Abstract
Microbial RNA is an important stimulator of innate immune responses. Differences in posttranscriptional RNA modification profiles enable the immune system to discriminate between self and non-self nucleic acids. This principle may be exploited by certain bacteria to circumvent immune cell activation. In this regard, 2'-O-methylation of Escherichia coli tRNATyr at position 18 (Gm18) has recently been described to inhibit TLR7-mediated IFN-a production in human plasmacytoid dendritic cells (pDCs). Extending these findings, we now demonstrate that Gm18 also potently inhibits TLR7-independent human monocyte activation by RNA derived from a variety of bacterial strains. The half minimal inhibitory concentration values were similar to those found for IFN-a inhibition in pDCs. Mechanistically, 2'-O-methylated RNA impaired upstream signalling events, including MAP kinase and NFκB activation. Our results suggest that antagonizing effects of Gm18-modified RNA are due to competition with stimulatory RNA for receptor binding. The antagonistic effect was specific for RNA because the small molecule TLR7/8 agonist R848 was not inhibited. Despite the striking phenotype in human cells, 2'-O-methylated RNA did not interfere with TLR13 activation by bacterial 23S rRNA in murine DC and BMDM. Thus, we identify here Gm18 in E. coli tRNATyr as a universal suppressor of innate immune activation in the human but not the murine system.
Key Words: Bacterial RNA, RNA modification, 2ߣ-O-methylation, Immunostimulation, Innate immunity, Peripheral blood mononuclear cells
Introduction
The innate immune system serves an important function in the early sensing and clearance of pathogens which is achieved by the recognition of conserved pathogen-associated molecular patterns by different pattern recognition receptors. Nucleic acids of both viral and bacterial origin constitute an important group of pathogen-associated molecular patterns that trigger a variety of cytosolic (RIG-I, MDA5, NLRP3, AIM2, cGAS, IFI16) and endosomal receptors (TLR3, 7, 8, 9 and in the murine system TLR13) [1, 2, 3, 4, 5, 6, 7, 8, 9]. Due to the pronounced chemical and structural similarities of host and microbial nucleic acids, discrimination between self and non-self is a fundamental but challenging task [5, 10, 11]. Three main principles for distinction of self/non-self nucleic acids have been deciphered, including subcellular compartmentalization, sequence composition and specific nucleotide modifications [12]. In the case of RNA, more than 100 different modifications have been identified that are all introduced posttranscriptionally [13]. Importantly, the extent and kind of nucleotide modifications incorporated varies significantly depending on the RNA species and its evolutionary origin. Eukaryotic RNA is more abundantly modified than prokaryotic RNA, and tRNAs contain most modifications [13]. Initial studies investigating the influence of RNA modifications on immunostimulation discovered that the incorporation of base modifications (m5C, m5U, s2U, m6A, pseudouridine) or 2'-O-methylation at the ribose into in vitro transcripts or siRNAs efficiently prevented TLR activation [14, 15, 16, 17]. While the influence of RNA modifications on immunostimulation was mostly investigated using artificial oligonucleotides with random incorporation of different modifications [15, 16], 2 recent studies addressed the role of RNA modifications in naturally occurring bacterial tRNA species. They demonstrated that most bacterial tRNAs can trigger the production of IFN-α in human pDCs in a TLR7-dependent manner [12, 18]. By contrast, a few bacterial tRNA species including Escherichia coli tRNATyr failed to do so. Further analysis identified a single 2'-O-methylation of guanosine at position 18 (Gm18) as a structural determinant being necessary and sufficient for immunosilencing [12]. This modification occurs regularly in eukaryotic RNA and is only rarely found in bacterial RNA (bRNA). Notably, Gm18 did not only render tRNAs non-stimulatory, but also acted as a TLR7 antagonist, i.e. it actively suppressed immune activation by an otherwise stimulatory tRNA species [12, 18]. The exact mechanism underlying the dominant inhibitory effect of Gm18 in E. coli tRNATyr remains poorly defined. Moreover, most studies investigating the effect of 2'-O-methylation on innate immune responses in human peripheral blood mononuclear cells (PBMCs) relied on the measurement of IFN-α as a commonly used read-out for TLR7-dependent pDC activation [12, 15, 19]. It is as yet unclear if the 2'-O-methyl modification also exerts a dominant inhibitory effect on other immune cells including human monocytes that do not express TLR7 [20] but are known to produce high amounts of pro-inflammatory cytokines (e.g. IL-12p40 and TNF) in response to microbial RNA.
In the present study we demonstrate that immunosilencing by naturally occurring 2'-O-methylated RNA is not restricted to TLR7 in pDCs, but also inhibits the secretion of pro-inflammatory cytokines in human monocytes. By contrast, 2'-O-methylated RNA did not impair activation of TLR13 in murine cells, indicating strictly species-specific inhibitory patterns. Notably, the antagonistic effect of 2'-O-methylation was stimulus specific as only bRNA, but no further TLR ligands, including the small molecule TLR7/8 agonist R848, could be suppressed. Further experiments suggest a direct binding of 2'-O-methylated RNA to TLR7, thus disrupting its interaction with stimulatory RNA.
Materials and Methods
Reagents
RPMI 1640 containing stable glutamine and DMEM were purchased from Biochrom (Berlin, Germany), FCS was obtained from Gibco (Darmstadt, Germany), Ficoll (1.078 g/ml) from Pan Biotech (Aidenbach, Germany), Lipofectamine 2000 and TRIzol from Invitrogen Life Technologies (Darmstadt, Germany), Pam3CSK4, R848 and CpG 2216 from Invivogen (San Diego, Calif., USA) and DOTAP [N-(1-[2,3-dioleoyloxy]propyl)N,N,N-trimethylammonium methylsulphate] from Roth (Karlsruhe, Germany). Ultrapure lipopolysaccharide (LPS) from Salmonella minnesota was a gift from U. Seydel (Borstel, Germany). HEK293-TLR7HA cells were purchased from Invivogen (Toulouse, France), anti-haemagglutinin (HA) beads from Sigma-Aldrich (Saint-Louis, Mo., USA), streptavidin beads from Thermo Scientific (Rockford, Ill., USA) and anti-HA antibody from Santa Cruz (Heidelberg, Germany). Primer sequences for quantitative real-time PCR were as follows: β-actin, fw 5′-CCC TGT GCT TGG CTT CAC CGA-3′, rev 5′-ACA GTG TGG GTG ACC CCG TTC-3′; TNF, fw 5′-GCC CAG GCA GTC AGA TCA TCT TC-3′, rev 5′-TGA GGT ACA GGC CCT CTG ATG G-3′; IL-12p40, fw 5′-GCG GAG CTG CTA CAC TCT C-3′, rev 5′-CCA TGA CCT CCA TGG GCA GAC-3′, and IRF7, fw 5′-CCC AGC AGG TAG CAT TCC C-3′, rev 5′-GCA GCA GTT CCT CCG TGT AG-3′. All primers were custom synthesized by MWG-Biotech (Ebersberg, Germany).
Oligoribonucleotides Used in the Study
RNA sequences used for stimulation experiments were the 2'-O-methylated tRNA fragment 5′-GGU GGG GUU CCC GAG CGmG CCA AAG GGA-3′ and the unmodified control tRNA fragment 5′-GGU GGG GUU CCC GAG CGG CCA AAG GGA-3′ (custom synthesized from Biomers, Ulm, Germany). RNA sequences used for co-immunoprecipitation experiments were biotinylated RNA 5′-GCA AGC UGA CCC UGA AGU UCA U-Biotin-3′ (Biomers, Ulm, Germany) and RNA corresponding in sequence to unmodified E. coli tRNATyr or its Gm18 variant 5′-GGU GGG GUU CCC GAG CG(m)G CCA AAG GGA GGG AGC AGA CUG UAA AUC UGC CGU CAC UCA CAG ACU UCG AAG GUU CGA AUC CUU CCU UCC CCC ACC ACC A-3′. Splinted ligation was performed by annealing 3 synthetic fragments of this RNA (IBA, Göttingen, Germany) as described previously [21]. Where indicated, RNA was biotinylated at the 3′ end. The ORN ssRNA40 (5′-GCC CGU CUG UUG UGU GAC UC-3′) was custom made by MWG-Biotech (Ebersberg, Germany).
Isolation and Stimulation of Human Immune Cells
Human PBMCs were isolated from heparinized blood of healthy donors upon informed consent by standard Ficoll-Hypaque density gradient centrifugation (Ficoll 1.078 g/ml) [15]. PBMCs were resuspended in RPMI 1640 supplemented with 10% heat-inactivated FCS. CD14-positive monocytes were isolated by positive selection using immunomagnetic cell separation (MACS-Microbeads, Miltenyi Biotech, Germany) according to the manufacturer's instructions in a semi-automated manner via an AutoMACS device. The purity of the CD14-positive cells exceeded 95% as confirmed by FACS analysis. For transfection experiments, bRNA was encapsulated with DOTAP at a ratio of 3 µl DOTAP/1 µg RNA in serum-free medium according to the manufacturer's protocol. Where indicated, 2'-O-methylated inhibitory RNA was mixed with bRNA prior to encapsulation with DOTAP. Reverse transfection was performed at a density of 4 × 105 cells/well (PBMCs) or 2 × 105 cells/well (CD14 positive cells) in a 96-well flat bottom plate. Cells were incubated overnight in a humidified 5% CO2 atmosphere at 37°C. As positive control, PBMCs were stimulated with TLR9-specific stimulus CpG2216 (1 µM) or TLR7/8-agonist R848 (1 μg/ml).
Bacteria Strains
The following microbial strains were used: Staphylococcus aureus (ATCC 25923), erythromycin-resistant S. aureus (patient isolate), Streptococcus agalactiae (ATCC 27956), Bacillus subtilis (ATCC 6051), Bifidobacterium bifidum (ATCC 29521), Helicobacter pylori (patient isolate), Campylobacter jejuni (patient isolate).
Murine Cell Isolation, Differentiation and Transfection
Bone marrow GM-CSF-derived myeloid DCs and BMDMs were prepared from 8- to 12-week-old mice as described previously [22]. DCs and BMDM were stimulated at a density of 2 × 105/well in antibiotic-free RPMI 1640 or DMEM, respectively, supplemented with 10% FCS. Cells were transfected with total RNA derived from S. aureus alone or together with 2'-O-methylated RNA complexed with Lipofectamine 2000 at a ratio of 1 µl Lipofectamine 2000/1 µg RNA. Cell culture supernatants were harvested after overnight incubation for cytokine measurement.
HEK-BlueTM Cells
HEK-Blue™ cells stably co-expressing human TLR8 together with an NF-κB-inducible secreted embryonic alkaline phosphatase (SEAP) reporter gene (InvivoGen, Toulouse, France) were cultured in DMEM supplemented with 10% heat-inactivated FCS. Cells were stimulated with ssRNA40 (1.5 µg/ml) in the presence or absence of the tRNA fragment (1.5 µg/ml) encapsulated with DOTAP. Stimulation was performed in HEK-Blue™ detection medium according to the manufacturer's instructions. HEK-Blue™ detection medium contains a specific SEAP substrate that produces a blue colour upon hydrolysis by the secreted SEAP reporter protein. The optical density at 620 nm was measured after overnight incubation using a microplate reader.
Preparation of Total Bacterial RNA and Bacterial tRNA
The different bacterial strains were grown in Luria-Bertani medium or brain heart infusion broth (Merck, Darmstadt, Germany) and harvested within the mid-log phase growth. After a digestion step with lysozyme (1 h at 40 mg/ml), total bRNA was isolated using TRIzol reagent according to the manufacturer's protocol. The obtained RNA underwent a further purification step using the RNeasy mini kit (QIAGEN, Hilden, Germany), including an on-column DNA digestion step. The purity of the RNA preparations was validated by determining the 260/230 nm and 260/280 nm extinction ratio by NanoDrop (Thermo Scientific). The tRNA was purified from total RNA preparations by denaturing PAGE, excised, eluted from the gel overnight in 0.3 M sodium acetate and precipitated with ethanol. The tRNA was resuspended in water, and the concentration was determined using NanoDrop [23].
Isolation of Mammalian RNA for Quantitative Real-Time PCR
Total RNA from cells was isolated using the peqGold Total RNA kit, including on-column DNA digestion (Peqlab, Erlangen, Germany). The RNA was transcribed into cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Woolston, UK) according to the manufacturer's instructions. Quantitative real-time PCR was performed with SYBR Green (Applied Biosystems) using a standard protocol. The identity of amplicons was verified by melting curve analysis, and no reverse transcriptase and no template controls were included. Analyses were performed in duplicates.
Co-Immunoprecipitation
For immunoprecipitation, HEK cells stably expressing human TLR7 fused at the C-terminus to an HA tag were transfected with RNA as indicated and incubated for 1 h. The cells were lysed using lysis buffer (136 mM NaCl, 20 mM Tris HCl, 10% glycerol, 2 mM EDTA, 1% Triton X100, pH 7.4, plus proteinase inhibitors) for 10 min on ice. Equilibrated streptavidin or HA beads as indicated were added to the samples and incubated for 3 h on an overhead rotator at 4°C. The samples were washed 3 times and resuspended in SDS-containing loading buffer. Immunoprecipitated proteins were separated by 6% SDS-PAGE and membranes were probed with an anti-HA tag antibody.
Immunoblotting and Cytokine Measurements
For the Western blot analysis of signalling cascade, cells were lysed in RIPA buffer, supplemented with the protease inhibitors leupeptin, aprotinin, pepstatin A (1 mg/ml) and 4-(2-aminoethyl)-benzenesulfonyl fluoride (1 mM). Proteins were separated by 12% SDS-PAGE and blotted on a nitrocellulose membrane. For the Western blot analysis of MAP kinase and NF-κB activation, membranes were probed with antibodies against IκBα, phosphorylated IκBα, phosphorylated p38, phosphorylated ERK1/2 and β-actin (all from Cell Signaling Technology). For cytokine measurement, the levels of TNF, IL-6, IL-12p40 (BD, Heidelberg, Germany) and IFN-α (Bender Med Systems, Vienna, Austria) were determined in cell-free supernatants by ELISA.
Statistical Analysis
The statistical significance between the groups was determined by the 2-tailed Student's t test. Differences were considered significant for p < 0.05, p < 0.01, p < 0.001. The half minimal inhibitory concentration (IC50) values for inhibitory RNA were calculated with R software using the 4-parameter Weibull function.
Results
2'-O-Methylated RNA Inhibits IFN-α Production by Total Bacterial RNA and tRNA in Human PBMC
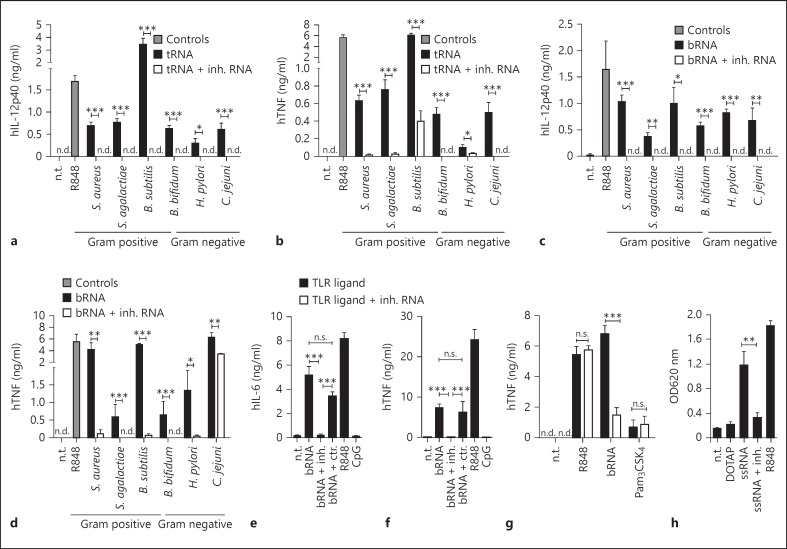
tRNA bearing a Gm18 2'-O-methylation has previously been shown to inhibit TLR7-mediated immunostimulation in human pDCs by single bacterial tRNAs and synthetic ssRNA40 [12, 18]. We now asked whether the inhibitory, antagonistic action of 2'-O-methylation would also affect other bacterial RNA species. Therefore, we made use of a short tRNA fragment derived from E. coli tRNATyr that incorporated a single 2'-O-modified guanosine only (fig. 1a). This fragment can be synthesized more conveniently than the full-length tRNA and was able to mimic the previously published inhibitory effects of modified, full-length tRNA (fig. 1b) [12, 21]. In a first approach, total tRNA and total bRNA were purified from a variety of different bacterial strains including Gram-positive and Gram-negative, pathogenic as well as commensal bacteria. As depicted in figure 1c, tRNA preparations from all tested bacteria induced IFN-α secretion. Secretion was abolished by the concomitant application of the 2'-O-methylated RNA fragment, indicating a dominant negative activity. The same was observed when PBMCs were treated with whole bRNA preparations (fig. 1d). Again, 2'-O-methylated RNA completely inhibited IFN-α secretion. By contrast, no antagonistic effect on pDC-derived IFN-α production was observed upon stimulation with CpG-DNA, a TLR9 ligand. Of note, stimulation by R848, a synthetic small molecule TLR7/8 agonist commonly used to mimic RNA-mediated responses, could not be inhibited by the modified RNA fragment (fig. 1e). Thus, 2'-O-methylated RNA is a potent and specific inhibitor of RNA-induced TLR7-responses in pDCs.
Fig. 1.
2'-O-methylated RNA specifically inhibits IFN-α production in pDCs induced by bRNA. a Schematic picture of the Gm18 tRNATyr fragment used in subsequent experiments. b Human PBMCs were transfected overnight with 0.5 µg/ml E. coli tRNATyr transcript and 1 µg/ml native full-length E. coli tRNATyr (containing the Gm18 modification) or the short tRNATyr fragment indicated in a. c-e Human PBMCs were transfected overnight with 0.5 µg/ml total tRNA (c) or total bRNA (d) purified from the indicated bacteria or were stimulated with R848 (0.5 µg/ml) or CpG (0.5 µM) (e). Where indicated, cells were transfected simultaneously with 1 µg/ml 2'-O-methylated RNA (inh. RNA). Cell-free supernatants were analysed for IFN-α production by ELISA. The results represent mean data + SEM of 2 (b) or 3 (c-e) independent experiments with different donors. E. faecalis = Enterococcus faecalis; n.d. = not detected; n.t. = non-treated negative control. * p < 0.05; ** p < 0.01; *** p < 0.001.
Inhibition of bRNA-Induced Cytokine Production by 2'-O-Methylated RNA Is also Operative in Human Monocytes
So far, most studies investigating immunosuppression by 2'-O-methylated RNA have been performed in human PBMCs using IFN-α production as read-out for TLR7-mediated pDC activation [12, 15]. However, pDCs constitute <1% of total PBMC and produce only a very limited panel of inflammatory mediators apart from IFN-α. By contrast, monocytes are a major source of TNF and IL-12p40 in response to TLR ligation [24, 25]. In order to investigate if the antagonistic effect of 2'-O-methylated RNA is restricted to TLR7/pDCs or also occurs in monocytes that are lacking TLR7 [20], PBMCs were cotransfected with total tRNA or bRNA preparations from a range of different bacteria together with inhibitory RNA. Similarly to the results obtained for type I IFN, 2'-O-methylated RNA greatly diminished TNF and IL-12p40 secretion induced by either tRNA or whole bRNA in PBMCs (fig. 2a-d). Suppression of cytokine secretion was nearly complete, except for total RNA purified from C. jejuni (fig. 2b). The remaining activity for this preparation might be explained by a contamination of this sample with other pathogen-associated molecular patterns, possibly LPS that is found in the cell wall of Gram-negative bacteria. Preparations of tRNA and bRNA varied in their immunostimulatory activity between the tested bacteria. To confirm that inhibition by 2'-O-methylated RNA was indeed operative in monocytes, we next specifically purified CD14+ monocytes to purities exceeding 95%. As for PBMCs, 2'-O-methylated RNA inhibited IL-6 and TNF secretion induced by bRNA (fig. 2e, f). The cotransfection of stimulatory bRNA with an unmodified but sequence-identical tRNA fragment served as specificity control and did not attenuate cytokine production (fig. 2e, f). In line with our previous results (fig. 1d), the silencing effect of 2'-O-methylated RNA on monocyte-derived pro-inflammatory cytokines was specific for bRNA as stimulation with R848 (triggering TLR8 in monocytes) and TLR2 ligand Pam3CSK4 was not impaired (fig. 2g). As monocytes do not express TLR7, these results indicate that the immune silencing property of 2'-O-methylated RNA is not restricted to TLR7 although the receptor sensing bRNA in monocytes is as yet poorly defined. However, TLR8 is a likely candidate receptor as it is expressed in high levels in human monocytes [20]. Indeed, Gm18 was able in suppress TLR8 activation in a HEK overexpression system, providing proof-of-principle that Gm18 can act on this receptor as well (fig. 2h).
Fig. 2.
Antagonistic effect of 2'-O-methylation on bRNA-induced cytokine production in human monocytes. a-d Human PBMCs were transfected overnight with 0.5 µg/ml total tRNA (a,b) or total bRNA (c,d) isolated from the indicated bacterial strains in the presence or absence of 2'-O-methylated RNA (inh. RNA, 1 µg/ml). e-g CD14+ sorted monocytes (e,f) or PBMCs (g) were transfected with total RNA purified from S. aureus (0.5 µg/ml) or were stimulated with R848 (0.5 µg/ml) or Pam3CSK4 (1 µg/ml) alone or with 1 µg/ml 2'-O-methylated RNA (inh. RNA). a-g Production of TNF, IL-12p40 and IL-6 was analysed in cell-free supernatants by ELISA. h HEK-Blue™ cells stably expressing human TLR8 together with an NF-κB-inducible SEAP reporter gene were transfected with ssRNA40 (1.5 µg/ml) in the presence or absence of 2'-O-methylated RNA (inh. RNA, 1.5 µg/ml) complexed with DOTAP. Stimulation was performed in HEK-Blue™ detection medium that allows measurement of SEAP activity by colour development upon substrate cleavage. The optical density at 620 nm was measured after overnight incubation. The data represent the mean values + SEM of 3 independent experiments (a-g) or 4 (h) independent experiments. n.d. = Not detectable; n.t. = non-treated.* p < 0.05; ** p < 0.01; *** p < 0.001.
2'-O-Methylated RNA Inhibits IFNα Secretion from pDCs as well as Monocyte-Derived Cytokine Production with Similar Efficiencies
Since 2'-O-methylated RNA attenuated pDCs as well as monocyte-derived cytokine production, we next addressed the question whether all cytokines were inhibited with similar efficiencies. Therefore, the inhibitory RNA was titrated to a fixed concentration of bRNA to determine the IC50 using the 4-parameter Weibull function (fig. 3a-d). The IC50 of the inhibitory RNA differed slightly for the tested cytokines and varied from 0.009 µg/ml for TNF and 0.019 µg/ml for IL-12p40 to 0.06 µg/ml for IFN-α (fig. 3a-d). Titration curves with unmodified control RNA were performed in parallel as control and showed approximately 10-fold higher IC50 values than for the inhibitory RNA (data not shown).
Fig. 3.
2'-O-methylated RNA inhibits IFN-α, TNF and IL-12p40 production with similar efficiency. a-c PBMCs were cotransfected overnight with 1 µg/ml bRNA purified from S. aureus and different ratios of 2'-O-methylated inhibitory RNA (inh. RNA). Levels of IFN-α (a), TNF (b) and IL-12p40 (c) were analysed in cell-free supernatants by ELISA. Data were normalized to cytokine production induced by bRNA alone to account for donor variation. Curve fit and IC50 (d) were calculated with R software using the 4-parameter Weibull function. Each data point represents the average value of 2–6 independent experiments with different donors.
2'-O-Methylation Does Not Affect Immunostimulation of TLR13 in the Murine System
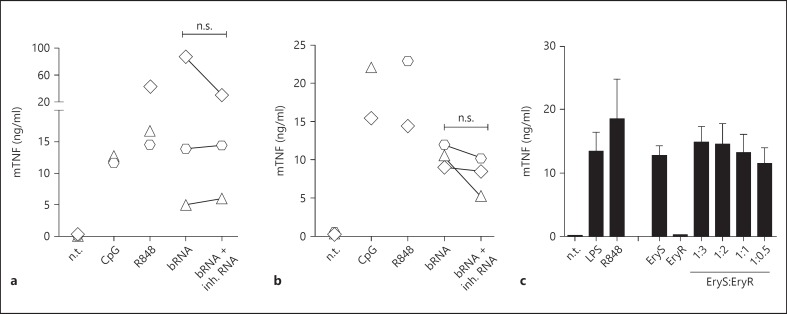
In murine BMDMs and DCs, TLR13, which is not expressed in human cells, has recently been identified as a sensor for bRNA. TLR13 recognizes a short but highly conserved region within the bacterial 23S rRNA. This raised the question if 2'-O-methylated RNA could also attenuate bRNA-induced activation of TLR13. However, as opposed to the striking phenotype in human cells, the dominant immunosilencing effect of 2'-O-methylation could not be observed in murine BMDMs and GM-CSF derived DCs (fig. 4a, b). Interestingly, it has been reported that another type of modification within the target sequence of 23S rRNA, i.e. a N6-dimethylation of adenosine at position A2058, abolishes TLR13 stimulatory properties [9, 26]. Notably, the same modification also renders bacteria resistant to macrolide antibiotics. We therefore decided to evaluate if N6-dimethylation not only renders 23S rRNA non-stimulatory but might also act as a dominant inhibitor of RNA-induced TLR13 responses in analogy to 2'-O-methylation in human cells. To this end, murine BMDMs were transfected in a titration experiment with a constant amount of total RNA derived from an erythromycin-sensitive S. aureus strain together with increasing concentrations of N6-dimethylated bRNA purified from an erythromycin-resistant S. aureus strain. Confirming previous findings, the modified RNA from erythromycin-resistant S. aureus lacked immunostimulation, whereas RNA from a susceptible strain induced TNF. However, N6-dimethylated bRNA did not attenuate cytokine production induced by stimulatory RNA, even when applied in 3-fold excess. Thus, naturally occurring 2'-O-methyl modifications are specific antagonists of RNA signalling in human but not murine immune cells.
Fig. 4.
2'-O-methylated RNA does not block TLR13 stimulation in murine cells. a-c Cell-free supernatants were analysed for TNF secretion by ELISA. Murine BMDCs (a) and murine BMDMs (b) were transfected overnight with bRNA derived from S. aureus (bRNA, 2 µg/ml) in the presence or absence of 2'-O-methylated inhibitory RNA (inh. RNA, 4 µg/ml). Stimulation with CpG or R848 served as positive control. Each symbol represents the mean value of 1 experiment. c Murine BMDMs were transfected overnight with total RNA (bRNA) derived from an erythromycin-sensitive S. aureus strain in the presence of different ratios of total RNA isolated from an erythromycin-resistant S. aureus clinical isolate. LPS, R848 and imiquimod were used as positive controls. Data represent mean values + SEM of 3 independent experiments. n.s. = Not significant; n.t. = non-treated negative control.
Inhibition by 2'-O-Methylated RNA Occurs at Proximal Levels of the TLR Signalling Cascade
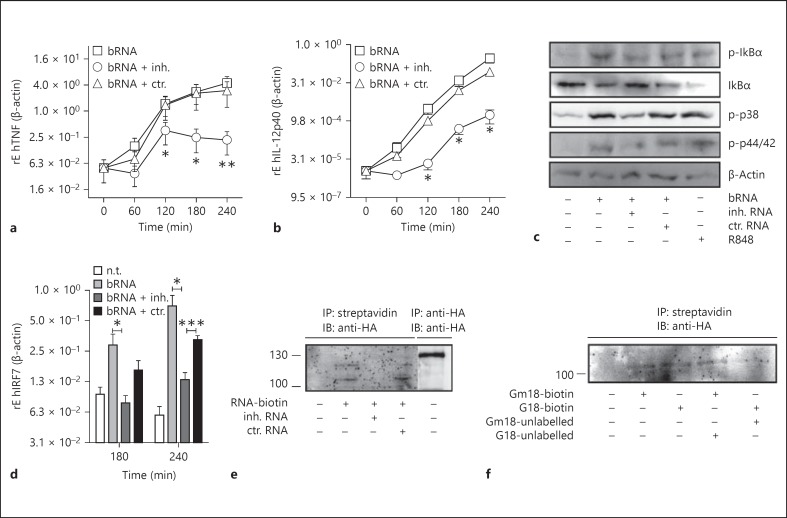
To determine the mechanisms underlying the suppression of immune activation by 2'-O-methylated RNA in human cells, we first evaluated if cytokine production was also inhibited on the transcriptional level. Expression of TNF and IL-12p40 mRNA in human PBMCs could be detected as early as 60 min after stimulation with lbRNA and substantially increased during the following hours. At all time points investigated, costimulation with inhibitory but not control RNA greatly diminished the induction of these cytokines (fig. 5a, b). In a next step, we investigated whether 2'-O-methylated RNA interfered with the 3 main signalling pathways activated by TLR ligation, i.e. activation of NF-κB, MAP kinases and IRFs. To this end, purified CD14+ monocytes as main source of TNF and IL-12p40 were stimulated with bRNA in the presence or absence of 2'-O-methylated RNA. Cell lysates were immunoblotted with antibodies recognizing activated forms of NF-κB, p38 and ERK. bRNA caused a rapid phosphorylation and degradation of Iκ-Bα as well as a phosphorylation of the tested MAP kinases (fig. 5c). The activation of all investigated signalling pathways was impaired in the presence of 2'-O-methylated RNA. The specificity of the inhibitory effect was again confirmed using an unmodified control oligonucleotide. To study the influence of inhibitory RNA on the IRF pathway, the induction of IRF7 was analysed by qPCR in PBMCs. The expression of IRF7 increased 3 h after stimulation with bRNA and was significantly suppressed by costimulation with 2'-O-methylated but not control RNA (fig. 5d). As inhibitory RNA attenuated bRNA-mediated immune responses at proximal levels of the signalling cascade, we speculated that 2'-O-methylated RNA competed with stimulatory RNA for receptor binding without inducing its activation. To test this hypothesis, HEK293 cells stably expressing human TLR7 fused with an HA tag at the C terminus were transfected with a biotinylated stimulatory RNA oligonucleotide in the presence or absence of inhibitory or control RNA. Cells were lysed, and biotinylated RNA was isolated with streptavidin beads. Co-immunoprecipitation of TLR7 was analysed by Western blot using an HA-specific antibody. When HEK cells were transfected with the biotinylated RNA alone or together with control RNA, coprecipitation of TLR7 could be detected, while the signal vanished upon costimulation with inhibitory RNA (fig. 5e). However, the detected bands appeared at a lower molecular weight than expected for the full-length TLR7 protein, presumably indicating the C-terminal cleaved fragment of TLR7 [27, 28]. Despite various modifications, we were not able to increase the signal strength with this assay. In a confirmatory approach, cells were cotransfected with equal amounts of biotinylated, Gm18-methylated and unlabelled, unmodified tRNATyr (fig. 5f, lane 4). Biotinylated RNA was again isolated using streptavidin beads. Under these conditions, the signal detected for RNA-bound TLR7 showed a similar intensity as observed for the transfection with Gm18 tRNATyr alone (fig. 5f, compare lanes 2 and 4). By contrast, when cells were cotransfected with unmodified, biotinylated tRNA plus unlabelled, Gm18-modified tRNA, only a weak band for TLR7 could be detected by Western blot analysis (fig. 5f, compare lanes 4 and 5). Together, these data indicate that 2'-O-methylated RNA can bind to TLR7 with a higher affinity than the unmodified counterpart, thus disrupting the interaction of stimulatory RNA with its receptor.
Fig. 5.
2'-O-methylated RNA impairs TLR activation at proximal levels. a,b,d Human PBMCs were transfected with bRNA (1 µg/ml) isolated from S. aureus in the presence or absence of 2'-O-methylated inhibitory (inh.) or unmethylated control (ctr.) RNA (2 µg/ml) or were stimulated with R848 (1 μg/ml) as positive control. Expression of TNF (a), IL-12p40 (b) and IRF7 (d) was detected by quantitative real-time PCR at the indicated time points. The data represent the mean values of 3 independent experiments normalized to the non-treated control. * p < 0.05; ** p < 0.01; *** p < 0.001. c Human CD14+ monocytes were stimulated for 30 min as in a. Cell extracts were immunoblotted with antibodies recognizing activated forms of NF-κB, p38 and ERK. One representative experiment of 1 is shown. e HEK-TLR7HA cells were transfected with an immunostimulatory biotin-labelled oligonucleotide with or without unlabelled inhibitory (inh.) or control (ctr.) RNA. After 1 h, the cells were lysed, and immunoprecipitation with streptavidin or anti-HA agarose beads was performed. The samples were washed, and Western blot analysis was performed using an anti-HA antibody. One representative experiment of 5 is shown. f HEK-TLR7HA cells were transfected with the indicated tRNATyr full-length in vitro transcripts. tRNA was immunoprecipitated using streptavidin beads, and co-immunoprecipitation of hTLR7 was detected by Western blot analysis.
Discussion
It has been hypothesized that 2'-O-methylation of bacterial tRNA may constitute a relevant immune escape mechanism for certain bacteria, similarly to what has recently been discovered regarding antiviral immunity. In corona- and flavivirus, 2'-O-methylation of the viral mRNA cap impaired the activation of MDA-5 and mediated the evasion of host restriction by IFIT members, respectively [29, 30]. In this study, we further characterized the modulatory effects of 2'-O-methylated bacterial tRNA on innate immune responses in human and murine immune cells. A major strength of the present investigation is the use of a naturally occurring E. coli tRNATyr sequence with the incorporation of a single Gm18 2'-O-methylation in its physiological context. By contrast, most previous studies in this field were performed with short synthetic oligoribonucleotides bearing multiple, randomly incorporated nucleotide modifications [15, 31, 32]. E. coli tRNATyr Gm18 has recently been demonstrated to be necessary and sufficient not only for silencing of human pDC activation via TLR7, but also to act as a dominant inhibitor resulting in inhibition of otherwise stimulatory RNA preparations [12, 18]. Extending previous findings, we demonstrate here that the inhibitory effect of Gm18 is not restricted to specific RNA species [12, 18] but that TLR7-mediated IFN-α production by a wide range of bacterial tRNA and whole bRNA preparations is abolished by concomitant application of 2'-O-methylated tRNA. Of note, a short Gm18 tRNA fragment composed of 27 nucleotides was as efficient in suppressing cytokine production as the full-length Gm18 tRNA, indicating that the classical 3-dimensional tRNA structure is irrelevant for the antagonistic effect. Mechanistically, 2'-O-methylated RNA competed with unmodified RNA for TLR7 binding, thus possibly displacing non-methylated stimulatory RNA from its receptor. Despite direct receptor binding, methylated RNA did not elicit cell activation, indicating that conformational changes of the receptor, which are required for initiating downstream signalling, are not induced. Competition experiments further revealed that the Gm18 tRNA fragment bound with a higher affinity to TLR7 than its unmodified counterpart. The higher binding affinity of modified RNA is supported by the low IC50 values required for the IFN-α inhibitory effect, with concentrations of 2'-O-methylated RNA corresponding to 1/15 of stimulatory bRNA. However, TLR7 appeared at a lower molecular weight in the co-immunoprecipitation experiments with RNA than expected for the full-length protein. It is probable that this band corresponds to the C-terminal cleaved fragment of activated TLR7 [27, 33]. Indeed, it has been described that uncleaved TLR7 resides in the ER under steady state conditions. Upon TLR7 ligand stimulation, TLR7 translocates from the ER to the endolysosome where it is cleaved into a C-terminal signalling competent fragment [28, 33]. Yet, the N-terminal part of TLR7 remains associated with the C-terminal fragment via a disulphide bond and also seems to be important for RNA recognition [34]. It is thus conceivable that full-length TLR7 is detected in unstimulated cells, while only the cleaved, active form coprecipitates with bRNA and appears under reducing Western Blot conditions as HA-tagged C-terminal fragment. Our results are also in line with a report of Hamm et al. [19] who analysed the binding of 2'-O-methylated siRNA to a purified TLR7-Fc fusion protein by AlphaScreen™. In contrast to our approach, Hamm et al. [19] used heavily modified siRNAs with 2'-O-methylations incorporated at every other base.
While the concept of TLR7 antagonistic properties of 2'-O-methylated RNA in pDCs is well accepted [12, 18], much less is known about its impact on other receptors and cell types. We demonstrate here that Gm18 also potently suppresses monocyte-derived pro-inflammatory cytokines including TNF, IL-6 and IL-12p40 with IC50 values similar to those found for IFN-α suppression. As TLR7 is not expressed in monocytes [20], these data indicate that the immunosilencing properties of Gm18 are not restricted to TLR7. Instead, monocytes express TLR8, an endosomal receptor reported to recognize AU-rich ssRNA of viral origin [5]. Although not formally proven, it is thus likely that bRNA can trigger TLR8 activation in human monocytes. In principle, Gm18 is able to suppress the activation of this receptor, as demonstrated in a HEK-TLR8 overexpression system. In contrast, Eberle et al. [35] did not observe a suppression of total bRNA-induced TNF secretion by 2'-O-methylated siRNA and whole tRNA preparations derived from T. thermophilus that contain a high proportion of Gm18-modified tRNA isoacceptors [18]. The discrepancies between different studies may be explained by relevant contaminations of certain bRNA preparations with cell wall components like LPS and lipopeptides that can efficiently trigger monocyte activation via TLR4 and TLR2, respectively [36, 37, 38]. Unlike monocytes, pDCs do not express TLR2 and TLR4 and are therefore more resistant to contamination. Indeed, an inhibitory effect of 2'-O-methylation on IL-6 production was observed when cells were stimulated with unmodified siRNA that should be free of contaminants due to its chemical synthesis [19]. However, the authors concluded that at least 3 modified residues were required for immunosilencing, while our data clearly demonstrate that a single modification is sufficient. It is likely that the sequence context in which the modification occurs is crucial for inhibition of TLR8, as previously demonstrated for TLR7. For the latter, the relevant sequence motif has been identified as [DmR], with D reflecting all bases except of cytosine and R all purines [21]. The pronounced antagonistic effect of Gm18 on monocyte activation might even be of greater importance for the subversion of innate immune responses than previously speculated for TLR7 suppression as the relevance of IFN-α in bacterial infections remains controversial [39, 40, 41]. Apart from a potential immune-evasive function, it was speculated that Gm18 might be responsible for the beneficial, immunomodulating properties of probiotic bacteria as tRNA from E. coli Nissle 1917 did not trigger TLR7 activation [18]. Yet, in this study, RNA preparations from probiotic B. bifidum were as stimulatory as RNA from other bacteria, challenging the hypothesis that non-stimulatory RNA is a hallmark of probiotic bacteria.
Notably, the inhibitory effect of 2'-O-methylated RNA on all tested cytokines was specific for RNA, as immune activation by CpG-DNA and synthetic TLR ligands including TLR7/8 agonist R848 was not affected. The differential inhibition of bRNA versus small molecule-induced TLR activation might be due to differences in the TLR7/8 binding sites of R848 and RNA, as recently described by Colak et al. [42]. They identified overlapping but also non-overlapping binding sites for R848 and RNA40 in the TLR7 and TLR8 ectodomains, resulting in subtle differences in the downstream signalling cascade.
Surprisingly, despite the striking phenotype in human cells, the antagonistic effect of 2'-O-methylation could not be reproduced in murine macrophages and DCs where TLR13 has recently been described as a sensor for bRNA that recognizes a highly conserved region within the 23S rRNA [9, 26]. Although we could reproduce the recently published finding that N6-dimethylation at a distinct position within this sequence renders RNA non-stimulatory, we demonstrate here that this modification does not possess dominant negative activity on TLR13 activation. This mode of action indicates a different mechanism of silencing at TLR13 in murine cells compared to TLR7/TLR8 in human cells, i.e. N6-dimethylation might simply destroy the recognition motif thereby impairing binding of N6-dimethylated RNA to TLR13. Thus, our data unambiguously reveal the existence of species-specific inhibitory patterns acting differentially on human versus murine receptors.
In summary, we identify here 2'-O-methylated E. coli tRNATyr as a universal inhibitor of immune activation by bRNA in the human but not the murine system. In future experiments, it will be interesting to determine the impact of 2'-O-methylation on immune responses by entire bacteria and to elucidate if the methylation status of bRNA correlates with virulence and infection outcome. However, the strict species specificity of Gm18-mediated immunosilencing will challenge further research in this field as commonly used murine infection models will not appropriately reflect the situation in humans.
Acknowledgements
We thank Suzan Leccese for her excellent technical support.
This work was supported by the German Research Foundation grant DA592/5 (to A.H.D.) and HE 3397/9 (to M.H.).
References
- 1.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 2.Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann K, Schlee M, Endres S, Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 3.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 4.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 5.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 6.Diebold SS, Kaisho T, Hemmi H, Akira S, Reise Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 7.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, Horiuchi T, Tomizawa H, Takeda K, Akira S. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 8.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Chen ZJ. Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. Elife. 2012;1:e00102. doi: 10.7554/eLife.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forsbach A, Nemorin J, Montino C, Müller C, Samulowitz U, Vicari AP, Jurk M, Mutwiri GK, Krieg AM, Lipford GB, Vollmer J. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J Immunol. 2008;180:3729–3738. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- 11.Vollmer J, Tluk S, Schmitz C, Hamm S, Jurk M, Forsbach A, Akira S, Kelly KM, Reeves WH, Bauer S, Krieg AM. Immune stimulation mediated by autoantigen binding sites within small nuclear RNAs involves Toll-like receptors 7 and 8. J Exp Med. 2005;202:1575–1585. doi: 10.1084/jem.20051696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gehrig S, Eberle M, Botschen F, Rimbach K, Eberle F, Eigenbrod T, Kaiser S, Holmes WM, Erdmann VA, Sprinzl M, Bec G, Keith G, Dalpke AH, Helm M. Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J Exp Med. 2012;209:225–233. doi: 10.1084/jem.20111044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37:D118–D121. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koski GK, Karikó K, Xu S, Weissman D, Cohen PA, Czerniecki BJ. Cutting edge: innate immune system discriminates between RNA containing bacterial versus eukaryotic structural features that prime for high-level IL-12 secretion by dendritic cells. J Immunol. 2004;172:3989–3993. doi: 10.4049/jimmunol.172.7.3989. [DOI] [PubMed] [Google Scholar]
- 15.Eberle F, Giessler K, Deck C, Heeg K, Peter M, Richert C, Dalpke AH. Modifications in small interfering RNA that separate immunostimulation from RNA interference. J Immunol. 2008;180:3229–3237. doi: 10.4049/jimmunol.180.5.3229. [DOI] [PubMed] [Google Scholar]
- 16.Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 17.Karikó K, Weissman D. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Curr Opin Drug Discov Devel. 2007;10:523–532. [PubMed] [Google Scholar]
- 18.Jöckel S, Nees G, Sommer R, Zhao Y, Cherkasov D, Hori H, Ehm G, Schnare M, Nain M, Kaufmann A, Bauer S. The 2′-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J Exp Med. 2012;209:235–241. doi: 10.1084/jem.20111075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamm S, Latz E, Hangel D, Müller T, Yu P, Golenbock D, Sparwasser T, Wagner H, Bauer S. Alternating 2′-O-ribose methylation is a universal approach for generating non-stimulatory siRNA by acting as TLR7 antagonist. Immunobiology. 2010;215:559–569. doi: 10.1016/j.imbio.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Hornung V, Rothenfusser S, Britsch S, Krug A, Jahrsdörfer B, Giese T, Endres S, Hartmann G. Quantitative expression of toll-like receptor 1–10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J Immunol. 2002;168:4531–4537. doi: 10.4049/jimmunol.168.9.4531. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser S, Rimbach K, Eigenbrod T, Dalpke AH, Helm M. A modified dinucleotide motif specifies tRNA recognition by TLR7. RNA. 2014;20:1351–1355. doi: 10.1261/rna.044024.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eigenbrod T, Franchi L, Muñoz-Planillo R, Kirschning CJ, Freudenberg MA, Núñez G, Dalpke A. Bacterial RNA mediates activation of caspase-1 and IL-1β release independently of TLRs 3, 7, 9 and TRIF but is dependent on UNC93B. J Immunol. 2012;189:328–336. doi: 10.4049/jimmunol.1103258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hengesbach M, Kobitski A, Voigts-Hoffmann F, Frauer C, Nienhaus GU, Helm M. RNA intramolecular dynamics by single-molecule FRET. Curr Protoc Nucleic Acid Chem. 2008 doi: 10.1002/0471142700.nc1112s34. chapter 11:unit 11.12. [DOI] [PubMed] [Google Scholar]
- 24.Schildberger A, Rossmanith E, Eichhorn T, Strassl K, Weber V. Monocytes, peripheral blood mononuclear cells, and THP-1 cells exhibit different cytokine expression patterns following stimulation with lipopolysaccharide. Mediators Inflamm. 2013;2013:697972. doi: 10.1155/2013/697972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watford WT, Moriguchi M, Morinobu A, O'Shea JJ. The biology of IL-12: coordinating innate and adaptive immune responses. Cytokine Growth Factor Rev. 2003;14:361–368. doi: 10.1016/s1359-6101(03)00043-1. [DOI] [PubMed] [Google Scholar]
- 26.Oldenburg M, Krüger A, Ferstl R, Kaufmann A, Nees G, Sigmund A, Bathke B, Lauterbach H, Suter M, Dreher S, Koedel U, Akira S, Kawai T, Buer J, Wagner H, Bauer S, Hochrein H, Kirschning CJ. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science. 2012;337:1111–1115. doi: 10.1126/science.1220363. [DOI] [PubMed] [Google Scholar]
- 27.Ewald SE, Engel A, Lee J, Wang M, Bogyo M, Barton GM. Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J Exp Med. 2011;208:643–651. doi: 10.1084/jem.20100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maschalidi S, Hässler S, Blanc F, Sepulveda FE, Tohme M, Chignard M, van Endert P, Si-Tahar M, Descamps D, Manoury B. Asparagine endopeptidase controls anti-influenza virus immune responses through TLR7 activation. PLoS Pathog. 2012;8:e1002841. doi: 10.1371/journal.ppat.1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin T, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Shi P, Diamond MS. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Züst R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson BR, Muramatsu H, Jha BK, Silverman RH, Weissman D, Karikó K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sioud M, Furset G, Cekaite L. Suppression of immunostimulatory siRNA-driven innate immune activation by 2′-modified RNAs. Biochem Biophys Res Commun. 2007;361:122–126. doi: 10.1016/j.bbrc.2007.06.177. [DOI] [PubMed] [Google Scholar]
- 33.Sepulveda FE, Maschalidi S, Colisson R, Heslop L, Ghirelli C, Sakka E, Lennon-Duménil A, Amigorena S, Cabanie L, Manoury B. Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity. 2009;31:737–748. doi: 10.1016/j.immuni.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 34.Kanno A, Yamamoto C, Onji M, Fukui R, Saitoh S, Motoi Y, Shibata T, Matsumoto F, Muta T, Miyake K. Essential role for Toll-like receptor 7 (TLR7)-unique cysteines in an intramolecular disulfide bond, proteolytic cleavage and RNA sensing. Int Immunol. 2013;25:413–422. doi: 10.1093/intimm/dxt007. [DOI] [PubMed] [Google Scholar]
- 35.Eberle F, Sirin M, Binder M, Dalpke AH. Bacterial RNA is recognized by different sets of immunoreceptors. Eur J Immunol. 2009;39:2537–2547. doi: 10.1002/eji.200838978. [DOI] [PubMed] [Google Scholar]
- 36.Takeuchi O, Kawai T, Mühlradt PF, Morr M, Radolf JD, Zychlinsky A, Takeda K, Akira S. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int Immunol. 2001;13:933–940. doi: 10.1093/intimm/13.7.933. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, Modlin RL, Akira S. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol. 2002;169:10–14. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- 38.Poltorak A, He X, Smirnova I, Liu MY, van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 39.Bogdan C, Mattner J, Schleicher U. The role of type I interferons in non-viral infections. Immunol Rev. 2004;202:33–48. doi: 10.1111/j.0105-2896.2004.00207.x. [DOI] [PubMed] [Google Scholar]
- 40.Schiavoni G, Mauri C, Carlei D, Belardelli F, Pastoris MC, Proietti E. Type I IFN protects permissive macrophages from Legionella pneumophila infection through an IFN-gamma-independent pathway. J Immunol. 2004;173:1266–1275. doi: 10.4049/jimmunol.173.2.1266. [DOI] [PubMed] [Google Scholar]
- 41.Decker T, Müller M, Stockinger S. The yin and yang of type I interferon activity in bacterial infection. Nat Rev Immunol. 2005;5:675–687. doi: 10.1038/nri1684. [DOI] [PubMed] [Google Scholar]
- 42.Colak E, Leslie A, Zausmer K, Khatamzas E, Kubarenko AV, Pichulik T, Klimosch SN, Mayer A, Siggs O, Hector A, Fischer R, Klesser B, Rautanen A, Frank M, Hill AV, Manoury B, Beutler B, Hartl D, Simmons A, Weber AN. RNA and imidazoquinolines are sensed by distinct TLR7/8 ectodomain sites resulting in functionally disparate signaling events. J Immunol. 2014;192:5963–5973. doi: 10.4049/jimmunol.1303058. [DOI] [PMC free article] [PubMed] [Google Scholar]