Abstract
The Drosophila immune deficiency pathway defends many bacterial pathogens and bears striking molecular similarities to the mammalian tumor necrosis factor signal transduction pathway. Orthologous inhibitors of apoptosis ubiquitin ligases act at a proximal stage of both responses to coordinate the assembly of signal transduction platforms that shape host immune responses. Despite the importance of inhibitor of apoptosis proteins within evolutionarily conserved innate immune responses, we know relatively little about the cellular machinery that controls inhibitor of apoptosis activity. In this study, we examined the molecular basis for inhibitor of apoptosis 2 protein regulation in the immune deficiency pathway. Our studies identified two distinct proteolytic events that determine the stability and composition of cellular inhibitor of apoptosis 2 protein pools. We found that apoptotic caspase activity cleaves inhibitor of apoptosis 2 at an N-terminal aspartate to generate a truncated protein that retains the ability to interact with immune deficiency pathway members. We also showed that a C-terminal ubiquitin ligase activity within inhibitor of apoptosis 2 directs the proteasomal destruction of full-length and truncated inhibitor of apoptosis 2 isoforms. These studies add to our appreciation of the regulation of innate immunity and suggest potential links between apoptotic caspases and innate defenses.
Key Words: Dredd, Dronc, IAP2, Immune deficiency, TNF pathway, Tumor necrosis factor
Introduction
The Drosophila melanogaster immune deficiency (IMD) pathway orchestrates a rapid immunological response upon detection of a bacterial diaminopimelic peptidoglycan (PGN) [1]. The IMD pathway has striking molecular similarities and critical physiological differences to the mammalian tumor necrosis factor (TNF) pathway. TNF signals through a homotrimeric TNF receptor to drive a number of context-dependent events that may include leukocyte extravasation, differentiation, proliferation and caspase-8-mediated cell death [2]. In contrast, the IMD pathway signals through a PGN recognition protein (PGRP) receptor to coordinate a nonapoptotic defense that includes elevated levels of circulating antimicrobial peptides.
Despite the overt differences between the IMD and the TNF pathway at the levels of ligand-receptor interactions and host response, both pathways share an evolutionarily conserved core of signal transduction molecules. In both cases, the receptor recruits a death domain-bearing adaptor that signals through inhibitor of apoptosis protein (IAP)-type ubiquitin ligases to a Tak1-associated binding protein (TAB)-TGF-β-activated kinase 1 (TAK1)-I-κB kinase (IKK) axis that activates nuclear factor (NF)-κB transcription factors. Many components of the IMD pathway were identified in a series of genetic and molecular studies that spanned a decade. Subsequent work delineated a complex arrangement of biochemical events that define IMD signals. While some details require clarification in animal models, available data suggest that the PGRP-LC receptor binds extracellular PGN [3, 4, 5, 6, 7] and nucleates a proximal complex that contains the death domain-bearing adaptor molecule Imd [8, 9, 10], the Drosophila Fas-associated death domain (FADD) ortholog [11, 12, 13] and the caspase-8 ortholog Dredd [14, 15, 16, 17]. Dredd proteolytically removes the N-terminal 30 amino acids of Imd [18] and cleaves between the N-terminal NF-κB domain and the cytosolic ankyrin domain of the p105 ortholog Relish (Rel) [17, 19].
The N-terminal residues of cleaved Imd constitute an IAP binding motif that recruits the RING domain-bearing Drosophila E3 ubiquitin ligase IAP2. Early studies implicated IAP2 in the control of programmed cell death in Drosophila [20, 21]. Later mutant and biochemical studies revealed an essential requirement for IAP2 in the IMD response [22, 23, 24, 25]. Biochemical data suggest that IAP2 is required for the conjugation of K63-linked ubiquitin chains to Dredd and Imd [18, 26]. By analogy to events in the TNF pathway, it is likely that ubiquitylated Imd recruits the Drosophila TAB2 ortholog [22, 24, 27] to initiate a kinase relay via the Drosophila TAK1 ortholog [28, 29], which simultaneously activates Drosophila c-Jun N-terminal kinase (JNK) [30, 31, 32] and IKK modules [33, 34, 35]. The IKK complex phosphorylates the NF-κΒ domain of Rel to establish a nuclear pool of phosphorylated NF-κΒ that promotes a rapid and comprehensive antibacterial transcriptional response.
Despite the apparent similarities between vertebrate TNF and invertebrate IMD signals, there are profound differences at the level of caspase activity. Whereas Dredd is a caspase-8 ortholog with an essential role in IMD responses and no known roles in cell death, mammalian caspase-8 acts at a later stage of the TNF pathway to induce an apoptotic response [36]. Active caspase-8 engages mitochondrial apoptotic factors to induce proteolytic activation of the initiator caspase-9, which stimulates a complex feed-forward loop that results in disassembly of the responding cell by apoptosis. In contrast to mammalian systems, there are no established interactions between the IMD response and the Drosophila caspase-9 ortholog Dronc [37, 38, 39, 40].
The remarkable differences between caspase involvement in the TNF and IMD pathways prompted us to explore the proximal, caspase-dependent events in the IMD pathway. We were particularly interested in relationships between Drosophila caspases and IAP2, as there are comparatively few data on the cellular control of IAP2 protein levels. In this study, we identified the interaction domains between IAP2 and Dredd, and produced cell culture and whole animal data that validated an N-terminal cleavage of IAP2 by apoptotic caspases. We demonstrated that cleaved IAP2 retains the ability to interact with IMD signal transduction molecules and suggest that Dronc is the principal caspase required for the cleavage of IAP2. We also uncovered a caspase-independent, proteasomal regulation of total IAP2 protein levels. Combined, our data expand our appreciation of the molecular regulation of an evolutionarily ancient and essential immune response pathway, and suggest possible connections between apoptotic caspases and immune signaling in Drosophila.
Materials and Methods
S2 Cell Culture
S2 cells were cultured at 25°C in HyQ TNM-FH medium (HyClone) with 10% heat-inactivated fetal bovine serum, 50 units/ml penicillin and 50 μg/ml streptomycin (Invitrogen). For transient transfections, cells were plated at 1 × 106 cells/ml and transfected with 2 μg/ml of plasmid DNA with Cellfectin II (Invitrogen) according to the manufacturer's recommendations. Expression analysis and immunoprecipitations were performed with 1 ml of cells in 12-well tissue culture plates or 3 ml of cells in 6-well plates, respectively. To monitor protein stability, S2 cells were incubated with 2.5 μg/ml of the translation inhibitor cycloheximide (Sigma) for the indicated times. To activate the IMD pathway, cells were treated with 5 μg/ml PGN (InvivoGen). To block proteasomal activity, cells were incubated with MG132 in DMSO (Sigma) for 3 h. RNAi was performed as described previously [41]. To inhibit caspase function, S2 cells were incubated with 100 μM of the general caspase inhibitor zVAD-FMK in DMSO (R&D Systems) for 3 h. To induce apoptosis, S2 cells were incubated with 25 μg/ml cycloheximide in DMSO for 8 h.
Generation of Expression Plasmids
The IAP2, Dredd, DreddCA and FADD expression constructs used in this study have been described elsewhere [41]. The Dredd variant used is the γ isoform of the Dredd* described previously [42]. The remainder of the expression constructs were generated by PCR from the respective cDNAs using the primers indicated in table 1. The individual PCR products were cloned into pENTR/D-TOPO (Invitrogen). Epitope-tagged expression constructs were generated by LR recombination with pAMW (6 × Myc) or pAHW (3 × HA) in a gateway LR clonase reaction according to the manufacturer's instructions (Invitrogen). To generate UAS-IAP2 transgenic flies, pENTR/D-TOPO IAP2 was recombined with pTMW in a gateway LR clonase reaction. Transgenic flies were made by Bestgene Inc.
Table 1.
Primers for expression constructs Name Forward Reverse
| IAP2 Δ 75 | CACCATGCTGGCTCCCAATCACTGC | TCACGAAAGGAACGTGCGC |
|---|---|---|
| IAP2 180–450 | CACCATGCAAATGGGCCCCCTTATAGAG | TCATAAGCGTGCATCCTTTAG |
| IAP2 1–450 | CACCATGACGGAGCTGGG | TCATAAGCGTGCATCCTTTAG |
| Dronc | CACCATGCAGCCGCCGGA | CTATTCGTTGAAAAACCCGGGATTG |
| Dredd - prodomain | CACCATGTCAGCGAGTGC | TCAATCCGTGCCATCCCG |
| Dredd - catalytic subunit | CACCGTGGATAAAGAACGACTAATCG | TCACAGACGAGGTGG |
Table 2 lists the primers used for site-directed mutagenesis of the respective cDNAs. To eliminate the parental (methylated) plasmid DNA, the newly synthesized nonmethylated DNA product was digested with 20 U Dpn1 (New England Biolabs) for 1 h at 37°C. Of the final reaction, 3 μl were transformed into 50 μl competent DH5α Escherichia coli.
Table 2.
Primers for site-directed mutagenesis
| Name | Forward | Reverse |
|---|---|---|
| IAP2D92A | CCCAGGAGCCAGGAGAGCGCCAACGAGGGAA | CGCTGTTTCCCTCGTTGGCGCTCTCCTGGCT |
| ACAGCG | CCTGGG | |
| IAP2D100A | CGAGGGAAACAGCGTAGTGGCCAGCCCGGA | GCAGGACTCCGGGCTGGCCACTACGCTGTTT |
| GTCCTGC | CCCTCG | |
| IAP2C472Y | GGCGTAGTGTTCCTGCCCTATGGCCACTTGGCC | GCAGGTGGCCAAGTGGCCATAGGGCAGGAACAC |
| ACCTGC | TACGCC | |
| DroncC318S | GGTGCTTATGTTTCCCTTTTCCCGCGGCGATG | CATATTCATCGCCGCGGGAAAAGGGAAACATA |
| AATATG | AGCACC |
The sequence of each expression construct was verified before use.
Immunoprecipitation
Cells were centrifuged at 1,000 g for 3 min and lysed in 200 μl lysis buffer [50 mM HEPES (pH 7.5), 10 mM EDTA (pH 8), 100 mM NaCl, 0.2% Nonidet P-40, protease inhibitors (Roche Applied Science), phosphatase inhibitors (Sigma, phosphatase inhibitor mixture)] for 10 min at 4°C. Samples were cleared by centrifugation at 21,000 g for 10 min at 4°C and then incubated with mouse anti-HA (Sigma, 1:400) at 4°C overnight. A 1:1 slurry of BSA-blocked protein G-Sepharose beads in lysis buffer (Amersham Biosciences) was added and samples were incubated for an additional hour at 4°C. Beads were pelleted three times by centrifugation briefly at 300 g and washed in lysis buffer. After the final wash, the supernatant was discarded and beads were resuspended in 2× sample buffer for analysis by Western blot.
Western Blots
Western blots were performed as described [39]. The antibodies used in this study were: rabbit anti-Myc (Sigma, 1:5,000); mouse anti-tubulin (DSHB E7, 1:1,000); mouse monoclonal anti-HA (Sigma, 1:5,000); rabbit anti-JNK (Santa Cruz Biotechnology, 1:4,000) and mouse anti-phospho-JNK (Cell Signaling, 1:2,000).
Results
Structural Evaluation of Interactions between Dredd and IAP2
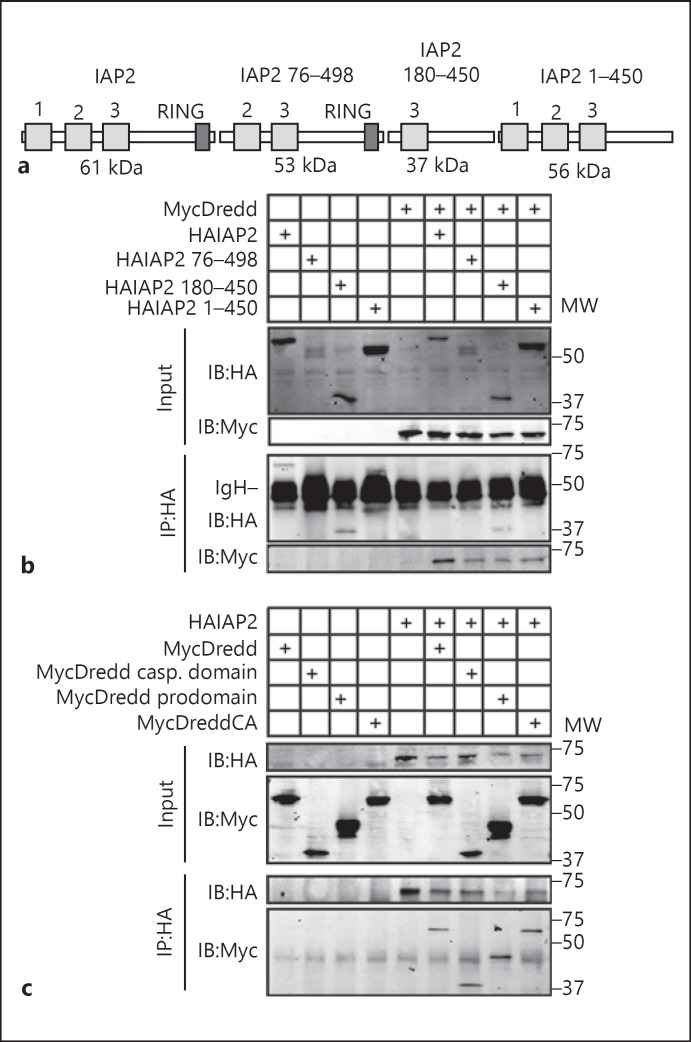
To identify the structural determinants of interactions between Dredd and IAP2, we monitored the ability of truncated IAP2 and Dredd variants to interact. For these studies, we generated an N-terminally HA-tagged full-length IAP2 expression construct, as well as HA-tagged expression constructs that lacked the N-terminal baculovirus IAP repeat (BIR) motif 1 (IAP2 76–498); BIR motifs 1 and 2 and the RING domain (IAP2 180–450), or the RING domain (IAP2 1–450; fig. 1a). We tested each IAP2 construct for its ability to precipitate N-terminally Myc-tagged Dredd in co-immunoprecipitation assays. We found that the HA antibody alone failed to immunoprecipitate MycDredd (fig. 1b, lane 5). In contrast, we readily detected a co-precipitation of MycDredd with all IAP2 expression constructs tested (fig. 1b, lanes 6–9). We interpret these data to suggest that the central 180–450 amino acids of IAP2 are sufficient to sustain interactions with Dredd.
Fig. 1.
a Schematic illustration of the IAP2 expression constructs used in this study. Numbers indicate BIR motifs in the respective constructs and a dark grey box indicates the RING domain. MW of the respective expression constructs including the 3HA tag are indicated below each protein. b, c Co-immunoprecipitation studies of lysates from S2 cells that were transfected with the expression constructs indicated at the top of the gel, and immunoprecipitated with a monoclonal anti-HA antibody. Input and immunoprecipitate fractions were visualized with anti-HA and anti-myc antibodies as indicated. MW is shown for each blot in kilodaltons. MycDreddCA indicates a proteolytically inactive variant of MycDredd. IgH indicates the Ig heavy chain from immunoprecipitations (b).
In reverse experiments, we tested the ability of HA-tagged IAP2 to precipitate MycDredd, the caspase domain of Dredd, the prodomain of Dredd or a catalytically inactive Dredd. While the HA antibody failed to immunoprecipitate any Dredd variant (fig. 1c, lanes 1–4), we observed a robust co-precipitation of the respective Dredd variants with IAP2 (fig. 1c, lanes 6–9). These data suggest that the prodomain and catalytic subunits of Dredd form separate interactions with IAP2.
Caspase Activity Modifies IAP2 Protein Levels
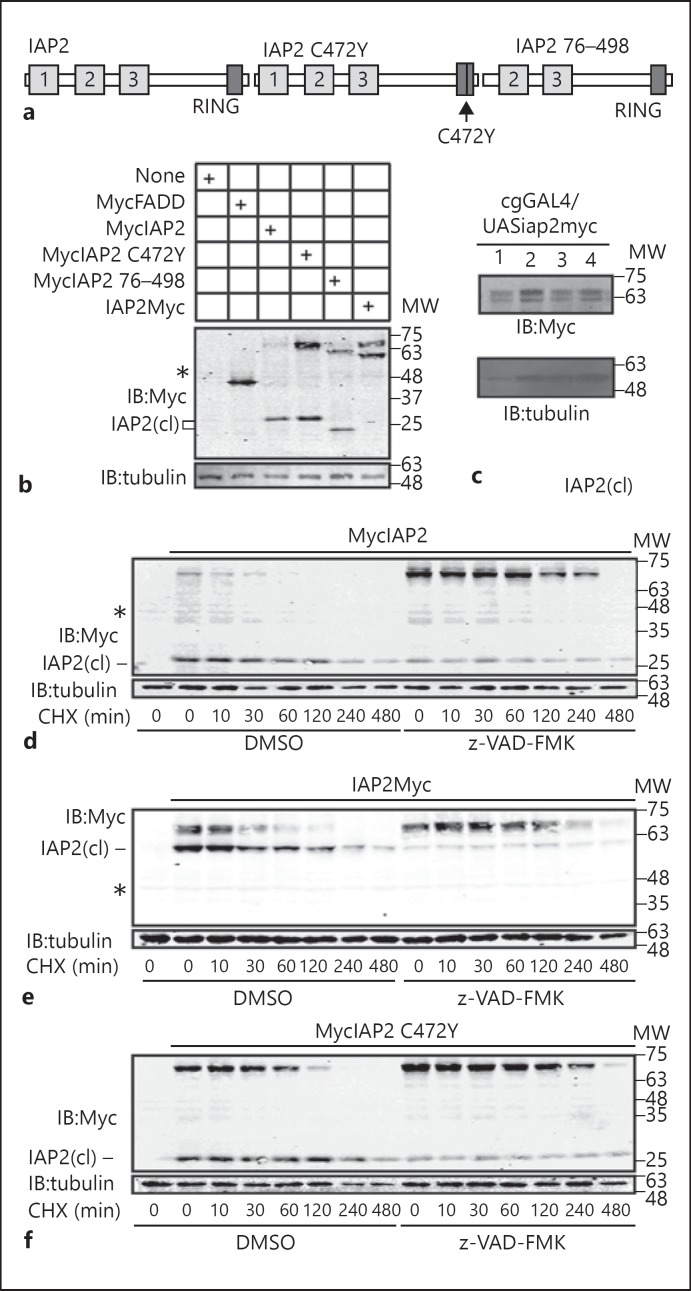
Our studies of Dredd-IAP2 interactions consistently revealed the appearance of two polypeptides of distinct molecular weights (MW) upon expression of epitope-tagged IAP2 constructs in S2 cells. We observed a comparatively faint protein with an MW consistent with full-length IAP2 and a second protein of considerably shorter length that suggests N-terminal processing of IAP2 in S2 cells [fig. 2b, marked as IAP2(cl)]. We observed a similar cleavage product upon expression of MycIAP2 with an inactivating point mutation in the RING domain (IAP2C472Y); MycIAP2 with a deletion of the N-terminal 75 amino acids (MycIAP2 76–498), or C-terminally Myc-tagged IAP2 (IAP2Myc; fig. 2b). We also detected N-terminal processing of IAP2 upon expression of a C-terminally Myc-tagged IAP2 transgenic construct in the fat bodies of adult flies (fig. 2c). Combined, our data suggest a RING domain-independent N-terminal processing of IAP2 in cell culture and in vivo, and they are consistent with a previous report of caspase-mediated cleavage of IAP2 in S2 cells [21].
Fig. 2.
a Schematic illustration of IAP2 expression constructs used in this study. b Western blot analysis of lysates from S2 cells that were transfected with the expression constructs indicated in the table. Cleaved IAP2 isoforms are indicated with IAP2(cl). c Western blot analysis of lysates from adult flies of the genotype cgGAL4/UASiap2. Four separate UASiap2 transgenic lines were used for these assays. d-f S2 cells were transfected with the expression constructs indicated at the top of each gel, and incubated with the translation inhibitor cycloheximide (CHX) for the indicated times. Samples were pre-incubated with DMSO or z-VAD-FMK as shown. Total cell lysates were immunoblotted with anti-myc antibodies where indicated. Cleaved IAP2 isoforms are indicated with IAP2(cl) and asterisks indicate a cross-reacting sample in Myc blots. For each blot, tubulin was measured as a loading control and MW is shown in kilodaltons.
To confirm caspase involvement in the processing of IAP2, we incubated S2 cells that expressed IAP2 or IAP2C472Y with the caspase inhibitor z-VAD-FMK. We treated each cell with the translation inhibitor cycloheximide and monitored the relative stability of the individual proteins. We noticed a substantial stabilization of full-length IAP2 in S2 cells treated with z-VAD-FMK irrespective of the location of the myc tag (fig. 2d, e). When we treated S2 cells with nonlethal doses of the translation inhibitor cycloheximide, we noticed that the cleaved IAP2 product persisted longer than the full-length isoform (fig. 2e, lanes 2–8). Inactivation of the RING domain catalytic activity stabilized full-length IAP2 (compare fig. 2f, lanes 2–8, to fig. 2d, lanes 2–8). However, we still detected a z-VAD-sensitive processing of RING domain mutant IAP2 (fig. 2f). These data suggest two proteolytic regulations of the IAP2 protein: caspase-dependent generation of an N-terminal cleavage product and RING domain-mediated destabilization of the full-length protein.
Synergistic Control of IAP2 Protein Levels
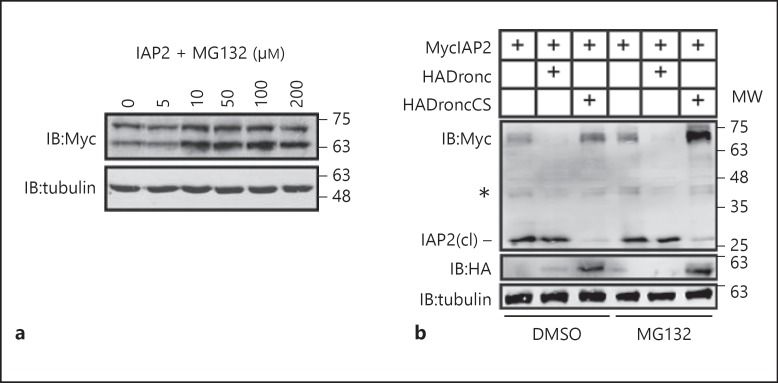
As RING domains are frequently implicated in the ubiquitin-mediated destruction of substrate proteins via the proteasome, we explored the impact of proteasomal inhibition on IAP2 protein levels. For these assays, we transfected S2 cells with an IAP2Myc expression plasmid and incubated the cells with increasing doses of the proteasomal inhibitor MG132. We found that all concentrations above 10 μM MG132 stabilized both the full-length and cleaved IAP2 isoforms (fig. 3a). The data in figure 3a suggest that proteasome inhibition does not affect the N-terminal cleavage of IAP2. These observations led us to ask if caspase activity and the proteasome synergistically control the levels of full-length IAP2. To test this hypothesis, we monitored IAP2 in S2 cells with independent or parallel inhibition of the effector caspase Dronc and the proteasome. Consistent with an involvement of caspases in the processing of IAP2, we found that expression of wild-type Dronc accelerated cleavage of IAP2, and expression of a dominant inhibitory Dronc variant (DroncCS) prevented processing of IAP2 (fig. 3b, lanes 2 and 3, respectively). Inhibition of the proteasome with MG132 also resulted in moderate stabilization of full-length IAP2 (fig. 3b, lane 4). In contrast, simultaneous inhibition of the proteasome and expression of DroncCS greatly increased the levels of full-length IAP2 (fig. 3b, lane 6). These findings uncover two separate proteolytic controls of IAP2 protein levels. Caspase activity generates a truncated protein independent of RING domain activity, and the IAP2 RING domain induces proteasomal degradation of both IAP2 isoforms.
Fig. 3.
Western blot of lysates from S2 cells transfected with the expression constructs indicated in the table at the top of the gel. a Cells were incubated with varying doses of the proteasomal inhibitor MG132 as indicated. b Cells were incubated with the proteasomal inhibitor MG132 or the MG132 solvent DMSO as indicated. Total cell lysates were immunoblotted with anti-myc and anti-HA antibodies where indicated. Tubulin levels were visualized for each membrane as a loading control. Cleaved IAP2 is indicated with IAP2(cl); asterisk indicates a cross-reacting species in Myc blots. MW is shown in kilodaltons.
Identification of the Caspase That Cleaves IAP2
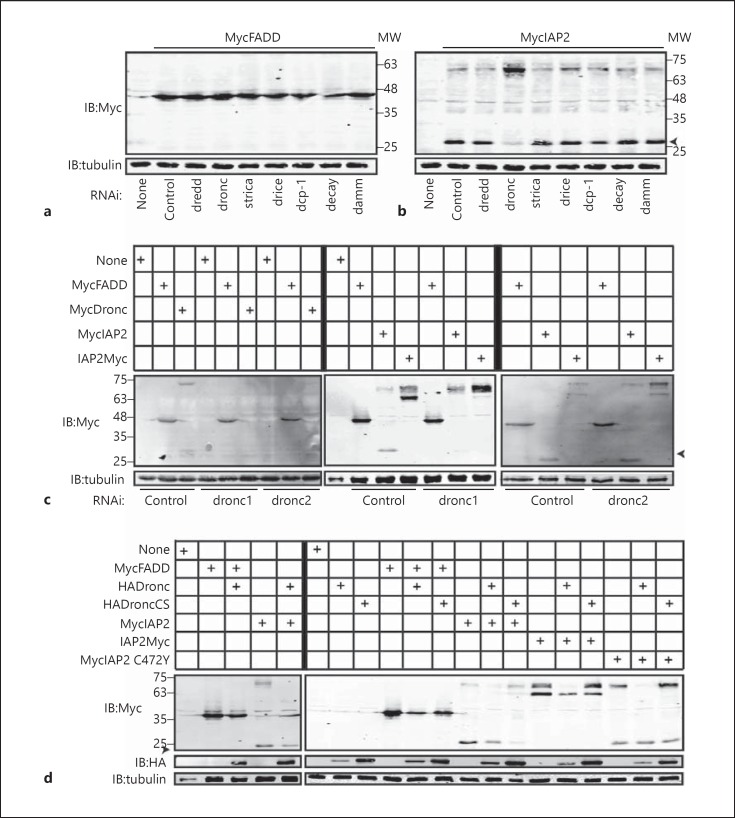
A previous study demonstrated cleavage of IAP2 by Drice in vitro [21]. To examine potential effects of additional caspases on IAP2, we tested all known Drosophila caspases for effects on the N-terminal proteolysis of IAP2. We used RNAi to deplete S2 cells of the seven annotated Drosophila caspases and measured the consequences for IAP2 cleavage. Caspase RNAi did not impair the expression or stability of the control Drosophila protein FADD (fig. 4a). Depletion of Drice had minimal effects on IAP2 cleavage. In contrast, loss of the initiator caspase Dronc greatly impaired cleavage of IAP2 and stabilized the full-length version (fig. 4b). To test requirements for Dronc in IAP2 cleavage and exclude potential off-target effects from our initial dronc dsRNA, we generated a second, nonoverlapping dronc dsRNA. Both dsRNA species prevented expression of Dronc and did not alter the expression of the control protein FADD (fig. 4c). Likewise, both dsRNA molecules blocked cleavage of IAP2 and stabilized the full-length protein (fig. 4c). We observed identical results for IAP2 proteins with Myc tags at the N or C terminus (fig. 4c).
Fig. 4.
Western blot analysis of lysates prepared from S2 cells that were transfected with the expression constructs indicated at the top of each gel. a-d Cells were incubated with the dsRNA indicated at the bottom of the gel prior to transfection. Total cell lysates were immunoblotted with anti-myc and anti-HA antibodies where indicated. Tubulin levels were visualized for each membrane as a loading control. Cleaved IAP2 is indicated with IAP2(cl). MW is shown for each blot in kilodaltons.
To validate a requirement for Dronc in the cleavage of IAP2, we monitored the effects of Dronc expression on IAP2 protein levels. Overexpression of Dronc did not alter the stability of the control protein FADD, but resulted in a complete loss of full-length IAP2 with a persistent expression of the N-terminal cleavage product (fig. 4d). Overexpression of Dronc enhanced IAP2 processing regardless of the location of the Myc tag or the presence of an active RING domain on IAP2 (fig. 4d). Finally, we found that expression of DroncCS stabilized full-length IAP2 (fig. 4d). Combined, these data suggest a novel role for Dronc in the N-terminal cleavage of IAP2.
Dronc Is Required for IAP2 Cleavage
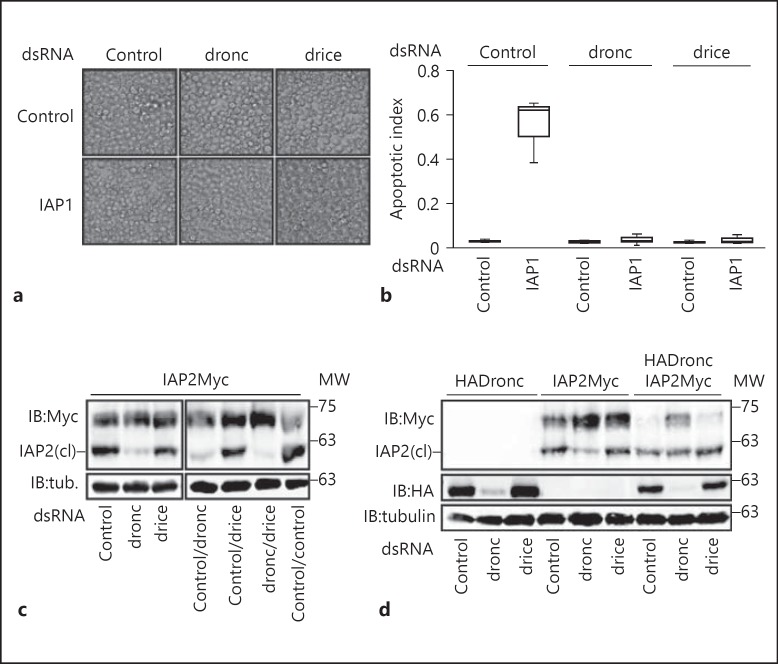
The data presented in figure 4 implicate Dronc in the proteolysis of IAP2. However, as Dronc is an initiator caspase in the Drosophila apoptotic pathway, we cannot exclude indirect effects of Dronc on the processing of IAP2 via additional proteases, such as the effector caspase Drice. To explore this possibility, we examined the consequences of simultaneous depletion of Dronc and Drice for IAP2. We first validated the efficacy of our Dronc and Drice RNAi regime by inhibition of apoptosis in S2 cells. IAP1 is an essential inhibitor of apoptosis in Drosophila, and loss of IAP1 results in widespread apoptosis (fig. 5a, b). Dual depletion of Dronc with IAP1, or Drice with IAP1, completely reverted the induction of apoptosis by depletion of IAP1 alone, indicating an efficient knockdown of both caspases.
Fig. 5.
a Visualization of the induction of apoptosis in S2 cells treated with the indicated combinations of dsRNAs. b Quantification of the relative amount of apoptosis in a. Cells were defined as apoptotic if they displayed a visible amount of membrane blebbing. The apoptotic index represents the ratio of apoptotic cells to total cells. c, d Western blot analysis of lysates prepared from S2 cells that were transfected with the expression constructs indicated at the top of each gel, and with the dsRNA indicated at the bottom of the gel. Total cell lysates were immunoblotted with anti-myc and anti-HA antibodies where indicated. Tubulin levels were visualized for each membrane as a loading control. Cleaved IAP2 is indicated with IAP2(cl). MW is shown for each blot in kilodaltons.
We also looked at IAP2 processing in cells treated with the same dsRNA at the same time as the assays performed in figure 5a, b. While inactivation of Dronc greatly diminished cleavage of IAP2, depletion of Drice had minimal effects (fig. 5c, lanes 2 and 3). Moreover, dual depletion of Dronc and Drice had nearly identical effects on IAP2 processing as depletion of Dronc alone (fig. 5c, lanes 4 and 6). Finally, we confirmed that expression of Dronc increased the extent of IAP2 processing (fig. 5d, lanes 4 and 7). As expected, Dronc RNAi partially reverted the accelerated processing observed upon Dronc expression (fig. 5d, lane 8). In contrast, Drice RNAi did not appear to diminish IAP2 processing (fig. 5d, lane 9). Thus, depletion of Drice to levels sufficient to prevent the genetic induction of apoptosis does not have appreciable effects on the extent of IAP2 processing. While we cannot exclude an involvement of residual Drice protein in IAP2 cleavage, we believe our data are more consistent with a prominent role for Dronc in the N-terminal cleavage of IAP2.
Identification of IAP2 Cleavage Sites
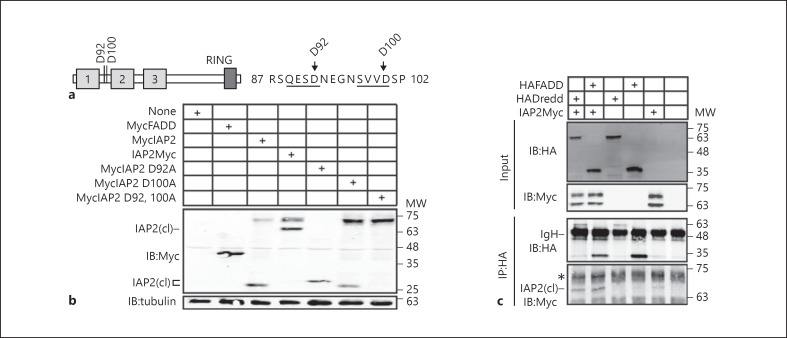
We then asked which residues of IAP2 are sensitive to caspase activity. Analysis of the primary amino acid sequence of IAP2 identified two prospective N-terminal caspase cleavage sites within IAP2 at residues 92 and 100 (fig. 6a). We converted the corresponding aspartates to alanines at both residues independently, or in tandem, and visualized the cleavage of the respective proteins. Introduction of an alanine at residue 92 (IAP2D92A) enhanced the cleavage of IAP2 and mutation of residue 100 (IAP2D100A) visibly impaired IAP2 cleavage, suggesting a critical role for D100 in the regulation of IAP2 stability (fig. 6b). In contrast, mutation of both residues to alanine (IAP2D92, 100A) completely blocked IAP2 cleavage (fig. 6b). These data suggest that IAP2 is cleaved at two aspartates in S2 cells. However, examination of figure 6b reveals that the cleavage product for wild-type IAP2 migrates the same distance as the D100A mutant, and thus corresponds to the shorter 92-residue peptide. Thus, we cannot exclude the possibility that aspartate 92 is the primary caspase substrate in S2 cells.
Fig. 6.
a Schematic illustration of IAP2 with putative caspase cleavage sites at residues 92 and 100. The primary amino acid sequences of residues 87–102 are also shown with putative caspase recognition sites underlined. b Western blot analysis of lysates from S2 cells transfected with the expression constructs indicated in the tables at the top of each gel. Total cell lysates were immunoblotted with anti-myc antibodies where indicated, and tubulin levels were visualized as a loading control. N-terminal IAP2 cleavage products are indicated with IAP2(cl). c Co-immunoprecipitation studies of lysates from S2 cells that were transfected with the expression constructs indicated at the top of the gel, and immunoprecipitated with a monoclonal anti-HA antibody. Input and immunoprecipitate fractions were visualized with anti-HA and anti-myc antibodies as indicated. MW is shown for each blot in kilodaltons.
To date, all studies of interactions between IAP2 and IMD pathway members examined physical interactions with the full-length IAP2 isoform. As our studies suggest that N-terminal cleavage of IAP2 is a constitutive event in cell culture and in vivo, we wanted to determine if cleaved IAP2 interacts with IMD partner proteins. For these assays, we expressed HA-tagged variants of FADD and Dredd alone, or together with IAP2Myc in S2 cells, and determined the ability of the respective IMD elements to co-immunoprecipitate cleaved IAP2. The HA antibody failed to immunoprecipitate the cleaved isoform of IAP2Myc (fig. 6c, lane 5). A cross-reacting band obscured the presence of full-length IAP2 in our immunoprecipitation assays (fig. 6c, indicated with an asterisk). However, we successfully co-precipitated the cleaved IAP2 isoform with HA-tagged FADD and Dredd [fig. 6c, lanes 1 and 2, cleaved IAP2 indicated with IAP2(cl)]. These findings indicate a novel involvement of apoptotic caspases in the generation of a truncated IAP2 variant that retains the ability to interact with immune response proteins.
Induction of Apoptosis Enhances Cleavage of IAP2
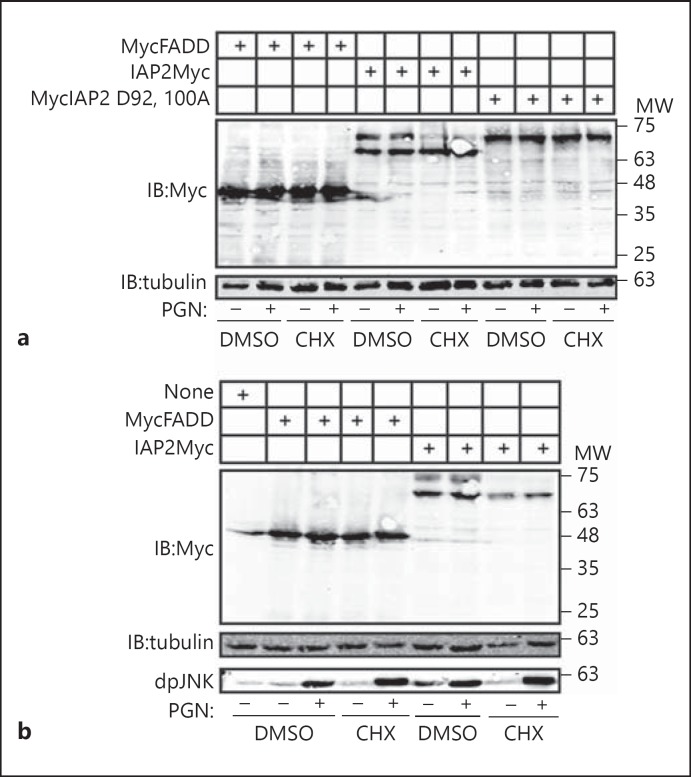
As Dronc is an essential initiator caspase in apoptosis with no known roles in the IMD pathway, we aimed to determine if induction of apoptosis enhances the proteolytic cleavage of wild-type IAP2. To test this hypothesis, we expressed IAP2 and IAP2D92, 100A in S2 cells and induced apoptosis through the addition of toxic doses of cycloheximide. Induction of apoptosis did not alter the expression or processing of the control protein FADD, and the cycloheximide solvent DMSO did not affect IAP2 cleavage (fig. 7a, lanes 1–4). We found that activation of the apoptotic pathway resulted in a complete N-terminal processing of IAP2 (fig. 7a, lanes 7–8) but failed to induce cleavage of the D92, 100A mutant (fig. 7a, lanes 11 and 12). Simultaneous stimulation of the IMD pathway did not alter the enhanced cleavage of IAP2 in apoptotic cells. We then asked if signals through the IMD pathway alter the N-terminal cleavage of IAP2 in the absence of proapoptotic signals. Incubation of S2 cells with PGN induced a robust IMD response as measured by the phosphorylation of Drosophila JNK (fig. 7b) without any measurable effect on IAP2 cleavage (fig. 7). Our findings suggest that Dronc is required for constitutive N-terminal cleavage of IAP2 and that apoptotic signals enhance IAP2 cleavage, while immunological signals do not.
Fig. 7.
a, b Western blot analysis of lysates from S2 cells transfected with the expression constructs indicated in the tables at the top of each gel. Cells were incubated with DMSO, cycloheximide (CHX) or PGN as shown. Total cell lysates were immunoblotted with anti-myc antibodies where indicated. For each membrane, tubulin levels were visualized as a loading control. Phosphorylated JNK (dpJNK) levels were examined to monitor activation of the IMD pathway. MW is shown for each membrane.
Discussion
The modules that govern the invertebrate IMD pathway and vertebrate TNF death receptor signals show striking similarities, suggesting that both evolved from an ancient signal transduction cassette. In Drosophila, the IMD pathway initiates an antimicrobial and prosurvival pathway via JNK and NF-κB responses to an exogenous signal, bacterial PGN. The vertebrate TNFR1 and DR3 receptors engage similar adaptor molecules and enzymatic relays to activate JNK and NF-κB responses. In contrast to Drosophila, prolonged signaling through the TNF pathway induces apoptosis. A key step in the initiation of the apoptotic pathway is the release of the signaling complex from the receptor and recruitment of FADD and caspase-8 [36]. The Drosophila orthologs FADD and Dredd are integral components of a proximal IMD signal transduction complex, but their presence does not result in apoptosis. Instead, it seems that Dronc is the sole initiator caspase in Drosophila apoptosis. In this study, we examined the relationship between the apoptotic caspase Dronc and the IMD pathway adaptor IAP2. Our data uncovered two independent proteolytic modifiers of IAP2 protein and implicate Dronc in the production of a cleaved IAP2 isoform that retains the ability to interact with IMD constituents.
Altered molecular associations accompany the apparent functional distinctions between Dredd and caspase-8. Whereas caspase-8 does not appear to interact physically with the Drosophila IAP2 ortholog cIAP1, Dredd forms a molecular complex with IAP2. In agreement with a previous identification of the BIR3 of IAP2 as the mediator of Dredd-IAP2 interactions [43], we identified residues 180–450 of IAP2 as sufficient for the association of IAP2 with Dredd. However, in contrast to the earlier report, our study suggests that the prodomains or catalytic subunits of Dredd alone are sufficient to mediate Dredd-IAP2 interactions.
In our exploration of Dredd-IAP2 interactions, we invariably detected N-terminal processing of IAP2 in lysates of S2 cells. Our findings partially overlap with a report that Drice cleaves IAP2 in S2 cells at aspartate 100 [21]. We found that site-directed mutagenesis of aspartate 100 prevented most but not all cleavage of IAP2. In contrast, mutations of aspartates 92 and 100 completely prevented the cleavage of IAP2. These observations indicate that aspartate 92 may act as an alternative target for caspase activity, and our studies of the N-terminal cleavage product suggest that aspartate 92 is a substrate in the wild-type protein. However, our sequence analysis of orthologous mammalian IAP proteins suggests a greater conservation of aspartate 100 than aspartate 92, raising the possibility that D100 is of greater relevance as a caspase substrate in vertebrates. Further studies are required to determine if mammalian caspases cleave pro-immune IAP orthologs. Our studies also suggest important roles for Dronc in the cleavage of IAP2. We found that RNAi-based inactivation of Drice did not prevent IAP2 processing, whereas inactivation of Dronc did. We also found that overexpression of Dronc accelerated cleavage of IAP2, while expression of a dominant inhibitory Dronc point mutant blocked IAP2 cleavage. Additional biochemical studies are required to test if IAP2 is a direct substrate for Dronc. However, we believe our studies point at a substantial role for Dronc in the cleavage of IAP2. In support of a requirement for Dronc in the proteolysis of IAP2, a previous study established that induction of apoptosis leads to IAP2 cleavage in vivo at a time that precedes Drice activation and overlaps with Dronc activation [21].
Our extended studies of IAP2 proteolysis identified two separate controls on the stability of the IAP2 protein. We found that proteasomal inhibitors or inactivating mutations in the RING domain stabilize IAP2, suggesting RING-dependent autoregulation, presumably through K48 conjugation of polyubiquitin chains. In addition, we showed that the RING domain acts in synergy with Dronc to control total levels of full-length IAP2. Despite the synergistic relationship between the RING domain and Dronc in the regulation of IAP2 stability, we believe that the phenotypic impacts of the two proteolytic events are distinct. We propose that RING domain-mediated proteolysis depletes the cellular pool of total and cleaved IAP2, effectively attenuating the strength of IMD activity. In contrast, Dronc generates a truncated IAP2 that retains the ability to associate with the IMD partner proteins FADD and Dredd. Cleaved IAP2 also has the RING domain required for ubiquitylation of Imd. Our observations suggest that Dronc activity does not diminish signal transduction through the IMD pathway. When we treated S2 cells with nonlethal doses of the translation inhibitor cycloheximide, we observed moderate stabilization of cleaved IAP2 relative to the full-length isoform (fig. 2e). These observations raise the possibility that Dronc may augment signal transduction through the IMD pathway through the generation of a functional and more stable IAP2 isoform, although additional molecular studies are required to validate this hypothesis. Consistent with a possible role for moderate levels of apoptotic signals in the potentiation of IMD responses, a previous report noted that mild increases in proapoptotic signals increase antimicrobial peptide production in vivo [23]. In addition, our previous search for quantitative modifiers of the IMD response found that loss of Dronc increased the strength of signal transduction through the IMD-JNK module [44]. This finding overlaps with an established role for the IMD-Rel module in the attenuation of the JNK component [32].
In summary, we believe our findings point at two independent proteolytic mechanisms that tune the cellular pools of IAP2 isoforms. We consider Dronc of particular interest, as it is exclusively known for its role as an apoptotic initiator. We speculate that our findings position Dronc as a sensor that tunes responses to physiological insults that typically warrant immunological or apoptotic responses. In this model, moderate stress may increase the level of Dronc activity to an extent that augments immune responses in preparation for microbial assault. In cases where the stress signal is simply too high, the extent of Dronc signaling diverts the primary response to activation of an apoptotic response and the induction of cell death.
Acknowledgments
Dronc cDNA was provided by Rollie Clem and Dredd cDNA was provided by John Abrams. S.G. held a studentship award from Alberta Innovates - Health Solutions (AIHS) and E.F. holds a Canada Research Chair (CRC) in innate immunity. This work was supported by the Natural Sciences and Engineering Research Council (RGPIN 371188 to E.F.).
References
- 1.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 2.Wajant H, Pfizenmaier K, Scheurich P. Tumor necrosis factor signaling. Cell Death Differ. 2003;10:45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- 3.Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- 4.Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- 5.Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, Harley W, Fox A, Golenbock D, Silverman N. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity. 2004;20:637–649. doi: 10.1016/s1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- 6.Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, Lee WJ, Mengin-Lecreulx D, Lemaitre B. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol. 2003;4:478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- 7.Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- 8.Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, Kopczynski C, Duyk G, Reichhart JM, Hoffmann JA. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev Cell. 2001;1:503–514. doi: 10.1016/s1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- 9.Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, Oshima Y, Peach C, Erturk-Hasdemir D, Goldman WE, Oh BH, Kurata S, Silverman N. PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol. 2006;7:715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- 10.Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart JM, Hoffmann JA. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc Natl Acad Sci U S A. 1995;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu S, Yang X. dFADD, a novel death domain-containing adapter protein for the Drosophila caspase DREDD. J Biol Chem. 2000;275:30761–30764. doi: 10.1074/jbc.C000341200. [DOI] [PubMed] [Google Scholar]
- 12.Leulier F, Vidal S, Saigo K, Ueda R, Lemaitre B. Inducible expression of double-stranded RNA reveals a role for dFADD in the regulation of the antibacterial response in Drosophila adults. Curr Biol. 2002;12:996–1000. doi: 10.1016/s0960-9822(02)00873-4. [DOI] [PubMed] [Google Scholar]
- 13.Naitza S, Rosse C, Kappler C, Georgel P, Belvin M, Gubb D, Camonis J, Hoffmann JA, Reichhart JM. The Drosophila immune defense against Gram-negative infection requires the death protein dFADD. Immunity. 2002;17:575–581. doi: 10.1016/s1074-7613(02)00454-5. [DOI] [PubMed] [Google Scholar]
- 14.Chen P, Rodriguez A, Erskine R, Thach T, Abrams JM. Dredd, a novel effector of the apoptosis activators reaper, grim, and hid in Drosophila. Dev Biol. 1998;201:202–216. doi: 10.1006/dbio.1998.9000. [DOI] [PubMed] [Google Scholar]
- 15.Leulier F, Rodriguez A, Khush RS, Abrams JM, Lemaitre B. The Drosophila caspase Dredd is required to resist Gram-negative bacterial infection. EMBO Rep. 2000;1:353–358. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoven S, Ando I, Kadalayil L, Engstrom Y, Hultmark D. Activation of the Drosophila NF-kappaB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 2000;1:347–352. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoven S, Silverman N, Junell A, Hedengren-Olcott M, Erturk D, Engstrom Y, Maniatis T, Hultmark D. Caspase-mediated processing of the Drosophila NF-kappaB factor Relish. Proc Natl Acad Sci U S A. 2003;100:5991–5996. doi: 10.1073/pnas.1035902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paquette N, Broemer M, Aggarwal K, Chen L, Husson M, Erturk-Hasdemir D, Reichhart JM, Meier P, Silverman N. Caspase-mediated cleavage, IAP binding, and ubiquitination: linking three mechanisms crucial for Drosophila NF-kappaB signaling. Mol Cell. 2010;37:172–182. doi: 10.1016/j.molcel.2009.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erturk-Hasdemir D, Broemer M, Leulier F, Lane WS, Paquette N, Hwang D, Kim CH, Stoven S, Meier P, Silverman N. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc Natl Acad Sci U S A. 2009;106:9779–9784. doi: 10.1073/pnas.0812022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leulier F, Ribeiro PS, Palmer E, Tenev T, Takahashi K, Robertson D, Zachariou A, Pichaud F, Ueda R, Meier P. Systematic in vivo RNAi analysis of putative components of the Drosophila cell death machinery. Cell Death Differ. 2006;13:1663–1674. doi: 10.1038/sj.cdd.4401868. [DOI] [PubMed] [Google Scholar]
- 21.Ribeiro PS, Kuranaga E, Tenev T, Leulier F, Miura M, Meier P. DIAP2 functions as a mechanism-based regulator of drICE that contributes to the caspase activity threshold in living cells. J Cell Biol. 2007;179:1467–1480. doi: 10.1083/jcb.200706027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gesellchen V, Kuttenkeuler D, Steckel M, Pelte N, Boutros M. An RNA interference screen identifies Inhibitor of Apoptosis Protein 2 as a regulator of innate immune signalling in Drosophila. EMBO Rep. 2005;6:979–984. doi: 10.1038/sj.embor.7400530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huh JR, Foe I, Muro I, Chen CH, Seol JH, Yoo SJ, Guo M, Park JM, Hay BA. The Drosophila inhibitor of apoptosis (IAP) DIAP2 is dispensable for cell survival, required for the innate immune response to Gram-negative bacterial infection, and can be negatively regulated by the Reaper/Hid/Grim family of IAP-binding apoptosis inducers. J Biol Chem. 2007;282:2056–2068. doi: 10.1074/jbc.M608051200. [DOI] [PubMed] [Google Scholar]
- 24.Kleino A, Valanne S, Ulvila J, Kallio J, Myllymaki H, Enwald H, Stoven S, Poidevin M, Ueda R, Hultmark D, Lemaitre B, Ramet M. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. EMBO J. 2005;24:3423–3434. doi: 10.1038/sj.emboj.7600807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leulier F, Lhocine N, Lemaitre B, Meier P. The Drosophila inhibitor of apoptosis protein DIAP2 functions in innate immunity and is essential to resist gram-negative bacterial infection. Mol Cell Biol. 2006;26:7821–7831. doi: 10.1128/MCB.00548-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meinander A, Runchel C, Tenev T, Chen L, Kim CH, Ribeiro PS, Broemer M, Leulier F, Zvelebil M, Silverman N, Meier P. Ubiquitylation of the initiator caspase DREDD is required for innate immune signalling. EMBO J. 2012;31:2770–2783. doi: 10.1038/emboj.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhuang ZH, Sun L, Kong L, Hu JH, Yu MC, Reinach P, Zang JW, Ge BX. Drosophila TAB2 is required for the immune activation of JNK and NF-kappaB. Cell Signal. 2006;18:964–970. doi: 10.1016/j.cellsig.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 28.Silverman N, Zhou R, Stoven S, Pandey N, Hultmark D, Maniatis T. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vidal S, Khush RS, Leulier F, Tzou P, Nakamura M, Lemaitre B. Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKS in the control of rel/NF-kappaB-dependent innate immune responses. Genes Dev. 2001;15:1900–1912. doi: 10.1101/gad.203301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boutros M, Agaisse H, Perrimon N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev Cell. 2002;3:711–722. doi: 10.1016/s1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- 31.Geuking P, Narasimamurthy R, Lemaitre B, Basler K, Leulier F. A non-redundant role for Drosophila Mkk4 and hemipterous/Mkk7 in TAK1-mediated activation of JNK. PLoS One. 2009;4:e7709. doi: 10.1371/journal.pone.0007709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park JM, Brady H, Ruocco MG, Sun H, Williams D, Lee SJ, Kato T, Jr, Richards N, Chan K, Mercurio F, Karin M, Wasserman SA. Targeting of TAK1 by the NF-kappa B protein Relish regulates the JNK-mediated immune response in Drosophila. Genes Dev. 2004;18:584–594. doi: 10.1101/gad.1168104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y, Wu LP, Anderson KV. The antibacterial arm of the Drosophila innate immune response requires an IkappaB kinase. Genes Dev. 2001;15:104–110. doi: 10.1101/gad.856901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutschmann S, Jung AC, Zhou R, Silverman N, Hoffmann JA, Ferrandon D. Role of Drosophila IKK gamma in a Toll-independent antibacterial immune response. Nat Immunol. 2000;1:342–347. doi: 10.1038/79801. [DOI] [PubMed] [Google Scholar]
- 35.Wu LP, Anderson KV. Regulated nuclear import of Rel proteins in the Drosophila immune response. Nature. 1998;392:93–97. doi: 10.1038/32195. [DOI] [PubMed] [Google Scholar]
- 36.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 37.Dorstyn L, Colussi PA, Quinn LM, Richardson H, Kumar S. DRONC, an ecdysone-inducible Drosophila caspase. Proc Natl Acad Sci U S A. 1999;96:4307–4312. doi: 10.1073/pnas.96.8.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hawkins CJ, Yoo SJ, Peterson EP, Wang SL, Vernooy SY, Hay BA. The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. J Biol Chem. 2000;275:27084–27093. doi: 10.1074/jbc.M000869200. [DOI] [PubMed] [Google Scholar]
- 39.Meier P, Silke J, Leevers SJ, Evan GI. The Drosophila caspase DRONC is regulated by DIAP1. EMBO J. 2000;19:598–611. doi: 10.1093/emboj/19.4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinn LM, Dorstyn L, Mills K, Colussi PA, Chen P, Coombe M, Abrams J, Kumar S, Richardson H. An essential role for the caspase Dronc in developmentally programmed cell death in Drosophila. J Biol Chem. 2000;275:40416–40424. doi: 10.1074/jbc.M002935200. [DOI] [PubMed] [Google Scholar]
- 41.Guntermann S, Foley E. The protein Dredd is an essential component of the c-Jun N-terminal kinase pathway in the Drosophila immune response. J Biol Chem. 2011;286:30284–30294. doi: 10.1074/jbc.M111.220285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams JM. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat Cell Biol. 1999;1:272–279. doi: 10.1038/12984. [DOI] [PubMed] [Google Scholar]
- 43.Meinander A, Runchel C, Tenev T, Chen L, Kim CH, Ribeiro PS, Broemer M, Leulier F, Zvelebil M, Silverman N, Meier P. Ubiquitylation of the initiator caspase DREDD is required for innate immune signalling. EMBO J. 2012;31:2770–2783. doi: 10.1038/emboj.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bond D, Foley E. A quantitative RNAi screen for JNK modifiers identifies Pvr as a novel regulator of Drosophila immune signaling. PLoS Pathog. 2009;5:e1000655. doi: 10.1371/journal.ppat.1000655. [DOI] [PMC free article] [PubMed] [Google Scholar]