Abstract
Since the discovery of the DNA double helix, the main molecular repository of genetic information, scientists have been struggling to find ways to efficiently manipulate genes. The ability to mark, modify, or regulate specific sequences of DNA in a controlled fashion is of key importance because of the ways that gene editing could be used to improve human life. For example, genetic therapies are being developed to permanently cure cancer and other life-threatening diseases.
In 2012 a breakthrough in biological research led to the discovery of a facile genome-editing technology, now commonly referred to as CRISPR-Cas9, that can be easily programmed to cleave and modify specific genes in living organisms.1,2 Because of its unprecedented versatility, precision, and cost-effectiveness, CRISPR-Cas9 is rapidly paving the way for revolutionary discoveries in biosciences, medicine, and biotechnology.
Today new genetic experiments performed with the technology are vastly improving our understanding of human health and disease. In biotechnology, CRISPR-Cas9 is being used to grow drought-resistant crops and driving advances in biofuel production. It also represents a new frontier in medicine. Genetic tools based on CRISPR-Cas9 will likely be used to design new drugs and revolutionary gene therapies. Physicians can now envision curing severe genetic diseases at their source. That capability could also provide new hope for people suffering from life-threatening illnesses such as cancer and cardiovascular diseases.
Even as CRISPR-Cas9 gains widespread use in the lab, biochemists’ understanding of how it works at the molecular level has remained opaque. Although experimental observations provided glimpses of those inner workings, molecular dynamics simulations have recently brought them into sharper focus. The simulations reveal an intricate biomolecular dance whose key steps—the recognition, binding, and cleavage of nucleic acids—must be performed with exquisite timing and precision.
A genetic breakthrough
The CRISPR-Cas9 technology is fundamentally a bacterial defense system against viral infections. When a virus invades a bacterium, parts of the foreign DNA are inserted between peculiar genetic sequences, called clustered regularly interspaced short palindromic repeats, or CRISPR, in the bacterial DNA. The DNA “spacer” sequences are markers of a viral invasion and are transcribed into complementary sequences of RNA. The RNA transcripts then bind with a specific enzyme called Cas9 and form the CRISPR-Cas9 complex. (For a glossary of genetic-engineering terms, see the box below.)
Because of its base-pair complementarity with viral DNA, the guide RNA segment in the CRISPR-Cas9 complex leads and docks the Cas9 enzyme at precise regions of the foreign genetic material. Once there, Cas9 cleaves the viral sequences and neutralizes the viral invasion, as shown in figure 1.After the infection, the spacer DNA remains stored between CRISPR sequences as a “memory” that immunizes against past infections.
FIGURE 1. CRISPR-CAS9 IMMUNE DEFENSE MECHANISM.
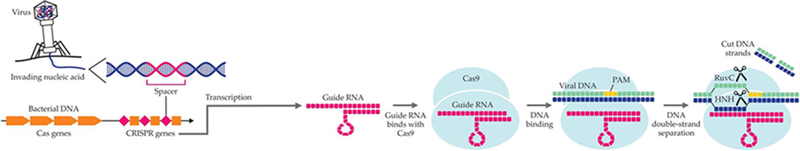
In an infection, segments of the viral nucleic acids (spacers) are inserted between clustered regularly interspaced short palindromic repeats (CRISPR) in a cell’s genetic material and form an array of CRISPR genes. The array is then transcribed into an RNA segment that functions as a guide for the Cas9 protein. The guide RNA binds to the protein to form a CRISPR-Cas complex. Because CRISPR-Cas9 contains a guide RNA segment that is complementary to viral DNA, it recognizes and binds foreign viral DNA as long as the DNA contains an adjacent PAM segment. Once bound to viral DNA, CRISPR-Cas9 separates and cleaves the two viral DNA strands, thus making them inactive.
The great breakthrough in CRISPR biology came with the realization that Cas9 could be reprogrammed to cleave not only viral DNA but also other DNA sequences2 by changing the guide RNA filament associated with Cas9. The enzyme is thus able to remove any undesired fragment of DNA and leave a tailor-made fragment in its place. Moreover, CRISPR-Cas9 is able to recognize DNA at specific sites by the presence of a short sequence known as protospacer adjacent motif (PAM), which consists of a few nucleotides and lies adjacent to the sequence to be cleaved.
In viral infections, PAM recognition is the first step to binding and subsequent cleavage of the adjacent DNA sequence by Cas9. If PAM is not present, CRISPR-Cas9 does not bind or cleave any DNA sequence, even if it perfectly matches the guide RNA segment. Thus CRISPR-Cas9 can be programmed only to cleave DNA sequences that are preceded by an appropriate PAM sequence. But not all DNA sequences are naturally preceded by a recognizable PAM sequence.
One of the most valuable goals in genome editing is the biomolecular engineering of Cas9-like enzymes that recognize any desired PAM sequence. Achieving that goal would expand the targeting capability of the technology.3 And it’s an example of how Cas9 can be further improved for genetic engineering. But to intelligently manipulate Cas9, biologists need a detailed understanding of how CRISPR-Cas9 recognizes, binds, and cleaves DNA.
Over the past five years, scientists have identified the fundamental biophysical aspects of CRISPR-Cas9. They’ve used state-of-the-art biochemical experiments and emerging electron-microscopy techniques to discover the intricate mechanism by which Cas9 edits genes.4 We now know, for instance, that upon PAM recognition, the complementary strand of the target DNA interacts with the RNA filament and produces a hybrid DNA–RNA double helix,3 as shown in figure 2.
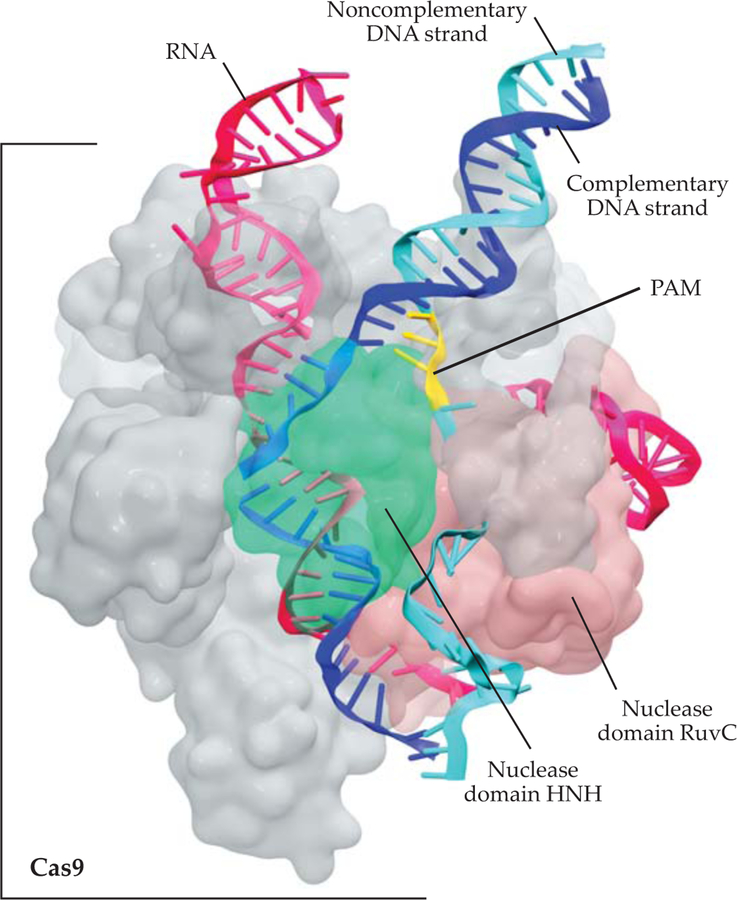
FIGURE 2. MOLECULAR ARCHITECTURE OF THE CRISPR-CAS9 COMPLEX,
from cryoelectron microscopy experiments and computational modeling.4,10 The Cas9 protein (gray) is bound to a guide RNA segment (magenta) and a matching DNA sequence (the complementary strand is dark blue; the noncomplementary strand, light blue; and the PAM segment, yellow). The Cas9 protein contains two nuclease domains, HNH (green) and RuvC (pink), which cleave the complementary and noncomplementary DNA strands, respectively.
X-ray crystallography reveals the overall architecture of Cas9, which is formed by several domains with specialized functions.4 When ready for catalysis, Cas9 positions its two nuclease domains—those specialized in cuing DNA—in close proximity to the two DNA strands. In that conformation, Cas9 can simultaneously cleave the two DNA strands to produce the characteristic double-strand breaks.
Sophisticated experimental studies have led to our current understanding of the biological function of CRISPR-Cas9. Our collection of experimentally obtained snapshots of Cas9 has given us a peek into the invisible dance that it performs as it binds to and cleaves nucleic acids. But to actually watch the conformational changes that compose the dance and understand how they are related to function is an extremely challenging experimental task. Here is where the power of computer simulations comes into play. They provide a dynamic, microscopic view that is out of reach when using experimental techniques.
The power of physical simulations
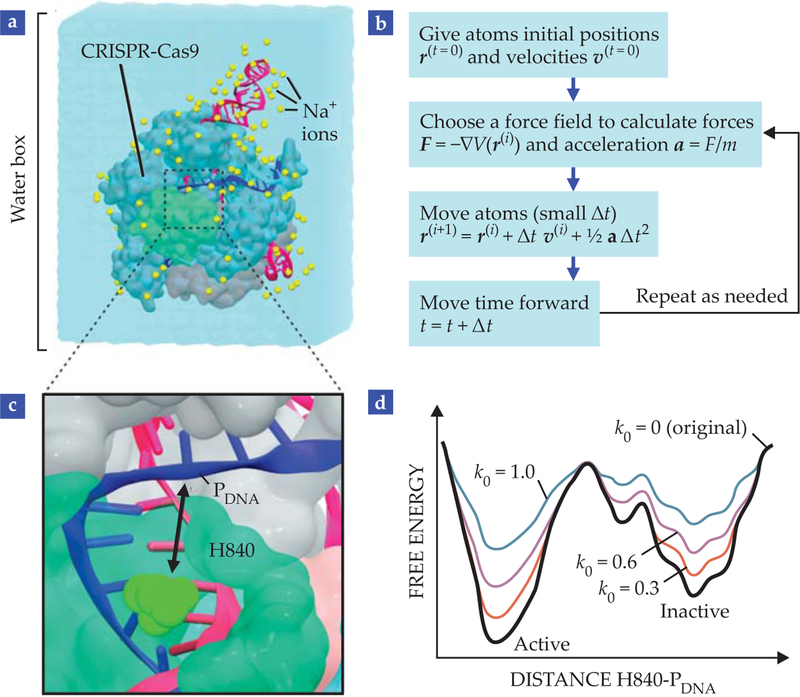
Molecular dynamics (MD) simulations, which use classical Newtonian physics to follow the motions of atoms through time, precisely capture the dynamics of bio-molecules.5 In classical MD, atoms are approximated as spheres, chemical bonds are approximated as springs, and the interactions are modeled by a set of parameterized functions, commonly referred to as force fields. Figure 3a illustrates the atomic-scale simulations.
FIGURE 3. MOLECULAR DYNAMICS (MD) SIMULATIONS.
produce a temporal trajectory of (a) CRISPR-Cas9 embedded in a water box that contains sodium ions at physiological concentration. (b) In a nutshell, MD consists of giving atoms initial positions and velocities, choosing a set of functions and parameters to describe the forces acting on each atom, and advancing time using Newton’s equations of motion. The resulting trajectory of atomic coordinates can be used to track the system’s properties over time. (c) Gaussian accelerated molecular dynamics6 (GaMD) describes the movement of histidine H840 (the catalytic residue in HNH) to the cleavage site in the target strand (PDNA) for catalysis.10 (d) In GaMD, quadratic functions modify the original potential energy of the system in order to overcome the barrier between active and inactive states. The extent of acceleration is controlled by parameters of the Gaussian function. The greater the value of k0, the greater the acceleration and the easier the system overcomes the barrier between states.
In the past few decades, progress in computational capability has made MD simulations significantly more powerful—and inexpensive, compared with any experimental method. A desktop computer can simulate biomolecular processes on the nanosecond time scale. Supercomputers and sophisticated methodologies can control the speed of the dynamics and allow scientists to observe processes that occur over milliseconds.
The state-of-the-art “accelerated” MD methodology uses quadratic functions to effectively decrease the potential energy barriers and accelerate the transitions between low-energy states,6 as shown in figure 3d. The simulations sample a wider configuration space and capture biological processes that cannot be described via conventional MD simulations. The advances have made it possible to simulate complex and slow biophysical events, such as folding, binding, and large-scale conformational transitions in biomolecules.6,7
At the same time, advances in the description of nucleic acids have enabled accurate simulations of proteins bound to DNA or RNA.8 Given the complexity of the CRISPR-Cas9 system, MD simulations are only meaningful over very long time scales. Accelerated MD methods therefore represent the best approach available to access biophysical processes that are representative of CRISPR-Cas9 biology. Indeed, accelerated simulations can access more than the conformational changes observed by conventional MD (see figure 3c, 3d).
MD techniques can also support experiments by revealing CRISPR-Cas9 at work on the molecular level and unveiling specific interactions and forces that are behind the Cas9 function. In the next section, we summarize the most exciting outcomes of computer simulations of CRISPR-Cas9 and the ongoing challenges in the field of genetic engineering.
Watching CRISPR-Cas9 at work
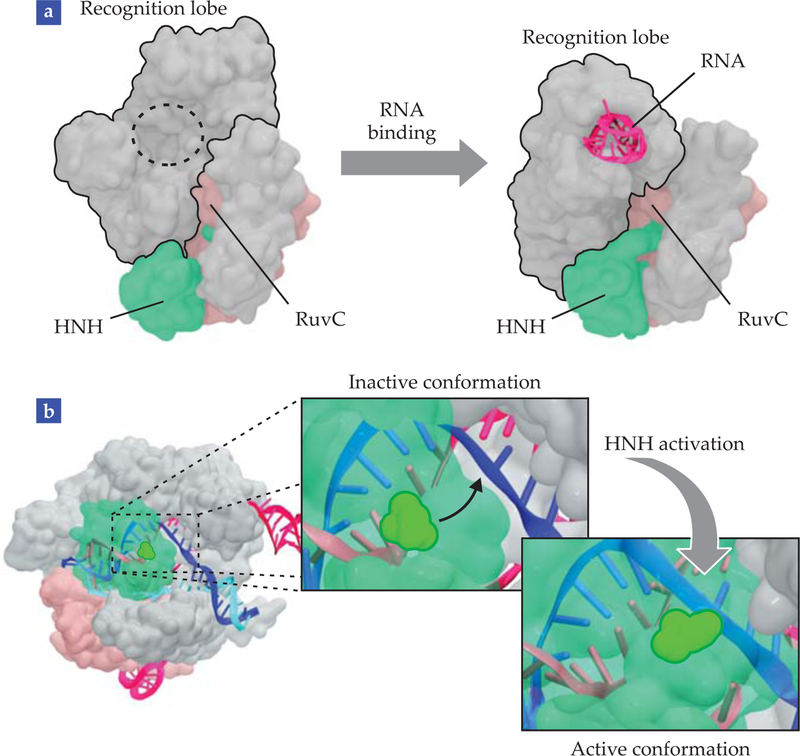
RNA binding is a critical step for Cas9 activation because it primes the protein to bind to DNA.4 It is therefore of paramount importance to understand the mechanism by which Cas9 binds with RNA. To that end, computational scientists have recently used enhanced-sampling MD techniques to simulate the recruitment of RNA by Cas9. Those simulations offered the first direct observation of domain movements that reshape the unbound Cas9 architecture into its RNA-bound state,9 illustrated in figure 4a.
FIGURE 4. MOLECULAR MECHANISMS OF CRISPR-CAS9,
revealed by molecular dynamics simulations. (a) Rearrangements made to the recognition lobe reshape the molecular architecture of unbound Cas9 into its RNA-bound conformation. (b) The movement of HNH into its final catalytic state prepares it to cleave the DNA. The conformational change, shown as insets, brings the HNH catalytic residue close to the cleavage site in the DNA complementary strand. In all structures, Cas9 nuclease domains HNH and RuvC are green and pink, respectively. RNA is magenta; DNA is dark blue (complementary strand) and light blue (noncomplementary strand).
During the conformational changes, Cas9 temporarily exposes basic residues to the solvent and creates a “positive cavity” that attracts and accommodates the negatively charged RNA filament. The observation of that positively charged cavity revealed the exact way in which electrostatic forces facilitate RNA–Cas9 binding.
Even after Cas9 is bound to its guide RNA, the CRISPR-Cas9 complex is not yet ready to catalyze the cleavage of DNA. Cas9 must first undergo a sequence of gradual conformational transitions and eventually relocate the two catalytic domains, HNH and RuvC, to optimal positions in order to cleave the complementary and noncomplementary DNA strands.
Using a combination of “steered” and accelerated MD simulations, computational scientists observed those conformational transitions and were able to identify the main players during Cas9 activation.9 In particular, the simulations revealed a striking plasticity of the HNH domain, which appears to be the main switch controlling the conformational dynamics. Prior to activation, the catalytic residues of HNH point away from the DNA cleavage site and in the opposite direction. To become active, HNH therefore needs to rotate by 180° and approach the site—a process captured by simulations and illustrated in figure 4b.
Starting from preactivated structures, MD simulations also enabled the first a empt to determine the final, completely activated structure of Cas9, where both HNH and RuvC domains are well positioned to cleave the DNA strands.9 Independent computational studies have confirmed and reproduced those results.10,11 In 2017 electron microscopy revealed an active structure of CRISPR-Cas9 in remarkably good agreement with the computational structure.12
MD simulations have also been used to perform a computational experiment designed to understand how the nucleic acids are involved in Cas9 activation. Although it’s known that the two DNA strands must be properly positioned for cleavage to occur, little was understood about how the noncomplementary strand—the one not hybridized with the guide RNA—functions in Cas9 activation. In the computational experiment, researchers simulated the CRISPR-Cas9 complex both in the presence and in the absence of the noncomplementary DNA strand.13 In the noncomplementary strand’s absence, HNH moved away from the cleavage site and adopted a conformation that made it inactive. But when the complementary strand was present, it facilitated the 180° rotation of HNH toward the catalytic site.
Scientists at the University of California, Berkeley, followed those simulations with sophisticated spectroscopic techniques to distinguish the conformational states of Cas9 in bulk and as a single molecule.14 The experiments confirmed the early computational outcome: They showed that the repositioning and docking of HNH at the cleavage site indeed requires the presence of the noncomplementary DNA strand.
Allosteric communications
One interesting aspect of the CRISPR-Cas9 complex is the ability of its domains to communicate with each other through a process known as allostery.15 Such communications are an important feature of many biological systems, and they allow perturbations at spatially distant regions of the protein to affect how the active site functions. The perturbation can be the binding of a ligand to an allosteric site, whose local effect is some-how transmitted all the way to functional regions where the ligand interferes with conformation and dynamics. Ligands that can exert remote control over a protein’s active region are commonly referred to as allosteric effectors.
Intriguingly, allostery often does not involve huge (or even obvious) conformational changes, which makes it difficult to understand how the information travels throughout the protein. In fact, because of the subtle nature of allosteric signaling, experiments o en fail to provide a full description of their effects. MD simulations, on the other hand, are valuable tools for studying allosteric effects. They provide a rich description of atomic fluctuations, from which correlated motions can be extracted.
To study allosteric communications in the CRISPR-Cas9 complex, computational scientists have used a mathematical approach based on Shannon’s entropy to identify pairs of atoms that display interdependent motions during the simulations. The approach produces pairwise correlation coefficients that dictate how much the position of one atom restricts the position of another, and vice versa.
With those coefficients at hand, it is possible to build network graphs by applying the same algorithms that Facebook and other media outlets use to describe social networks. The algorithms organize individuals into different communities and identify the most efficient communication pathways between thousands of people—or atoms, in the case of MD simulations. By describing Cas9 as a network of interactions, scientists can track the main communication pathway in the CRISPR-Cas9 complex.16 It turns out that PAM—the small sequence of DNA that initiates recognition and binding—works as an allosteric effector and facilitates communication between the two catalytic domains, RuvC and HNH.
Importantly, the cross talk between RuvC and HNH is what allows Cas9 to cleave the two DNA strands in a concerted fashion. Thus PAM is required not only to ensure DNA recognition by Cas9 but also to activate the two domains for catalysis.
The future of CRISPR-Cas9
The studies discussed in this article are all based on classical MD simulations and focus on the sequence of conformational changes that prepare Cas9 for DNA cleavage. The actual cleavage mechanism, however, cannot be simulated by classical MD. That’s because it requires a proper description of the electronic effects in chemical reactions.
To understand the catalytic mechanism by which Cas9 cuts the DNA, computational scientists need to employ high-level quantum mechanics simulations, which describe the formation and breakup of chemical bonds. Such simulations have recently determined the structure of the reactant state of Cas9.17 Building on that structure, which depicts a fully reactive CRISPR-Cas9 catalytic complex, we expect future quantum mechanics simulations to shed light on the catalytic mechanism of the system and yield crucial information on how to optimize or tune Cas9’s activity.
We hope to have convinced you of the power of molecular simulations for understanding how CRISPR-Cas9 edits genes. Gaining that knowledge is the first step to improving the technology for genome-editing purposes. Despite the remarkable advantages of CRISPR-Cas9 relative to other such systems, some issues still need to be addressed before it can be considered a safe genetic therapy. One concern is the occurrence of so-called off-target cleavages, which occur when CRISPR-Cas9 mistakenly cuts DNA sequences that are similar but not identical to the target sequence.
Because off-target cleavages can produce unpredictable and detrimental mutations, the specificity of CRISPR-Cas9 must be improved before it can be safely used for clinical purposes. In that respect, recent computational simulations promise to provide valuable insights on the molecular determinants of off-target effects.18 They are sure to help in designing novel and highly specific Cas9-like enzymes.
Acknowledgments
Our work is supported in part by the National Institutes of Health, the National Biomedical Computation Resource, the San Diego Supercomputer Center, and the Extreme Science and Engineering Discovery Environment, which provided computer time. We thank Ying-long Miao for discussions about accelerated MD simulations.
A GENETIC-ENGINEERING GLOSSARY
- Catalysis
The breaking of a chemical bond, facilitated by an enzyme.
- CRISPR
An acronym for clustered regularly interspaced short palindromic repeats. Peculiar genetic sequences in bacteria, between which viral DNA segments are inserted, serve as markers of an infection.
- CRISPR-Cas9
The complex formed by a CRISPR RNA transcript and a Cas9 protein.
- DNA
Deoxyribonucleic acid, the molecular repository of genetic information for all cellular life forms and many viruses. It is located in the nucleus and is normally found as a double helix, two intertwined strands.
- Enzyme
A biomolecule, normally a protein, that catalyzes a specific chemical reaction by lowering the activation energy.
- Gene
A segment of DNA that encodes the genetic information required for the synthesis of functional biological products. Mostly proteins, the products could also be some types of RNA.
- Genome
The entire genetic information in a living organism, encoded in DNA (or in RNA in some viruses).
- HNH and RuvC
Nuclease domains of the Cas9 protein, responsible for cleaving the complementary and noncomplementary DNA strands, respectively.
- Mutation
An alteration in DNA structure that produces permanent changes in the genetic information encoded therein. Detrimental mutations are associated with aging and cancer.
- Nuclease
An enzyme that cleaves the internucleotide (phosphodiester) linkages in the strands of nucleic acids.
- PAM
An acronym for protospacer adjacent motif, a short segment of a few nucleotides that occurs in the viral DNA adjacent to the sequence that is cleaved by Cas9.
- Protein (or enzyme) domain
A compact unit within a protein chain that can exist and function independently of the rest of the chain.
- RNA
Ribonucleic acid, the molecular carrier of genetic information for all cellular life forms. It is normally found as a single strand that can adopt widely different and complex three-dimensional structures, but it can also form hybrid double helices with a complementary DNA strand, as in CRISPR.
- Transcript
The RNA product of a DNA transcription.
- Transcription
The synthesis of an RNA segment complementary to a DNA template.
Biography
Giulia Palermo

Clarisse G. Ricci

J. Andrew McCammon

Contributor Information
Giulia Palermo, University of California, Riverside..
Clarisse G. Ricci, University of California, San Diego..
J. Andrew McCammon, UCSD.
REFERENCES
- 1.Doudna JA, Charpentier E, Science 346, 1258096 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Jinek M et al. , Science 337, 816 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anders C et al. , Nature 513, 569 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang F, Doudna JA, Annu. Rev. Biophys 46, 505 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Karplus M, McCammon JA, Nat. Struc. Biol 9, 646 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Miao Y, Feher VA, McCammon JA, Chem J. Theory Comput 11, 3584 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miao Y, McCammon JA, Proc. Natl. Acad. Sci. USA 115, 3036 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palermo G et al. , Acc. Chem. Res 48, 220 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Palermo G et al. , Proc. Natl. Acad. Sci. USA 114, 7260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palermo G et al. , Q. Rev. Biophys 51, e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuo Z, Liu J, Sci. Rep 7, 17271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huai C et al. , Nat. Commun 8, 1375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palermo G et al. , ACS Cent. Sci 2, 756 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dagdas YS et al. , Sci. Adv 3, eaao0027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sternberg SH et al. , Nature 527, 110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palermo G et al. , J. Am. Chem. Soc 139, 16028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palermo G, J. Chem. Inf. Model (2019), doi:/ 10.1021/acs.jcim.8b00988. [DOI] [PubMed] [Google Scholar]
- 18.Ricci CG et al. , ACS Cent. Sci (2019), doi:/ 10.1021/acscentsci.9b00020. [DOI] [PMC free article] [PubMed] [Google Scholar]