Abstract
The interferon (IFN) pathway plays an essential role in the innate immune response following viral infections and subsequent shaping of adaptive immunity. Infections with influenza A viruses (IAV) activate the IFN pathway after the recognition of pathogen-specific molecular patterns by respective pattern recognition receptors. The IFN regulatory factors IRF3 and IRF7 are key players in the regulation of type I and III IFN genes. In this study, we analyzed the role of IRF3 and IRF7 for the host response to IAV infections in Irf3−/−, Irf7−/−, and Irf3−/−Irf7−/− knockout mice. While the absence of IRF3 had only a moderate impact on IFN expression, deletion of IRF7 completely abolished IFNα production after infection. In contrast, lack of both IRF3 and IRF7 resulted in the absence of both IFNα and IFNβ after IAV infection. In addition, IAV infection of double knockout mice resulted in a strong increase of mortality associated with a massive influx of granulocytes in the lung and reduced activation of the adaptive immune response.
Keywords: Irf3-deficient mice, Irf7-deficient mice, Influenza A virus, Host response, Interferon pathway
Introduction
Influenza A virus (IAV) infections represent a threat to both human and animal health [1]. In addition to virulence factors of IAV, recent studies in animal models [2] and humans [3, 4] revealed that host genetic factors are strong determinants for the severity of influenza disease. The interferon (IFN) system is a major player in the host pathogen defense, especially against viruses. Type I and type III IFN are induced if pathogens are recognized by cell surface and endosomal pattern recognition receptors and intracellular viral sensors in infected cells [5, 6, 7, 8]. These receptors in turn activate the IFN regulatory factors (IRF) that are transcription factors inducing the expression of IFN genes [9, 10]. IFNs are subsequently secreted from infected cells and initiate an antiviral state in neighboring cells and systemically by inducing a large number of IFN-induced genes [11, 12, 13]. The IRF gene family consists of 9 members, and Irf3, Irf5, Irf7, and Irf9 have been shown to be important for viral infections [9, 10, 14]. The systemic expression of IFNA genes was completely dependent on Irf7 after murine cytomegalovirus (MCMV) infection in mice but independent of Irf3, whereas IFNB production was not affected in single knockout (KO) or double KO (DKO) Irf3/Irf7 mice. However, single KO or DKO Irf3/Irf7 mice were only modestly more susceptible than wild-type (WT) mice to MCMV infections [15]. Mice lacking Irf3 and Irf7 as single KO or DKO still produced reduced levels of Ifnb1 in myeloid dendritic cells after West Nile virus infection, but only additional deletion of Irf5 resulted in increased susceptibility to West Nile virus and murine norovirus in triple KO mice [16]. Robust induction of type I IFNs after herpes simplex virus type 1, encephalomyocarditis virus or vesicular stomatitis virus infection of embryonic fibroblast cells or dendritic cells from mutant mice was entirely dependent on Irf7, whereas Irf3 contributed only little in the absence of Irf7[17]. IFN serum levels were only slightly reduced after herpes simplex virus or vesicular stomatitis virus infection in Irf3 mutant mice, but strongly reduced in Irf7 mutant mice [17]. Irf7 mutant mice were highly susceptible to infections with these viruses. Mice deficient in Irf3 were more susceptible to ECMV infections and produced reduced levels of type I IFNs [18]. The infection of Irf3/Irf7 double-mutant mice with Chikungunya virus was highly lethal and no type I IFNs were detected in the blood [19]. After the infection of neonatal mice with metapneumovirus, Irf3 and Irf3/Irf7 double-mutant mice exhibited higher viral loads in their lungs compared to WT mice but not Irf7 single-mutant mice [20].
In IAV-infected murine trachea epithelial cells, Irf3 deficiency had only a minor impact on IFN type I and III expression, whereas the lack of Irf7 reduced IFN production significantly and no IFN induction was seen in DKO Irf3/Irf7 mice [21]. These data indicate that the role and importance of Irf3 and Irf7 differs depending on the pathogen and whether cell cultures or in vivo experiments are used.
Therefore, we investigated the role of IRF3 and IRF7 for the host defense to IAV in vivo by infecting single KO and DKO mice. We demonstrate here that the respective single KO resulted in a less pronounced increase of susceptibility to IAV, whereas DKO exhibited a strongly increased mortality and an aberrant immune response.
Materials and Methods
Ethics Statement
The mice experiments were approved by the Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit, Oldenburg, Germany (permit No. 3392 42502-04-136/1234) according to the guidelines of the German animal welfare law.
Virus and Mice
Original stocks of mouse-adapted A/Puerto Rico/8/34 (H1N1, PR8M) virus were obtained from Stefan Ludwig, University of Münster [22]. Virus stocks were propagated in the chorioallantoic cavity of 10-day-old pathogen-free embryonated chicken eggs for 48 h at 37°C as described previously [23]. The viral titer was determined by focus-forming unit (FFU) assay as described previously [23]. Mutant B6;129S6-Irf3tm1Ttg (Irf3−/−) and B6;129P2-Irf7tm1Ttg (Irf7−/−) mice were kindly provided by Tadatsugu Taniguchi [17, 18] and backcrossed to C57BL/6J for at least 10 generations. The background was confirmed by mouse universal genotyping array. The analysis revealed 99.86% of the single nucleotide polymorphisms matched to the C57BL/6J background containing an 18.66-Mb congenic region on chromosome 7 in Irf3−/− mice and 99.89% C57BL/6J background containing a 6.12-Mb congenic region on chromosome 7 in Irf7−/− mice, respectively. B6.129-Irf3tm1TtgIrf7tm1Ttg (Irf3−/−Irf7−/−) DKO mice were generated by breeding Irf3−/− with Irf7−/− mice. As controls, C57BL/6J mice were obtained from Janvier (Le Genest-Saint-Isle, France). All mice were maintained under specific pathogen-free conditions at the animal facilities of the Helmholtz Centre for Infection Research (HZI). For genotyping, genomic DNA was extracted from mouse tails and the DNA concentration was quantified with a Nano Drop 100 (Thermo Scientific) spectrophotometer. A total of 100 ng of DNA and 10 pmol of primer oligonucleotides were used for PCR. Primers were designed for sequences flanking the targeted Irf3 locus (Irf3-common 5′-GAA CCT CGG AGT TAT CCC GAA GG-3′, Irf3-ds 5′-GTT TGA GTT ATC CCT GCA CTT GGG-3′, Irf3-neo 5′-TCG TGC TTT ACG CTA TCG CCG CTC CCG ATT-3′) and Irf7 locus (Irf7-uzu 5′-GTG GTA CCC ACT CCT GCC CTC TTT ATA ATC T-3′, Irf7-long 5′-AGT AGA TCC AAG CTC CCG GCT AAG TTC GTA C-3′, Irf7-neo 5′-TCG TGC TTT ACG GTA TCG CCG CTC CCC ATT C-3′).
Infection of Mice
Female mice at the age of 8–12 weeks were anesthetized by intraperitoneal injection of ketamine/xylazine (10% [v/v] of 100 mg/ml ketamine and 5% (v/v) of 20 mg/ml xylazine in 0.9% [w/v] NaCl) with a dose adjusted to the individual body weight (200 µl/20 g body weight). Infection was attained by the intranasal application of virus solution in 20 µl of sterile phosphate-buffered saline (PBS). Subsequently, survival and body weight loss were monitored until day 14 postinfection (pi). In addition to mice that were found dead, mice with a weight loss of more than 30% of their initial body weight were euthanized and recorded as dead.
Determination of Infectious Viral Particles
For determining the viral load in lungs, organs were extracted and placed into 2 ml of PBS containing 0.1% BSA. Lung tissue was subsequently homogenized using the Polytron 2100 homogenizer. Debris was removed by centrifugation, and aliquots were stored at −70°C. Virus titers were determined on Madin-Darby Canine Kidney (MDCK) cells as FFUs as described previously [22].
Hematology
For the monitoring of hematological parameters, hearts were punctured and blood was collected in EDTA tubes. Absolute numbers of white blood cells, lymphocytes, granulocytes, and monocytes in the blood were determined immediately using the hematologic system VetScan HM5 (Abaxis, Griesheim, Germany). The ratio of granulocytes to lymphocytes were calculated and normalized to the values from noninfected animals for each respective mouse line.
ELISA
Mice were infected with 2 × 105 FFU PR8M (H1N1) virus and PBS for mock controls. They were anesthetized by isoflurane inhalation and euthanized by retro-orbital bleeding on day 1 and 2 pi. A sterile, 22-gauge catheter was inserted into the exposed tracheal lumen. Bronchoalveolar lavage (BAL) fluid was collected by the instillation and subsequent aspiration of 1 ml of PBS into the lung. Supernatants were aliquoted, stored at −70°C and used to detect the IFNA, IFNB1, and IFNL2/3 (also referred to as IL-28A/B) protein concentration by the respective ELISA kits from PBL Assay Science (Piscataway, N.J., USA). Standard curves were performed according to the manufacturer's instructions. Quantification of the respective proteins was based on the standard curves determined.
Histological and Immunohistochemical Analyses
Ten-week-old female mice were infected with 2 × 105 FFU A/Puerto Rico/8/34 (H1N1, PR8M) virus and their lungs were prepared on day 3 pi. Subsequently tissues were immersion-fixed for 24 h in 4% buffered formaldehyde solution (pH 7.4), dehydrated in a series of graded ethanol and embedded in paraffin. Sections were cut from 3 evenly distributed levels and stained with hematoxylin and eosin (HE). For immunohistochemical studies, sections were stained overnight at 4°C with a primary antibody against IAV nucleoprotein (clone hb65; ATCC, Wessel, Germany), incubated for 30 min with HRP-labeled goat-anti-mouse IgG2a (Biozol, Eching, Germany) and counterstained with hematoxylin. Analysis of HE-stained lung sections from 5 WT and 4 Irf3−/−Irf7−/− DKO mice on day 3 pi was performed using a semiquantitative scoring system for increase in cellular infiltration: 0 = none, 1 = mild, 2 = moderate, 3 = severe. The degree of monocytic and granulocytic infiltration was evaluated separately on 3 HE sections per animal and ratios of mean values were calculated. In addition, bronchiolar structures with and without necrotic changes of the epithelial cells were counted on 3 complete lung sections per animal to determine epithelial damage. Influenza virus-infected cells were counted on 3 immunohistochemically stained sections of the complete lung per animal.
Preparation of RNA
Eight- to 12-week-old female mice were intranasally infected with 2 × 103 FFU A/Puerto Rico/8/34 (H1N1, PR8M) virus or mock infected with PBS. On day 3 pi, mice were euthanized by CO2 asphyxiation, lungs were extracted and immediately stored in RNAlater solution (Qiagen, Hilden, Germany). RNA was isolated using Qiagen RNeasyMidi Kit according to the manufacturer's instructions. RNA was eluted in RNase-free water, measured by NanoDrop spectrophotometer and quality controlled on a 2100 Bioanalyzer Instrument (Agilent Technologies, Waldbronn, Germany).
Microarray Analysis
All samples had an RNA integrity number of ≥9.7. One hundred nanograms of total RNA were applied for Cy3-labelling reaction using a one color Quick Amp Labeling protocol (Agilent Technologies). Labeled cRNA was hybridized to Agilentʼs mouse microarrays (Agilent 4×44k Mouse V2 Design ID 026655) for 16 h at 68°C and scanned using the Agilent DNA Microarray Scanner. Expression values were calculated by the software package Feature Extraction 10.5.1.1 (Agilent Technologies). Microarray data were then analyzed using the R software package [24]. Preprocessing steps included background correction, quantile normalization and annotation using the MmAgilentDesign026655.db [25], LIMMA [26], and Agi4×44PreProcess [27] packages. Multigroup comparisons and the identification of DEPS (differentially expressed probe sets) were performed with the LIMMA package [26] using BH correction for multiple testing [28]. DEPS were identified based on an adjusted p value <0.05 and exhibiting more than a 2-fold difference in expression levels (abs[log2] >1). Reactome enrichment analysis and cluster profiling was performed with the package clusterProfiler [29]. KEGG pathway analysis was performed using the pathview package in R [30]. Digital cell quantification (DCQ) analysis was performed according to the algorithm described by Altboum et al. [31]. Gene expression values were preprocessed by subtracting the mean of the mock-infected mice from the infected mice for each mouse strain and by selecting the most variant probe sets for genes with multiple probe sets based on the standard deviation across all groups. DCQ analysis was then performed at the public server provided by the University of Tel Aviv (http://dcq.tau.ac.il/) using the immune cell database at http://dcq.tau.ac.il/. The results were then visualized by heat maps using the R software [24]. The raw data has been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE77600.
Real-Time Reverse Transcription Polymerase Chain Reaction
Aliquots from the same RNA preparations that were used for array analysis were subsequently analyzed by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). One microgram of lung-extracted RNA from each sample was used for cDNA synthesis (SuperScripIII; Invitrogen, Karlsruhe, Germany) using a 1:1 mixture of oligodT and random hexamers (Invitrogen). Specific primers were designed for the respective genes (see online suppl. table 1; for all online suppl. material, see www.karger.com/doi/10.1159/000450705). The cDNA was diluted into a final volume of 50 µl, and RT-PCR was carried out with the DNA Master SYBR Green I kit (Roche, Mannheim, Germany) using a LightCycler 480 apparatus (Multi-Well Plate 96; Roche). For relative quantification of specific host genes, standard curves were generated from a serial 10-fold dilution of cDNA of 1 infected sample using the respective specific primers. Gene induction was normalized to the ribosomal protein L4 (Rpl4) housekeeping gene using the ΔΔCT method [32].
Data Analysis
Means ± standard error of the mean (SEM) were calculated and the data for percent body weights, viral titers, hematology, epithelial damage, cellular infiltration, influenza virus-infected cells, and cytokine/chemokine expression were evaluated for statistically significant differences by the nonparametric Mann-Whitney U test using GraphPad Prism version 5.04 and 6.05 for Windows (GraphPad Software, San Diego, Calif., USA). p values ≤0.05 were considered significant. The log-rank Mantel-Cox test was used to calculate the significance of differences in survival rates between KO and WT mice.
Flow Cytometry
For the measurements of lymphocytes, lungs were rinsed and passed through a 70-µm cell strainer. Subsequently, lymphocytes were separated by a density gradient centrifugation using Easycoll (Biochrom, Berlin, Germany). The single cell suspensions were stained with αCD3-V500 (BD Biosciences, Heidelberg, Germany), αCD4-PE-Cy7 (eBioscience, Frankfurt, Germany), αCD8-BV650 (BioLegend, London, UK), αIL-10-BV421 (BioLegend), αIL-17-FITC (eBioscience), and LIVE/DEAD Fixable Blue Dead cell stain kit (UV excitation, ThermoFisher, Schwerte, Germany) to exclude dead cells. Intracellular staining for interleukin (IL)-10 and IL-17 was performed using the Cytofix/Cytoperm system (BD Biosciences). To analyze granulocytes, lungs were isolated and passed through a 40-µm cell strainer and a density centrifugation using Lympholyte M (Biozol) was performed. After washing the cells twice with PBS/2% fetal calf serum, Fc receptors were blocked with αCD16/32 and subsequently stained with αCD45-FITC, αCD11b-APC and αLy-6G/Ly-6C-PerCp (Gr-1; eBioscience). To exclude dead cells, propidium iodide was added to each sample. Samples were measured using an Accuri C6 or LSR Fortessa flow cytometer (BD Biosciences) and data were analyzed using FlowJo version 7.6.5 (Tree Star, Ashland, Oreg., USA).
SDS-PAGE and Western Blot Analysis
Eight- to 12-week-old female mice were intranasally infected with 2 × 103 FFU PR8M virus or mock infected with PBS. On day 3 pi, mice were euthanized by CO2 asphyxiation, and their lungs were extracted and immediately stored in RIPA buffer (1× PBS, 1% NP-40, 0.5% sodium deoxycholate, 1% SDS and Halt phosphatase/protease inhibitor cocktail; ThermoFisher, Grand Island, Nebr., USA). The lungs were then homogenized in Fastprep homogenizer (MP Biomedicals, LLC, Santa Ana, Calif., USA) for 60 s, centrifuged (1 min, 1,000 × g), and debris was discarded. Approximately 20 μg of protein homogenate was diluted in 2× SDS loading buffer (125 mM Tris-HCl, pH 6.8, 5% SDS, 20% glycerol, 0.004% bromophenol blue, and 10% β-mercaptoethanol), boiled for 5 min, and the proteins were resolved by SDS-PAGE on a 10% Tris-Glycine SDS gel. The proteins were transferred to a nitrocellulose membrane. The membrane was blocked with 5% BSA in 1× TBS for 1 h at room temperature. Phosphorylated and total JNK proteins were detected by incubating the membrane overnight at 4°C with rabbit anti-JNK-pT183/Y185 (1:2,000) and mouse anti-JNK antibodies (1:1,000; Cell Signaling Technologies, Danvers, Mass., USA) diluted in 5% BSA in 1× TBST (TBS + 0.05% Tween-20). The membranes were washed 5 times in 1× TBST and then incubated with donkey anti-mouse infrared dye (IRDye) 680RD and donkey anti-rabbit IRDye 800CW secondary antibodies (1:10,000, LI-COR; Lincoln, Nebr., USA) diluted in 5% BSA-TBST for 1 h at room temperature. After washing the membrane 5 times in 1× TBST and once in 1× TBS, the bands were visualized and fluorescent intensities were quantified using an Odyssey CLx imager and Image Studio Software, respectively (LI-COR).
Results
Deletion of Irf3 and Irf7 Results in Increased Susceptibility to Influenza A Infection
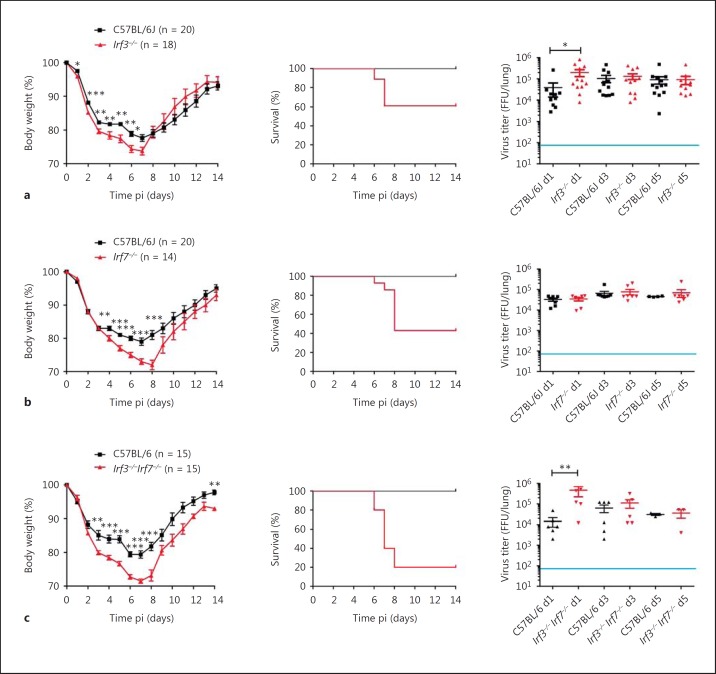
IRF genes are important mediators of the host response to infections. Here, we examined the function of both Irf3 and Irf7 and their interaction for the immune response to IAV infection using Irf3−/− and Irf7−/− single KO and Irf3−/−Irf7−/− DKO mice. Eight- to 11-week-old female WT, KO and DKO mice were infected with mouse-adapted PR8M (A/Puerto/Rico/8/34, H1N1). Irf3−/− and Irf7−/− single KO and the DKO mice lost significantly more body weight after infection with PR8M and revealed significantly increased mortality rates compared to WT mice (fig. 1). Whereas 60% of Irf3−/− mice survived the infection, only 40% of Irf7−/− and 20% of Irf3−/−Irf7−/− KO mice survived. Furthermore, a significantly higher viral load was detected on day 1 in Irf3−/− and Irf3−/−Irf7−/− DKO mice compared to WT mice, whereas no differences were observed at other time points with respect to viral loads in WT mice (fig. 1a, c, right panel). Irf7−/−-infected mice did not have a significantly different viral load compared to WT mice on any day tested pi (fig. 1b, right panel).
Fig. 1.
Body weight loss, survival, and viral load of Irf3−/−, Irf7−/−, and Irf3−/−Irf7−/− mutant mice after IAV infection. Irf3−/− (a), Irf7−/− (b) and Irf3−/−Irf7−/− (c) female mice at the age of 8–12 weeks were infected intranasally with 2 × 105 FFU PR8M (H1N1) IAV. C57BL/6J (WT) female mice at the same age were used as controls. Body weight (left panel) and survival (middle panel) was monitored until day 14 pi. Mice reaching more than 30% of their initial body weight had to be euthanized and recorded as dead. Weight loss data represent mean values ± SEM. Group sizes were the same for body weight and survival measurements (as indicated in the body weight loss curves). Homozygous KO and DKO mice revealed significantly higher weight loss early after infection (* p < 0.05, ** p < 0.01, *** p < 0.001) and significantly lower survival rates (p < 0.01 for Irf3−/− and p < 0.001 for Irf7−/− and Irf3−/−Irf7−/−, respectively). The viral load (right panel) was determined in lung homogenates. The detection limit of the assay was 80 infectious particles per lung, as indicated by the blue line. Significances were calculated using the Mann-Whitney U test (body weight loss and viral load) and log-rank test (survival). The results on infections of Irf7 mutant mice include data from our previous publication [44] plus additional mice.
Global Analysis of Gene Expression Revealed Qualitative Differences of the Host Response in Dependence of Irf3 and Irf7
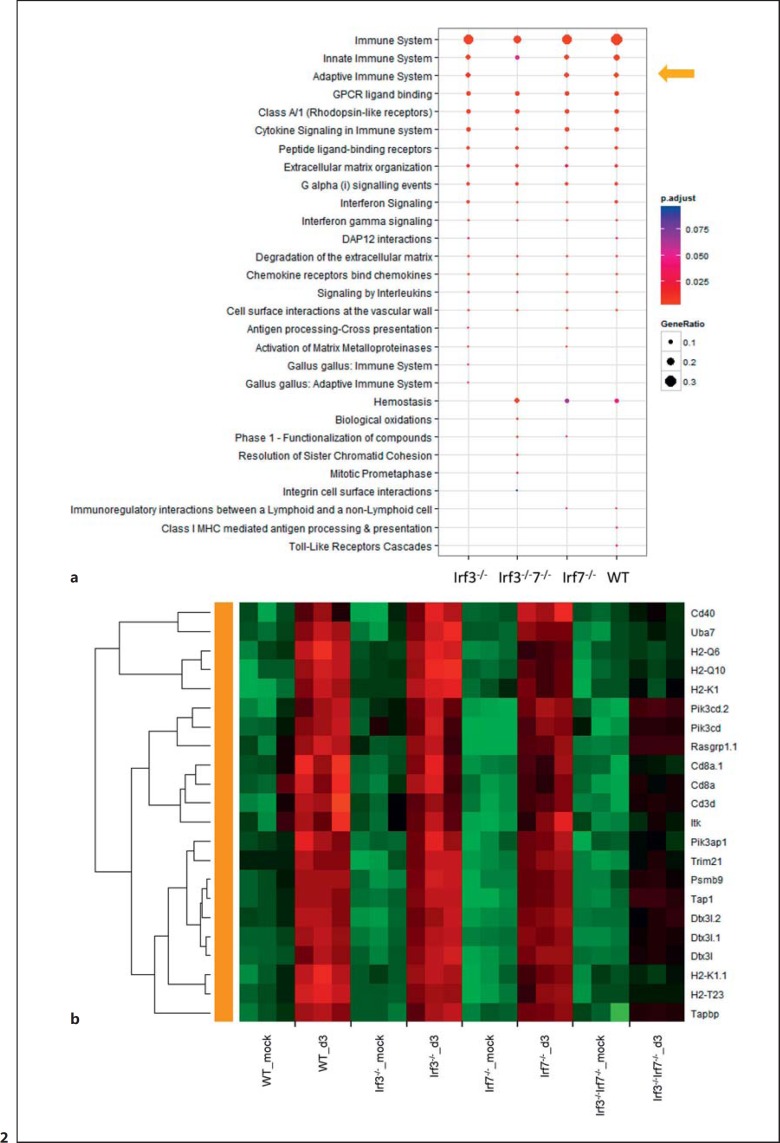
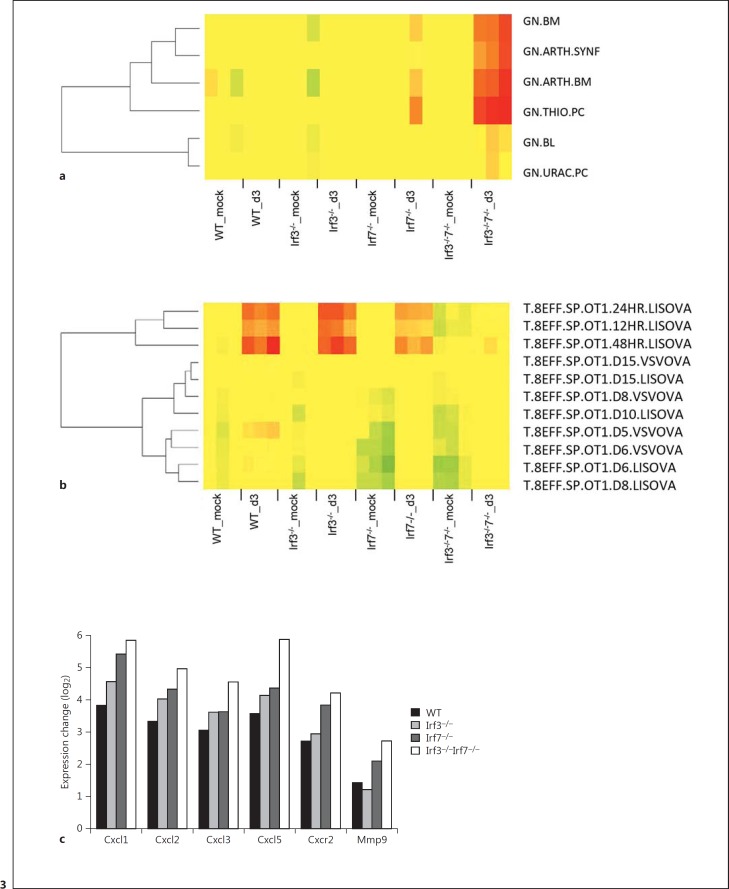
To characterize the host response in more detail, we analyzed global changes of the transcriptome in PR8M-infected lungs 3 days pi in the lungs of Irf3 and Irf7 single KO as well as DKO mice in comparison to WT mice. A similar number of differentially expressed probe sets (defined as more than a 2-fold change, adjusted p < 0.05) was found to be regulated after IAV infection compared to the respective mock controls in WT as well as single and DKO mice (online suppl. fig. 1). Functional analysis of differentially expressed genes using the Reactome pathway database revealed a group of genes related to the ‘adaptive immune response’ that was strongly reduced in DKO mice (fig. 2a). A heat map showing expression levels from the genes that belong to this pathway revealed 22 probesets with decreased levels in DKO mice (fig. 2b). Next, we used a DCQ analysis based on the Immunological Genome Project (ImmGen) database as described in Altboum et al. [31] to determine the relative number of different immune cells in infected lungs (fig. 3a, b). We found that Irf3−/−Irf7−/− DKO mice exhibited a pronounced increase of gene expression signals that are specific for granulocytes (fig. 3a). In addition, T cell-specific gene expression signals were much lower in DKO mice (fig. 3b). Furthermore, expression levels of neutrophil attracting chemokines (Cxcl1, Cxcl2, Cxcl3, Cxcl5) as well as the respective Cxcr2 receptor and the neutrophil-specific Mmp9 gene were higher in DKO compared to WT and single KO mice (fig. 3c).
Fig. 2.
Functional analysis of differentially expressed genes. a Pathway enrichment analysis for differentially expressed genes (DEGs) from the lungs of WT, KO, and DKO mice (n = 3) infected with 2 × 103 FFU PR8M virus using cluster profiler. The x-axis indicates mouse groups (Irf3−/−, Irf7−/−, Irf3−/−Irf7−/−). The y-axis lists pathway descriptions from the Reactome pathway database. The sizes of filled circles reflect the ratio between the number of differentially expressed genes in the group and the total number of genes that belong to the respective pathway. The color indicates p values for the probability that the observed number of DEGs for the respective pathway occurs by chance. The cut-off for adjusted p values was set to 0.1 and the maximum number of displayed pathways was set to 20. DEGs from DKO mice were not enriched for the ‘adaptive immune system’ pathway (yellow arrow) as it was seen for WT and single KO mice. b Heat map of log2-transformed normalized expression values from the lungs of individual mice for DEGs that belonged to the ‘adaptive immune system’ pathway and which showed lower expression levels in DKO compared to WT and single KO mice. Green indicates lower (lowest values are bright green), red higher (highest values are bright red) and black mean (at or close to mean value) expression. Rows represent expression values per gene and columns represent individual samples. Rows were clustered based on Pearson correlations.
Fig. 3.
Changes in the relative numbers of immune cells in the lungs of WT, single KO, and DKO mice. The relative number of immune cells in the lungs of WT, single KO, and DKO mice (n = 3 for all groups) on day 3 pi after infection with 2 × 103 FFU PR8M was determined by DCQ, based on gene expression values. Results from the analysis were visualized as heat maps for selected immune cell populations: granulocytes (a), T cells (b). A detailed description for the designation of the different immune cell types can be found at https://www.immgen.org/ListPDFs/Myeloid.pdf. c log2 gene expression values of the differences between infected mice and respective mock-infected controls (mean infected mice minus mean mock-treated controls) for Cxcl1, Cxcl2, Cxcl3, Cxcl5, Cxcr2, and Mmp9 from the lungs of WT, single KO, and DKO mice that were significantly regulated (based on ANOVA, p <0.001).
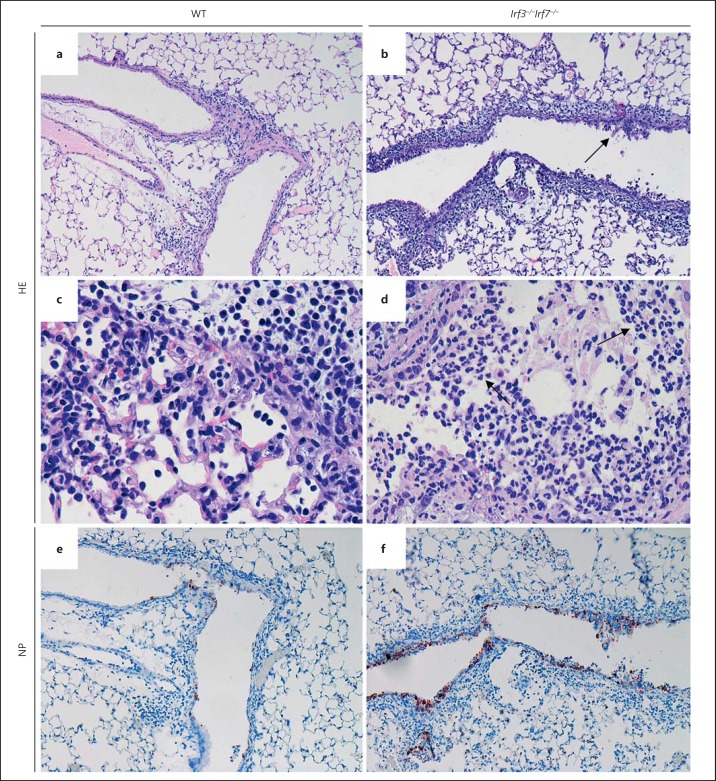
Increased Epithelial Cell Damage and Granulocyte Infiltration in Irf3−/−Irf7−/− DKO Mutant Mice
Histopathological analysis of infected lungs revealed increased epithelial damage in Irf3−/−Irf7−/− DKO mice (fig. 4a, b; online suppl. fig. 2). A more pronounced infiltration of granulocytes was found in the infected lungs on day 3 pi of the DKO mice compared to WT controls (fig. 4c, d; online suppl. fig. 2). Immunohistochemical analyses of viral antigen expression revealed increased numbers of infected cells in the bronchiolar epithelium of DKO lungs compared to WT mice (fig. 4e, f; online suppl. fig. 2). Since the total amount of infectious particles was not significantly different after day 3 and 5 pi (fig. 1) in DKO versus WT mice, these observations suggest a faster spreading of the virus in the lungs of DKO mice.
Fig. 4.
Histological differences in lungs of IAV-infected C57BL/6J and Irf3−/−Irf7−/− mice. Serial lung sections of 10-week-old mice were stained with HE (a–d) and anti-influenza NP antibody (e, f) on day 3 pi with 2 × 105 FFU PR8M. Original magnification, ×10 (a, b, e, f), ×40 (c, d). Both WT and mutant mice revealed strong infiltrations with more densely consolidated tissue and increased epithelial damage (arrow in b) in DKO mice. Many more infiltrating polymorphonuclear cells were observed in the infected lungs of DKO mice compared to WT controls (c, arrows in d). Virus-infected cells were observed mainly in bronchiolar regions of WT (e) and DKO (f) mice.
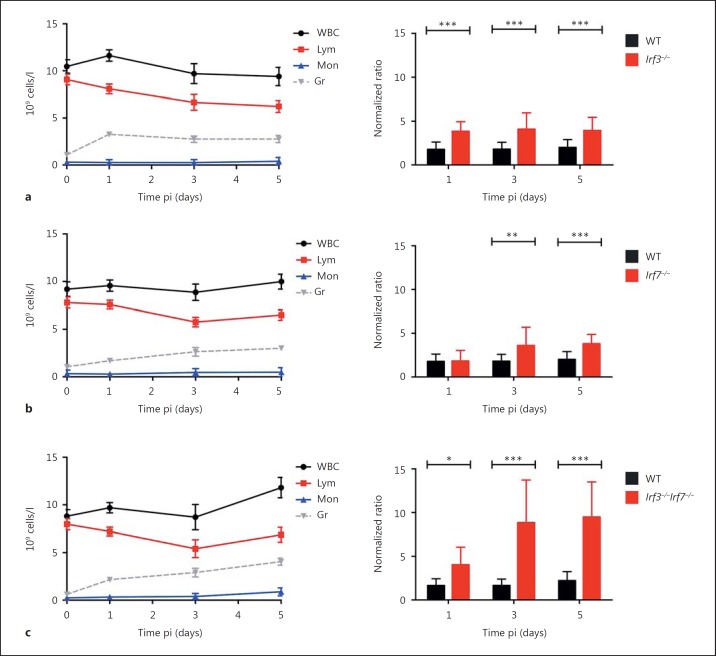
In addition, we investigated the inflammatory response by monitoring the composition and absolute numbers of white blood cell lymphocytes, monocytes, and granulocytes in the peripheral blood between day 1 and day 5 pi (fig. 5, left panel). During the course of infection, a slight increase of granulocytes and monocytes was observed in single KO mice. It is noteworthy that a maximum of granulocyte infiltration was already seen in Irf3−/− mice on day 1 pi, which remained at that level until day 5 pi. In DKO mice, the number of granulocytes increased steadily and was 6.8 times higher at 5 days pi compared to day 0, whereas Irf3−/− and Irf7−/− mice reached 2.6-fold and 2.8-fold higher levels compared to day 0, respectively. The absolute amount of monocytes was 3.7 times higher on day 5 in DKO mice compared to a 1.3-fold and 1.4-fold increase in Irf3−/− and Irf7−/−, respectively. The ratio of granulocyte to lymphocyte numbers, which is an indicator for the severity of IAV infectious disease [33], was significantly increased in Irf3−/− compared to WT mice on all of the days tested (fig. 5a, right panel). In comparison to WT infected mice, this ratio increased in Irf7−/− mice on days 3 and 5 pi (fig. 5b, right panel). The granulocyte to lymphocyte ratio in Irf3−/−Irf7−/− DKO mice rose considerably from day 1 to day 5 pi (fig. 5c, right panel).
Fig. 5.
Hematological analysis of influenza-infected WT, KO, and DKO mice. Eight- to 12-week-old female Irf3−/− (n = 14–19; a), Irf7−/− (n = 16–18; b), and Irf3−/−Irf7−/− (n = 11–21; c) mice were infected with 2 × 105 FFU PR8M. Hematological parameters in blood were measured using the VetScan HM5 system and absolute numbers of white blood cells (WBC), lymphocytes (Lym), monocytes (Mon), and granulocytes (Gr) were determined at different days pi. Error bars represent ±1 SEM. The ratio of granulocyte to lymphocyte cell counts was calculated and then normalized to uninfected values (right panel). Significances were calculated using the Mann-Whitney U test (* p < 0.05, ** p < 0.01, *** p < 0.001). Note that there was also an increase in the granulocyte to lymphocyte ratio in infected C57BL/6J mice. However, this increase is not apparent because of the scale used for the y-axis.
In summary, these results indicated a qualitative and quantitative difference in the host innate immune response of Irf3−/−Irf7−/− DKO mutant mice after IAV infection characterized by increased numbers of infiltrating granulocytes and a lack of T cell activation.
Impaired Induction of IFN Response in Irf3−/−, Irf7−/−, and Irf3−/−Irf7−/− Mutant Mice
Irf3 and Irf7 are important regulators of the host IFN response to infections. We therefore analyzed changes in the expression levels of IFN and IFN response genes in WT, single KO, and DKO mice after infection with PR8M. Only genes that exhibited a significant change with an adjusted p value <0.05 between groups using ANOVA were considered. Of the type I IFNs, Ifna1 and Ifna4 were solely upregulated in WT mice whereas Ifna7 was significantly increased in WT (1.3-fold) and Irf3−/− mice (1.7-fold; online suppl. fig. 3a). Ifnb1 was strongly elevated (56-fold) in infected WT mice and to a lesser extend in the single mutants Irf3−/− and Irf7−/− (9- and 29-fold, respectively; online suppl. fig. 3b). In DKO mice, no significant regulation of Ifnb1 after infection was observed. The expression of type III IFN Ifnl3 (also referred to as Il28b, probeset A_52_P254088) was more strongly induced in Irf3−/− mice (13-fold) compared to WT mice (7-fold), whereas Irf7−/− mice revealed only a slight increase (3-fold). DKO presented no significant regulation of Ifnl3. Furthermore, we found type II IFN Ifng gene to be upregulated in all mutant and WT mice to similar levels. Whereas the single KO mice exhibited a pronounced upregulation of the IFN-stimulated Mx1 gene, which was even higher than in WT mice, DKO mice revealed a more than 4-fold lower expression level although an activation compared to mock-infected controls could still be observed (online suppl. fig. 3c). Similar results were obtained for Oas encoding genes (online suppl. fig. 3d) as well as other IFN-stimulated genes (online suppl. fig. 3e, f). The results from the array analysis were confirmed by qRT-PCR for 9 genes (online suppl. fig. 4).
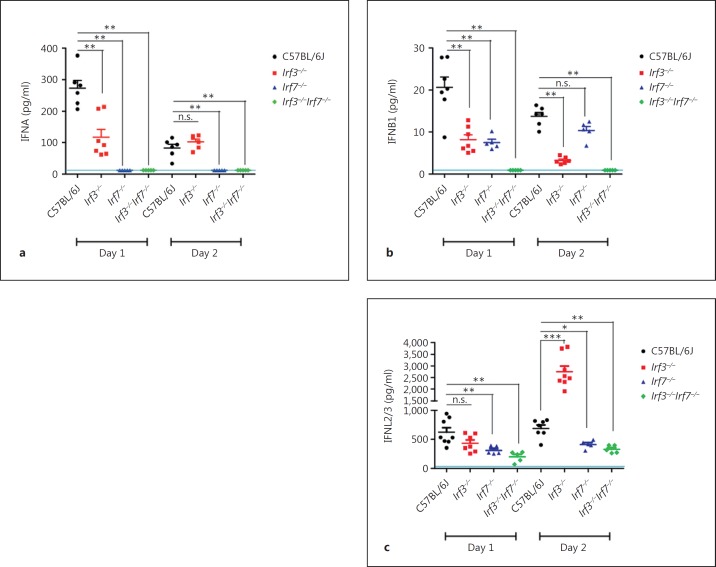
To also verify the regulation of IFNs at the protein level, we analyzed BAL from infected Irf3−/−, Irf7−/−, DKO, and WT mice on day 1 and 2 pi (fig. 6). Whereas Irf3−/− mice revealed a decrease of IFNA and IFNB1 proteins accompanied by strongly elevated levels of IFNL2/3 proteins on day 2 pi, Irf7−/− mice showed no detectable IFNA protein expression, reduced levels of IFNB1 and lower IFNL2/3 protein synthesis compared to WT mice. In DKO mice no induction of IFNA and IFNB proteins was observed in BAL, and only small amounts of IFNL2/3 proteins were detectable. Most notably, single Irf7−/− KO and DKO mice did not exhibit an increase in IFNL2/3 production.
Fig. 6.
Quantification of IFN type I and type III protein levels in KO and WT mice infected with PR8M IAV. After intranasal infection of female mice at the age of 8–11 weeks (n = 5–8) with PBS or 2 × 105 FFU PR8M, BAL was collected at the indicated days and used for IFNA (a), IFNB1 (b) or IFNL2/3 (IL-28A/B; c) detection by ELISA. Each sample was measured in technical duplicates. Values from individual mice, the mean and SEM are presented. The blue line indicates the detection limit of each specific assay. Protein measurements that were below the detection limit were set to the detection limit. The significance of differences in protein concentrations between mutant mice and WT controls were calculated at each time point by the two-tailed Mann-Whitney U test. n.s. = No significance. * p < 0.05, ** p < 0.01, *** p < 0.001.
In summary, Irf7 was crucial for the induction of IFNA. The expression of IFNB1 was more affected in the absence of Irf3 than Irf7. Interestingly, IFNL2/3 reached a significantly higher level in Irf3−/− mice, whereas a defect in Irf7 caused reduced transcription and translation of IFNL2/3.
Altered IFN Induction in Mutant Mice Impacts Downstream Pathways and Activation of Adaptive Immune Response
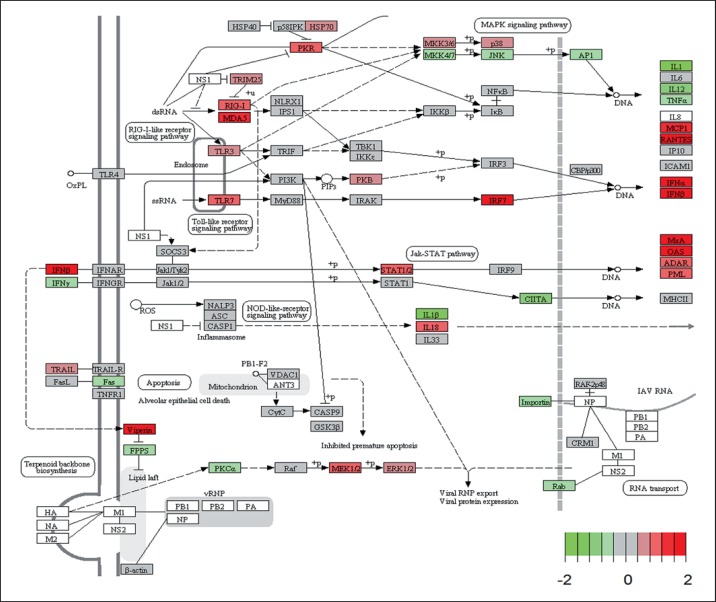
KEGG pathway analysis of transcriptome differences in DKO versus WT infected mice revealed an increased activation of the MKK/JNK pathway in DKO mice (fig. 7). This activation resulted in higher expression levels of the downstream proinflammatory cytokines IL-1 and TNF (fig. 7). Signaling via IFNAR and the JAK/STAT pathway induces the expression of antiviral genes including Mx and Oas. However, the activation of the IFN pathway and downstream IFN response genes was strongly reduced in DKO mice compared to WT mice (fig. 7). The results from the array analysis were confirmed by qRT-PCR for 10 genes (online suppl. fig. 4).
Fig. 7.
KEGG pathway view for influenza-activated genes. Mean differences between gene expression changes in Irf3−/−Irf7−/− mice (mean infected minus mean mock-infected controls) to WT mice (mean infected minus mean mock-infected controls) were calculated and visualized on the KEGG pathway ‘influenza A pathway: mmu05164’ using the ‘pathview’ package. The image was slightly modified (e.g. trimmed) for better visualization. Red indicates higher expression in WT mice and green indicates higher expression levels in DKO mice. The range for the color scale was set from −2 to 2 to increase sensitivity for small differences (total range was 6 to −3.6, 1st quantile −0.183, 3rd quantile 0.149).
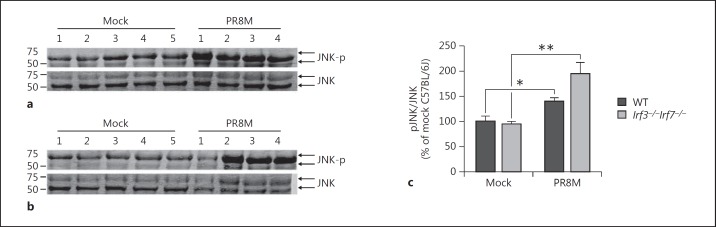
This pathway analysis revealed that genes of the JNK pathway were upregulated in DKO mice and a concomitant increase in granulocyte infiltration was observed while induction of T cell responses were significantly reduced. Thus, to confirm the gene expression data, we directly assayed MKK/JNK pathway activation by Western blot analysis of phosphorylated JNK protein (JNK-pT183/Y185) in infected mouse lungs. On day 3 pi, we observed a marked increase (∼50%) in JNK activation (pJNK/JNK) in PR8M-infected WT mice compared to mock-infected mice (fig. 8a, c). Consistent with our gene expression data, the virus-induced activation was stronger in DKO mice (fig. 8b, c) suggesting increased activation of the MKK/JNK pathway at the protein level in these mice compared to WT mice.
Fig. 8.
Western blot analysis of active JNK proteins in infected mouse lungs. Eight- to 12-week-old female WT (a) and DKO (b) mice were intranasally infected with 2 × 105 FFU PR8M (n = 4 per mouse strain) or mock infected (n = 5 per mouse strain) with PBS. On day 3 pi, the lungs were harvested from euthanized mice, homogenized, and proteins were separated by SDS-PAGE and immunoblotted using primary antibodies for total JNK (JNK) or active JNK phosphorylated at T183 and Y185 (JNK-p), and IRDye-conjugated secondary antibodies. The bands were visualized using the LI-COR Odyssey CLx imager. c LI-COR Image Studio software was used to quantify band intensities. The data are presented as the increase of active to total JNK ratio (pJNK/JNK) relative to that observed in mock-infected WT mice. Error bars represent ±1 SEM. Significance was calculated by an unpaired t test (* p < 0.05, ** p < 0.01).
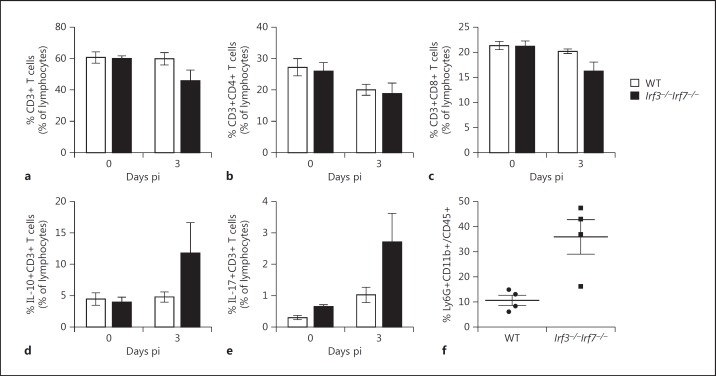
Furthermore, we performed flow cytometry studies of immune cells in the lung of DKO and WT infected mice to further substantiate these observations. IAV infection of WT mice did not reveal any differences in the frequency or functionality of lung CD3+ T cells (60%; fig. 9a). However, in the DKO mice IAV infection resulted in a decreased frequency of CD3+ T cells on day 3 (∼46%) as compared to day 0 (∼60%), although these differences were not significant (fig. 9a). The detailed analysis of CD4+ and CD8+ T cells showed that this observation might be due to reduced frequencies of CD8+ T cells (∼16%) in the lungs of DKO mice as compared to uninfected mice (∼21%; fig. 9b, c). With respect to their functionality, increased frequencies of IL-10- (∼12%) and IL-17- (∼2.7%) secreting CD3+ T cells were detected in DKO mice as compared to the lungs from mice prior to infection (approx. 3 and 0.6%; fig. 9d, e). These findings revealed phenotypical and functional differences of T cells in the lungs of DKO mice after infection. Analysis of granulocyte markers confirmed the above observations from gene expression arrays and histological studies of an increased infiltration of granulocytes in DKO compared to WT mice at day 3 pi (fig. 9f).
Fig. 9.
Flow cytometry of lung cells in WT and DKO mice. The frequencies of CD3+, CD4+, and CD8+ T cells in the lungs of WT and DKO mice (n = 3–5) on day 3 pi after infection with 2 × 103 FFU PR8M were determined by flow cytometry staining of cells ex vivo: CD3+ T cells (a), CD3+CD4+ T cells (b), CD3+CD8+ T cells (c), IL-10+CD3+ T cells (d), IL-17+CD3+ T cells (e), and granulocytes (f). Differences between WT and DKO in a–e were not significant; differences between WT and DKO in f were significant (p < 0.05, Mann-Whitney U test).
Discussion
We recently described the susceptible phenotype of Irf7−/− mice [34], a key player of host IFN response to IAV infections. In this study, we investigated the roles of both Irf3 and Irf7 in the host response early after IAV using single KO and DKO mice. In line with previous studies, the absence of Irf3 and particularly Irf7 genes resulted in a significant reduction of type I IFN expression in mutant mouse lungs after viral infection [16, 17, 18, 19, 21, 35, 36, 37]. This correlated with the higher susceptibility of the Irf3−/−, Irf7−/− and Irf3−/−Irf7−/− mice, underlining the importance of the IFN response to fight IAV infections.
A previous publication studied the role of Irf3 and Irf7 for IFN induction after IAV infection using epithelial cell cultures from the respective KO mice [21]. Cultured murine tracheal epithelial cells were able to produce type I and type III IFN in a RIG-I/MAVS-dependent manner. Crotta et al. [21] found a significantly reduced production of IFN type I in Irf7−/− epithelial cells. Irf7 was also identified as a highly regulated gene in an RNAseq transcriptome analysis from IAV-infected lungs of susceptible and resistant mouse strains [34]. This study also demonstrated that Irf7−/− mice were more susceptible than WT mice to IAV infections [34].
Here, we report that the deletion of Irf3 also had a significant impact on the host response since KO mice exhibited increased susceptibility to PR8 infection whereas Irf3 was not required for IFN induction in primary epithelial cells [21]. Notably, we found an increased level of IFNL2/3 in BAL of Irf3−/− mice after infection compared to WT mice. These results suggest a compensatory mechanism in Irf3−/− KO mice which has also been suggested by Crotta et al. [21]. However, the increased IFNL2/3 was not able to compensate for the lower type I IFN levels in Irf3−/− mice, resulting in a higher viral load on day 1, which is most likely the cause for increased mortality. It is important to note that the differences between the in vitro and in vivo findings that were observed underscore the necessity to validate gene functions in in vivo models.
A case report from a 2.5-year-old child suffering from severe influenza revealed heterozygous mutations (F410V and Q421X) in IRF7 and a remarkable reduction in the production of IFN type I (except IFNβ) and type III [38]. This observation supports our findings that, despite a certain redundancy of the IFN system, the effect of a loss-of-function may be much more severe in the context of the whole organism.
In line with the data from Crotta et al. [21], we found that type I IFN was completely absent in Irf3−/−/Irf7−/− DKO and only a slight production of IFNL2/3 was observed. Using a 100-fold lower dose of PR8M, DKO mice showed a survival rate of 75% (data not shown), demonstrating that DKO mice are still able to clear the infection and that the alternative pathways are functional, albeit to a very limited degree.
MX1 is not functional in C57BL/6 mice but RNA is still expressed and can be used to monitor the activation of IFN-stimulated genes. In DKO mice, Mx1 expression was strongly reduced as well as other antiviral genes like the Oas family. From these results, we conclude that alternative pathways can still be activated even in the absence of Irf3 and Irf7, but have limited protection from severe disease.
The genes of the JNK pathway were upregulated in DKO mice and, in addition, an increased activation of the JNK pathway was observed. A pronounced infiltration of granulocytes was observed in DKO mice whereas induction of a T cell response was missing and the numbers of infiltrating T cells were decreased in DKO mice. The genes encoding the proinflammatory cytokines TNF and IL-1 were found to be more highly expressed in DKO compared to WT mice, as well as other granulocyte-attracting chemokines like Cxcl1, Cxcl2, Cxcl3, and Cxcl5. The role of granulocytes in viral infections is dichotomous, since they can have beneficial and detrimental effects [39, 40, 41, 42]. The release of their toxic substances might be responsible for massive cell damage and tissue necrosis leading to severe pneumonia and acute respiratory distress syndrome. Brandes et al. [39] revealed qualitative differences in the lung transcriptome between lethal versus nonlethal influenza infections. They identified a feedforward inflammatory loop that was driven by accumulation of neutrophils in infected lungs. Their study indicated a narrow therapeutic window. Moderate neutrophil depletion increased survival of IAV-infected mice by reducing the damaging effects of these cells, whereas the total depletion of granulocytes resulted in increased mortality. Our results indicate that upregulation of the Irf3- and Irf7-independent pathways, namely the JNK pathway, and the resulting higher infiltration of granulocytes may be an important driver of a severe outcome. This result is in line with the findings of Börgeling et al. [43], who presented p38 MAPK as a potential target for antiviral strategies since an inhibitor was able to protect mice from lethal influenza.
In conclusion, this study adds to our understanding of the role of IRF genes and downstream IFN pathway activation for the host defense to IAV infection. Deletion of both Irf3 and Irf7 resulted in the absence of induction of IFNA and IFNB. Concomitantly, we observed a wider spread of virus in the lungs, a larger degree of tissue damage, an increase in granulocyte infiltration and reduced activation of the adaptive immune response in DKO mice. Together, these factors contribute to the observed higher susceptibility of DKO mice to IAV infection.
Author Contributions
B.H., E.W. and K.S. conceived the experiments. B.H., H.H., I.G., P.R., H.E., R.G., E.W. performed the experiments. B.H., H.H., E.W., P.R., H.E. and K.S. analyzed the data. B.H., E.W., H.H. and K.S. contributed to writing of the manuscript.
Disclosure Statement
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare that they do not have any conflicts of interests.
Supplementary Material
Supplementary data
Acknowledgements
We wish to thank Karin Lammert for excellent technical assistance, and Irit Gat-Viks and Yael Steuerman (Cell Research and Immunology Department, Tel Aviv University, Tel Aviv, Israel) for help with digital cell quantification. Part of this work was for a PhD thesis (H.H.) at the University of Veterinary Medicine, Hannover.
This work was supported by intramural grants from the Helmholtz-Association (Program Infection and Immunity), a FluResearchNet research grant (No. 01KI07137) from the German Ministry of Education and Research, and an ‘Infection challenge in the German Mouse Clinic’ research grant from the German Ministry of Education and Research to K.S. The PhD work of H.H. was partially financed by a scholarship from the Tierärztlichen Hochschule Hannover and a grant from the Vietnamese Government (Program Biotechnology and Aquaculture - Ministry of Education and Training and Ministry of Agriculture and Rural Development).
References
- 1.Fauci AS. Seasonal and pandemic influenza preparedness: science and countermeasures. J Infect Dis. 2006;194((suppl 2)):S73–S76. doi: 10.1086/507550. [DOI] [PubMed] [Google Scholar]
- 2.Kollmus H, Wilk E, Schughart K. Systems biology and systems genetics - novel innovative approaches to study host-pathogen interactions during influenza infection. Curr Opin Virol. 2014;6:47–54. doi: 10.1016/j.coviro.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Ciancanelli MJ, Abel L, Zhang SY, Casanova JL. Host genetics of severe influenza: from mouse Mx1 to human IRF7. Curr Opin Immunol. 2016;38:109–120. doi: 10.1016/j.coi.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.To KK, Zhou J, Chan JF, Yuen KY. Host genes and influenza pathogenesis in humans: an emerging paradigm. Curr Opin Virol. 2015;14:7–15. doi: 10.1016/j.coviro.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 6.Kawasaki T, Kawai T, Akira S. Recognition of nucleic acids by pattern-recognition receptors and its relevance in autoimmunity. Immunol Rev. 2011;243:61–73. doi: 10.1111/j.1600-065X.2011.01048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem Biophys Res Commun. 2009;388:621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- 8.Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 9.Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Honda K, Taniguchi T. IRFs: master regulators of signalling by toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 11.Haller O, Kochs G, Weber F. The interferon response circuit: induction and suppression by pathogenic viruses. Virology. 2006;344:119–130. doi: 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Page C, Genin P, Baines MG, Hiscott J. Interferon activation and innate immunity. Rev Immunogenet. 2000;2:374–386. [PubMed] [Google Scholar]
- 13.Malmgaard L. Induction and regulation of IFNs during viral infections. J Interferon Cytokine Res. 2004;24:439–454. doi: 10.1089/1079990041689665. [DOI] [PubMed] [Google Scholar]
- 14.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 15.Steinberg C, Eisenacher K, Gross O, Reindl W, Schmitz F, Ruland J, Krug A. The IFN regulatory factor 7-dependent type I IFN response is not essential for early resistance against murine cytomegalovirus infection. Eur J Immunol. 2009;39:1007–1018. doi: 10.1002/eji.200838814. [DOI] [PubMed] [Google Scholar]
- 16.Lazear HM, Lancaster A, Wilkins C, Suthar MS, Huang A, Vick SC, Clepper L, Thackray L, Brassil MM, Virgin HW, Nikolich-Zugich J, Moses AV, Gale M, Jr, Fruh K, Diamond MS. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog. 2013;9:e1003118. doi: 10.1371/journal.ppat.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 18.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 19.Rudd PA, Wilson J, Gardner J, Larcher T, Babarit C, Le TT, Anraku I, Kumagai Y, Loo YM, Gale M, Jr, Akira S, Khromykh AA, Suhrbier A. Interferon response factors 3 and 7 protect against chikungunya virus hemorrhagic fever and shock. J Virol. 2012;86:9888–9898. doi: 10.1128/JVI.00956-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spann KM, Loh Z, Lynch JP, Ullah A, Zhang V, Baturcam E, Werder RB, Khajornjiraphan N, Rudd P, Loo YM, Suhrbier A, Gale M, Jr, Upham JW, Phipps S. IRF-3, IRF-7, and IPS-1 promote host defense against acute human metapneumovirus infection in neonatal mice. Am J Pathol. 2014;184:1795–1806. doi: 10.1016/j.ajpath.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 21.Crotta S, Davidson S, Mahlakoiv T, Desmet CJ, Buckwalter MR, Albert ML, Staeheli P, Wack A. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 2013;9:e1003773. doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blazejewska P, Koscinski L, Viegas N, Anhlan D, Ludwig S, Schughart K. Pathogenicity of different PR8 influenza A virus variants in mice is determined by both viral and host factors. Virology. 2011;412:36–45. doi: 10.1016/j.virol.2010.12.047. [DOI] [PubMed] [Google Scholar]
- 23.Wilk E, Schughart K. The mouse as model system to study host-pathogen interactions in influenza A infections. Curr Protoc Mouse Biol. 2012;2:177–205. doi: 10.1002/9780470942390.mo110173. [DOI] [PubMed] [Google Scholar]
- 24.R_Core_Team: R: A language and environment for statistical computing Vienna, R Foundation for Statistical Computing. 2013 http://www.R-project.org/ [Google Scholar]
- 25.Carlson M. Agilent chips that use Agilent design number 026655 annotation data (chip MmAgilentDesign026655) R package version 2.14.0. 2014 http://bioconductor.org/packages/release/data/annotation/html/MmAgilentDesign026655.db.html. [Google Scholar]
- 26.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. article3. [DOI] [PubMed] [Google Scholar]
- 27.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful apporach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 29.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29:1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altboum Z, Steuerman Y, David E, Barnett-Itzhaki Z, Valadarsky L, Keren-Shaul H, Meningher T, Mendelson E, Mandelboim M, Gat-Viks I, Amit I. Digital cell quantification identifies global immune cell dynamics during influenza infection. Mol Syst Biol. 2014;10:720. doi: 10.1002/msb.134947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Dengler L, Kuhn N, Shin DL, Hatesuer B, Schughart K, Wilk E. Cellular changes in blood indicate severe respiratory disease during influenza infections in mice. PLoS One. 2014;9:e103149. doi: 10.1371/journal.pone.0103149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilk E, Pandey AK, Leist SR, Hatesuer B, Preusse M, Pommerenke C, Wang J, Schughart K. RNAseq expression analysis of resistant and susceptible mice after influenza A virus infection identifies novel genes associated with virus replication and important for host resistance to infection. BMC Genomics. 2015;16:655. doi: 10.1186/s12864-015-1867-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christensen JE, Fenger C, Issazadeh-Navikas S, Krug A, Liljestrom P, Goriely S, Paludan SR, Finsen B, Christensen JP, Thomsen AR. Differential impact of interferon regulatory factor 7 in initiation of the type I interferon response in the lymphocytic choriomeningitis virus-infected central nervous system versus the periphery. J Virol. 2012;86:7384–7392. doi: 10.1128/JVI.07090-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daffis S, Samuel MA, Suthar MS, Keller BC, Gale M, Jr, Diamond MS. Interferon regulatory factor IRF-7 induces the antiviral alpha interferon response and protects against lethal West Nile virus infection. J Virol. 2008;82:8465–8475. doi: 10.1128/JVI.00918-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daffis S, Suthar MS, Szretter KJ, Gale M, Jr, Diamond MS. Induction of IFN-β and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent signal that does not require IRF-3 and IRF-7. PLoS Pathog. 2009;5:e1000607. doi: 10.1371/journal.ppat.1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciancanelli MJ, Huang SX, Luthra P, Garner H, Itan Y, Volpi S, Lafaille FG, Trouillet C, Schmolke M, Albrecht RA, Israelsson E, Lim HK, Casadio M, Hermesh T, Lorenzo L, Leung LW, Pedergnana V, Boisson B, Okada S, Picard C, Ringuier B, Troussier F, Chaussabel D, Abel L, Pellier I, Notarangelo LD, Garcia-Sastre A, Basler CF, Geissmann F, Zhang SY, Snoeck HW, Casanova JL. Infectious disease: life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 2015;348:448–453. doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brandes M, Klauschen F, Kuchen S, Germain RN. A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell. 2013;154:197–212. doi: 10.1016/j.cell.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galani IE, Andreakos E. Neutrophils in viral infections: current concepts and caveats. J Leukoc Biol. 2015;98:557–564. doi: 10.1189/jlb.4VMR1114-555R. [DOI] [PubMed] [Google Scholar]
- 41.Tate MD, Brooks AG, Reading PC, Mintern JD. Neutrophils sustain effective CD8+ T-cell responses in the respiratory tract following influenza infection. Immunol Cell Biol. 2012;90:197–205. doi: 10.1038/icb.2011.26. [DOI] [PubMed] [Google Scholar]
- 42.Tate MD, Ioannidis LJ, Croker B, Brown LE, Brooks AG, Reading PC. The role of neutrophils during mild and severe influenza virus infections of mice. PLoS One. 2011;6:e17618. doi: 10.1371/journal.pone.0017618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Börgeling Y, Schmolke M, Viemann D, Nordhoff C, Roth J, Ludwig S. Inhibition of p38 mitogen-activated protein kinase impairs influenza virus-induced primary and secondary host gene responses and protects mice from lethal H5N1 infection. J Biol Chem. 2014;289:13–27. doi: 10.1074/jbc.M113.469239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hatesuer B, Bertram S, Mehnert N, Bahgat MM, Nelson PS, Pohlman S, Schughart K. Tmprss2 is essential for influenza H1N1 virus pathogenesis in mice. PLoS Pathog. 2013;9:e1003774. doi: 10.1371/journal.ppat.1003774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data