Abstract
In Drosophila, peptidoglycan (PGN) is detected by PGN recognition proteins (PGRPs) that act as pattern recognition receptors. Some PGRPs such as PGRP-LB or PGRP-SCs are able to cleave PGN, therefore reducing the amount of immune elicitors and dampening immune deficiency (IMD) pathway activation. The precise role of PGRP-SC is less well defined because the PGRP-SC genes (PGRP-SC1a, PGRP-SC1b and PGRP-SC2) lie very close on the chromosome and have been studied using a deletion encompassing the three genes. By generating PGRP-SC-specific mutants, we reevaluated the roles of PGRP-LB, PGRP-SC1 and PGRP-SC2, respectively, during immune responses. We showed that these genes are expressed in different gut domains and that they follow distinct transcriptional regulation. Loss-of-function mutant analysis indicates that PGRP-LB is playing a major role in IMD pathway activation and bacterial load regulation in the gut, although PGRP-SCs are expressed at high levels in this organ. We also demonstrated that PGRP-SC2 is the main negative regulator of IMD pathway activation in the fat body. Accordingly, we showed that mutants for either PGRP-LB or PGRP-SC2 displayed a distinct susceptibility to bacteria depending on the infection route. Lastly, we demonstrated that PGRP-SC1 and PGRP-SC2 are required in vivo for full Toll pathway activation by Gram-positive bacteria.
Key Words: Amidase, Drosophila, Immune deficiency, Innate immunity, Nuclear factor-κB, Peptidoglycan, Peptidoglycan recognition proteins, Toll signaling
Introduction
Peptidoglycan (PGN) and PGN recognition proteins (PGRPs) are the main microbe-associated molecular patterns and pattern recognition receptors that regulate the antibacterial response in Drosophila, respectively [1, 2, 3, 4]. Some PGRP family members such as PGRP-LC, PGRP-SA, PGRP-SD or PGRP-LE have the ability to bind PGN and are therefore essential sentinels upstream of the two NF-κB-dependent Drosophila signaling cascades, called Toll and immune deficiency (IMD) [4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15]. Recognition of lysine (Lys)-type PGN by PGRP-SA is sufficient to trigger the Toll/Dorsal/Dif signaling, whereas detection of diaminopimelic (DAP)-type PGN (either membrane-associated via PGRP-LC or intracellularly via PGRP-LE) promotes IMD/Relish signaling activation [16, 17].
Biochemical experiments have demonstrated that other PGRP family members, such as PGRP-LB, PGRP-SB and PGRP-SC, are not only able to bind PGN but also display an amidasic activity that allows them to cleave PGN into smaller nonimmunogenic muropeptides [18, 19, 20]. In the case of PGRP-LB, in vivo experiments have clearly shown that by degrading PGN, PGRP-LB provides a negative feedback regulation that allows a tight adjustment of the immune activation to the intensity of the infection [18, 21]. In the absence of PGRP-LB, flies overrespond to bacteria and eventually die for unknown reasons. Although the ability of PGRP-SC proteins to cleave PGN is clearly documented, published results on the in vivo role of PGRP-SC in immune system activation are difficult to reconcile into a coherent model.
The PGRP-SC1 protein is coded by two genes, PGRP-SC1a and PGRP-SC1b, which both produce the same polypeptide. This amidase was initially identified as a scavenger receptor that, by cleaving Staphylococcus aureus Lys-type PGN, reduces its immune-stimulatory activity on the IMD pathway in cultured cells [22]. Later on, using an RNAi-mediated approach, it was shown that simultaneous inactivation of PGRP-SC1a/PGRP-SC1b and PGRP-SC2 in the gut induces ectopic expression of immune-inducible genes in the fat body following Escherichia coli ingestion [23]. This role of PGRP-SCs as negative regulators of IMD pathway activation was later confirmed using a deletion removing PGRP-SC1a/ PGRP-SC1b and PGRP-SC2 [21]. This study also revealed that PGRP-SC-dependent negative regulation takes place in the fat body during the systemic response and not in the gut itself. PGRP-SC1 was independently identified through an EMS genetic screen as a protein required for Toll pathway activation and for phagocytosis [24]. Surprisingly, while the PGN-cleaving activity is required to mediate S. aureus phagocytosis, it is dispensable for Toll activation. Finally, a recent report proposed that by reducing IMD/Relish signaling in the gut, PGRP-SC2 is preventing commensal dysbiosis, stem cell hyperproliferation and epithelial dysplasia, and, in turn, prevents gut aging [25]. The different conclusions drawn from these studies could be explained, at least partly, either by the different techniques used to inactivate the genes (RNAi, KO or EMS, for example) or by the fact that while some studies analyzed the effect of removing one PGRP-SC (PGRP-SC1 or PGRP-SC2), others described the phenotype of Drosophila mutant affecting both PGRP-SC1 and PGRP-SC2.
To clarify the respective role of PGRP-SCs and PGRP-LB in immune response modulation, we generated specific KO for each of the PGRP-SC genes and analyzed their immune phenotypes. Our results failed to identify any clear IMD-dependent function for PGRP-SC1, although its transcriptional induction is the highest of the entire genome after gut bacterial colonization. We demonstrated that although PGRP-SC2 and PGRP-LB are both strong negative regulators of IMD, they act in different tissues. Whereas PGRP-LB is needed in the gut to cleave PGN and prevent both local gut activation and PGN dissemination into the hemolymph, PGRP-SC2 is mainly required in the fat body to control systemic immune response. Rescue experiments also show that PGRP-SC2 and PGRP-LB are not functionally equivalent. Finally, mutant phenotype analysis indicated that both PGRP-SC1 and PGRP-SC2 are positive regulators of the Toll signaling cascade.
Materials and Methods
Bacterial Strains
The following microorganisms were used: Lactobacillus plantarumWJL, Erwinia carotovora carotovora 15 2141 (Ecc), Micrococcus luteus and Enterococcus faecalis. All strains were cultured in Luria-Bertani medium, except L. plantarum (MRS medium). L. plantarum and E. faecalis were cultured at 37°C; Ecc and M. luteus at 30°C.
D. melanogaster Strains and Maintenance
The following strains were used in this work: Oregon-R (WT), SC1a-Gal4 (this work), SC1b-mCherry (this work), UAS-nlsGFP BL No. 4775, imdshadok[15], PGRP-LE112[7], PGRP-LCΔE12[15], SC2-Gal4 (this work), PGRP-SC1a/1b−/− (this work) and PGRP-SC2−/− KOs (this work).
Flies were grown at 25°C on a yeast/cornmeal medium. For 1 liter of food, 8.2 g of agar (VWR, cat. No. 20768.361), 80 g of cornmeal flour (Westhove, Farigel Maize H1) and 80 g of yeast extract (VWR, cat. No. 24979.413) were cooked for 10 min in boiling water; 5.2 g of methylparaben sodium salt (Merck, cat. No. 106756) and 4 ml of 99% propionic acid (Carlo Erba, cat. No. 409553) were added when the food had cooled down. For antibiotic treatment, standard medium was supplemented with ampicillin, kanamycin, tetracycline and erythromycin at final concentrations of 50 μg/ml. For all tests, the adult flies used were exclusively 6-day-old females.
Mutant Generation
PGRP-SC1a/1b −/− and PGRP-SC2−/− KO lines were generated by homologous recombination. Gene clusters were replaced by a mini-white gene. DNA flanking the 5′ and 3′ ends used were 3,041 and 2,982 bp for the PGRP-SC1a/1b locus, and 3,002 and 2,980 bp for the PGRP-SC2 locus, respectively. Sequences were cloned into the pW25 vector [26].
Monoassociation of Germ-Free Flies with L. plantarum
Germ-free embryos laid on standard culture medium by germ-free females were covered with 150 ml L. plantarum suspension with an optical density (OD) of 2. Emerging larvae were allowed to develop on the contaminated media. Third-instar larvae (96 h after egg laying) were then dissected.
Natural Infection of Adults by Ecc-15
Overnight bacterial cultures were centrifuged at 2,500 g for 8 min at room temperature and resuspended in fresh Luria-Bertani medium. Cells were serially diluted in PBS and their concentration was determined by OD measurement at 600 nm. For oral infection, flies were first incubated for 2 h at 29°C in empty vials and then placed in a fly vial with food. The food solution was obtained by mixing a pellet of an overnight culture of bacterial Ecc-15 (OD = 200) with a solution of 5% sucrose (50/50) and added to a filter disk that completely covered the agar surface of the fly vial. Septic injuries were performed by pricking adult females with a thin needle contaminated with Ecc-15.
Bacterial Loads
The bacterial load of surface-sterilized individuals was quantified by plating serial dilutions of lysates obtained from 10 individuals on nutrient agar plates (MRS for L. plantarum). Homogenization of individuals was performed using the Precellys 24-tissue homogenizer (Bertin Technologies, France) and 0.75-/1-mm glass beads in 800 ml of the appropriate bacterial culture medium. (Bacterial loads were analyzed 72 h after monoassociation in larval flies.)
Survival Tests with Bacterial Infection
Orally induced bacterial infections were performed with Pseudomonas entomophila. An overnight culture was centrifuged at 2,500 g for 5 min at room temperature and resuspended at OD = 1 in fresh Luria-Bertani medium with 2.5% of sucrose. This bacterial solution was deposited on a filter disk that completely covered the agar surface of the fly vial and flies were added.
Systemic infections (septic injury) were performed with P. entomophila or E. faecalis by pricking adult females in the thorax with a thin needle previously dipped into a concentrated pellet of the bacterial culture. Infected flies were subsequently maintained at 29°C. At least two tubes of 20 flies were used for each survival assay and three replicates of this experiment were done.
Mutants and control populations are compared using the log-rank and the Wilcoxon test (χ2 distribution and p values). All analyses were performed using GraphPad Prism 6 statistical software.
Imaging
Larval or adult tissues were dissected in PBS and fixed for 20 min in 4% paraformaldehyde on ice. After several rinses in PBT (PBS + 0.1% Triton X-100), the tissues were mounted in Vectashield (Vector Laboratories) fluorescent mounting medium with DAPI. Images were captured with either a Stereo Discovery V12 microscope or an LSM 780 Zeiss confocal microscope.
Quantitative Real-Time PCR
RNAs from entire flies (n = 12), guts (n = 12), fat bodies (n = 12) or cultured S2 cells (300 μl) were extracted with the RNeasy mini kit (Qiagen, cat. No. 74106). Quantitative real-time PCR, TaqMan and SYBR Green analyses were performed as previously described [27]. Information on primers can be obtained upon request. The amount of mRNA detected was normalized to control rp49 mRNA values. Normalized data were used to quantify the relative levels of a given mRNA according to cycle threshold analysis (ΔCT).
Drosophila S2 Cells for PGRP-SC Expression
cDNA fragments corresponding to the whole coding sequences of PGRP-SC1a and PGRP-SC2 were amplified using specific oligonucleotides. The PCR-amplified DNAs were digested by the appropriate restriction enzymes and then inserted at the specific sites into the vector pMT/BIP/V5-HisA (Life Technologies, cat. No. V4120-20). The resulting recombinant plasmids encode for V5-His-tagged proteins. The integrity of all constructs was assessed by DNA sequencing. The recombinant plasmids encoding PGRP-SC1 and PGRP-SC2 were cotransfected with the pAc5C-pac vector (an actin5C-driven expression vector for puromycin acetyltransferase) into Drosophila S2 cells according to the protocol from Life Technologies. Stable clones were obtained using puromycin selection. Cells were grown in suspension at 23°C at a cell density of 3-4 × 106 cells/ml and kept under selection in Schneider's medium (Sigma) containing 0.5 μg/ml puromycin (Life Technologies), 50 μg/ml streptomycin (Gibco), 50 µg/ml penicillin (Gibco) and 10% heat-inactivated fetal bovine serum (Gibco). For immune tests with bacteria, cells were diluted at a cell density of 106 cells/ml on day 1. On day 2, expression of the proteins was induced by addition of 0.05 mM CuSO4, and the contact with dead bacteria was done on day 3 for 24 h.
Results
PGRP-SC1a and PGRP-SC1b Are Expressed in Overlapping Domains in the Larval Gut
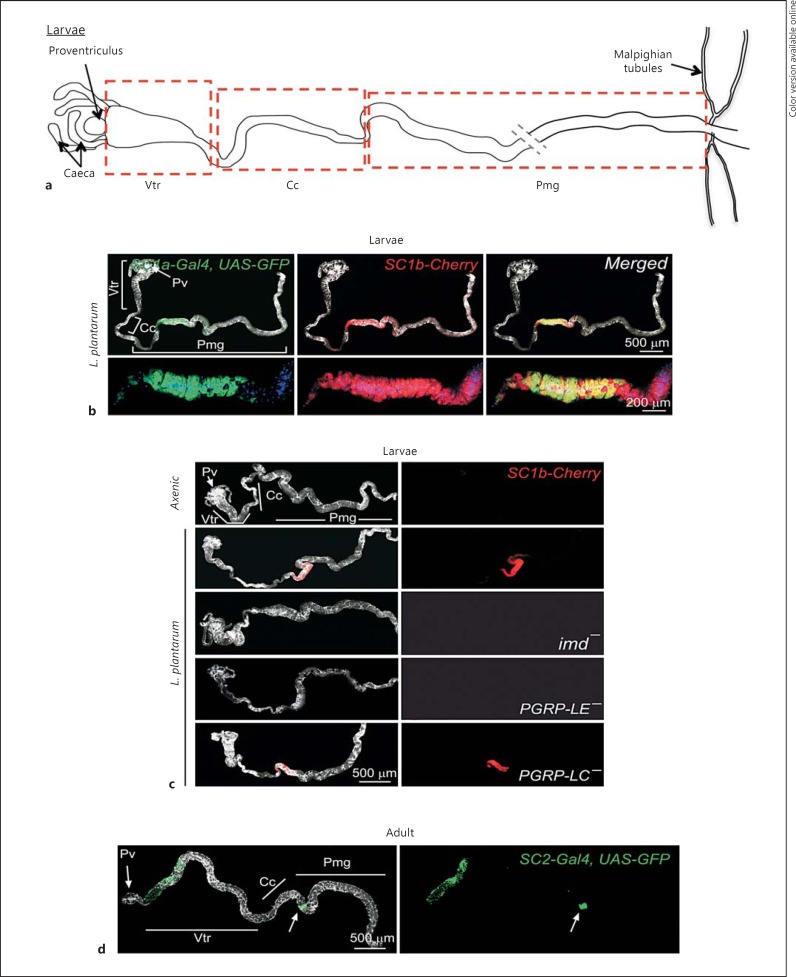
The PGRP-SC1a and PGRP-SC1b genes lie 3 kb apart on the second chromosome and code for two identical polypeptides [28]. To reveal their expression pattern, we generated reporter lines in which 1.5 kb of genomic DNA 5′ of each coding region was cloned upstream to either mCherry (for PGRP-SC1b)- or Gal4-coding sequences (for PGRP-SC1a). When raised in axenic condition, transgenic PGRP-SC1a-mCherry or PGRP-SC1b-Gal4, UAS-nlsGFP larvae did not display fluorescent signals in the intestinal tract (data not shown). When larvae of the same genotypes were fed with the commensal strain, L. plantarum, both transgenes were highly and specifically expressed in the anterior part of the posterior midgut (Pmg) in almost completely overlapping domains (fig. 1a, b). Very similar expression patterns were observed for the PGRP-SC1 transgenes in adult infected guts (data not shown). This indicated that these two genes that code for the same protein and are expressed in very similar domains are transcriptionally regulated by two different promoter regions. To insure that the lines were faithfully reporting endogenous gene expression patterns, PGRP-SC1a-mCherry flies were crossed to IMD pathway mutants known to regulate its transcription. As expected from previous work [29], PGRP-SC1 gene activation upon L. plantarum gut colonization required a functional PGRP-LE receptor and an IMD adaptor but was PGRP-LC independent (fig. 1c).
Fig. 1.
Expression pattern of PGRP-SC1a, PGRP-SC1b and PGRP-SC2 in the larval gut. Colors refer to the online version only. a Schematic representation of a dissected larval gut. b PGRP-SC1a and PGRP-SC1b expression pattern in the larval gut of flies associated with L. plantarum. PGRP-SC1a-Gal4, UAS-nlsGFP (green) and PGRP-SC1b-mCherry (red) are coexpressed in the anterior part of the midgut. c IMD and PGRP-LE, but not PGRP-LC, are required for PGRP-SC1 expression in the gut. Confocal pictures of axenic or L. plantarum-associated larval guts of the following genotypes: SC1b-Cherry(PGRP-SC1b-mCherry), imd-(PGRP-SC1b-Cherry, imdshadok), PGRP-LE-(PGRP-SC1b-mCherry; PGRP-LE112) and PGRP-LC-(PGRP-SC1b-mCherry; PGRP-LCE12). d PGRP-SC2 expression in adult guts of axenic flies. PGRP-SC2-Gal4, UAS-nlsGFP (green) is expressed in Vtr and in a narrow domain of the Pmg. Pv = Proventriculus.
PGRP-SC1, PGRP-SC2 and PGRP-LB Have Different Expression Patterns in the Gut
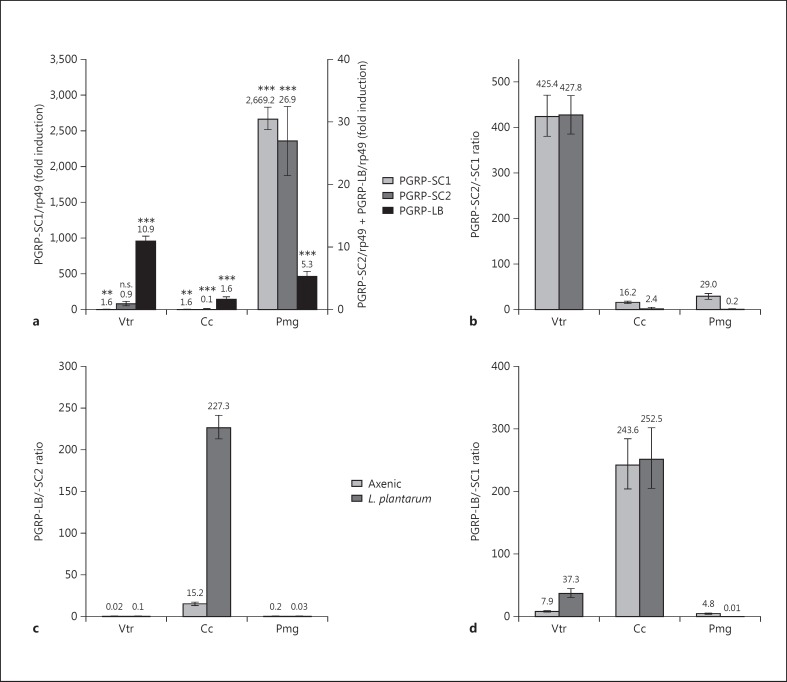
Previous work has shown that PGRP-SC1, PGRP-SC2 and PGRP-LB are all expressed in the gut [21]. We then decided to quantify the relative expression levels of PGRP-SC1a/PGRP-SC1b, PGRP-SC2 and PGRP-LB mRNAs in different domains of naive and L. plantarum-colonized midguts (fig. 2a). While PGRP-SC1a/PGRP-SC1b and PGRP-SC2 transcription were not modified upon L. plantarum recolonization in ventriculus (Vtr) and copper cells (Cc), it was strongly up-regulated in the Pmg of bacterium-colonized guts. PGRP-SC2 induction was, however, a hundred times lower than that of PGRP-SC1 (fig. 2a). This contrasted with PGRP-LB mRNAs, whose levels were only moderately affected by the presence of bacteria with a 10 and 5 times increase in the Vtr and Pmg, respectively. We then compared the relative abundance of the three amidase mRNAs in the different gut domains contaminated or not with bacteria (fig. 2b, c, d). In axenic larvae, PGRP-SC2 mRNAs were more abundant than PGRP-SC1 mRNAs in all gut domains, but most dramatically in the anterior Vtr, in which the PGRP-SC2/PGRP-SC1 ratio can be increased up to 400 times (fig. 2b). These results indicated that the pattern and dynamics of PGRP-SC1a/PGRP-SC1b and PGRP-SC2 expression are very different. Whereas PGRP-SC1a/PGRP-SC1b are transcribed at very low levels in axenic guts and are strongly up-regulated in the Pmg after bacterial colonization, PGRP-SC2 expression is constitutive in anterior domains and moderately modified by bacteria in the Pmg. A similar pattern of expression was observed at the adult stage using a PGRP-SC2-Gal4, UAS-nlsGFP reporter line (fig. 1d). In contrast to PGRP-SC1a-mCherry lines in which the signal was only detected in the presence of bacteria, the GFP signal in the PGRP-SC2-Gal4, UAS-nlsGFP line was already observed in axenic conditions (fig. 1d) and not strongly modified by bacteria (not shown). These data indicated that although the three amidases are expressed in the gut, their specific domain of expression, their relative ratio and their induction upon bacterial infection are unique to a given amidase. In other words, each gut domain contains a specific cocktail of amidases that is either constitutively expressed or induced by the presence of microorganisms.
Fig. 2.
PGRP mRNA levels in L. plantarum-colonized gut domains. a Induction levels of PGRP-SC1, PGRP-SC2 and PGRP-LB mRNAs in gut domains of L. plantarum-colonized larvae compared to axenic larvae (= control value). Means ± SD of three independent experiments. Statistical significance of the results with Student's t test analysis is included: p > 0.05 (n.s.), ** p < 0.05 and *** p < 0.01. b-d Relative ratios between PGRP-SC2 and PGRP-SC1 (b), between PGRP-LB and PGRP-SC2 (c) and between PGRP-LB and PGRP-SC1 (d) mRNAs in different gut domains of axenic and L. plantarum-colonized larvae.
Generating Single PGRP-SC1 and PGRP-SC2 Mutants
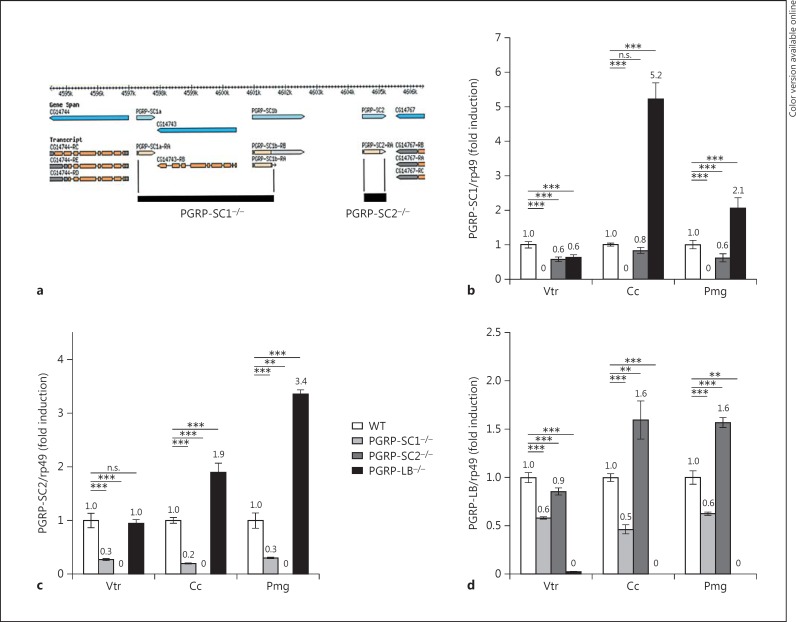
The fact that each amidase displays a unique transcription pattern suggested that they might play a distinct role in the gut immune response. In order to further dissect their respective contribution to immune responses in the gut but also in other immune tissues, we took advantage of a previously generated PGRP-LB−/− mutant and have generated PGRP-SC1−/−- and PGRP-SC2−/−-specific KOs through homologous recombination (fig. 3a) [21]. Since the CG14743 gene is inserted in between PGRP-SC1a and PGRP-SC1b, the PGRP-SC1−/− construct also removed it (fig. 3a). Our results demonstrated that this gene is not expressed in the gut (data not shown), confirming previously published data showing that its inactivation does not affect immune responses [21]. As expected, quantitative real-time PCR on dissected adult gut domains showed that PGRP-SC1 and PGRP-SC2 mRNAs were absent in their respective KO mutant, thereby validating the tools generated (fig. 3b, c). However, the proximity of the two genes on the chromosome led us to test whether genome editing performed at one locus could impact gene expression at the neighboring locus. In axenic guts, basal PGRP-SC1 transcription was slightly affected by the absence of PGRP-SC2 (fig. 3b). The effects were stronger with the reciprocal combination with a 70–80% decrease in the PGRP-SC2 mRNA level in PGRP-SC1−/−. However, since PGRP-SC2 is a gene whose basal expression is high in the gut, the PGRP-SC1−/− mutant cannot be considered as a double mutant. Moreover, in other tissues such as the fat body, PGRP-SC1 inactivation had no influence on PGRP-SC2 expression and vice versa (online suppl. fig. S1; see www.karger.com/doi/10.1159/437368 for all online suppl. material). Finally, PGRP-LB inactivation was associated with a slight increase in PGRP-SC1 and PGRP-SC2 mRNA levels (fig. 3b, c); online suppl. fig. S1). In contrast, PGRP-SC elimination had only minor effects on PGRP-LB expression (fig. 3d).
Fig. 3.
Generation and characterization of PGRP-SC1- and PGRP-SC2-specific mutants. a Schematic representation of PGRP-SC1a/b and PGRP-SC2 mutants. The gene map was adapted from FlyBase. The deleted segment replaced by the mini-white gene (black box) is indicated. For the PGRP-SC1a/b locus, the deletion starts at position 2R: 8,709,733 and ends at position 2R: 8,715,094 and also includes the CG14743 gene; for the PGRP-SC2 locus, the deletion starts at position 2R: 8,716,950 and ends at position 2R: 8,717,695. b-d Relative gene expression of PGRP-SC1 (b), PGRP-SC2 (c) and PGRP-LB (d) in different gut domains of axenic larvae of the three specific mutants compared to controls (WT). Means ± SD of three independent experiments. p > 0.05 (n.s.), ** p < 0.05 and *** p < 0.01 (Student's t test).
Amidase Inactivation Alters Gut IMD Pathway Activation by L. plantarum
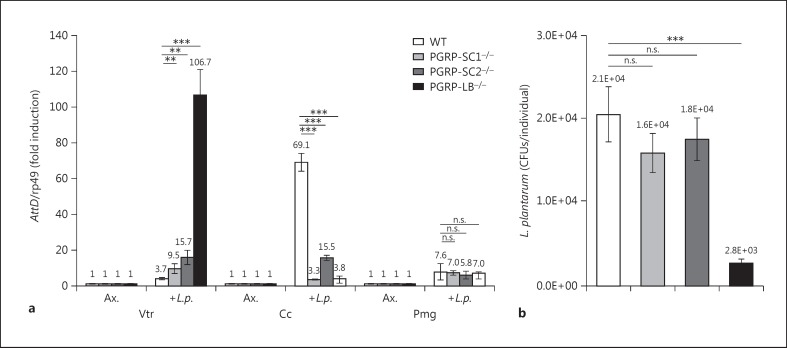
Taking advantage of the newly generated mutants, we tested the implication of each amidase for gut immune responses following colonization with L. plantarum, bacteria previously shown to be tolerated by the gut immune system [30]. Using attacin D (AttD) as a molecular readout of the IMD pathway, we showed that amidase inactivation had domain-specific effects (fig. 4a). Although inactivation of any of the three amidases provoked an up-regulation of AttD expression in the Vtr, the strongest effects were seen in PGRP-LB−/−. Since PGRP-SC1 mRNAs were almost absent in the Vtr and since PGRP-SC1−/− strongly reduced the PGRP-SC2 mRNA level, the effects seen in PGRP-SC1−/− could be secondary to a reduction in PGRP-SC2 mRNAs. Conversely, AttD expression was decreased in the Cc region soon after removal of one of the three amidases. Finally, inactivating any of the three amidases had no real impact on the intensity of the immune response to the colonization of the Pmg with L. plantarum. These data indicated that although PGRP-SC2 is expressed at very high levels in the gut, its absence had only limited effects on regulating IMD pathway activation after bacterial ingestion. They also highlighted the essential role of PGRP-LB in regulating IMD pathway activation in the anterior part of the gut. Since the Vtr is part of the gut that hosts gut-associated bacteria, we tested the ability of the amidase mutants to control bacterial load in the intestinal tract [29]. While mutants for either PGRP-SC1 or PGRP-SC2 had no effect on bacterial load of L. plantarum-recolonized guts, a very strong bacterial reduction was observed in PGRP-LB mutants (fig. 4b). This confirmed that PGRP-LB is the main IMD pathway regulator in the gut and that PGRP-SCs do not play a role in regulating bacterial load and intensity of immune responses in the intestinal tract.
Fig. 4.
PGRP-SC1, PGRP-SC2 and PGRP-LB mutants have specific effects on AMP expression and bacterial load in the gut. a AttD expression in gut domains of controls (WT) and PGRP mutants recolonized by L. plantarum (L.p.).b Bacterial loads of larval guts recolonized by L. plantarum. Means ± SD of three independent experiments. p > 0.05 (n.s.), ** p < 0.05 and *** p < 0.01 (Student's t test). Ax. = Axenic.
Tissue-Specific Role of PGRP-SC2 and PGRP-LB in Dampening IMD Activation following Ecc Infection
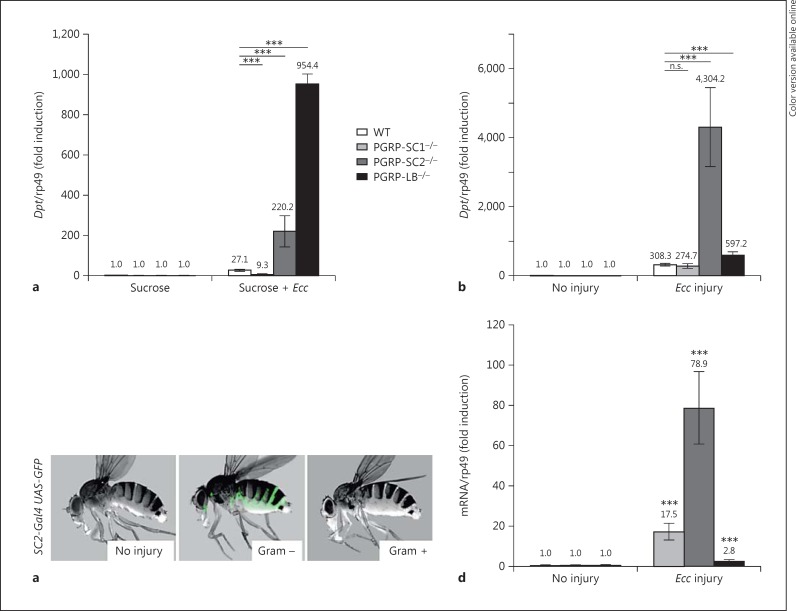
The fact that PGRP-SC1 and PGRP-SC2 inactivation had only minor effects on local gut immune responses prompted us to test their putative implication for other host immune tissues such as the fat body. For that purpose, we turned to Ecc, which has the ability to activate both local gut but also systemic immune responses when present in the gut, probably by releasing PGN that reaches the hemolymph [31, 32, 33]. Diptericin (Dpt) transcription, which is a molecular readout for IMD activation in the fat body, was monitored in the three amidase mutants and compared to WT controls. Whereas feeding with Ecc had only a moderate effect on Dpt induction in WT and PGRP-SC1 mutants, elimination of PGRP-SC2 or PGRP-LB mRNAs provokes its strong up-regulation (fig. 5a). These effects could either reflect the action of PGN-degrading enzymes in the intestinal tract or in the circulating hemolymph. To distinguish between these possibilities, we monitored Dpt expression in flies infected with Ecc by pricking. Using such a protocol, the inactivation of PGRP-SC2 had a much stronger effect on Dpt expression than the inactivation of PGRP-LB, which had only mild consequences (fig. 5b). Here again, the absence of PGRP-SC1 was without any consequences. These results are well correlated with the fact that PGRP-SC2 but not PGRP-LB transcription is strongly up-regulated in the fat body of infected flies (fig. 5c, d). These results showed that while the absence of PGRP-SC1 mRNA has no effect on the level of activation of systemic immune responses, both PGRP-SC2 and PGRP-LB act as negative regulators of the IMD pathway. Interestingly, however, their effects seem to depend on the inoculation route. While PGRP-LB is needed to degrade Ecc PGN in the gut, our results suggest that PGRP-SC2 is doing so within the body cavity. To further challenge this hypothesis in vivo, we decided to compare the ability of specific amidase mutants to resist infection with the same pathogenic bacterial species inoculated via different routes.
Fig. 5.
The impact of PGRP-SC2 and PGRP-LB on IMD pathway repression after Ecc infection is tissue specific. Color refers to the online version only. a, b Dpt expression in WT and PGRP mutant flies 24 h after Ecc feeding (a) and 24 h after septic injury with Ecc (b). c PGRP SC2 expression in adult flies 48 h after septic injury. PGRP-SC2-Gal4, UAS-nlsGFP (green) is mainly expressed in the fat body after injury with Ecc. d Induction levels of PGRP-SC1, PGRP-SC2 and PGRP-LB mRNAs in the fat body of adult flies 24 h after Ecc injury. Means ± SD of three independent experiments. p > 0.05 (n.s.) and *** p < 0.01 (Student's t test).
PGRP-LB and PGRP-SC2 Mutants Display Different Susceptibility to P. entomophila Infection Depending on the Inoculation Route
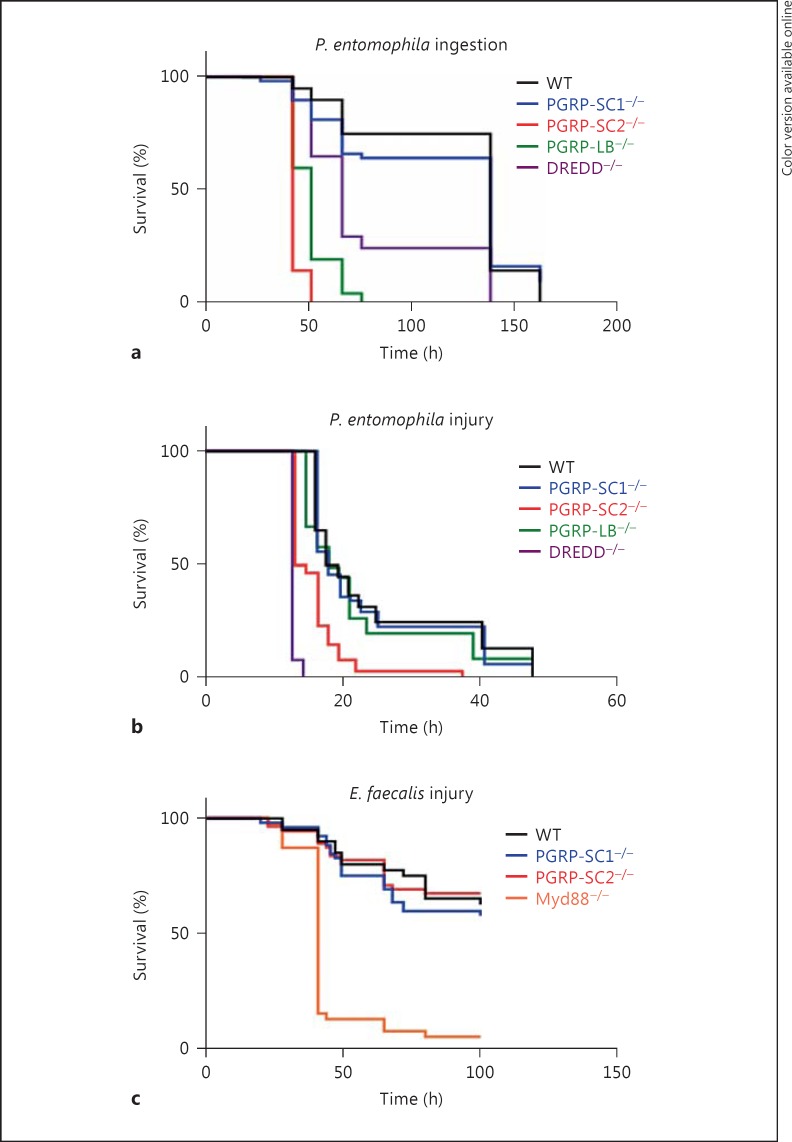
Others and we have shown that uncontrolled IMD pathway activation is detrimental to the fly [21, 25]. The present data indicate that while PGRP-LB mutants present an exacerbated immune response when bacteria are fed to the flies, this is not the case if bacteria are inoculated in the body cavity by pricking. This contrasted with PGRP-SC2mutants in which IMD pathway overactivation is observed with both modes of infection, although more strongly after cuticle injury. In order to test whether these differences could impact the ability of a fly to survive the detrimental effects of IMD pathway overactivation, we used P. entomophila spp. In contrast to Ecc,P. entomophila is pathogenic to Drosophila but is also able to trigger AMP production in the fat body when present in the gut [34, 35, 36, 37]. We then compared the ability of WT and amidase-mutant flies to survive to P. entomophila infection (fig. 6a, b). Using both infection modes (oral or pricking), PGRP-SC1 mutants behave as WT controls confirming the results obtained with Ecc. Interestingly, PGRP-LB and PGRP-SC2 mutants behave differently when infected with P. entomophila. While both mutants showed a high mortality rate when infected orally with P. entomophila, only the PGRP-SC2 mutant died quicker than controls when injured with P. entomophila. This suggested that when PGN is present in the hemolymph, it is degraded by PGRP-SC2 to reduce IMD pathway activation. If it is in the gut, PGN is taken care of mainly by PGRP-LB.
Fig. 6.
Survival rates of PGRP-SC1 and PGRP-SC2 mutants after infection with bacterial pathogens. a Survival analysis of WT (Oregon-R), PGRP-SC1 mutant, PGRP-SC2 mutant, PGRP-LB−/− and DREDD−/− after ingestion with P. entomophila. PGRP-SC2−/−, PGRP-LB−/− and DREDD−/− mutants are more susceptible than controls (p < 0.001). b Survival analysis of WT (Oregon-R), PGRP-SC1−/−, PGRP-SC2−/−, PGRP-LB−/− and DREDD−/− after infection with P. entomophila. PGRP-SC2−/− and DREDD−/− mutants are more susceptible than controls (p < 0.001). c Survival analysis of WT (Oregon-R), PGRP-SC1−/−,PGRP-SC2−/− and Myd88−/− mutants after infection with E. faecalis. Myd88−/− mutants are more susceptible than controls (p < 0.001). Survival curves are representative of at least three independent trials. p values were calculated with the log-rank and the Wilcoxon test.
PGRP-SC1 and PGRP-SC2 Are Required for Full Toll Pathway Activation following Infection with Gram-Positive Bacteria
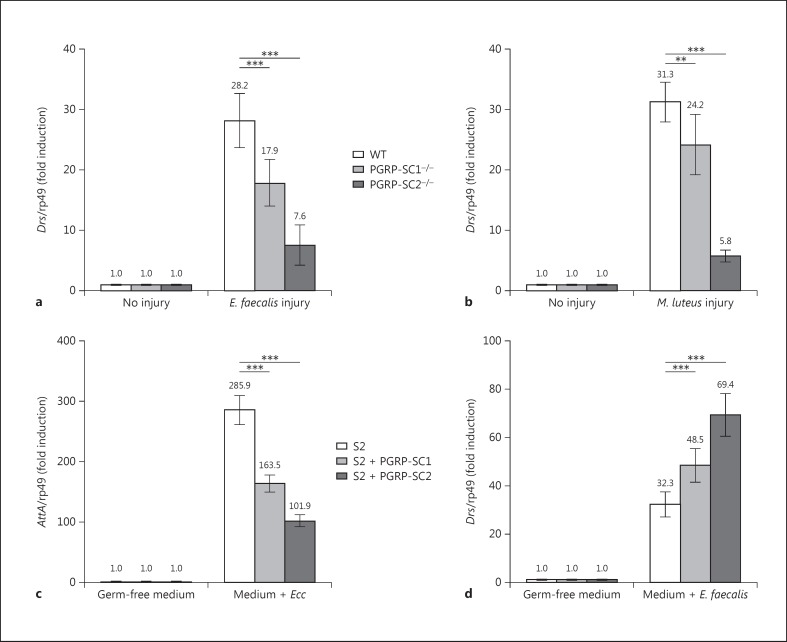
In all the above tests, elimination of PGRP-SC1 had no consequence on IMD pathway activation. Taking advantage of the mutant generated, we tested whether PGRP-SC1 could influence, as previously proposed [24], the activation of the Toll pathway, the second NF-κB signaling cascade involved in fly innate immunity. Inactivation of PGRP-SC2, and to a lesser extent of PGRP-SC1, provoked a marked reduction in transcriptional activation of the Toll pathway target gene drosomycin (Drs) after infection with Gram-positive bacteria compared to WT controls (fig. 7a, b). This result was rather unexpected since Drosophila amidases have been proposed to cleave DAP-type PGN rather than Lys-type PGN. In addition, amidases seem here to act as facilitators of Toll pathway activation whereas they are repressors of IMD signaling. To test this hypothesis in a simplified system, we analyzed whether the same amidases could display antagonistic effects on Toll and IMD pathway activation in S2 cells (online suppl. fig. S2). For that purpose, we compared the ability of naïve and PGRP-SC1- or PGRP-SC2-expressing S2 cells to activate the Toll and IMD pathways, respectively, after incubation with Gram-positive or -negative bacteria. Consistent with previous data, amidase-producing cells had a reduced ability to activate the IMD pathway compared to controls when incubated with heat-inactivated Ecc (fig. 7c). Interestingly, when the same PGRP-SC2- or PGRP-SC1-producing cells were incubated with dead E. faecalis, they showed an enhanced ability to activate Drs, the Toll target gene (fig. 7d). Reduction in Toll pathway activation in PGRP-SC mutants was, however, not sufficient to impair the ability of the flies to resist to E. faecalis infection (fig. 6c). These in vitro and in vivo results show that PGRP-SC1 and PGRP-SC2 exert an opposite effect on the activation of the two NF-κB signaling cascades. While they reduced the ability of DAP-type PGN to activate the IMD pathway, they increased the elicitor activity of Lys-type PGN on Toll pathway activation. Here, again, the effects seen with PGRP-SC2 were stronger than with PGRP-SC1.
Fig. 7.
Inactivation of PGRP-SC1 and PGRP-SC2 reduce Toll pathway activation. a, b Drs expression in controls and PGRP-SC1 and PGRP-SC2 mutants 24 h after infection with E. faecalis (a) and M. luteus (b). c AttA expression in S2 cells 24 h after addition of dead Ecc. S2 cells overexpressing PGRP-SC1 or PGRP-SC2 show decreased AttA expression. d Drs expression in S2 cells 24 h after addition of dead E. faecalis. S2 cells overexpressing PGRP-SC1 or PGRP-SC2 show increased Drs expression. Means ± SD of three independent experiments. ** p < 0.05 and *** p < 0.01 (Student's t test).
Discussion
The results presented here demonstrate that PGRP-LB, PGRP-SC1 and PGRP-SC2 have different spatiotemporal expression patterns and play specific roles in regulating Drosophila immune responses. As far as the IMD pathway is concerned, PGRP-SC1a/PGRP-SC1b elimination did not provoke any modification in immune pathway activation. This was rather unexpected, since PGRP-SC1 is the Drosophila most induced gene following bacterial colonization. In contrast, our data showed that both PGRP-SC2 and PGRP-LB are strong dampeners of the IMD pathway. In accordance with previous work, we demonstrated that PGRP-LB is the essential amidase in the gut. However, it remained unclear why the PGRP-SC2 amidase which is highly expressed in the gut has such a minor role in regulating IMD pathway activation or bacterial load in this organ. Some kind of functional redundancy could explain the lack of effect. However, previous work did not report a very strong IMD pathway up-regulation in the double PGRP-SC mutant [21]. In addition, whereas PGRP-LB and PGRP-SC2 are both expressed in the Vtr, removing the PGRP-LB gene had a clear phenotype, speaking against functional redundancy between these two PGN-cleaving enzymes. In addition, using ectopic expression tools, we were able to show that while ectopic PGRP-LB expression can rescue the PGRP-LB-mutant phenotype, PGRP-SC1 and PGRP-SC2 cannot (online suppl. fig. S3). This clearly demonstrated that in addition to being expressed in different spatiotemporal patterns, amidases are not functionally equivalent. In this respect, it is interesting to note that PGRP-LB is functionally important in the gut and PGRP-SC2 in the circulating hemolymph. Indeed, we have previously shown that the mode of bacterial detection in the gut and in the fat body are different [29]. While most enterocytes rely on the intracellular PGRP-LE for PGN detection, fat body cells detect PGN mainly via PGRP-LC. It is well possible that these two receptors are activated in vivo by different ligands. A possible model could be that PGRP-LB is preventing the production of PGRP-LE-activating ligands (such as TCT) whereas PGRP-SC2 is preventing accumulation of PGRP-LC ligands. Further experiments will be needed to test this hypothesis.
Using PGRP-SC mutants, we also showed that amidases are not only required to dampen the IMD pathway but also to facilitate Toll signaling activation. This indicated that, surprisingly, the action of amidases had opposite effects on Toll and IMD signaling activation. Since the activation of both pathways depends on PGN recognition by PGRP family members, one can postulate that while a PGRP-SC-digested DAP-type PGN will be a weaker IMD pathway activator and therefore probably a weak PGRP-LC ligand, a PGRP-SC-digested Lys-type PGN will be a good inducer of Toll signaling and therefore strongly recognized by PGRP-SA. This antagonistic effect correlates well with the fact that while the IMD cascade strongly needs to be down-regulated to prevent flies from dying of infection, this is not at all the case for the Toll pathway whose constitutive activation has no effect on the flies' viability but is probably more efficient to fight infection.
The data presented here demonstrated the complexity and interdependence of the interactions that are occurring to adapt the immune responses towards bacteria entering the body cavity of Drosophila. Analyzing immune responses in Vtr, Cc and Pmg separately, we have demonstrated that different gut domains produce different amidase cocktails and display specific responses. However, gut dissection has shown that the gut can be anatomically subdivided into more than ten subdomains [38]. This could potentially greatly increase the complexity of the regulation. In addition, one also cannot exclude the possibility that amidases are acting successively to degrade PGN. If such a PGN will be first cleaved by a given amidase before being a target for another PGN-cleaving enzyme, the interpretation of the mutant phenotype will be even more complicated. This could be the case for Ecc PGN that could first be modified in the gut lumen by PGRP-LB before being digested in the hemolymph by PGRP-SC2. One should also keep in mind that PGRPs with amidase activity are potentially secreted proteins and could therefore act distant from the site where they are produced. They could eventually travel together with the bacteria from one gut domain to another. Finally, our data showed that mutations in a given amidase can have opposite effects on the regulation of the two main signaling immune pathways, IMD and Toll. This could potentially be explained with two biological roles of PGRP-SC, an amidase-dependent and an amidase-independent function. Consistently, Garver at al. [24] demonstrated that a noncatalytic cysteine-serine PGRP-SC1a transgene is able to rescue a PGRP-SC1a mutant as far as Toll pathway activation is concerned. Knowing that some immune genes are specifically activated by one cascade whereas others depend on both signaling pathways, one should interpret the immunomodulation and immune phenotypes observed in amidase mutants with caution.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
We thank Fatoumata Djitte for technical assistance, and the members of Royet's laboratory for comments and discussion. This work was supported by the Equipe FRM DEQ20140329541 to Julien Royet, and by the Centre National de la Recherche Scientifique and the Agence Nationale pour la Recherche.
References
- 1.Royet J, Gupta D, Dziarski R. Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nat Rev Immunol. 2011;11:837–851. doi: 10.1038/nri3089. [DOI] [PubMed] [Google Scholar]
- 2.Royet J, Dziarski R. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol. 2007;5:264–277. doi: 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- 3.Buchon N, Silverman N, Cherry S. Immunity in Drosophila melanogaster - from microbial recognition to whole-organism physiology. Nat Rev Immunol. 2014;14:796–810. doi: 10.1038/nri3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurata S. Extracellular and intracellular pathogen recognition by Drosophila PGRP-LE and PGRP-LC. Int Immunol. 2010;22:143–148. doi: 10.1093/intimm/dxp128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol. 2006;7:715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- 6.Choe KM, Lee H, Anderson KV. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc Natl Acad Sci USA. 2005;102:1122–1126. doi: 10.1073/pnas.0404952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takehana A, Yano T, Mita S, Kotani A, Oshima Y, et al. Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in Drosophila immunity. EMBO J. 2004;23:4690–4700. doi: 10.1038/sj.emboj.7600466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Werner T, Borge-Renberg K, Mellroth P, Steiner H, Hultmark D. Functional diversity of the Drosophila PGRP-LC gene cluster in the response to lipopolysaccharide and peptidoglycan. J Biol Chem. 2003;278:26319–26322. doi: 10.1074/jbc.C300184200. [DOI] [PubMed] [Google Scholar]
- 9.Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- 10.Leone P, Bischoff V, Kellenberger C, Hetru C, Royet J, et al. Crystal structure of Drosophila PGRP-SD suggests binding to DAP-type but not lysine-type peptidoglycan. Mol Immunol. 2008;45:2521–2530. doi: 10.1016/j.molimm.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 11.Bischoff V, Vignal C, Boneca IG, Michel T, Hoffmann JA, et al. Function of the Drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat Immunol. 2004;5:1175–1180. doi: 10.1038/ni1123. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Gilbert RJ, Atilano ML, Filipe SR, Gay NJ, et al. Peptidoglycan recognition protein-SD provides versatility of receptor formation in Drosophila immunity. Proc Natl Acad Sci USA. 2008;105:11881–11886. doi: 10.1073/pnas.0710092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- 14.Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- 15.Gottar M, Gobert V, Michel T, Belvin M, Duyk G, et al. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- 16.Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol. 2003;4:478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- 17.Stenbak CR, Ryu JH, Leulier F, Pili-Floury S, Parquet C, et al. Peptidoglycan molecular requirements allowing detection by the Drosophila immune deficiency pathway. J Immunol. 2004;173:7339–7348. doi: 10.4049/jimmunol.173.12.7339. [DOI] [PubMed] [Google Scholar]
- 18.Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, et al. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 2006;24:463–473. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 19.Zaidman-Remy A, Poidevin M, Herve M, Welchman DP, Paredes JC, et al. Drosophila immunity: analysis of PGRP-SB1 expression, enzymatic activity and function. PLoS One. 2011;6:e17231. doi: 10.1371/journal.pone.0017231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellroth P, Steiner H. PGRP-SB1: an N-acetylmuramoyl L-alanine amidase with antibacterial activity. Biochem Biophys Res Commun. 2006;350:994–999. doi: 10.1016/j.bbrc.2006.09.139. [DOI] [PubMed] [Google Scholar]
- 21.Paredes JC, Welchman DP, Poidevin M, Lemaitre B. Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity. 2011;35:770–779. doi: 10.1016/j.immuni.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 22.Mellroth P, Karlsson J, Steiner H. A scavenger function for a Drosophila peptidoglycan recognition protein. J Biol Chem. 2003;278:7059–7064. doi: 10.1074/jbc.M208900200. [DOI] [PubMed] [Google Scholar]
- 23.Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, et al. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2006;2:e14. doi: 10.1371/journal.ppat.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garver LS, Wu J, Wu LP. The peptidoglycan recognition protein PGRP-SC1a is essential for Toll signaling and phagocytosis of Staphylococcus aureus in Drosophila. Proc Natl Acad Sci USA. 2006;103:660–665. doi: 10.1073/pnas.0506182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo L, Karpac J, Tran SL, Jasper H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell. 2014;156:109–122. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong WJ, Golic KG. Genomic deletions of the Drosophila melanogaster Hsp70 genes. Genetics. 2004;168:1467–1476. doi: 10.1534/genetics.104.030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charroux B, Royet J. Elimination of plasmatocytes by targeted apoptosis reveals their role in multiple aspects of the Drosophila immune response. Proc Natl Acad Sci USA. 2009;106:9797–9802. doi: 10.1073/pnas.0903971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Werner T, Liu G, Kang D, Ekengren S, Steiner H, et al. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97:13772–13777. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosco-Drayon V, Poidevin M, Boneca IG, Narbonne-Reveau K, Royet J, et al. Peptidoglycan sensing by the receptor PGRP-LE in the Drosophila gut induces immune responses to infectious bacteria and tolerance to microbiota. Cell Host Microbe. 2012;12:153–165. doi: 10.1016/j.chom.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Storelli G, Defaye A, Erkosar B, Hols P, Royet J, et al. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 2011;14:403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 31.Quevillon-Cheruel S, Leulliot N, Muniz CA, Vincent M, Gallay J, et al. Evf, a virulence factor produced by the Drosophila pathogen Erwinia carotovora, is an S-palmitoylated protein with a new fold that binds to lipid vesicles. J Biol Chem. 2009;284:3552–3562. doi: 10.1074/jbc.M808334200. [DOI] [PubMed] [Google Scholar]
- 32.Acosta Muniz C, Jaillard D, Lemaitre B, Boccard F. Erwinia carotovora Evf antagonizes the elimination of bacteria in the gut of Drosophila larvae. Cell Microbiol. 2007;9:106–119. doi: 10.1111/j.1462-5822.2006.00771.x. [DOI] [PubMed] [Google Scholar]
- 33.Basset A, Khush RS, Braun A, Gardan L, Boccard F, et al. The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc Natl Acad Sci USA. 2000;97:3376–3381. doi: 10.1073/pnas.070357597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haller S, Limmer S, Ferrandon D. Assessing Pseudomonas virulence with a nonmammalian host: Drosophila melanogaster. Methods Mol Biol. 2014;1149:723–740. doi: 10.1007/978-1-4939-0473-0_56. [DOI] [PubMed] [Google Scholar]
- 35.Liehl P, Blight M, Vodovar N, Boccard F, Lemaitre B. Prevalence of local immune response against oral infection in a Drosophila/Pseudomonas infection model. PLoS Pathog. 2006;2:e56. doi: 10.1371/journal.ppat.0020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vodovar N, Vallenet D, Cruveiller S, Rouy Z, Barbe V, et al. Complete genome sequence of the entomopathogenic and metabolically versatile soil bacterium Pseudomonas entomophila. Nat Biotechnol. 2006;24:673–679. doi: 10.1038/nbt1212. [DOI] [PubMed] [Google Scholar]
- 37.Vodovar N, Vinals M, Liehl P, Basset A, Degrouard J, et al. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc Natl Acad Sci USA. 2005;102:11414–11419. doi: 10.1073/pnas.0502240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchon N, Osman D, David FP, Fang HY, Boquete JP, et al. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 2013;3:1725–1738. doi: 10.1016/j.celrep.2013.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data