Abstract
Since its discovery in 2003, the type III interferon-λ (IFN-λ) family has been found to contribute significantly to the host response to infection. Whilst IFN-λ shares many features with type I IFN induction and signalling pathways, the tissue-specific restricted expression of its receptor, IL28RA, makes IFN-λ a major mediator of host innate immunity in tissues and organs with a high epithelial cell content. Host susceptibility and responses to infection are known to be heterogeneous, and the identification of common genetic variants linked to disease outcome by genome-wide association studies (GWAS) has underscored the significance of host polymorphisms in responses to infection. Several such GWAS have highlighted the IFN-λ locus on chromosome 19q13 as an area of genetic variation significantly associated with hepatitis C virus (HCV) infection, and the rs12979860 genotype can be used in clinical practice as a biomarker for predicting a successful response to treatment with pegylated IFN and ribavarin. Here, we discuss IFN-λ genetic polymorphisms and their role in HCV and other infectious diseases as well as their potential impact on clinical diagnostics, patient stratification and therapy. Finally, the broader role of IFN-λ in the immunopathogenesis of non-infectious inflammatory diseases is considered.
Key Words: Interferon-λ, IL28B, Polymorphism, Single-nucleotide polymorphism, Hepatitis C virus, Autoimmunity, Cytokines, Epithelium, Host defence, Immune response, Virology
Introduction
The host response to infection is complex, and whilst the virulence of the infecting pathogen plays a major role in the establishment and severity of disease, the contribution of host genetic variations to disease susceptibility and the outcome of infection are significant. Several diseases and disorders are known to be caused by genetic variation within components of the immune system, for example in interferon (IFN)-mediated host immunity pathways [1, 2]. Genome-wide association studies (GWAS) provide a powerful tool to link genetic polymorphisms to specific diseases and have enabled the identification of markers associated with pathogens including the human immunodeficiency virus (HIV), hepatitis C virus (HCV), dengue virus, malaria, and Mycobacterium tuberculosis[3].
In 2009, three independent GWAS found a single-nucleotide polymorphism (SNP), rs12979860, in the promoter region of IFN-λ3 (IL28B) to be significantly associated with spontaneous virus clearance and a sustained virological response (SVR) to HCV infection following treatment [4, 5, 6]. The identification of this SNP has led to considerable efforts to understand the IFN-λ locus and how its polymorphisms influence HCV infection, as well as other diseases, with the number of publications increasing from around 40 in 2010 to almost 300 in 2012 and up to 200 to date in 2014 (search terms ‘IL28B polymorphism’, ‘rs12979860’ and ‘rs8099917’).
This review provides a brief overview of the IFN-λ family, the numerous genetic polymorphisms found at this locus and the relationship between these variants and clinical infection medicine. The importance of ethnic and genetic considerations when studying disease association of SNPs will be discussed. Finally, we will explore the role of IFN-λ and its polymorphisms in other non-infectious diseases to highlight the wide-reaching implications of IFN-λs and their underlying genetics.
IFN-λ: Production, Signalling and Biological Effects
IFNs provide the first line of innate immune defence against viruses and intra-cellular bacteria and are classified into families based upon sequence homology, receptor specificity and the responses they initiate [7]. The type I IFN family (13 subtypes of IFN-α, IFN-β and the minor IFNs IFN-ε, IFN-κ and IFN-ω) bind the ubiquitously expressed IFNAR1/2 receptor complex and induce expression of IFN-stimulated genes (ISGs) via STAT1/STAT2 heterodimers or STAT1 homodimers, while the sole type II IFN, IFN-γ, produced largely by activated T cells and natural killer cells, binds the IFNGR1/2 receptor complex to induce ISGs via STAT1 homodimers only. The type III IFNs [IFN-λ1 (IL29), IFN-λ2 (IL28A), IFN-λ3 (IL28B) and the most recently discovered IFN-λ4] are a relatively new addition to the IFN family and bind a complex of the ubiquitous IL10RB receptor and the less common IL28RA receptor to induce ISGs via ISGF3 [8, 9, 10, 11].
Type III IFNs, like type I IFNs, are expressed in response to viral infection but, despite this and the commonality of activated anti-viral signal transduction pathways, IFN-λs display distinct functional roles [8, 12]. Type I IFNs cause a rapid peak and fall of ISG expression, whereas IFN-λs induce a steady increase and prolonged expression of ISGs due in part to their induction of the constituents of the transactivator ISGF3 (STAT1, STAT2 and IRF9) [13, 14, 15, 16]. As IFN-λ subtypes are differentially expressed (with IFN-λ1 requiring IRF1 and NF-κB in addition to IRF3 and IRF7), the variation in ISG activation between the subtypes is likely attributable to relative levels of IFN-λ subtypes as well as receptor binding affinities and tissue-specific expression of transcription activators (e.g. IRF1 and Med23), and repressors (e.g. Zeb1) [12, 17, 18, 19, 20]. These differences in IFN expression, receptor utilisation and subsequent ISG induction between and within the 3 IFN families suggest that they are likely to have complementary roles in host defence against invading pathogens.
Anti-Viral Effects of IFN-λ
The IFN-λ family exerts anti-viral activity against a range of RNA and DNA viruses responsible for diverse infections, including hepatitis B (HBV) and C viruses (HCV) [21], herpes simplex virus types 1 and 2 (HSV-1/2) [20, 22], cytomegalovirus [23], human herpesvirus 6B [24] and HIV [25]. As the predominant IFN induced in response to respiratory viruses [26], IFN-λ contributes considerably to the control of viral infections of the respiratory tract, including influenza A virus [27, 28, 29], human metapneumovirus and the severe acute respiratory syndrome (SARS) coronavirus [30]. Although only expressed in a minority of individuals, IFN-λ4 exerts anti-viral activities against HCV and 2 human coronaviruses, HCoV-229E and the Middle East respiratory syndrome (MERS)-CoV, despite its relatively low level of expression [9, 31]. The capacity of cells to respond to IFN-λ is determined by the expression of the IL28RA receptor subunit which is predominantly expressed on hepatocytes and cells of epithelial origin [8, 10, 32, 33]. This restricted receptor expression and the consequent tissue-specific anti-viral effects suggest a major role for IFN-λ in the protection of the epithelium from viral infection [34, 35, 36].
Anti-Tumorigenic Effects of IFN-λ
Type I IFNs are used therapeutically in the treatment of some cancers [37], and due to the overlap in their downstream signalling pathways there is mounting evidence that the IFN-λ family shares these anti-tumorigenic properties [15, 38]. IFN-λs have been demonstrated to possess pro-apoptotic and anti-metastatic effects in mouse models [39, 40] as well as in human lung cancer cell lines in vitro [41]. These findings highlight the significance of IFN-λ in the cellular defence against both invading viral pathogens and malignant host cells.
IFN-λ Polymorphisms
IFN-λ Polymorphisms and Viral Hepatitis
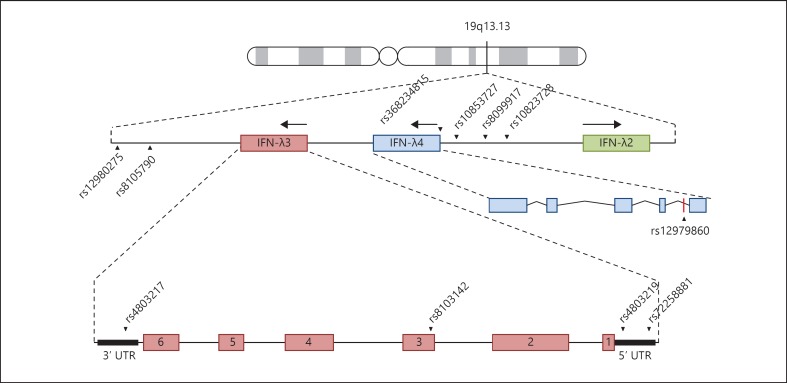
In 2009, GWAS aiming to identify genetic determinants of therapy response in HCV patients identified multiple SNPs within the IFN-λ locus on chromosome 19q13 that were linked to disease and treatment outcome [4, 5, 6]. Since then, numerous studies have verified these associations, discovered correlations with other diseases and identified more SNPs within this locus (table 1). The rs12979860 SNP is located 3 kb upstream of IFN-λ3 (fig. 1), and a CC genotype at this location is associated with both SVR to pegylated IFN-α and ribavirin treatment, and spontaneous viral clearance in chronic HCV infection [4, 42]. A similar study found a haplotype with 6 polymorphisms [rs12980275, rs8105790, rs8103142, rs10853727, rs8109886 and rs8099917 (GCCTAG)] to be overrepresented in patients who failed to respond to treatment, and in particular the minor G allele at both rs12980275 and rs8099917 was found to be strongly linked to a failure to clear infection (null virological response; NVR) [5, 6].
Table 1.
Interferon-λ polymorphisms and disease association
| SNP | Gene | Minor/major allele | Effects of the minor allele | Ethnicities implicated | References |
|---|---|---|---|---|---|
| rs12979860a | IFN-λ3 (3 kb upsteam) | T/C | HCV: decreased effectiveness of PegIFN-α/ribavirin treatment | AA, C, H, LA | 4, 103 |
| Decreased rates of SVC | A, Eu | 42 | |||
| HSV: higher rates of recurrent/more severe herpes labialis | I | 20 | |||
| Allergic disease: greater chance of being affected by allergies | C | 87 | |||
| rs8099917 | IFN-λ3 (8.9 kb upstream) | G/T | HCV: decreased effectiveness of PegIFN-α/ribavirin treatment | Au (Eu), LA | 5, 103 |
| Decreased rates of SVC in HCV | Eu, Ch | 61, 104 | |||
| HTLV-1: development of HAM/TSP | B | 81 | |||
| CMV: lower viral replication in transplant recipients | Na | 67 | |||
| rs28416813 | IFN-λ3 (distal promoter region) | G/C | HCV: decreased effectiveness of PegIFN-α/ribavirin treatment | J | 6 |
| rs8103142 | IFN-λ3 (third exon) | C/T | HCV: decreased effectiveness of PegIFN-α/ribavirin treatment | J, Au | 5, 6 |
| rs4803217 | IFN-λ3 (3′ UTR) | A/C | HCV: decreased SVC in HIV/HCV co-infected patients | Eu | 105 |
| rs368234815 (ss469415590) | Frame shift expresses IFN-X4 | AG/TT | HCV: poor response to PegIFN-α/ribavirin treatment (associated with lower rates of viral clearance) | AA, S | 9, 45 |
| rs11881222 | IFN-λ3 (7.5 kb upstream) | G/A | HCV: decreased effectiveness of PegIFN-a and ribavirin treatment | J | 6 |
| rs4803219 | IFN-λ3 (5′ UTR) | T/C | HCV: decreased effectiveness of PegIFN-α/ribavirin treatment | J | 6 |
| rs72258881 | IFN-λ3 (promoter region) | TA repeat | Number of repeats correlated with gene expression | J | 70 |
| rs8105790 | IFN-λ3 | T/C | HCV: decreased effectiveness of PegIFN-α/ribavirin treatment | J, Au | 5, 6 |
| Decreased rates of SVC | Ch | 104 | |||
| rs7248668 | Upstream of IFN-λ3 | G/A | HCV: decreased effectiveness of PegIFN-α/ribavirin treatment | J | 6 |
| rs12980275 | Downstream of IFN-λ3 | G/A | HCV: associated with NVR with PegIFN-α/ribavirin treatment | J, L | 6, 103 |
| Decreased rates of SVC | Ch | 104 | |||
| rs10853727a | IFN-λ3 (promoter region) | G/A | HCV: associated with NVR with PegIFN-α/ribavirin treatment | Au | 5 |
| rs8109886 | IFN-λ3 (promoter region) | A/C | HCV: associated with NVR with PegIFN-α/ribavirin treatment | Au | 5 |
PegIFN-a = Pegylated interferon-a; AA = African American; C = Caucasian; H = Hispanic; LA = Latin American; Eu = European; Ch = Chinese; J = Japanese; Au = Australian; I = Italian; B = Brazilian; NA = North American; A = African; S = Swiss.
Originally identified in the IFN-λ3 promoter.
Fig. 1.
Location of SNPs in the IFN-λ gene locus. Schematic representation of the IFNL locus in chromosome 19q13, with key SNPs within non-coding and coding regions highlighted. SNPs discussed throughout this review are highlighted.
Following the identification of rs12979860 as a strong predictor of therapy outcome in HCV patients, studies in other cohorts found alternative SNPs to be better predictors of treatment success. rs8099917 is the strongest predictor of disease outcome in the Japanese population, and an increased number of TA dinucleotide repeats at rs72258881, in the promoter region of IFN-λ3, is also strongly linked to a successful viral response in Japanese individuals [43, 44]. The dinucleotide variant (TT/ΔG) rs368234815 (or ss469415590) has recently been identified, and the ΔG frame shift allele, which leads to expression of IFN-λ4, is the major variant in Africans and the strongest known host factor for predicting failure to clear HCV in this population [9]. The homozygous TT genotype at this locus also has a strong positive correlation with HCV clearance and treatment outcome [45, 46, 47].
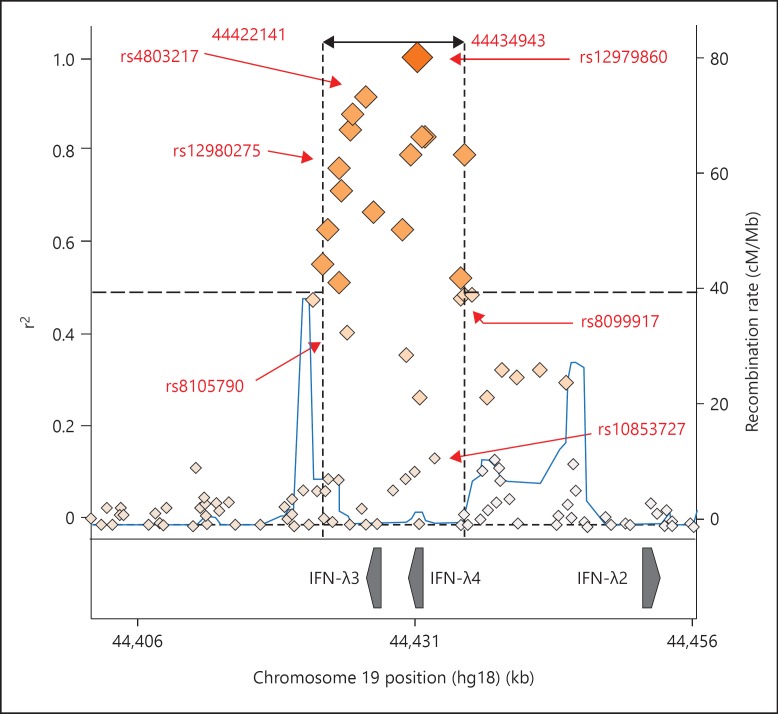
Linkage disequilibrium (LD) between SNPs found within the IFN-λ region is a prominent feature and makes it difficult to assign a causative role to individual SNPs, particularly if they are far apart. In sickle cell anaemia, for example, a functional SNP has been found in high LD with SNPs up to 2.5 Mbp away [48]. In the context of IFN-λ3 polymorphisms, the high LD observed between rs12979860 and rs368234815 (r2 = 0.988) gives the latter SNP limited use as a marker in this population (fig. 2) [49]. However, the evidence that the rs368234815 genotype actually provides a better prediction of therapy outcome in Caucasians co-infected with HIV-1 and HCV highlights the difficulties in forming reliable associations between disease and SNPs [50].
Fig. 2.
Regional LD plot with rs12979860. An LD plot was generated which highlights all SNPs within 50 bp up or downstream and in LD (r2 > 0.5) with rs12979860. SNPs discussed in this review, for which LD data is available, are highlighted. The plot was generated with SNP Annotation and Proxy Search (SNAP) using the 1000 Genomes Pilot 1 data set [101].
The ability of cells to respond to IFN-λ is dependent on the expression of the receptor IL28RA, and there is some evidence that SNPs on this gene are linked to chronic HCV infection. Whilst the major allele at rs10903035 (AA) was found to be overrepresented in a persistently infected, albeit untreated, at-risk group in the Chinese population, the minor G allele has been linked to PEG-IFN-α treatment failure in chronic HCV [51, 52]. Cui et al. [51] also found the rs11249006 GG allele to be linked to a reduced susceptibility to persistent HCV; however, a later study in the Chinese Han population found no such link [53]. Further investigation in much larger sample groups is required to fully understand the role of IL28RA polymorphisms, if any, in disease.
Whilst the impact of IFN-λ and its SNPs on HCV is clear, its role in HBV infection remains controversial despite increasing global interest [reviewed in [54]]. Several SNPs have been studied in HBV patient cohorts. The CG rs12979860/rs8099917 haplotype block was found to be associated with HBV seroclearance [55], and both rs12979860 and rs1298025 have been positively linked to response to pegylated IFN-α therapy [56, 57]. In addition, the rs10853728 SNP [in low LD with rs8105790 (68%), rs12979860 (70%) and rs8099917 (68%)] has been linked to chronic hepatitis in HBeAg-negative patients (p = 0.032), suggesting a potential role for IFN-λ in hepatic inflammation in HBV patients [58]. However, as many other reports have found no links between IFN-λ and chronic HBV infection, a consensus on its role is yet to be reached [59, 60].
Mechanisms of Action
In the area of HCV, significant associations have been made between SNPs at the IFN-λ locus, disease susceptibility and treatment outcome; however, the mechanisms via which these polymorphisms functionally influence innate immunity remain ill defined. Of the numerous SNPs within the IFN-λ locus (fig. 1), many are located in the promoter region and could directly influence protein expression by altering the transcription factor binding affinity or chromatin accessibility [61, 62]. For example, SNPs rs12978960, rs4803221 and rs368234815 are proposed to be located within a CpG island [17, 45, 62, 63]. Methylation and deamination at CpG islands cause epigenetic changes promoting the recruitment of proteins associated with less transcriptionally active heterochromatic DNA and thus may alter IFN-λ3 expression. Whilst a recent investigation into the transcriptional regulation of IFN-λ suggested that the majority of the control occurs within the first kilo base pair upstream of IL28B, excluding both rs12979860 and rs8099917, it is well established that the risk alleles at these SNPs (T and G, respectively) alter the mRNA and protein expression in peripheral blood mononuclear cells, sera and the liver in HCV patients as well as healthy individuals [5, 6, 64, 65, 66, 67, 68, 69].
In addition to being located within a CpG island, the ΔG allele at rs368234815 is associated with expression of IFN-λ4 and a reduced expression of IFN-λ3 mRNA. The high LD between this and SNP rs12979860, and the combined physiological effects of reduced IFN-λ3, increased IFN-λ4 expression and the associated feedback mechanisms remains unclear. The promoter polymorphism rs72258881 consists of a TA nucleotide repeat, and whilst evidence suggests that the increasing length of TA repeats results in a higher level of transcriptional activity of the IFN-λ3, the mechanism is undefined [70].
Recently, host polymorphism-dependent expression of IFN-λ has been linked to the HCV-induced micro-RNA (miRNA) miR-122, which is highly expressed in the liver. High levels of miR-122 are linked to a protective CC genotype in rs12979860 and SVR in chronic HCV [71, 72]. Although miR-122 increases HCV RNA replication in vitro, a correlation has been found between high miR-122 and a reduced HCV viral load in the liver [71]. The favourable G allele at a second polymorphism in the 3′ untranslated region (UTR) of IL28B, rs4803217, was found to be resistant to post-transcriptional regulation and AU-rich element-mediated decay via HCV-induced miR-208b and miR-499a-5p [73]. These studies provide enticing evidence for the interplay between host regulatory elements, genetic polymorphisms and disease control.
IFN-λ Polymorphisms and HCV Treatment
Following the original GWAS studies, and numerous subsequent independent re-validations, a patient's IL28B genotype is known to be associated with their likelihood of achieving an SVR to pegylated IFN and ribavarin therapy. Current guidelines recommend IL28B genotype testing if the results would influence management plans [74]. Considering the substantial side effect burden of IFN therapy, patients may be reluctant to start treatment, but knowledge of their IL28B genotype and likely response to treatment can facilitate a more informed decision. However, the truly remarkable efficacy of new direct-acting anti-viral agents for the treatment of genotype 1, 2 and 3 HCV infections means that the pegylated IFN and ribavarin therapy regime is now all but obsolete [75, 76]. It remains to be seen whether IL28B genotype will also be predictive of the responsiveness to these new direct-acting anti-viral agents.
IFN-λ Polymorphisms and Other Infectious Diseases
In addition to their role in HCV infection, IFN-λ SNPs have been investigated in the context of other infectious diseases. Following the identification of Med23 as an anti-viral host factor in HSV-1 infection in vitro, the IL28B genotype was explored in patients suffering recurrent orofacial herpes. The minor T allele at rs12979860 was found to be associated with the severity and frequency of oral herpes labialis recurrence [20]. A more recent study in HIV-infected patients found that individuals homozygous for the ΔG mutation at SNP rs368234815 had a higher incidence of developing CMV retinitis [77]. Infection of retinal epithelial cells by CMV causes retinal damage resulting in blindness, and the reduced IFN-λ3 expression as a result of this ΔG mutation may contribute to higher replication levels of CMV and an increased likelihood of developing retinitis. IFN-λ SNPs have also been investigated in the context of HIV ‘controllers’ who suppress the HIV viral load in the absence of therapy. One such study found the protective rs12979860 CC genotype to be associated with spontaneous HIV control; however, in addition to having very low numbers in comparison to non-controllers (53 vs. 389), this group also had a high rate of HCV co-infection, making firm conclusions difficult [78].
Genetic variation in IFN-λs has also been linked to infection with 2 potentially oncogenic viruses: human papilloma virus and human T-lymphotropic leukaemia virus (HTLV) type I. A genetic association study found SNPs in IFN-λ2 and IFN-λ3 to be linked to human papilloma virus persistence and cervical cancer, albeit with a low significance [79]. The risk alleles at rs12979860 (CT) and rs8099917 (GG) have been linked to the incidence of HTLV-1-associated myelopathy/tropical spastic paraparesis [80, 81]. However, another study failed to link the rs8099917 genotype and disease, even though the protective allele (TT) was associated with higher IL28B levels in HTLV-1 mono-infected patients in comparison to HTLV-1/HCV co-infected patients [82]. As a virus predominantly infecting CD4+ T-lymphocytes, the relevance of IFN-λ polymorphisms and generally a type III IFN response in HTLV-1 infection and associated inflammatory diseases is not clear, and further studies in larger cohorts are required to fully understand this and the role of IFN-λ in the development of virus-induced cancers.
Despite being the major IFN expressed in airway cells in response to infection [26, 27, 28], surprisingly little is known about the effect of IFN-λ SNPs on respiratory infectious diseases. One study exploring the SNPs rs12979860 and rs8099917 in infants hospitalised with RSV-associated bronchiolitis found no link to the course of clinical infection or viral load even though a significant association between raised IFN-λ1 levels in airway secretions and the severity of disease in infants hospitalised due to RSV bronchiolitis has been observed [83, 84]. This apparent negative link between IFN-λ and disease prognosis raises the possibility that overexpression of IFN-λ may actually be detrimental to patient health, particularly in the context of respiratory infections. In such infections, the predominant induction of IFN-λ may contribute significantly to the phenomenon of hypercytokinaemia, acute respiratory distress syndrome and respiratory failure, necessitating mechanical ventilation in life-threatening disease [85].
IFN-λ Polymorphisms, Inflammation and Allergic Disease
IFN-λs contribute to inflammatory responses, and thus polymorphisms which alter IFN-λ expression may have a role in the immunopathogenesis of inflammatory diseases [86]. IFN-λ suppresses allergic asthma in mouse models as a result of promoting Th1 and suppressing Th2 responses [88, 89], and bronchial epithelial cells from asthmatic patients are impaired in their ability to express IFN-λ in response to infection [90]. It is possible, therefore, that polymorphisms that reduce IFN-λ levels are responsible for a dominance of Th2 cells and the resultant inflammatory and allergic diseases, and one study found the risk T allele at rs12979860 to be overrepresented in a cohort of allergy sufferers [87]. However, contradictory reports of elevated IFN-λ levels in asthma patients [91, 92] indicate the need for further studies to elucidate the role of IFN-λ and its polymorphisms in inflammatory disease.
IFN-λ expression and signalling are also linked to the autoimmune disease systemic lupus erythematosus (SLE), with higher levels of IFN-λ (mRNA and protein), and its subsequent stimulation of cytokine expression, in SLE patients thought to contribute to the observed inflammation and associated organ damage in SLE disease [93, 94]. Interestingly, the IL28RA SNP rs4649203 minor allele is linked to an increased risk of SLE in the Chinese Han population [96] and, whilst the effect of this polymorphism is not known, its location in the 3′ UTR suggests that it may influence receptor expression. Whilst the underlying cause of SLE remains undefined, an infectious agent has long been suspected [reviewed in [96]]. It is unsurprising, therefore, to find links between host polymorphisms influencing IFN-λ pathways and SLE, and the role of IFN-λ in inflammatory and autoimmune diseases warrants further investigation.
IFN-λ Polymorphisms and Disease Association: Ethnicity and Gender Considerations
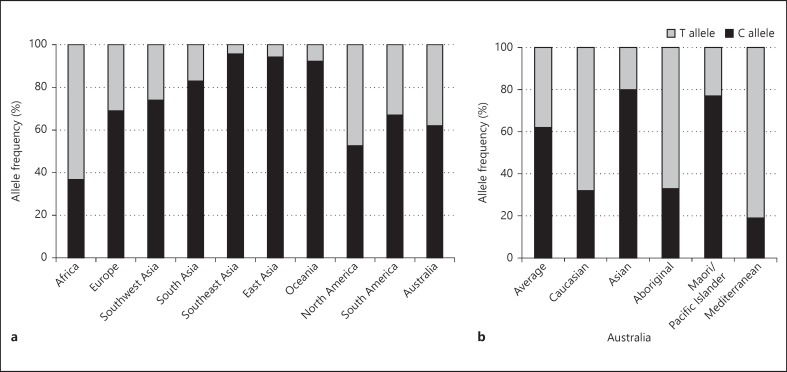
The most studied SNP in the IFN-λ locus, rs12979860, shows extensive geographical variation in allele frequencies, with the C allele, associated with clearance of HCV, present at very high (>90%) rates in East-Asian populations and at low rates (<40%) in African populations (fig. 3a). This disparity in C allele frequency correlates with the observed clinical differences between these populations, where East Asians have a more than 3 times greater rate of HCV clearance [42, 97]. The ethnicity-dependent distribution of the protective CC genotype also partially explains the differences in SVR between African Americans and European Americans, where chronic infection occurs more often in people of African descent (76.4%) compared to Caucasian populations (44%) [4]. Such variation in allele frequencies is also likely to contribute to the global variations in ‘predictor’ genotypes, such as the SNPs rs8099917 and rs368134815 being the strongest predictors of HCV therapy outcome in Japanese and African populations, respectively [9, 43]. Intra-regional variation in SNP allele frequencies is also noted, with CC frequencies ranging from 80% in Asian Australians to 19% in Mediterranean Australians (fig. 3b) [98], and this high level of variation even within the relatively small population of Australians (∼23 million) should be considered when utilising genotypes for patient stratification.
Fig. 3.
Geographical variations in rs12979860 allele frequency. Comparison of the relative frequency of C and T alleles in SNP rs12979860 in distinct worldwide regions (a) and in specific ethnic groups within Australia (b). Africa includes African Americans; Europe includes European Americans, and South America excludes the Ticuna population [adapted from [42, 98, 102]].
The observed correlation between geographically distinct genotypes and chronic disease prevalence provides evolutionary evidence for maintenance of the protective alleles in the IFN-λ SNPs, i.e. maintenance of a CC at rs12989760 in areas endemic for chronic HCV. However, this does not explain the high prevalence of the so-called ‘risk’ T allele at this SNP in populations such as the Australian Aboriginal and Mediterranean populations. In some well-known genetic disorders, such as sickle cell anaemia, individuals homozygous for the mutation in question suffer severe disease, whilst the heterozygosity actually confers a level of protection against malaria. It is possible, therefore, that the risk allele at rs12979860 may confer some as yet unidentified genetic advantage to some populations whilst simultaneously rendering others susceptible to chronic diseases such as HCV.
Interestingly, there is also evidence for a gender bias with respect to the association between IL28B SNPs and HCV. The rs12979860 genotype has been found to be a more reliable predictor of a favourable outcome of HCV infection in the female gender [42]. This gender bias was supported by a later study showing a similar rate of spontaneous viral clearance in women with TT/CT alleles and men with the favourable CC genotype [99]. Such a gender bias has also been observed in allergic disease, where a study of the rs12979860 SNP found that female children with the ‘protective’ CC genotype had decreased rates of allergy-associated epidermis symptoms [87].
Concluding Remarks
Since its discovery in 2003, there has been an explosion of data describing the possible roles type III IFN in infectious and non-infectious human disease. Whilst the assessment of host genetics contributes to diagnostic and therapy options in oncology, this is surprisingly underexploited in other fields of medicine. The IL28B polymorphism rs12979860 is the only host SNP to be used as a genetic biomarker for infectious diseases, and genotyping at this locus is incorporated into patient stratification for the clinical management of HCV infection [100]. Whilst the link between IFN-λ polymorphisms and HCV infection is the most robust and well described, this review has highlighted a number of other infectious and inflammatory diseases in which polymorphisms affecting IFN-λ expression or signalling have been implicated. The data for many of these diseases, however, remain unconvincing. Many studies have been limited by often very small sample sizes and small effect sizes, and variations in patient characteristics make inter-study comparisons difficult. Given the levels of intra-regional genotype variation within some populations, caution should be used when interpreting the population stratification in such association studies, particularly in those with relatively high p values. Finally, the existence of LD between SNPs within the IFN-λ locus makes it difficult to assign causality to an individual polymorphism. However, it seems likely that host polymorphisms affecting the IFN-λ signalling axis will play a key role in the advancing field of novel host biomarkers for patient management and differential therapy.
References
- 1.Thio CL, Thomas DL, Carrington M. Chronic viral hepatitis and the human genome. Hepatology. 2000;31:819–827. doi: 10.1053/he.2000.4316. [DOI] [PubMed] [Google Scholar]
- 2.Zhang SY, Boisson-Dupuis S, Chapgier A, Yang K, Bustamante J, Puel A, Picard C, Abel L, Jouanguy E, Casanova JL. Inborn errors of interferon (IFN)-mediated immunity in humans: insights into the respective roles of IFN-alpha/beta, IFN-gamma, and IFN-lambda in host defense. Immunol Rev. 2008;226:29–40. doi: 10.1111/j.1600-065X.2008.00698.x. [DOI] [PubMed] [Google Scholar]
- 3.Chapman SJ, Hill AV. Human genetic susceptibility to infectious disease. Nat Rev Genet. 2012;13:175–188. doi: 10.1038/nrg3114. [DOI] [PubMed] [Google Scholar]
- 4.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 5.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E, Riordan S, Sheridan D, Smedile A, Fragomeli V, Muller T, Bahlo M, Stewart GJ, Booth DR, George J. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, Ito Y, Mita E, Tanaka E, Mochida S, Murawaki Y, Honda M, Sakai A, Hiasa Y, Nishiguchi S, Koike A, Sakaida I, Imamura M, Ito K, Yano K, Masaki N, Sugauchi F, Izumi N, Tokunaga K, Mizokami M. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 7.Taniguchi T, Takaoka A. The interferon-alpha/beta system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr Opin Immunol. 2002;14:111–116. doi: 10.1016/s0952-7915(01)00305-3. [DOI] [PubMed] [Google Scholar]
- 8.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. ITN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 9.Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I, Chen S, Brand N, Tarway M, Liu L, Sheikh F, Astemborski J, Bonkovsky HL, Edlin BR, Howell CD, Morgan TR, Thomas DL, Rehermann B, Donnelly RP, O'Brien TR. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, Ostrander C, Dong D, Shin J, Presnell S, Fox B, Haldeman B, Cooper E, Taft D, Gilbert T, Grant FJ, Tackett M, Krivan W, McKnight G, Clegg C, Foster D, Klucher KM. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Z, Hamming OJ, Ank N, Paludan SR, Nielsen AL, Hartmann R. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J Virol. 2007;81:7749–7758. doi: 10.1128/JVI.02438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Onoguchi K, Yoneyama M, Takemura A, Akira S, Taniguchi T, Namiki H, Fujita T. Viral infections activate types I and III interferon genes through a common mechanism. J Biol Chem. 2007;282:7576–7581. doi: 10.1074/jbc.M608618200. [DOI] [PubMed] [Google Scholar]
- 13.Diegelmann J, Beigel F, Zitzmann K, Kaul A, Goke B, Auernhammer CJ, Bartenschlager R, Diepolder HM, Brand S. Comparative analysis of the lambda-interferons IL-28A and IL-29 regarding their transcriptome and their antiviral properties against hepatitis C virus. PLoS One. 2010;5:e15200. doi: 10.1371/journal.pone.0015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, Rice CM. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 15.Maher SG, Sheikh F, Scarzello AJ, Romero-Weaver AL, Baker DP, Donnelly RP, Gamero AM. IFNalpha and IFNlambda differ in their antiproliferative effects and duration of JAK/STAT signaling activity. Cancer Biol Ther. 2008;7:1109–1115. doi: 10.4161/cbt.7.7.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu BS, Janssen HL, Boonstra A. IL-29 and IFNalpha differ in their ability to modulate IL-12 production by TLR-activated human macrophages and exhibit differential regulation of the IFNgamma receptor expression. Blood. 2011;117:2385–2395. doi: 10.1182/blood-2010-07-298976. [DOI] [PubMed] [Google Scholar]
- 17.Lee HC, Narayanan S, Park SJ, Seong SY, Hahn YS. Transcriptional regulation of IFN-lambda genes in hepatitis C virus-infected hepatocytes via IRF-3·IRF-7·NF-κB complex. J Biol Chem. 2014;289:5310–5319. doi: 10.1074/jbc.M113.536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel R, Eskdale J, Gallagher G. Regulation of IFN-lambda1 promoter activity (IFN-lambda1/IL-29) in human airway epithelial cells. J Immunol. 2011;187:5636–5644. doi: 10.4049/jimmunol.1003988. [DOI] [PubMed] [Google Scholar]
- 19.Swider A, Siegel R, Eskdale J, Gallagher G. Regulation of interferon lambda-1 (IFNL1/IFN-lambda1/IL-29) expression in human colon epithelial cells. Cytokine. 2014;65:17–23. doi: 10.1016/j.cyto.2013.09.020. [DOI] [PubMed] [Google Scholar]
- 20.Griffiths SJ, Koegl M, Boutell C, Zenner HL, Crump CM, Pica F, Gonzalez O, Friedel CC, Barry G, Martin K, Craigon MH, Chen R, Kaza LN, Fossum E, Fazakerley JK, Efstathiou S, Volpi A, Zimmer R, Ghazal P, Haas J. A systematic analysis of host factors reveals a Med23-interferon-lambda regulatory axis against herpes simplex virus type 1 replication. PLoS Pathog. 2013;9:e1003514. doi: 10.1371/journal.ppat.1003514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79:3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006;80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brand S, Beigel F, Olszak T, Zitzmann K, Eichhorst ST, Otte JM, Diebold J, Diepolder H, Adler B, Auernhammer CJ, Goke B, Dambacher J. IL-28A and IL-29 mediate antiproliferative and antiviral signals in intestinal epithelial cells and murine cmv infection increases colonic IL-28A expression. Am J Physiol Gastrointest Liver Physiol. 2005;289:G960–G968. doi: 10.1152/ajpgi.00126.2005. [DOI] [PubMed] [Google Scholar]
- 24.Nordstrom I, Eriksson K. HHV-6B induces IFN-lambda1 responses in cord plasmacytoid dendritic cells through TLR9. PLoS One. 2012;7:e38683. doi: 10.1371/journal.pone.0038683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou W, Wang X, Ye L, Zhou L, Yang ZQ, Riedel E, Ho WZ. Lambda interferon inhibits human immunodeficiency virus type 1 infection of macrophages. J Virol. 2009;83:3834–3842. doi: 10.1128/JVI.01773-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okabayashi T, Kojima T, Masaki T, Yokota S, Imaizumi T, Tsutsumi H, Himi T, Fujii N, Sawada N. Type-III interferon, not type-I, is the predominant interferon induced by respiratory viruses in nasal epithelial cells. Virus Res. 2011;160:360–366. doi: 10.1016/j.virusres.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 27.Jewell NA, Cline T, Mertz SE, Smirnov SV, Flano E, Schindler C, Grieves JL, Durbin RK, Kotenko SV, Durbin JE. Lambda interferon is the predominant interferon induced by influenza a virus infection in vivo. J Virol. 2010;84:11515–11522. doi: 10.1128/JVI.01703-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khaitov MR, Laza-Stanca V, Edwards MR, Walton RP, Rohde G, Contoli M, Papi A, Stanciu LA, Kotenko SV, Johnston SL. Respiratory virus induction of alpha-, beta- and lambda-interferons in bronchial epithelial cells and peripheral blood mononuclear cells. Allergy. 2009;64:375–386. doi: 10.1111/j.1398-9995.2008.01826.x. [DOI] [PubMed] [Google Scholar]
- 29.Mordstein M, Kochs G, Dumoutier L, Renauld JC, Paludan SR, Klucher K, Staeheli P. Interferon-lambda contributes to innate immunity of mice against influenza a virus but not against hepatotropic viruses. PLoS Pathog. 2008;4:e1000151. doi: 10.1371/journal.ppat.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, Sorgeloos F, Ehl S, Mayer D, Kochs G, Schwemmle M, Gunther S, Drosten C, Michiels T, Staeheli P. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol. 2010;84:5670–5677. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamming OJ, Terczynska-Dyla E, Vieyres G, Dijkman R, Jorgensen SE, Akhtar H, Siupka P, Pietschmann T, Thiel V, Hartmann R. Interferon lambda 4 signals via the IFNlambda receptor to regulate antiviral activity against HCV and coronaviruses. EMBO J. 2013;32:3055–3065. doi: 10.1038/emboj.2013.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB, Dagnaes-Hansen F, Thomsen AR, Chen Z, Haugen H, Klucher K, Paludan SR. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J Immunol. 2008;180:2474–2485. doi: 10.4049/jimmunol.180.4.2474. [DOI] [PubMed] [Google Scholar]
- 33.Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pott J, Mahlakoiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, Staeheli P, Hornef MW. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci U S A. 2011;108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russell CD, Griffiths SJ, Haas J. Interferon lambda genetic polymorphisms and viral infection: the tip of the iceberg? DNA Cell Biol. 2014;33:60–63. doi: 10.1089/dna.2013.2261. [DOI] [PubMed] [Google Scholar]
- 36.Pica F, Volpi A, Gaziano R, Garaci E. Interferon-lambda in immunocompetent individuals with a history of recurrent herpes labialis. Antivir Ther. 2010;15:737–743. doi: 10.3851/IMP1610. [DOI] [PubMed] [Google Scholar]
- 37.Pasquali S, Mocellin S. The anticancer face of interferon alpha (IFN-alpha): from biology to clinical results, with a focus on melanoma. Curr Med Chem. 2010;17:3327–3336. doi: 10.2174/092986710793176393. [DOI] [PubMed] [Google Scholar]
- 38.Dumoutier L, Tounsi A, Michiels T, Sommereyns C, Kotenko SV, Renauld JC. Role of the interleukin (IL)-28 receptor tyrosine residues for antiviral and antiproliferative activity of IL-29/interferon-lambda 1: similarities with type I interferon signaling. J Biol Chem. 2004;279:32269–32274. doi: 10.1074/jbc.M404789200. [DOI] [PubMed] [Google Scholar]
- 39.Ding S, Khoury-Hanold W, Iwasaki A, Robek MD. Epigenetic reprogramming of the type III interferon response potentiates antiviral activity and suppresses tumor growth. PLoS Biol. 2014;12:e1001758. doi: 10.1371/journal.pbio.1001758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Numasaki M, Tagawa M, Iwata F, Suzuki T, Nakamura A, Okada M, Iwakura Y, Aiba S, Yamaya M. IL-28 elicits antitumor responses against murine fibrosarcoma. J Immunol. 2007;178:5086–5098. doi: 10.4049/jimmunol.178.8.5086. [DOI] [PubMed] [Google Scholar]
- 41.Tezuka Y, Endo S, Matsui A, Sato A, Saito K, Semba K, Takahashi M, Murakami T. Potential anti-tumor effect of IFN-lambda2 (IL-28A) against human lung cancer cells. Lung Cancer. 2012;78:185–192. doi: 10.1016/j.lungcan.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 42.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito K, Higami K, Masaki N, Sugiyama M, Mukaide M, Saito H, Aoki Y, Sato Y, Imamura M, Murata K, Nomura H, Hige S, Adachi H, Hino K, Yatsuhashi H, Orito E, Kani S, Tanaka Y, Mizokami M. The rs8099917 polymorphism, when determined by a suitable genotyping method, is a better predictor for response to pegylated alpha interferon/ribavirin therapy in Japanese patients than other single nucleotide polymorphisms associated with interleukin-28B. J Clin Microbiol. 2011;49:1853–1860. doi: 10.1128/JCM.02139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masaki N, Sugiyama M, Shimada N, Tanaka Y, Nakamuta M, Izumi N, Watanabe S, Tsubota A, Komatsu M, Masaki T, Enomoto N, Yoneda M, Murata K, Ito K, Koike K, Mizokami M. Pretreatment prediction of the outcome of response-guided peginterferon-alpha and ribavirin therapy for chronic hepatitis C. J Gastroenterol Hepatol. 2014;29:1996–2005. doi: 10.1111/jgh.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bibert S, Roger T, Calandra T, Bochud M, Cerny A, Semmo N, Duong FH, Gerlach T, Malinverni R, Moradpour D, Negro F, Mullhaupt B, Bochud PY. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med. 2013;210:1109–1116. doi: 10.1084/jem.20130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujino H, Imamura M, Nagaoki Y, Kawakami Y, Abe H, Hayes CN, Kan H, Fukuhara T, Kobayashi T, Masaki K, Ono A, Nakahara T, Honda Y, Naeshiro N, Urabe A, Yokoyama S, Miyaki D, Murakami E, Kawaoka T, Hiraga N, Tsuge M, Hiramatsu A, Hyogo H, Aikata H, Takahashi S, Miki D, Ochi H, Ohishi W, Chayama K. Predictive value of the IFNL4 polymorphism on outcome of telaprevir, peginterferon, and ribavirin therapy for older patients with genotype 1B chronic hepatitis C. J Gastroenterol. 2014;49:1548–1556. doi: 10.1007/s00535-013-0924-9. [DOI] [PubMed] [Google Scholar]
- 47.Meissner EG, Bon D, Prokunina-Olsson L, Tang W, Masur H, O'Brien TR, Herrmann E, Kottilil S, Osinusi A. IFNL4-ΔG genotype is associated with slower viral clearance in hepatitis C, genotype-1 patients treated with sofosbuvir and ribavirin. J Infect Dis. 2014;209:1700–1704. doi: 10.1093/infdis/jit827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stattermayer AF, Strassl R, Maieron A, Rutter K, Stauber R, Strasser M, Beinhardt S, Datz C, Scherzer TM, Steindl-Munda P, Gschwantler M, Trauner M, Hofer H, Ferenci P. Polymorphisms of interferon-λ4 and IL28B - effects on treatment response to interferon/ribavirin in patients with chronic hepatitis C. Aliment Pharmacol Ther. 2014;39:104–111. doi: 10.1111/apt.12547. [DOI] [PubMed] [Google Scholar]
- 50.Franco S, Aparicio E, Parera M, Clotet B, Tural C, Martinez MA. IFNL4 ss469415590 variant is a better predictor than rs12979860 of pegylated interferon-alpha/ribavirin therapy failure in hepatitis C virus/HIV-1 coinfected patients. AIDS. 2014;28:133–136. doi: 10.1097/QAD.0000000000000052. [DOI] [PubMed] [Google Scholar]
- 51.Cui Q, Zhang YX, Su J, Chen X, Ding K, Lei N, Liu Y, Li J, Zhang Y, Yu RB. Genetic variation in IL28RA is associated with the outcomes of HCV infection in a high-risk Chinese population. Infect Genet Evol. 2011;11:1682–1689. doi: 10.1016/j.meegid.2011.06.016. [DOI] [PubMed] [Google Scholar]
- 52.Jimenez-Sousa MA, Berenguer J, Rallon N, Guzman-Fulgencio M, Lopez JC, Soriano V, Fernandez-Rodriguez A, Cosin J, Restrepo C, Garcia-Alvarez M, Miralles P, Benito JM, Resino S. IL28RA polymorphism is associated with early hepatitis C virus (HCV) treatment failure in human immunodeficiency virus-/HCV-coinfected patients. J Viral Hepat. 2013;20:358–366. doi: 10.1111/jvh.12041. [DOI] [PubMed] [Google Scholar]
- 53.Jin G, Kang H, Chen X, Dai D. Evaluation of the relationship between IL28B, IL10RB and IL28RA single-nucleotide polymorphisms and susceptibility to hepatitis C virus in Chinese Han population. Infect Genet Evol. 2014;21:8–14. doi: 10.1016/j.meegid.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi T. Interleukin 28B genetic polymorphism and hepatitis B virus infection. World J Gastroenterol. 2014;20:12026–12030. doi: 10.3748/wjg.v20.i34.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seto WK, Wong DK, Kopaniszen M, Proitsi P, Sham PC, Hung IF, Fung J, Lai CL, Yuen MF. HLA-DP and IL28B polymorphisms: Influence of host genome on hepatitis B surface antigen seroclearance in chronic hepatitis B. Clin Infect Dis. 2013;56:1695–1703. doi: 10.1093/cid/cit121. [DOI] [PubMed] [Google Scholar]
- 56.Sonneveld MJ, Wong VW, Woltman AM, Wong GL, Cakaloglu Y, Zeuzem S, Buster EH, Uitterlinden AG, Hansen BE, Chan HL, Janssen HL. Polymorphisms near IL28B and serologic response to peginterferon in HBeAg-positive patients with chronic hepatitis B. Gastroenterology. 2012;142:513–520. doi: 10.1053/j.gastro.2011.11.025. e1. [DOI] [PubMed] [Google Scholar]
- 57.Lampertico P, Vigano M, Cheroni C, Facchetti F, Invernizzi F, Valveri V, Soffredini R, Abrignani S, De Francesco R, Colombo M. IL28B polymorphisms predict interferon-related hepatitis B surface antigen seroclearance in genotype D hepatitis B e antigen-negative patients with chronic hepatitis B. Hepatology. 2013;57:890–896. doi: 10.1002/hep.25749. [DOI] [PubMed] [Google Scholar]
- 58.Lee IC, Lin CH, Huang YH, Huo TI, Su CW, Hou MC, Huang HC, Lee KC, Chan CC, Lin MW, Lin HC, Lee SD. IL28B polymorphism correlates with active hepatitis in patients with HBeAg-negative chronic hepatitis B. PLoS One. 2013;8:e58071. doi: 10.1371/journal.pone.0058071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Galmozzi E, Vigano M, Lampertico P. Systematic review with meta-analysis: Do interferon lambda 3 polymorphisms predict the outcome of interferon-therapy in hepatitis B infection? Aliment Pharmacol Ther. 2014;39:569–578. doi: 10.1111/apt.12631. [DOI] [PubMed] [Google Scholar]
- 60.Lee DH, Lee JH, Kim YJ, Park NH, Cho Y, Lee YB, Yoo JJ, Lee M, Cho Y, Choi WM, Yu SJ, Yoon JH, Kim CY, Lee HS. Relationship between polymorphisms near the IL28B gene and spontaneous HBsAg seroclearance: a systematic review and meta-analysis. J Viral Hepat. 2014;21:163–170. doi: 10.1111/jvh.12193. [DOI] [PubMed] [Google Scholar]
- 61.Rauch A, Rohrbach J, Bochud PY. The recent breakthroughs in the understanding of host genomics in hepatitis C. Eur J Clin Invest. 2010;40:950–959. doi: 10.1111/j.1365-2362.2010.02337.x. [DOI] [PubMed] [Google Scholar]
- 62.Smith KR, Suppiah V, O'Connor K, Berg T, Weltman M, Abate ML, Spengler U, Bassendine M, Matthews G, Irving WL, Powell E, Riordan S, Ahlenstiel G, Stewart GJ, Bahlo M, George J, Booth DR. Identification of improved IL28B SNPs and haplotypes for prediction of drug response in treatment of hepatitis C using massively parallel sequencing in a cross-sectional European cohort. Genome Med. 2011;3:57. doi: 10.1186/gm273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayes CN, Imamura M, Aikata H, Chayama K. Genetics of IL28B and HCV - response to infection and treatment. Nat Rev Gastroenterol Hepatol. 2012;9:406–417. doi: 10.1038/nrgastro.2012.101. [DOI] [PubMed] [Google Scholar]
- 64.Langhans B, Kupfer B, Braunschweiger I, Arndt S, Schulte W, Nischalke HD, Nattermann J, Oldenburg J, Sauerbruch T, Spengler U. Interferon-lambda serum levels in hepatitis C. J Hepatol. 2011;54:859–865. doi: 10.1016/j.jhep.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 65.Fukuhara T, Taketomi A, Motomura T, Okano S, Ninomiya A, Abe T, Uchiyama H, Soejima Y, Shirabe K, Matsuura Y, Maehara Y. Variants in IL28B in liver recipients and donors correlate with response to peg-interferon and ribavirin therapy for recurrent hepatitis C. Gastroenterology. 2010;139:1577–1585. doi: 10.1053/j.gastro.2010.07.058. 1585.e1-3. [DOI] [PubMed] [Google Scholar]
- 66.Duggal P, Thio CL, Wojcik GL, Goedert JJ, Mangia A, Latanich R, Kim AY, Lauer GM, Chung RT, Peters MG, Kirk GD, Mehta SH, Cox AL, Khakoo SI, Alric L, Cramp ME, Donfield SM, Edlin BR, Tobler LH, Busch MP, Alexander G, Rosen HR, Gao X, Abdel-Hamid M, Apps R, Carrington M, Thomas DL. Genome-wide association study of spontaneous resolution of hepatitis C virus infection: data from multiple cohorts. Ann Intern Med. 2013;158:235–245. doi: 10.7326/0003-4819-158-4-201302190-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Egli A, Levin A, Santer DM, Joyce M, O'Shea D, Thomas BS, Lisboa LF, Barakat K, Bhat R, Fischer KP, Houghton M, Tyrrell DL, Kumar D, Humar A. Immunomodulatory function of interleukin 28B during primary infection with cytomegalovirus. J Infect Dis. 2014;210:717–727. doi: 10.1093/infdis/jiu144. [DOI] [PubMed] [Google Scholar]
- 68.Honda M, Shirasaki T, Shimakami T, Sakai A, Horii R, Arai K, Yamashita T, Sakai Y, Okada H, Murai K, Nakamura M, Mizukoshi E, Kaneko S. Hepatic interferon-stimulated genes are differentially regulated in the liver of chronic hepatitis C patients with different interleukin-28B genotypes. Hepatology. 2014;59:828–838. doi: 10.1002/hep.26788. [DOI] [PubMed] [Google Scholar]
- 69.Shi X, Pan Y, Wang M, Wang D, Li W, Jiang T, Zhang P, Chi X, Jiang Y, Gao Y, Zhong J, Sun B, Xu D, Jiang J, Niu J. IL28B genetic variation is associated with spontaneous clearance of hepatitis C virus, treatment response, serum IL-28B levels in Chinese population. PLoS One. 2012;7:e37054. doi: 10.1371/journal.pone.0037054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sugiyama M, Tanaka Y, Wakita T, Nakanishi M, Mizokami M. Genetic variation of the IL-28B promoter affecting gene expression. PLoS One. 2011;6:e26620. doi: 10.1371/journal.pone.0026620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Estrabaud E, Lapalus M, Broet P, Appourchaux K, De Muynck S, Lada O, Martinot-Peignoux M, Bieche I, Valla D, Bedossa P, Marcellin P, Vidaud M, Asselah T. Reduction of microRNA 122 expression in IFNL3 CT/TT carriers and during progression of fibrosis in patients with chronic hepatitis C. J Virol. 2014;88:6394–6402. doi: 10.1128/JVI.00016-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kamo Y, Ichikawa T, Miyaaki H, Uchida S, Yamaguchi T, Shibata H, Honda T, Taura N, Isomoto H, Takeshima F, Nakao K. Significance of miRNA-122 in chronic hepatitis C patients with serotype 1 on interferon therapy. Hepatol Res. 2014 doi: 10.1111/hepr.12317. DOI: 10.1111/hepr.12317. [DOI] [PubMed] [Google Scholar]
- 73.McFarland AP, Horner SM, Jarret A, Joslyn RC, Bindewald E, Shapiro BA, Delker DA, Hagedorn CH, Carrington M, Gale M, Jr, Savan R. The favorable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus-induced microRNAs. Nat Immunol. 2014;15:72–79. doi: 10.1038/ni.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yee HS, Chang MF, Pocha C, Lim J, Ross D, Morgan TR, Monto A. Update on the management and treatment of hepatitis C virus infection: recommendations from the Department of Veterans Affairs Hepatitis C Resource Center Program and the National Hepatitis C Program Office. Am J Gastroenterol. 2012;107:669–689. doi: 10.1038/ajg.2012.48. quiz 690. [DOI] [PubMed] [Google Scholar]
- 75.Zeuzem S, Dusheiko GM, Salupere R, Mangia A, Flisiak R, Hyland RH, Illeperuma A, Svarovskaia E, Brainard DM, Symonds WT, Subramanian GM, McHutchison JG, Weiland O, Reesink HW, Ferenci P, Hezode C, Esteban R. Sofosbuvir and ribavirin in HCV genotypes 2 and 3. N Engl J Med. 2014;370:1993–2001. doi: 10.1056/NEJMoa1316145. [DOI] [PubMed] [Google Scholar]
- 76.Afdhal N, Zeuzem S, Kwo P, Chojkier M, Gitlin N, Puoti M, Romero-Gomez M, Zarski JP, Agarwal K, Buggisch P, Foster GR, Brau N, Buti M, Jacobson IM, Subramanian GM, Ding X, Mo H, Yang JC, Pang PS, Symonds WT, McHutchison JG, Muir AJ, Mangia A, Marcellin P. Ledipasvir and sofosbuvir for untreated HCV genotype 1 infection. N Engl J Med. 2014;370:1889–1898. doi: 10.1056/NEJMoa1402454. [DOI] [PubMed] [Google Scholar]
- 77.Bibert S, Wojtowicz A, Taffe P, Manuel O, Bernasconi E, Furrer H, Gunthard HF, Hoffmann M, Kaiser L, Osthoff M, Cavassini M, Bochud PY. The IFNL3/4 ΔG variant increases susceptibility to cytomegalovirus retinitis among HIV-infected patients. AIDS. 2014;28:1885–1889. doi: 10.1097/QAD.0000000000000379. [DOI] [PubMed] [Google Scholar]
- 78.Machmach K, Abad-Molina C, Romero-Sanchez MC, Abad MA, Ferrando-Martinez S, Genebat M, Pulido I, Viciana P, Gonzalez-Escribano MF, Leal M, Ruiz-Mateos E. IL28B single-nucleotide polymorphism rs12979860 is associated with spontaneous HIV control in white subjects. J Infect Dis. 2013;207:651–655. doi: 10.1093/infdis/jis717. [DOI] [PubMed] [Google Scholar]
- 79.Safaeian M, Hildesheim A, Gonzalez P, Yu K, Porras C, Li Q, Rodriguez AC, Sherman ME, Schiffman M, Wacholder S, Burk R, Herrero R, Burdette L, Chanock SJ, Wang SS. Single nucleotide polymorphisms in the PRDX3 and RPS19 and risk of HPV persistence and cervical precancer/cancer. PLoS One. 2012;7:e33619. doi: 10.1371/journal.pone.0033619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trevino A, Lopez M, Vispo E, Aguilera A, Ramos JM, Benito R, Roc L, Eiros JM, de Mendoza C, Soriano V. Development of tropical spastic paraparesis in human T-lymphotropic virus type 1 carriers is influenced by interleukin 28B gene polymorphisms. Clin Infect Dis. 2012;55:e1–e4. doi: 10.1093/cid/cis343. [DOI] [PubMed] [Google Scholar]
- 81.Assone T, Souza FV, Gaester KO, Fonseca LA, Luiz Odo C, Malta F, Pinho JR, Goncalves Fde T, Duarte AJ, de Oliveira AC, Casseb J. IL28B gene polymorphism SNP rs8099917 genotype GG is associated with HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) in HTLV-1 carriers. PLoS Negl Trop Dis. 2014;8:e3199. doi: 10.1371/journal.pntd.0003199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kamihira S, Usui T, Ichikawa T, Uno N, Morinaga Y, Mori S, Nagai K, Sasaki D, Hasegawa H, Yanagihara K, Honda T, Yamada Y, Iwanaga M, Kanematu T, Nakao K. Paradoxical expression of IL-28B mRNA in peripheral blood in human T-cell leukemia virus type-1 mono-infection and co-infection with hepatitis C virus. Virol J. 2012;9:40. doi: 10.1186/1743-422X-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scagnolari C, Midulla F, Riva E, Monteleone K, Solimini A, Bonci E, Cangiano G, Papoff P, Moretti C, Pierangeli A, Antonelli G. Evaluation of interleukin 28B single nucleotide polymorphisms in infants suffering from bronchiolitis. Virus Res. 2012;165:236–240. doi: 10.1016/j.virusres.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Selvaggi C, Pierangeli A, Fabiani M, Spano L, Nicolai A, Papoff P, Moretti C, Midulla F, Antonelli G, Scagnolari C. Interferon lambda 1–3 expression in infants hospitalized for RSV or HRV associated bronchiolitis. J Infect. 2014;68:467–477. doi: 10.1016/j.jinf.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wei H, Wang S, Chen Q, Chen Y, Chi X, Zhang L, Huang S, Gao GF, Chen JL. Suppression of interferon lambda signaling by SOCS-1 results in their excessive production during influenza virus infection. PLoS Pathog. 2014;10:e1003845. doi: 10.1371/journal.ppat.1003845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng YW, Li H, Yu JP, Zhao H, Wang SE, Ren XB. Interferon-lambdas: special immunomodulatory agents and potential therapeutic targets. J Innate Immun. 2013;5:209–218. doi: 10.1159/000345365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gaudieri S, Lucas M, Lucas A, McKinnon E, Albloushi H, Rauch A, di Iulio J, Martino D, Prescott SL, Tulic MK. Genetic variations in IL28B and allergic disease in children. PLoS One. 2012;7:e30607. doi: 10.1371/journal.pone.0030607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jordan WJ, Eskdale J, Srinivas S, Pekarek V, Kelner D, Rodia M, Gallagher G. Human interferon lambda-1 (IFN-lambda1/IL-29) modulates the Th1/Th2 response. Genes Immun. 2007;8:254–261. doi: 10.1038/sj.gene.6364382. [DOI] [PubMed] [Google Scholar]
- 89.Koltsida O, Hausding M, Stavropoulos A, Koch S, Tzelepis G, Ubel C, Kotenko SV, Sideras P, Lehr HA, Tepe M, Klucher KM, Doyle SE, Neurath MF, Finotto S, Andreakos E. IL-28A (IFN-lambda2) modulates lung DC function to promote Th1 immune skewing and suppress allergic airway disease. EMBO Mol Med. 2011;3:348–361. doi: 10.1002/emmm.201100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Contoli M, Message SD, Laza-Stanca V, Edwards MR, Wark PA, Bartlett NW, Kebadze T, Mallia P, Stanciu LA, Parker HL, Slater L, Lewis-Antes A, Kon OM, Holgate ST, Davies DE, Kotenko SV, Papi A, Johnston SL. Role of deficient type III interferon-lambda production in asthma exacerbations. Nat Med. 2006;12:1023–1026. doi: 10.1038/nm1462. [DOI] [PubMed] [Google Scholar]
- 91.Bullens DM, Decraene A, Dilissen E, Meyts I, De Boeck K, Dupont LJ, Ceuppens JL. Type III IFN-lambda mRNA expression in sputum of adult and school-aged asthmatics. Clin Exp Allergy. 2008;38:1459–1467. doi: 10.1111/j.1365-2222.2008.03045.x. [DOI] [PubMed] [Google Scholar]
- 92.He S, Li T, Chen H, Ma W, Yao Q, Yang H, Wang H, Wang F, Zhao C, Yang P. Cd14+ cell-derived IL-29 modulates proinflammatory cytokine production in patients with allergic airway inflammation. Allergy. 2011;66:238–246. doi: 10.1111/j.1398-9995.2010.02455.x. [DOI] [PubMed] [Google Scholar]
- 93.Lin SC, Kuo CC, Tsao JT, Lin LJ. Profiling the expression of interleukin (IL)-28 and IL-28 receptor alpha in systemic lupus erythematosus patients. Eur J Clin Invest. 2012;42:61–69. doi: 10.1111/j.1365-2362.2011.02557.x. [DOI] [PubMed] [Google Scholar]
- 94.Wu Q, Yang Q, Lourenco E, Sun H, Zhang Y. Interferon-lambda1 induces peripheral blood mononuclear cell-derived chemokines secretion in patients with systemic lupus erythematosus: its correlation with disease activity. Arthritis Res Ther. 2011;13:R88. doi: 10.1186/ar3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li Y, Cheng H, Zuo XB, Sheng YJ, Zhou FS, Tang XF, Tang HY, Gao JP, Zhang Z, He SM, Lv YM, Zhu KJ, Hu DY, Liang B, Zhu J, Zheng XD, Sun LD, Yang S, Cui Y, Liu JJ, Zhang XJ. Association analyses identifying two common susceptibility loci shared by psoriasis and systemic lupus erythematosus in the Chinese Han population. J Med Genet. 2013;50:812–818. doi: 10.1136/jmedgenet-2013-101787. [DOI] [PubMed] [Google Scholar]
- 96.Rigante D, Mazzoni MB, Esposito S. The cryptic interplay between systemic lupus erythematosus and infections. Autoimmun Rev. 2014;13:96–102. doi: 10.1016/j.autrev.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 97.Liu CH, Liu CJ, Lin CL, Liang CC, Hsu SJ, Yang SS, Hsu CS, Tseng TC, Wang CC, Lai MY, Chen JH, Chen PJ, Chen DS, Kao JH. Pegylated interferon-alpha-2a plus ribavirin for treatment-naive Asian patients with hepatitis C virus genotype 1 infection: a multicenter, randomized controlled trial. Clin Infect Dis. 2008;47:1260–1269. doi: 10.1086/592579. [DOI] [PubMed] [Google Scholar]
- 98.Roberts SK, Mitchell J, Leung R, Booth D, Bollipo S, Ostapowicz G, Sloss A, McCaughan GW, Dore GJ, Thompson A, Crawford DH, Sievert W, Weltman M, Cheng W, George J. Distribution of interferon lambda-3 gene polymorphisms in Australian patients with previously untreated genotype 1 chronic hepatitis. C analysis from the PREDICT and CHARIOT studies. J Gastroenterol Hepatol. 2014;29:179–184. doi: 10.1111/jgh.12424. [DOI] [PubMed] [Google Scholar]
- 99.Grebely J, Page K, Sacks-Davis R, van der Loeff MS, Rice TM, Bruneau J, Morris MD, Hajarizadeh B, Amin J, Cox AL, Kim AY, McGovern BH, Schinkel J, George J, Shoukry NH, Lauer GM, Maher L, Lloyd AR, Hellard M, Dore GJ, Prins M. The effects of female sex, viral genotype, and IL28B genotype on spontaneous clearance of acute hepatitis C virus infection. Hepatology. 2014;59:109–120. doi: 10.1002/hep.26639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Russell CD, Griffiths SJ, Baillie JK, Haas J. Stratified infection medicine: a call to arms. Lancet Infect Dis. 2014;14:451. doi: 10.1016/S1473-3099(14)70774-1. [DOI] [PubMed] [Google Scholar]
- 101.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. Snap: A web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Indolfi G, Mangone G, Bartolini E, Nebbia G, Calvo PL, Moriondo M, Tovo PA, de Martino M, Azzari C, Resti M. Comparative analysis of rs12979860 SNP of the IFNL3 gene in children with hepatitis C and ethnic matched controls using 1000 Genomes Project data. PLoS One. 2014;9:e85899. doi: 10.1371/journal.pone.0085899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Venegas M, Villanueva RA, Gonzalez K, Brahm J. IL28B polymorphisms associated with therapy response in Chilean chronic hepatitis C patients. World J Gastroenterol. 2011;17:3636–3639. doi: 10.3748/wjg.v17.i31.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rao HY, Sun DG, Jiang D, Yang RF, Guo F, Wang JH, Liu F, Zhang HY, Zhang HH, Du SC, Jin Q, Qin H, Lok AS, Wei L. IL28B genetic variants and gender are associated with spontaneous clearance of hepatitis C virus infection. J Viral Hepat. 2012;19:173–181. doi: 10.1111/j.1365-2893.2011.01497.x. [DOI] [PubMed] [Google Scholar]
- 105.di Iulio J, Ciuffi A, Fitzmaurice K, Kelleher D, Rotger M, Fellay J, Martinez R, Pulit S, Furrer H, Gunthard HF, Battegay M, Bernasconi E, Schmid P, Hirschel B, Barnes E, Klenerman P, Telenti A, Rauch A. Estimating the net contribution of interleukin-28B variation to spontaneous hepatitis C virus clearance. Hepatology. 2011;53:1446–1454. doi: 10.1002/hep.24263. [DOI] [PMC free article] [PubMed] [Google Scholar]