Abstract
Proline-rich tyrosine kinase 2 (Pyk2) is a member of the focal adhesion kinase (FAK) family and is mainly expressed in neuronal and hematopoietic cells. As FAK family members are involved in signaling connections downstream of integrins, we studied the role of Pyk2 in complement-receptor 3 (CR3, also known as Mac-1, integrin αMβ2, CD11b/CD18)-mediated phagocytosis, a key process in innate immunity. Using 3 independent approaches, we observed that Pyk2 contributes to CR3-dependent phagocytosis by RAW 264.7 macrophages, but is dispensable for Fcγ receptor (FcγR)-mediated uptake. Reduction of Pyk2 expression levels via siRNA, the pharmacological inhibition of Pyk2 kinase activity as well as macrophage treatment with a cell permeable TAT fusion protein containing the C-terminus of Pyk2 (TAT-PRNK) significantly impaired CR3-mediated phagocytosis without affecting FcγR-mediated uptake. In addition, Pyk2 was strongly recruited to complement opsonized Escherichia coli and the pharmacological inhibition of Pyk2 significantly decreased uptake of the bacteria. Finally, CRISPR/Cas-mediated disruption of the pyk2 gene in RAW 264.7 macrophages confirmed the role of this protein tyrosine kinase in CR3-mediated phagocytosis. Together, our data demonstrate that Pyk2 selectively contributes to the coordination of phagocytosis-promoting signals downstream of CR3, but is dispensable for FcγR-mediated phagocytosis.
Key Words: Pyk2, Phagocytosis, CR3, Bacteria, Opsonization, Fcγ receptor, Macrophages
Introduction
Phagocytosis is an important process for both unicellular and multicellular eukaryotes. In Metazoa, internalization of apoptotic bodies contributes to tissue homeostasis. Moreover, the phagocytosis of microorganisms like fungi or bacteria limits infections [1]. Therefore, phagocytosis is a major part of the first-line defense against pathogens and plays a pivotal role in the innate immune response. Moreover, phagocytosis is linked to the release of proinflammatory cytokines and is a prerequisite for antigen presentation. It is thus also instrumental in orchestrating the adaptive immunity [2].
Professional phagocytes, namely granulocytes, macrophages and dendritic cells, are able to recognize and ingest pathogenic microorganisms via specific phagocytic receptors. Some phagocytic receptors, such as Dectin1 or CEACAM3, directly recognize characteristic structures on bacterial or fungal cells [3, 4, 5] whereas other phagocytic receptors require prior opsonization of the particles. For example, Fc receptors (FcRs) mediate the uptake of pathogens decorated by specific immunoglobulins via recognition of the Fc part of antibodies (Abs) [6]. A further mechanism, which does not require specific Abs, is the opsonization of pathogens with factors of the complement system, mainly iC3b. Phagocytosis of complement-opsonized particles occurs via binding and signaling through complement receptors (CRs) [7]. The 2 main complement receptors, CR3 (also known as Mac-1, CD11b/CD18, integrin αMβ2) and CR4 (also known as gp150/95, CD11c/CD18, integrin αXβ2), are integrin family members with a common β2 subunit. While CR4 is mainly expressed in tissue macrophages, CR3 expression is predominant in blood monocytes and granulocytes [8].
A family of cytoplasmic nonreceptor protein tyrosine kinases (PTKs), which is connected to integrin signaling in many cell types, is the focal adhesion kinase (FAK) family, consisting of FAK and proline-rich kinase 2 (Pyk2, also known as FAKB, CAK beta, RAFTK or FAK2). Pyk2 and FAK have a common domain structure with the central kinase showing a sequence similarity of 60% [9, 10, 11, 12]. However, while FAK is ubiquitously expressed, Pyk2 expression is restricted to cells of the central nervous system and hematopoietic cells. Both kinases have been linked to integrin-mediated cell adhesion and migration [13]. For example, macrophages with decreased Pyk2 expression or genetic deficiency in Pyk2 are significantly impaired in their migratory potential [14, 15, 16]. In line with this functional impairment, Pyk2-deficient macrophages show a reduced contractile activity of lamellipodia and diminished membrane ruffling [16]. Importantly, the stimulation of integrins, such as αVβ3 or αMβ2 (CR3), results in the activation of Pyk2 and its colocalization with integrins [14, 17, 18, 19]. The functional connection between integrins and Pyk2 in hematopoietic cells is in line with the observed functional role of Pyk2 in migration and the spreading of phagocytes. These observations also raise the question, if additional integrin-dependent processes in phagocytic cells, such as complement receptor-mediated phagocytosis, might be influenced by Pyk2. Interestingly, Pyk2-deficient mice show an impaired immune response to Staphylococcus aureus infection in vivo [20]. However, in vitro experiments with serum-opsonized bacteria or polystyrene beads do not reveal an altered uptake or reduced bacterial killing in the case of Pyk2-deficient murine granulocytes [20].
To resolve these apparent discrepancies, we have directly addressed the function of Pyk2 in CR3-mediated phagocytosis. By using pharmacological inhibition, protein transduction with dominant-negative mutants, siRNA-mediated knockdown as well as CRISPR/Cas-mediated knockout of Pyk2, we observe a significant contribution of Pyk2 to CR3-mediated but not FcR-mediated phagocytosis. In murine macrophages, Pyk2 is recruited to complement-opsonized but not to IgG-opsonized particles, and bacteria opsonized by IgG-depleted serum are internalized in a Pyk2-dependent manner. Together, our results demonstrate that Pyk2 positively contributes to integrin-mediated phagocytosis in macrophages.
Materials and Methods
Cell Culture and Transfection
RAW 264.7 macrophages were cultured in DMEM/10% heat-inactivated fetal bovine serum (hiFBS) at 37°C, 5% CO2. Pyk2 siRNA or siGLO control siRNA (Dharmacon, Lafayette, Colo., USA) were delivered using INTERFERin® transfection reagent (Polyplus-Transfection, Illkirch, France) according to the manufacturer's recommendations.
Reagents, Antibodies and Coupling of Polystyrene Beads
Pyk2/FAK inhibitor PF431396 was purchased from Tocris Bioscience (Wiesbaden-Nordenstadt, Germany). Pervanadate (PV) was freshly prepared by incubation of 50 mM H2O2 and 10 mM sodium orthovanadate for 5 min and the subsequent addition of 1 mg catalase. Carboxylated polystyrene microbeads (Polysciences, Eppelheim, Germany) were covalently coupled to proteins according to the manufacturer's protocol. Beads were coupled to 400 µg murine IgG1, ovalbumin or recombinant protein G (Sigma Aldrich, Hamburg, Germany). Protein G-coupled beads were incubated with rat monoclonal Ab against CD11b, purified from hybridoma cell supernatants (Clone M1/70.15.11.5.2, Developmental Studies Hybridoma Bank, University of Iowa) or the isotype-matched control rat Ab (IgG2b, ImmunoTools, Friesoythe, Germany). During protein coupling, the beads were also biotin-labeled with 0.5 mg/ml sulfo-NHS-LC Biotin (Thermo Fisher Scientific, Bonn, Germany). For Western blot analysis, rabbit polyclonal Pyk2 Ab (H-102, Santa Cruz Biotechnology, Heidelberg, Germany), rabbit polyclonal phospho-Pyk2 [pY402] Ab (17HCLC, Life Technologies, Darmstadt, Germany), monoclonal tubulin Ab (clone E7), polyclonal C3 Ab (MP Biomedicals, Eschwege, Germany) were used.
Cloning and Purification of TAT-GST Fusion Proteins
The sequence of the transactivator protein (TAT) was inserted via complementary oligonucleotides (5′-TACCGCGGCGGCGCTGGCGGCGTTTCTTGCGGCCGTACAT-3′ and 5′-TAATGTACGGCCGCAAGAAACGCCGCCAGCGCCGCCGCGG-3′) in the NdeI site of pET-42a(+) (Merck Millipore, Darmstadt, Germany) resulting in pET-42-TAT. The C-terminal part of human Pyk2, hPRNK, or the fluorescent protein EGFP were amplified by PCR (PRNK sense: 5′-TGCATGACTAGTAGTGACGTTTATCAGATGG-3′, PRNK antisense: 5′-TGACAGGTCGACTCACTCTGCAGGTGG-3′; EGFP sense: 5′-ATGCTGACTAGTAGCAAGGGCGAGGAG-3′, EGFP antisense: 5′-ACTGCTGTCGACGTTAATTAAGTTTGTGCCCCAG-3′) and inserted via SalI and SpeI restriction sites in pET-42-TAT. The TAT-fusion proteins were expressed in Escherichia coli BL-21 DE3 and purified using a GSTrapFF column (GE Healthcare, Freiburg, Germany). TAT-GST fusion proteins were applied 30 min before the actual start of the experiment.
Microbead Phagocytosis Assay
1 × 105 RAW 264.7 macrophages were seeded on gelatine-coated coverslips in 24-well plates and then serum-starved for 16 h (0.5% BSA in DMEM). The cells were stimulated for 10 min with DMEM/10% hiFBS prior to the addition of the biotinylated, protein-coupled beads. In some cases, the Pyk2 inhibitor PF431396 or DMSO were added 10 min before bead addition. Protein-coupled and biotinylated beads [30 beads/cell; multiplicity of infection (MOI) 30] were centrifuged onto the cells (for 3 min at 500 g). After 2 h, the cells were fixed and extracellular beads were stained using rhodamine-streptavidin (Jackson ImmunoResearch Laboratories, Newmarket, UK). Samples were analyzed using wide-field microscopy (Leica AF6000 LX) and the ratio of intracellular versus total cell-associated beads was determined.
Pyk2 Immunostaining
For immunostaining of Pyk2, 1 × 105 cells were seeded as indicated on coverslips coated with gelatine (0.1%) or poly-L-lysine (10 μg/ml) in 24-well plates, and then serum-starved for 16 h. The cells were activated with growth medium or growth medium containing 200 ng/ml PMA 10 min before infection with protein-coupled microbeads or E. coli for the indicated times and at the indicated MOI. Microbeads and bacteria were labeled with 0.05 mg/ml fluorescein isothiocyanat (FITC; Sigma Aldrich, Hamburg, Germany) in PBS for 1 h at 37°C prior to infection. After fixation and permeabilization, samples were stained with polyclonal anti-Pyk2 Ab and fluorescently labeled secondary anti-rabbit IgG Ab (Jackson ImmunoResearch Laboratories). If indicated, cells were additionally stained with Phalloidin-Cy5 (Life Technologies). For fluorescence intensity profiles, line plots with the same distances were created for each fluorescence channel at sites of bead or bacteria attachment using ImageJ software. Intensities of Pyk2-Cy3 and Phalloidin-Cy5 (when indicated) were normalized to the maximal fluorescence intensity of beads or bacteria, respectively. Mean values of normalized data of 7-10 attachment sites in different cells were combined.
Scanning Electron Microscopy
1 × 105 cells were seeded and treated as indicated. After incubation with microbeads or bacteria, fixation buffer (3% formaldehyde, 3% glutaraldehyde in sodium cacodylate buffer containing 0.09 M sucrose, 0.01 M CaCl2 and 0.01 M MgCl2, pH 6.9) was added. After 5 min, the fixation buffer, together with the medium, was aspirated, fresh buffer was then added and incubation took place for 30 min. Subsequently, the fixation buffer was changed again and incubated overnight at 4°C before the cells were washed 3 times with sodium cacodylate buffer. After stepwise dehydration to 70%, critical-point drying with CO2 was performed, and the samples were sputter-coated with 5 nm gold-palladium in a BAL-TEC SCD 030. The preparations were analyzed on a Zeiss Auriga CrossBeam Workstation at 15 kV, using the secondary electron detector at the Electron Microscopy Center, of the University of Konstanz.
Serum Preparation and Opsonization of E. coli
Fresh murine blood samples were allowed to clot at room temperature for at least 30 min. The clotted material was removed by centrifugation for 15 min at 3,000 rpm and the serum was snap-frozen in liquid N2 and stored at −80°C. IgG depletion of the serum was accomplished by incubating 500 µl serum with 2 ml formaldehyde-fixed Streptococcus dysgalactiae suspension in PBS for 16 h at 4°C. IgG depletion was confirmed via Western blot. For heat-inactivation, the IgG-depleted serum was incubated for 30 min at 65°C. Opsonization of E. coli with serum samples was achieved by incubation of 1 × 108 bacteria with 30% serum in PBS for 30 min at 37°C.
Flow Cytometry-Based Phagocytosis Assay
1 × 106 RAW 264.7 macrophages were seeded in 6-well plates and serum-starved for 16 h. Cells were activated with 200 ng/ml PMA 10 min prior to infection with FITC-labeled and opsonized E. coli at a MOI of 100 in presence or absence of 10 µM PF431396. Bacteria were gently centrifuged onto the cells and samples were incubated for 1 h at 37°C. Cells were washed 3 times with PBS, detached, resuspended in PBS and analyzed via flow cytometry. Overall FITC fluorescence of cell-associated bacteria was measured before extracellular signals were quenched with trypan blue to selectively detect fluorescence from internalized bacteria. The uptake index was calculated by multiplication of the percentage of FITC-positive cells with the mean fluorescence.
CRISPR/Cas-Mediated Knockout of Pyk2 in RAW 264.7 Cells
Transfection of RAW 264.7 macrophages was performed using the Nucleofector™ kit V for the Nucleofector device (Lonza Group Ltd, Basel, Switzerland) according to the manufacturer's protocol. Cells were cotransfected with plasmids containing gRNAs targeting exon 1 (gRNA sequence: 5′-GGGCCCCCCAGAGCCCATGG-3′) and exon 2 (gRNA sequence: 5′-GCTGCACCCACAGATGACCG-3′) of the pyk2 gene; and a plasmid encoding for the Cas9 enzyme together with a puromycin resistance cassette (pX459 provided by Feng Zhang via www.addgene.org; plasmid No. 48139). Two days after transfection, cells harboring the Cas9-encoding vector were selected, using 1.5 µg/ml puromycin for 3 days, and single-cell clones were propagated. Complete knockout of Pyk2 in selected clonal lines was verified via immunoblotting against Pyk2 protein.
Statistical Analysis
For microbead phagocytosis assays, data are shown as medians, with boxes representing the 25th to 75th percentiles and error bars the 10th to 90th percentiles of 3 independent experiments. For the flow cytometry-based invasion assay, data are shown as means ± SD. Differences between sample means were assessed using the Mann-Whitney U test (* p < 0.05; ** p < 0.01; *** p < 0.001).
Results
CD11b Ab-Coated Beads Trigger CR3-Mediated Phagocytosis
To analyze the function of Pyk2 during opsono-phagocytosis, we used Ab-coupled microbeads, which selectively address either CR3-mediated or FcγR-mediated uptake. Thus, carboxylated polystyrene microbeads were first covalently coupled with protein G and then incubated with a rat monoclonal Ab directed against the murine CD11b subunit of CR3 (anti-CR3 beads) or with an isotype-matched rat control Ab (rat IgG beads), respectively. In addition, some beads were directly coupled to murine IgG (Fc beads) to trigger phagocytic processes via FcγR, and a further batch of beads was coated with ovalbumin (albumin beads) to serve as an additional control. During coupling, the beads were also biotinylated by the addition of sulfo-NHS-LC-biotin. The different protein-coated, biotinylated bead preparations were incubated for 120 min with RAW 264.7 macrophages. Following fixation, streptavidin-rhodamine was added to selectively stain extracellular beads. Microscopic evaluation by phase-contrast and fluorescence microscopy allowed the visualization and quantification of extra- and intracellular beads (online suppl. fig. 1A; for all online suppl. material, see www. karger.com/doi/10.1159/000442944). Albumin bead internalization occurred, albeit at a low percentage of approximately 10%. A slightly greater uptake of rat IgG beads, around 20%, was observed; in contrast, 40–50% of the cell-associated particles were intracellular for Fc or anti-CR3 beads (online suppl. fig. 1A, C). To demonstrate the selective uptake of anti-CR3 beads via CR3, parallel samples were incubated with beads in the presence of a CR3-blocking Ab (online suppl. fig. 1B). Clearly, blocking of CR3 by Abs reduced the internalization of anti-CR3 beads down to the background level of control Ab uptake whereas blocking of the CR3 receptor had no effect on the uptake of Fc, control or albumin beads (online suppl. fig. 1B, C). These results demonstrate that the microbead phagocytosis assay is able to discriminate between FcγR- and CR3-mediated phagocytosis, and this allowed us to analyze the role of Pyk2 during these processes.
Pyk2 Is Recruited to Sites of CR3-Mediated Phagocytosis
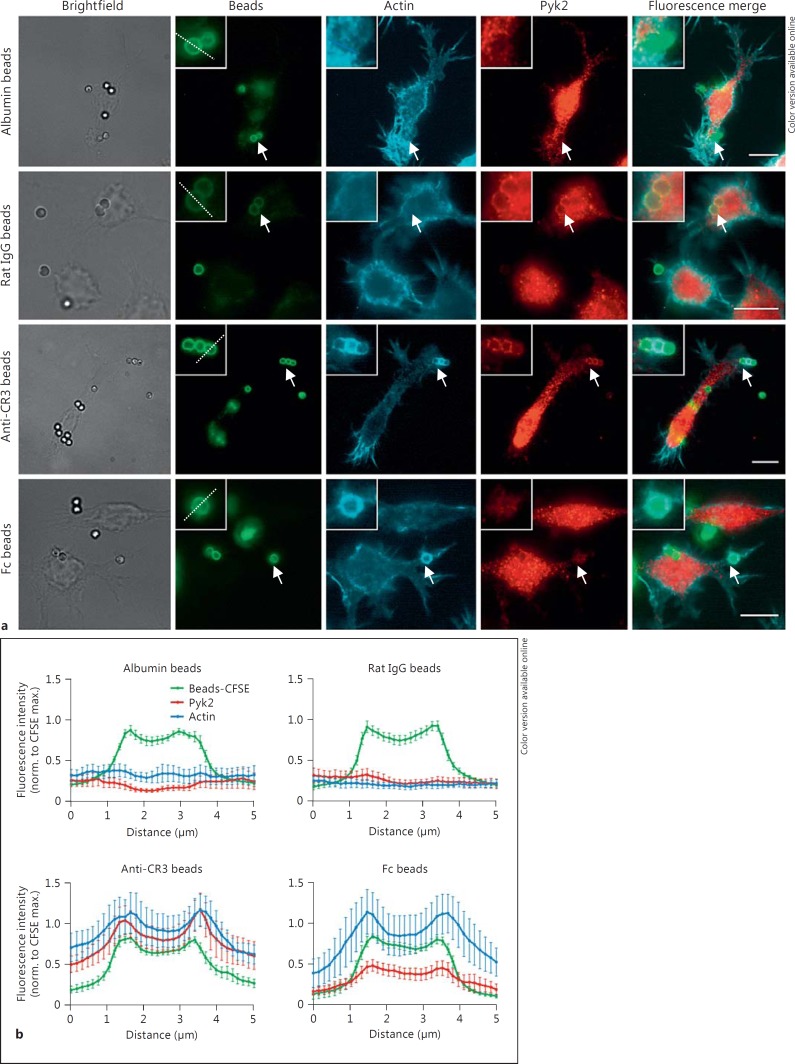
In the podosomes of migrating macrophages, a colocalization of Pyk2 with integrin αMβ2 has been observed [14]. To elucidate whether Pyk2 localizes to sites of CR3-mediated phagocytosis, RAW 264.7 cells were incubated with anti-CR3, control, Fc or albumin beads for 60 min and Pyk2 was visualized by immunostaining (fig. 1a). The additional staining of F-actin revealed a ring-like accumulation of actin at the sites of internalization for Fc and anti-CR3 beads confirming the involvement of the actin cytoskeleton in both processes [21]. A comparable F-actin accumulation could not be observed in the isotype control or albumin beads (fig. 1a). Despite an equivalent uptake of Fc and anti-CR3 beads by the macrophages (online suppl. fig. 1), a clear enrichment of Pyk2 at sites of internalization could only be observed for anti-CR3-coupled beads (fig. 1a). The recruitment of Pyk2 to CR3 was clearly visible in intensity profiles through attachment sites of beads coated with anti-CR3 Ab whereas control, albumin or Fc beads did not exhibit a local enrichment of Pyk2 staining (fig. 1b). The selective recruitment of Pyk2 during the uptake of anti-CR3 beads suggests a function of Pyk2 during integrin αMβ2-mediated phagocytosis.
Fig. 1.
Pyk2 is recruited to cell-associated anti-CR3 beads. a Serum-starved RAW 264.7 cells were activated with DMEM/10% hiFBS for 10 min and infected with CFSE-labeled beads coupled to albumin, rat IgG2b (control), α-CD11b Ab (anti-CR3) or IgG (MOI 15) for 30 min. After infection, the cells were fixed and immunostained with α-Pyk2 (1:50) and α-rabbit-Cy3 Ab. In addition, actin was stained using Phalloidin-Cy5. The regarded beads are indicated with arrows. Scale bars: 10 μm. b Normalized fluorescence intensity profiles of sites of bead attachment over a 5-µm distance (dashed lines in a). Intensities were normalized to maximal CFSE fluorescence and mean values of 10 intensity profiles are shown with error bars representing SEM.
Knockdown and Inhibition of Pyk2 Impairs CR3-Mediated Phagocytosis
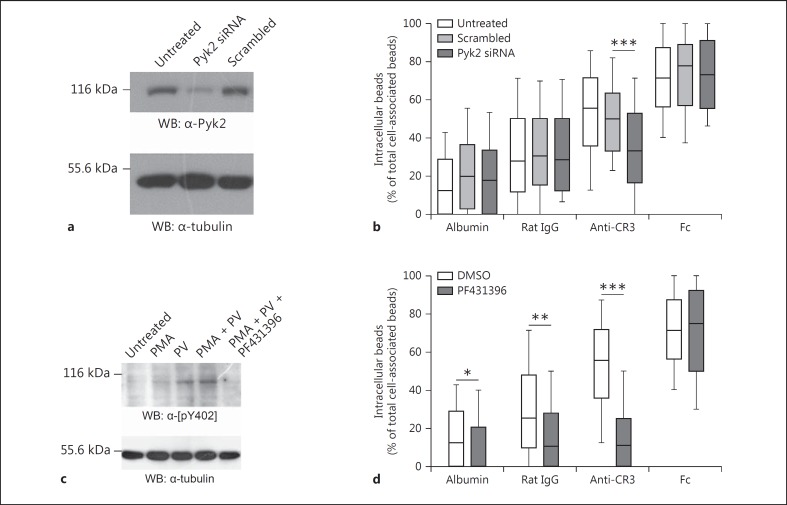
To specifically interfere with Pyk2 function in CR3-mediated phagocytosis, we used 3 independent experimental approaches for the microbead phagocytosis assay. First, the expression level of Pyk2 in macrophages was reduced via siRNA treatment. Western blot analysis revealed an efficient knockdown of Pyk2 in RAW 264.7 cells compared to scrambled siRNA or untreated cells (fig. 2a). Macrophages with a decreased level of Pyk2 were further used in microbead phagocytosis assays to analyze their phagocytic potential. For control and albumin beads, the percentages of internalized beads, 30 and 20%, respectively, were not altered by treatment with scrambled or Pyk2 siRNA (fig. 2b). For Fc beads, the ratio of approximately 70% intracellular beads was also not affected in cells treated with siRNA. However, Pyk2 siRNA significantly reduced the uptake of anti-CR3 beads to the levels of the rat IgG control beads (fig. 2b). Thus, the decrease of Pyk2 expression selectively interfered with the uptake of anti-CR3 beads, suggesting a specific functional role for Pyk2 during complement-mediated phagocytosis.
Fig. 2.
Pyk2 expression and activity are essential for CR3-mediated phagocytosis. a Whole-cell lysates of RAW 264.7 macrophages treated with 100 nM Pyk2 siRNA or 100 nM scrambled siRNA were analyzed by Western blot using α-Pyk2 Ab (1:400) and compared with untreated cells. b Pyk2 siRNA- or scrambled siRNA-treated and -untreated RAW 264.7 macrophages were seeded on gelatine-coated coverslips and serum-starved before activation with DMEM/10% hiFBS and infection (for 2 h at MOI 30) with biotinylated polystyrene beads coupled to albumin, rat IgG2b (control), α-CD11b Ab (anti-CR3) or IgG. After infection, cells were fixed and extracellular beads were stained with streptavidin-rhodamine. The ratio of internalized beads was calculated by counting extra- and intracellular beads per cell. c Cells were treated for 10 min with either 200 ng/ml PMA or 100 µM pervanadate (PV) or PMA + PV together in the presence or absence of PF431396 (10 μM). After incubation, whole-cell lysates were immunoblotted with α-pY402-Pyk2 specific Ab (1:100). Tubulin served as a loading control. d Serum-starved macrophages were incubated with polystyrene beads (as in b) in the presence or absence of PF431396 (10 μM). b, d Whisker plots are derived from 150 cells/sample from 3 independent experiments. Groups were compared by Mann-Whitney U test. * p < 0.05; ** p < 0.01; *** p < 0.001.
In a second approach, we analyzed whether Pyk2 kinase activity is relevant for complement-mediated phagocytosis. To this end, we interfered with Pyk2 function by pharmacological inhibition of its kinase activity, using the Pyk2/FAK inhibitor PF431396 [22]. To analyze the potency of the inhibitor, macrophages were treated either with PMA to stimulate Pyk2 autophosphorylation or with pervanadate (PV) to maintain the phosphorylation state, or with both PMA and PV for 30 min. Western blot analysis of Pyk2 autophosphorylation revealed enhanced phosphorylation of Pyk2 in macrophages treated with PV, which increased slightly in the cells cotreated with PMA (fig. 2c). However, when cells were treated with PMA and PV together with PF431396, Pyk2 autophosphorylation strongly decreased (fig. 2c), confirming the ability of the inhibitor to efficiently block Pyk2 kinase activity. Next, macrophages were treated or not treated with PF431396 and microbead phagocytosis assays were performed. In line with the previous Pyk2 siRNA results, phagocytosis of Fc beads was not influenced by the Pyk2 inhibitor and resulted in about 70% intracellular beads, both in the presence and absence of PF431396 (fig. 2d). In contrast, the treatment with PF431396 led to a strong decrease (approx. 40%) in the internalization of anti-CR3 beads (fig. 2d). Furthermore, the uptake of control or albumin beads was slightly, but significantly, affected by PF431396 treatment (fig. 2d). Together, these data show that the inhibition of Pyk2 kinase activity or the specific reduction of its expression level significantly reduce the phagocytosis of anti-CR3 beads, suggesting an important role for Pyk2 in the process of complement-mediated phagocytosis.
TAT-PRNK Reduces CR3-Mediated Phagocytosis
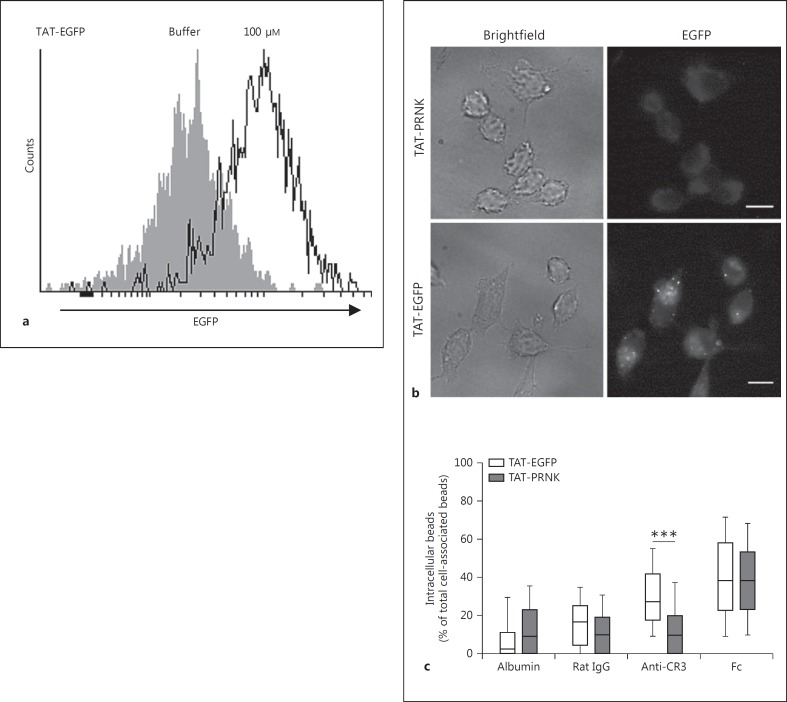
In a third approach, we used the Pyk2 related non-kinase (PRNK), resembling the C-terminus of Pyk2, which is known to interfere with the subcellular localization of Pyk2 [23, 24] (online suppl. fig. 2E). To apply PRNK during CR3-mediated phagocytosis, we fused PRNK with a short peptide taken from the HIV transactivator protein (TAT-peptide). A recombinant TAT-EGFP fusion protein was used to establish the efficient protein transduction into murine macrophages. Incubation of macrophages with 100 µM of purified TAT-EGFP resulted in significant intracellular EGFP fluorescence, as confirmed via flow cytometry and microscopy (fig. 3a, b). Next, macrophages were preincubated for 30 min with TAT-PRNK or the TAT-EGFP control protein, and were subsequently incubated with microbeads for 2 h. The basal uptake of albumin or control beads as well as the internalization of Fc beads was not altered in the presence of TAT-PRNK compared to TAT-EGFP (fig. 3c). However, the internalization of anti-CR3 beads was significantly reduced, and only about 10% of the anti-CR3 beads were intracellular in TAT-PRNK-treated cells (fig. 3b). As PRNK interferes with the proper subcellular localization of Pyk2, this result indicates that the localization of Pyk2 to sites of infection is required for CR3-mediated uptake. Together, our combined results suggest an important role for Pyk2 in complement-dependent phagocytosis in macrophages.
Fig. 3.
PRNK blocks CR3-mediated phagocytosis. a Serum-starved RAW 264.7 macrophages seeded on gelatine-coated coverslips were incubated for 90 min with DMEM/10% hiFBS medium containing 100 μM of TAT-EGFP or buffer. After washing with acetic wash buffer (0.5% acetic acid and 0.5% NaCl in PBS), cells were analyzed by flow cytometry to measure efficiency of protein transduction. b Cells were incubated (as in a) with TAT-EGFP or TAT-PRNK. After incubation, the cells were washed once with acetic wash buffer prior to fixation and analyzed on fluorescence microscopy. Scale bars: 10 μm. c Serum-starved cells were incubated (as in a) with 100 μM TAT-EGFP or TAT-PRNK. Next, polystyrene beads coupled to albumin, rat IgG2b (control), α-CD11b Ab (anti-CR3) or IgG were added (MOI 30). After 2 h, cells were fixed and extracellular beads were stained with streptavidin-rhodamine. The ratio of internalized beads was calculated by counting extra- and intracellular beads per cell. Microscopic quantification was achieved by counting nonstained intracellular and rhodamine-stained extracellular beads. The whisker plots are derived from 3 independent experiments (150 cells/sample). Groups were compared with the Mann-Whitney U test. *** p < 0.001.
Inhibition of Pyk2 Does Not Alter the Formation of Membrane Protrusions
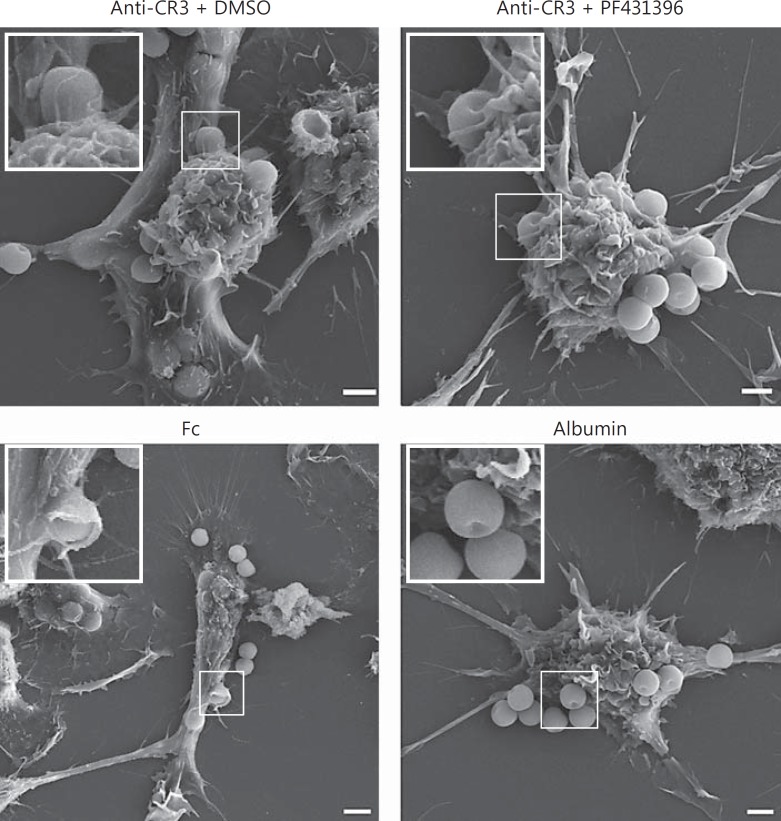
The impaired uptake of anti-CR3 beads upon Pyk2 inhibition might be due to a role of Pyk2 in the formation of membrane protrusions. To test this hypothesis, we analyzed the morphology of macrophages incubated with microbeads using scanning electron microscopy (SEM). SEM analysis revealed the formation of distinct membrane protrusions upon incubation with Fc and anti-CR3 beads, while the incubation with albumin beads did not induce membrane ruffles (fig. 4). Interestingly, the inhibition of Pyk2 with PF431396 did not prevent the formation of membrane ruffles in macrophages incubated with anti-CR3 beads. This observation indicates that Pyk2 is not involved in the formation of membrane protrusions.
Fig. 4.
Pyk2 is not involved in the formation of membrane protrusions during phagocytosis. SEM analysis of macrophages incubated with albumin, anti-CR3 or Fc beads. RAW 264.7 macrophages were seeded on gelatine-coated coverslips and serum-starved before the addition of DMEM/10% hiFBS containing albumin, anti-CR3 or Fc beads, respectively (20 beads/cell). In the case of the anti-CR3 beads, cells were preincubated with DMSO (control) or 10 μM of PF431396 as indicated for 10 min before addition of beads. After incubation for 2 h, the samples were fixed, dehydrated and analyzed via SEM. Images show overviews of whole cells with insets highlighting magnified details.
Pyk2 Is Essential for Efficient Phagocytosis of Complement-Opsonized Bacteria
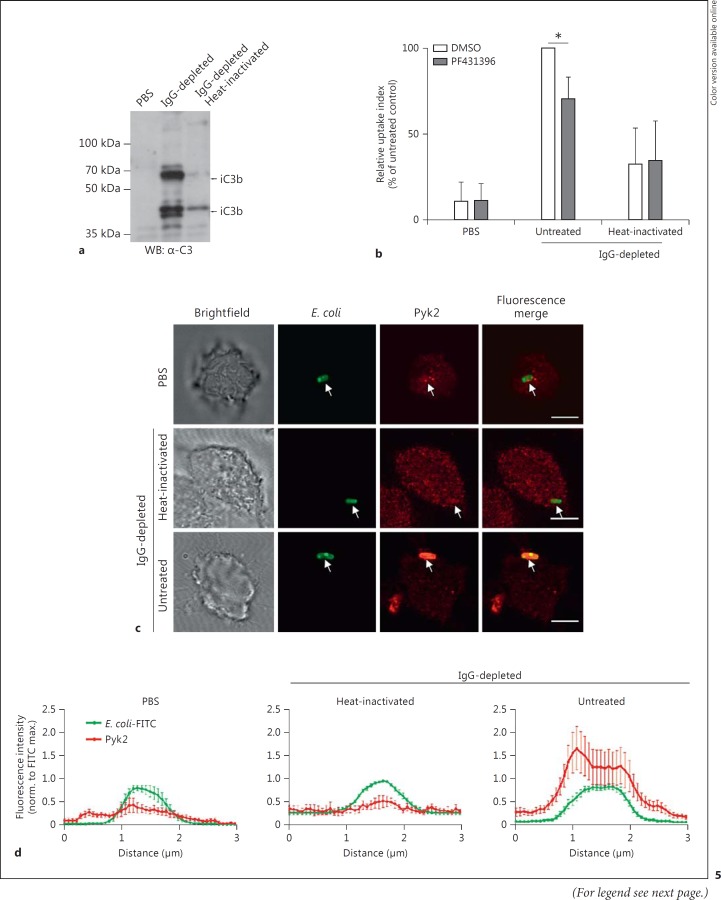
Analyzing phagocytosis by means of Ab-coated microbeads affords precise control over the phagocytic receptors involved. However, it might not reflect the physiological situation, where multiple receptors, having both particulate and soluble ligands, might be stimulated simultaneously. To analyze the role of Pyk2 in a more relevant context, E. coli bacteria were opsonized with either normal or IgG-depleted mouse serum. IgG was removed through the incubation of serum samples with an excess of inactivated, protein G-expressing S. dysgalactiae. Immunoblotting of the resulting serum with an anti-IgG/IgM Ab demonstrated the successful depletion of IgG by S. dysgalactiae (online suppl. fig. 2A). Importantly, the serum levels of C3 were only marginally affected by the IgG depletion (online suppl. fig. 2B). When E. coli was incubated with IgG-depleted serum, immunoblotting with anti-IgG Ab revealed the lack of IgG opsonization by the IgG-depleted serum whereas the normal serum led to IgG deposition on the bacteria (online suppl. fig. 2C). Therefore, to selectively opsonize bacteria with complement, we further used the IgG-depleted serum. As controls, bacteria were incubated in PBS or heat-inactivated serum. Western blot analysis of bacterial pellets showed a strong opsonization of bacteria with C3-derived peptides, when incubated with IgG-depleted serum (fig. 5a). In contrast, incubation with PBS or the heat-inactivated serum resulted in no/very poor opsonization (fig. 5a).
Fig. 5.
Inhibition of Pyk2 reduces CR3-mediated phagocytosis of E. coli. a E. coli were incubated with PBS, IgG-depleted or heat-inactivated serum for 30 min at 37°C and analyzed for efficient opsonization via Western blot with an anti-C3 Ab (iC3b). b Serum-starved macrophages were activated with 200 ng/ml PMA and infected with FITC-labeled E. coli for 60 min (MOI 100) in the presence or absence of 10 µM PF431396. Cells were resuspendend and analyzed via flow cytometry. Uptake indices (% positive cells × mean fluorescence) were measured in the presence of trypan blue and are presented as relative values (normalized to mean of DMSO-treated control). Groups were compared by the Mann-Whitney U test. * p < 0.05. c Serum-starved RAW 264.7 cells seeded on poly-L-lysine-coated coverslips were activated with 200 ng/ml PMA and incubated for 30 min with FITC-labeled E. coli incubated with PBS, IgG-depleted or heat-inactivated serum. Cells were fixed and stained for Pyk2. Scale bars: 10 μm. d Normalized fluorescence intensity profiles of sites of E. coli infection over a 3-µm distance in samples of c. Intensities were normalized to maximal FITC fluorescence of bacteria and mean values of 7-10 intensity profiles are shown with error bars representing SEM.
Complement-opsonized E. coli bacteria were labeled with FITC and then used to study the phagocytosis of bacteria via CR3. For this, macrophages were infected for 60 min with FITC-labeled E. coli, which were either opsonized with complement via the IgG-depleted serum, or incubated with PBS or the heat-inactivated serum, respectively. There was only a moderate uptake of bacteria preincubated with heat-inactivated serum or with PBS, and the low levels of internalization were not influenced by the presence of the Pyk2 inhibitor PF431396 (fig. 5b). As expected, complement opsonization of E. coli allowed increased uptake by the macrophages compared to the control samples (fig. 5b). Strikingly, the internalization of complement-opsonized E. coli by macrophages was significantly reduced in the presence of PF431396, demonstrating the importance of Pyk2 kinase activity during this process (fig. 5b). To rule out that the reduced cell uptake of complement-opsonized bacteria is based on an altered surface expression of CR3 upon treatment with the Pyk2 inhibitor, we analyzed the surface levels of CR3 by Ab staining and flow cytometry. Clearly, treatment with PF431396 did not cause any alterations in CR3 surface expression (online suppl. fig. 2D). In contrast to complement-opsonized bacteria, only a few microbes, which had been incubated with PBS or heat-inactivated serum, were associated with macrophages 1 h after infection. Moreover, immunostaining of Pyk2 showed no colocalization of these macrophage-bound bacteria with the kinase (fig. 5c). However, when E. coli were opsonized with complement containing IgG-depleted serum, a strong recruitment of Pyk2 to the cell-associated bacteria could be observed (fig. 5c). Intensity profiles through attachment sites of complement-opsonized E. coli highlight the local enrichment of Pyk2 around bacteria-engaged CR3 whereas bacteria incubated with the heat-inactivated, IgG-depleted serum do not induce recruitment of Pyk2 (fig. 5d). These findings confirm the positive contribution of Pyk2 to the complement-mediated phagocytosis of bacteria and suggest that, in macrophages, efficient integrin-mediated uptake of bacteria requires the activity of this nonreceptor tyrosine kinase.
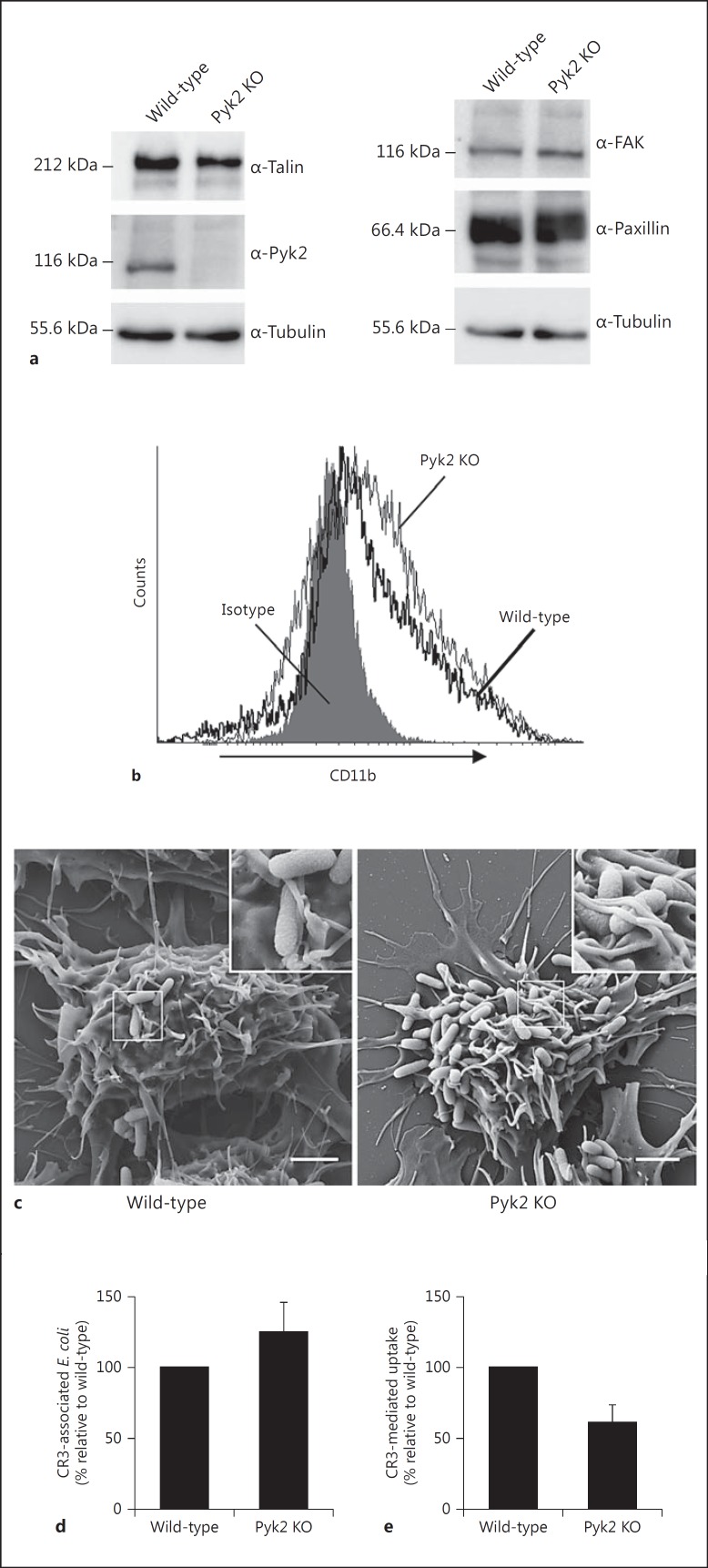
Knockout of Pyk2 Impairs CR3-Mediated Phagocytosis without Affecting Binding Affinity to Opsonized E. coli
To provide unambiguous confirmation of Pyk2's contribution to CR3-mediated phagocytosis, we established a Pyk2-deficient RAW 264.7 cell line (Pyk2 knockout, KO) using the CRISPR/Cas system. Lack of Pyk2 expression in the obtained clonal cell line was confirmed by Western blot analysis (fig. 6a). To rule out that genetic deletion of Pyk2 alters the expression of the Pyk2-associated proteins paxillin and talin, or the expression of the second family member FAK, we tested Pyk2 KO cells for the expression of these proteins (fig. 6a). Immunoblotting against paxillin, talin and FAK revealed no change in their expression in Pyk2 KO in comparison to wild-type cells. Furthermore, flow cytometry measurements confirmed that the expression of the CR3 receptor is not reduced upon genetic deletion of Pyk2 but is rather slightly increased (fig. 6b). We next employed the Pyk2 KO macrophages to analyze their ability to form membrane protrusions upon infection with opsonized E. coli. Accordingly, cells were infected for 30 min with E. coli, which had been preincubated with IgG-depleted serum and processed for SEM. No difference in the formation of membrane protrusions at sites of infection could be observed between Pyk2 KO and wild-type cells (fig. 6c). In line with the observation that the surface expression of CR3 is slightly increased following disruption of the pyk2 gene, flow cytometry-based measurements also showed a minor increase in total cell-associated, opsonized E. coli after 30 min of infection of Pyk2 KO when compared to wild-type cells (fig. 6d). There was, however, an approximate 40% decrease in intracellular bacteria in Pyk2 KO cells, again pointing to an important contribution of Pyk2 to CR3-mediated phagocytosis (fig. 6e). The combined results obtained with Pyk2 KO cells demonstrate that Pyk2 is not necessary for the initial binding of complement-opsonized bacteria or for the formation of membrane protrusions at the sites of infection. However, Pyk2 clearly contributes to the downstream signaling upon CR3 engagement to promote the phagocytic process.
Fig. 6.
CRISPR/Cas-mediated knockout of Pyk2 impairs CR3-mediated phagocytosis of E. coli. a Whole-cell lysates of wild-type and Pyk2 KO RAW 264.7 macrophages were used for Western blot analysis against Pyk2, talin, paxillin and FAK. Tubulin served as a loading control. b Surface localization of CR3 in wild-type (solid black line) or Pyk2 KO (gray line) macrophages was detected by staining with anti-CD11b Ab followed by flow cytometry. The signal derived from cells stained with an isotype-matched control Ab is indicated by the gray shaded area. c Serum-starved RAW 264.7 cells seeded on gelatine-coated coverslips were infected (MOI 100) for 30 min with FITC-labeled E. coli opsonized with IgG-depleted serum. After incubation, samples were fixed, dehydrated and analyzed via SEM. Images show overviews of whole cells, with insets highlighting details of phagocytic processes. Scale bars: 2 μm. d Serum-starved wild-type or Pyk2 KO macrophages were infected with FITC-labeled complement-opsonized E. coli for 30 min (MOI 100). Cells were resuspendend and analyzed via flow cytometry in the absence of trypan blue to measure total associated bacteria. e Samples (as in d) were analyzed in the presence of trypan blue to detect intracellular bacteria. Mean ± SEM of 3 independent experiments.
Discussion
In this work, we studied the contribution of Pyk2 to complement-mediated phagocytosis by murine macrophages. Our results show that Pyk2 is recruited to sites of complement-mediated internalization. Furthermore, we observed that the loss of proper Pyk2 function strongly interferes with complement receptor-mediated uptake of opsonized particles.
We employed a microscope-based assay that enabled discrimination between FcγR- and CR3-mediated phagocytosis. With this assay, we first analyzed the localization of Pyk2 during phagocytosis via immunofluorescence staining. Staining of the actin cytoskeleton revealed a ring-like actin organization at the sites of internalization for IgG and anti-CR3 beads, confirming that, in both processes, an active actin assembly occurs [21]. Such an active actin reorganization was absent in the macrophages in contact with control or albumin beads. Interestingly, despite the similarity in actin rearrangements induced by IgG and anti-CR3 beads, we could observe a selective recruitment of Pyk2 to anti-CR3 beads only. In addition, Pyk2 was selectively recruited to sites of internalization of complement-opsonized E. coli and was not recruited in cells incubated with nonopsonized bacteria. The association of Pyk2 with sites of CR3-mediated phagocytosis is in line with studies showing a colocalization of Pyk2 with integrin β2 in the podosomes of migrating macrophages [14]. Our novel findings extend the functional association of Pyk2 and integrin β2 to the process of phagocytosis.
The siRNA-mediated knockdown of Pyk2 resulted in a significant reduction of internalized anti-CR3 beads, with no influence on the phagocytosis of control, albumin or IgG beads. However, the pharmacological inhibition also affected the uptake of the control and albumin beads, albeit to a much lesser extent than the anti-CR3 beads. In addition, the internalization of IgG beads was not influenced at all by the pharmacological Pyk2 inhibitor. The effect on the uptake of control beads could be due to the blockage of additional kinases by PF431396 [25]. Indeed, the Pyk2-related kinase FAK is also inhibited by this compound [22]. Background levels of control bead uptake might be due to serum components, such as fibronectin or vitronectin, sticking to the microbeads. These plasma proteins are recognized and endocytosed by integrins α5β1 or αvβ3 via FAK-dependent pathways, explaining the reduced uptake of control beads in the presence of PF431396 [26, 27, 28].
For competitive inhibition of Pyk2 localization and function, we used PRNK. To enable transport of recombinant PRNK into the cytosol of macrophages, PRNK was fused to the TAT-peptide derived from HIV. The presence of TAT-PRNK resulted in a significant decrease in CR3-mediated phagocytosis, which is consistent with the results obtained after treatment with the Pyk2 siRNA or with the Pyk2 kinase inhibitor. TAT-PRNK only had a negative effect on the internalization of anti-CR3 beads, without affecting the uptake of IgG-coupled beads or the background uptake observed in the negative controls. Together, our combined results hint at an important and selective role of Pyk2 in complement-mediated phagocytosis in macrophages. In line with our results for the CR3-dependent uptake, previous reports indicate a role for both Pyk2 and FAK in the integrin β1-dependent uptake of Yersinia pseudotuberculosis. In this regard, Pyk2 is mainly important for the uptake of Yersinia via YadA, an extracellular matrix- and complement factor-binding protein [29, 30]. However, which of the known YadA ligands triggers uptake in this scenario has not been analyzed in detail.
In addition to the microbead assay, we also analyzed complement-mediated phagocytosis in a more physiological context. We analyzed the phagocytosis of complement-opsonized E. coli in the presence or absence of the Pyk2 inhibitor PF431396. This analysis revealed a significant decrease in the uptake of complement-opsonized bacteria when Pyk2 is inhibited. Importantly, the surface expression of CR3 was not influenced upon treatment with PF431396. Again, these findings strongly argue for an involvement of Pyk2 in the CR3-mediated phagocytosis of microorganisms.
SEM analysis of macrophages revealed no difference in the formation of membrane protrusions around macrophage-associated Fc- or anti-CR3 beads. This observation confirms recent data that CR3-mediated phagocytosis also involves membrane ruffling [31, 32]. Strikingly, the inhibition of Pyk2 had no effect on the formation of membrane protrusions during CR3-mediated phagocytosis. This might indicate that Pyk2 is not involved in the actin reorganization occurring in CR3-dependent phagocytosis. This suggestion is further supported by the analysis of Pyk2 KO macrophages with respect to their association with complement-opsonized E. coli. Flow cytometry and SEM analysis of infected Pyk2 KO versus wild-type cells showed no difference in their ability to bind the bacteria and form membrane protrusions at the sites of infection. Although the Pyk2 KO macrophages are able to bind complement-opsonized bacteria in a manner comparable to wild-type cells, they show defects in CR3-mediated phagocytosis. These effects are unlikely to be a result of altered expression of proteins that might be involved in the same signaling pathway because the expression pattern of FAK, talin, paxillin and CR3 in Pyk2 KO cells is unchanged. In summary, interfering with Pyk2 signaling via its inhibition, knockdown or genetic deletion consistently leads to a clear reduction in phagocytosis of anti-CR3 beads or complement-opsonized E. coli. These findings place Pyk2 downstream of the integrin-initiated signals necessary for efficient CR3-phagocytosis.
There are several possibilities as to how Pyk2 could be integrated in the CR3-initiated signaling cascade. Although the inhibition of Pyk2 does not impair the formation of membrane protrusions at sites of CR3-mediated uptake, Pyk2 could contribute in other ways to CR3 downstream signaling. For example, Pyk2 was shown to associate with phosphatidylinositol-3′ kinase (PI3K) and to get activated upon integrin β1 engagement and PI3K activation [33]. PI3K catalyzes the production of phosphatidylinositol(3,4,5)trisphosphate, which is necessary for the closure of phagosomes [34, 35]. As Pyk2-deficient macrophages and platelets are impaired in the activation of PI3K [16, 36], this could explain the observed reduction of CR3-mediated uptake. Besides several common downstream signaling molecules, such as the small GTPase RhoG [37] and the protein tyrosine kinase Syk [38, 39, 40], FcγR- and CR3-mediated phagocytosis triggers distinct molecular mechanisms of uptake. For example, FcγR-mediated phagocytosis depends on the small GTPases Rac2 and Cdc42 whereas phagocytosis via CR3 involves RhoA [41]. Our study further emphasizes that these two opsonin-dependent routes of phagocytosis rely on distinct signaling modules to mediate the internalization of particles. It could be speculated that these different uptake routes also dictate the distinct fates of the internalized microbes, an area of research that warrants further exploration.
Taken together, we could show that Pyk2 activity and its localization at sites of internalization are crucial for efficient CR3-mediated phagocytosis. These results highlight an additional function of Pyk2 in phagocytic cells. It remains to be seen how Pyk2 is linked to the CR3 upon receptor engagement. Given the known interaction of Pyk2 with several integrin-associated proteins such as paxillin and Hic-5, Pyk2 might be closely connected to the integrin β2 subunit. Further analysis of the spatial and temporal regulation of this kinase in CR3-mediated signaling will clarify the contribution of Pyk2 to this important process of innate immunity.
Disclosure Statement
The authors declare no conflicts of interest.
Supplementary Material
Supplementary data
Supplementary data
Acknowledgement
We thank C. Hentschel, S. Feindler-Boeckh and P. Zoll-Kiewitz for expert technical assistance as well as Ann-Kathrin Fuchs, Julia Nagel and Benjamin Frommeyer for analyzing samples. We are also grateful for the expert SEM support supplied by J. Hentschel and L. Kling at the Electron Microscopy Center of the University of Konstanz. We acknowledge support with flow cytometry analysis by the FlowCon facility of the University of Konstanz. A.B. was the recipient of a Fellowship according to the LGFG, Baden-Württemberg and the Zukunftskolleg at the University of Konstanz. This study was supported by funds from the Ministry of Science, Research and the Arts of Baden-Württemberg (CAP17) to C.R.H.
References
- 1.Desjardins M, Houde M, Gagnon E. Phagocytosis: the convoluted way from nutrition to adaptive immunity. Immunol Rev. 2005;207:158–165. doi: 10.1111/j.0105-2896.2005.00319.x. [DOI] [PubMed] [Google Scholar]
- 2.Flannagan RS, Jaumouille V, Grinstein S. The cell biology of phagocytosis. Annu Rev Pathol. 2012;7:49–86. doi: 10.1146/annurev-pathol-011811-132445. [DOI] [PubMed] [Google Scholar]
- 3.Herre J, Marshall AS, Caron E, Edwards AD, Williams DL, Schweighoffer E, Tybulewicz V, Reis e Sousa C, Gordon S, Brown GD. Dectin-1 uses novel mechanisms for yeast phagocytosis in macrophages. Blood. 2004;104:4038–4045. doi: 10.1182/blood-2004-03-1140. [DOI] [PubMed] [Google Scholar]
- 4.Goodridge HS, Underhill DM, Touret N. Mechanisms of Fc receptor and dectin-1 activation for phagocytosis. Traffic. 2012;13:1062–1071. doi: 10.1111/j.1600-0854.2012.01382.x. [DOI] [PubMed] [Google Scholar]
- 5.Buntru A, Roth A, Nyffenegger-Jann NJ, Hauck CR. Hemitam signaling by ceacam3, a human granulocyte receptor recognizing bacterial pathogens. Arch Biochem Biophys. 2012;524:77–83. doi: 10.1016/j.abb.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 6.Anderson CL, Shen L, Eicher DM, Wewers MD, Gill JK. Phagocytosis mediated by three distinct Fc gamma receptor classes on human leukocytes. J Exp Med. 1990;171:1333–1345. doi: 10.1084/jem.171.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groves E, Dart AE, Covarelli V, Caron E. Molecular mechanisms of phagocytic uptake in mammalian cells. Cell Mol Life Sci. 2008;65:1957–1976. doi: 10.1007/s00018-008-7578-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myones BL, Dalzell JG, Hogg N, Ross GD. Neutrophil and monocyte cell surface p150,95 has iC3b-receptor (CR4) activity resembling CR3. J Clin Invest. 1988;82:640–651. doi: 10.1172/JCI113643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanner SB, Aruffo A, Chan P-Y. Lymphocyte antigen receptor activation of a focal adhesion kinase-related tyrosine kinase substrate. Proc Natl Acad Sci USA. 1994;91:10484–10487. doi: 10.1073/pnas.91.22.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasaki H, Nagura K, Ishino M, Tobioka H, Kotani K, Sasaki T. Cloning and characterization of cell adhesion kinase beta, a novel protein-tyrosine kinase of the focal adhesion kinase subfamily. J Biol Chem. 1995;270:21206–21219. doi: 10.1074/jbc.270.36.21206. [DOI] [PubMed] [Google Scholar]
- 11.Avraham S, London R, Fu Y, Ota S, Hiregowdara D, et al. Identification and characterization of a novel related adhesion focal tyrosine kinase (RAFTK) from megakaryocytes and brain. J Biol Chem. 1995;270:27742–27751. doi: 10.1074/jbc.270.46.27742. [DOI] [PubMed] [Google Scholar]
- 12.Lev S, Moreno H, Martinez R, Canoll P, Peles E, Musacchio JM, Plowman GD, Rudy B, Schlessinger J. Protein tyrosine kinase Pyk2 involved in Ca(2+)-induced regulation of ion channel and map kinase functions. Nature. 1995;376:737–745. doi: 10.1038/376737a0. [DOI] [PubMed] [Google Scholar]
- 13.Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 14.Duong LT, Rodan GA. PYK2 is an adhesion kinase in macrophages, localized in podosomes and activated by beta(2)-integrin ligation. Cell Motil Cytoskeleton. 2000;47:174–188. doi: 10.1002/1097-0169(200011)47:3<174::AID-CM2>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 15.Guinamard R, Okigaki M, Schlessinger J, Ravetch JV. Absence of marginal zone B cells in Pyk-2-deficient mice defines their role in the humoral response. Nat Immunol. 2000;1:31–36. doi: 10.1038/76882. [DOI] [PubMed] [Google Scholar]
- 16.Okigaki M, Davis C, Falasca M, Harroch S, Felsenfeld DP, Sheetz MP, Schlessinger J. Pyk2 regulates multiple signaling events crucial for macrophage morphology and migration. Proc Natl Acad Sci USA. 2003;100:10740–10745. doi: 10.1073/pnas.1834348100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butler B, Blystone SD. Tyrosine phosphorylation of beta3 integrin provides a binding site for Pyk2. J Biol Chem. 2005;280:14556–14562. doi: 10.1074/jbc.M411765200. [DOI] [PubMed] [Google Scholar]
- 18.Gao C, Blystone SD. A Pyk2-Vav1 complex is recruited to beta3-adhesion sites to initiate Rho activation. Biochem J. 2009;420:49–56. doi: 10.1042/BJ20090037. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Learoyd J, Duan Y, Leff AR, Zhu X. Hematopoietic Pyk2 regulates migration of differentiated hl-60 cells. J Inflamm (Lond) 2010;7:26. doi: 10.1186/1476-9255-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamen LA, Schlessinger J, Lowell CA. Pyk2 is required for neutrophil degranulation and host defense responses to bacterial infection. J Immunol. 2011;186:1656–1665. doi: 10.4049/jimmunol.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.May RC, Machesky LM. Phagocytosis and the actin cytoskeleton. J Cell Sci. 2001;114:1061–1077. doi: 10.1242/jcs.114.6.1061. [DOI] [PubMed] [Google Scholar]
- 22.Buckbinder L, Crawford DT, Qi H, Ke HZ, Olson LM, Long KR, Bonnette PC, Baumann AP, Hambor JE, Grasser WA, Pan LC, Owen TA, Luzzio MJ, Hulford CA, Gebhard DF, Paralkar VM, Simmons HA, Kath JC, Roberts WG, Smock SL, Guzman-Perez A, Brown TA, Li M. Proline-rich tyrosine kinase 2 regulates osteoprogenitor cells and bone formation, and offers an anabolic treatment approach for osteoporosis. Proc Natl Acad Sci USA. 2007;104:10619–10624. doi: 10.1073/pnas.0701421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong WC, Macklem M, Parsons JT. Expression and characterization of splice variants of Pyk2, a focal adhesion kinase-related protein. J Cell Sci. 1998;111:1981–1991. doi: 10.1242/jcs.111.14.1981. [DOI] [PubMed] [Google Scholar]
- 24.Sun CK, Man K, Ng KT, Ho JW, Lim ZX, Cheng Q, Lo CM, Poon RT, Fan ST. Proline-rich tyrosine kinase 2 (Pyk2) promotes proliferation and invasiveness of hepatocellular carcinoma cells through c-SRC/ERK activation. Carcinogenesis. 2008;29:2096–2105. doi: 10.1093/carcin/bgn203. [DOI] [PubMed] [Google Scholar]
- 25.Han S, Mistry A, Chang JS, Cunningham D, Griffor M, Bonnette PC, Wang H, Chrunyk BA, Aspnes GE, Walker DP, Brosius AD, Buckbinder L. Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design. J Biol Chem. 2009;284:13193–13201. doi: 10.1074/jbc.M809038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bretscher MS. Endocytosis and recycling of the fibronectin receptor in CHO cells. EMBO J. 1989;8:1341–1348. doi: 10.1002/j.1460-2075.1989.tb03514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caswell P, Norman J. Endocytic transport of integrins during cell migration and invasion. Trends Cell Biol. 2008;18:257–263. doi: 10.1016/j.tcb.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann C, Berking A, Agerer F, Buntru A, Neske F, Chhatwal GS, Ohlsen K, Hauck CR. Caveolin limits membrane microdomain mobility and integrin-mediated uptake of fibronectin-binding pathogens. J Cell Sci. 2010;123:4280–4291. doi: 10.1242/jcs.064006. [DOI] [PubMed] [Google Scholar]
- 29.Bruce-Staskal PJ, Weidow CL, Gibson JJ, Bouton AH. Cas, Fak and Pyk2 function in diverse signaling cascades to promote Yersinia uptake. J Cell Sci. 2002;115:2689–2700. doi: 10.1242/jcs.115.13.2689. [DOI] [PubMed] [Google Scholar]
- 30.Owen KA, Thomas KS, Bouton AH. The differential expression of Yersiniapseudotuberculosis adhesins determines the requirement for Fak and/or Pyk2 during bacterial phagocytosis by macrophages. Cell Microbiol. 2007;9:596–609. doi: 10.1111/j.1462-5822.2006.00811.x. [DOI] [PubMed] [Google Scholar]
- 31.Hall AB, Gakidis MA, Glogauer M, Wilsbacher JL, Gao S, Swat W, Brugge JS. Requirements for Vav guanine nucleotide exchange factors and Rho GTpases in FcgammaR- and complement-mediated phagocytosis. Immunity. 2006;24:305–316. doi: 10.1016/j.immuni.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Patel PC, Harrison RE. Membrane ruffles capture C3bi-opsonized particles in activated macrophages. Mol Biol Cell. 2008;19:4628–4639. doi: 10.1091/mbc.E08-02-0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melikova S, Dylla SJ, Verfaillie CM. Phosphatidylinositol-3-kinase activation mediates proline-rich tyrosine kinase 2 phosphorylation and recruitment to beta1-integrins in human CD34+ cells. Exp Hematol. 2004;32:1051–1056. doi: 10.1016/j.exphem.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 34.Araki N, Johnson MT, Swanson JA. A role for phosphoinositide 3-kinase in the completion of macropinocytosis and phagocytosis by macrophages. J Cell Biol. 1996;135:1249–1260. doi: 10.1083/jcb.135.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura N, Hazeki K, Okazaki N, Kametani Y, Murakami H, Takaba Y, Ishikawa Y, Nigorikawa K, Hazeki O. Specific role of phosphoinositide 3-kinase p110alpha in the regulation of phagocytosis and pinocytosis in macrophages. Biochem J. 2009;423:99–108. doi: 10.1042/BJ20090687. [DOI] [PubMed] [Google Scholar]
- 36.Consonni A, Cipolla L, Guidetti G, Canobbio I, Ciraolo E, Hirsch E, Falasca M, Okigaki M, Balduini C, Torti M. Role and regulation of phosphatidylinositol 3-kinase beta in platelet integrin alpha2beta1 signaling. Blood. 2012;119:847–856. doi: 10.1182/blood-2011-07-364992. [DOI] [PubMed] [Google Scholar]
- 37.Tzircotis G, Braga VM, Caron E. RhoG is required for both FcgammaR- and CR3-mediated phagocytosis. J Cell Sci. 2011;124:2897–2902. doi: 10.1242/jcs.084269. [DOI] [PubMed] [Google Scholar]
- 38.Crowley MT, Costello PS, Fitzer-Attas CJ, Turner M, Meng F, Lowell C, Tybulewicz VL, DeFranco AL. A critical role for Syk in signal transduction and phagocytosis mediated by Fcgamma receptors on macrophages. J Exp Med. 1997;186:1027–1039. doi: 10.1084/jem.186.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiefer F, Brumell J, Al-Alawi N, Latour S, Cheng A, Veillette A, Grinstein S, Pawson T. The Syk protein tyrosine kinase is essential for Fcgamma receptor signaling in macrophages and neutrophils. Mol Cell Biol. 1998;18:4209–4220. doi: 10.1128/mcb.18.7.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Y, Tohyama Y, Kadono T, He J, Miah SM, Hazama R, Tanaka C, Tohyama K, Yamamura H. Protein-tyrosine kinase Syk is required for pathogen engulfment in complement-mediated phagocytosis. Blood. 2006;107:4554–4562. doi: 10.1182/blood-2005-09-3616. [DOI] [PubMed] [Google Scholar]
- 41.Caron E, Hall A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTpases. Science. 1998;282:1717–1721. doi: 10.1126/science.282.5394.1717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data