Abstract
Natural killer (NK) cells are innate lymphocytes that aid in self-nonself discrimination by recognizing cells undergoing pathological alterations. The NKR-P1B inhibitory receptor recognizes Clr-b, a self-encoded marker of cell health downregulated during viral infection. Here, we show that Clr-b loss during mouse cytomegalovirus (MCMV) infection is predicated by a loss of Clr-b (Clec2d) promoter activity and nascent transcripts, driven in part by MCMV ie3 (M122) activity. In contrast, uninfected bystander cells near MCMV-infected fibroblasts reciprocally upregulate Clr-b expression due to paracrine type-I interferon (IFN) signaling. Exposure of fibroblasts to type-I IFN augments Clec2d promoter activity and nascent Clr-b transcripts, dependent upon a cluster of IRF3/7/9 motifs located ∼200 bp upstream of the transcriptional start site. Cells deficient in type-I IFN signaling components revealed IRF9 and STAT1 as key transcription factors involved in Clr-b upregulation. In chromatin immunoprecipitation experiments, the Clec2d IRF cluster recruited STAT2 upon IFN-α exposure, confirming the involvement of ISGF3 (IRF9/STAT1/STAT2) in positively regulating the Clec2d promoter. These findings demonstrate that Clr-b is an IFN-stimulated gene on healthy bystander cells, in addition to a missing-self marker on MCMV-infected cells, and thereby enhances the dynamic range of innate self-nonself discrimination by NK cells.
Keywords: Natural killer cell, Missing-self recognition, NKR-P1B, Clr-b, Clec2d, Paracrine, Interferon, Mouse cytomegalovirus
Introduction
Natural killer (NK) cells are an important component of innate immune responses to pathological target cells, including tumor, virus-infected, antibody-coated, transplanted, and “stressed” cells. NK cell effector functions include cellular cytotoxicity, mediated by exocytosis of preformed granules (containing perforin, granzymes, and granulysin), expression of apoptosis-inducing surface molecules (such as Fas-L and TRAIL), cytokine secretion [most notably interferon (IFN)-γ, TNF-α, and GM-CSF], and chemokine responses [1]. Recently, NK cells have been reclassified as a subset of group-1 innate lymphoid cells (ILC) that share with ILC1 both T-bet (Tbx21) expression and IFN-γ secretion yet also possess cytotoxic function and enhanced expression of eomesodermin (Eomes) [2, 3]. NK cells distinguish between healthy and pathological target cells through a complex integration of signaling events mediated by inhibitory and stimulatory cell surface receptors, which in turn recognize cognate ligands either downregulated or induced on target cells under surveillance [1].
In mice, self-specific NK cell receptors (NKR) include the Ly49 (Klra) family of inhibitory and activating receptors that mainly recognize classical MHC-I molecules, the bifunctional CD94/NKG2 (Klrd1/Klrc) family that recognizes nonclassical MHC-I molecules, the stimulatory NKG2D (Klrk1) receptor that recognizes MHC-I-related stress ligands, and the NKR-P1 (Klrb) family of inhibitory and stimulatory receptors that recognize C-type-lectin-related (Clr/Clec2) glycoproteins [4]. All of these receptors are encoded within the NK gene complex (NKC) located on mouse chromosome 6, rat chromosome 4, and human chromosome 12 [5], yet a number of loci encoding other NKR, including NKp46 (Ncr1) and 2B4 (Cd244), are located outside the NKC [reviewed in [6]].
Among this diversity, the NKR-P1 receptor family is somewhat conspicuous in being genetically linked to its cognate Clr ligand family, akin to one other MHC-independent recognition system, i.e., CD244:CD48; this arrangement may ensure co-inheritance of self-specific receptor-ligand interactions [7, 8, 9]. In the mouse, 5 functional NKR-P1 receptors have been identified, including the activating receptors NKR-P1A (Klrb1a; unknown ligand), NKR-P1C (NK1.1;Klrb1c; unknown ligand), and NKR-P1F (Klrb1f; recognizes Clr-c,d,g), as well as the inhibitory isoforms NKR-P1B/D (Klrb1b; recognizes Clr-b) and NKR-P1G (Klrb1g; recognizes Clr-d,f,g) [10]. At least one other receptor pseudogene locus is annotated (NKR-P1E; Klrb1-ps1), while other Clr loci with unknown or pseudogene function also exist (Clr-a,e,h,i,j) [11, 12]. In humans, only a single inhibitory NKR-P1A receptor exists (CD161/KLRB1), genetically linked to its cognate ligand (LLT1/CLEC2D); however, related stimulatory receptors are also encoded within the NKC, tightly linked to their respective ligand loci, including NKp80 (KLRF1; ligand, AICL/CLEC2B) and NKp65 (KLRF2; ligand, KACL/CLEC2A) [13].
The inhibitory NKR-P1B: Clr-b interaction is currently the most well-characterized recognition pair. Like the MHC-I-specific Ly49 receptors, NKR-P1B: Clr-b interactions are involved in both NK cell education and target cell recognition [14, 15]. Clr-b (Clec2d) is a type-II transmembrane C-type lectin-like glycoprotein expressed on most hematopoietic and some nonhematopoietic cells in a pattern similar to that of MHC-I molecules [8]. However, under pathological conditions such as oncogenesis, virus infection, and genotoxic stress, Clr-b is rapidly downregulated at both the cell surface and steady-state transcript levels, rendering these cells more sensitive to NK cell-mediated “missing-self” recognition [8, 16, 17, 18, 19]. The regulatory mechanisms governing the modulation of Clr-b expression remain to be elucidated, although previous work has suggested roles for transcriptional and posttranscriptional, as well as ubiquitin-proteasomal and endolysosomal, control [16, 17, 18, 19].
To investigate the genomic control of the Clr-b/Clec2d gene at the promoter and nascent transcript levels in healthy versus virus-infected cells, we used MCMV as a model pathogen. MCMV is a β-herpesvirus with a large double-stranded DNA genome capable of accommodating numerous immunoevasin genes that subvert host immune responses. Previous studies have shown that MCMV, RCMV-E, and vaccinia virus infections promote a rapid loss of mouse Clr-b/Clec2d and rat Clr-11/Clec2d11 on fibroblasts [16, 18, 19]. Interestingly, at early time points during MCMV infection in vitro, uninfected fibroblasts actually upregulate Clr-b expression, as do cells exposed to passaged viral supernatants. This reciprocal regulation may represent a means to ensure optimal self-nonself discrimination between uninfected “bystander” cells in the vicinity of infected “missing-self” targets.
Here, we demonstrate that MCMV-mediated downregulation of Clr-b steady-state transcripts is controlled by disruption of Clec2d promoter activity, mediated at least in part by the cell-autonomous action of the MCMV ie3 gene product in trans. In contrast, Clr-b upregulation on uninfected bystander cells is driven by paracrine type-I interferon (IFNαβ) in a manner that is dependent upon IFNAR1 signaling and occupancy of the Clec2d promoter in cis by a complex containing IRF9, STAT1, and STAT2, most likely the ISGF3 heterotrimer (IRF9/STAT1/STAT2). Discerning how NKR ligands are regulated on both healthy and pathological target cells is an important facet in further understanding NK recognition and harnessing NK cell activity in disease therapy.
Materials and Methods
Animals
Stat1 −/− and wild-type (WT) control mice (129S6 strain) were purchased from Taconic Biosciences and handled in accordance with approved animal care protocols at Sunnybrook Research Institute.
Cell Culture and Reagents
NIH3T3 cells were purchased from the American Type Culture Collection and maintained in DMEM (HyClone) with 10% FCS (Gibco) supplemented with 10 mM HEPES (HyClone), 1 mM sodium pyruvate (HyClone), 50 μg/mL gentamicin sulfate (GIBCO), 100 U/mL penicillin (HyClone), 100 μg/mL streptomycin (HyClone), 2 mM GlutaMAX (Gibco), and 50 μM 2-mercaptoethanol (Gibco). To generate stable NIH3T3 cell lines, cells were electroporated with Amaxa Nucleofector II (see below) with linearized plasmid and selected using 2.5 μg/mL puromycin (Life Technologies). Puromycin was removed for 24 h prior to the experiments. Primary mouse embryonic fibroblasts (MEF) were generated in the lab of Dr. Karen Mossman (McMaster University) and grown in αMEM with 15% FCS and supplemented with 1 mM sodium pyruvate (HyClone) and 2 mM GlutaMAX (Gibco). MEF cells were validated for knockout status by PCR following Jackson Laboratory protocols. Primary splenocytes and bone marrow cells were harvested from Stat1−/− and WT mice, red blood cells were lysed using ACK lysis buffer, and cells were analyzed immediately following a 6 h IFN-α4 treatment (103 units/mL). Adult ear fibroblasts (AEF) were generated from minced ear tissue, dissociated, and cultured in 10% supplemented DMEM. Primary AEF cells were treated with or without the STAT1 inhibitors, nifuroxazide or fludarabine (Cedarlane Labs), at titrated doses (50 μM is shown) during IFN-α4 treatment. All cells were maintained in a subconfluent state and grown at 37°C and 5% CO2.
Murine IFN-α4 was provided by Dr. Eleanor N. Fish (University of Toronto). MCMV-Smith and MCMV-GFP were described previously [20, 21] and in vitro passaged in MEF cells in our laboratory. For most MCMV infections or IFN-α4 treatments, 2 × 105 cells were plated in 1 mL medium and infected with MCMV (multiplicity of infection ∼0.5 PFU/cell, spin-fected by centrifugation at 800 g for 30 min at 37°C) or exposed to IFN-α4 (103 U/mL) for 24 h, unless otherwise indicated.
A piggyBac tetracycline-inducible system [22] was modified to replace the β-geo cassette with a puromycin resistance gene (PuroR); this vector was then used to generate doxycycline (Dox)-inducible NIH3T3 stable transfectants. Briefly, NIH3T3 cells were transiently transfected with the modified PB-TET vector containing viral ORF of interest, plus PB transposase and reverse transactivator (rtTA) vectors at a 1:1:1 ratio. Dox was added at a concentration of 1.5 μg/mL the next day, and then the cells were selected in 2.5 μg/mL puromycin plus 1.5 μg/mL Dox for 5 days and allowed to recover for 2 days in 10% dMEM before being used in the experiments.
Flow Cytometry
Surface Clr-b was detected using biotinylated Clr-b mAb (4A6) [8], and IFNAR1 was detected using biotinylated mAb (MAR1-5A3) (BioLegend) plus secondary streptavidin-allophycocyanin (Life Technologies). Cells were stained, washed, and analyzed [23] using BD FACSCalibur and FlowJo software (TreeStar). All flow plots show cells gated by forward scatter, side scatter, lack of propidium iodide uptake, and GFP expression, where necessary.
Vector Construction
Respective B6-strain Clec2d upstream regulatory regions were cloned from BAC RP24-384I3 (BacPac Resources) using specific primers (online suppl. Table 1; for all online suppl. material, see www.karger.com/doi/10.1159/000454926) and AccuPrime HiFi Taq (Life Technologies). The mutated 500-bp promoter PCR product was generated by GeneSOE using the indicated primers (online suppl. Table 1) and ExpandPLUS High-Fidelity enzyme (Roche). The mutated sequence was validated by 2 independent transcription factor search algorithms to be devoid of transcription factor binding sites [24] (http://diyhpl.us/∼bryan/irc/protocol-online/protocol-cache/TFSEARCH.html). PCR products were cloned into the luciferase vectors pGL3-Basic or pGL4.22 (Promega) and then sequenced (Macrogen Inc., South Korea, or TCAG Sequencing, Hospital for Sick Children, Toronto, ON, Canada). The pGL4.22 reporter vector was modified to contain a puromycin resistance cassette. The pRL-TK vector was used as a control for transfection efficiency (Promega).
To overexpress IRF3/7/9, their respective coding sequences were PCR amplified from MCMV-infected NIH3T3 cDNA using the Q5 enzyme (New England Biolabs) and the primers listed in online supplementary Table 1. The sequences were ligated into pcDNA3.1 (Life Technologies) and sequenced. M27 and the immediate early genes were amplified as described above from MCMV-infected NIH3T3 cells and ligated into pIRES2-GFP for transient transfectants and the modified piggyBac tetracycline-inducible system [22] for stable transfectants.
DNA Transfections
NIH3T3 cells were electroporated using program U-030 on a Nucleofector II machine with the Nucleofector® Kit R (Lonza) according to the manufacturer's protocol. MEF cells were electroporated using a 10-μL Neon Transfection Kit (Life Technologies) with buffer R and the following program: 1,350 V, 30 ms, and 1 pulse. The cells received a total of 1 μg DNA with a molar ratio of 10:1 (pGL3:pRL-TK) for transient luciferase reporter assays. BWZ.36 reporter cells were transfected with the 10-μL Neon Transfection Kit with buffer R and the following program: 1,400 V, 20 ms, and 2 pulses.
Intronic qRT-PCR
Total RNA was extracted from NIH3T3 cells using an RNeasy Plus Mini Kit (Qiagen) following the manufacturer's instructions. The RNA was digested with DNaseI (Life Technologies) and reverse transcribed using a Super-Script III Kit, with random hexamer primers (Life Technologies). qRT-PCR was performed using SsoFast EvaGreen Supermix (Bio-Rad) or PerfeCTa SYBR Green Supermix (Quanta BioSciences) on a CFX96 (Bio-Rad) instrument. For SsoFast EvaGreen, the cycling conditions were 95°C for 30 s and 40 cycles of 95°C for 5 s and 60°C for 5 s. For PerfecTa, the cycling conditions were 95°C for 3 min and 40 cycles of 95°C for 10 s and 60°C for 30 s. PCR products were confirmed by melting curve analysis. The primers used for intronic qRT-PCR are listed in online supplementary Table 1. For all qRT-PCR experiments, primers targeting 5 independent housekeeping genes were tested for each condition, with the least variable gene(s) being selected as reference gene(s) for those conditions. Analysis was done using CFX Manager software (Bio-Rad) and validated by manual calculations.
Luciferase Bioluminescence and Protein Assays
Following transfection, NIH3T3 or MEF cells were plated in 6-well plates or 24-well plates, respectively. After overnight incubation, cells were treated with IFN-α4 or infected with MCMV for 24 h. Transiently transfected cells were assayed using the Dual Luciferase reporter system (Promega) and a Varioskan microplate reader (Thermo Scientific). Transient transfection pGL3 vector results represent the averaged ratios of firefly (Photinus) to Renilla (sea pansy) relative light units (RLU), subsequently normalized to values obtained using the pGL3 empty vector, where indicated. Stably transfected cells were assayed using the Luciferase Reporter Assay System (Promega) and results represent the average firefly RLU values normalized to total protein per sample and indexed relative to empty vector values, where indicated.
Chromatin Immunoprecipitation
NIH3T3 cells (10 × 106) were treated with IFN-α4 (103 U/mL) or left untreated for 1.5 h. ChIP was performed as described previously [25]. Briefly, cells were cross-linked with 1% formaldehyde at room temperature for 10 min, washed twice with ice-cold PBS, collected in 1 mL PBS, and spun down. Cells were resuspended in 1 mL lysis buffer [1% SDS, 10 mM EDTA, and 50 mM Tris-HCl (pH 8)] plus proteinase inhibitors, incubated on ice for 10 min, and sonicated to an average size of 500 bp. Chromatin was precleared with 25 μL Pansorbin (Merck Millipore) at 4°C for 15 min. A 100-μL aliquot of sonicated chromatin was immunoprecipitated with 2 μg STAT2 mAb (D9J7L; Cell Signaling Technology) or normal rabbit IgG (Santa Cruz Biotechnology) at 4°C overnight. Immunoprecipitated samples were centrifuged at 13,200 rpm and supernatants were incubated with 10 μL Pansorbin at room temperature for 15 min. Precipitates were washed twice sequentially for 3 min in 1× dialysis buffer [2 mM EDTA, 50 mM Tris-HCl (pH 8), and 0.2% sarkosyl] and then in IP wash buffer 4 times [1% Nonidet P-40, 100 mM Tris-HCl (pH 9), 500 mM LiCl 1%, and deoxycholic acid]. Samples were extracted twice with 150 μL elution buffer (1% SDS and 50 mM NaHCO3), heated at 65°C overnight to reverse cross-linking, and then DNA fragments were purified with a QIAEX II Gel Extraction Kit (Qiagen). For ChIP analysis, qPCR amplification was performed on the CFX Connect Real-Time PCR Detection System (Bio-Rad). Amplicons were detected using SYBR Green (Invitrogen) and the primers listed in online supplementary Table 1.
BWZ Reporter Cell Assays
BWZ.36 cells expressing the CD3ζ/NKR-P1B fusion receptor (BWZ.P1B) were used as reporter cells [8], with the following modification: the Ifnar1 receptor gene was targeted using CRISPR-Cas9 technology. Oligonucleotide pairs 5′-caccg GCT GGT GGC CGG GGC GCC TT-3′, or 5′-aaac AAG GCG CCC CGG CCA CCA GC c-3′, were designed following the Zhang lab protocol (http://crispr.mit.edu) and cloned into the pSpCas9-E2A-EGFP (PX458) plasmid. Plasmids were transiently transfected into BWZ(-) or BWZ.P1B cells using the Neon Transfection System (Life Technologies). Three days after transfection, the cells were single-cell sorted on a FACSDiva for a GFP+ and IFNAR1− phenotype. Clones were subsequently validated by flow cytometry for a lack of IFNAR expression.
For reporter cell assays, NIH3T3 or MEF cells were used as stimulator cells and plated in 3-fold serial dilutions in flat-bottomed 96-well plates. Modified BWZ.P1B reporter cells (Ifnar1−/−) were added at a density of 5 × 104 cells/well. Half of the co-cultures were treated with 103 U/mL IFN-α4 and the other half were left untreated. For a positive control, BWZ cells were stimulated with 10 ng/mL PMA and 0.5 μM ionomycin. Purified blocking Clr-b mAb (4A6) antibody was added at 10 μg/mL. Co-cultures were incubated overnight at 37°C. Cells were washed with PBS and developed using 100 μL 1× CPRG buffer [90 mg/L chlorophenol-red-β-D-galactopyranoside (Roche), 9 mM MgCl2, 0.1% NP-40, in PBS] at room temperature. Optical densities (OD) were measured with a Varioskan microplate reader (Thermo Scientific) using differential ΔOD595–655 values.
Western Blots
Proteins were extracted from NIH3T3 cells using RIPA buffer (50 mM Tris-HCl pH8.0, 150 mM NaCl, 0.02% sodium azide, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate) in the presence of complete protease inhibitor cocktail (Roche). Protein concentrations were quantitated using Bio-Rad protein assay reagent (Bio-Rad). Proteins were resolved using SDS-PAGE and transferred to PVDF membranes. Membranes were preblocked with 5% milk powder or 5% BSA in TBS-T and blotted using STAT2 mAb (D9J7L; Cell Signaling Technology) plus secondary anti-rabbit HRP-conjugated antibody. Signals were detected using ECL (Immobilon Western HRP substrate; EMD Millipore) and a MicroChem 4.2 imager (DNR Bio-imaging Systems).
Statistical Analysis
Data were analyzed using GraphPad Prism 7 with a paired Student's t test or ANOVA. All graphs show means ± SEM. Only significant differences are annotated.
Results
MCMV Infection Reciprocally Modulates Clr-b Levels on Infected and Bystander Cells
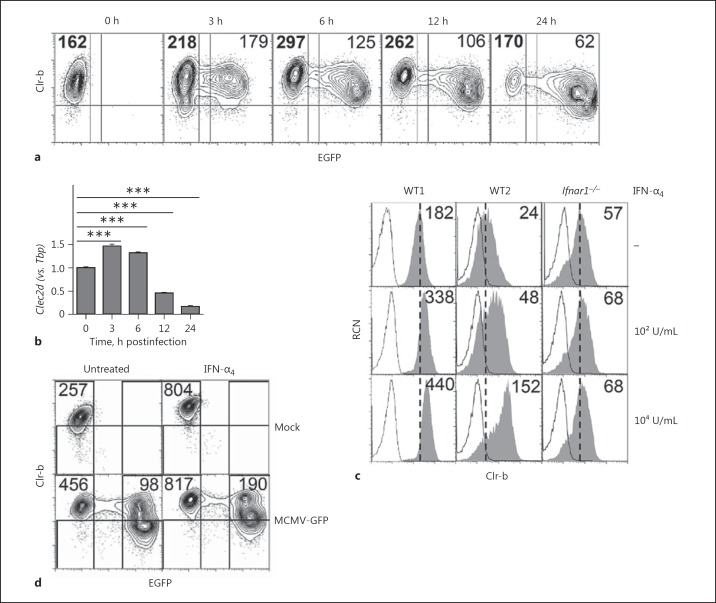
Previous studies have shown that infection of various mouse and rat cells with a number of viruses (MCMV, RCMV-E, vaccinia, ectromelia) promotes a rapid loss of mouse Clr-b (Clec2d) and the rat Clr-b homolog rClr-11 (Clec2d11) at both the steady-state transcript and cell surface levels [6, 16, 18, 19]. To distinguish between infected and uninfected (bystander) cells at the single cell level, we infected mouse NIH3T3 fibroblasts over an early time course using a modified MCMV-GFP reporter virus in which an enhanced GFP transgene is driven by an immediate early gene (ie1/3) promoter in the MCMV-Smith (VR-194) strain [19, 20, 21]. While we consistently observed a loss of Clr-b surface expression on the infected population at later time points (GFP+, 12–24 h postinfection), at early time points the uninfected bystander population expressed elevated Clr-b surface levels relative to mock-infected parental cells (GFP−, 3–12 h postinfection; Fig. 1a). Notably, exposure of NIH3T3 cells to UV-irradiated viral supernatants still promoted early Clr-b upregulation (data not shown), while infection using highly purified MCMV viral particles did not cause significant bystander Clr-b upregulation [19]. This suggested that a soluble mediator in viral supernatants may promote bystander Clr-b upregulation.
Fig. 1.
Reciprocal modulation of Clr-b expression on mouse cytomegalovirus (MCMV)-infected and interferon (IFN)-stimulated bystander cells. NIH3T3 cells were infected with MCMV-GFP supernatants at a multiplicity of infection of 0.5 PFU/cell over an extended time course. Bold numbers represent the Clr-b median fluorescence intensity (MFI) for the uninfected cells. a Cells were analyzed by flow cytometry for Clr-b surface expression. Numbers represent median fluorescence intensities. b Quantitation of steady-state Clec2d transcripts in a by qRT-PCR normalized to Tbp expression and 0 h. Significance was determined by a 2-tailed t test (n = 3). c Flow cytometric analysis of wild-type (WT) and Ifnar1−/− primary mouse embryonic fibroblasts cells with or without IFN-α4 treatment for 24 h. The shaded histogram represents Clr-b expression. Dotted lines represent the secondary reagent alone. Vertical dotted lines show median fluorescence intensities of untreated cells. Numbers represent median fluorescence intensities. d NIH3T3 cells were exposed to medium alone (untreated) or 104 U/mL IFN-α4 overnight, followed by MCMV-GFP infection at a multiplicity of infection of 1 PFU/cell or mock infection for 24 h. The results are representative of at least 3 independent experiments. ***p < 0.001.
Time course analysis of total steady-state mRNA levels by qRT-PCR confirmed (at the population level) that MCMV-infected NIH3T3 cells initially induced and then downregulated Clr-b transcripts relative to mock-infected cells (Fig. 1b). Collectively, this suggests that MCMV infection may differentially modulate Clr-b levels in infected and bystander cells, whereby early induction of Clr-b may be due to exogenous factors produced upon MCMV infection acting in trans.
Bystander Clr-b Induction during MCMV Infection Is Type-I IFN Dependent
We hypothesized that the induced levels of Clr-b protein and Clec2d transcripts in uninfected bystander cells might be due to paracrine type-I IFN cytokine stimulation. To test this, we exposed primary murine embryonic fibroblasts (MEF) from both WT B6 and Ifnar1−/− mice to IFN-α4 (or IFN-β; data not shown) and then examined Clr-b levels by flow cytometry. Treatment of WT MEF with IFN-α4 upregulated Clr-b (∼2-fold at a low dose and ∼6-fold at a high dose), while Ifnar1−/− MEF showed no change in Clr-b levels (Fig. 1c). Clr-b expression also remained elevated for ∼2–4 h after IFN was removed from the culture medium (data not shown). Thus, Clr-b levels are upregulated on primary MEF cells in response to type-I IFN in an IFNAR1-dependent manner.
To test whether pre-conditioning cells with type-I IFN could block MCMV infection-mediated Clr-b loss, NIH3T3 cells were pre-treated overnight with a high dose (104 U/mL) of IFN-α4 prior to infection. Notably, IFN-α4 pre-exposure did not prevent MCMV-GFP virus infection (Fig. 1d). In addition, while an IFN-mediated induction of Clr-b was observed on all cells, infected (GFP+) cells still displayed reduced Clr-b levels in comparison to uninfected (GFP−) cells. This suggests that the IFN-induced antiviral state is insufficient to prevent WT MCMV infection in vitro, and that infection-mediated Clr-b loss is IFN independent. To probe this reciprocal regulation further, we examined genomic control at the level of the Clr-b (Clec2d) promoter.
Reciprocal Modulation of Clec2d Promoter Activity by MCMV Infection and Type-I IFN
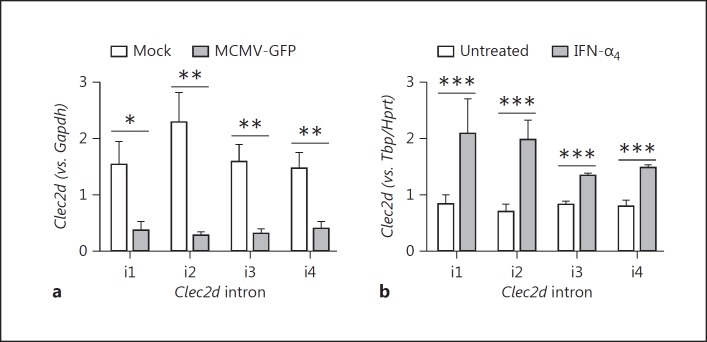
To elucidate the role of the Clec2d promoter, independently of mRNA stability, we performed intronic qRT-PCR to assess nascent (pre-mRNA) Clec2d transcript levels, since intron removal during mRNA splicing usually occurs co-transcriptionally. Clec2d is a relatively compact gene, consisting of 5 known exons separated by 4 introns; therefore, the nascent abundance of introns 1–4 (i1-i4) was quantitated during MCMV infection relative to healthy cells. Following overnight MCMV-GFP infection, nascent Clec2d transcript levels were reduced ∼5-fold relative to mock-infected cells (Fig. 2a; MCMV-GFP vs. mock). In contrast, following overnight IFN-α4 treatment, a ∼2- to 3-fold increase in nascent Clec2d transcripts was observed (Fig. 2b; IFN-α4 vs. untreated). Thus, MCMV infection promotes a significant decrease in nascent Clr-b transcript levels, while type-I IFN treatment causes a significant increase, suggesting that Clec2d promoter activity is reciprocally modulated in infected and bystander cells.
Fig. 2.
Mouse cytomegalovirus (MCMV) infection and type-I interferon (IFN) oppositely regulate Clr-b nascent transcript levels. NIH3T3 cells were infected with MCMV-GFP at a multiplicity of infection of 0.5 PFU/cell (a) or treated with 103 U/mL IFN-α4 for 24 h (b). Levels of Clec2d nascent pre-mRNA transcripts across introns 1–4 were quantified by qRT-PCR and normalized relative to Gapdh (a) or Hprt and Tbp (b). Significance was determined by a 2-tailed t test (n = 3–5 experiments). * p < 0.05, ** p < 0.01, *** p < 0.001.
The Clec2d Promoter Is Regulated by a Functional Cluster of IRF Binding Sites (IRFC)
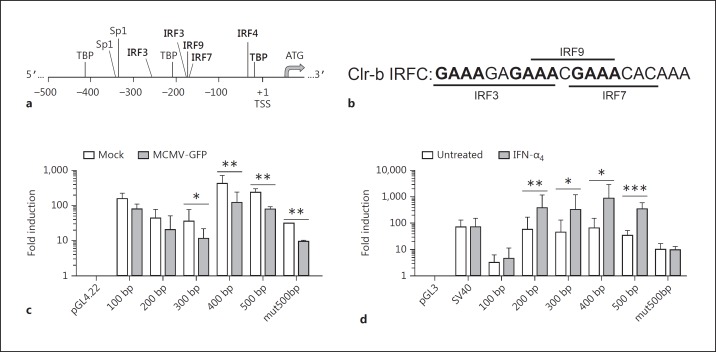
To decipher whether infected and bystander cells display differential Clec2d promoter activity, luciferase reporter assays were employed. To this end, Clec2d promoter fragments of various sizes were subcloned from a BAC vector into luciferase reporter vectors (pGL3 for transient and pGL4.22 for stable transfectants) (Fig. 3a-d). NIH3T3 transfectants were subsequently exposed to either MCMV or IFN-α4 overnight and assayed for luciferase activity. As observed at the transcript level, MCMV infection caused a ∼4- to 5-fold decrease in Clec2d promoter activity, relative to the empty pGL4.22 vector, in transfectants containing ≥300 bp upstream of the Clec2d transcriptional start site (TSS) (Fig. 3c). In contrast, IFN-α4 treatment resulted in a ∼3-fold increase in Clec2d promoter activity, relative to the empty pGL3 vector or the SV40 promoter, in transfectants containing ≥200 bp upstream of the Clec2d TSS (Fig. 3d). Collectively, this suggests that MCMV-mediated downregulation and IFN-mediated induction of Clec2d are regulated by distinct regulatory regions, and that DNA element(s) within the first ∼200 bp upstream of the Clec2d TSS are required for IFN-mediated Clr-b induction.
Fig. 3.
Mouse cytomegalovirus (MCMV) infection and type-I interferon (IFN) reciprocally regulate Clec2d promoter activity. a Schematic representation of the Clec2d upstream regulatory region showing selected consensus motifs; ATG, translational start site; +1, transcriptional start site (TSS); TBP, TATA binding protein; IRF, interferon regulatory factor; Sp, specificity protein. b Nucleotide sequence of the cluster of predicted IRF-binding sites (IRF cluster) within the first 200 bp of the Clec2d promoter. The upstream GAAA motifs in bold may enhance STAT1 (and thus ISGF3) recruitment to the IRF9 motif, in turn minimizing the requirement for the IRF3/7 motifs in response to type-I IFN [30]. c, d NIH3T3 cells were transfected with luciferase reporter plasmids containing varying sizes of the Clec2d promoter region as outlined in a and b. c Stable transfectants were infected with MCMV at a multiplicity of infection of 0.5 PFU/cell for 24 h. d Transient transfectants were treated with 103 U/mL IFN-α4 for 24 h the next day. Promoter activity was assayed by luciferase reporter assay relative to total protein and empty vector (pGL4.22) (b) or Renilla and empty vector (pGL3) (c). Significance was determined by a 2-tailed t test on log-transformed values (n = 3–7 experiments). * p < 0.05, ** p < 0.01, *** p < 0.001.
We next analyzed the Clec2d promoter region to identify putative transcription factor binding sites responsible for Clr-b regulation. Several IFN regulatory factor (IRF) binding sites were predicted within the ∼200- to 300-bp promoter responsible for IFN-mediated induction (Fig. 3a, b, d). A putative IRF4 binding site was identified within the first 100 bp, although this region is not sufficient to augment Clec2d promoter activity (Fig. 3a, d). Importantly, the 200-bp promoter fragment, which is sufficient to induce Clec2d in response to IFN-α4, contains a cluster of overlapping binding sites for IRF3, IRF7, and IRF9 [see Fig. 3a and b for the IRF cluster (IRFC) nucleotide sequence]; in addition, another IRF3 binding site was predicted within the 300-bp fragment (Fig. 3a). To investigate whether the ∼200-bp IRFC was necessary for type-I IFN induction, we mutated the IRF3/7/9 consensus motifs within the 500-bp promoter fragment (mut500bp) to sequences devoid of transcription factor activity (as determined by MatInspector and TfSearch). In contrast to the intact 500-bp promoter fragment, the mut500bp fragment failed to augment Clec2d promoter activity in response to IFN-α4, indicating that this region is necessary for Clr-b induction (Fig. 3d). However, reduced Clec2d promoter activity was still observed during MCMV infection using the mut500bp fragment (Fig. 3c), suggesting that virus infection more broadly affects the Clec2d promoter. Interestingly, a recent study examined whole-genome RNA polymerase II (RNAPII) occupancy by ChIP-Seq in MEF cells during HSV-1 infection [26]; analysis of the Clec2d promoter from these data revealed that RNAPII was essentially absent following HSV-1 infection (online suppl. Fig. 1). Thus, the loss of Clec2d promoter activity and nascent Clr-b transcripts may be a generalized response to herpesvirus infection, and perhaps other viruses [16, 18, 26].
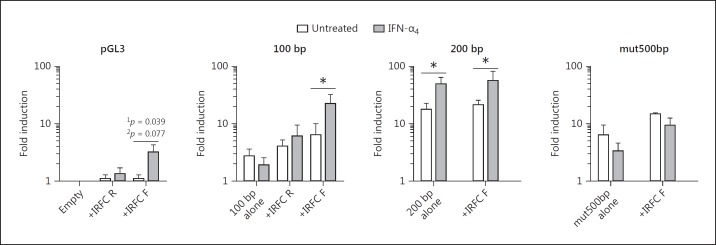
Next, we examined whether the IRFC was sufficient to mediate IFN-inducible expression in a minimal promoter/enhancer assay. In transient NIH3T3 transfectants, the IRFC alone provided limited IFN-dependent induction in the forward orientation only, although this was only significant using a 1-tailed t test (Fig. 4). More importantly, the IFN-nonresponsive 100-bp promoter fragment became IFN responsive upon upstream addition of a forward-oriented IRFC (Fig. 4). However, providing an additional IRFC to the IFN-responsive 200-bp promoter fragment, or adding an intact IRFC upstream of the mut500bp promoter fragment, did not further increase the expression (Fig. 4). Taken together, these orientation/proximity-dependent results suggest that the IRFC acts as part of an IFN-inducible promoter but not an independent enhancer. It remains possible that the additional IRF3 site may contribute to this context dependence.
Fig. 4.
An overlapping IRF3/7/9 consensus cluster (IRF cluster; IRFC) is required for type-I interferon (IFN)-dependent promoter activity. NIH3T3 cells were transfected with luciferase reporter vectors and then treated with 103 U/mL IFN-α4 for 24 h the next day. Promoter activity was assayed by dual luciferase reporter assay relative to Renilla luciferase and normalized to the pGL3 empty vector. Significance was determined by a 1-tailed (1p) or 2-tailed (2p) t test on log-transformed values (n = 3 experiments). * p < 0.05.
Requirement for IFNAR1, IRF9, STAT1, and STAT2 in Clec2d Induction by Type-I IFN
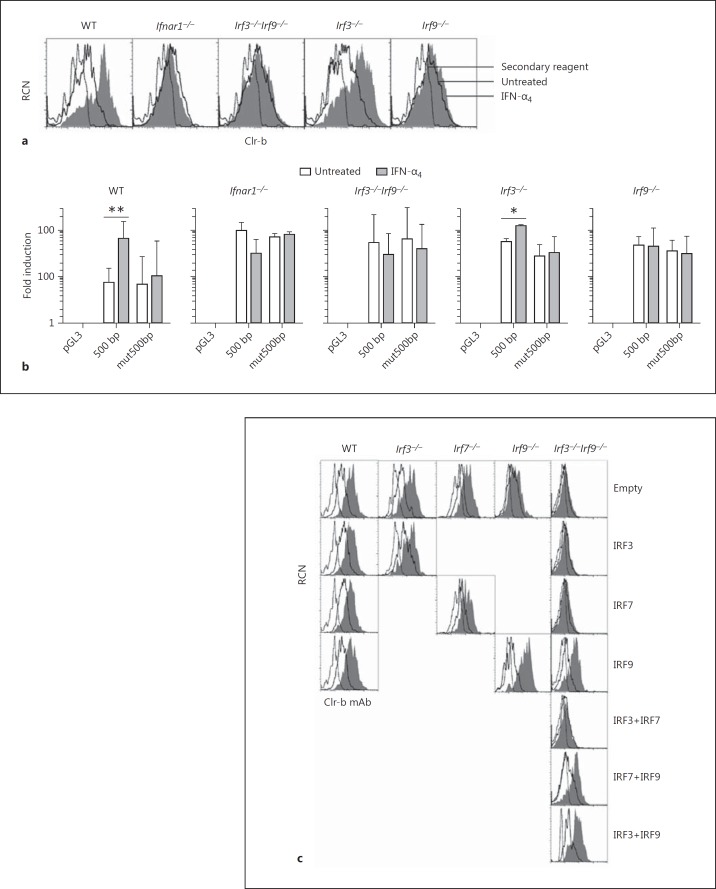
To examine the mechanism of IFN-induced Clr-b induction further, we utilized MEF cells deficient in type-I IFN signaling pathway components. To this end, MEF cells from WT, Ifnar1−/−, and various IRF-deficient mice were exposed to IFN-α4 or MCMV and examined for Clr-b levels by flow cytometry. As shown previously, IFN-α4 upregulated Clr-b on WT but not Ifnar1−/− MEF (Fig. 5a), demonstrating a requirement for IFNAR1 signaling. IFN-dependent Clr-b induction was also observed on Irf1−/−Irf3−/−,Irf3−/−Irf7−/−,Irf3−/−, and Irf7−/− MEF, but not on Irf3−/−Irf9−/− or Irf9−/− MEF, demonstrating a requirement for IRF9 function (Fig. 5a and data not shown).
Fig. 5.
Type-I interferon (IFN)-dependent induction of Clr-b is IRF9 dependent. a Primary mouse embryonic fibroblast cells derived from wild-type (WT), Ifnar1−/−,Irf3−/−Irf9−/−,Irf3-/−, and Irf9−/− mice were treated with 103 U/mL IFN-α4 for 24 h and analyzed for Clr-b cell surface expression by flow cytometry. The dotted line represents the secondary reagent alone; solid lines represent untreated cells; shaded histograms represent treated cells. b Mouse embryonic fibroblast cells as in a were transfected with luciferase reporter constructs and analyzed by dual luciferase reporter assay. The graphs show fold luciferase induction relative to Renilla and normalized to the pGL3 empty vector. Significance was determined by a 2-tailed t test on log-transformed values (n = 3–5 experiments). c Mouse embryonic fibroblast cells were transfected with overexpression vectors containing IRF3, IRF7, and/or IRF9 for 24 h and then left untreated or treated with 103 U/mL IFN-α4 for an additional 24 h before analysis of Clr-b expression by flow cytometry. Histograms are gated on GFP+ cells. Dotted lines represent the secondary reagent alone; solid lines represent untreated cells; shaded histograms represents IFN-treated cells. The results are representative of at least 4 independent experiments.
On the other hand, upon MCMV-GFP exposure, infected (GFP+) MEF cells downregulated Clr-b independently of genotype, and WT bystander (GFP−) MEF cells routinely induced Clr-b, yet Irf3−/−Irf9−/− and Irf9−/− bystander (GFP−) MEF cells did not upregulate Clr-b (online suppl. Fig. 2). Results using Irf3−/− and Irf7−/− MEF were not as clear, with bystander cells inducing Clr-b in some but not all experiments (dependent upon resting Clr-b levels and viral multiplicity of infection; online suppl. Fig. 2). Taken together, this suggests that IFN-mediated Clr-b induction requires IRF9, while MCMV-mediated Clr-b downregulation is more complex and appears to be IRF1/3/7/9 independent.
We next used mutant MEF cells to reanalyze Clec2d promoter activity via luciferase reporter assays. As shown previously for NIH3T3, the 500-bp Clec2d promoter fragment augmented the luciferase activity in response to IFN-α4 in WT and Irf3−/− MEF cells, while the mut500bp fragment did not (Fig. 5b). In contrast, neither the intact 500-bp nor the mut500bp promoter fragments augmented luciferase activity in response to IFN-α4 treatment in Ifnar1−/−,Irf3−/−Irf9−/−, or Irf9−/− MEF transfectants (Fig. 5b). This suggests that both IRF9 and the IRF3/7/9 cluster, in addition to intact IFNAR1 signaling, are required for IFN-mediated Clec2d induction. Importantly, complementation studies using mutant MEF cells and overexpression of their deleted gene products demonstrated that re-introduction of IRF9 into Irf3−/−Irf9−/− and Irf9−/− MEF cells re-established IFN-dependent Clr-b induction, further confirming the importance of IRF9 (Fig. 5c).
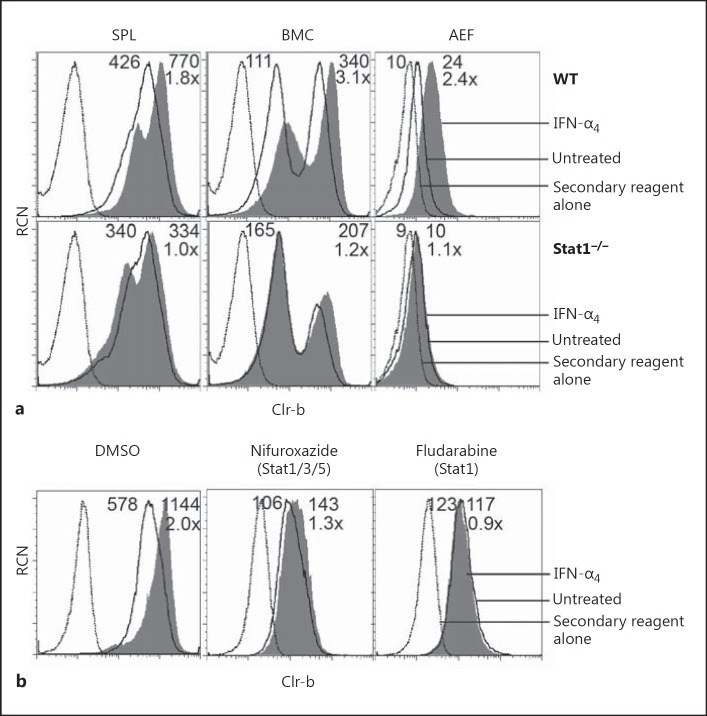
IRF9 commonly activates transcription of ISG by forming a complex with STAT1 and STAT2 (termed ISGF3). To dissect this pathway further, we analyzed the IFN-α4 responses of primary cells from Stat1−/− and WT control (129S6 strain) mice. Notably, Clr-b induction on Stat1−/− splenocytes, bone marrow cells, and primary AEF was abrogated in response to IFN-α4 treatment, while the same tissues from strain-matched (129S6) control WT mice upregulated Clr-b in response to IFN-α4 (1.8-, 3.1-, and 2.4-fold, respectively; Fig. 6a). Furthermore, 2 independent STAT1 inhibitors, i.e., nifuroxazide (which inhibits STAT1/3/5 phosphorylation by Jak2/Tyk2) and fludarabine (which specifically depletes STAT1 protein and mRNA) [27, 28, 29], also blocked Clr-b upregulation upon IFN-α4 treatment of NIH3T3 cells (Fig. 6b). Taken together, these results suggest that type-I IFN-mediated Clr-b upregulation is dependent upon STAT1. Notably, it has been previously suggested that GAAA sequences localized upstream of IRF9 binding sites help to facilitate the recruitment of STAT1 and IRF9 to ISGF3-dependent IFN-stimulated response elements [30]; thus, the enrichment of GAAA sequences in the IRFC (Fig. 3a, b) may explain both the STAT1 dependence and IRF3/7 independence of this cluster.
Fig. 6.
Type-I interferon (IFN)-mediated Clr-b induction is dependent on STAT1. a Splenocytes (SPL), bone marrow cells (BMC), and primary adult ear fibroblasts (AEF) were harvested from wild-type (WT) and Stat1−/− mice, treated with 103 U/mL IFN-α4 for 6 h, and analyzed by flow cytometry for Clr-b expression. b NIH3T3 cells were treated with vehicle alone (DMSO), 50 μM nifuroxazide, or 50 μM fludarabine for 22 h, at which point 103 U/mL IFN-α4 was added for 3 h prior to flow cytometric analysis of Clr-b expression. Dotted lines represent the secondary reagent alone; solid lines represent cells not treated with IFN-α4; shaded histograms represent IFN-α4-treated cells. Numbers reflect the median fluorescence intensity of untreated cells (left) or IFN-α4-treated cells (right); the lower number (e.g. 8×) represents the fold change in Clr-b MFI (IFN-α4 treated/untreated).
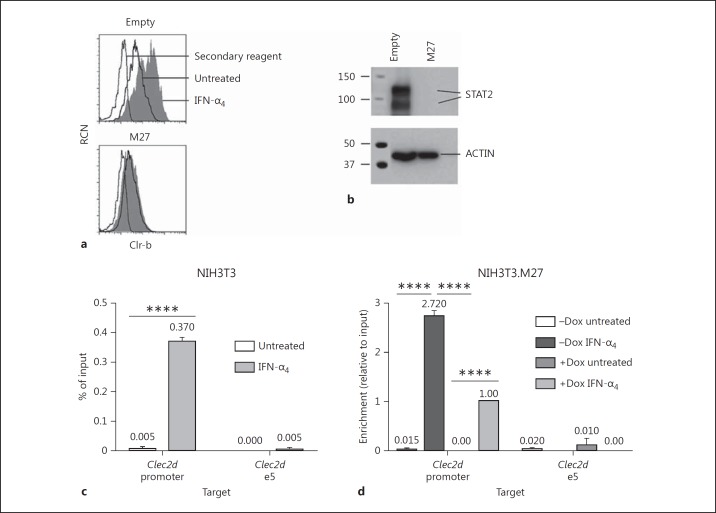
To demonstrate a role for STAT2 in type-I IFN-mediated Clec2d induction, we overexpressed the MCMV immunoevasin M27, known for its ability to target and degrade STAT2 [31], in NIH3T3 cells. Strikingly, M27 overexpression resulted in a complete loss of STAT2 protein, in turn preventing Clr-b upregulation in response to IFN treatment; thus, STAT2 is necessary for IFN-dependent Clr-b induction (Fig. 7a, b). To formally show recruitment of STAT2 to the Clec2d promoter, we performed chromatin immunoprecipitation (ChIP) experiments on IFN-treated NIH3T3 cells. Here, the IFN-α4 treatment was reduced to ∼1.5 h, a time point reflecting an optimal increase in nascent Clr-b transcript levels prior to protein induction (online suppl. Fig. 3a, b). Interestingly, an enrichment of Clec2d promoter gDNA (containing the IRFC) was observed upon ChIP of IFN-treated NIH3T3 cells using a STAT2 mAb (Fig. 7c), while this was not observed for downstream Clec2d exon-5 gDNA. In addition, similar results were observed using stable Dox-inducible NIH3T3.M27 transductants [22], whereby Dox induction of M27 expression mitigated the IFN-dependent recruitment of STAT2 to the Clec2d promoter containing the IRFC (Fig. 7d). This demonstrates that STAT2 is directly and specifically recruited to the Clec2d promoter IRFC element upon type-I IFN treatment. Notably, attempts to perform ChIP on mouse cells using multiple anti-IRF9 antibodies and Flag-tagged IRF9 failed due to specificity and sensitivity problems (data not shown). Nonetheless, the above results collectively demonstrate that ISGF3 heterotrimer (IRF9/STAT2/STAT1) recruitment to the IRFC is likely responsible for Clr-b induction.
Fig. 7.
STAT2 is required for Clr-b induction following type-I interferon (IFN) treatment. a NIH3T3 cells were transfected with a vector encoding the mouse cytomegalovirus M27 immunoevasin or pIRES2-GFP empty vector and then treated with 103 U/mL IFN-α4 for 24 h the next day. Clr-b expression was analyzed by flow cytometry, gated on GFP+ cells. Dotted lines represent the secondary reagent alone; solid lines represent untreated cells; shaded histograms represent IFN-treated cells. b Cells were transfected as in a and then examined by Western blot for STAT2 expression the next day. c ChIP using an anti-STAT2 mAb was performed on NIH3T3 cells treated for 1.5 h with 103 U/mL IFN-α4, using qRT-PCR primers spanning the IRF cluster of the Clec2d promoter or exon 5 (n = 2 experiments). d NIH3T3 cells stably expressing M27 using a modified Dox-inducible piggyBac vector (NIH3T3.M27) were used to perform STAT2 ChIP as in c (n = 2 experiments). **** p < 0.0001.
IFN-Dependent Clr-b Induction Augments NKR-P1B-Ligand Function
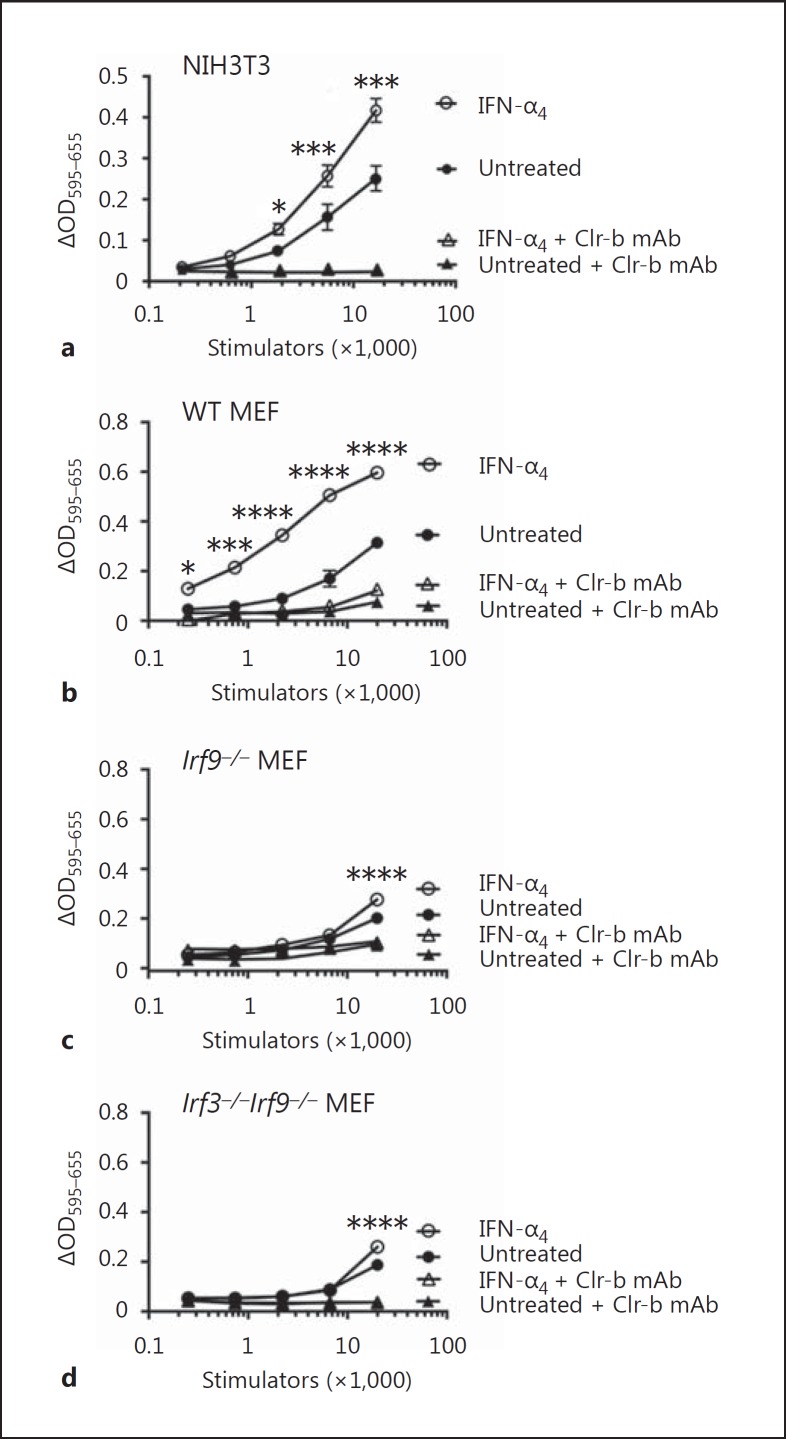
To assess the functional consequences of Clr-b upregulation by type-I IFN, we utilized BWZ reporter cell assays. Here, BWZ.36 reporter cells expressing a CD3ζ-fusion receptor of the NKR-P1B ectodomain (BWZ.P1B cells) were used to specifically quantitate NKR-P1B-ligand (Clr-b) function on various stimulator cells. Preliminary experiments using native BWZ.P1B cells suggested that reporter cell signaling was partially reduced by type-I IFN in a dose-dependent fashion (online suppl. Fig. 4a, c), possibly due to altered signaling [32, 33] or threshold effects on the BWZ.36 (T-lineage) cell line. Therefore, we utilized CRISPR-Cas9 gene-editing technology to generate Ifnar1−/− BWZ.P1B and control BWZ(-) cells. IFNAR1 deficiency was confirmed for multiple BWZ clones by flow cytometry using IFNAR1 mAb (online suppl. Fig. 4b). Notably, IFN-α4 treatment of NIH3T3 stimulator cells augmented NKR-P1B-ligand function using IFNAR1-deficient BWZ.P1B versus BWZ(-) cells (Fig. 8a; online suppl. Fig. 4c). This signal was Clr-b-specific, as it was blocked using Clr-b mAb.
Fig. 8.
BWZ.NKR-P1B reporter cell analysis of Clr-b ligand function on type-I interferon (IFN)-treated stimulator cells. NIH3T3 cells (a), primary wild-type mouse embryonic fibroblast cells (b), primary Irf9−/− mouse embryonic fibroblast cells (c), or primary Irf3−/−Irf9−/− mouse embryonic fibroblast cells (d) were used as stimulator cells for Ifnar1−/− CD3ζ/NKR-P1B receptor-bearing BWZ.36 reporter cells, either untreated or treated overnight with 103 U/mL IFN-α4, in the absence or presence of blocking Clr-b mAb. Control Ifnar1−/− BWZ.36 cells are not shown. Experiments were analyzed using ANOVA with Bonferroni's post hoc analysis comparing treated to untreated. The results are representative of at least 4 independent experiments, and the graphs show means ± SEM. OD, optical density. * p < 0.05, *** p < 0.001, **** p < 0.0001.
To confirm a role for IRF9 in Clr-b induction, we repeated these assays using primary MEF from WT, Irf9−/−, and Irf3−/−Irf9−/− mice as stimulator cells. Notably, treatment of WT MEF with IFN-α4 greatly augmented BWZ.P1B reporter activity, which was blocked using Clr-b mAb (Fig. 8b). In contrast, treatment of Irf9−/− or Irf3−/−Irf9−/− MEF with IFN-α4 yielded almost no BWZ.P1B responses, with minimal dose-dependent induction (Fig. 8c, d). Taken together, these results confirm that Irf9−/− MEF cells possess an intrinsic functional defect in IFN-dependent induction of the NKR-P1B-ligand Clr-b.
The MCMV ie3 Gene Product Cell-Autonomously Represses Clec2d Expression
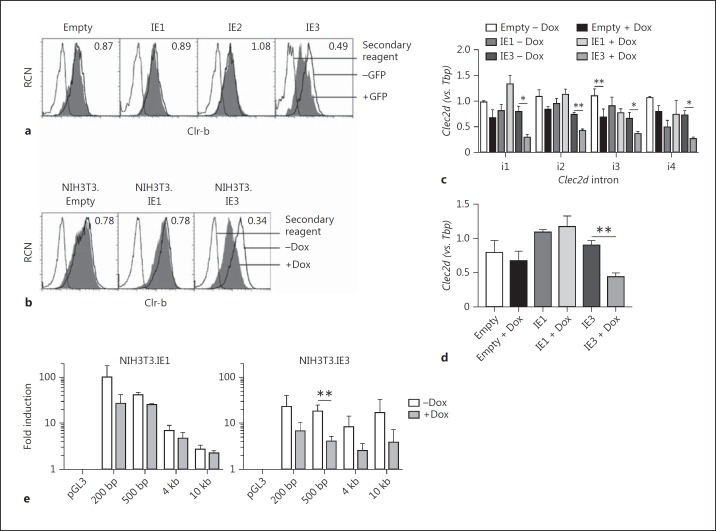
In an attempt to understand mechanistically how MCMV infection downregulates Clr-b, we have tested a number of MCMV large genomic-deletion mutants and cloned numerous MCMV gene products for independent overexpression. Importantly, neither m02 or m145 family immunoevasin genes nor several core genes were found to negatively regulate Clr-b levels in isolation. However, overexpression of a single immediate-early gene product, ie3, but not ie1 or ie2, consistently promoted a significant (≥50%) Clr-b downregulation when transiently transfected into NIH3T3 cells (Fig. 9a).
Fig. 9.
Immediate early gene 3 (ie3) cell autonomously promotes downregulation of Clr-b levels by repressing Clec2d promoter activity. a Vectors encoding the MCMV immediate early gene ORF (ie1,2,3) or the empty pIRES2-GFP vector were transiently transfected into NIH3T3 cells and then analyzed for GFP and Clr-b expression after 24 h. Dotted lines represent the secondary reagent alone; solid black lines represent Clr-b expression gated on GFP− (untransfected) cells, and shaded histrograms represent Clr-b expression gated on GFP+ (transfected) cells. Numbers represent the fold change in Clr-b expression (GFP+/GFP− cells). b NIH3T3 stable transfectants of Dox-inducible piggyBac vectors encoding the ie gene products or empty vector controls were induced for 2 days in 1.5 μg/mL Dox and then analyzed by flow cytometry. Dotted lines represent the secondary reagent alone; solid black lines represent Clr-b expression on untreated cells (-Dox); shaded histograms represent Clr-b expression on Dox-treated cells (+Dox). Numbers represent the fold change in Clr-b expression (+Dox/-Dox). c RNA from the cells in a was analyzed by qRT-PCR for Clec2d nascent pre-mRNA transcripts of introns 1–4 normalized relative to Tbp (n = 3 experiments). Significance was determined by a 2-tailed t test. d RNA from the cells in b was analyzed for Clec2d steady-state levels by qRT-PCR relative to Tbp (n = 3–4 experiments). e Cells as in b were transfected with luciferase-reporter vectors containing various Clec2d promoter fragments and then promoter activity was assayed by luciferase reporter assay relative to Renilla luciferase and normalized to the empty vector control (pGL3). Significance was determined by a 2-tailed t test on log-transformed values (n = 3 experiments). * p < 0.05, ** p < 0.01.
To further assess the effects of ie3 on Clr-b levels, we generated stable Dox-inducible NIH3T3 transfectants of the alternately spliced ie1/3 genes [22]. Following Dox induction, ie3 was found to promote a significant (≥66%) loss of surface Clr-b, with minimal downregulation observed for ie1 and empty vector controls (Fig. 9b). To examine the effect of ie3 on Clec2d gene expression, we next quantitated Clr-b nascent and steady-state transcript levels by qRT-PCR. Here, Dox-induced ie3 expression promoted a significant decrease in both Clr-b nascent and steady-state transcript levels (≥2- to 3-fold; Fig. 9c, d). To determine whether the ie3 gene product directly repressed Clec2d promoter activity, dual-luciferase constructs of the promoter fragments were transfected into the Dox-inducible stable NIH3T3.IE1/3 transfectants. In line with Clr-b protein and transcript data, overexpression of ie3 resulted in significantly decreased Clec2d promoter activity using the intact 500-bp promoter construct, while ie1 had no significant effect (Fig. 9e). Taken together, these findings suggest that the ie3 gene product cell-autonomously represses the Clec2d promoter, in turn partially extinguishing Clr-b nascent transcripts and downregulating surface Clr-b protein early during MCMV infection.
Discussion
This study provides mechanistic evidence that Clr-b (Clec2d) is an inducible IFN-stimulated gene (ISG), in addition to its demonstrated function as a “missing-self” marker of cell health that interacts with the NKR-P1B inhibitory receptor on NK cells and ILC subsets [7]. In keeping with this dual function, we propose that Clr-b enhances self-nonself discrimination by NK cells during viral infection by two means: (i) MCMV-infected cells downregulate resting levels of Clr-b via a host pattern recognition mechanism, in order to render the “missing-self” cells more susceptible to clearance via NK cell disinhibition, while (ii) normal “bystander” cells in the vicinity of infected cells induce Clr-b as a marker of cell health in response to paracrine type-I IFN, thereby reinforcing NK cell inhibition via NKR-P1B. The net effect is that NK cells encounter a broader dynamic range of Clr-b-mediated inhibition/disinhibition during self-nonself discrimination, in turn rendering them better able to integrate signaling inputs and redirect cytotoxic machinery to recognize pathological versus healthy target cells in a battlefield of infection.
This work has shown that both of the above responses are facilitated at the genomic level, where significant changes in Clec2d promoter activity lead to direct and immediate changes in nascent Clr-b transcript and protein levels. In this model, infected cells lose Clr-b in a manner partially attributable to ie3-mediated Clec2d repression, presumably involving a loss of transcription factors and/or RNAPII occupancy at the Clec2d promoter (see below). Meanwhile, infected cells secreting type-I IFN may be sensed in a paracrine manner by uninfected bystander cells via the IFNAR1 receptor. Subsequently, canonical IFNAR1 signaling activates JAK1/TYK2 to phosphorylate and activate STAT1 and STAT2 to heterotrimerize in a complex with IRF9 as ISGF3. An ISGF3-containing transcription factor complex is then recruited to the Clec2d promoter at the proximal ∼200-bp IRFC element (the IRF3/7/9 cluster, which acts as an IFN-stimulated response element), in turn augmenting Clr-b nascent transcripts and protein levels. This combined regulation mechanism is significant because it creates a greater disparity between the infected and uninfected cell populations, facilitating enhanced NK cell recognition.
On the other hand, in infected cells, pattern recognition receptor (PRR) signaling events following the detection of MCMV-specific pathogen-associated molecular patterns (such as cytosolic nucleic acids), in combination with the effects of ie3 on host gene transcription (and/or unidentified immunoevasins), lead directly to the loss of Clec2d promoter occupancy, Clr-b transcripts, and surface protein. While the complexity of this “missing-self” host response mechanism remains to be elucidated, the direct repressive effects of ie3 on host gene expression in general and Clec2d in particular follow the tenets of a pattern recognition system, implicating the NKR-P1B: Clr-b interaction as a pattern recognition receptor axis where the pattern itself is host transcriptional integrity.
In support of this, previous studies have shown that ie3 (M122) is critical for MCMV fitness, such that without it infectious viral progeny can only be produced by cellular ie3 complementation [34, 35, 36]. Thus, MCMV (the pathogen) cannot easily mutate ie3 (the molecule) to avoid cellular detection of its effects on host cells (the pattern - shutting down host gene expression and turning on early viral genes) without compromising viral fitness. In turn, the Clec2d gene serves by proxy as an innate detector (the recognition axis) of the active repression of cellular host gene expression by diverse viruses. Following the shut-off of Clec2d, this signal is relayed to NK cells via the “missing-self” loss of Clr-b, which acts as a marker of cell health and the loss of which disinhibits (activates) NK cells via NKR-P1B (the receptor). In contrast, normal bystander cells augment Clec2d and Clr-b expression in response to type-I IFN as a mechanism to relay to NK cells that they are indeed healthy yet responding to paracrine IFN as a byproduct of pathological infected cells in the nearby vicinity.
One remaining unanswered question is how infection with diverse viruses (CMV, HSV, and poxviruses) mechanistically promotes a rapid loss of Clec2d transcription in infected cells. One possibility is a general viral shut-off and subversion of host mRNA transcription. Another possibility is that MCMV may encode an immunoevasin that directly modulates Clr-b levels by design. Interestingly, we have previously shown that both rat CMV and MCMV encode decoy (Clr-b-surrogate) immunoevasins to elude missing-self recognition via NKR-P1B [16, 19], perhaps suggesting that the viruses cannot otherwise subvert general host mRNA loss and instead directly target host Clr-b and/or NKR-P1B function. Notably, while previous attempts have failed to identify CMV genes that directly lead to the loss of Clr-b expression, here we show that exogenous expression of ie3 autonomously promotes Clr-b downregulation in the absence of infection. Since ie3 is required for productive virus infection and early CMV gene expression [34, 35, 36], the virus cannot easily circumvent this requirement, making Clr-b a pattern recognition axis and the NKR-P1B receptor a self-specific pattern recognition receptor. IE3 is known to promote cell cycle arrest in G1/G₀ and interact in MCMV replication compartments with PML, Daxx, and ND10 complex proteins involved in viral restriction, and the HCMV homolog IE2 functions to repress important host transcription factors (including HDAC and TBP/TFIID) [37, 38, 39, 40] and it is thought to function as a transcription-associated factor [41]. This is interesting because the Clr-b promoter contains an inverted TATA motif and may function as a TBP-dependent promoter. Future studies will elucidate ie3-interacting partners in host gene transcriptional regulation. It is also possible that ie3 is not the sole gene involved in Clr-b downregulation during infection. Rather, since it is an immediate early gene, ie3 likely acts as an immediate repressor of Clec2d, whereas early or late gene products may then contribute to the complete loss of surface Clr-b and/or counteract the loss of Clr-b ligand or NKR-P1B inhibition specifically. Indeed, our current work implicates an m02 family member as a decoy ligand for the NKR-P1B receptor, and other ongoing work implicates an m145 family member in moderately antagonizing Clr-b downregulation during MCMV infection.
In conclusion, this work has demonstrated that Clr-b, a marker of cell health and a self ligand for the NKR-P1B inhibitory receptor, is reciprocally regulated on MCMV-infected and bystander cells. While the mechanism of “missing-self” Clr-b downregulation remains to be fully characterized, Clr-b induction on bystander cells occurs via a canonical paracrine type-I IFN signaling mechanism, making Clr-b a bona fide ISG. This dual tuning mechanism involving the inhibitory NKR-P1B: Clr-b axis facilitates NK cell self-nonself discrimination during viral infection by increasing the recognition threshold between pathological (missing-self) targets and normal (healthy-self) bystander cells.
Disclosure Statement
No competing financial interests exist.
Supplementary Material
Supplementary data
Supplementary data
Acknowledgements
We thank Dr. A. Makrigiannis and Dr. S. Vidal for providing us with the MCMV viruses, Dr. E. Fish and Beata Majchrzak for the IFN-α4 supernatant, and Dr. Geneve Awong, Courtney McIntosh, and Vincent Cheng for cell sorting. We also thank Drs. J.C. Züñiga-Pflücker, A. Martin, A. Nagy, and T. Schrader for plasmids. C.L.K. was funded by a CGS-M Award from the Natural Sciences and Engineering Research Council of Canada (NSERC) and Ontario Graduate Scholarship (OGS) Awards. A.M. was funded by a Vanier Scholarship from the Canadian Institutes of Health Research (CIHR), and O.A.A. and J.H.F. were funded by PGS-D3 scholarships from NSERC. This work was funded by an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund and grants from the CIHR (106491 to J.R.C.).
References
- 1.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 2.Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, Lindsten T, Reiner SL. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36:55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, Powrie F, Vivier E. Innate lymphoid cells - a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 4.Kirkham CL, Carlyle JR. Complexity and diversity of the NKR-P1: Clr (Klrb1: Clec2) recognition systems. Front Immunol. 2014;5:214. doi: 10.3389/fimmu.2014.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hao L, Klein J, Nei M. Heterogeneous but conserved natural killer receptor gene complexes in four major orders of mammals. Proc Natl Acad Sci USA. 2006;103:3192–3197. doi: 10.1073/pnas.0511280103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrillo-Bustamante P, Kesmir C, de Boer RJ. The evolution of natural killer cell receptors. Immunogenetics. 2016;68:3–18. doi: 10.1007/s00251-015-0869-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iizuka K, Naidenko OV, Plougastel BF, Fremont DH, Yokoyama WM. Genetically linked C-type lectin-related ligands for the NKRP1 family of natural killer cell receptors. Nat Immunol. 2003;4:801–807. doi: 10.1038/ni954. [DOI] [PubMed] [Google Scholar]
- 8.Carlyle JR, Jamieson AM, Gasser S, Clingan CS, Arase H, Raulet DH. Missing self-recognition of Ocil/Clr-b by inhibitory NKR-P1 natural killer cell receptors. Proc Natl Acad Sci USA. 2004;101:3527–3532. doi: 10.1073/pnas.0308304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlyle JR, Mesci A, Fine JH, Chen P, Belanger S, Tai LH, Makrigiannis AP. Evolution of the Ly49 and Nkrp1 recognition systems. Semin Immunol. 2008;20:321–330. doi: 10.1016/j.smim.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Chen P, Belanger S, Aguilar OA, Zhang Q, St-Laurent A, Rahim MM, Makrigiannis AP, Carlyle JR. Analysis of the mouse 129-strain Nkrp1-Clr gene cluster reveals conservation of genomic organization and functional receptor-ligand interactions despite significant allelic polymorphism. Immunogenetics. 2011;63:627–640. doi: 10.1007/s00251-011-0542-8. [DOI] [PubMed] [Google Scholar]
- 11.Plougastel B, Dubbelde C, Yokoyama WM. Cloning of Clr, a new family of lectin-like genes localized between mouse Nkrp1a and Cd69. Immunogenetics. 2001;53:209–214. doi: 10.1007/s002510100319. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Q, Rahim MM, Allan DS, Tu MM, Belanger S, Abou-Samra E, Ma J, Sekhon HS, Fairhead T, Zein HS, Carlyle JR, Anderson SK, Makrigiannis AP. Mouse Nkrp1-Clr gene cluster sequence and expression analyses reveal conservation of tissue-specific MHC-independent immunosurveillance. PLoS One. 2012;7:e50561. doi: 10.1371/journal.pone.0050561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogler I, Steinle A. Vis-a-vis in the NKC: genetically linked natural killer cell receptor/ligand pairs in the natural killer gene complex (NKC) J Innate Immun. 2011;3:227–235. doi: 10.1159/000324112. [DOI] [PubMed] [Google Scholar]
- 14.Chen P, Aguilar OA, Rahim MM, Allan DS, Fine JH, Kirkham CL, Ma J, Tanaka M, Tu MM, Wight A, Kartsogiannis V, Gillespie MT, Makrigiannis AP, Carlyle JR. Genetic investigation of MHC-independent missing-self recognition by mouse NK cells using an in vivo bone marrow transplantation model. J Immunol. 2015;194:2909–2918. doi: 10.4049/jimmunol.1401523. [DOI] [PubMed] [Google Scholar]
- 15.Rahim MM, Chen P, Mottashed AN, Mahmoud AB, Thomas MJ, Zhu Q, Brooks CG, Kartsogiannis V, Gillespie MT, Carlyle JR, Makrigiannis AP. The mouse NKR-P1B: Clr-b recognition system is a negative regulator of innate immune responses. Blood. 2015;125:2217–2227. doi: 10.1182/blood-2014-02-556142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voigt S, Mesci A, Ettinger J, Fine JH, Chen P, Chou W, Carlyle JR. Cytomegalovirus evasion of innate immunity by subversion of the NKR-P1B: Clr-b missing-self axis. Immunity. 2007;26:617–627. doi: 10.1016/j.immuni.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Fine JH, Chen P, Mesci A, Allan DS, Gasser S, Raulet DH, Carlyle JR. Chemotherapy-induced genotoxic stress promotes sensitivity to natural killer cell cytotoxicity by enabling missing-self recognition. Cancer Res. 2010;70:7102–7113. doi: 10.1158/0008-5472.CAN-10-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams KJ, Wilson E, Davidson CL, Aguilar OA, Fu L, Carlyle JR, Burshtyn DN. Poxvirus infection-associated downregulation of C-type lectin-related-b prevents NK cell inhibition by NK receptor protein-1B. J Immunol. 2012;188:4980–4991. doi: 10.4049/jimmunol.1103425. [DOI] [PubMed] [Google Scholar]
- 19.Aguilar OA, Mesci A, Ma J, Chen P, Kirkham CL, Hundrieser J, Voigt S, Allan DS, Carlyle JR. Modulation of Clr ligand expression and NKR-P1 receptor function during murine cytomegalovirus infection. Journal of Innate Immunity. 2015;7:584–600. doi: 10.1159/000382032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith MG. Propagation in tissue cultures of a cytopathogenic virus from human salivary gland virus (SGV) disease. Proc Soc Exp Biol Med. 1956;92:424–430. doi: 10.3181/00379727-92-22498. [DOI] [PubMed] [Google Scholar]
- 21.Henry SC, Schmader K, Brown TT, Miller SE, Howell DN, Daley GG, Hamilton JD. Enhanced green fluorescent protein as a marker for localizing murine cytomegalovirus in acute and latent infection. J Virol Methods. 2000;89:61–73. doi: 10.1016/s0166-0934(00)00202-0. [DOI] [PubMed] [Google Scholar]
- 22.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung HK, Nagy A. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlyle JR, Martin A, Mehra A, Attisano L, Tsui FW, Zuniga-Pflucker JC. Mouse NKR-P1B, a novel NK1.1 antigen with inhibitory function. J Immunol. 1999;162:5917–5923. [PubMed] [Google Scholar]
- 24.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 25.Ni Z, Bremner R. Brahma-related gene 1-dependent STAT3 recruitment at IL-6-inducible genes. J Immunol. 2007;178:345–351. doi: 10.4049/jimmunol.178.1.345. [DOI] [PubMed] [Google Scholar]
- 26.Abrisch RG, Eidem TM, Yakovchuk P, Kugel JF, Goodrich JA. Infection by herpes simplex virus type-1 causes near-complete loss of RNA polymerase II occupancy on the host cell genome. J Virology. 2015;90:2503–2513. doi: 10.1128/JVI.02665-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frank DA, Mahajan S, Ritz J. Fludarabine-induced immunosuppression is associated with inhibition of STAT1 signaling. Nat Med. 1999;5:444–447. doi: 10.1038/7445. [DOI] [PubMed] [Google Scholar]
- 28.Torella D, Curcio A, Gasparri C, Galuppo V, De Serio D, Surace FC, Cavaliere AL, Leone A, Coppola C, Ellison GM, Indolfi C. Fludarabine prevents smooth muscle proliferation in vitro and neointimal hyperplasia in vivo through specific inhibition of STAT-1 activation. Am J Physiol Heart Circ Physiol. 2007;292:H2935–H2943. doi: 10.1152/ajpheart.00887.2006. [DOI] [PubMed] [Google Scholar]
- 29.Nelson EA, Walker SR, Kepich A, Gashin LB, Hideshima T, Ikeda H, Chauhan D, Anderson KC, Frank DA. Nifuroxazide inhibits survival of multiple myeloma cells by directly inhibiting STAT3. Blood. 2008;112:5095–5102. doi: 10.1182/blood-2007-12-129718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenoever BR, Ng SL, Chua MA, McWhirter SM, Garcia-Sastre A, Maniatis T. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science. 2007;315:1274–1278. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- 31.Zimmermann A, Trilling M, Wagner M, Wilborn M, Bubic I, Jonjic S, Koszinowski U, Hengel H. A cytomegaloviral protein reveals a dual role for STAT2 in IFN-(gamma) signaling and antiviral responses. J Exp Med. 2005;201:1543–1553. doi: 10.1084/jem.20041401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petricoin EF, 3rd, Ito S, Williams BL, Audet S, Stancato LF, Gamero A, Clouse K, Grimley P, Weiss A, Beeler J, Finbloom DS, Shores EW, Abraham R, Larner AC. Antiproliferative action of interferon-alpha requires components of T-cell-receptor signalling. Nature. 1997;390:629–632. doi: 10.1038/37648. [DOI] [PubMed] [Google Scholar]
- 33.Sun S, Zhang X, Tough DF, Sprent J. Type I interferon-mediated stimulation of T cells by CpG DNA. J Exp Med. 1998;188:2335–2342. doi: 10.1084/jem.188.12.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buhler B, Keil GM, Weiland F, Koszinowski UH. Characterization of the murine cytomegalovirus early transcription unit e1 that is induced by immediate-early proteins. J Virol. 1990;64:1907–1919. doi: 10.1128/jvi.64.5.1907-1919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Messerle M, Buhler B, Keil GM, Koszinowski UH. Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate-early gene 3. J Virol. 1992;66:27–36. doi: 10.1128/jvi.66.1.27-36.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angulo A, Ghazal P, Messerle M. The major immediate-early gene ie3 of mouse cytomegalovirus is essential for viral growth. J Virol. 2000;74:11129–11136. doi: 10.1128/jvi.74.23.11129-11136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hagemeier C, Walker S, Caswell R, Kouzarides T, Sinclair J. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J Virol. 1992;66:4452–4456. doi: 10.1128/jvi.66.7.4452-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caswell R, Hagemeier C, Chiou CJ, Hayward G, Kouzarides T, Sinclair J. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J Gen Virol. 1993;74:2691–2698. doi: 10.1099/0022-1317-74-12-2691. [DOI] [PubMed] [Google Scholar]
- 39.Martinez FP, Cosme RS, Tang Q. Murine cytomegalovirus major immediate-early protein 3 interacts with cellular and viral proteins in viral DNA replication compartments and is important for early gene activation. J Gen Virol. 2010;91:2664–2676. doi: 10.1099/vir.0.022301-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez KJ, Martinez FP, Cosme-Cruz R, Perez-Crespo NM, Tang Q. A short cis-acting motif in the M112–113 promoter region is essential for IE3 to activate M112-113 gene expression and is important for murine cytomegalovirus replication. J Virol. 2013;87:2639–2647. doi: 10.1128/JVI.03171-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukac DM, Harel NY, Tanese N, Alwine JC. TAF-like functions of human cytomegalovirus immediate-early proteins. J Virol. 1997;71:7227–7239. doi: 10.1128/jvi.71.10.7227-7239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data