitor) for 15 min on ice and were vortexed for 10 s. Nuclei were pelleted by centrifugation at 13,000 g for 1 min and the pellets and cytosolic protein-containing supernatants were collected. The protein concentration of each sample was determined using a Bradford assay kit (Bio-Rad, Hercules, CA, USA). A 100-μg aliquot of each sample was subjected to 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis and was transferred to a nitrocellulose membrane (Amersham). Blots were blocked with nonfat dry milk prior to incubation with primary and secondary antibodies. Bands were detected using the Super Signal chemiluminescent reagent (Pierce, Rockford, IL, USA).
Standard ELISA kits were used to quantify HBV E and S antigen (HBeAg, HBsAg) levels (Shanghai Kehua Biotechnology, Shanghai, China). Standard ELISA kits were used to quantify IFNα, β and λ1 protein levels (Neobioscience, Shenzhen, China).
Reverse-Transcription Reaction and Quantitative Real-Time PCR Analysis
Total cellular RNA was extracted with TRIzol (Invitrogen). Cellular RNA samples were reverse transcribed with random primers at 37°C for 1 h. Relative quantitative real-time PCR was performed using SYBR Select Master Mix (Applied Biosystems, Waltham, MA, USA) and a LightCycler 480 system (Roche) with primers for the indicated target genes. The primers are listed in online supplementary Table 1 (for all online suppl. material, see www.karger.com/doi/10.1159/000460294).
Measurement of IAV Replication
A549 cells were infected with IAV at an MOI of 1 as previously described [35]. Viral titers were measured by a hemagglutination assay in U-shaped plates at various time points postinfection, as previously described [36]. IAV nucleoprotein (NP) RNA was reverse transcribed with the following primers: for specific viral RNA (NP-vRNA), 5′-CTCACCGAGTGACATCAACATCATG- 3′; for NP-cRNA, 5′-AGTAGAAACAAGGGTATTTTTCTTTAATTGTCAT-3′, and for NP mRNA, oligo(dT). Relative levels of IAV RNA were measured by quantitative real-time PCR as previously described [37].
Immunofluorescence
After transfection, cells were fixed with 4% paraformaldehyde for 15 min, washed 3 times with PBS, permeabilized with PBS containing 0.5% Triton X-100 for 5 min, washed 3 times with PBS, and blocked with PBS containing 4% BSA for 1 h at room temperature. Next, the cells were incubated with the primary antibody overnight at 4°C, followed by incubation with secondary antibodies (ProteinTech Group) for 1 h. Mounting was done with vectashield mounting medium with DAPI (Vector Laboratories), and the cells were visualized by confocal laser microscopy (FLUOVIEW FV1000; Olympus, Tokyo, Japan).
Nuclear Extraction
Cells were incubated in serum-free media for 24 h, washed twice with cold PBS, and scraped into 1 mL of cold PBS, followed by centrifugation at 12,000 g for 10 min in a microcentrifuge. The cell pellet was then resuspended in 2 packed cell volumes of buffer A (10 mM HEPES, pH 8, 0.5% NP-40, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, and 200 mM sucrose) and incubated for 10 min on ice with flipping of the tube. Nuclei were collected by centrifugation (12,000 g for 15 s). Pellets were rinsed with buffer A, resuspended in buffer B (20 mM HEPES, pH 7.9, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, and 1.0 mM DTT), and incubated on a rocking platform for 30 min (4°C). Nuclei were clarified by centrifugation (12,000 rpm for 15 min), and the supernatants were diluted 1:1 with buffer C (20 mM HEPES, pH 7.9, 100 mM KCl, 0.2 mM EDTA, 20% glycerol, and 1 mM DTT). Cocktail protease inhibitor was added to each type of buffer. Nuclear extracts were stored at −70°C until use.
Coimmunoprecipitation
Coimmunoprecipitation analysis was performed as previously described [38]. Briefly, after cell treatment, cells were lysed using lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.5% (vol/vol) Nonidet-P40, 1 mM EDTA, 30 mM NaF, 2 mM sodium pyrophosphate, 10 mg/mL aprotinin, and 10 mg/mL leupeptin). Lysates were mixed and precipitated overnight at 4°C with antibodies or IgG and protein G-agarose beads. Beads were washed 5 times with lysis buffer and bound proteins were separated by SDS-PAGE with subsequent immunoblotting analysis.
Chromatin Immunoprecipitation Assay
A549 cells were incubated for 24 h after transfection (if transfection was necessary), after which they were serum starved for an additional 24 h. One percent formaldehyde was added to the culture medium, and the cells were washed twice with PBS, scraped, and lysed in buffer containing 1% SDS, 10 mM Tris-HCl, pH 8.0, and protease inhibitor cocktail for 10 min on ice. The lysates were sonicated on ice and the debris was removed by centrifugation at 12,000 g for 15 min at 4°C. One-fourth of the supernatant was used as a DNA input control. The remaining sample was diluted 10-fold with dilution buffer (0.01% SDS, 1% Triton X-100, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0, and 150 mM NaCl) followed by incubation with antibodies against NF-κB/p50, CREB1, or IgG overnight at 4°C. Immunoprecipitated complexes were collected using protein A/G-sepharose. The pellets were washed 4 times with dialysis buffer containing 2 mM EDTA and 50 mM Tris-HCl, pH 8.0. After washing, the precipitates were incubated in an elution buffer (1% SDS and 0.1 M NaHCO3) at room temperature. Supernatants were transferred to clean tubes, and RNase A was added to destroy bound RNA in the sample. Samples were incubated at 65°C for 5 h to reverse the formaldehyde crosslinks, and DNA was precipitated with ethanol and extracted twice with phenol/chloroform. Pellets were suspended in TE buffer and subjected to PCR amplification using the promoter detection primers. The primers are listed in online supplementary Table 2.
Statistical Analysis
The relationship between GBP5 expression and the IAV gene NP level in the clinical samples was analyzed by the Pearson correlation coefficient. All other experiments were performed in triplicate. Two group comparisons were analyzed by the Student t test, and the data are expressed as means with standard deviations. p < 0.05 was considered statistically significant.
Results
GBP5 Expression Is Elevated in Patients Infected with IAV
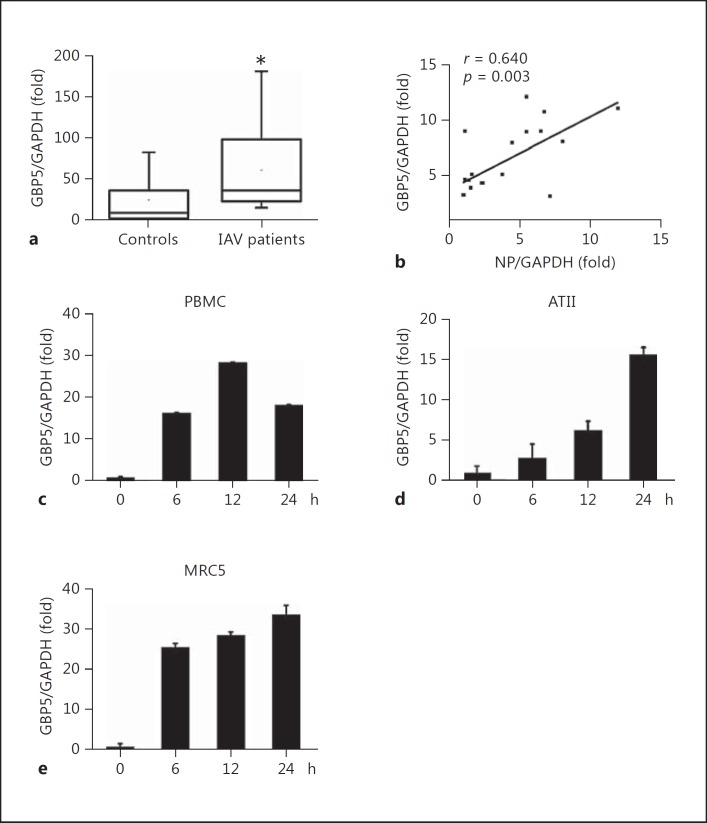
In order to investigate the potential role of GBP5 during IAV infection, we first determined whether GBP5 expression was induced by IAV infection by isolating total mRNA from throat swab samples of patients infected with IAV (n = 19) and healthy individuals (n = 18). GBP5 mRNA expression levels were significantly higher in IAV patients than in healthy individuals (60.08 ± 45.38 vs. 25.87 ± 31.25, respectively, p < 0.05; Fig. 1a).
Fig. 1.
Determination of GBP5 levels in patients infected with IAV and healthy individuals. a Total RNA was extracted from freshly isolated throat swabs from healthy individuals (n = 18) or IAV patients (n = 19). GBP5 mRNA was quantified by qRT-PCR. The sample with the lowest GBP5 expression level in healthy individuals served as a reference value. b Correlation analysis between NP mRNA and GBP5 mRNA (n = 19) levels in throat swabs from IAV patients. Pearson correlation analysis was used to determine R values; Student t test was used to determine p values. c PBMCs were infected with IAV A/Hong Kong/498/97 (H3N2) at an MOI of 1 for the indicated times. GBP5 mRNA was quantified by qRT-PCR. d Human primary alveolar epithelial type II (ATII) cells were infected with A/Hong Kong/498/97 (H3N2; MOI = 1) for the indicated times. e MRC5 cells were infected with A/Hong Kong/498/97 (H3N2; MOI = 1) for the indicated times. GBP5 mRNA was quantified by qRT-PCR. Bar graphs represent means ± SD; n = 3.
Furthermore, to verify whether the GBP5 expression was related to the virus infection, the relationship between the IAV NP mRNA and GBP5 mRNA levels was determined by Pearson correlation analysis. A statistically significant correlation was observed between NP mRNA and GBP5 expression in patients with seasonal IAV (NP and GBP5: r = 0.6397, p < 0.01; Fig. 1b). Thus, GBP5 expression was positively correlated with IAV infection in the IAV patients.
Next, primary cultured cells were tested. Freshly isolated PBMCs, ATII and MRC5 cells were infected with influenza virus A/HongKong/498/97 (H3N2) at an MOI of 1 and harvested at the indicated time points, and the GBP5 mRNA level was measured by qRT-PCR. The results showed that IAV infection induced GBP5 expression in primary cultured cells (Fig. 1c-e).
GBP5 Is Upregulated by IAV Infection
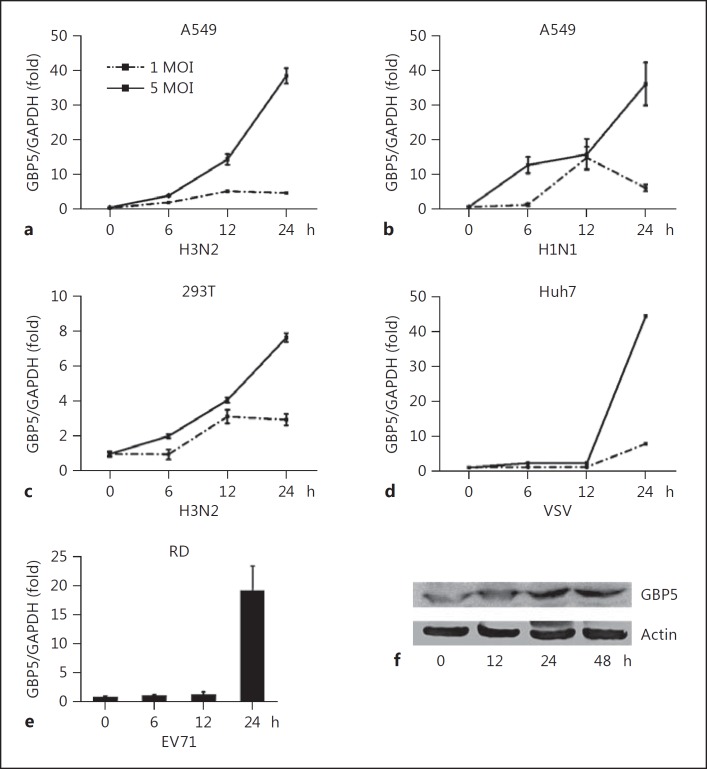
Because GBP5 expression correlated with IAV infection in influenza patients and primary cultured cells, we investigated the expression of GBP5 in different cell cultures stimulated by viral infection. The A549 human lung epithelial cell line was infected with H3N2 at an MOI of 1 or 5 and harvested at the indicated time points, and GBP5 mRNA expression levels were assessed by qRT-PCR. As shown in Figure 2a, GBP5 mRNA expression was induced by IAV infection in a time-dependent manner (Fig. 2a). Then, the A549 was infected with H1N1 at an MOI of 1 or 5 and harvested at the indicated time point, and GBP5 mRNA expression levels were assessed by qRT-PCR. The result also showed that GBP5 mRNA expression was induced by H1N1 infection in a time-dependent manner (Fig. 2b).
Fig. 2.
Induction of GBP5 by IAV in different cultured cells. a A549 cells were infected with H3N2 at an MOI of 1 or 5, and the cells were harvested at the indicated time points. GBP5 mRNA was quantified by qRT-PCR. b A549 cells were infected with H1N1 at an MOI of 1 or 5 for the indicated times. GBP5 mRNA was quantified by qRT-PCR. c 293T cells were infected with H3N2 with an MOI of 1 or 5 for the indicated times. GBP5 mRNA was quantified by qRT-PCR. d Huh7 cells were infected with VSV with an MOI of 1 or 5 for the indicated times. GBP5 protein was quantified by Western blot. e RD cells were infected with EV71 at an MOI of 1, and the cells were harvested at the indicated time points. GBP5 mRNA was quantified by qRT-PCR. f A549 cells were infected with H3N2 at an MOI of 1, and the cells were harvested at the indicated time points. The GBP5 protein level was quantified by Western blot. All experiments were repeated at least 3 times with similar results. Bar graphs represent means ± SD; n = 3 (** p < 0.01; * p < 0.05).
The same experiments were also conducted in the 293T cells and a similar expression pattern was observed (Fig. 2c). While in Huh7, vesicular stomatitis virus (VSV) also showed this kind of expression pattern (Fig. 2d). Since a high concentration of EV71 would lead to the apoptosis of RD cells, so only 1 MOI EV71 were used to infected the RD cell, and turned to induce GBP5 in a time-depend manner (Fig. 2e). GBP5 protein levels were also measured by Western blot and followed the same trend in A549 cells infected by H3N2 (Fig. 2f).
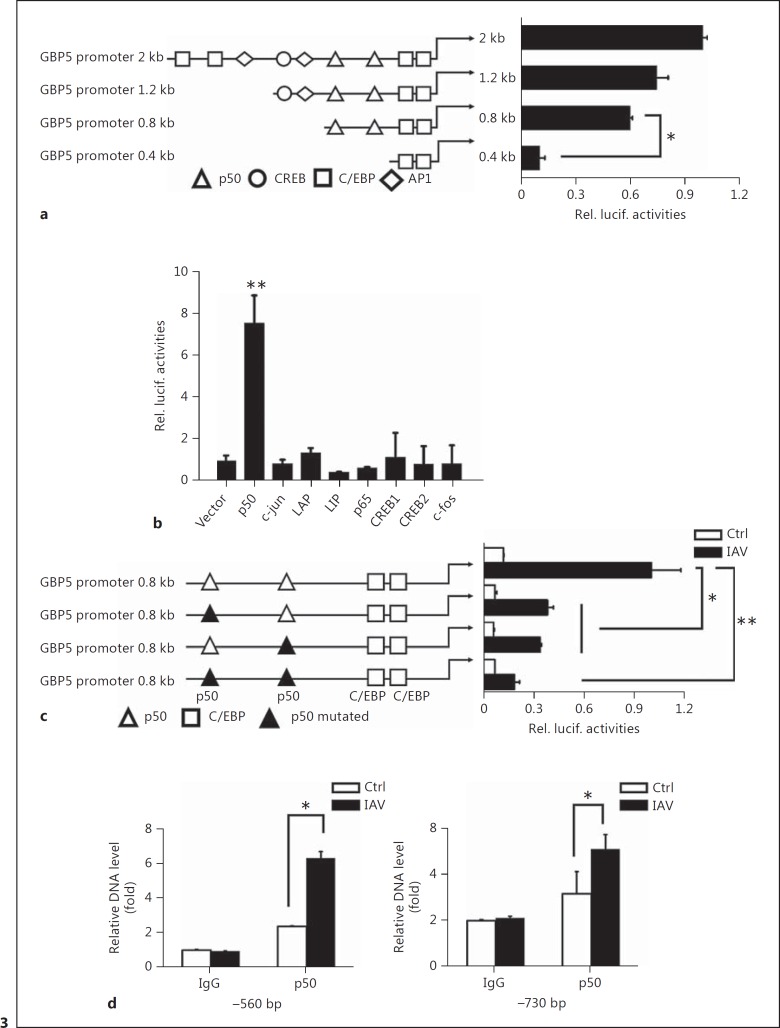
NF-κB/p50 Plays a Role in IAV-Induced GBP5 Promoter Activation
To further investigate the mechanism that regulates GBP5 expression, we generated truncation promoter reporters in which different truncated promoters were cloned upstream of the luciferase gene. To determine promoter activity, the reporter constructs were transfected into A549 cells following IAV infection. However, no significant change in activity was observed until the 2 p50 binding sites were deleted (Fig. 3a), suggesting that p50 may be an important transcription factor that regulates GBP5 expression.
Fig. 3.
NF-κB/p50 plays a role in IAV-induced GBP5 promoter activation. a A549 cells were transfected with luciferase reporter plasmids containing the wild-type GBP5 promoter or with plasmids containing truncation mutants of the GBP5 promoter, along with IAV infection (MOI = 1). Luciferase activities were measured in each sample after serum starvation for 24 h and were compared to those obtained from control cells transfected with empty vector. All data were normalized to the Renilla luciferase value. b A549 cells were transfected with luciferase reporter plasmids containing the wild-type GBP5 promoter along with the different transcription factor expression plasmids. c A549 cells were transfected with luciferase reporter plasmids containing the wild-type GBP5 promoter or with plasmids containing mutants of the GBP5 promoter, along with IAV infection (MOI = 1). Luciferase activities were measured in each sample after serum starvation for 24 h and were compared to those obtained from control cells transfected with empty vector. All data were normalized to the Renilla luciferase value. d A549 cells were infected with IAV (MOI = 1) for 12 h and ChIP assays were performed using 2 μg of anti-p50. Normal rabbit IgG was used as a control. Immunoprecipitated DNA or control DNA was collected and amplified using specific primers for the NF-κB binding site in the GBP5 promoter. All experiments were repeated at least 3 times with similar results. Bar graphs represent means ± SD; n = 3 (** p < 0.01; * p < 0.05).
In order to test which transcription factor correlated with the expression of GBP5, a GBP5 full promoter reporter was cotransfected along with different transcription factor expression plasmids or a control vector into 293T cells. A series of transcription factors were tested, including p50, c-Jun, LAP, LIP, p65, CREB1, CREB2, and c-Fos. The results showed that GBP5 promoter activity can be significantly enhanced by p50 (Fig. 3b). Furthermore, p50 shRNA or control shRNA were transfected into A549 cells to verify the effect of p50 on endogenous GBP5 expression. The results showed that knockdown of p50 by specific shRNA decreased the expression of GBP5 (online suppl. Fig. S1A)
Then, to confirm whether the NF-κB binding sites play an important role in the regulation of GBP5 production upon IAV infection, 2 possible NF-κB binding sites were mutated individually and the mutant constructs were transfected into A549 cells as described above to examine the promoter activity in response to IAV infection. The data indicated that each single site mutant could decrease the promoter activity significantly and the dual mutant totally suppressed the promoter activity. These results prove that both of the NF-κB binding sites on the promoter contribute to the induction of GBP5 (Fig. 3c).
Next, we performed chromatin immunoprecipitation (ChIP) in A549 cells to verify the binding of NF-κB to the GBP5 promoter. We found that p50 bound to both sites in the GBP5 promoter. Furthermore, more p50 was bound to these sites in IAV-infected cells than in control cells. Together, our data demonstrate that NF-κB is an important cis-regulatory element required for the activation of the GBP5 promoter by IAV (Fig. 3d).
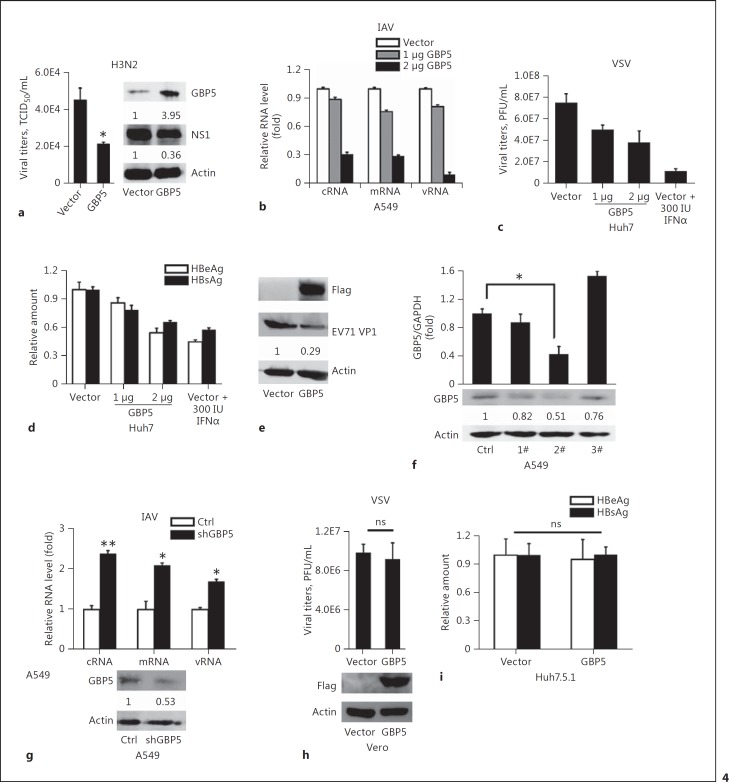
Expression of GBP5 Inhibits Virus Replication
Since the production of GBP5 can be induced by IAV, we asked whether GBP5 influences IAV replication. A549 cells were transfected with the GBP5 expression plasmid or the control plasmid, and 12 h later the cells were infected with IAV (MOI = 1). The cells were then harvested 24 hpi, and the protein expression was detected by Western blot. The results showed that the decrease of viral titer was observed (Fig. 4a, graph) and the IAV NS1 was reduced significantly following GBP5 overexpression (Fig. 4a, blot). The viral RNA were then tested. A549 cells were transfected with the GBP5 expression plasmid or the control plasmid, and 12 h later the cells were infected with IAV (MOI = 1). The cells were harvested for RNA extraction 24 hpi. Viral NP gene expression levels, including vRNA, cRNA, and mRNA, were measured by qRT-PCR. The expression of GBP5 led to significant decreases in the expression of all 3 types of viral RNA examined (Fig. 4b).
Fig. 4.
Antiviral activity of GBP5. a A549 cells were transfected with vector or GBP5 expression plasmid for 24 h, then the cells were infected with IAV (MOI = 1) and harvested at 24 hpi. Viral titers in the culture supernatants were quantified by a TCID50 test (left). Protein level of GBP5 and NS1 were measured by Western blot (right). b A549 cells were transfected with the GBP5 expression plasmid with 1 or 2 μg for 24 h, then the cells were infected with IAV (MOI = 1) and harvested at 24 hpi. Relative levels of NP-specific mRNA, cRNA, and vRNA were measured by qRT-PCR. c A549 cells were transfected with the p3×FLAG-GBP5 plasmid with 1 or 2 μg for 24 h, then the cells were infected with VSV (MOI = 1), and treated with IFNα (300 IU/ml) as a positive control. After 24 h, the supernatant was harvested for plaque experiment in Vero cells. d Huh7 cells were transfected with pHBV1.3 along with the GBP5 expression plasmid with 1 or 2 μg for 48 h, and treated with IFNα (300 IU/mL) as a positive control. HBeAg and HBsAg in the supernatant were then measured by ELISA. e RD cells were transfected with the GBP5 expression plasmid for 24 h, then the cells were infected with EV71 (MOI = 1) and harvested at 8 hpi. EV71 VP1 protein was then measured by Western blot. f A549 cells were transfected with shNC or specific shRNA for GBP5, and GBP5 levels were measured by qRT-PCR and Western blot. g A549 cells were transfected with shGBP5 2# for 24 h, then the cells were infected with IAV (MOI = 1) and harvested at 6 hpi. Relative levels of NP-specific mRNA, cRNA, and vRNA were measured by qRT-PCR and the protein level of GBP5 was measured by Western blot. h Vero cells were transfected with the p3×FLAG-GBP5 plasmid for 24 h, then the cells were infected with VSV (MOI = 1). After 24 h, the supernatant was harvest for plaque experiments in Vero cells. i Huh7.5.1 cells were transfected with pHBV1.3 along with the GBP5 expression plasmid for 48 h. HBeAg and HBsAg in the supernatant were then measured by ELISA. All experiments were repeated at least 3 times with similar results. Bar graphs represent means ± SD; n = 3 (** p < 0.01; * p < 0.05).
To test whether GBP5 exerts a universal antiviral function, we also assessed the effects of GBP5 on the replication of different viruses, including VSV, HBV, and EV71. VSV replication was determined by plaque formation after virus infection in Huh7 cells. The results showed that viral titers were significantly lower in GBP5-overexpressing cells than that in control cells (Fig. 4c). HBV1.3 was cotransfected with the GBP5 expression plasmid or the control plasmid into Huh7 cells to assess HBV protein (HBeAg and HBsAg) expression in the culture supernatant. The result showed that GBP5 also inhibited HBV expression (Fig. 4d). Furthermore, RD cells were infected by EV71 after transfection with the GBP5 expression plasmid or the control plasmid. The cells were then harvested 12 hpi, and the protein expression was detected by Western blot. The results showed that the viral protein VP1 was reduced significantly following GBP5 overexpression (Fig. 4e).
Next, specific shRNAs for GBP5 were constructed, and shGBP5 2# had a high efficiency for knocking down GBP5 at both the mRNA and protein levels (Fig. 4f). Then, shGBP5 2# was used in antiviral assays against IAV infection in A549 cells. The results showed that higher levels of viral NP mRNA, cRNA, and vRNA were observed after knockdown of GBP5 (Fig. 4g).
To test whether the antiviral function of GBP5 depends on the presence of IFN, Vero cells, a cell line lacking functional type I IFN genes [39, 40], were chosen for investigation of the antiviral function of GBP5 against VSV. VSV is a pathogen that is extremely sensitive to the action of type I IFN. When Vero cells were transfected with the GBP5 expression plasmid or the control plasmid in the presence of VSV, no significant difference in viral titer was observed (Fig. 4g). Similarly, in Huh 7.5.1, a cell line derived from Huh7 cells with a point mutation in the dsRNA sensor RIG-I [41, 42, 43], transfected with the HBV1.3 plasmid, the expression of the viral HBeAg and HBsAg proteins was not changed in the presence of GBP5 (Fig. 4h). Overall, our data indicate that GBP5 exerts a universal antiviral function in response to infection by different viruses and it is likely that the antiviral function of GBP5 depends on the presence of IFN.
GBP5 Activates IFN-Related Pathways
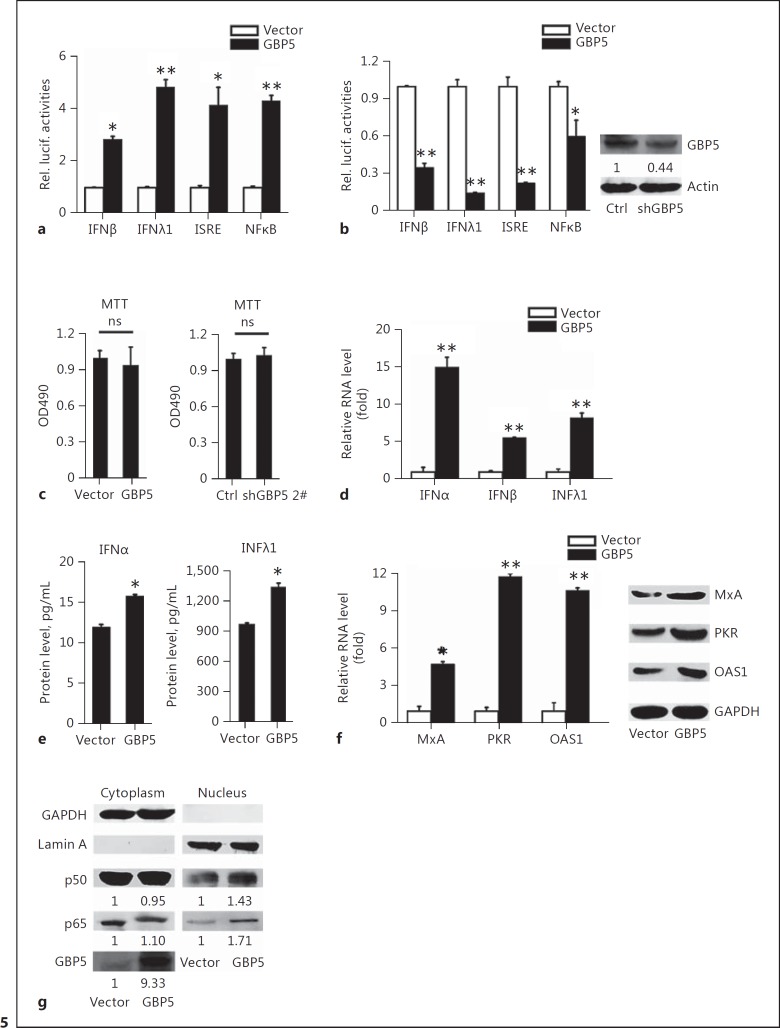
Since our results demonstrated that GBP5 can inhibit viral replication and that its antiviral activity may depend on the presence of IFN, we sought to determine the relationship between GBP5 and IFN. In IFN pathway-related reporter assays, GBP5 significantly promoted IAV-induced activation of the IFNβ, IFNλ1, ISRE, and NF-κB promoters in 293T cells (Fig. 5a). In contrast, the promoter activities of IFNβ, IFNλ1, ISRE, and NF-κB were downregulated by GBP5 shRNA (Fig. 5b). The data showed that GBP5 does not affect cell viability (Fig. 5c). The RNA levels of IFNα, IFNβ, and IFNλ1 were then detected by qRT-PCR. The data showed that GBP5 activates IFN expression in A549 cells (Fig. 5d). ELISAs of culture supernatants demonstrated that GBP5 activate the production of secreted IFNs in A549 cells (Fig. 5e). We next assessed whether GBP5 induced IFN downstream effectors. A549 cells were transfected with the GBP5 expression plasmid followed by IAV infection, and qRT-PCR and Western blot were performed to determine the levels of MxA, PKR, and OAS1 induced by IFN. As expected, GBP5 increased the mRNA and protein levels of each IFN effector examined (Fig. 5f). Furthermore, the cellular localization of transcription factors were assessed in A549 cells transfected with the GBP5 expression plasmid for 24 h. Cytosolic and nuclear fractions were prepared from GBP5-transfected cells, and Western blot analysis was performed to detect the protein distribution. Our data revealed that the levels of p65 and p50 protein within the nucleus increased in the GBP5-transfected cells (Fig. 5g). Together, our results suggest that GBP5 not only contributes to the activation of IFNs and downstream IFN effectors, but also activates the NF-κB pathway.
Fig. 5.
GBP5 activates the expression of IFNs. a The indicated luciferase promoter plasmids were transfected into 293T cells along with the GBP5 expression plasmid or a control plasmid. Luciferase assays were performed 24 h after H3N2 (MOI = 1) infection. b The indicated luciferase promoter plasmids were transfected into 293T cells along with the shGBP5 expression plasmid or a negative control. Luciferase assays were performed 24 h after H3N2 (MOI = 1) infection (left). Cells were harvested for Western blot analysis with the indicated antibodies (right). c A549 cells were transfected with the GBP5 expression plasmid or a control vector for 24 h (left), and transfected with the shGBP5 expression plasmid or a negative control vector for 24 h (right), then the cell viability was measured by the MTT assay. d A549 cells were transfected with the indicated plasmids and infected with IAV (MOI = 1) for 24 h, then the mRNA levels of IFNα, IFNβ, and IFNλ1 were measured by qRT-PCR. e A549 cells were transfected with the indicated plasmids and infected with IAV (MOI = 1) for 24 h, then the protein levels of IFNα and IFNλ1 were measured by ELISA. f A549 cells were transfected with the indicated plasmids and infected with IAV (MOI = 1) for 24 h. The protein levels of OAS1, PKR, and IRF3 were measured by Western blot and mRNA levels were measured by qRT-PCR. g A549 cells were transfected with the GBP5 expression plasmid or a control plasmid. Then, cytosolic and nuclear extracts were prepared at the indicated time points and were subjected to Western blot analysis. GAPDH and lamin A were used as markers for the cytosolic and nuclear fractions, respectively. Protein levels were measured by Western blot analysis with the indicated antibodies. All experiments were repeated at least 3 times with similar results. Bar graphs represent means ± SD; n = 3 (** p < 0.01; * p < 0.05).
GBP5 Interacts with the NEMO Complex
In humans, the innate immune system recognizes pathogens via PRRs, such as Toll-like receptors, to initiate IFN induction [44, 45]. So far, 3 types of IFNs have been identified [46]. Type I IFNs include IFN-α and IFN-β, which play an important role in limiting viral replication [47, 48]. Type I IFNs secreted by infected cells act on IFN-α/β receptors (IFN-α/βR) of the same cell or neighboring cells, and activate the transcription factor NF-κB through the IKK complex, which is composed of IKKa, IKKb, and NEMO, resulting in the induction of IFN and inflammatory cytokines [48, 49].
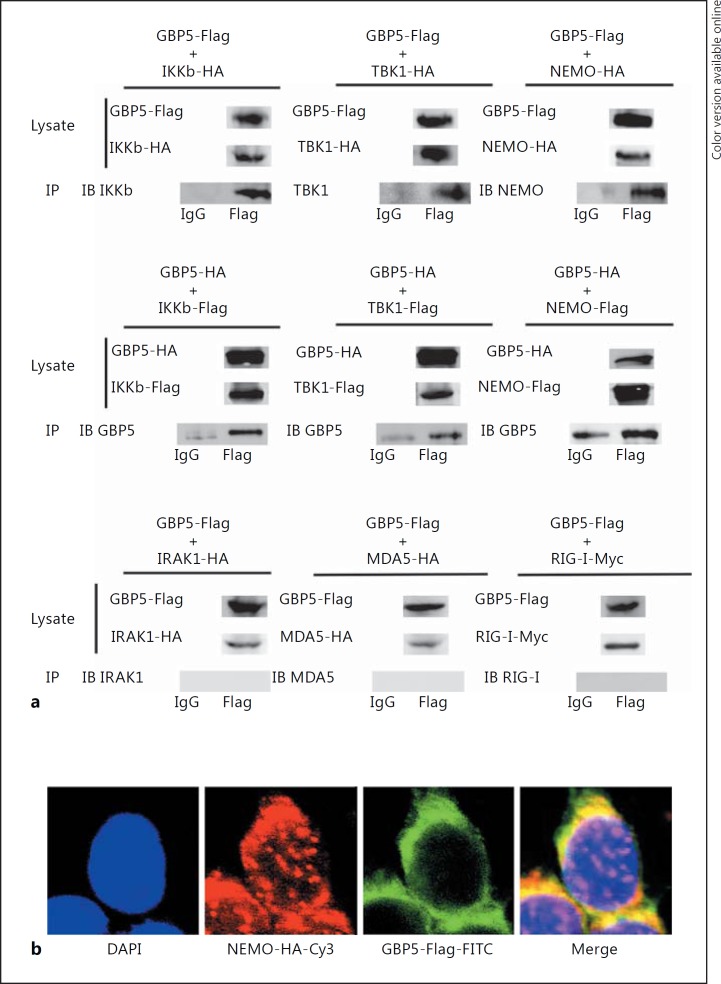
To further investigate the mechanism of GBP5-induced IFN production, a series of IFN-related factors, including NEMO, IKKβ, TBK1, RIG-I, MDA5, and IRAK1, were cloned into the indicating plasmid and screened for their interaction with GBP5 by coimmunoprecipitation. The results indicated that GBP5 interacted with NEMO, IKKβ, and TBK1, but not with IRAK1, MDA5, and RIG-I (Fig. 6a). Immunofluorescent staining experiments suggested that GBP5 and NEMO had a similar distribution pattern (Fig. 6b).
Fig. 6.
GBP5 interacts with the NEMO protein complex. a 293T cells were transfected with FLAG-tagged or HA-tagged GBP5 and the indicated plasmids tagged with HA or Flag. At 48 h posttransfection, coimmunoprecipitation and immunoblot analysis were performed with the indicated antibodies. b 293T cells were transfected with FLAG-GBP5 and HA-NEMO for 48 h prior to immunofluorescence assays.
GBP5 Induces COX-2 Activation through CREB1
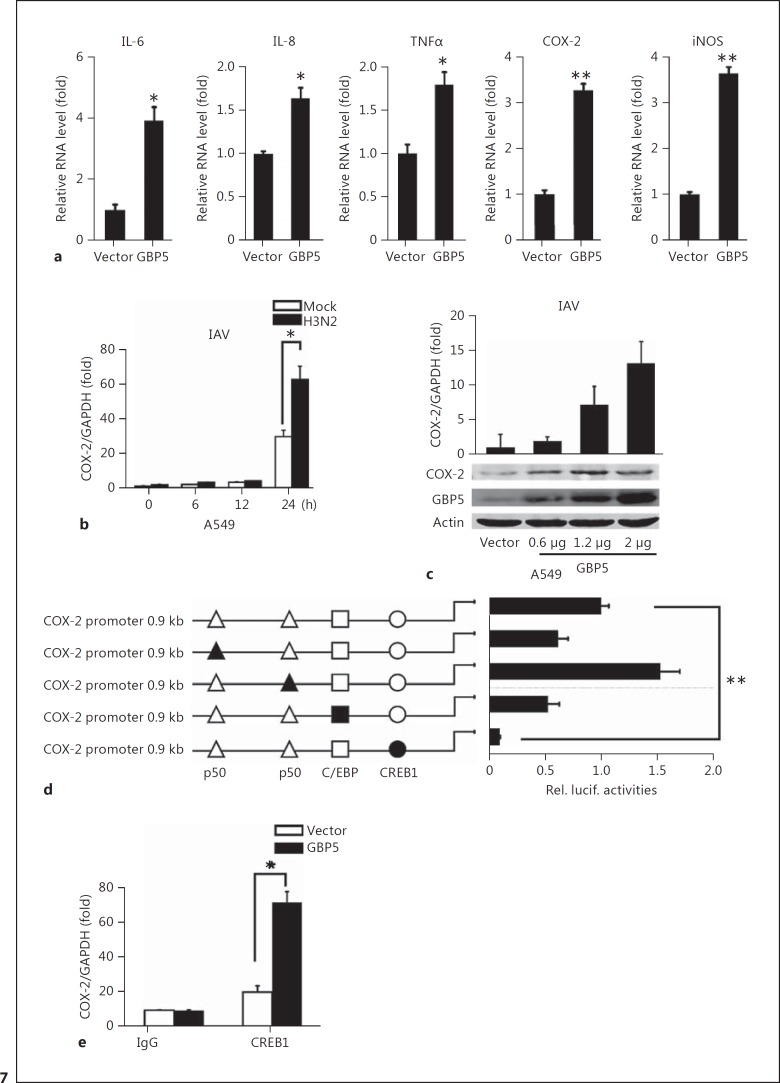
In human influenza infection, a phenomenon called “cytokine storm” can lead to acute lung injury or acute respiratory distress syndrome [50, 51]. During the storm, a large amount of inflammatory factors (e.g., TNFα, IL-6, IL-8, IFN-γ, etc.) are produced [52]. Therefore, we asked whether GBP5 affects the expression of inflammatory factors. Five proinflammatory factors, including IL-6, IL-8, TNF, COX-2, and iNOS, were examined. The results showed that all of these cellular factors were induced in the presence of GBP5 (Fig. 7a). Because COX-2 is a critical inflammatory factor related to virus infection and IFN production [53, 54], we sought to determine how GBP5 regulates COX-2 expression. First, COX-2 expression was tested at the indicated time points after transfection with the GBP5 expression plasmid. The result showed that COX-2 expression was dramatically induced by GBP5 at 24 h (Fig. 7b). Next, GBP5 was transfected in a dose gradient in A549 cells, and the expression of COX-2 was assessed after 48 h by qPCR and Western blot. The result showed that COX-2 was induced by GBP5 in a dose-dependent manner (Fig. 7c). Furthermore, to find the mechanism by which COX-2 was activated, a series of mutant COX-2 promoters were used to test the cis-regulatory elements involved. Our results showed that mutation of the CREB1 site significantly decreased COX-2 promoter activity (Fig. 7d). A549 cells were transfected with specific siRNA for CREB1 or irrelevant control siRNA to verify the effect of CREB1 on endogenous COX-2 expression. The results indicated that knockdown of CREB1 by specific siRNA decreased the expression of COX-2 (online suppl. Fig. S1B). In addition, ChIP assays confirmed that CREB1 bound to the COX-2 promoter in the presence of GBP5 (Fig. 7e).
Fig. 7.
GBP5 stimulates the expression of COX-2 during IAV infection. a A549 cells were transfected with 2 μg of GBP5 expression plasmid for 24 h, then the cells were infected with IAV (MOI = 1) and harvested at 24 hpi. Relative mRNA levels of the indicated genes were measured by qRT-PCR. b A549 cells were transfected with vector or GBP5 for 24 h, then the cells were infected with IAV (MOI = 1) and harvested at the indicated time. Relative levels of COX-2 mRNA were measured by qRT-PCR. c A549 cells were transfected with the GBP5 expression plasmid in a dose gradient for 24 h, then the cells were infected with IAV (MOI = 1) and harvested at 24 hpi. The relative level of COX-2 mRNA was measured by qRT-PCR and the protein level was measured by Western blot. d 293T cells were transfected with luciferase reporter plasmids containing the wild-type COX-2 promoter or mutant COX-2 promoter, along with IAV (MOI = 1) infection. Luciferase activities were measured in each sample after serum starvation for 24 h, and were compared to those obtained from control cells transfected with empty vector. e A549 cells were transfected with the GBP5 expression plasmid and then infected with IAV (MOI = 1) for 12 h and ChIP assays were performed using 2 μg of anti-CREB1. Normal rabbit IgG was used as a control. Immunoprecipitated DNA or control DNA was collected and amplified using specific primers for the CREB1 binding site in the COX-2 promoter. Bar graphs represent means ± SD; n = 3 (** p < 0.01; * p < 0.05).
Discussion
Here, we delineated a previously unrecognized function of GBP5 and identified cellular signaling pathways that contributed to GBP5 expression during IAV infection, as well as the antiviral mechanism of GBP5. Our data demonstrate that IAV infection induces GBP5 expression through NF-κB, which then activates the expression of IFNs, IFN effectors, and proinflammatory factors.
PBMCs are involved mainly in immune responses and play a major role in regulating host defense against microbial infections [55]. In this study, PBMCs were used to evaluate the expression of GBP5 during viral infection. These cells are widely used to research the immediate immune responses to viral and other microbial infections [55, 56, 57]. In fact, our previous study demonstrated that PBMCs infected by influenza virus are a clean and efficient model for studying the antiviral immune response [56].
GBPs are mostly reported as ISGs (IFN-stimulated genes) to exert an antibacterial protein, but there are also reports of hGBP1 interacting with IAV NS1 to antagonize antiviral activity, and also to antagonize the dengue virus [58, 59]. Murine GBP2 was reported to antagonize VSV and encephalomyocarditis virus [60]. As a member in the GBP family, it was reported that pyroptosis induced by the Salmonella enterica serovar Typhimurium can be positively regulated by prior IFN-γ stimulation, and this process is mediated by IFN-γ-induced GBP5 [30]. However, there have been few studies regarding GBP5 induction by viruses. Our results suggest that multiple viruses are able to activate the expression of GBP5. To investigate the mechanism by which GBP5 is induced by IAV, we tested the effects of 8 different transcription factors on GBP5 promoter activity. The results show that GBP5 is mainly induced by NF-κB/p50, and the binding of NF-κB to the GBP5 promoter is necessary for this induction. However, if some inhibitors exist on the GBP5 promoter, they will interfere with the result and some important transcriptional factors may not be found.
Since the induction of GBP5 is related to the viral infection, we also examined IAV replication in the presence of GBP5. Three different viruses other than IAV were tested, and the replication of all 3 viruses was suppressed. These results reveal a critical role for GBP5 in the innate immune response against viral infection. We also confirmed that GBP5 increased the expression of type I IFN downstream effectors, including OAS, PKR, and Mx [61]. Thus, GBP5 can enhance the production of IFNs and their downstream effectors. To determine the mechanism of crosstalk between GBP5 and IFN signaling, IFN pathway-related cellular proteins were screened for interaction with GBP5 by coimmunoprecipitation. Among them, the NEMO complex was shown to associate with GBP5. Because the rapid induction of type I IFN expression is key in initiating the innate antiviral response, clarification of NEMO-mediated antiviral signaling is important for understanding innate immune signaling; however, NEMO-mediated antiviral signaling is not well elucidated. Recent studies found that several ubiquitin E3 ligases are involved in the regulation of innate immune signaling [62, 63, 64, 65, 66, 67]. For this reason, it would be interesting to determine whether NEMO and GBP5 are involved in ubiquitination. More experiments need to be conducted to explore the mechanism of GBP5 ubiquitination in the future.
Recently, GBP5 was reported as a critical activator of AIM2 during infection with Francisella novicida; GBP2 and GBP5 targeted cytosolic F. novicida and promoted bacteriolysis. Thus, in addition to their roles in host defense against vacuolar pathogens, GBPs also facilitate the presentation of ligands by directly attacking cytosolic bacteria [28]. These results imply that GBP5 may be involved in the cytokine burst during inflammation. Furthermore, the inducible form of cyclooxygenase, COX-2, is induced by inflammatory stimuli, which quickly results in the biosynthesis of prostaglandins of the E-series, mainly PGE2 [53]. Reports show that COX-2 is activated by viruses such as EBV, IAV, HBV, HCV, and HSV-1 (herpes simplex virus type 1), and PGE2 has antiviral activity in the host response to these viruses [54, 68, 69, 70, 71, 72]. The induction of the COX-2 promoter is mainly facilitated by NF-κB and CREB1, which bind to and activate the promoter separately. Our results suggest that COX-2 induction is facilitated by GBP5 during IAV infection.
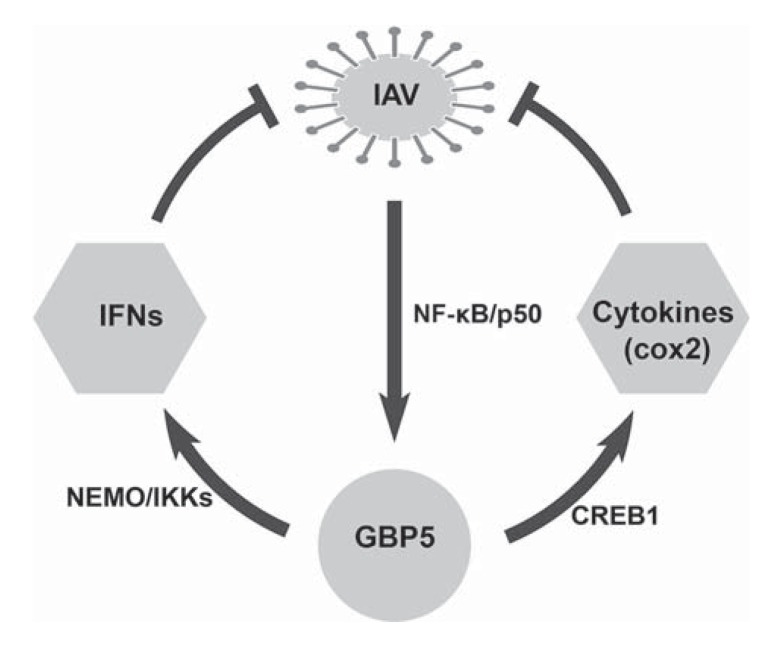
In conclusion, we demonstrated that GBP5 is a regulator of antiviral immunity that is induced in response to virus infection. GBP5 plays an important role in endogenous IFN production and exhibits antiviral activity toward viral infections in an IFN-dependent manner (Fig. 8).
Fig. 8.
A hypothetical model related to IAV infection and host response. Viral infection induces GBP5 expression through NF-κB/p50 signaling. The induction of GBP5 contributes to the activation of both IFNs and cytokine production, which in turn hamper viral replication.
Supplementary Material
Supplementary data
Supplementary data
Acknowledgments
This work was supported by research grants from the National Natural Science Foundation of China (31570870 and 81461130019), the Major State Basic Research Development Program of China (2013CB911102), Chinese 111 project (B06018). The funding agencies had no role in study design, data collection, or analysis, decision to publish, or preparation of the manuscript.
References
- 1.Noymer A, Garenne M. The 1918 influenza epidemic's effects on sex differentials in mortality in the United States. Popul Dev Rev. 2000;26:565–581. doi: 10.1111/j.1728-4457.2000.00565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annu Rev Pathol. 2008;3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stöhr K. Influenza - WHO cares. Lancet Infect Dis. 2002;2:517. doi: 10.1016/s1473-3099(02)00366-3. [DOI] [PubMed] [Google Scholar]
- 4.Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holt PG, Strickland DH, Wikström ME, Jahnsen FL. Regulation of immunological homeostasis in the respiratory tract. Nat Rev Immunol. 2008;8:142–152. doi: 10.1038/nri2236. [DOI] [PubMed] [Google Scholar]
- 6.Zhong B, Tien P, Shu HB. Innate immune responses: crosstalk of signaling and regulation of gene transcription. Virology. 2006;352:14–21. doi: 10.1016/j.virol.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 7.Sanders CJ, Doherty PC, Thomas PG. Respiratory epithelial cells in innate immunity to influenza virus infection. Cell Tissue Res. 2011;343:13–21. doi: 10.1007/s00441-010-1043-z. [DOI] [PubMed] [Google Scholar]
- 8.Chelbi-Alix MK, Wietzerbin J. Interferon, a growing cytokine family: 50 years of interferon research. Biochimie. 2007;89:713–718. doi: 10.1016/j.biochi.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 9.van de Sandt CE, Kreijtz JH, Rimmelzwaan GF. Evasion of influenza A viruses from innate and adaptive immune responses. Viruses. 2012;4:1438–1476. doi: 10.3390/v4091438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szretter KJ, Gangappa S, Belser JA, Zeng H, Chen H, Matsuoka Y, Sambhara S, Swayne DE, Tumpey TM, Katz JM. Early control of H5N1 influenza virus replication by the type I interferon response in mice. J Virol. 2009;83:5825–5834. doi: 10.1128/JVI.02144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 12.Turner SJ, Olivas E, Gutierrez A, Diaz G, Doherty PC. Disregulated influenza A virus-specific CD8+ T cell homeostasis in the absence of IFN-γ signaling. J Immunol. 2007;178:7616–7622. doi: 10.4049/jimmunol.178.12.7616. [DOI] [PubMed] [Google Scholar]
- 13.Mordstein M, Kochs G, Dumoutier L, Renauld J, Paludan S, Klucher K, Staeheli P. Interferon-λ contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 2008;4:e1000151. doi: 10.1371/journal.ppat.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogdan C. Nitric oxide synthase in innate and adaptive immunity: an update. Trends Immunol. 2015;36:161–178. doi: 10.1016/j.it.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Steer SA, Moran JM, Christmann BS, Maggi LB, Corbett JA. Role of MAPK in the regulation of double-stranded RNA- and encephalomyocarditis virus-induced cyclooxygenase-2 expression by macrophages. J Immunol. 2006;177:3413–3420. doi: 10.4049/jimmunol.177.5.3413. [DOI] [PubMed] [Google Scholar]
- 16.Smith WL, DeWitt DL, Garavito RM. Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem. 2000;69:145–182. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- 17.Inoue H, Yokoyama C, Hara S, Tone Y, Tanabe T. Transcriptional regulation of human prostaglandin-endoperoxide synthase-2 gene by lipopolysaccharide and phorbol ester in vascular endothelial cells: involvement of both nuclear factor for interleukin-6 expression site and cAMP response element. J Biol Chem. 1995;270:24965–24971. doi: 10.1074/jbc.270.42.24965. [DOI] [PubMed] [Google Scholar]
- 18.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 19.Martens S, Howard J. The interferon-inducible GTPases. Annu Rev Cell Dev Biol. 2006;22:559–589. doi: 10.1146/annurev.cellbio.22.010305.104619. [DOI] [PubMed] [Google Scholar]
- 20.Prakash B, Renault L, Praefcke GJ, Herrmann C, Wittinghofer A. Triphosphate structure of guanylate-binding protein 1 and implications for nucleotide binding and GTPase mechanism. EMBO J. 2000;19:4555–4564. doi: 10.1093/emboj/19.17.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tiwari S, Choi HP, Matsuzawa T, Pypaert M, MacMicking JD. Targeting of the GTPase Irgm1 to the phagosomal membrane via PtdIns(3,4)P2 and PtdIns(3,4,5)P3 promotes immunity to mycobacteria. Nat Immunol. 2009;10:907–917. doi: 10.1038/ni.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E, Ponpuak M, Master S, Pilli M, White E, Komatsu M, Deretic V. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol. 2010;12:1154–1165. doi: 10.1038/ncb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunn JP, Koenen-Waisman S, Papic N, Schroeder N, Pawlowski N, Lange R, Kaiser F, Zerrahn J, Martens S, Howard JC. Regulatory interactions between IRG resistance GTPases in the cellular response to Toxoplasma gondii. EMBO J. 2008;27:2495–2509. doi: 10.1038/emboj.2008.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olszewski MA, Gray J, Vestal DJ. In silico genomic analysis of the human and murine guanylate-binding protein (GBP) gene clusters. J Interferon Cytokine Res. 2006;26:328–352. doi: 10.1089/jir.2006.26.328. [DOI] [PubMed] [Google Scholar]
- 25.Cheng Y, Colonno RJ, Yin FH. Interferon induction of fibroblast proteins with guanylate binding activity. J Biol Chem. 1983;258:7746–7750. [PubMed] [Google Scholar]
- 26.Schwemmle M, Staeheli P. The interferon-induced 67-kDa guanylate-binding protein (hGBP1) is a GTPase that converts GTP to GMP. J Biol Chem. 1994;269:11299–11305. [PubMed] [Google Scholar]
- 27.Shenoy AR, Wellington DA, Kumar P, Kassa H, Booth CJ, Cresswell P, MacMicking JD. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 2012;336:481–485. doi: 10.1126/science.1217141. [DOI] [PubMed] [Google Scholar]
- 28.Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Ruhl S, Dussurgey S, Dick MS, Kistner A, Rigard M, Degrandi D, Pfeffer K, Yamamoto M, Henry T, Broz P. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol. 2015;16:476–484. doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krapp C, Hotter D, Gawanbacht A, McLaren PJ, Kluge SF, Sturzel CM, Mack K, Reith E, Engelhart S, Ciuffi A, Hornung V, Sauter D, Telenti A, Kirchhoff F. Guanylate binding protein (GBP) 5 is an interferon-inducible inhibitor of HIV-1 infectivity. Cell Host Microbe. 2016;19:504–514. doi: 10.1016/j.chom.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 30.Rupper AC, Cardelli JA. Induction of guanylate binding protein 5 by gamma interferon increases susceptibility to Salmonella enterica serovar Typhimurium-induced pyroptosis in RAW 264.7 cells. Infect Immun. 2008;76:2304–2315. doi: 10.1128/IAI.01437-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S, Peng NF, Xie J, Hao Q, Zhang M, Zhang Y, Xia Z, Xu G, Zhao F, Wang Q, Han T, Zhu Y. Human hepatitis B virus S and E antigens inhibit major vault protein signaling in interferon induction pathways. J Hepatol. 2015;62:1015–1023. doi: 10.1016/j.jhep.2014.11.035. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q, Chen X, Feng J, Cao Y, Song Y, Wang H, Zhu C, Liu S, Zhu Y. Soluble interleukin-6 receptor-mediated innate immune response to DNA and RNA viruses. J Virol. 2013;87:11244–11254. doi: 10.1128/JVI.01248-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi HJ, Dinarello CA, Shapiro L. Interleukin-18 inhibits human immunodeficiency virus type 1 production in peripheral blood mononuclear cells. J Infect Dis. 2001;184:560–568. doi: 10.1086/322805. [DOI] [PubMed] [Google Scholar]
- 34.Yue X, Yang F, Yang Y, Mu Y, Sun W, Li W, Xu D, Wu J, Zhu Y. Induction of cyclooxygenase-2 expression by hepatitis B virus depends on demethylation-associated recruitment of transcription factors to the promoter. Virol J. 2011;8:118. doi: 10.1186/1743-422X-8-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou F, Liu Y, Liu L, Wu K, Wei W, Zhu Y, Wu J. Retinoic acid activates human inducible nitric oxide synthase gene through binding of RARα/RXRα heterodimer to a novel retinoic acid response element in the promoter. Biochem Biophys Res Commun. 2007;355:494–500. doi: 10.1016/j.bbrc.2007.01.178. [DOI] [PubMed] [Google Scholar]
- 36.Mukhtar MM, Li S, Li W, Wan T, Mu Y, Wei W, Kang L, Rasool ST, Xiao Y, Zhu Y, Wu J. Single-chain intracellular antibodies inhibit influenza virus replication by disrupting interaction of proteins involved in viral replication and transcription. Int J Biochem Cell Biol. 2009;41:554–560. doi: 10.1016/j.biocel.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Li W, Liu Y, Mukhtar MM, Gong R, Pan Y, Rasool ST, Gao Y, Kang L, Hao Q, Peng G, Chen Y, Chen X, Wu J, Zhu Y. Activation of interleukin-32 pro-inflammatory pathway in response to influenza A virus infection. PLoS One. 2008;3:e1985. doi: 10.1371/journal.pone.0001985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shu H, Takeuchi M, Goeddel DV. The tumor necrosis factor receptor 2 signal transducers TRAF2 and c-IAP1 are components of the tumor necrosis factor receptor 1 signaling complex. Proc Natl Acad Sci USA. 1996;93:13973–13978. doi: 10.1073/pnas.93.24.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pauli EK, Schmolke M, Wolff T, Viemann D, Roth J, Bode JG, Ludwig S. Influenza A virus inhibits type I IFN signaling via NF-κB-dependent induction of SOCS-3 expression. PLoS Pathog. 2008;4:e1000196. doi: 10.1371/journal.ppat.1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prescott J, Hall P, Acuna-Retamar M, Ye C, Wathelet MG, Ebihara H, Feldmann H, Hjelle B. New world hantaviruses activate IFNλ production in type I IFN-deficient vero E6 cells. PLoS One. 2010;5:e11159. doi: 10.1371/journal.pone.0011159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sumpter R, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 45.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science. 2004;303:566. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 46.Kawagoe T, Sato S, Matsushita K, Kato H, Matsui K, Kumagai Y, Saitoh T, Kawai T, Takeuchi O, Akira S. Sequential control of Toll-like receptor-dependent responses by IRAK1 and IRAK2. Nat Immunol. 2008;9:684–691. doi: 10.1038/ni.1606. [DOI] [PubMed] [Google Scholar]
- 47.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel D. TRAF6 is a signal transducer for interleukin-1. Nature. 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 48.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 49.Iwasaki H, Takeuchi O, Teraguchi S, Matsushita K, Uehata T, Kuniyoshi K, Satoh T, Saitoh T, Matsushita M, Standley DM, Akira S. The IκB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-1. Nat Immunol. 2011;12:1167–1175. doi: 10.1038/ni.2137. [DOI] [PubMed] [Google Scholar]
- 50.Kaiser L, Fritz RS, Straus SE, Gubareva L, Hayden FG. Symptom pathogenesis during acute influenza: interleukin-6 and other cytokine responses. J Virol. 2001;64:262–268. doi: 10.1002/jmv.1045. [DOI] [PubMed] [Google Scholar]
- 51.Eliopoulos AG, Gallagher NJ, Blake SMS, Dawson CW, Young LS. Activation of the p38 mitogen-activated protein kinase pathway by Epstein-Barr virus-encoded latent membrane protein 1 coregulates interleukin-6 and interleukin-8 production. J Biol Chem. 1999;274:16085–16096. doi: 10.1074/jbc.274.23.16085. [DOI] [PubMed] [Google Scholar]
- 52.Finco D, Grimaldi C, Fort M, Walker M, Kiessling A, Wolf B, Salcedo T, Faggioni R, Schneider A, Ibraghimov A, Scesney S, Serna D, Prell R, Stebbings R, Narayanan PK. Cytokine release assays: current practices and future directions. Cytokine. 2014;66:143–155. doi: 10.1016/j.cyto.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 53.Harris RE, Casto BC, Harris ZM. Cyclooxygenase-2 and the inflammogenesis of breast cancer. World J Clin Oncol. 2014;5:677–692. doi: 10.5306/wjco.v5.i4.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu W, Fan Z, Han Y, Xu L, Wang M, Zhang D, Mao Y, Li J, Wang H. Activation of NF-κB signaling pathway in HSV-1-induced mouse facial palsy: possible relation to therapeutic effect of glucocorticoids. Neuroscience. 2015;289:251–261. doi: 10.1016/j.neuroscience.2014.12.062. [DOI] [PubMed] [Google Scholar]
- 55.Ronni T, Sareneva T, Pirhonen J, Julkunen I. Activation of IFN-alpha, IFN-gamma, MxA, and IFN regulatory factor 1 genes in influenza A virus-infected human peripheral blood mononuclear cells. J Immunol. 1995;154:2764–2774. [PubMed] [Google Scholar]
- 56.Liu L, Cao Z, Chen J, Li R, Cao Y, Zhu C, Wu K, Wu J, Liu F, Zhu Y. Influenza A virus induces interleukin-27 through cyclooxygenase-2 and protein kinase A signaling. J Biol Chem. 2012;287:11899–11910. doi: 10.1074/jbc.M111.308064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Netea MG, Azam T, Lewis EC, Joosten LA, Wang M, Langenberg D, Meng X, Chan ED, Yoon DY, Ottenhoff T, Kim SH, Dinarello CA. Mycobacterium tuberculosis induces interleukin-32 production through a caspase-1/IL-18/interferon-γ-dependent mechanism. PLoS Med. 2006;3:e277. doi: 10.1371/journal.pmed.0030277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu Z, Shi Z, Yan W, Wei J, Shao D, Deng X, Wang S, Li B, Tong G, Ma Z. Nonstructural protein 1 of influenza A virus interacts with human guanylate-binding protein 1 to antagonize antiviral activity. PLoS One. 2013;8:e55920. doi: 10.1371/journal.pone.0055920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan W, Zuo X, Feng T, Shi X, Dai J. Guanylate-binding protein 1 participates in cellular antiviral response to dengue virus. Virol J. 2012;9:292. doi: 10.1186/1743-422X-9-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carter CC, Gorbacheva VY, Vestal DJ. Inhibition of VSV and EMCV replication by the interferon-induced GTPase, mGBP-2: differential requirement for wild-type GTP binding domain. Arch Virol. 2005;150:1213–1220. doi: 10.1007/s00705-004-0489-2. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Q, Gong R, Qu J, Zhou Y, Liu W, Chen M, Liu Y, Zhu Y, Wu J. Activation of the Ras/Raf/MEK pathway facilitates hepatitis C virus replication via attenuation of the interferon-JAK-STAT pathway. J Virol. 2012;86:1544–1554. doi: 10.1128/JVI.00688-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gack MU, Shin YC, Joo C, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 63.Arimoto K, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci USA. 2007;104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-β induction during the early phase of viral infection. J Biol Chem. 2009;284:807–817. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- 65.Chuang TH, Ulevitch RJ. Triad3A, an E3 ubiquitin-protein ligase regulating Toll-like receptors. Nat Immunol. 2004;5:495–502. doi: 10.1038/ni1066. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka T, Grusby MJ, Kaisho T. PDLIM2-mediated termination of transcription factor NF-κB activation by intranuclear sequestration and degradation of the p65 subunit. Nat Immunol. 2007;8:584–591. doi: 10.1038/ni1464. [DOI] [PubMed] [Google Scholar]
- 67.Lin R, Yang L, Nakhaei P, Sun Q, Sharif-Askari E, Julkunen I, Hiscott J. Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. J Biol Chem. 2006;281:2095–2103. doi: 10.1074/jbc.M510326200. [DOI] [PubMed] [Google Scholar]
- 68.Li W, Yang F, Liu Y, Gong R, Liu L, Feng Y, Hu P, Sun W, Hao Q, Kang L, Wu J, Zhu Y. Negative feedback regulation of IL-32 production by iNOS activation in response to dsRNA or influenza virus infection. Eur J Immunol. 2009;39:1019–1024. doi: 10.1002/eji.200838885. [DOI] [PubMed] [Google Scholar]
- 69.Murono S, Inoue H, Tanabe T, Joab I, Yoshizaki T, Furukawa M, Pagano JS. Induction of cyclooxygenase-2 by Epstein-Barr virus latent membrane protein 1 is involved in vascular endothelial growth factor production in nasopharyngeal carcinoma cells. Proc Natl Acad Sci USA. 2001;98:6905–6910. doi: 10.1073/pnas.121016998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nunez O, Fernandez-Martinez A, Majano PL, Apolinario A, Gomez-Gonzalo M, Benedicto I, Lopez-Cabrera M, Bosca L, Clemente G, Garcia-Monzon C, Martin-Sanz P. Increased intrahepatic cyclooxygenase 2, matrix metalloproteinase 2, and matrix metalloproteinase 9 expression is associated with progressive liver disease in chronic hepatitis C virus infection: role of viral core and NS5A proteins. Gut. 2004;53:1665–1672. doi: 10.1136/gut.2003.038364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng CY, Huang WR, Chi PI, Chiu HC, Liu HJ. Cell entry of bovine ephemeral fever virus requires activation of Src-JNK-AP1 and PI3K-Akt-NF-κB pathways as well as Cox-2-mediated PGE2/EP receptor signalling to enhance clathrin-mediated virus endocytosis. Cell Microbiol. 2015;17:967–987. doi: 10.1111/cmi.12414. [DOI] [PubMed] [Google Scholar]
- 72.He L, Zhang Y, Fang Y, Liang W, Lin J, Cheng M. Classical swine fever virus induces oxidative stress in swine umbilical vein endothelial cells. BMC Vet Res. 2014;10:279. doi: 10.1186/s12917-014-0279-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data