Abstract
Lipoteichoic acid (LTA) is a component of the cell wall of Gram-positive bacteria and induces a toll-like receptor 2 (TLR2)-mediated inflammatory response upon initial binding to lipopolysaccharide-binding protein (LBP) and subsequent transfer to CD14. In this study, we identified a novel role for the nuclear protein high-mobility group box 1 (HMGB1) in LTA-mediated inflammation. Results of ELISA, surface plasmon resonance and native PAGE electrophoretic mobility shift analyses indicated that HMGB1 binds to LTA in a concentration-dependent manner and that this binding is inhibited by LBP. Native PAGE, fluorescence-based transfer and confocal imaging analyses indicated that HMGB1 catalytically disaggregates LTA and transfers LTA to CD14. NF-κB p65 nuclear transmigration, degradation of IκBa and reporter assay results demonstrated that NF-κB activity in HEK293-hTLR2/6 cells is significantly upregulated by a mixture of LTA and soluble CD14 in the presence of HMGB1. Furthermore, the production of TNF-a and IL-6 in J774A.1 and RAW264.7 cells increased significantly following treatment with a mixture of LTA and HMGB1 compared with treatment with LTA or HMGB1 alone. Thus, we propose that HMGB1 plays an important role in LTA-mediated inflammation by binding to and transferring LTA to CD14, which is subsequently transferred to TLR2 to induce an inflammatory response.
Key Words: HMGB1, Gram-positive bacteria, Lipoteichoic acid, TLR2, Inflammation
Introduction
Approximately 50% of bacterial sepsis or septic shock cases are caused by Gram-positive bacteria [1]. Lipoteichoic acid (LTA), a component of the Gram-positive bacterial cell wall, is a membrane-associated amphiphile adhesin. LTA is considered a major virulence factor and has immunostimulatory activity. It is composed of repeating polymer and glycolipid units and shares many of its pathophysiological properties with lipopolysaccharide (LPS), which is composed of lipid A and polysaccharides [2, 3]. In Gram-negative bacterial infections, LPS-binding protein (LBP) binds to LPS and catalyzes the transfer of LPS monomers from LPS aggregates to CD14 [4], resulting in increased NF-κB signaling through the activation of the TLR4-MD2 receptor [5]. LTA in the circulation forms a complex with LBP and then binds to CD14, activating signaling via TLR2 in a mechanism similar to the LPS-mediated activation of TLR4 signaling [6]. LTA-mediated TLR2 activation triggers the MyD88-dependent signaling pathway via MyD88, IL-1 receptor-associated kinase and tumor necrosis factor receptor-associated factor 6, leading to NF-κB activation [7, 8]. However, the presence of LTA alone is insufficient to cause the type of severe inflammation associated with septic shock and multiple organ failure [9, 10]. LBP plays an important role in protecting the host by sensing and enhancing the LPS- and LTA-mediated inflammation associated with Gram-negative and Gram-positive bacterial infections.
The nuclear protein high-mobility group box 1 (HMGB1) is a well-known danger-associated molecular pattern molecule that stabilizes the nucleosome and regulates gene transcription [11]. The release of HMGB1 into the extracellular space as a result of modification [12, 13, 14, 15] or cell necrosis [16] can trigger inflammation and sepsis [17, 18]. In addition, HMGB1 can form complexes with CpG DNA [19], IL-1β [20] and CXCL12 [21], thus exacerbating inflammation [22]. Previously, we reported that HMGB1 binds to and facilitates the transfer of LPS to CD14, ultimately upregulating LPS-mediated cytokine production [23, 24]. Our results identified HMGB1 as a novel LPS-sensing and LPS-transferring molecule. Considering that LBP plays an important role in binding to and transferring LTA to CD14 to initiate TLR2-mediated proinflammatory responses [6], we hypothesized that HMGB1 also binds to LTA and facilitates LTA-stimulated TLR2 signaling.
In this study, we demonstrate that HMGB1 interacts with LTA, the primary component of the Gram-positive bacterial cell wall, and transfers LTA to CD14, ultimately resulting in the upregulation of TNF-α and IL-6 production via LTA-stimulated TLR2 signaling in macrophages. Our results suggest that HMGB1 plays an important role in TLR-mediated signaling associated with Gram-positive bacterial infections.
Materials and Methods
LTA and Proteins
LTAs from Staphylococcus aureus (sLTA) and Bacillus subtilis (bLTA; endotoxin levels, 0.01 EU/μg) were purchased from InvivoGen (San Diego, Calif., USA). Recombinant human LBP (R&D Systems, Minneapolis, Minn., USA) and soluble CD14 protein (sCD14; amino acids, aa 1–352) were also purchased from R&D Systems. LPS extracted from Escherichia coli was purchased from Sigma-Aldrich (St. Louis, Mo., USA). Wild-type recombinant human HMGB1, acidic tail-deleted HMGB1 (ΔC-HMGB1, aa 1–185), HMGB1 A box (aa 1–79) and HMGB1 B box (aa 88–162) incorporating six His-tags were expressed in E. coli BL21 (DE3) pLysE and purified using Ni2+-NTA, Sephadex G75 and ion-exchange columns [13, 23]. Wild-type HMGB1 produced in baculovirus-infected Spodoptera frugiperda 9 insect cells was used to confirm the results of the study. Endotoxin was removed by phase separation using Triton X-114 [25]. The absence of endotoxin in HMGB1 and ΔC-HMGB1 preparations was confirmed using the Limulus amebocyte lysate assay (Lonza, Basel, Switzerland), with the level being under 1.0 EU/μg protein.
Analysis of HMGB1 Binding to LTA Using ELISA and Competition ELISA
To observe the binding of HMGB1 to LTA, 96-well PolySorp microtiter plates (Nunc, Rochester, N.Y., USA) were coated with 1 μg/ml of LBP or HMGB1. The plates were washed with PBS (0.137 M NaCl, 0.005 M KCl, 0.009 M Na2HPO4, and 0.001 M KH2PO4, pH 7.4) containing 0.01% Tween 20 (v/v, PBST) and blocked with 2% BSA in PBST. Various concentrations (101-5 ng/ml) of LTA were added and incubated for 1 h. After washing, mouse anti-LTA antibody (Ab; Abcam, Cambridge, Mass., USA) was added and incubated. HRP-conjugated anti-mouse IgGAM (Sigma-Aldrich) was used as a secondary Ab. TMB solution was added for color development and the optical density of each well was measured at 450 nm. For the reverse assay, microtiter plate wells were coated with 1 μg/ml of LTA and incubated with various concentrations of HMGB1. The ELISA was performed using anti-HMGB1 Ab (Abcam).
A competition ELISA was performed to confirm whether LBP inhibits the binding of HMGB1 to LTA. Microtiter plate wells were coated with 0.1 μg/ml of LTA and then blocked with 2% BSA in PBST. HMGB1 (0.1 μg/ml) was added to the wells in the presence of various concentrations of LBP. After washing with PBST, rabbit anti-HMGB1 Ab (Abcam) was added and incubated. HRP-conjugated anti-rabbit IgGAM (Sigma-Aldrich) was used as the secondary Ab.
Surface Plasmon Resonance Assay
The binding of LTA to HMGB1 was analyzed in real time by means of surface plasmon resonance using a BIAcore 2000 instrument (BIAcore AB, Uppsala, Sweden). For surface preparation, wild-type HMGB1, ΔC-HMGB1 or HMGB1 A and B boxes at a concentration of 100 µg/ml were immobilized as the ligand on a carboxymethyl dextran (CM5) sensor chip in 10 mM sodium acetate buffer (pH 4). The surface of the CM5 dextran sensor chip was activated with 0.2 M N-ethyl-N'-(3-diethylamino-propyl)-carbodiimide and 0.05 M N-hydroxysuccinimide. HBS running buffer (10 mM HEPES, 150 mM NaCl, 3.4 mM EDTA, and 0.005% Tween 20, pH 7.4) was used for sample dilution and analysis. To evaluate binding, sLTA was diluted in HBS buffer at various concentrations (0.25-4 µg/ml), and samples were passed over the sensor chip at a flow rate of 10 µl/min. Response unit values for samples were calculated by subtracting the control values using BIAevaluation 3.0 software (BIAcore).
Native PAGE Gel Mobility Shift Assay and Western Blotting
Native PAGE was used to demonstrate the electrophoretic mobility shift resulting from the HMGB1-mediated transfer of LTA to CD14. A mixture of sCD14 and LTA was incubated with different amounts of HMGB1 or LBP for 12 h at 37°C and then electrophoresed on a 4–15% polyacrylamide gradient gel (Bio-Rad, Hercules, Calif., USA) containing no reducing agent. The gels were run at 20 mA under cold conditions with native electrophoresis buffer (24 mM Tris base, 192 mM glycine, pH 8.3). After electrophoresis, the gels were silver-stained using a SilverQuest™ staining kit (Invitrogen). For Western blotting, electrophoresed proteins were transferred onto polyvinylidene fluoride membranes (GE Bioscience, Uppsala, Sweden), which were incubated in blocking solution (5% nonfat milk in Tris-buffered saline containing 0.05% Tween 20) for 1 h at room temperature. A rabbit anti-CD14 Ab (Abcam) was used to label CD14, and HMGB1 was labeled with a rabbit anti-HMGB1 Ab (Abcam). The membranes were then incubated with an HRP-labeled secondary Ab for 1 h at room temperature for subsequent development and visualization.
Cell Culture and NF-κB Reporter Assay
Human embryonic kidney (HEK) 293 cells stably expressing TLR2 and TLR6 (HEK293-hTLR2/6) were purchased from InvivoGen. The cells were maintained in DMEM supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10 µg/ml blasticidin (InvivoGen). HEK293-hTLR2/6 cells were plated at 2 × 105 per well in 24-well culture plates 24 h before transfection. The seeded cells were transiently transfected with 250 ng each of NF-κB-luc and β-galactosidase plasmids using PolyExpress (Excellgen, Rockville, Md., USA). At 24 h after transfection, the cells were washed with serum-free medium to remove the FBS and stimulated with a preincubated mixture including sCD14, HMGB1 or LBP, and LTA for 5 h. Luciferase activity was measured using a luciferase reporter assay system (Promega, Madison, Wisc., USA) according to the manufacturer's instructions. Luminescence was quantified using a Multilabel Reader (Perkin Elmer, Wellesley, Mass., USA). Results are shown as relative luciferase activity, defined as the ratio of activity stimulated by test proteins to the activity of β-galactosidase. Transfection of the β-galactosidase gene was performed to calculate transfection efficiency.
IκBα Degradation and Western Blotting
Activation of NF-κB was assessed by measuring IκB degradation. J774A.1 cells were seeded at 5 × 105 per well in a 6-well plate and incubated for 24 h in 10% FBS RPMI1640 medium, after which the cells were washed with serum-free medium to remove the FBS. The cells were stimulated with sLTA (60 ng/ml), HMGB1 (1 μg/ml), LBP (1 μg/ml), or a mixture of HMGB1 (1 or 5 μg/ml) and sLTA and maintained in medium for 30 min. A mixture of LBP (1 μg/ml) and sLTA was used as a positive control. The cells were lysed with RIPA buffer supplemented with protease and phosphatase inhibitor cocktails. Total cell extracts were prepared, and 15 μg of each extract was electrophoresed on an SDS-PAGE gel and immunoblotted with rabbit anti-IκBα or rabbit anti-actin Ab (Cell Signaling). An HRP-labeled secondary Ab was applied, and the blot was developed using the ECL system.
Measurement of BODIPY® FL-LTA Fluorescence
sLTA was labeled using a BODIPY FL (boron-dipyrromethene fluorescein leakage) labeling kit (Molecular Probes) according to the manufacturer's procedure. Briefly, sLTA was suspended in 0.5 M sodium bicarbonate buffer (pH 9.0) containing labeling reagent and incubated for 30 min with frequent sonication. BODIPY FL-conjugated LTA was quickly separated from free BODIPY by gel filtration on a PD10 column (GE Healthcare). Attachment of BODIPY FL to sLTA was evidenced by an increase in the fluorescence of BODIPY FL-LTA following solubilization by the addition of 2% SDS. BODIPY FL-sLTA (1 μg/ml) and sCD14 (5 μg/ml) were added to PBS lacking Ca2+ and Mg2+ in the presence or absence of 1 or 5 μg/ml of HMGB1. LBP at 2 μg/ml was used as a positive control. Fluorescence was measured using a Varioskan™ Flash Multimode Reader (Thermo Scientific) after incubation for 10 h at 25°C. The fluorescence of BODIPY FL-LTA was recorded at a wavelength of 525 nm with excitation at 488 nm.
Cytokine Production
Cytokine production was measured in J774A.1 and RAW264.7 murine macrophage cells. Cells were cultured in 24-well plates in OPTI-MEM medium (Invitrogen) at a density of 3 × 105 cells/ml for J774A.1 cells and 5 × 105 cells/ml for RAW264.7 cells. The J774A.1 and RAW264.7 cells were treated with sLTA and bLTA in the presence or absence of HMGB1 or LBP for 20 h at 37°C, after which the culture supernatants were collected by centrifugation at 5,000 rpm at 4°C. HMGB1 was degraded by incubation in the presence of 50 ng/ml of proteinase K for 2 h at 37°C, after which the proteinase K was denatured by boiling the sample for 30 min. The concentrations of TNF-α and IL-6 were determined using a sandwich ELISA (R&D Systems) according to the manufacturer's procedure.
Confocal Microscopy
The HMGB1-mediated transfer of BODIPY FL-LTA to TLR2 was assessed using immunofluorescent staining. HEK293-hTLR2/6 cells were cultured in LabTek II chambers (Nalgene) and washed with serum-free medium to remove the FBS. The cells were incubated with a mixture of LTA (1 μg/ml) and sCD14 (0.5 μg/ml) in the presence or absence of HMGB1 (1 μg/ml) or LBP (1 μg/ml) in serum-free medium for 4 h. After treatment, the cells were fixed in 4% paraformaldehyde in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, and 4 mM MgSO4, pH 7.0) for 20 min at room temperature. The cells were washed with PBS, blocked with 1% BSA in PBS for 10 min and incubated with mouse anti-TLR2 Ab (eBioscience) overnight at 4°C. After three washes, Alexa594-conjugated goat anti-mouse Ig (Invitrogen) was added. The localization of BODIPY FL-LTA and TLR2 was visualized using a FluoView FV1000 confocal microscope (Olympus, Tokyo, Japan).
To investigate whether HMGB1-mediated LTA transfer enhances the nuclear translocation of NF-κB p65, J774A.1 cells were seeded in LabTek II chambers, and the medium was exchanged with serum-free medium after 24 h. The cells were treated with a mixture of sLTA (60 ng/ml) and HMGB1 (1 μg/ml). A mixture of sLTA and LBP (1 μg/ml) was used as a positive control. The cells were washed with PBS and fixed with 4% paraformaldehyde for 30 min at room temperature. The fixed cells were permeabilized by incubation in 0.5% Triton X-100 in PBS for 20 min at room temperature. The cells were then washed three times with PBS, blocked with 1% BSA in PBS for 10 min and incubated with mouse anti-p65 Ab (Santa Cruz Biotech) overnight at 4°C. After washing with PBS, the cells were incubated with Alexa594-conjugated goat anti-mouse Ig (Invitrogen). The cells were mounted using VECTASHIELD with DAPI (Vector Laboratories) and examined using confocal microscopy.
Statistics
A one-way ANOVA test was used for the analysis with Bonferroni correction.
Results
Binding of HMGB1 to LTA
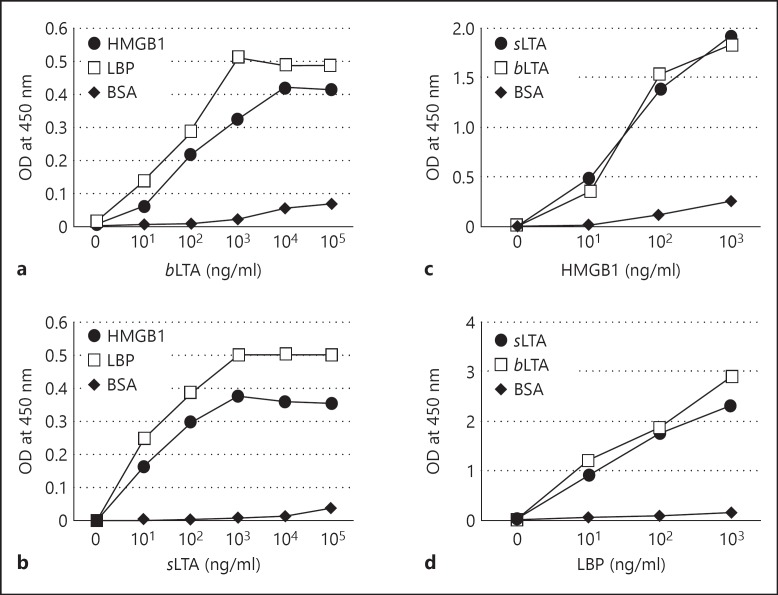
Previous studies have shown that HMGB1 binds to the Gram-negative bacteria endotoxin LPS, enhancing LPS-mediated cytokine production through the HMGB1-mediated transfer of LPS to CD14 [23, 24]. In this study, we found that HMGB1 binds to LTA and mediates the transfer of LTA to CD14-TLR2, enhancing the release of proinflammatory cytokines. To characterize this phenomenon, microtiter EIA plates were coated with 1 μg/ml of HMGB1, after which sLTA and bLTA were added to the wells and the binding was examined by ELISA. Both different LTAs isolated from S. aureus and B. subtilis were used because of the heterogeneity of LTA. Both LTAs bound to HMGB1 in a concentration-dependent manner (fig. 1a, b). LBP, which is known to bind LTA [6], was used as a positive control. In the reverse experiment, microtiter plates were coated with 1 μg/ml of sLTA or bLTA; the added HMGB1 and LBP bound to both LTAs in a concentration-dependent manner (fig. 1c, d).
Fig. 1.
Binding of HMGB1 to LTA. OD = Optical density. a, b HMGB1 (1 μg/ml) was immobilized in wells of microtiter plates; various concentrations of bLTA (a) or sLTA (b) were added and binding was evaluated by ELISA. LBP was used as a positive control. c, d Either bLTA or sLTA (1 μg/ml) was immobilized in wells of a microtiter plate; various concentrations of HMGB1 (c) or LBP (d) were added and binding was evaluated by ELISA. Data represent 1 of 2 similar independent experiments.
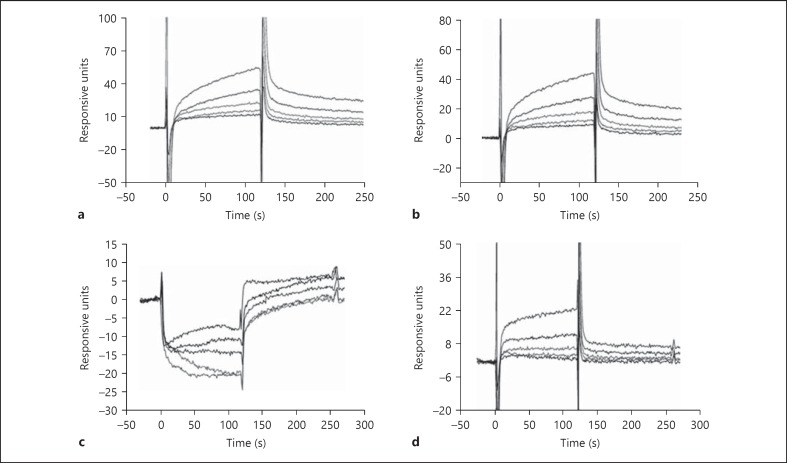
The binding of LTA to HMGB1 was analyzed in real time using surface plasmon resonance. Wild-type HMGB1, ΔC-HMGB1 and HMGB1 A and B boxes were immobilized onto CM5 dextran sensor chips, and then different concentrations of sLTA as a represensative were flowed over the chips. sLTA bound to both HMGB1 and ΔC-HMGB1 in a concentration-dependent manner (fig. 2a, b). LTA bound to ΔC-HMGB1 in a similar manner as wild-type HMGB1. When washing buffer was applied, the bound sLTA slowly dissociated. The result of surface plasmon resonance studies showed that LTA binds to the HMGB1 B box in a dose-dependent manner, but there was no correlative binding to the A box protein (fig. 2c, d).
Fig. 2.
Surface plasmon resonance analysis of HMGB1 binding to LTA. Wild-type HMGB1 (a), ΔC-HMGB1 (b), HMGB1 A box (c), and HMGB1 B box (d) proteins were immobilized on a CM5 dextran sensor chip and a solution of sLTA at concentrations of 0.25, 0.5, 1, 2, or 4 µg/ml was flowed over the chip at time 0 and dissociated sLTA with washing buffer at the time of 125 s. The binding of LTA to wild-type HMGB1 or mutant HMGB1 proteins was increased in a dose-dependent manner in a, b, d. An activated and blocked flow cell without immobilized ligand was used to evaluate nonspecific binding.
Formation of HMGB1-LTA Complexes as Determined by Gel Mobility Shift Assay
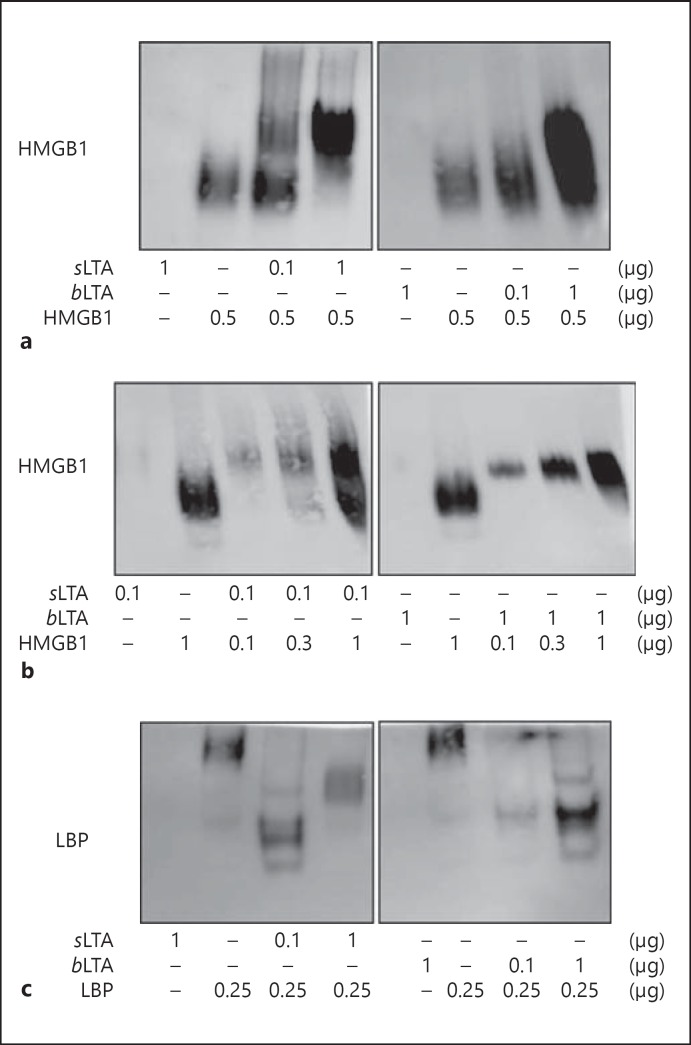
To confirm the formation of HMGB1-LTA complexes, HMGB1 was incubated with sLTA or bLTA overnight at 37°C, and a shift in the electrophoretic mobility of HMGB1 was observed by means of native PAGE followed by immunoblotting analysis. Both sLTA and bLTA bound to HMGB1, and the HMGB1-LTA complexes migrated more slowly than HMGB1 alone; band intensity also increased with increasing LTA concentration (fig. 3a). When the concentration of LTA was fixed, the formation of HMGB1-LTA complexes increased in an HMGB1 concentration-dependent manner (fig. 3b). When LBP was used as a positive control, the migration position of LBP-LTA was shifted relative to LBP alone (fig. 3c).
Fig. 3.
Native PAGE mobility shift assay of LTA and HMGB1 complexes. a, bsLTA and bLTA were incubated with HMGB1 for 12 h at 37°C and subjected to native PAGE on a 4–15% gradient gel. A fixed concentration of HMGB1 was incubated with the indicated amount of LTA (a) and a fixed concentration of LTA was incubated with the indicated amount of HMGB1 to assess complex formation (b). Membranes were immunoblotted with an anti-HMGB1 Ab. c The formation of a complex between LTA and LBP was analyzed as a positive control, and the membrane was immunoblotted using an anti-LBP Ab.
HMGB1 Competes with LBP for Binding to LTA
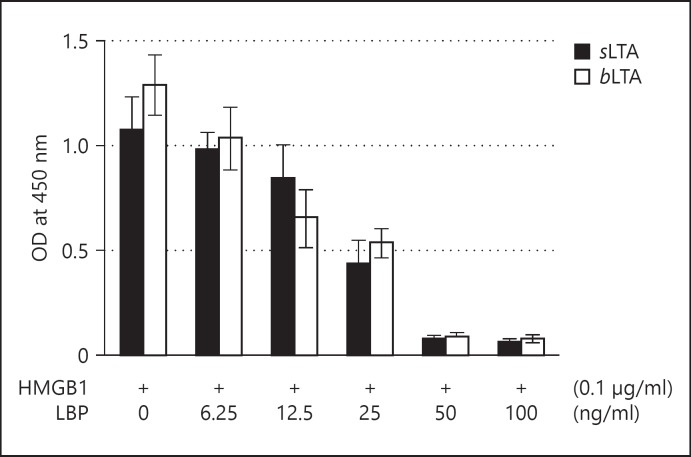
LBP rapidly binds to LTA during invasion by Gram-positive bacteria and catalyzes the transfer of LTA [6, 7]. An inhibition ELISA was used to investigate whether HMGB1 and LBP compete for binding to LTA. Microtiter plate wells were coated with 0.1 μg/ml sLTA or bLTA, and HMGB1 at a concentration of 0.1 μg/ml was added in the presence of various concentrations of LBP. The binding of HMGB1 to both LTAs was inhibited by LBP in a concentration-dependent manner, suggesting that both HMGB1 and LBP bind to a similar region of LTA (fig. 4).
Fig. 4.
Competition between HMGB1 and LBP for binding to LTA. OD = Optical density. Either bLTA or sLTA (0.1 μg/ml) was immobilized in wells of a microtiter plate, and HMGB1 (0.1 µg/ml) was added to the wells in the presence of various concentrations of LBP. Binding of HMGB1 to LTA was evaluated by ELISA using an anti-HMGB1 Ab and measurement of absorbance.
HMGB1 Facilitates Formation of LTA-CD14 Complexes
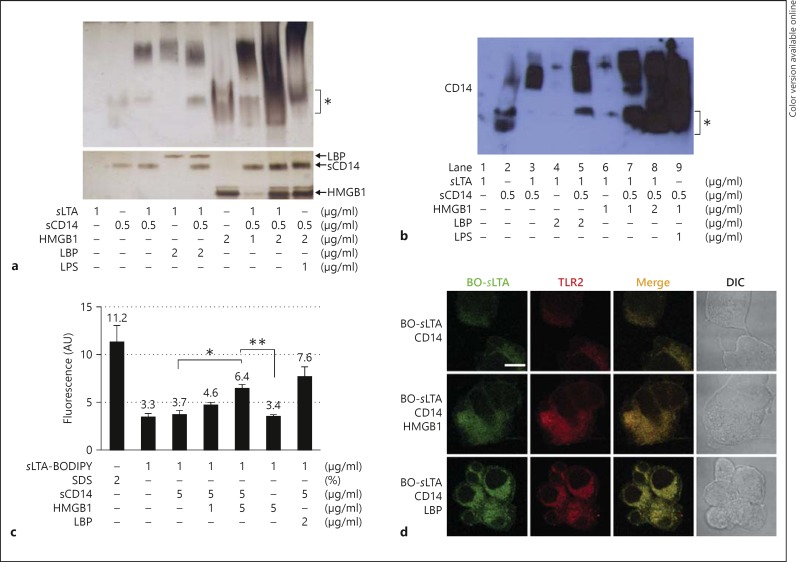
Upon entering the bloodstream, LPS is rapidly bound by the serum protein LBP, and the resulting LPS-LBP complexes readily bind to CD14 to induce proinflammatory responses via TLR4-mediated NF-κB activation and TNF-α production [26]. Because LTA also binds to LBP and is transferred to CD14 to induce TLR2-mediated inflammatory responses [6], we investigated whether HMGB1 plays a role in the transfer of LTA to CD14 to initiate proinflammatory cytokine production. In the native PAGE electrophoretic mobility shift assay, HMGB1 formed complexes with both LTA and CD14, resulting in a shift in mobility to a position similar to the LTA-LBP-CD14 complex positive control (fig. 5a). The lower panel in figure 5a depicts the SDS-PAGE loading control. A mixture of sLTA, HMGB1 and sCD14 preincubated prior to electrophoresis displayed a retarded shift compared with uncomplexed sCD14 or HMGB1 alone and showed a similar pattern to the positive control mixture consisting of LTA, LBP and sCD14. We confirmed the migration of the LTA-sCD14 complex formed by the transfer of LTA by HMGB1 using Western blot analysis with an anti-CD14 Ab after native PAGE (fig. 5b). The LTA-sCD14 complex migrated similarly to the complex formed by the transfer of LTA by LBP.
Fig. 5.
HMGB1 interacts with LTA and mediates the transfer of LTA to sCD14. a Native PAGE mobility shift assay of the transfer of LTA to sCD14 following HMGB1-LTA binding. A mixture of sLTA and sCD14 was incubated with HMGB1 for 12 h at 37°C and then subjected to native PAGE analysis on a 4–15% gradient gel, with subsequent silver staining (upper panel). LBP was used as a positive control. Lower panel represents SDS-PAGE analysis as a protein loading control. The position of uncomplexed sCD14 and HMGB1 is indicated by the asterisk. b Western blot analysis using an anti-CD14 Ab after native PAGE. Asterisk indicates uncomplexed sCD14. c HMGB1-mediated transfer of BODIPY FL-LTA to CD14. A mixture of BODIPY FL-sLTA and sCD14 was incubated in the presence or absence of HMGB1 for 10 h at 25°C and the fluorescence level was measured. LBP was used as a positive control, and 2% SDS was used to solubilize the disaggregated BODIPY FL-sLTA completely to maximize the fluorescence. * p < 0.05, ** p < 0.01 (n = 4). d Transfer of BODIPY FL-sLTA to TLR2. BO = BODIPY FL. HEK293-hTLR2/6 cells were stimulated with a mixture of 1 μg/ml of BODIPY FL-sLTA and 0.5 μg/ml of sCD14 in the presence or absence of 1 μg/ml of HMGB1 for 4 h. LBP (1 μg/ml) was used as a positive control. The cells were then fixed and stained with an anti-TLR2 Ab. Scale bar = 10 μm.
To confirm the transfer of LTA by HMGB1, sLTA was labeled with BODIPY FL and then incubated with sCD14 in the presence or absence of HMGB1, after which dissociation of BODIPY FL was monitored. The fluorescence of sLTA-BODIPY FL was quenched in the resting state. The fluorescence of BODIPY FL increased significantly - from 3.7 to 6.4 absorbance units - when HMGB1 was added to a mixture of sLTA-BODIPY FL and sCD14 (fig. 5c).
We next investigated the transfer of LTA to TLR2 by HMGB1. HEK293-hTLR2/6 cells were incubated with a mixture of sLTA-BODIPY FL and sCD14 for 4 h in the presence or absence of HMGB1. sLTA-BODIPY FL was observed on the cell surface and in the cytoplasm of HEK293-hTLR2/6 cells in the presence of HMGB1, similar to cells treated with LBP, whereas the fluorescence level was low in the absence of HMGB1 (fig. 5d). These results indicate that HMGB1 catalyzes the transfer of LTA to CD14 and TLR2.
HMGB1 Facilitates LTA-Mediated TLR2 Activation and TNF-α and IL-6 Production
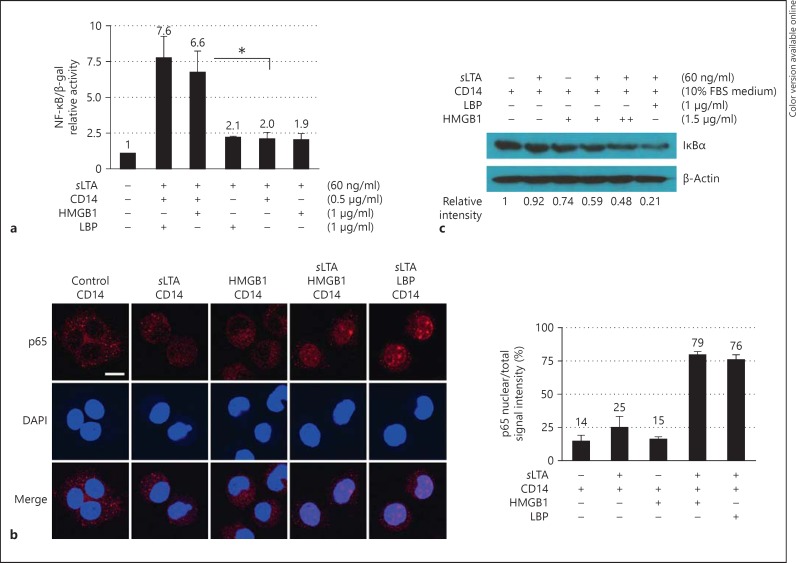
The initiation of proinflammatory signaling was monitored in order to evaluate the effect of sLTA transfer by HMGB1 to CD14, which is subsequently transferred to TLR2. HEK293-hTLR2/6 cells were transiently transfected with NF-κB reporter and β-galactosidase plasmids and then stimulated with a mixture of sLTA and sCD14 in the presence or absence of HMGB1 (fig. 6a). LBP was used as a positive control. The addition of HMGB1 to the sLTA and sCD14 mixture induced a significant 3.3-fold increase in NF-κB activity relative to samples without HMGB1 (fig. 6a). To explore these results further, the migration of NF-κB p65 to the nucleus of J774A.1 cells was observed 2 h after treatment with a mixture of sLTA and HMGB1 or sLTA or HMGB1 alone. Treating cells with a mixture of sLTA and HMGB1 enhanced the migration of p65 to the nucleus more so than treatment with either sLTA or HMGB1 alone, as determined by confocal microscopy and signal localization (fig. 6b).
Fig. 6.
HMGB1 enhances LTA-mediated induction of NF-κB activity. β-gal = β-Galactosidase. a HEK293-hTLR2/6 cells were stimulated with sLTA in the presence or absence of CD14 and HMGB1, and NF-κB activity was measured using an NF-κB reporter plasmid. A β-galactosidase plasmid was used for normalization of the transfection efficiency. * p < 0.01 (n = 3). b Subcellular localization of NF-κB p65 in J774A.1 cells. J774A.1 cells were stimulated with 1 μg/ml of sLTA and 0.5 μg/ml of CD14 in the presence or absence of 1 μg/ml of HMGB1 for 2 h. LBP (1 μg/ml) was used as a positive control. Cells were permeabilized and incubated with an anti-NF-κB p65 Ab overnight at 4°C prior to analysis by confocal microscopy (left panels). Scale bar = 10 μm. The localization of p65-associated fluorescence in nuclei and in whole cells was examined in more than 10 cells using ImageJ software (NIH), and the ratio of mean fluorescence intensity in the nuclei to that in whole cells is shown (right panel). c Western blot analysis of IκBα degradation. J774A.1 cells were stimulated with sLTA and HMGB1 for 30 min in 10% FBS RPMI1640, which contained CD14, and whole cell lysates were analyzed for IκBα degradation after SDS-PAGE. The ratio of the intensity of IκBα band to that of the β-actin band was determined.
In addition, IκB degradation was measured 30 min after LTA stimulation. Degradation of IκBα, which is an inhibitor of NF-κB, enables NF-κB release and nuclear transmigration. As shown in figure 6c, IκBα was significantly degraded when the cells were treated with a mixture of sLTA and HMGB1. These data show that stimulation with a mixture of LTA, HMGB1 and sCD14 initiates proinflammatory signaling and subsequent NF-κB activation.
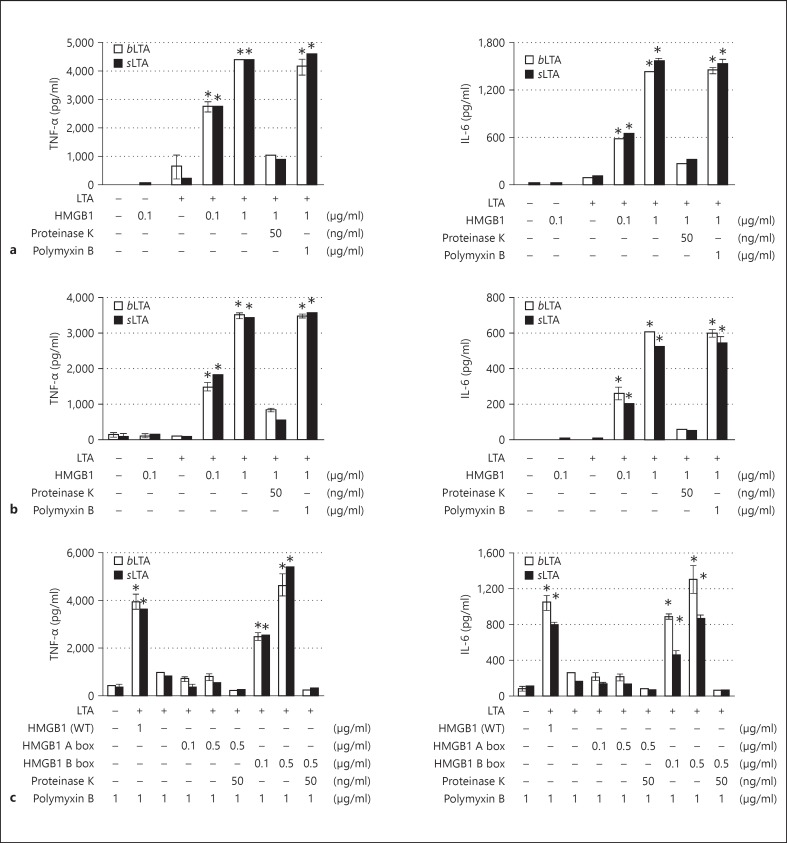
Finally, we evaluated LTA-mediated TNF-α and IL-6 production in J774A.1 and RAW264.7 cells. When cells of both lines were treated with a mixture of sLTA or bLTA and HMGB1, TNF-α production increased significantly in an HMGB1 dose-dependent manner, whereas treatment with LTA or HMGB1 alone resulted in only a minimal increase in TNF-α production (fig. 7a, b). A similar pattern was observed with IL-6 production in cells treated with a mixture of LTA and HMGB1 (fig. 7a, b). The stimulatory effect on cytokine production observed in the present study by cells treated with 10 ng/ml of bLTA was similar to that of cells treated with sLTA at 60 ng/ml, which is consistent with a previous report [27]. In cells treated with polymyxin B, which inhibits LPS-mediated stimulation but only partially inhibits LTA-mediated stimulation at high concentration [27], there was almost no change in TNF-α and IL-6 production by cells treated with a mixture of LTA and HMGB1. The degradation of HMGB1 in the LTA/HMGB1 mixture via the addition of proteinase K followed by the inactivation of proteinase K by boiling resulted in a dramatic decrease in the production of both TNF-α and IL-6, suggesting that HMGB1 plays the role of a transfer molecule (fig. 7a, b). When we evaluated the LTA-mediated transfer of TNF-α and IL-6 using each domain of HMGB1 A and B box proteins in LTA transfer in J774A.1 cells, both TNF-α and IL-6 productions were significantly increased in an HMGB1 B box dose-dependent manner, whereas there was no change by HMGB1 A box protein (fig. 7c). These results suggest that HMGB1 transfers LTA to CD14, which is subsequently transferred to TLR2 to initiate the inflammatory response.
Fig. 7.
TNF-α and IL-6 production in J774A.1 and RAW264.7 cells after HMGB1-mediated transfer of LTA. WT = Wild-type. J774A.1 (a, c) and RAW264.7 cells (b) were treated with LTA in the presence or absence of wild-type HMGB1 or HMGB1 A and B box proteins in OPTI-MEM for 20 h, after which the levels of TNF-α and IL-6 in the supernatants were measured. The experiment was repeated more than 3 times, and the mean ± SEM are shown. * p < 0.005 vs. LTA only: one-way ANOVA test with Bonferroni correction.
Discussion
HMGB1 is known to exert proinflammatory effects by stimulating the production of a diverse array of cytokines [28]. In addition, HMGB1 directly binds to pattern-associated molecular pattern molecules such as LPS, DNA, RNA, and CpG, synergistically enhancing the proinflammatory effect [19, 20, 23, 29, 30]. Here, we report the novel finding that HMGB1 binds to and catalyzes the transfer of LTA monomers from LTA micelles to CD14, synergistically enhancing cytokine production via TLR2-mediated signaling.
There are many LTA-binding proteins, as follows: LBP, CD14, mannose-binding protein, L-ficolin, albumin [6, 31, 32, 33], apolipoproteins A and C [34], transferrin and hemoglobin [35, 36], high- and low-density lipoproteins [37], hemoglobin subunits α and β and S100A9 and SPLUNC2 [38]. Until now, however, LBP was the only molecule known to mediate the transfer of LTA to CD14 to enhance the inflammatory response. Recently, it has been shown that hemoglobin synergistically activates macrophages with LTA, and hemoglobin alone can induce secretion of HMGB1, which is in synergy with LTA in IL-6 production [36]. The present report is the first to demonstrate that HMGB1 also binds to LTA and mediates its transfer to CD14, thus enhancing the inflammatory response. Previously, we reported that, similar to LBP, HMGB1 plays an important role in LPS-mediated proinflammatory signaling by catalyzing the transfer of LPS monomers from LPS micelles to CD14, initiating TLR4-mediated inflammatory responses [23]. We propose, here, that HMGB1 plays an important synergistic role in proinflammatory cytokine production in LTA-mediated TLR2 signaling. Our results show that HMGB1 acts as a PAMP-sensing molecule in infections with Gram-positive and Gram-negative bacteria.
HMGB1 is composed of two homologous DNA-binding domains (the A and B boxes) and a negatively charged C-terminal tail. HMGB1 has a proline-rich linker between the internal repeats of the A and B boxes and a disulfide bond between Cys23 and Cys45. There is no significant difference between ΔC-HMGB1 and wild-type HMGB1 in terms of LTA binding affinity, suggesting that the acidic tail of HMGB1 is not involved in binding. The redox status of HMGB1 influences its cytokine production activity. Native HMGB1, that is, disulfide-HMGB1 at Cys23 and Cys45, induced cytokine production, but the all-thiol form of HMGB1 at Cys23, 45 and 106 showed no significant effect on cytokine production [39]. In our data, native HMGB1 and the all-thiol form of HMGB1 showed no significant altered binding affinity, which suggests that HMGB1 oxidation may not be involved in LTA-mediated TLR2 signaling in the HMGB1-LTA interaction (data not shown). Interestingly, our surface plasmon resonance analyses showed that LTA binds to the B box not the A box, which is similar to LPS binding of the HMGB1 B box [23]. LPS consists of repeating units of polysaccharide and lipid A regions with multiple acyl chains, and LTA similarly consists of a hydrophilic region of repeating polymer units and a hydrophobic glycolipid or phosphoglycolipid region of diacylglycerol units substituted by a di- or triglycoside with five different types based on the chemical structure of the repeating polymer [3, 40]. Based on our data, HMGB1, especially the B box protein, shows high-affinity binding to lipid anchor regions of both macroamphiphiles of LPS and LTA and enhances the inflammatory response. HMGB1 has two LPS-binding motifs, which are located in the A and B box domains and are involved in binding to the polysaccharide and lipid A regions, respectively [24]. Synthetic peptides containing these two LPS-binding motifs inhibit LPS-stimulated TNF-α production [24]. Identification of the LTA-binding peptide region of the HMGB1 B box may enable the development of a peptide useful for neutralizing LTA-stimulated TNF-α production, since the B box itself exhibits proinflammatory effects. Thus, further study of the LTA-binding amino acid residue of HMGB1 is needed to better understand both the molecular interaction between HMGB1 and LTA and the mechanism of the HMGB1-mediated transfer of LTA to CD14.
LPS contamination is common in LTA preparations [10]. However, our LTA transfer assay using silver-stained native PAGE gels gave no indication of LPS contamination, as the migration pattern of a mixture of LTA, sCD14 and HMGB1 differed from that of a mixture of LPS, sCD14 and HMGB1. In our cytokine production assay, treating J774A.1 and RAW264.7 cells with a mixture of LTA and HMGB1, with the addition of the potent LPS antagonist polymyxin B, had no effect on TNF-α and IL-6 production compared with the same treatment without polymyxin B, suggesting that the effects associated with LTA were not due to contamination with LPS.
In conclusion, we demonstrate for the first time that HMGB1 acts as a sensor molecule that binds to and mediates the transfer of LTA to CD14, resulting in the enhancement of LTA-mediated signaling leading to proinflammatory cytokine production. HMGB1-mediated LTA signaling may, therefore, serve as a therapeutic target in the treatment of Gram-positive bacterial sepsis.
Acknowledgment
This work was supported by grants from the National Research Foundation of Korea (NRF) funded by the Korean government (MEST; grant No. 2011-0017611 and 2014R1A4A1008625).
References
- 1.Martin GS, Mannino DM, Eaton S, Moss M. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med. 2003;348:1546–1554. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- 2.Ginsburg I. Role of lipoteichoic acid in infection and inflammation. Lancet Infect Dis. 2002;2:171–179. doi: 10.1016/s1473-3099(02)00226-8. [DOI] [PubMed] [Google Scholar]
- 3.Percy MG, Grundling A. Lipoteichoic acid synthesis and function in Gram-positive bacteria. Annu Rev Microbiol. 2014;68:81–100. doi: 10.1146/annurev-micro-091213-112949. [DOI] [PubMed] [Google Scholar]
- 4.Tobias PS, Mathison JC, Ulevitch RJ. A family of lipopolysaccharide binding proteins involved in responses to Gram-negative sepsis. J Biol Chem. 1988;263:13479–13481. [PubMed] [Google Scholar]
- 5.Gioannini TL, Weiss JP. Regulation of interactions of Gram-negative bacterial endotoxins with mammalian cells. Immunol Res. 2007;39:249–260. doi: 10.1007/s12026-007-0069-0. [DOI] [PubMed] [Google Scholar]
- 6.Schroder NW, Morath S, Alexander C, Hamann L, Hartung T, Zahringer U, Gobel UB, Weber JR, Schumann RR. Lipoteichoic acid (LTA) of Streptococcus pneumoniae and Staphylococcus aureus activates immune cells via Toll-like receptor (TLR)-2, lipopolysaccharide-binding protein (LBP), and CD14, whereas TLR-4 and MD-2 are not involved. J Biol Chem. 2003;278:15587–15594. doi: 10.1074/jbc.M212829200. [DOI] [PubMed] [Google Scholar]
- 7.Buckley JM, Wang JH, Redmond HP. Cellular reprogramming by Gram-positive bacterial components: a review. J Leukoc Biol. 2006;80:731–741. doi: 10.1189/jlb.0506312. [DOI] [PubMed] [Google Scholar]
- 8.Beutler B, Rietschel ET. Innate immune sensing and its roots: the story of endotoxin. Nat Rev Immunol. 2003;3:169–176. doi: 10.1038/nri1004. [DOI] [PubMed] [Google Scholar]
- 9.De Kimpe SJ, Kengatharan M, Thiemermann C, Vane JR. The cell wall components peptidoglycan and lipoteichoic acid from Staphylococcus aureus act in synergy to cause shock and multiple organ failure. Proc Natl Acad Sci U S A. 1995;92:10359–10363. doi: 10.1073/pnas.92.22.10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao JJ, Xue Q, Zuvanich EG, Haghi KR, Morrison DC. Commercial preparations of lipoteichoic acid contain endotoxin that contributes to activation of mouse macrophages in vitro. Infect Immun. 2001;69:751–757. doi: 10.1128/IAI.69.2.751-757.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SA, Kwak MS, Kim S, Shin JS. The role of high mobility group box 1 in innate immunity. Yonsei Med J. 2014;55:1165–1176. doi: 10.3349/ymj.2014.55.5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonaldi T, Talamo F, Scaffidi P, Ferrera D, Porto A, Bachi A, Rubartelli A, Agresti A, Bianchi ME. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 2003;22:5551–5560. doi: 10.1093/emboj/cdg516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Youn JH, Shin JS. Nucleocytoplasmic shuttling of HMGB1 is regulated by phosphorylation that redirects it toward secretion. J Immunol. 2006;177:7889–7897. doi: 10.4049/jimmunol.177.11.7889. [DOI] [PubMed] [Google Scholar]
- 14.Oh YJ, Youn JH, Ji Y, Lee SE, Lim KJ, Choi JE, Shin JS. HMGB1 is phosphorylated by classical protein kinase C and is secreted by a calcium-dependent mechanism. J Immunol. 2009;182:5800–5809. doi: 10.4049/jimmunol.0801873. [DOI] [PubMed] [Google Scholar]
- 15.Ito I, Fukazawa J, Yoshida M. Post-translational methylation of high mobility group box 1 (HMGB1) causes its cytoplasmic localization in neutrophils. J Biol Chem. 2007;282:16336–16344. doi: 10.1074/jbc.M608467200. [DOI] [PubMed] [Google Scholar]
- 16.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, Frazier A, Yang H, Ivanova S, Borovikova L, Manogue KR, Faist E, Abraham E, Andersson J, Andersson U, Molina PE, Abumrad NN, Sama A, Tracey KJ. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999;285:248–251. doi: 10.1126/science.285.5425.248. [DOI] [PubMed] [Google Scholar]
- 18.Sunden-Cullberg J, Norrby-Teglund A, Rouhiainen A, Rauvala H, Herman G, Tracey KJ, Lee ML, Andersson J, Tokics L, Treutiger CJ. Persistent elevation of high mobility group box-1 protein (HMGB1) in patients with severe sepsis and septic shock. Crit Care Med. 2005;33:564–573. doi: 10.1097/01.ccm.0000155991.88802.4d. [DOI] [PubMed] [Google Scholar]
- 19.Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, Parroche P, Drabic S, Golenbock D, Sirois C, Hua J, An LL, Audoly L, La Rosa G, Bierhaus A, Naworth P, Marshak-Rothstein A, Crow MK, Fitzgerald KA, Latz E, Kiener PA, Coyle AJ. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 20.Sha Y, Zmijewski J, Xu Z, Abraham E. HMGB1 develops enhanced proinflammatory activity by binding to cytokines. J Immunol. 2008;180:2531–2537. doi: 10.4049/jimmunol.180.4.2531. [DOI] [PubMed] [Google Scholar]
- 21.Schiraldi M, Raucci A, Munoz LM, Livoti E, Celona B, Venereau E, Apuzzo T, De Marchis F, Pedotti M, Bachi A, Thelen M, Varani L, Mellado M, Proudfoot A, Bianchi ME, Uguccioni M. HMGB1 promotes recruitment of inflammatory cells to damaged tissues by forming a complex with CXCL12 and signaling via CXCR4. J Exp Med. 2012;209:551–563. doi: 10.1084/jem.20111739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bianchi ME. HMGB1 loves company. J Leukoc Biol. 2009;86:573–576. doi: 10.1189/jlb.1008585. [DOI] [PubMed] [Google Scholar]
- 23.Youn JH, Oh YJ, Kim ES, Choi JE, Shin JS. High mobility group box 1 protein binding to lipopolysaccharide facilitates transfer of lipopolysaccharide to CD14 and enhances lipopolysaccharide-mediated TNF-α production in human monocytes. J Immunol. 2008;180:5067–5074. doi: 10.4049/jimmunol.180.7.5067. [DOI] [PubMed] [Google Scholar]
- 24.Youn JH, Kwak MS, Wu J, Kim ES, Ji Y, Min HJ, Yoo JH, Choi JE, Cho HS, Shin JS. Identification of lipopolysaccharide-binding peptide regions within HMGB1 and their effects on subclinical endotoxemia in a mouse model. Eur J Immunol. 2011;41:2753–2762. doi: 10.1002/eji.201141391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aida Y, Pabst MJ. Removal of endotoxin from protein solutions by phase separation using Triton X-114. J Immunol Methods. 1990;132:191–195. doi: 10.1016/0022-1759(90)90029-u. [DOI] [PubMed] [Google Scholar]
- 26.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 27.Zidek Z, Farghali H, Kmonickova E. Intrinsic nitric oxide-stimulatory activity of lipoteichoic acids from different Gram-positive bacteria. Nitric Oxide. 2010;23:300–310. doi: 10.1016/j.niox.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Andersson U, Wang H, Palmblad K, Aveberger AC, Bloom O, Erlandsson-Harris H, Janson A, Kokkola R, Zhang M, Yang H, Tracey KJ. High mobility group 1 protein (HMG-1) stimulates proinflammatory cytokine synthesis in human monocytes. J Exp Med. 2000;192:565–570. doi: 10.1084/jem.192.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urbonaviciute V, Furnrohr BG, Meister S, Munoz L, Heyder P, De Marchis F, Bianchi ME, Kirschning C, Wagner H, Manfredi AA, Kalden JR, Schett G, Rovere-Querini P, Herrmann M, Voll RE. Induction of inflammatory and immune responses by HMGB1-nucleosome complexes: implications for the pathogenesis of SLE. J Exp Med. 2008;205:3007–3018. doi: 10.1084/jem.20081165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hreggvidsdottir HS, Ostberg T, Wahamaa H, Schierbeck H, Aveberger AC, Klevenvall L, Palmblad K, Ottosson L, Andersson U, Harris HE. The alarmin HMGB1 acts in synergy with endogenous and exogenous danger signals to promote inflammation. J Leukoc Biol. 2009;86:655–662. doi: 10.1189/jlb.0908548. [DOI] [PubMed] [Google Scholar]
- 31.Long EM, Millen B, Kubes P, Robbins SM. Lipoteichoic acid induces unique inflammatory responses when compared to other toll-like receptor 2 ligands. PLoS One. 2009;4:e5601. doi: 10.1371/journal.pone.0005601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch NJ, Roscher S, Hartung T, Morath S, Matsushita M, Maennel DN, Kuraya M, Fujita T, Schwaeble WJ. L-ficolin specifically binds to lipoteichoic acid, a cell wall constituent of Gram-positive bacteria, and activates the lectin pathway of complement. J Immunol. 2004;172:1198–1202. doi: 10.4049/jimmunol.172.2.1198. [DOI] [PubMed] [Google Scholar]
- 33.Polotsky VY, Fischer W, Ezekowitz RA, Joiner KA. Interactions of human mannose-binding protein with lipoteichoic acids. Infect Immun. 1996;64:380–383. doi: 10.1128/iai.64.1.380-383.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jang KS, Baik JE, Kang SS, Jeon JH, Choi S, Yang YH, Kim BG, Yun CH, Han SH. Identification of staphylococcal lipoteichoic acid-binding proteins in human serum by high-resolution LTQ-Orbitrap mass spectrometry. Mol Immunol. 2012;50:177–183. doi: 10.1016/j.molimm.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 35.Triantafilou M, Mouratis MA, Lepper PM, Haston RM, Baldwin F, Lowes S, Ahmed MA, Schumann C, Boyd O, Triantafilou K. Serum proteins modulate lipopolysaccharide and lipoteichoic acid-induced activation and contribute to the clinical outcome of sepsis. Virulence. 2012;3:136–145. doi: 10.4161/viru.19077. [DOI] [PubMed] [Google Scholar]
- 36.Cox KH, Cox ME, Woo-Rasberry V, Hasty DL. Pathways involved in the synergistic activation of macrophages by lipoteichoic acid and hemoglobin. PLoS One. 2012;7:e47333. doi: 10.1371/journal.pone.0047333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levels JH, Abraham PR, van Barreveld EP, Meijers JC, van Deventer SJ. Distribution and kinetics of lipoprotein-bound lipoteichoic acid. Infect Immun. 2003;71:3280–3284. doi: 10.1128/IAI.71.6.3280-3284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong SW, Seo DG, Baik JE, Cho K, Yun CH, Han SH. Differential profiles of salivary proteins with affinity to Streptococcus mutans lipoteichoic acid in caries-free and caries-positive human subjects. Mol Oral Microbiol. 2014;29:208–218. doi: 10.1111/omi.12057. [DOI] [PubMed] [Google Scholar]
- 39.Yang H, Lundback P, Ottosson L, Erlandsson-Harris H, Venereau E, Bianchi ME, Al-Abed Y, Andersson U, Tracey KJ, Antoine DJ. Redox modification of cysteine residues regulates the cytokine activity of high mobility group box-1 (HMGB1) Mol Med. 2012;18:250–259. doi: 10.2119/molmed.2011.00389. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Neuhaus FC, Baddiley J. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in Gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]